Abstract
A durum wheat consensus linkage map was developed by combining segregation data from six mapping populations. All of the crosses were derived from durum wheat cultivars, except for one accession of T. ssp. dicoccoides. The consensus map was composed of 1,898 loci arranged into 27 linkage groups covering all 14 chromosomes. The length of the integrated map and the average marker distance were 3,058.6 and 1.6 cM, respectively. The order of the loci was generally in agreement with respect to the individual maps and with previously published maps. When the consensus map was aligned to the deletion bin map, 493 markers were assigned to specific bins. Segregation distortion was found across many durum wheat chromosomes, with a higher frequency for the B genome. This high-density consensus map allowed the scanning of the genome for chromosomal rearrangements occurring during the wheat evolution. Translocations and inversions that were already known in literature were confirmed, and new putative rearrangements are proposed. The consensus map herein described provides a more complete coverage of the durum wheat genome compared with previously developed maps. It also represents a step forward in durum wheat genomics and an essential tool for further research and studies on evolution of the wheat genome.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-012-1939-y) contains supplementary material, which is available to authorized users.
Introduction
Durum wheat [Triticum turgidum (L.) subsp. turgidum (L.) convar. durum (Desf.)] is characterized by a large allotetraploid genome (AABB genome, 2n = 4x = 28, seven homoeologous groups—13,000 Mbp). Although durum wheat accounts for about 10 % of the total wheat production (World Grain Statistic, http://www.igc.org.uk), it is particularly important for its end products, which are mainly pasta, couscous and bulgur. Intense breeding activities have been carried out over the past century to improve the durum wheat varieties in terms of grain yield and quality, disease resistance, and drought tolerance (De Vita et al. 2007). Plant breeding is a long-term process and molecular tools can be used to overcome difficulties and to open the way for more rapid and efficient breeding strategies (Gupta et al. 2008; Tester and Langridge 2010). The phenotypic variation of many complex traits of agricultural or evolutionary importance is influenced by quantitative trait loci (QTL), their interactions, the environment, and the interactions between the QTL and the environment. Linkage mapping has been largely adopted in wheat to identify genomic regions that are involved in the control of complex traits (Breseghello and Sorrells 2006; Kuchel et al. 2007; Gupta et al. 2008), and many genetic maps of durum wheat have been published (Blanco et al. 1998; Nachit et al. 2001; Elouafi and Nachit 2004; Zhang et al. 2008; Peleg et al. 2008; Mantovani et al. 2008; Gadaleta et al. 2009). The early maps were based on restriction fragment length polymorphism (RFLP) markers (Blanco et al. 1998), while later the polymerase chain reaction (PCR)-based markers became dominant for genetic map construction, e.g. amplified fragment length polymorphisms (AFLPs) (Nachit et al. 2001) and simple sequence repeats (SSRs) (Peleg et al. 2008; Gadaleta et al. 2009). More recently, single-nucleotide polymorphisms (SNPs) have been included in durum wheat genetic maps (Zhang et al. 2008; Trebbi et al. 2011). The availability of SSR markers for durum wheat (Eujayl et al. 2002) and the development of high-throughput systems such as diversity array technology (DArT) (Jaccoud et al. 2001) have overcome the difficulties of genotyping large panels of genotypes with many loci. DArT technology in particular provides a highly multiplexed platform, which allows for rapid and cost-effective genome-wide genotyping (Wenzl et al. 2004; Akbari et al. 2006).
The construction of integrated maps provides the opportunity to increase the marker coverage with respect to individual maps. Consensus maps have been developed in many plant species: bread wheat (Somers et al. 2004), barley (Wenzl et al. 2006), rye (Gustafson et al. 2009), soybean (Hwang et al. 2009), red clover (Isobe et al. 2009), and ryegrass (Studer et al. 2010). In Vitis Vinifera L. (Vezzulli et al. 2008) and durum wheat (Trebbi et al. 2011), integrated maps have allowed new SNP markers to be mapped (501 and 157, respectively). The importance of the construction of consensus maps relies on the development of genetic tools that provide an essential basis for further genomic research.
Structural rearrangements revealed by colinearity failures among homoeologs can be genetically characterized with linkage maps. In wheat specific chromosome rearrangements have been documented in the A, B, and D genomes, e.g. the cyclic translocation involving chromosomes 4A, 5A, and 7B (Blanco et al. 1998). An integrated genetic map with high marker density can be useful to scan the whole genome for different kinds of chromosomal rearrangements as translocations, inversions, and duplications.
The main aim of the present study was to develop a high-density durum wheat consensus map derived from the integration of six individual maps, as a reference resource for durum wheat scientists in molecular breeding programs, as well as for comparative genomics within grass species. Along with the consensus map, the assignment to deletion bin map of many markers is herein reported. Regions with segregation distortion were identified by combining data from the single populations. Finally, an extensive analysis of multi-locus markers has allowed the identification of numerous chromosomal rearrangements.
Materials and methods
Segregating populations and genetic maps
A total of six mapping populations, developed to serve specific needs for qualitative and quantitative trait analysis, were used to integrate nearly 2,000 unique loci into a single consensus map: ‘Creso’ × ‘Pedroso’ [CP, 123 recombinant inbred lines (RILs) F8–F9], ‘Ofanto’ × ‘Cappelli’ (OC, 161 RILs F8–F9), ‘Cirillo’ × ‘Neodur’ (CN, 178 RILs F8–F9), ‘Ciccio’ × ‘Svevo’ (CS, 120 RILs F7–F8), ‘Latino’ × ‘Primadur’ (LP, 121 F2–F3 families), and ‘Messapia’ × ‘MG4343’ (MM, 65 RILs F7–F8). All of these genotypes are durum wheat varieties, except MG4343, which is an accession of Triticum turgidum (L.) sub-species dicoccoides.
The genetic map obtained from the MM population represented the first map to be constructed in tetraploid wheat (Blanco et al. 1998). This was achieved mainly with RFLP markers, then the map was enriched with SSRs (Blanco et al. 2004), and this version was used for the development of the consensus map. The CP population was used to study the genetic basis of durable leaf rust resistance of the cultivar Creso (Marone et al. 2009) and then implemented with additional 75 microsatellite markers (Marone et al., personal communication). The map derived from the OC population was developed to find chromosomal regions involved in the response to drought stress, as these two cultivars have different water-use efficiencies (Rizza et al. 2012). Furthermore, both CP and OC were used to map genes coding for different lipoxygenase isoforms on chromosome 4B (Verlotta et al. 2010). The CN map allowed the identification of a major and some minor QTL that explain the resistance against soil-borne cereal mosaic virus in the Neodur variety (Russo et al. 2011). As the CN map was implemented after the beginning of the work on consensus map, the dataset used in the present study is smaller than the one used by Russo et al. (2011) (290 vs. 426 markers). The CS map was developed for genetic and physical mapping of new expressed sequence tag (EST)-SSRs (Gadaleta et al. 2009), and it was then enriched with DArT markers to identify loci that are involved in seed protein content (Blanco et al. 2012). Finally, the LP population was used to determine the genetic basis of yellow pigment content and carotenoid accumulation (Blanco et al. 2011).
The main features of the segregating populations and the corresponding genetic maps are reported in Table 1, whereas Online resource 1 reports the different sources of markers used for the construction of the maps.
Table 1.
Summary of the six mapping populations used to construct the consensus map of durum wheat
| Parents | Population size | Markers | Total markers | Map length (cM) | Marker density (cM/marker) | Reference | |||
|---|---|---|---|---|---|---|---|---|---|
| SSR | EST-derived (SSR,STS) | DArT | Other markersa | ||||||
| ‘Creso’ × ‘Pedroso’ | 123 | 191 | 44 | 340 | – | 575 | 2,221.3 | 3.8 | Marone et al. (2009) |
| ‘Ofanto’ × ‘Cappelli’ | 161 | 154 | 23 | 437 | 4 | 618 | 1,649.4 | 2.6 | Verlotta et al. (2010) |
| ‘Cirillo’ × ‘Neodur’ | 178 | 71 | 7 | 212 | – | 290 | 1,568.5 | 5.4 | Russo et al. (2011) |
| ‘Ciccio’ × ‘Svevo’ | 120 | 132 | 110 | 584 | 4 | 830 | 1,765.8 | 2.1 | Gadaleta et al. (2009) |
| ‘Latino’ × ‘Primadur’ | 121 | 96 | 22 | 322 | – | 440 | 1,066.2 | 2.4 | Blanco et al. (2011, 2012) |
| ‘Messapia’ × ‘MG4343’ | 65 | 84 | – | – | 356 | 440 | 2,913.2 | 6.6 | Blanco et al. (2004) |
aRFLP, TRAP, biochemical and morphological markers
Construction of the consensus map
JoinMap 4.0 software (Van Ooijen and Voorips 2004) was used to reproduce the six durum wheat genetic maps and to generate the consensus map. The significance of deviations of the observed allelic frequencies from the expected ratios (1:1 or 1:2:1) (P < 0.01) was tested using Chi-squared analysis. The segregation data of each mapping population were first analyzed chromosome by chromosome, using a minimum logarithm of odds (LOD) score of 4 for grouping. The Kosambi mapping function (Kosambi 1943) and the “fixed order” of marker loci were used to reproduce linkage groups that correspond to the single maps previously developed. Subsequently, the linkage groups for each chromosome derived from the six mapping populations were joined using the “combine groups for map integration” function within the JoinMap software. When necessary, markers were removed from the analysis (i.e. markers with too much missing data), and the order was recalculated, until a stable and consistent order was obtained with respect to the single genetic maps. For some markers on chromosome 7B the order of consensus loci reproduced by the software was not consistent with that observed in individual maps. In this case, a fixed order was imposed based on data consistency in more than one individual map. The centromeres were positioned onto the consensus map at the midpoints between the most proximal markers on the short and long arms, according to common markers between this map and those of Röder et al. (1998), Somers et al. (2004) and Gadaleta et al. (2009).
