Abstract
Constitutional deletions of distal 9q34 encompassing the EHMT1 (euchromatic histone methyltransferase 1) gene, or loss-of-function point mutations in EHMT1, are associated with the 9q34.3 microdeletion, also known as Kleefstra syndrome [MIM#610253]. We now report further evidence for genomic instability of the subtelomeric 9q34.3 region as evidenced by copy number gains of this genomic interval that include duplications, triplications, derivative chromosomes and complex rearrangements. Comparisons between the observed shared clinical features and molecular analyses in 20 subjects suggest that increased dosage of EHMT1 may be responsible for the neurodevelopmental impairment, speech delay, and autism spectrum disorders revealing the dosage sensitivity of yet another chromatin remodeling protein in human disease. Five patients had 9q34 genomic abnormalities resulting in complex deletion-duplication or duplication-triplication rearrangements; such complex triplications were also observed in six other subtelomeric intervals. Based on the specific structure of these complex genomic rearrangements (CGR) a DNA replication mechanism is proposed confirming recent findings in C elegans telomere healing. The end-replication challenges of subtelomeric genomic intervals may make them particularly prone to rearrangements generated by errors in DNA replication.
Keywords: chromosome 9q34.3, duplication, triplication, molecular mechanism, subtelomeric rearrangements, genomic disorder, telomere stabilization
Introduction
The subtelomeric regions of human chromosomes are frequently involved in rearrangements including deletions, duplications, triplications, translocations, and complex chromosome rearrangements (Ledbetter and Martin 2007; Shao et al. 2008; Yatsenko et al. 2009a). Screening large patient cohorts with clinically relevant phenotypes, such as intellectual disability (ID) and multiple congenital anomalies (MCA), by array comparative genomic hybridization (aCGH) has lead to the discovery and characterization of many novel microdeletion and microduplication syndromes and other genomic disorders (Lupski 1998; 2009). Most of these aberrations arise sporadically and vary in size and breakpoint(s) locations.
Previous genetic studies in patients with terminal subtelomeric deletions suggested that the majority of aberrations are the result of double strand DNA (dsDNA) breakage, followed by telomere loss, and repair processes (Varga et al. 2005). Diverse molecular mechanisms that generate and stabilize broken chromosomes were proposed. Terminal deletions can be stabilized by telomere healing, the addition of a telomere (TTAGGG)n repeat sequences by the enzyme telomerase (Bonaglia et al. 2006; Bonaglia et al. 2011; Flint et al. 1994; Lamb et al. 1993; Varley et al. 2000; Verdun and Karlseder 2007; Wilkie et al. 1990), by a telomerase-independent mechanism mediated by nonallelic homologous recombination (NAHR) in which telomere repeats are acquired from homologous chromosome or a sister chromatid (Stankiewicz and Lupski, 2002), or by a nonhomologous end-joining (NHEJ) recombination mechanism with another chromosome resulting in the formation of a derivative chromosome (Flint et al. 1996; Ballif et al. 2004, D’Angelo et al. 2009). In addition, numerous studies indicate that some subtelomeric rearrangements may be mediated by interspersed repetitive elements such as Alu, LINE, long-terminal repeats and simple tandem repeats. Such repetitive elements may play an important role by formation of a specific DNA secondary structure that stabilizes broken chromosomes or assist in DNA repair (Ballif et al. 2004; Yatsenko et al. 2009a). Alternatively, repetitive sequences may facilitate a replication fork template stalling and switching (Boone et al. 2011; Moser et al. 2009). These processes can differ significantly from chromosome to chromosome, likely reflecting specific complex DNA organization at each subtelomere (Dsosopoulos et al. 2012). Repetitive sequences and transposable elements have recently been found to play a prominent role in the structural variation of human personal genomes (Lupski 2010; Beck et al. 2010; Huang et al. 2010; Iskow et al. 2010; Ewing and Kazazian 2010).
