Abstract
Type 2 diabetes has profound effects on metabolism that can be detected in plasma. While increases in circulating non-esterified fatty acids (NEFA) are well-described in diabetes, effects on signaling lipids have received little attention. Oxylipins and endocannabinoids are classes of bioactive fatty acid metabolites with many structural members that influence insulin signaling, adipose function and inflammation through autocrine, paracrine and endocrine mechanisms. To link diabetes-associated changes in plasma NEFA and signaling lipids, we quantitatively targeted >150 plasma lipidome components in age- and body mass index-matched, overweight to obese, non-diabetic (n = 12) and type 2 diabetic (n = 43) African-American women. Diabetes related NEFA patterns indicated ∼60% increase in steroyl-CoA desaturase activity and ∼40% decrease in very long chain polyunsaturated fatty acid chain shortening, patterns previously associated with the development of nonalcoholic fatty liver disease. Further, epoxides and ketones of eighteen carbon polyunsaturated fatty acids were elevated >80% in diabetes and strongly correlated with changes in NEFA, consistent with their liberation during adipose lipolysis. Endocannabinoid behavior differed by class with diabetes increasing an array of N-acylethanolamides which were positively correlated with pro-inflammatory 5-lipooxygenase-derived metabolites, while monoacylglycerols were negatively correlated with body mass. These results clearly show that diabetes not only results in an increase in plasma NEFA, but shifts the plasma lipidomic profiles in ways that reflect the biochemical and physiological changes of this pathological state which are independent of obesity associated changes.
Introduction
Obesity is a risk factor for the development of Type 2 diabetes, a disease which chronically increases circulating non-esterified fatty acids (NEFA) [1], dampens the pulsatile secretion of insulin [2], [3], and diminishes tissue glucose uptake while promoting hepatic glucose output [2], [4], [5]. Peripheral insulin resistance and fuel partitioning in type 2 diabetes are well-studied with respect to glucose, yet impacts on many metabolic domains remain to be assessed. Investigations of diabetes employing global metabolomics in plasma have reported changes in numerous metabolites including lipids, carbohydrates and amino acids, highlighting the fact that type 2 diabetes elicits broad perturbations of energy metabolism [6]–[8]. For example, diabetes increases circulating medium- and long-chain acylcarnitines [6] and branched-chain amino acids [7], [9], suggesting broad dysfunctions in fuel catabolism and mitochondrial function [6]. In contrast, studies addressing the impact of diabetes on circulating levels of low abundance signaling lipids including oxylipins (OxL) and endocannabinoids (eCBs) are less common. Here, we quantified many of these potent mediators along with NEFA to assess the covariant behavior of these molecules in obese diabetic and non-diabetic cohorts.
OxL and eCBs are structurally diverse groups of molecules with broad effects on cellular function acting through receptor- and ion channel-mediated processes [10], [11]. The arachidonic acid-derived OxLs (i.e. eicosanoids) are known to influence insulin signaling, inflammation and vascular function with mechanistic implications at the tissue level in diabetes and associated pathologies [10]. However, little is known regarding the impact of diabetes on plasma concentrations of eicosanoids and other polyunsaturated fatty acid (PUFA)-derived OxLs. Molecules with eCB properties include the N-acylethanolamides (NAEs), monoacylglycerols (MAGs) and lipoamino acids (LAA). Obesity and diabetes increase plasma and tissue arachidonate-derived eCBs, N-arachidonoylethanolamide (A-EA) and 2-arachidonylglycerol (2-AG) levels [11]. These changes are hypothetically linked to dysfunctions in eCB regulation involving dietary fat intake [12], leptin [13] and/or insulin signaling [14]. While distinct signaling and significant cross-talk exists between individual eCBs and their structural analogs [15] little is known regarding the impact of diabetes on the eCB array beyond A-EA and 2-AG. Moreover, while NEFA, OxL and eCB metabolism are linked [16], coordinated changes of these three pathways have not been previously characterized.
To identify linkages between changes in NEFA and circulating signaling lipids, we performed a quantitative metabolomic investigation targeting over 150 plasma lipids including NEFA, OxLs and eCBs in overweight to obese, age and BMI matched non-diabetic and diabetic women. Multivariate analysis methods were used to identify key changes in biosynthetic relationships which are predictive of the diabetic phenotype.
Methods
Subjects and Study Design
Study volunteers were recruited in the Project SuGAR study, described in detail elsewhere [17]. This group displays an extraordinarily low genetic admixture, lives in a relatively small geographical space, and has a common dietary intake pattern which is uniformly rich in animal fats. There were 1279 registered participants in Project SuGAR, of which 290 were genotyped to identify persons with a missense uncoupling protein 3 (UCP3) G304A polymorphism. Of 52 subjects positive for the UCP3 g/a polymorphism (43 females, 9 males), complete datasets for clinical chemistries (blood lipids, glucose, lactate, HbA1c) and oral glucose tolerance test (OGTT) were available for 28 women (22 T2Ds, 6 non-diabetics). These subjects were thus chosen for subsequent metabolomics analyses, along with an age- and BMI-matched set of 28 women without the polymorphism (22 T2Ds, 6 non-diabetics) for comparison. A 2-way ANOVA was used to evaluate the association and interactions between the primary metabolic discriminates of type 2-diabetes reported below, and the UCP3 genotype. Neither significant associations with the UCP3 polymorphism nor diabetes x polymorphism interactions were detected (Table S1). A comparison of clinical parameters for subjects with and without type 2 diabetes is shown in Table 1 . One diabetic subject was omitted due to insufficient sample volume. Of the 43 diabetic study participants included, 34 (79%) were on insulin and or a combination of oral anti-hyperglycemic and lipid lowering medications (insulin, n = 21; insulin and biguanide, n = 2; insulin, fibrate and thiazoladendiones, n = 1; biguanide, n = 2; sulfonylurea, n = 7; biguanide and sulfonylurea, n = 1).
Table 1. Characteristics of overweight diabetic and non-diabetic African American Gullah-speaking female study participants.
| Clinical Parameter | non-diabetic(n = 12) | type 2 diabetic(n = 43) |
| Body Mass (kg) | 89.3±17 | 92.7±17 |
| BMI, kg/m2 (range) | 33±6 (24,43) | 36±6 (26,47) |
| Age,yrs (range) | 49±17 (21,69) | 55±14 (19,87) |
| Glucose, mg/dL | 92±10 | 210±79* |
| HbA1c (%) | 5.4±1 | 9.1±2* |
| Lactate, mmol/L | 1.1±0 | 1.2±0 |
| Triglycerides, mg/dL | 100±61 | 120±75 |
| Cholesterol, mg/dL | 200±52 | 210±47 |
| HDL cholesterol, mg/dL | 45±12 | 44±13 |
| LDL cholestrol, mg/dL | 140±42 | 140±41 |
| VLDL cholesterol, mg/dL | 20±12 | 23±15 |
p<0.0001 by Mann-Whitney U-test.
The Institutional Review Boards of the University of California, Davis, Medical University of South Carolina, and the University of Alabama at Birmingham approved the studies and participants provided informed consent. Sample collection and plasma clinical chemistry analyses are described elsewhere [6]. Volunteers were instructed to eat their regular diets and avoid unusual activity or exercise for 3d prior to blood collection. Diabetic patients were instructed to refrain from oral medications the evening before and morning of the study. Patients treated with insulin (n = 23) were instructed to take regular or rapid-acting insulin at dinner the night before the study, but withhold intermediate- or long-acting insulin the evening before or morning of the blood collection. Blood was obtained between ∼0800 hrs and 0900 hrs by arm venipuncture into EDTA-treated tubes from fasted subjects: no food or drink since 2000 hrs the previous night. Plasma was frozen at −20°C for 1–7 d before transport to −80°C freezers. Plasma aliquots were used to carry out the current analysis, acylcarnitine profiling [6], and global metabolomics [7].