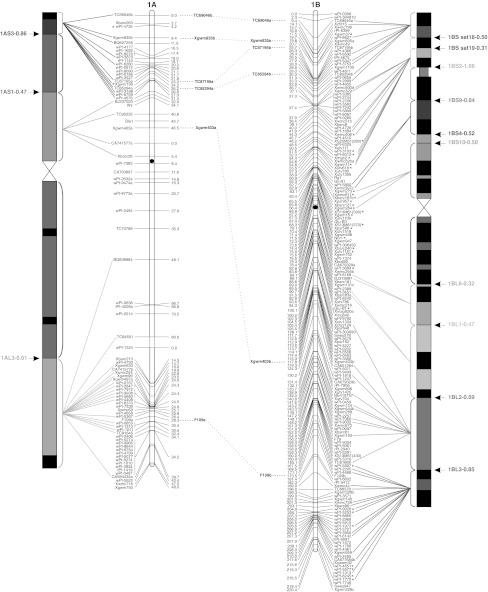
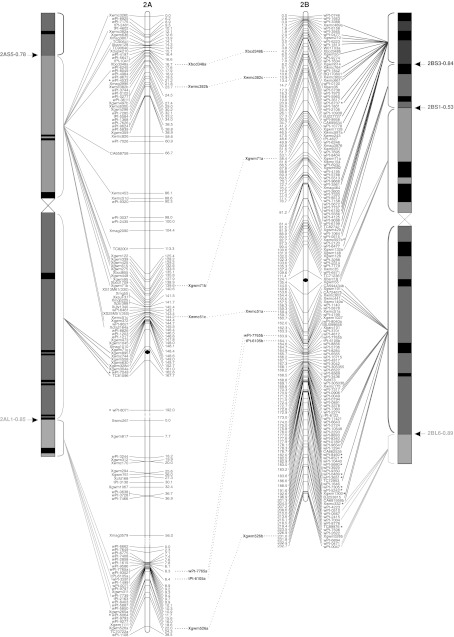
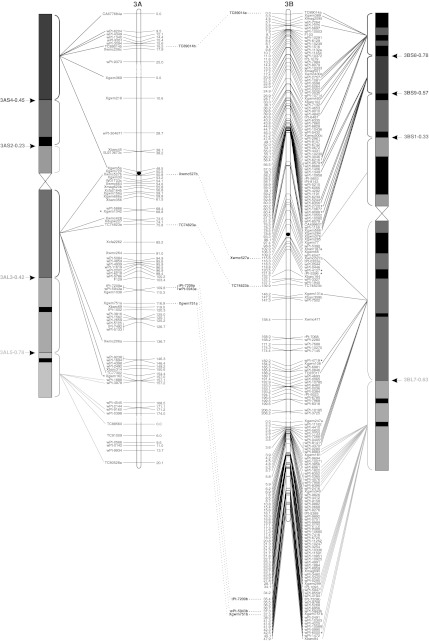
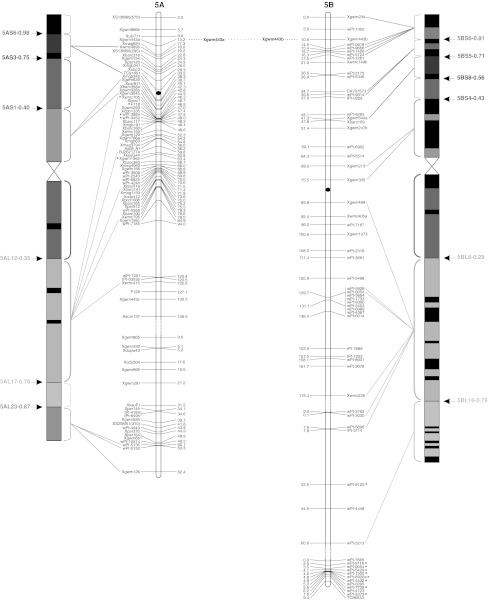
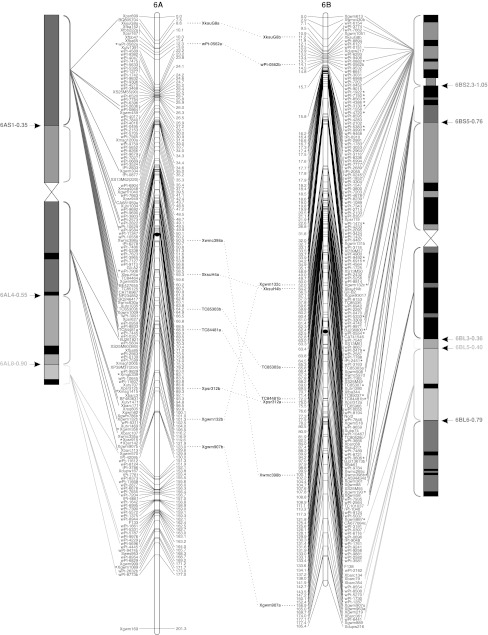
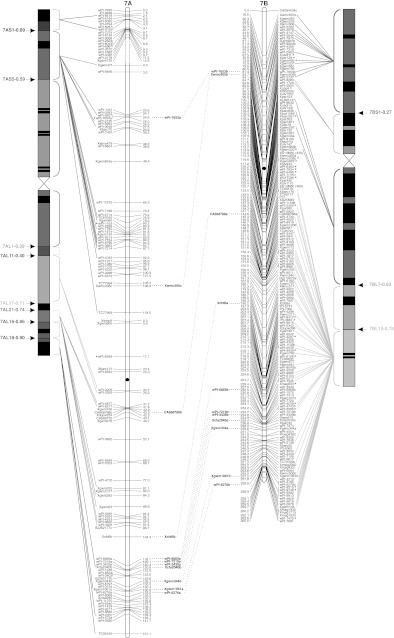
To validate the marker order of the consensus map, genomic SSR, EST-SSR, and DArT markers were assigned to specific deletion bins when possible, using the resources available. The physical positions of genomic SSR and EST-SSR markers were obtained by Francki et al. (2008) and Gadaleta et al. (2009), whereas for DArT markers this was determined by Francki et al. (2008) and by the deletion bin maps available at http://www.cereals.uk.net. Figure 1 reports the physical map described by Gadaleta et al. (2009) based on a set of 58 common wheat deletion lines dividing the A and B genome chromosomes into 94 bins (Endo and Gill 1996), in which physical mapping data derived from different sources were integrated.
Fig. 1.
The durum wheat consensus linkage map. The deletion bin map as reported by Gadaleta et al. (2009) was aligned with the consensus map and the colored lines show the genetic/physical relationships for each marker. Straight lines connect markers to specific bins, characterized by different colors. Dotted lines connect homoeologous loci that are reported between the two chromosomes of each pair. Markers with segregation distortion (P < 0.01) are marked with an asterisk. Markers identifying two or more mapped loci have the suffix “.a”, “.b”, and so on. The centromeres are indicated by black circles. Dashed lines on chromosomes indicate break points (color figure online)
Once developed, the consensus map was compared with the six individual maps and with the bread and durum wheat maps available in the literature (Röder et al. 1998; Somers et al. 2004; Francki et al. 2008; Gadaleta et al. 2009; Trebbi et al. 2011), in terms of marker order and genetic marker distance.
Analysis of colinearity between homoeologous chromosomes and structural rearrangements
Multi-locus markers were considered, and loci revealed by the same marker were inspected to identify homoeologous and paralogous loci. Chromosomal rearrangements were analyzed by studying orthologous loci mapped on non-homoeologous groups and by evaluating the information available in the literature on the position of all markers included in the putative translocated regions.
Results
Overview of the individual linkage maps
The six individual genetic maps differ according to the types and the numbers of markers, the lengths, and the marker densities of the linkage groups; their main features are summarized in Table 1.
The individual maps carried between 290 (CN) and 830 (CS) loci that are assembled into a number of linkage groups, ranging from 19 (LP) to 37 (CS). The marker density was from 2.1 cM/marker (CS) to 6.6 (MM) cM/marker. The highest proportion of markers (24 %) was positioned on chromosome 3B in CS, whereas the lowest (1.3 %) was mapped on chromosome 5A in CN. Generally, the homoeologous groups 3, 6, and 7 contained higher percentages of loci in most of the analyzed populations. On the contrary, the homoeologous groups 1 and 5 were characterized by a lower marker density. The percentages of mapped markers were higher on the B than the A genome (A genome: 38–45 % vs. B genome: 55–62 %), except for MM, where 55 % of the markers were mapped on the A genome. The lengths of the resulting maps varied between 1,066 cM (LP) and 2,913 cM (MM). Gaps with genetic distances greater than 20 cM were found in all of these linkage maps.
Construction of the consensus map
The durum wheat consensus map is composed of 1,898 loci (1,185 DArT, 388 genomic SSR, 166 EST-derived loci, and 159 other markers, e.g. RFLP, TRAP, biochemical and morphological loci) arranged into 27 linkage groups covering all 14 chromosomes (Table 2; see Online resource 2). A total of 650 mapped markers (216 PCR-based and 434 DArT) were common among at least two of the mapping populations (Table 3), while a total of 1,397 markers were unique to a specific mapping population. The total length of the integrated map was 3,058.6 cM. The mean length of the 27 linkage groups was 218.4 cM, although the chromosome size ranged from 142.7 cM (4B) to 286.5 cM (2A) (Table 2). The number of markers for each chromosome ranged from 58 (4B) to 261 (3B), with an average of 136.8. The average marker distance per chromosome was between 0.9 cM (4A and 3B) and 3.9 cM (5B), with an average density of one marker per 1.6 cM for the whole genome.
Table 2.
Density and distribution of markers in the consensus map of durum wheat
| Chromosome | Markers | Total markers | Map length (cM) | Marker density (cM/marker) | |||
|---|---|---|---|---|---|---|---|
| DArT | SSR | EST-derived (SSR, STS) | Othera | ||||
| 1A | 50 | 13 | 15 | 2 | 80 | 174.5 | 2.2 |
| 2A | 61 | 44 | 12 | 10 | 127 | 286.5 | 2.3 |
| 3A | 37 | 23 | 11 | 1 | 72 | 219.1 | 3.0 |
| 4A | 105 | 27 | 8 | 14 | 154 | 147.2 | 0.9 |
| 5A | 16 | 34 | 10 | 23 | 83 | 200.9 | 2.4 |
| 6A | 96 | 21 | 19 | 27 | 163 | 201.3 | 1.2 |
| 7A | 80 | 22 | 9 | – | 111 | 283.2 | 2.5 |
| Genome A | 445 | 184 | 84 | 77 | 790 | 1,512.7 | 1.9 |
| 1B | 84 | 45 | 17 | 27 | 173 | 220.4 | 1.2 |
| 2B | 112 | 35 | 15 | 2 | 164 | 232.7 | 1.4 |
| 3B | 229 | 24 | 8 | – | 261 | 254.1 | 0.9 |
| 4B | 22 | 21 | 6 | 9 | 58 | 142.7 | 2.4 |
| 5B | 48 | 12 | 2 | – | 62 | 245.3 | 3.9 |
| 6B | 119 | 25 | 16 | 16 | 176 | 185.4 | 1.0 |
| 7B | 126 | 42 | 18 | 28 | 214 | 265.5 | 1.2 |
| Genome B | 740 | 204 | 82 | 82 | 1,108 | 1,545.9 | 1.4 |
| Total | 1,185 | 388 | 166 | 159 | 1,898 | 3,058.6 | 1.6 |
aRFLP, TRAP, biochemical and morphological markers
Table 3.
Common markers across mapping populations
| Common markers between | Markers | Total common markers between populations | |||
|---|---|---|---|---|---|
| DArT | PCR-based | ||||
| Genome A | Genome B | Genome A | Genome B | ||
| Two populations | 102 | 171 | 77 | 70 | 420 |
| Three populations | 64 | 70 | 16 | 27 | 177 |
| Four populations | 12 | 15 | 12 | 12 | 51 |
| Five populations | – | – | 2 | – | 2 |
| Total | 178 | 256 | 107 | 109 | 650 |
The approximate location of the centromere was identified (Fig. 1) based on the integration of bread and durum wheat linkage (Röder et al. 1998; Somers et al. 2004; Gadaleta et al. 2009) and physical maps (Francki et al. 2008; http://www.cerealsdb.uk.net). Most markers were relatively evenly distributed along the chromosomes in terms of their genetic distances, although some regions were characterized by higher or lower marker densities (Fig. 1). The largest gap (that is a big genetic distance between two adjacent markers of the same linkage group) was 26.4 cM, between wPt-7185 (94.0 cM) and wPt-7201 (120.4 cM) on chromosome 5A of the consensus map. Additional gaps (more than 20 cM) were present on chromosomes 2A, 6A, 7A, 4B, and 5B. Some of the gaps of the individual maps were also present in the consensus map, whereas others were filled by integrating the information derived from different populations. For example, the 25 cM gap in CN on chromosome 4A (between wPt-6330 and wPt-1262) is well covered by markers in the consensus map.
The order of loci of the consensus map was in good agreement with the corresponding orders of the individual linkage maps. Some exceptions concerned changes that occurred within a small interval (<10 cM). Considering the relative marker distances, the largest discrepancy was observed between markers Xwmc716 and Xbarc213 on chromosome 1A, where the genetic distance was 30 cM in the consensus, while it was only 7 cM in CP.
The position of the loci mapped in this study was compared with recently published maps of bread and durum wheat. The ITMI map (Song et al. 2005) and the consensus map developed by Somers et al. (2004), which represent two well-saturated bread wheat maps, were considered for SSRs, while the bread and durum wheat maps described by Crossa et al. (2007), Peleg et al. (2008), and Trebbi et al. (2011) were taken into account for comparisons of both the SSR and DArT marker positions. The genetic positions of most SSR and DArT loci in the durum wheat consensus map showed consistency with their positions in the reference maps with few exceptions. In some cases, differences in the relative distances between two markers were found, although these were not considered as real discrepancies as they involved markers that identified multiple loci, with paralogous loci mapped on the same chromosome. As an example, the markers Xgwm443 and Xgwm666 were positioned on chromosome 5A at a distance of about 120 cM by Somers et al. (2004) and Song et al. (2005). Three loci were mapped in the durum wheat consensus map for Xgwm443: the Xgwm443b locus was located on chromosome 5B, while the Xgwm443a and Xgwm443c loci were located on chromosome 5A. Xgwm443a was 13.2 cM from Xgwm666, but Xgwm443c was positioned at 130.5 cM according to the bread wheat consensus map.
The marker order is in agreement with the maps published by Somers et al. (2004), Song et al. (2005) and Trebbi et al. (2011) for ten chromosomes (1B, 2A, 3A, 3B, 4A, 4B, 5A, 5B, 6A, and 7A), while some inconsistencies were seen for the remaining chromosomes.
A distance of about 40 cM was reported between the markers Xgwm497 and Xgwm99 by Somers et al. (2004) and Song et al. (2005) on the long arm of chromosome 1A, while the two markers were co-segregating on our consensus map. Nevertheless, the positions of these two markers in two individual maps (CS, 3 cM, and LP, 1 cM) supported the distances found in the consensus map, which was also confirmed by the durum wheat map reported by Elouafi and Nachit (2004), where the two markers were positioned 6 cM apart.
On the long arm of chromosome 1A, a group of DArT markers (which contained wPt-6754 and wPt-8644) were positioned more than 40 cM distal from the marker Xwmc716 by Trebbi et al. (2011), while in our consensus map, the marker Xwmc716 was 13 cM distal from the same group of DArT markers. Even if this region was contributed only by the CP individual map, the distances in our consensus map for the markers Xwmc716 and wPt-8644 agree with findings reported by Peleg et al. (2008).
On chromosome 2B, an inconsistency was found for the region between markers Xwmc149 and wPt-6643, which were localized in the telomeric region of the long arm by Trebbi et al. (2011), and in the pericentromeric region in our map. However, the detailed analysis of genetic marker distances within this interval in the individual maps validated the orientation reported here.
Another discrepancy in the order of the loci was observed between our durum wheat consensus map and previous reports for chromosome 6B for DArT markers. The marker wPt-7935 was mapped near the centromere, at <2 cM from the marker Xgwm193, whereas Trebbi et al. (2011) mapped the two markers at a distance of 60 cM; however, the two markers were placed 2 cM apart by Peleg et al. (2008). Finally, the marker wPt-0530 co-segregated with Xgwm344 in the map of Trebbi et al. (2011) on the long arm of chromosome 7B, while a distance of 20 cM was found between the two markers in our consensus map, and a similar distance (13 cM) was reported by Crossa et al. (2007).