Oxylipins, Acylamides, Acylglycerols, and Lipoamino Acids Measurements
Oxylipins and eCBs were isolated and quantified using modifications of published protocols [18]. Briefly, plasma aliquots (100 µL) were spiked with deuterated OxL and eCB surrogates, and extracted with 60 mg Oasis HLB (Waters Corporation, Milford, MA) solid phase extraction cartridges. Solvents were removed under vacuum. Residues were reconstituted in methanol containing an internal standard, filtered at 0.1 µm and analyzed by UPLC-MS/MS. Analytes were separated with a 2.1×150 mm, 1.7 µm Acquity BEH column on an Acquity UPLC (Waters Inc, Milford MA), ionized by electrospray ionization and detected by multi-reaction monitoring on an API4000 QTRAP (AB-SCIEX, Foster City, CA). Oxylipins and NAEs/MAGs/LAAs were analyzed in independent injections and ionized in negative and positive modes, respectively. See Table S2 for NAE, MAG and LAA mass transitions.
Non-esterified Fatty Acids (NEFA) Measurements
Plasma NEFA were isolated and converted to fatty acid methyl esters using a modified extractive methylation procedure [19]. Samples aliquots (25 µL) were spiked with 30 µL of 20 µM 15∶1n5 fatty acid (Nu-chek Prep, Inc., Elysian MN) in methanol, mixed with 125 µL ethereal diazomethane, and incubated for 10 min at 25°C. Solvents were removed by vacuum and residues were reconstituted in hexane containing internal standards and aliquots (1 µL) were analyzed on an HP6890 GC-5973N MSD (Agilent Technologies, San Jose, CA) equipped with a 30 m×0.25 id×0.25 µm DB-225 ms column (Agilent Technologies) with electron impact ionization. Spectral data was acquired in simultaneous selected ion monitoring/full scan (SIM/Scan) mode. Analytes were quantified with ChemStation vE.02.14 (Agilent Technologies) using internal standard methodologies against a 5 to 7 pt calibration curves.
Data Quality Assurance and Control
Assay variability was assessed by analyzing sample replicates and laboratory reference materials in each batch and found to be stable across the study. Data was corrected for surrogate losses (see Table S3). Analytes are not reported if the signal to noise ratio is <2, calculated concentrations are below the lowest calibrant, or surrogate recoveries are below 40%. Of the ∼150 plasma lipids measured, 80 metabolites met these reporting criteria.
Univariate Statistics
Significant changes in metabolite concentrations and enzyme activity indices (i.e. product to substrate ratios) were assessed in R (version 2.13.1) [20] by Mann-Whitney U test. To control for the false discovery rate (FDR) associated with multiple hypothesis testing, the p<0.05 significance criterion was adjusted to allow a maximum 10% probability (q = 0.1) of false positive detection [21]. The percent change in geometric mean (GM) between diabetic and non-diabetic cohorts was calculated using the following equation:
Discriminant Modeling
Partial least squares projection to latent structures-discriminant analysis (PLS-DA) [22] and orthogonal PLS-DA (OPLS-DA) [23] multivariate classification models were developed for the study population. Model performance was optimized using a novel method for iterative variable selection (IVS). A metabolite-driven OPLS-DA model was compared to a model built with clinical variables alone [6]. The clinical parameters included plasma concentrations of glucose, lactate, HbA1c, triglycerides, and both total and lipoprotein cholesterol, as well as age, body mass, and BMI. Classification performance was determined by the goodness of the prediction statistic Q2 [23] and area under the receiver operator characteristic curve (AUROC).
The IVS feature selection approach belongs to a larger class of “wrapper methods” [24], and implements a heuristic (i.e. experience based) algorithm to accomplish a randomized, forward and backward variable subset selection. The full feature set is stochastically divided into subsets that undergo iterative backward variable deletion and forward variable addition until model performance converges to a local maxima, based on the cross-validated R2 or Q2. After each iteration the algorithm-optimized variable sets are combined and the procedure is repeated until the model performance converges to the global maxima. Due to the small study population feature selection was not included in the model validation procedures. However, the degree of model overfitting by this procedure was independently assessed.
IVS was implemented in R [package: pls [25]] using non-linear partial least squares [26] on log transformed and centered metabolite concentrations, scaled to unit variance. Using normality transformations optimized for each metabolite did not alter model results, as compared to uniformly log transformed data. The optimal model was chosen based on maximum Q2 from 100 independent IVS optimized PLS-DA models. To simplify model interpretation the selected model parameters were used to construct an OPLS-DA model, which collapses informative model aspects into a single latent variable. The degree of IVS optimized model overfitting was estimated by carrying out 7 independent IVS feature selection embedded model validations. The full data set was randomly split between training (2/3) and test sets (1/3). The training data was used to develop 100 IVS optimized sub-models. Comparison of these models’ predictive performances for the held out test sets to that of the training sets were used to estimate the degree of model overfitting for the reported model. Based on this investigation, the performance statistics of the final model shown in Table S4 is expected to be lowered by ∼4±10% due overfitting.
OPLS-DA model training and validation was conducted in SIMCA-P+ v 12.0.1 (Umetrics; Umeå/Malmö, Sweden) by randomly assigning 2/3 of samples to training and remaining 1/3 to test sets. External OPLS-DA model cross-validation was conducted by repeating the training/testing procedure 3 times, to ensure that each subject was represented among one of the three test sets. The model diagnostic and performance statistics were calculated as the mean ± standard deviation of the 3 independent training/testing procedures. A summary model, including all subjects, was constructed to evaluate model scores and loadings. As a baseline for comparison, a clinical parameter OPLS-DA model (no IVS optimization) was generated using identical training and testing procedures.
Parameter Connectivity Network Generation
Similarities in metabolite correlation patterns were translated into proximities within a visualized network using multi-dimensionally scaled Euclidean distances calculated from 1-|Spearman’s ρ|. Correlations between measurements (e.g. metabolites), represented by nodes (i.e. shapes), are designated by edges (i.e. connecting lines) whose characteristics are determined based on the Spearman’s rank-order coefficient of correlation (ρ) between two respective species. High density regions among biosynthetically or biologically related metabolites are highlighted by ellipses defined by Hotelling’s T2 (p<0.05) for group multivariate normal distributions within the coordinate space. Network visualizations were generated using imDEV v 1.4 [27].
Results
Plasma Free Fatty Acids
In subjects with Type 2 diabetes, increases in plasma NEFA and stearoyl-CoA desaturase (SCD) activity, and reductions in peroxisome-dependent synthesis of very long chain (≥22 carbon) PUFAs (VLCPUFAs) were observed. For instance, diabetic subjects showed a 114% increase in circulating NEFA ( Table 2 ) and total NEFA was positively correlated with fasting glucose (ρ = 0.68, p<0.0001), with monounsaturated fatty acids (MUFA) showing the greatest magnitude of changes. The ratio of the MUFA species, palmitoleate (16∶1n7) and oleate (18∶1n9), to their saturated fatty acid (SFA) precursors, palmitate (16∶0) and stearate (18∶0), were used to estimate SCD activity [28]. As seen in Table 3 and Figure 1 , diabetes was associated with increases in both measures of SCD activity. A similar strategy was used to assess changes in VLCPUFA biosynthesis [29]. While the Δ6-desaturase (D6D) activity was unchanged in diabetic subjects, an increase in the elongase of very long chain fatty acids 2 (ELOVL2) activity is suggested by the ratio of 22∶5n3 over 20∶5n3 ( Table 3 ). The conversions of 22∶4n6 to 22∶5n6 and 22∶5n3 to 22∶6n3 rely on the combined activities of ELOVL2, D6D and Sprecher pathway chain shortening (SPCS) via peroxisomal β-oxidation [30]. While not explicit defined within this calculation, this ratio may be further affected by other enzymes in the very long chain fatty acid biosynthesis which elongate 24 carbon fatty acids, including ELOVL1 and ELOVL4, to produce fatty acids up to 38 carbons in length. Since ELOVL2 increased and D6D was unchanged, changes in the ratios of 22∶5n6 over 22∶4n6 and 22∶6n3 over 22∶5n3 may suggest a reduction in SPCS activity, and/or a general reduction in very long chain biosynthesis in diabetic subjects. Changes in this potential marker of VLCPUFA biosynthesis activity were inversely correlated with the SCD activity index (ρ = −0.45; p = 0.002). Results for all measured plasma NEFA species can be found in Table S5.