Comparison of marker positions between the consensus and deletion bin maps
Four hundred and ninety-three markers (25.9 %; 256 DArTs, 153 gSSRs, and 83 EST-derived markers) were assigned to specific bins of the wheat deletion bin map (Fig. 1). Sixty-six of the 94 bins were covered by at least one marker, and the number of markers per bin ranged from 1 to 47, with a mean of 7.5. Except for the short arm of chromosome 4A, represented by only one molecular marker, the bin coverage with genetically mapped molecular markers was relatively good. For instance, all of the bins of chromosome 2B were covered by markers genetically mapped in the consensus map. These findings allowed anchoring between the consensus and physical maps of the durum wheat genome. A few discrepancies were noted. A group of nine DArTs (wPt-4533, wPt-4197, wPt-5647, wPt-0102, wPt-3611, wPt-0277, wPt-7626, wPt-9624, and wPt-5839) were genetically mapped on a region of chromosome 2AS between markers physically positioned in the pericentromeric region in the consensus map, while they were previously located to the short arm of the same chromosome (http://www.cerealsdb.uk.net). The marker wPt-3566, which was mapped to the centromeric region of chromosome 1B by Francki et al. (2008), was instead positioned on the long arm in the consensus map. On the same chromosome, based on the physical positions of the surrounding markers, the marker Xbarc8 was assigned to the bin 1BS2-1.06, while it was physically mapped to the bin 1BS10-0.00-0.50 by Gadaleta et al. (2009). In all of these examples, the map positions in the consensus map for these markers were in agreement with those reported by Trebbi et al. (2011).
The physical positions on chromosome 3A of DArTs wPt-5084, wPt-4859, wPt-1562, and wPt-2659, which were previously mapped to the bin 3AL5-0.78-1.00 (http://www.cerealsdb.uk.net), corresponded to the centromeric region in the consensus map. In this case, the genetic positions of these markers were supported by very good agreement between the CP and OC individual maps.
On chromosome 5A of the consensus map, the markers Xgwm186, Xbarc165, and Xbarc100 were on the long arm instead of the centromere, as described by Francki et al. (2008). Nevertheless, two loci were physically mapped by Sourdille et al. (2004) for the marker Xgwm186, in the centromeric region and the long arm, while the marker Xbarc100 was assigned to 5AL in the same study.
The marker Xbarc3, which was previously physically positioned in the bin 6AS1-0.35-1.00 (Francki et al. 2008), was genetically mapped in the pericentromeric region on 6AS in the consensus map, as according to Somers et al. (2004) and Goyal et al. (2005). Similarly, the marker CA668788b that was previously positioned in the pericentromeric region of chromosome 7AS (Gadaleta et al. 2009) was mapped on chromosome 7AL in the consensus map. This was supported by the genetic position of the marker, which was highly consistent in two individual maps (OC and CS).
Finally, Xwmc479 was previously mapped in the bin 7AS1-0.89 (Gadaleta et al. 2009), while in the consensus map its position is between Xgwm471 and Xgwm60, and it is physically mapped in the 7AS5-0.59 bin (Francki et al. 2008; Xue et al. 2008). This genetic position is as according to Somers et al. (2004) and Xue et al. (2008).
Of note, some genetically close markers were instead located physically in distant bins. For example, Xgwm5 and Xwmc527b were at a distance of only 2.3 cM on chromosome 3A, but they were physically mapped on the bins 3AS4-0.45-1.00 and 3AL3-0.00-0.42, respectively. Similar data were shown for this region by Somers et al. (2004). Analogous cases were observed on chromosome 4B for the markers CA663888 and Xgwm165, at a distance of only 0.3 cM, but mapped physically in the bins 4BS4-0.00-0.37 and 4BL5-0.86-1.00, respectively, and on chromosome 6A, where the co-mapping markers wPt-0357 and BJ261821 were instead mapped physically on the short and the long arms, respectively.
Segregation distortion
The percentage of skewed markers (P < 0.01) was different across the populations, varying from 0.6 to 11.8 % for CS and OC, respectively. Both co-dominant (SSR, STS, RFLP) and dominant (DArT) markers were subjected to deviation from the expected Mendelian 1:1 and 1:2:1 ratios. The distribution of the markers with segregation distortion was not uniform across chromosomes. Chromosomes 7B (OC) and 6B (LP) had 26 and 20 skewed markers, respectively, positioned in regions spanning less than 40 cM, and these were the chromosomes with the highest proportion of skewed markers. Chromosome 1A was the least affected by segregation distortion in all of the individual maps. Moreover, clusters of markers with skewed segregation were identified in all of the individual maps (data not reported).
A total of 149 markers (7.8 %) showed distorted segregation (P < 0.01) on the consensus map (Fig. 1). A similar ratio of skewed markers was found for DArT and SSR markers (7.3 and 7.9 %, respectively). Markers with segregation distortion were spread across all of the durum wheat chromosomes. Nevertheless, a statistically significant difference (P = 0.0032) was seen between the A and B genomes for the number of skewed markers: 120 markers on B and 29 on A (Fig. 1). The percentages with respect to the total number of markers positioned on each genome were 10.7 and 3.7 %, respectively. Considering all of the pairs of homoeologous chromosomes, the number of skewed markers was higher for those belonging to the B genome. The difference between homoeologs was low for groups 3 and 5, but generally high for the other pairs, e.g. on chromosome 1 there were two skewed markers (0.2 %) on 1A, compared with 30 skewed markers (17 %) on 1B.
Also in the consensus map skewed markers defined particular chromosome regions with distorted segregation, which therefore putatively contain loci involved in this phenomenon. In some cases, the skewed markers were spread over large chromosome regions, as seen for chromosomes 1A, 2A, 3A, 3B, and 7B. Single skewed regions were identified on chromosomes 4BS (pericentromeric region), 2BL and 5BL (telomeric region), while two regions were identified on chromosomes 1B (close to the centromere and close to the telomere of the long arm), and 6B (on the short arm and in the pericentromeric region). Finally, chromosome 7B showed a high number of skewed markers that were clustered in three regions all along the chromosome.
The clusters of skewed markers derived specifically from a single population, except for one region on chromosome 1B (from 44.3 to 100.1 cM), found to carry skewed markers in both MM and CP (see Online resource 3).
Analysis of colinearity between homoeologous chromosomes and structural rearrangements
Multi-locus markers were mapped in the present study, which we define as markers based on the same primer pair or clone that identified more than one locus.
A total of 214 loci were produced by 94 multi-locus markers, and out of these, 82 (56 SSRs, 20 DArTs and 6 RFLPs) were mapped on homoeologous chromosomes, whereas 132 (105 SSRs, 18 DArTs, 6 RFLPs, and 3 TRAPs) were assigned to intra-chromosome or non-homoeologous inter-chromosome positions (see Online resource 4). The colinearity between chromosomes within homoeologous groups was well conserved, as shown in Fig. 1, except for some markers. Six markers (4 EST-SSRs and 2 SSRs) that mapped to homoeologous sites were identified for homoeologous group 1. The same order and genetic positions characterized all of the loci, with the exception of Xgwm403, which showed a locus on the short arm of chromosome 1A and another one on the long arm of chromosome 1B.
Fourteen homoeologous loci (10 SSRs and 4 DArTs) were identified for group 2. Their order was highly consistent, although three markers (Xbcd348, Xwmc382, and Xgwm71) were mapped on the pericentromeric region of the short arm of chromosome 2A and on the telomeric region of the short arm of chromosome 2B. Furthermore, two DArT markers (wPt-7765 and tPt-6105) were located on the telomere of chromosome 2AL and on the centromeric area of chromosome 2BL.
A comparable number of homoeologous loci was found for group 3 (8 SSRs and 4 DArTs). There was consistent order and genetic positions of the markers along the chromosomes, except for the loci tPt-7209, wPt-5943, and Xgwm751, which mapped near to the centromere on chromosome 3AL and on the long arm of chromosome 3B. Only two homoeologous loci were mapped on both groups 4 and 5. While for group 5 the genetic position of the loci Xgwm443 on chromosomes 5AS and 5BS showed perfect correspondence, Xwmc617 was mapped in the pericentromeric region on chromosome 4AL and on the short arm of chromosome 4B. The same results were reported by Röder et al. (1998) for the marker Xgwm165, positioned on chromosomes 4AS and 4AL.
Nine markers (5 SSRs, 3 RFLPs, and 1 DArT) detected homoeologous loci on both chromosomes of group 6. The correspondence in terms of genetic position and marker order was good, except for Xpsr312, the loci of which were positioned around the centromere on chromosome 6AL and on the short arm of chromosome 6B, Xgwm132, which mapped on the long arm of chromosome 6A and on the short arm of chromosome 6B, and Xwmc398, for which two loci on chromosomes 6AS and 6BL were found. Finally, 22 homoeologous loci (12 SSRs and 10 DArTs) were identified on chromosomes 7A and 7B, showing colinearity between the chromosomes.
Different chromosomal rearrangements have occurred during wheat evolution, such as duplications, inversions, and translocations. The construction of the consensus map reported in the present study with high number of markers and marker density allowed the wheat genome to be scanned for identification of such rearrangements. Groups of multi-locus markers that had loci mapped on non-homoeologous positions were considered to be involved in putative translocations. Then the markers included in these regions were evaluated in terms of their genetic positions reported in the literature (Fig. 2).
Fig. 2.
Genetic mapping of putative translocations in durum wheat. For each translocation the chromosome pair is reported with the molecular markers that identified non-homoeologous loci (bold characters). Markers positioned on one chromosome in the durum consensus map are in normal characters, for which additional loci on the other chromosome are reported in literature
The markers wPt-7491 and Xgwm834 identified loci on chromosome 7AS and on the long arm of chromosome 4A, instead of chromosome 7B, as shown in Fig. 2a. Furthermore, Xbarc70, which mapped in the same region of chromosome 7AS, showed an additional locus on chromosome 4AL in OC (data not shown). Three additional markers (wPt-3648, wPt-3135, and Xgwm471), for which a single locus on chromosome 7AS was identified in the consensus map, were previously located on chromosome 4A (Song et al. 2005; Francki et al. 2008; Jing et al. 2009). Taken together, these data suggest that a translocation event took place between homoelogous groups 4 and 7. In addition, two markers (Xgwm274 and Xwmc232) located on the putative translocated region of chromosome 4AL were previously mapped on chromosome 7B in bread wheat (Somers et al. 2004; Semagn et al. 2006; Xue et al. 2008), which suggests that a segment of chromosome 7B moved to chromosome 4A.
The terminal portion of the long arm of chromosome 5A carries the locus Xgwm6b, which corresponds to the locus Xgwm6a on the long arm of chromosome 4B (Fig. 2b). The marker Xdupw43 was positioned in the same region of chromosome 5A, while it was previously mapped on chromosome 4B in hexaploid wheat by Semagn et al. (2006).
DArT marker wPt-8920 revealed two loci located on the long arm of chromosome 5B and on the short arm of chromosome 7B (Fig. 2c). In the same 7B region, there are two SSRs (Xgwm537 and Xwmc405) that were previously mapped on chromosome 5B in the durum × ssp. dicoccoides map Omrabi/600545//Omrabi (Xgwm537) (Elouafi and Nachit 2004) and in bread and durum wheat maps (Xwmc405) (Somers et al. 2004; Mantovani et al. 2008).
A putative translocation that occurred between chromosomes 3B and 7B is also shown in the present study (Fig. 2d). Three loci that correspond to the EST-SSR marker BJ213673 were mapped on the long arm of chromosome 7B and on the short arm of chromosomes 3A and 6B. Nevertheless, the same region of chromosome 7B carries three markers (Xgwm112, Xwmc540, and Xcfd6) that have already been shown to map on the corresponding region of chromosome 3B by Röder et al. (1998) and Somers et al. (2004), which suggests the occurrence of a T3B-7B translocation event.
Based on the same criteria adopted for previously described rearrangements, three more regions were identified that were characterized by a non-homoeologous relationship, even if two of them were based on only one marker. The segment of the short arm of chromosome 5B that contains the marker Xwmc149 corresponds to the distal region on the long arm of chromosome 2B (Fig. 2e). The loci Xgwm191 and Xwmc363 were also mapped in this region; they were previously positioned on chromosome 5B in hexaploid wheat (Röder et al. 1998; Somers et al. 2004; Song et al. 2005), which suggests that this chromosome region was affected by a translocation.
The regions on chromosomes 1A and 6A reported in Fig. 2f, were collinear for the markers wPt-9474, wPt-2632, and wPt-8773. The possibility that a rearrangement involved these chromosome segments is further supported by the marker Swes953, which is located near the locus wPt-9474b on the long arm of chromosome 6A, and was previously mapped in wheat on chromosome 1B (Li et al. 2007).
Finally, two paralogous loci for the marker Xgwm786 were mapped on chromosomes 5A and 6A (Fig. 2g). The short segment of chromosome 5A also includes Xwmc150, for which Somers et al. (2004) identified two loci on chromosomes 5A and 6A, in agreement with our results. A translocation T6B-5A can be hypothesized, as this region on chromosome 5A carries the locus Xbarc180, which was previously mapped on chromosome 6B in bread wheat (Song et al. 2005).
Discussion
Features of the durum wheat consensus map
The consensus map developed in the present study presents a much higher average density than that observed across the six individual maps. Values ranging from 11.8 cM/marker (Nachit et al. 2001) to 5.7 cM/marker (Mantovani et al. 2008) have been reported for published durum wheat individual linkage maps. Two consensus maps were previously developed for bread wheat by Somers et al. (2004) and Crossa et al. (2007). They mapped 1,235 and 1,644 markers, respectively, on the A, B, and D genomes. With respect to these maps, the durum consensus map herein described represents a large improvement. Due to the presence of regions which lack the statistical support for linking linkage groups belonging to the same chromosome, it was not possible to connect all the groups in a number corresponding to 14 chromosomes. The same feature characterized the map published by Trebbi et al. (2011). It is a durum wheat integrated map that they developed by merging two individual datasets. Even if the lengths of the two consensus maps were very similar, the consensus developed in the present study still has a greater number of markers (1,898 vs. 1,479), with better marker density (1.6 vs. 2.0 cM/marker). This is probably due to the integration of six individual maps that were derived from genotypes more genetically distant (11 durum wheat cultivars and one ssp. dicoccoides).
The high number of common markers, as well as the small differences in the recombination frequencies of the common markers across the different populations, can allow to position markers on a highly reliable reference map also in those regions that were poorly covered in the individual maps. Nevertheless, some regions with insufficient marker coverage (>20 cM) are still present in the consensus, which indicates a lack of polymorphism between specific parental pairs, due to recent co-ancestry, as suggested by pedigree data (data not shown). Alternatively, this can be due to the domestication bottle neck or a locally altered genetic versus physical distance ratio.
Segregation distortion
In the present study, a similar proportion of skewed markers was found for DArT and SSR markers, according to Akbari et al. (2006), Peleg et al. (2008) and Mantovani et al. (2008). The major occurrence of skewed markers on the B genome as compared with the A genome is a common feature for durum wheat linkage maps (Peleg et al. 2008; Mantovani et al. 2008). An opposite behavior was seen for the bread wheat map reported by Semagn et al. (2006), in which the chromosomes of the A genome always had many more skewed markers than the B counterpart, except for group 6.
As markers with distorted segregation were clustered into specific regions, and as the same regions were identified in different backgrounds, this indicates that this phenomenon is linked to genetic factors and is unlikely to be due to genotyping or scoring errors. Distorted segregation can be explained by reduced fitness of gametes and zygotes that is determined by loci with lethal or sub-lethal effects linked to molecular markers (Foolad et al. 1995; Blanco et al. 1998). In our study based on RIL populations, the gametophytic selection probably has a limited role in segregation distortion, compared with other studies based on double haploid populations (Cadalen et al. 1997). Furthermore, chromosomal rearrangements can also explain segregation distortion (Faure et al. 1993). Indeed, the region on chromosome 7BS putatively involved in the translocation described in the present study contained skewed markers (Fig. 1). Nearly half of the regions that included skewed markers in this consensus map were located around centromeres, which are regions that generally show reduced recombination (Faris et al. 2000), according to most of the aforementioned studies. Knowing the positions of the skewed regions is very important in plant breeding, as they can affect the association marker-QTL and the obtaining of the desired recombinants.
Analysis of colinearity between homoeologous chromosomes and structural rearrangements
Several of the 21 chromosomes of hexaploid wheat contain translocations of considerable sizes (Gale 1990), and the evolutionary evidence for translocations that have involved chromosome arms 4AL, 5AL, and 7BS has been firmly established (Chao et al. 1989; Naranjo 1990; Liu et al. 1992; Chen and Gustafson 1994, 1997; Blanco et al. 1998; Devos et al. 1995; Mickelson-Young et al. 1995; Nelson et al. 1995; Quarrie et al. 2005). This important cyclic translocation (4AL–5AL–7BS), evident also in our consensus map, has become an evolutionary signature of polyploidy wheat, which has conferred an adaptive advantage during the course of evolution (Devos et al. 1995). Possibly at the diploid level, chromosomes 4AL and 5AL exchanged terminal segments. Then, in tetraploid wheat, the distal portion of the chromosome 5A segment on chromosome 4AL was exchanged with a terminal segment from chromosome 7BS. Similar rearrangements have been documented in other grass genomes, such as rye, which has a translocation that corresponds to T4A–5A in wheat (King et al. 1994).