Table 2. Plasma non-esterified fatty acids (µM) in BMI-matched obese non-diabetic and type 2 diabetic African-American women.*.
| Lipid | non-diabetic(n = 12) | type 2 diabetic(n = 43) | ΔGM (%) |
| Σ NEFA | 290 [109, 653] | 621 [182, 1960] | 114 |
| Σ SFA | 135 [47.0, 341] | 281 [79.0, 787] | 109 |
| 14∶0 | 2.93 [1.00, 19.0] | 5.48 [1.00, 18.0] | 87 |
| 16∶0 | 92.7 [30.0, 260] | 196 [46.0, 582] | 111 |
| 18∶0 | 38.7 [14.0, 66.0] | 79.7 [30.0, 197] | 106 |
| 19∶0 | 0.05 [0.02, 0.13] | 0.09 [0.03, 0.37] | 85 |
| 20∶0 | 0.12 [0.05, 0.43] | 0.21 [0.01, 2.82] | 75 |
| Σ MUFA | 54.8 [21.6, 124] | 176 [48.0, 682] | 220 |
| Σ n7 fatty acids | 8.76 [3.00, 28.0] | 23.8 [8.00, 80.0] | 172 |
| Σ n9 fatty acids | 46.0 [18.0, 95.0] | 152 [40.0, 626] | 230 |
| 16∶1n7 | 3.08 [1.00, 17.0] | 8.78 [3.00, 38.0] | 185 |
| 18∶1n7 | 5.68 [3.00, 10.0] | 15.0 [5.00, 49.0] | 165 |
| 18∶1n9 | 45.1 [18.0, 95.0] | 150 [40.0, 620] | 231 |
| 20∶1n9 | 0.37 [0.10, 0.65] | 1.40 [0.14, 6.50] | 278 |
| Σ PUFA | 99.2 [41.0, 185] | 162 [33.0, 579] | 63 |
| 18∶2n6 | 54.8 [24.0, 102] | 103 [22.0, 363] | 87 |
| 9ct,11t-CLA | 0.85 [0.32, 2.00] | 1.29 [0.34, 4.00] | 52 |
| 18∶3n3 | 1.57 [0.34, 9.00] | 4.02 [1.00, 16.0] | 156 |
| 22∶4n6 | 0.42 [0.03, 2.00] | 0.74 [0.05, 3.00] | 76 |
| 22∶5n3 | 1.15 [0.12, 3.00] | 2.17 [0.28, 9.00] | 89 |
| Σ trans fatty acids | 44.7 [0.21, 3.65] | 92.1 [0.86, 13.0] | 106 |
| trans 16∶1n7 | 0.76 [0.19, 2.50] | 2.04 [0.50, 9.00] | 168 |
| trans 18∶2n6 | 0.28 [0.03, 1.00] | 1.17 [0.05, 4.00] | 211 |
Values are reported as geometric means [ranges] if changes in geometric means between groups are significant (Mann-Whitney U-test, p<0.05 with FDR adjustment at q = 0.1). For remaining measurements see Table S5.
Table 3. Estimated enzyme activities in BMI-matched obese non-diabetic and type 2 diabetic African-American women based on plasma NEFA product to substrate ratios.*.
| Activity Indices† | non-diabetic (n = 12) | type 2 diabetic (n = 43) | ΔGM (%) |
| SCD | 0.60 [0.45, 1.01] | 0.96 [0.29, 1.91] | 60 |
| 16∶1n7/16∶0 | 0.03 [0.02, 0.07] | 0.04 [0.02, 0.11] | 35 |
| 18∶1n9/18∶0 | 1.16 [0.88, 1.98] | 1.88 [0.56, 3.77] | 62 |
| D6D | 0.08 [0.05, 0.13] | 0.08 [0.05, 0.14] | – |
| 18∶3n6/18∶2n6 | 0.01 [ND, 0.02] | 0.01 [ND, 0.02] | – |
| 20∶3n3/20∶4n6 | 0.15 [0.08, 0.25] | 0.16 [0.10, 0.26] | – |
| ELOVL2 | 0.42 [0.24, 0.60] | 0.72 [0.23, 10.4] | 70 |
| 22∶4n6/20∶4n6 | 0.02 [ND, 0.04] | 0.03 [ND, 0.19] | – |
| 22∶5n3/20∶5n3 | 0.82 [0.47, 1.18] | 1.40 [0.46, 20.8] | 70 |
| ELOVL2/D6D/SPCS ‡ | 6.71 [3.99, 25.2] | 3.99 [1.48, 11.0] | −41 |
| 22∶5n6/22∶4n6 | 3.12 [0.96, 44.7] | 1.37 [0.23, 17.0] | −56 |
| 22∶6n3/22∶5n3 | 8.76 [5.76, 26.2] | 6.15 [2.71, 13.4] | −30 |
Values are reported as geometric means [ranges] if changes in geometric means between groups are significant (Mann-Whitney U-test, p<0.05 with FDR adjustment at q = 0.1).
SCD, steroyl-CoA desaturase; ELOVL2, elongase of very long chain fatty acids 2; D6D, delta 6 desaturase; SPCS, Sprecher pathway VLCPUFA chain shortening.
ELOVL2/D6D/SPCS has inherent dependencies on other enzymes in the very long chain fatty acid synthesis pathway including ELOVL1 and ELOVL4.
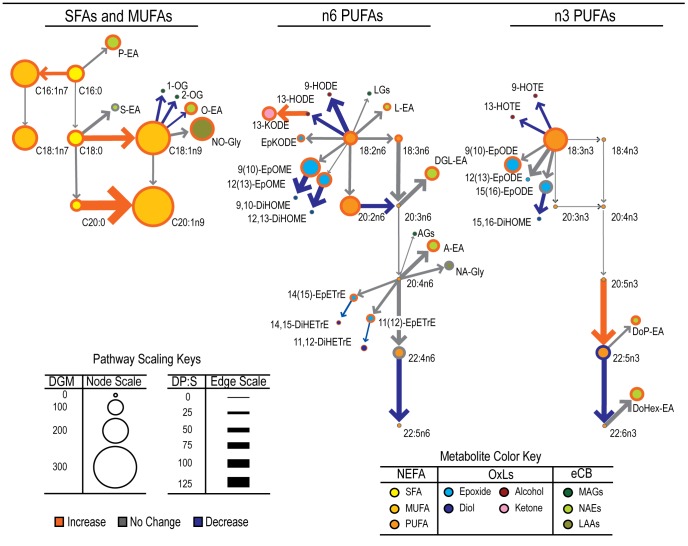
Figure 1. The type 2 diabetes-associated lipidomic changes projected in context of their biological relationships in obese African-American women.
Metabolites are represented by circular “nodes” linked by “edges” with arrows designating the direction of the biosynthetic gradient (i.e. substrate to product). Some metabolites are linked by more than one enzymatic step. Node sizes represent magnitudes of differences in plasma metabolite geometric means (ΔGM). Arrow widths represent magnitudes of changes in product over substrate ratios (ΔP:S). Colors of node borders and arrows represent the significance and direction of changes relative to non-diabetics as per the figure legend. Differences are significant at p<0.05 by Mann-Whitney U test adjusted for FDR (q = 0.1).