Furthermore, a pericentric inversion within chromosome 4A has also been reported in the literature (Miftahudin et al. 2004). Our results confirm this inversion (marker Xwmc617—Fig. 1) and suggest other inversions for the homoeologous groups 1 and 2 (markers Xgwm403 on group 1 and Xwmc51 on group 2—Fig. 1).
Moreover, the markers Xwmc51 and Xgwm71 show an additional locus on the same chromosome 2A, which suggests that a duplication of the region that comprises these markers has occurred during the evolution of the wheat genome. The presence of two distinct loci for the marker Xgwm71 was confirmed by Röder et al. (1998) and Somers et al. (2004).
The other translocation, identified with the consensus map, on the long arm of chromosome 7B (Fig. 2d) could correspond to the translocation T3B:7B described for T. dicoccoides by Badaeva et al. (2007). As the rearrangements reported by these authors were cytogenetically identified, the translocation identified in the present study represents the description of a translocation T3B–7B with the molecular markers genetically mapped.
Three putative translocations are herein suggested that have never been described before, for chromosomes 2B–5A, 1B–6A, 5A–6B (Fig. 2). Out of seven translocation events proposed in the present study, five are inter-genomic, which suggests that recombination between homoeologous chromosomes might be common in polyploids following interspecific wheat hybridization (Wendel and Wessier 2000). Transposable elements might represent one of the mechanisms that form the basis of intergenomic rearrangements. Indeed, the union of two genomes into a single nucleus can be perceived as the introduction of ‘foreign DNA’ and can induce the activation of transposable elements during polyploidization, as has been shown in cotton and wheat (Wendel and Wessier 2000; Chantret et al. 2005). Interestingly, the markers wPt-5964 and wPt-7214 that are positioned within and near to a region on chromosome 7A that is involved in a translocation, correspond to sequences that code for retrotransposons (data not shown).
It is reasonable to expect that the ongoing wheat sequencing projects (http://www.wheatgenome.org) will reveal the evolution of the chromosome structure and the distributions and physical locations of additional breakpoints and rearrangements within all of the chromosomes of both hexaploid and tetraploid wheat, as revealed by a recent analysis of the gene content in chromosome 5A (Vitulo et al. 2011).
The consensus map as a tool for advanced genetic studies in durum wheat
Consensus maps can be used to perform meta-QTL analysis (Haberle et al. 2009; Löffler et al. 2009), to combine genetic marker data and QTL characteristics (location, confidence interval, effects, and traits used for QTL detection) on a single map, as obtained from independent QTL mapping. This kind of study allows the optimal set of distinct consensus QTL (meta-QTL) to finally be estimated. Danan et al. (2011) clustered 144 QTL into 24 meta-QTL using a consensus map of potato that comprised 2,141 markers.
A limitation of QTL studies performed on individual biparental populations is often seen in the low number of molecular markers present in the region in which the QTL is identified. The projection onto high-density integrated maps of information derived from single population studies allows the QTL region to be enriched with many more molecular markers with respect to single populations. This thus represents an important advantage for fine QTL analysis, map-based gene cloning, transfer of QTL among different genetic backgrounds, and comparative studies between different genomes.
Linkage disequilibrium-based QTL analyses have been carried out for many agronomic traits related to grain yield and disease resistance in both common wheat (Crossa et al. 2007; Neumann et al. 2010) and durum wheat (Maccaferri et al. 2010). High-coverage integrated maps can have a positive effect on association mapping studies (Neumann et al. 2010; Trebbi et al. 2011). If unmapped markers have been used to genotype plants for association mapping, they can be tentatively ordered using the information provided by consensus maps. With more than 1,100 mapped DArT markers, the consensus map developed in the present study represents a valuable tool for association mapping of QTL on populations characterized with this high-throughput and cost-effective genotyping system.
The consensus map herein presented contains mapping data regarding 167 PCR-based molecular markers and 182 DArT markers for which the clone sequence is available (http://www.triticarte.com.au) and for which a match was found in the public databases using BLASTN and BLASTX searches (AM Mastrangelo, personal communication). Overall, a total of 349 markers related to expressed sequences were mapped in the present study, thus adding a functional value to the consensus map. The presence on a genetic map of markers derived from expressed sequences helps to associate candidate genes with identified QTL. Expressed-sequence-based molecular markers are also essential for colinearity studies among genomes, thus taking advantage of the available sequenced plant genome information, for faster fine mapping of QTL in species like wheat, for which the sequenced genome has not been released yet.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was supported by the Italian Ministry of Agriculture (MiPAAF), with the special grant AGRONANOTECH, MAPPA 5A, BIOMASSVAL and ESPLORA, and by the Ministry of Education, Universities and Research (MIUR), with the special grant AGROGEN. We are grateful to V. Giovanniello and P. Ferragonio for their technical support in the construction of three genetic maps, and to Dr. Christopher Berrie for scientific English language editorial assistance.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Footnotes
D. Marone and G. Laidò contributed equally to this study.
References
- Akbari M, Wenzl P, Caig V, Carlig J, Xia L, Yang S, Uszynski G, Mohler V, Lehmensiek A, Kuchel H, Hayden MJ, Howes N, Sharp P, Vaughan P, Rathmell B, Huttner E, Kilian A. Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet. 2006;113:1409–1420. doi: 10.1007/s00122-006-0365-4. [DOI] [PubMed] [Google Scholar]
- Badaeva ED, Dedkova OS, Gay G, Pukhalskyi VA, Zelenin AV, Berbard S, Berbard M. Chromosomal rearrangements in wheat: their types and distribution. Genome. 2007;50:907–926. doi: 10.1139/G07-072. [DOI] [PubMed] [Google Scholar]
- Blanco A, Bellomo MP, Cenci A, De Giovanni C, D’Ovidio R, Iacono E, Laddomada B, Pagnotta MA, Porceddu E, Sciancalepore A, Simeone R, Tanzarella OA. A genetic linkage map of durum wheat. Theor Appl Genet. 1998;97:721–728. doi: 10.1007/s001220050948. [DOI] [Google Scholar]
- Blanco A, Simeone R, Cenci A, Gadaleta A, Tanzarella OA, Porceddu E, Salvi S, Tuberosa R, Figliuolo G, Spagnoletti P, Röder MS, Korzun V. Extension of the Messapia × dicoccoides linkage map of Triticumturgidum (L.) Thell. Cell Mol Biol Letters. 2004;9:529–541. [PubMed] [Google Scholar]
- Blanco A, Mangini G, Giancaspro A, Giove S, Colasuonno P, Simeone R, Signorile A, De Vita P, Mastrangelo AM, Cattivelli L, Gadaleta A. Relationships between grain protein content and grain yield components through quantitative trait locus analyses in a recombinant inbred line population derived from two elite durum wheat cultivars. Mol Breed. 2012;30:79–92. doi: 10.1007/s11032-011-9600-z. [DOI] [Google Scholar]
- Blanco A, Colasuonno P, Gadaleta A, Mangini G, Schiavulli A, Simeone R, Digesù AM, De Vita P, Mastrangelo AM, Cattivelli L. Quantitative trait loci for yellow pigment concentration and individual carotenoid compounds in durum wheat. J Cereal Sci. 2011;54:255–264. doi: 10.1016/j.jcs.2011.07.002. [DOI] [Google Scholar]
- Breseghello F, Sorrells ME. Association analysis as a strategy for improvement of quantitative traits in plants. Crop Sci. 2006;46:1323–1330. doi: 10.2135/cropsci2005.09-0305. [DOI] [Google Scholar]
- Cadalen T, Boeuf C, Bernard S, Bernard M. An intervarietal molecular marker map in Triticumaestivum L. Em. Thell. and comparison with a map from a wide cross. Theor Appl Genet. 1997;94:367–377. doi: 10.1007/s001220050425. [DOI] [Google Scholar]
- Chantret N, Salse J, Sabot F, Rahman S, Bellec A, Laubin B, Dubois I, Dossat C, Sourdille P, Joudrier P, Gautier MF, Cattolico L, Beckert M, Aubourg S, Weissenbach J, Caboche M, Bernard M, Leroy P, Chalhoub B. Molecular basis of evolutionary events that shaped the Hardness locus in diploid and polyploidy wheat species (Triticum and Aegilops) Plant Cell. 2005;17:1043–1055. doi: 10.1105/tpc.104.029181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao S, Sharp PJ, Worland AJ, Warham EJ, Koebner RMD, Gale MD. RFLP-based genetic maps of wheat homoeologous group 7 chromosomes. Theor Appl Genet. 1989;78:495–504. doi: 10.1007/BF00290833. [DOI] [PubMed] [Google Scholar]
- Chen J, Gustafson JP. Physical mapping of genetically mapped molecular markers in homoeologous group 7 chromosomes of wheat by in situ hybridization. J Jiangsu Agric Coll. 1994;15:1–9. [Google Scholar]
- Chen J, Gustafson JP. Chromosomal rearrangement of wheat (T.aestivum) chromosome 4A by in situ hybridization. Chin J Genet. 1997;24:39–47. [Google Scholar]
- Crossa J, Burgueño J, Dreisigacker S, Vargas M, Herrera-Foessel SA, Lillemo M, Singh RP, Trethowan R, Warburton M, Franco J, Reynolds M, Crouch JH, Ortiz R. Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics. 2007;177:1889–1913. doi: 10.1534/genetics.107.078659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danan S, Veyrieras J-B, Lefebvre V. Construction of a potato consensus map and QTL meta-analysis offer new insights into the genetic architecture of late blight resistance and plant maturity traits. BMC Plant Biol. 2011;11:16. doi: 10.1186/1471-2229-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vita P, Matteu L, Mastrangelo AM, Di Fonzo N, Cattivelli L. Effetti del miglioramento genetico sul frumento duro prodotto in Italia nel XX secolo. Ital J Agron/Riv Agron. 2007;4:451–461. [Google Scholar]
- Devos KM, Dubcovsky J, Dvorak J, Chinoy CN, Gale MD. Structural evolution of wheat chromosomes 4A, 5A and 7B and its impact on recombination. Theor Appl Genet. 1995;91:282–288. doi: 10.1007/BF00220890. [DOI] [PubMed] [Google Scholar]
- Elouafi I, Nachit MM. A genetic linkage map of the Durum × Triticum dicoccoides backcross population based on SSRs and AFLP markers, and QTL analysis for milling traits. Theor Appl Genet. 2004;108:401–413. doi: 10.1007/s00122-003-1440-8. [DOI] [PubMed] [Google Scholar]
- Endo TR, Gill BS. The deletion stocks of common wheat. J Hered. 1996;87:295–307. doi: 10.1093/oxfordjournals.jhered.a023003. [DOI] [Google Scholar]
- Eujayl I, Sorrells ME, Baum M, Wolters P, Powell W. Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor Appl Genet. 2002;104:399–407. doi: 10.1007/s001220100738. [DOI] [PubMed] [Google Scholar]
- Faris JD, Haen KM, Gill BS. Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics. 2000;154:823–835. doi: 10.1093/genetics/154.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S, Noyer JL, Horry JP, Bakry F, Lanaud C, Gonzales de Leon D. A molecular marker-based linkage map of diploid bananas (Musa acuminata) Theor Appl Genet. 1993;87:517–526. doi: 10.1007/BF00215098. [DOI] [PubMed] [Google Scholar]
- Foolad MR, Arulsekar S, Becerra V, Bliss FA. A genetic map of Prunus based on an interspecific cross between peach and almond. Theor Appl Genet. 1995;91:262–269. doi: 10.1007/BF00220887. [DOI] [PubMed] [Google Scholar]
- Francki MG, Walker E, Crawford AC, Broughton S, Ohm HW, Barclay I, Wilson RE, McLean R. Comparison of genetic and cytogenetic maps of hexaploid wheat (Triticum aestivum L.) using SSR and DArT markers. Mol Genet Genomics. 2008;281:181–191. doi: 10.1007/s00438-008-0403-9. [DOI] [PubMed] [Google Scholar]
- Gadaleta A, Giancaspro A, Giove SL, Zacheo S, Mangini G, Simeone R, Signorile A, Blanco A. Genetic and physical mapping of new EST-derived SSRs on the A and B genome chromosomes of wheat. Theor Appl Genet. 2009;118:1015–1025. doi: 10.1007/s00122-008-0958-1. [DOI] [PubMed] [Google Scholar]
- Gale MD (1990) Comparative mapping in Triticeae genomes. In: McGuire PE, Corke H, Qualset CO (eds) Genome mapping of wheat and related species. Proceedings of the 1st public workshop international triticeae map initiative. University of California, West Sacramento, California, pp 17–19
- Goyal A, Bandopadhyay R, Sourdille P, Endo TR, Balyan HS, Gupta PK. Physical molecular maps of wheat chromosomes. Funct Integr Genomics. 2005;5:260–263. doi: 10.1007/s10142-005-0146-1. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Mir RR, Mohan A, Kumar J (2008) Wheat genomics: present status and future prospects. Intern J Plant Genom. ID 896451 [DOI] [PMC free article] [PubMed]
- Gustafson JP, Ma XF, Korzun V, Snape JW. A consensus map of rye integrating mapping data from five mapping populations. Theor Appl Genet. 2009;118:793–800. doi: 10.1007/s00122-008-0939-4. [DOI] [PubMed] [Google Scholar]
- Haberle J, Holzapfel J, Schweizer G, Hartl L. A major QTL for resistance against Fusarium head blight in European winter wheat. Theor Appl Genet. 2009;119:325–332. doi: 10.1007/s00122-009-1041-2. [DOI] [PubMed] [Google Scholar]
- Hwang TY, Sayama T, Takahashi M, Takada Y, Nakamoto Y, Funatsuki H, Hisano H, Sasamoto S, Sato S, Tabata S, Kono I, Hoshi M, Hanawa M, Yano C, Xia Z, Harada K, Kitamura K, Ishimoto M. High-density integrated linkage map based on SSR markers in soybean. DNA Res. 2009;16:213–225. doi: 10.1093/dnares/dsp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe S, Kolliker R, Hisano H, Sasamoto S, Wada T, Klimenko I, Okumura K, Tabata S. Construction of a consensus linkage map for red clover (Trifolium pratense L.) BMC Plant Biol. 2009;9:57. doi: 10.1186/1471-2229-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccoud D, Peng K, Feinstein D, Kilian A. Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001;29:e25. doi: 10.1093/nar/29.4.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing HC, Bayon C, Kanyuka K, Berry S, Wenzl P, Huttner E, Kilian A, Hammond-Kosack KE. DArT markers: diversity analyses, genomes comparison, mapping and integration with SSR markers in Triticum monococcum. BMC Genomics. 2009;10:458. doi: 10.1186/1471-2164-10-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JP, Purdie KA, Liu CJ, Reader SM, Orford SE, Pittaway TS, Miller TE. Detection of interchromosomal translocations within the Triticeae by RFLP analysis. Genome. 1994;37:882–887. doi: 10.1139/g94-125. [DOI] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1943;12:172–175. [Google Scholar]
- Kuchel H, Williams KJ, Langridge P, Eagles KA, Jefferies SP. Genetic dissection of grain yield in bread wheat. I. QTL analysis. Theor Appl Genet. 2007;115:1029–1041. doi: 10.1007/s00122-007-0629-7. [DOI] [PubMed] [Google Scholar]
- Li S, Jia J, Wei X, Zhang X, Li L, Chen H, Fan Y, Sun H, Zhao X, Lei T, Xu Y, Jiang F, Wang H, Li L. A intervarietal genetic map and QTL analysis for yield traits in wheat. Mol Breed. 2007;20:167–178. doi: 10.1007/s11032-007-9080-3. [DOI] [Google Scholar]
- Liu CJ, Devos KM, Chinoy CN, Atkinson MD, Gale MD. Non-homoeologous translocations between group 4, 5 and 7 chromosomes in wheat and rye. Theor Appl Genet. 1992;83:305–312. doi: 10.1007/BF00224276. [DOI] [PubMed] [Google Scholar]
- Löffler M, Schon CC, Miedaner T. Revealing the genetic architecture of FHB resistance in hexaploid wheat (Triticum aestivum L.) by QTL meta analysis. Mol Breed. 2009;23:473–488. doi: 10.1007/s11032-008-9250-y. [DOI] [Google Scholar]
- Maccaferri M, Sanguineti MC, Demontis A, El-Ahmed A, Garcia del Moral L, Maalouf F, Nachit M, Nserallah N, Ouabbou H, Rhouma S, Royo C, Villegas D, Tuberosa R. Association mapping in durum wheat grown across a broad range of water regimes. J Exp Bot. 2010;62:409–438. doi: 10.1093/jxb/erq287. [DOI] [PubMed] [Google Scholar]
- Mantovani P, Maccaferri M, Sanguineti MC, Tuberosa R, Catione I, Wenzl P, Thomson B, Carling J, Huttner E, De Ambrogio E, Kilian A. An integrated DArT-SSR linkage map of durum wheat. Mol Breed. 2008;22:629–648. doi: 10.1007/s11032-008-9205-3. [DOI] [Google Scholar]