Plasma Free Oxylipins
While the total OxL concentration were unchanged in the diabetic state, increases in the concentrations of some fatty acid epoxides, diols and ketones were detected (Table 4). Each of the three measured linoleic acid (18∶2n6)-derived epoxides and one of three α-linolenic acid (18∶3n3)-derived epoxides were elevated from 47 to 127% in diabetic subjects ( Table 4 ). In contrast two of the 3 measured arachidonic acid (20∶4n6)-derived epoxides, were elevated by ∼50%. The vicinal or 1,2-diol metabolites of the 18 carbon fatty acid epoxide were unchanged, while the arachidonate-derived 14,15- and 11,12-dihydroxyeicosatrieneoates (DiHETrE) were elevated by ∼35%. The fatty acid ketone, 13-KODE, an NAD+ dependent dehydrogenation product of the 18∶2n6 alcohol 13-HODE [31], was also elevated 86% in diabetic subjects. Analogous fatty acid ketones including 9-KODE and the arachidonate-derived KETEs either did not meet reporting criteria or did not display significant diabetes-associated effects (Table S6 and Table S7).
Table 4. Concentrations of selected plasma oxylipins (nM) in BMI-matched obese non-diabetic and type 2 diabetic African-American women.*.
| Lipid | non-diabetic(n = 12) | type 2 diabetic(n = 43) | ΔGM (%) |
| Σ oxylipins | 79.2 [43.4, 370] | 91.1 [15.3, 365] | – |
| ΣC18 Epoxides | 5.10 [2.12, 50.7] | 9.4 [2.27, 60.6] | 84 |
| 9(10)-EpODE | 0.32 [0.10, 4.00] | 0.66 [0.10, 6.00] | 106 |
| 9(10)-EpOME | 1.18 [0.30, 11.0] | 2.68 [0.70, 20.0] | 127 |
| 12(13)-EpOME | 1.43 [0.50, 12.0] | 2.76 [0.70, 17.0] | 93 |
| EKODE | 1.69 [0.80, 18.0] | 2.43 [0.20, 14.0] | 44 |
| ΣC20 Epoxides | 1.14 [0.57, 5.14] | 1.61 [0.48, 9.20] | 41 |
| 11(12)-EpETrE | 0.46 [0.20, 2.00] | 0.70 [0.10, 4.00] | 52 |
| 14(15)-EpETrE | 0.24 [0.10, 1.00] | 0.35 [0.10, 2.00] | 46 |
| Σ Diols | 23.4 [9.50, 66.4] | 22.4 [2.29, 71.9] | – |
| 11,12-DiHETrE | 0.44 [0.32, 0.70] | 0.62 [0.34, 1.24] | 41 |
| 14,15-DiHETrE | 0.52 [0.38, 0.72] | 0.67 [0.38, 1.44] | 29 |
| Σ Ketones | 7.94 [4.05, 30.4] | 13.2 [1.29, 75.7] | – |
| 13-KODE | 5.63 [3.00, 25.0] | 10.5 [0.80, 68.0] | 86 |
Plasma N-acylethanolamides and Lipoamino Acids
With the exception of stearoyl-ethanolamide, the measured NAEs were significantly elevated in diabetic subjects ( Table 5 ). Of the two measured LAAs only N-oleoylglycine (NO-Gly) was elevated in diabetic subjects ( Table 5 ). Mean MAG concentrations were unchanged in association with diabetes (Table S8) but showed negative correlations with BMI in these obese cohorts.
Table 5. Plasma N-acylamides and lipoamino acids (nM) in BMI-matched obese non-diabetic and type 2 diabetic African-American women.* .
| Lipid | non-diabetic (n = 12) | Diabetic (n = 43) | ΔGM (%) |
| Σ NAE | 67.4 [27.0, 124] | 106 [25.0, 440] | 57 |
| P-EA | 9.59 [5.00, 40.0] | 18.9 [6.00, 162] | 97 |
| O-EA | 20.3 [8.00, 33.0] | 36.2 [9.00, 175] | 78 |
| L-EA | 7.91 [4.00, 27.0] | 11.4 [5.00, 44.0] | 44 |
| DGL-EA | 0.5 [0.30, 1.00] | 0.93 [0.30, 4.00] | 86 |
| A-EA | 2.09 [1.00, 3.00] | 3.57 [1.00, 8.00] | 71 |
| DoP-EA | 1.29 [0.20, 2.76] | 1.97 [0.68, 4.66] | 52 |
| DoHex-EA | 0.55 [0.30, 1.00] | 1.02 [0.40, 2.00] | 85 |
| Σ LAA | 8.67 [2.50, 36.1] | 21.0 [4.50, 88.6] | 142 |
| NO-Gly | 8.15 [2.00, 34.0] | 20.2 [4.00, 86.0] | 148 |
Values are reported as geometric means [ranges] if changes in geometric means between groups are significant (Mann-Whitney U-test, p<0.05 with FDR adjustment at q = 0.1). For remaining measurements see Table S8.
Combined Analysis of NEFA, OxL and eCB Changes
The lipidomic changes associated with diabetes are projected in context of their biochemical relationships in Figure 1 . Metabolites are represented by circular “nodes” with colored borders linked with “edges” represented by arrows designating the enzyme-dependent substrate to product transformation. Node color indicates chemical class while node size represents percent changes in metabolite concentrations. Edge widths represent percent changes in product to substrate ratios. The colors of node borders and edges indicate direction of significant changes, with gray indicating p>0.05.
In Figure 1 , SFA and MUFA metabolites are displayed at the left, while omega-6 and omega-3 PUFA metabolites are displayed in the center and right. Orange circled nodes indicate increasing concentrations in diabetic subjects, with the low abundance eicosenoic acid (20∶1n9) showing the largest increase (276%; Table 2 ). As with 18∶1n9, N-oleoylethanolamide (O-EA) and NO-Gly also increased in diabetics. However, while the NO-Gly/18∶1n9 ratio was unchanged, the O-EA/18∶1n9 decreased as indicated by a blue connecting arrow. All NAEs (lime green nodes) matched the observed increases in their parent NEFA as shown by the linkage of these lipids by gray edges. This holds true for the epoxides (cyan nodes) as well. Unlike the diols derived from 20∶4n6 epoxides, diabetes-associated increases in the 18∶2n6- and 18∶3n3-derived epoxides are not matched by significant changes in their diols.
Changes in indices of enzymatic activity are also highlighted in this figure. Increased SCD activity is indicated by the orange arrows between all SCD-linked SFAs and MUFAs (e.g. 16∶0 to 16∶1n7 and 18∶0 to 18∶1n9 nodes), while the diabetes-associated decrease in SPCS activity is reflected in the blue arrows linking 22∶6n3 with 22∶5n3 and 22∶4n6 with 22∶5n6.
Predictive Models for Type 2 Diabetes
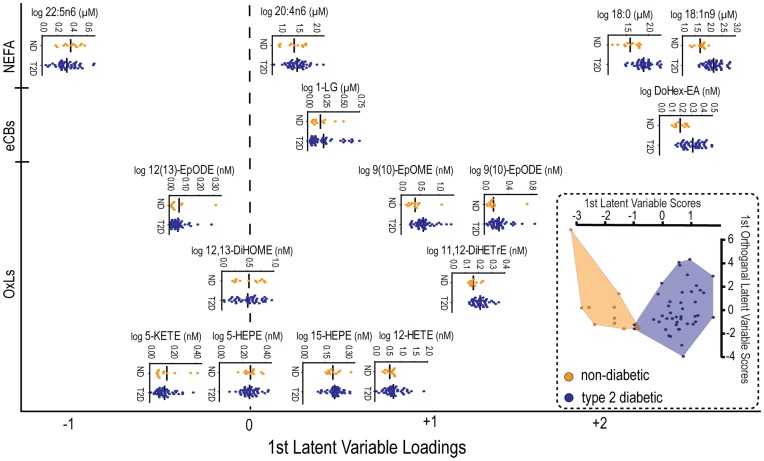
OPLS-DA models built either with metabolites or clinical parameters were excellent predictors of type 2 diabetes. However, the metabolite-based model had superior classification statistics (Q2 = 0.61 vs. 0.46; AUROC = 0.97 vs. 0.94; Table S4) and highlighted metabolic shifts associated with diabetes. OPLS-DA model scores for each subject ( Figure 2 ) correlate with both fasting glucose (r = 0.7, p<0.0001) and glycosylated hemoglobin (HbA1c; r = 0.5, p<0.05).
Figure 2. An OPLS-DA model built from 15 plasma lipids discriminates non-diabetic and diabetic cohorts.
Horizontal scatter plots of the log transformed concentrations for each model variable are shown. The horizontal arrangement of metabolite scatter plots is scaled to their loading in the discriminant model. A given species importance in the classification increases with increasing displacement from the origin (broken line). The direction of the displacement, left or right, designates whether the species was decreased (left) or increased (right) in the diabetic relative to the non-diabetic patients. The overall model discrimination performance is presented as a scatter plot of subject model scores (inset).
The model loading plot ( Figure 2 ) describes the distributions and intercorrelations for model components in the context of their importance in the classification model. Metabolites are arranged based on their loading on the models’ predictive latent variable. Species with the greatest displacement from the origin (hashed line) indicate the most influential model parameters; whereas, the direction of their displacement designates if the metabolite was decreased (left) or increased (right) in diabetic compared to non-diabetic cohorts. The loading plot indicates that 18∶1n9, its precursor 18∶0, the docosahexaenoyl-ethanolamide (DoHex-EA) and the VLCPUFA 22∶5n6 are the dominant discriminating variables. Together, 18∶0 and 18∶1n9 report on the increased SCD activity in diabetic subjects. Changes in SFA but not MUFA are correlated with the VLCPUFA model components, 20∶4n6 and docosapentaenoate (22∶5n6), which are themselves highly correlated (p<0.0001) and report on the diabetes-associated change in VLCPUFA biosynthesis. The elevation in 18∶2n6- and 18∶3n3-derived epoxides 9(10)-EpOMEs and 9(10)-EpODEs displayed positive correlation with SFA and MUFA. The retention of the 20∶4n6 metabolite 12-HETE in the model is also of particular interest. While the 12-HETE values between diabetic and non-diabetic cohorts did not reach significance (p = 0.3), ∼20% of the diabetic subjects showed substantially elevated 12-HETE concentrations.
Analysis of Intercorrelations Among Metabolites and Clinical Parameters
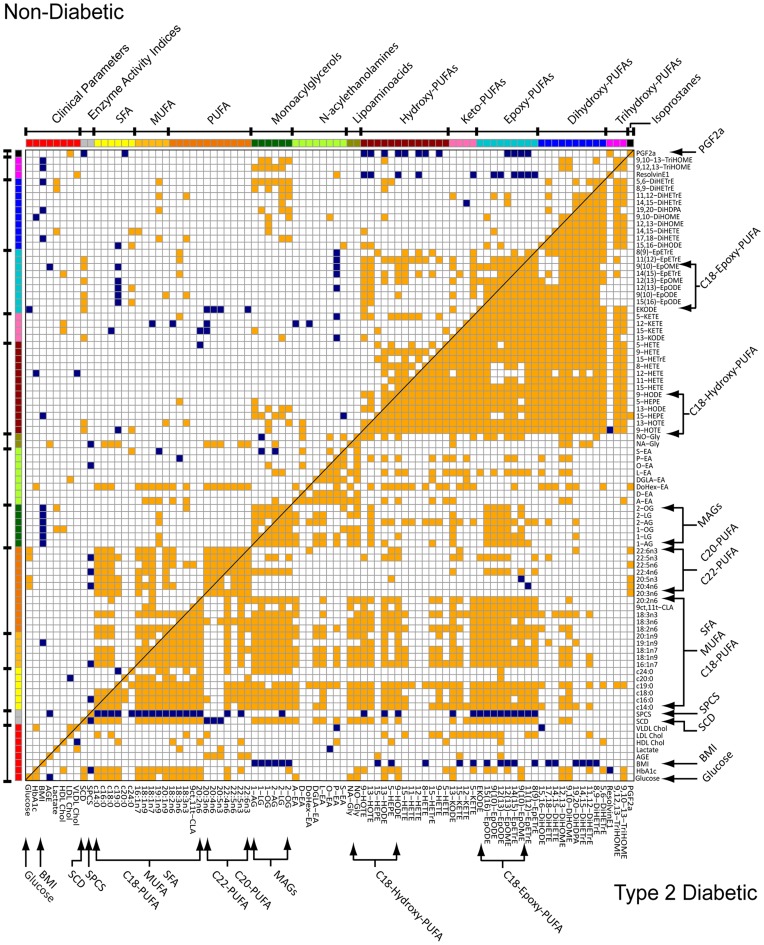
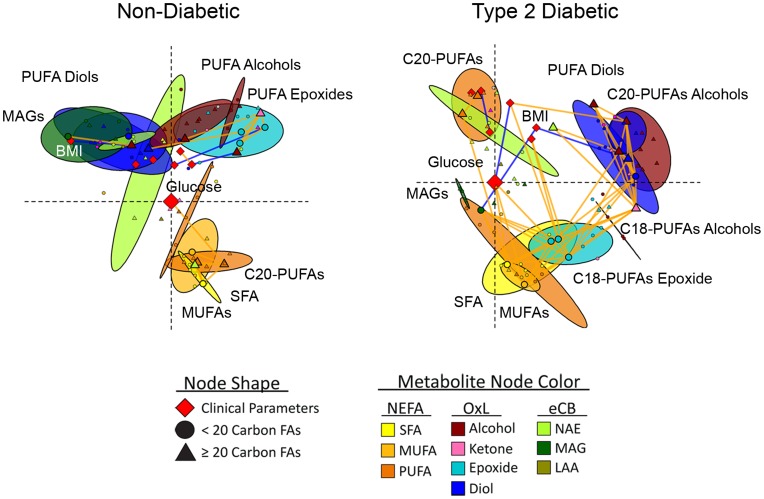
Correlation matrix heat maps ( Figure 3 ) and parameter connectivity networks (PCNs; Figure 4 ) are used to visualize diabetes-associated changes in parameter relationships. Diabetes was associated with the striking emergence of metabolite and pathway correlations not apparent in the non-diabetic condition. Correlations among variables are designated by orange (positive) or blue (negative) heat map intersections (p<0.05) and PCN connecting lines (p<0.05, non-diabetic; p<0.01, diabetic). In Figure 4 variables with similar intercorrelations are positioned closely, and metabolite classes are contained within ellipses representing their 95% probability regions. While powerful, this 3-dimensional projection can closely position uncorrelated compounds in the 2-dimensional display.
Figure 3. Analysis of correlations among all measured variables and estimated enzyme activities in non-diabetic and type 2 diabetic African-American women.
Significant (p<0.05) non-parametric Spearman’s correlations for non-diabetic (top left triangle) and type 2 diabetic (bottom right triangle) subjects are indicated by orange (positive) and blue (negative) intersections.
Figure 4. Parameter connectivity networks of metabolites and clinical parameters in African-American women with and without type 2 diabetes.