- Marone D, Del Olmo AI, Laidò G, Sillero JC, Emeran AA, Russo MA, Ferragonio P, Giovanniello V, Mazzucotelli E, De Leonardis AM, De Vita P, Blanco A, Cattivelli L, Rubiales D, Mastrangelo AM. Genetic analysis of durable resistance against leaf rust in durum wheat. Mol Breed. 2009;24:25–39. doi: 10.1007/s11032-009-9268-9. [DOI] [Google Scholar]
- Mickelson-Young L, Endo TR, Gill BS. A cytogenetic ladder-map of the wheat homoeologous group 4 chromosomes. Theor Appl Genet. 1995;90:1007–1011. doi: 10.1007/BF00222914. [DOI] [PubMed] [Google Scholar]
- Miftahudin RK, Ma XF, Mahmoud AA, Layton J, Milla MA, Chikmawati T, Ramalingam J, Feril O, Pathan MS, Momirovic GS, Kim S, Chema K, Fang P, Haule L, Struxness H, Birkes J, Yaghoubian C, Skinner R, McAllister J, Nguyen V, Qi LL, Echalier B, Gill BS, Linkiewicz AM, Dubcovsky J, Akhunov ED, Dvorák J, Dilbirligi M, Gill KS, Peng JH, Lapitan NL, Bermudez-Kandianis CE, Sorrells ME, Hossain KG, Kalavacharla V, Kianian SF, Lazo GR, Chao S, Anderson OD, Gonzalez-Hernandez J, Conley EJ, Anderson JA, Choi DW, Fenton RD, Close TJ, McGuire PE, Qualset CO, Nguyen HT, Gustafson JP. Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics. 2004;168:651–663. doi: 10.1534/genetics.104.034827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachit MM, Elouafi I, Pagnotta MA, El Saleh A, Iacono E, Labhilili M, Asbati A, Azrak M, Hazzam H, Benscher D, Khairallah M, Ribaut JM, Tanzarella OA, Porceddu E, Sorrells ME. Molecular linkage map for an intraspecific recombinant inbred population of durum wheat (Triticum turgidum L. var durum) Theor Appl Genet. 2001;102:177–186. doi: 10.1007/s001220051633. [DOI] [Google Scholar]
- Naranjo T. Chromosome structure of durum wheat. Theor Appl Genet. 1990;79:397–400. doi: 10.1007/BF01186085. [DOI] [PubMed] [Google Scholar]
- Nelson JC, Sorrels ME, Van Deynze AE, Lu YH, Atkinson M, Bernard M, Leroy P, Faris JD, Anderson JA. Molecular mapping of wheat: major genes and rearrangements in homoeologous groups 4, 5 and 7. Genetics. 1995;141:721–731. doi: 10.1093/genetics/141.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann K, Kobiljski B, Denčić S, Varshney RK, Borner A. Genome-wide association mapping: a case of study in bread wheat (Triticum aestivum L.) Mol Breed. 2010;27:37–58. doi: 10.1007/s11032-010-9411-7. [DOI] [Google Scholar]
- Peleg Z, Saranga Y, Suprunova T, Ronin Y, Röder MS, Kilian A, Korol AB, Fahima T. High-density genetic map of durum wheat x wild emmer wheat based on SSR and DArT markers. Theor Appl Genet. 2008;117:103–115. doi: 10.1007/s00122-008-0756-9. [DOI] [PubMed] [Google Scholar]
- Quarrie SA, Steed A, Calestani C, Semikhodskii A, Lebreton C, Chinoy C, Steele N, Pljevljakusic D, Waterman E, Weyen J, Schondelmaier J, Habash DZ, Farmer P, Saker L, Clarkson DT, Abugalieva A, Yessimbekova M, Turuspekov Y, Abugalieva S, Tuberosa R, Sanguineti MC, Hollington PA, Araguès R, Royo A, Dodig D. A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring × SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor Appl Genet. 2005;110:865–880. doi: 10.1007/s00122-004-1902-7. [DOI] [PubMed] [Google Scholar]
- Rizza F, Ghashghaie J, Meyer S, Matteu L, Mastrangelo AM, Badeck F-W. Constitutive differences in water use efficiency between two durum wheat cultivars. Field Crops Res. 2012;120:49–60. doi: 10.1016/j.fcr.2011.09.001. [DOI] [Google Scholar]
- Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW. A microsatellite map of wheat. Genetics. 1998;149:2007–2023. doi: 10.1093/genetics/149.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo MA, Ficco DBM, Marone D, De Vita P, Vallega V, Rubies-Autonell C, Ratti C, Ferragonio P, Giovanniello V, Pecchioni N, Cattivelli L, Mastrangelo AM. A major QTL for resistance to soil-borne cereal mosaic virus derived from an old Italian durum wheat cultivar. J Plant Inter. 2011 [Google Scholar]
- Semagn K, Bjornstad A, Skinnes H, Maroy AG, Tarkegne Y, William M. Distribution of DArT, AFLP, and SSR markers in a genetic linkage map of a double-haploid hexaploid wheat population. Genome. 2006;49:545–555. doi: 10.1139/G06-002. [DOI] [PubMed] [Google Scholar]
- Somers JD, Isaac P, Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.) Theor Appl Genet. 2004;109:1105–1114. doi: 10.1007/s00122-004-1740-7. [DOI] [PubMed] [Google Scholar]
- Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J, Gill BS, Ward R, Cregan PB. Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet. 2005;110:550–560. doi: 10.1007/s00122-004-1871-x. [DOI] [PubMed] [Google Scholar]
- Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M. Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.) Funct Integr Genomics. 2004;4:12–25. doi: 10.1007/s10142-004-0106-1. [DOI] [PubMed] [Google Scholar]
- Studer B, Kölliker R, Muylle H, Asp T, Frei U, Roldán-Ruiz I, Barre P, Barth S, Skøt L, Armstead IP, Dolstra O, Roulund N, Nielsen KK, Lübberstedt T. EST-derived SSR markers used as anchor loci for the construction of a consensus linkage map in ryegrass (Lolium spp.) BMC Plant Biol. 2010;10:177. doi: 10.1186/1471-2229-10-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Langridge P. Breeding technologies to increase crop production in a changing world. Science. 2010;317:818–822. doi: 10.1126/science.1183700. [DOI] [PubMed] [Google Scholar]
- Trebbi D, Maccaferri M, Heer P, Sørensen A, Giuliani S, Salvi S, Sanguineti MC, Massi A, Vossen EAG, Tuberosa R. High-throughput SNP discovery and genotyping in durum wheat (Triticum durum Desf.) Theor Appl Genet. 2011;123:555–569. doi: 10.1007/s00122-011-1607-7. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW, Voorips RE (2004) JoinMap Version 3.0, Software for the calculation of genetic linkage maps. Kyazma BV, Wageningen
- Verlotta A, De Simone V, Mastrangelo AM, Cattivelli L, Papa R, Trono D. Insight into durum wheat Lpx-B1: a small gene family coding for the lipoxygenase responsible for carotenoid bleaching in mature grains. BMC Plant Biol. 2010;10:263. doi: 10.1186/1471-2229-10-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli S, Troggio M, Coppola G, Jermakow A, Cartwright D, Zharkikh A, Stefanini M, Grando MS, Viola R, Adam-Blondon AF, Thomas M, This P, Velasco R. A reference integrated map for cultivated grapevine (Vitis vinifera L.) from three crosses, based on 283 SSR and 501 SNP-based markers. Theor Appl Genet. 2008;117:499–511. doi: 10.1007/s00122-008-0794-3. [DOI] [PubMed] [Google Scholar]
- Vitulo N, Albiero A, Forcato C, Campagna D, Dal Pero F, Bagnaresi P, Colaiacovo M, Faccioli P, Lamontanara A, Šimková H, Kubaláková M, Perrotta G, Facella P, Lopez L, Pietrella M, Gianese G, Doležel J, Giuliano G, Cattivelli L, Valle G, Stanca AM. First survey of the wheat chromosome 5A composition through a next generation sequencing approach. PLoS One. 2011;6:e26421. doi: 10.1371/journal.pone.0026421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Wessier SR. Retrotransposon-mediated genome evolution on a local ecological scale. Proc Natl Acad Sci USA. 2000;97:6250–6252. doi: 10.1073/pnas.97.12.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A. Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc Natl Acad Sci USA. 2004;101:9915–9920. doi: 10.1073/pnas.0401076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzl P, Li H, Carling J, Zhou M, Raman H, Paul E, Hearnden P, Maier C, Xia L, Caig V, Ovesná J, Cakir M, Poulsen D, Wang J, Raman R, Smith KP, Muehlbauer GJ, Chalmers KJ, Kleinhofs A, Huttner E, Kilian A. A high-density consensus map of barley linking DArT markers to SSR and RFLP loci and agronomic traits. BMC Genomics. 2006;7:206–228. doi: 10.1186/1471-2164-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Zhang Z, Lin F, Kong Z, Cao Y, Li C, Yi H, Mei M, Zhu H, Wu J, Xu H, Zhao D, Tian D, Zhang C, Ma Z. A high-density intervarietal map of the wheat genome enriched with markers derived from expressed sequence tags. Theor Appl Genet. 2008;117:181–189. doi: 10.1007/s00122-008-0764-9. [DOI] [PubMed] [Google Scholar]
- Zhang W, Chao S, Manthey F, Chicaiza O, Brevis JC, Echenique V, Dubcovsky J. QTL analysis of pasta quality using a composite microsatellite and SNP map of durum wheat. Theor Appl Genet. 2008;117:1361–1377. doi: 10.1007/s00122-008-0869-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.