Spearman’s correlations were used to generate multi-dimensionally scaled parameter connectivity networks for variable intercorrelations. Networks were oriented with fasting glucose at the origin and SFA in the lower right quadrant. Colored ellipses represent the 95% probability locations of metabolite classes (Hoettlings T2, p<0.05). Nodes indicate clinical parameters (diamonds), <20-carbon fatty acid metabolites (circles) and ≥20-carbon fatty acid metabolites (triangles), with discriminant model variables and glucose enlarged. Significant correlations between species are designated by orange (positive) or blue (negative) connecting lines (p<0.05, non-diabetic; p<0.01, diabetic participants).
In short, variable intercorrelations are dramatically shifted in the diabetic state. The positive correlation between fasting glucose and C20-C22 PUFAs in the non-diabetic group is replaced by correlations with SFA, MUFAs, A-EA, and a selection of OxLs in the diabetic group. Conversely BMI is negatively correlated to MAGs in both groups, but negatively correlated with the 18 carbon epoxides and diols only in the diabetic cohort. In diabetes, LDL cholesterol gains a positive correlation with EpODEs and PUFA ketones ( Figure 3 ). This display also highlights the shifts in the SCD and the VLCPUFA activity as reported by the SPCS activity indices. In the OPLS-DA model, DoHex-EA was the key acylethanolamide discriminant of diabetes. DoHex-EA is also positively correlated with SFA, MUFA, and PUFAs in the non-diabetic group only, a pattern not shared by A-EA ( Figure 3 ). Finally, while OxL correlations are increased in diabetic subjects, class specific associations change. Notably, the 5-lipoxygenase-dependent products, 5-KETE and 5-HEPE increase their correlations with all PUFA-metabolites, while remaining uncorrelated to their parent lipids. In contrast, the isoprostanes became positively correlated with the >20 carbon PUFA pools in diabetic participants. These patterns of changes are well summarized in Figure 4 , where the connectivity networks highlight the migration of epoxides (cyan ellipse) toward the bulk of the NEFAs (yellow and orange ellipses), while the >20 carbon PUFAs become dissociated from this group. Thus, the NEFA fine structure shifts from a highly intercorrelated matrix, having weaker correlations with the omega-3 PUFAs, to a clear subdivision between SFA/MUFA/18 carbon PUFAs and >20 carbon PUFAs.
Discussion
The diabetes-associated perturbations of circulating lipid mediators and their relationships to plasma fatty acids are largely unexplored. Plasma NEFA concentrations reflect shift in NEFA uptake and release by adipose, liver and muscle [32], being dominated by adipose lipolysis in the fasted state [33]. Not unexpectedly, increases in plasma SFA and MUFA dominated the diabetes-associated changes in NEFA and were strong predictors of type 2 diabetes. However our targeted lipidomic survey uncovered subtle relationships between freely circulating NEFA, oxylipins, and endocannabinoids.
Based on the array of biosynthetically-related MUFA to SFA ratios, a net increase in systemic SCD activity was apparent in the diabetic cohort ( Table 3 and Figure 1 ). Increased SCD activity and/or expression have been reported in hypertriglyceridemia, obesity, nonalcoholic fatty liver disease (NAFLD), and the metabolic syndrome [34]–[37]. As the current study cohorts were BMI-matched and not hypertriglyceridemic [6], it is apparent that systemic SCD activity was elevated in the type 2 diabetic cohort beyond that expected by obesity alone. Such an observation is consistent with the increase in available glucoses, increasing de novo lipogenesis resulting in an elevation in SCD activity [35]
Despite the observed type 2 diabetes-associated increases in plasma NEFA, long and very long chain polyunsaturated fatty acids were not significantly elevated in this disease. In fact plasma markers of peroxisome-dependent VLCPUFA chain-shortening in both omega-3 and omega-6 pathways were reduced in diabetes, which also inversely correlated with SCD activity. Impaired VLCPUFA synthesis have previously been reported in the retina of streptozotocin-induced diabetic rats [38] and in NAFLD [39]. In fact, insulin resistance accelerates NAFLD in rodents [40] and observed changes in the VLCPUFA indices suggest an increased prevalence of NAFLD among the diabetic cohort.Diabetes and obesity are associated with pancreatic β-cell dysfunction, chronic inflammation, and vascular complications, conditions directly influenced by oxylipin metabolism [10]. While hyperglycemia and diabetes increase oxidative stress [41], and the total OxL concentration was positively correlated with fasting glucose, it did not increase in the diabetic compared to the non-diabetic cohort. Moreover, oxidative stress markers including the F2 isoprostanes were equivalent between cohorts. However, the concentration of multiple fatty acid epoxides, as well as several diols and ketones increased in association with diabetes. In particular, the epoxides of 18∶2n6 and 18∶3n3, and the 18∶2n6 ketone 13-KODE, were increased 2-fold and positively correlated with changes in SFA and MUFA. Changes in levels of 20∶4n6 and 20∶5n3-derived oxylipins were more subtle with ∼50% increases. Type 2 diabetes-associated changes in these species may arise from increased biosynthesis, decreased degradation, and or increased release of preformed metabolites. However, since fasting increases adipose lipolysis [33] and in this cohort epoxides and ketones are strongly correlated with plasma NEFA, we hypothesize that these species are derived from lipolysis of adipose. In support of this hypothesis, we have found that the adipose triglycerides of hamsters fed differing lipids in the diet preferentially accumulate 18 carbon species, with EpOMEs >5-fold over DiHOMEs, and KODEs ∼2-fold over HODEs in this pool [42].
Endocannabinoids and their related metabolites are important regulators of inflammation and energy balance, functioning through interaction with cannabinoid type 1 and type 2 receptors, transient receptor potential vanilloid type 1, and peroxisome proliferator activated receptors [11], [43], [44]. Previous investigations have reported increases in eCBs among both obese and diabetic humans [11]. Here plasma MAGs were inversely proportional to BMI, but unaffected by diabetes status. Conversely, plasma NAEs, including A-EA, were elevated and its concentrations correlated with SFA and MUFA in the diabetic cohort ( Figure 3 ). Given that insulin dependent suppression of plasma A-EA is inversely correlated with liver fat [14], the higher A-EA in this studies diabetic cohort is consistent with insulin resistance and an increased prevalence of NAFLD. Similarly, increases in the omega-3 DoHex-EA were strong predictors for the type 2 diabetes-associate phenotype ( Figure 2 ) and correlated with two 5-lipooxygenase metabolites, 5-HEPE and 5-KETE among diabetic participants. The basis for higher NAE metabolites in diabetes remains to be established, but an anti-inflammatory role is plausible. Specifically, the diabetes-associated elevation in SFA can activate Toll-Like Receptor 4 (TLR4), which has been reported to initiate inflammatory signaling and enhance NAE, but not MAG synthesis [45]. The NAEs may therefore act in a feedback loop to suppress inflammatory signaling through cannabinoid type 2 receptors [46].
In conclusion, to determine impacts of type 2 diabetes on the non-esterified plasma lipidome we quantified NEFA, oxylipins, acylethanolamides, lipoaminoacids and monoacylglycerides in weight-matched obese diabetic and non-diabetic cohorts. Diabetes-associated NEFA patterns indicate increases in SCD activity and decreases in VLCPUFA chain shortening, which may indicate impaired hepatic insulin sensitivity and/or fatty liver disease. Among diabetic participants, increases in 18 carbon epoxides and ketones correlate strongly with changes in SFA and MUFA, consistent with an enhanced release from adipose stores and/or suppressed degradation of these oxylipins. Type 2 diabetes is also associated with increases in NAEs and LAAs, but not MAGs. While the increases in NAE tone may constitute an adaptive mechanism to suppress inflammation developed in response to increases in circulating SFAs, MAG concentrations provide a diabetes-independent metabolic marker of body mass. Together, the observed changes in the plasma lipidome describe type 2 diabetes as a state of imbalance with respect to metabolic processes associated with fatty acid desaturation, VLCPUFA systhesis, adipose lipolysis, endocannabinoid tone and inflammatory activation. Oxylipin analysis revealed that these changes occur in the absence of systemic oxidative stress greater than that seen in obesity. Through this targeted lipidomic analysis, the following plasma markers were found as predictors of the type 2 diabetes phenotype: 1) 18∶1n9/18∶0, a marker of increased SCD activity; 2) 22∶5n6/22∶4n6, a marker of suppressed VLCPUFA synthesis; 3) 9(10)-EpOME and 9(10)-EpODE, putative markers of adipose lipolysis; 4) DoHex-EA, a marker of increased eCB system tone; 5) 1-LG, a metabolic marker of obesity.
Supporting Information
Diabetes x UCP3 G304A polymorphism 2-way ANOVA p-values for Type 2 diabetes associated metabolic changes. Type 2 Diabetes associated changes were not significantly different between evaluated UCP3 genotypes and no interactions were identified.
(DOC)
NAE, MAG and LAA mass transitions. This table lists the positive mode electrospray ionization mass transitions used to detect and quantify the listed analytical targets.
(DOC)
Analytical surrogate recoveries. The recovery and precision of analytical surrogates associated with non-esterified fatty acids (n = 1), endocannabinoids (n = 8) and oxylipins (n = 6) are reported.
(DOC)
Average modeling statistics for type 2 diabetes predictive models. Orthogonal projection to latent structures discriminate analysis (OPLS-DA) models were constructed using either metabolites or clinical parameters. Both models accurately discriminated diabetic status, with the metabolite-based model slightly out-performing the clinical model in terms of model fit and the area under the receiver operator characteristic curve for model based prediction.
(DOC)
Plasma non-esterified fatty acids (µM) in obese African-American women. Geometric mean and ranges are listed for all measured metabolites in this class.
(DOC)
Plasma eighteen carbon oxylipins (nM) in obese African-American women. Geometric mean and ranges are listed for all measured metabolites in this class for experimental groups with and without Type 2 diabetes.
(DOC)
Plasma twenty and twenty-two carbon oxylipins (nM) in obese African-American women. Geometric mean and ranges are listed for all measured metabolites in this class for experimental groups with and without Type 2 diabetes.
(DOC)
Plasma N-acylethanolamides (nM), lipoamino acids (nM) and monoacylglycerols (µM) in obese African-American women. Geometric mean and ranges are listed for all measured metabolites in this class for experimental groups with and without Type 2 diabetes.
(DOC)
Acknowledgments
We would like to thank William R. Keyes of the USDA-ARS-WHNRC for technical assistance with GC-MS analysis protocols. The USDA is an equal opportunity employer.
Funding Statement
This work was funded in part by the following: National Institute of General Medical Sciences-National Institutes of Health (NIGMS-NIH) T32-GM008799 (D.G.); intramural United States Department of Agriculture-Agricultural Research Service (USDA-ARS) Project 5306-51530-019-00D (J.W.N. and S.H.A.); National Institute of Diabetes and Digestive and Kidney Diseases (NIH-NIDDK) R01DK078328-01 (S.H.A.); NIH grants DK-038764, DK-083562, and P01 HL-055782 and the Merit Review program of the Department of Veterans Affairs (W.T.G.). The authors also acknowledge support from the research core facilities of the University of Alabama (UAB) Center for Clinical and Translational Science (UL1 RR025777), the UAB Nutrition and Obesity Research Center (P30-DK56336), and the UAB Diabetes Research and Training Center (P60 DK079626). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bergman RN, Ader M (2000) Free fatty acids and pathogenesis of type 2 diabetes mellitus. Trends Endocrinol Metab 11: 351–356. [DOI] [PubMed] [Google Scholar]
- 2. Matthews DR, Naylor BA, Jones RG, Ward GM, Turner RC (1983) Pulsatile insulin has greater hypoglycemic effect than continuous delivery. Diabetes 32: 617–621. [DOI] [PubMed] [Google Scholar]
- 3. Roden M, Stingl H, Chandramouli V, Schumann WC, Hofer A, et al. (2000) Effects of free fatty acid elevation on postabsorptive endogenous glucose production and gluconeogenesis in humans. Diabetes 49: 701–707. [DOI] [PubMed] [Google Scholar]
- 4. Komjati M, Bratusch-Marrain P, Waldhausl W (1986) Superior efficacy of pulsatile versus continuous hormone exposure on hepatic glucose production in vitro. Endocrinology 118: 312–319. [DOI] [PubMed] [Google Scholar]
- 5. Paolisso G, Scheen AJ, Giugliano D, Sgambato S, Albert A, et al. (1991) Pulsatile insulin delivery has greater metabolic effects than continuous hormone administration in man: importance of pulse frequency. J Clin Endocrinol Metab 72: 607–615. [DOI] [PubMed] [Google Scholar]
- 6. Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, et al. (2009) Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr 139: 1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fiehn O, Garvey WT, Newman JW, Lok KH, Hoppel CL, et al. (2010) Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS One 5: e15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng M, Che2 Z, Liang Y, Wang B, Chen X, et al. (2009) GC–MS Based Plasma Metabolic Profiling of Type 2 Diabetes Mellitus. Chromatographia: 941–948.
- 9. She P, Van Horn C, Reid T, Hutson SM, Cooney RN, et al. (2007) Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am J Physiol Endocrinol Metab 293: E1552–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luo P, Wang MH (2011) Eicosanoids, beta-cell function, and diabetes. Prostaglandins Other Lipid Mediat 95: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Marzo V, Piscitelli F, Mechoulam R (2011) Cannabinoids and endocannabinoids in metabolic disorders with focus on diabetes. Handb Exp Pharmacol: 75–104. [DOI] [PubMed]
- 12. Dipatrizio NV, Astarita G, Schwartz G, Li X, Piomelli D (2011) From the Cover: Endocannabinoid signal in the gut controls dietary fat intake. Proc Natl Acad Sci U S A 108: 12904–12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Marzo V, Goparaju SK, Wang L, Liu J, Batkai S, et al. (2001) Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature 410: 822–825. [DOI] [PubMed] [Google Scholar]
- 14.Di Marzo V, Verrijken A, Hakkarainen A, Petrosino S, Mertens I, et al.. (2009) Role of insulin as a negative regulator of plasma endocannabinoid levels in obese and nonobese subjects. Eur J Endocrinol. [DOI] [PubMed]
- 15. Lambert DM, Di Marzo V (1999) The palmitoylethanolamide and oleamide enigmas: Are these two fatty acid amides cannabimimetic? Current Medicinal Chemistry 6: 757–773. [PubMed] [Google Scholar]
- 16. Woodward DF, Carling RW, Cornell CL, Fliri HG, Martos JL, et al. (2008) The pharmacology and therapeutic relevance of endocannabinoid derived cyclo-oxygenase (COX)-2 products. Pharmacol Ther 120: 71–80. [DOI] [PubMed] [Google Scholar]
- 17. Sale MM, Lu L, Spruill IJ, Fernandes JK, Lok KH, et al. (2009) Genome-wide linkage scan in Gullah-speaking African American families with type 2 diabetes: the Sea Islands Genetic African American Registry (Project SuGAR). Diabetes 58: 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shearer GC, Harris WS, Pedersen TL, Newman JW (2010) Detection of omega-3 oxylipins in human plasma and response to treatment with omega-3 acid ethyl esters. J Lipid Res 51: 2074–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pace-Asciak CR (1989) One-step rapid extractive methylation of plasma nonesterified fatty acids for gas chromatographic analysis. J Lipid Res 30: 451–454. [PubMed] [Google Scholar]
- 20.R Development Core Team (2011) R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
- 21. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 57: 289–300. [Google Scholar]
- 22. Wold S, Sjostrom M, Eriksson L (2001) PLS-regression: a basic tool of chemometrics. Chemometrics and Intelligent Laboratory Systems 58: 109–130. [Google Scholar]
- 23.Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikström C, et al.. (2006) Multi- and megavariate data analysis - principles and applications.: Umetrics Academy.
- 24. Saeys Y, Inza I, Larranaga P (2007) A review of feature selection techniques in bioinformatics. Bioinformatics 23: 2507–2517. [DOI] [PubMed] [Google Scholar]
- 25.Wehrens R, Mevik B-H (2007) pls: Partial Least Squares Regression (PLSR) and Principal Component Regression (PCR). R package version 2.1–0 ed.
- 26.Wold H, editor (1966) Estimation of principal components and related models by iterative least squares. New York: Academic Press. 391–420 p.
- 27.Grapov D, Newman JW (2012) imDEV: a Graphical User Interface to R Multivariate Analysis Tools in Microsoft Excel. Bioinformatics. [DOI] [PMC free article] [PubMed]
- 28. Mar-Heyming R, Miyazaki M, Weissglas-Volkov D, Kolaitis NA, Sadaat N, et al. (2008) Association of stearoyl-CoA desaturase 1 activity with familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol 28: 1193–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Voss A, Reinhart M, Sankarappa S, Sprecher H (1991) The metabolism of 7,10,13,16,19-docosapentaenoic acid to 4,7,10,13,16,19-docosahexaenoic acid in rat liver is independent of a 4-desaturase. J Biol Chem 266: 19995–20000. [PubMed] [Google Scholar]
- 30. Ferdinandusse S, Denis S, Mooijer PA, Zhang Z, Reddy JK, et al. (2001) Identification of the peroxisomal beta-oxidation enzymes involved in the biosynthesis of docosahexaenoic acid. J Lipid Res 42: 1987–1995. [PubMed] [Google Scholar]
- 31. Earles SM, Bronstein JC, Winner DL, Bull AW (1991) Metabolism of oxidized linoleic acid: characterization of 13-hydroxyoctadecadienoic acid dehydrogenase activity from rat colonic tissue. Biochim Biophys Acta 1081: 174–180. [DOI] [PubMed] [Google Scholar]
- 32. Lewis GF, Carpentier A, Adeli K, Giacca A (2002) Disordered fat storage and mobilization in the pathogenesis of insulin resistance and type 2 diabetes. Endocr Rev 23: 201–229. [DOI] [PubMed] [Google Scholar]
- 33. Jelic K, Hallgreen CE, Colding-Jorgensen M (2009) A model of NEFA dynamics with focus on the postprandial state. Ann Biomed Eng 37: 1897–1909. [DOI] [PubMed] [Google Scholar]
- 34. Attie AD, Krauss RM, Gray-Keller MP, Brownlie A, Miyazaki M, et al. (2002) Relationship between stearoyl-CoA desaturase activity and plasma triglycerides in human and mouse hypertriglyceridemia. J Lipid Res 43: 1899–1907. [DOI] [PubMed] [Google Scholar]
- 35. Hulver MW, Berggren JR, Carper MJ, Miyazaki M, Ntambi JM, et al. (2005) Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab 2: 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tomita K, Teratani T, Yokoyama H, Suzuki T, Irie R, et al. (2011) Plasma free myristic acid proportion is a predictor of nonalcoholic steatohepatitis. Dig Dis Sci 56: 3045–3052. [DOI] [PubMed] [Google Scholar]
- 37. Sethom MM, Fares S, Feki M, Hadj-Taieb S, Elasmi M, et al. (2011) Plasma fatty acids profile and estimated elongase and desaturases activities in Tunisian patients with the metabolic syndrome. Prostaglandins Leukot Essent Fatty Acids 85: 137–141. [DOI] [PubMed] [Google Scholar]
- 38. Tikhonenko M, Lydic TA, Wang Y, Chen W, Opreanu M, et al. (2010) Remodeling of retinal Fatty acids in an animal model of diabetes: a decrease in long-chain polyunsaturated fatty acids is associated with a decrease in fatty acid elongases Elovl2 and Elovl4. Diabetes 59: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Puri P, Wiest MM, Cheung O, Mirshahi F, Sargeant C, et al. (2009) The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology 50: 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ota T, Takamura T, Kurita S, Matsuzawa N, Kita Y, et al. (2007) Insulin resistance accelerates a dietary rat model of nonalcoholic steatohepatitis. Gastroenterology 132: 282–293. [DOI] [PubMed] [Google Scholar]
- 41. Inoguchi T, Li P, Umeda F, Yu HY, Kakimoto M, et al. (2000) High glucose level and free fatty acid stimulate reactive oxygen species production through protein kinase C–dependent activation of NAD(P)H oxidase in cultured vascular cells. Diabetes 49: 1939–1945. [DOI] [PubMed] [Google Scholar]
- 42.Grapov D (2012) Application of Targeted Metabolomic Profiling and Multivariate Data Analysis for the Study of Nutrition and Disease [Dissertation]. Davis: University of California Davis. 236 p.
- 43. Everaerts W, Nilius B, Owsianik G (2010) The vanilloid transient receptor potential channel TRPV4: from structure to disease. Prog Biophys Mol Biol 103: 2–17. [DOI] [PubMed] [Google Scholar]
- 44. O’Sullivan SE (2007) Cannabinoids go nuclear: evidence for activation of peroxisome proliferator-activated receptors. British Journal of Pharmacology 152: 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu J, Batkai S, Pacher P, Harvey-White J, Wagner JA, et al. (2003) Lipopolysaccharide induces anandamide synthesis in macrophages via CD14/MAPK/phosphoinositide 3-kinase/NF-kappa B independently of platelet-activating factor. Journal of Biological Chemistry 278: 45034–45039. [DOI] [PubMed] [Google Scholar]
- 46. Lunn CA, Reich EP, Bober L (2006) Targeting the CB2 receptor for immune modulation. Expert Opin Ther Targets 10: 653–663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diabetes x UCP3 G304A polymorphism 2-way ANOVA p-values for Type 2 diabetes associated metabolic changes. Type 2 Diabetes associated changes were not significantly different between evaluated UCP3 genotypes and no interactions were identified.
(DOC)
NAE, MAG and LAA mass transitions. This table lists the positive mode electrospray ionization mass transitions used to detect and quantify the listed analytical targets.
(DOC)
Analytical surrogate recoveries. The recovery and precision of analytical surrogates associated with non-esterified fatty acids (n = 1), endocannabinoids (n = 8) and oxylipins (n = 6) are reported.
(DOC)
Average modeling statistics for type 2 diabetes predictive models. Orthogonal projection to latent structures discriminate analysis (OPLS-DA) models were constructed using either metabolites or clinical parameters. Both models accurately discriminated diabetic status, with the metabolite-based model slightly out-performing the clinical model in terms of model fit and the area under the receiver operator characteristic curve for model based prediction.
(DOC)
Plasma non-esterified fatty acids (µM) in obese African-American women. Geometric mean and ranges are listed for all measured metabolites in this class.
(DOC)
Plasma eighteen carbon oxylipins (nM) in obese African-American women. Geometric mean and ranges are listed for all measured metabolites in this class for experimental groups with and without Type 2 diabetes.
(DOC)
Plasma twenty and twenty-two carbon oxylipins (nM) in obese African-American women. Geometric mean and ranges are listed for all measured metabolites in this class for experimental groups with and without Type 2 diabetes.
(DOC)
Plasma N-acylethanolamides (nM), lipoamino acids (nM) and monoacylglycerols (µM) in obese African-American women. Geometric mean and ranges are listed for all measured metabolites in this class for experimental groups with and without Type 2 diabetes.
(DOC)