Telomere loss can also generate complex chromosomal abnormalities through the breakage-fusion-bridge (BFB) mechanism, which was originally described in maize (McClintock 1941) and proposed as a molecular recombination mechanism in patients with constitutional duplication-deletion rearrangements of subtelomeric regions (Ballif et al. 2003; Zuffardi et al. 2009). BFB cycles are activated during DNA replication when the sister chromatids of a broken chromosome fuse at their ends forming a dicentric chromosome. During anaphase, the fused sister chromatids form a bridge when two centromeres are pulled in opposite directions. During cytokinesis, these dicentric chromosomes are broken unevenly producing a chromosome with a telomere deletion and an interrupted inverted duplication, a hallmark of BFB, and a chromosome with a terminal deletion. The chromatids may fuse and break again during the next nuclear division leading to the repeated BFB cycles until the chromosome eventually acquires a new telomere or becomes stable. End-to-end chromosome fusions that occur in the context of telomerase deficiency can trigger duplications. For more than 70 years these duplications, that are notable for an inverted, interrupted duplication structure, have been attributed to breakage-fusion-bridge (BFB) cycles.
Terminal chromosomal deletions, derivative chromosomes and duplication-deletion rearrangements can be caused by different molecular mechanisms (Hastings et al. 2009a); however, a variety of other rearrangement products can be detected from subtelomeric chromosome aberrations, including duplications, triplications, and complex rearrangements. BFB cycles may not be the only mechanism causing subtelomeric duplication-deletion events. Molecular studies using high resolution array CGH and analyses of breakpoint sequences demonstrate that many genomic rearrangements are complex events showing discontinuous duplications, interrupted by segments with deletions or triplications (Lee et al. 2007; Zhang et al. 2009a; Carvalho et al. 2009; Zhang et al. 2009b), such as DUP-TRP/INV-DUP structures, or other complexities (Carvalho et al. 2011; Chiang et al. 2012; Liu et al. 2011, Talkowski et al. 2012). Such complex rearrangements often do not show structures consistent with a BFB mechanism, such as inverted - interrupted duplications, but can be potentially generated by a DNA replication based mechanism such as “fork stalling and template switching” (FoSTeS) (Lee et al. 2007) or microhomology-mediated break-induced replication (MMBIR) mechanism (Hastings et al. 2009b). Recent studies from Caenorrhabditis elegans telomere replication mutants provide evidence that indeed DNA synthesis, via an apparent FoSTeS like mechanism, generates terminal duplications that seal end-to-end chromosome fusions (Lowden et al. 2011).
Whereas terminal 9q34 deletions have been studied recently in detail (Ballif et al. 2004; Yatsenko et al. 2009a), no systematic experimental investigations have examined gains at 9q34. To obtain further insight into the mechanisms of human subtelomeric rearrangements we studied 20 patients with duplications, triplications and complex alterations of the distal 9q34 region and also investigated samples from six subjects with subtelomeric triplications of other chromosomes. We propose that a replication mechanism, such as FoSTeS and/or MMBIR may play a central role in generating complex duplication-triplication-deletion aberrations at human subtelomeric regions.
Materials and Methods
Human Subjects
We studied 20 unrelated children with variable phenotypes who were found to have a gain in DNA copy number in the subtelomeric 9q34 region revealed by either array CGH, FISH or chromosome analysis performed at Baylor College of Medicine or elsewhere (Table 1). Whole genome oligonucleotide array CGH analysis has been performed at Baylor College of Medicine on all children using a custom-designed array with approximately 180,000 (60 mer) interrogating oligonucleotides, manufactured by Agilent Technologies, Inc. (Santa Clara, CA, USA) with exon coverage of ~1,700 known or candidate disease genes (average of 4.2 probes/exon). The entire genome is covered with an average probe spacing of 30 kb, excluding low-copy repeats and other repetitive sequences. In addition, two adult subjects (mothers of P33 and P46) were ascertained through abnormal FISH analyses performed because of an affected child; and these parents were found to be similarly affected. Blood samples were obtained from 20 individuals and their parents after informed consent for Institutional Review Board (IRB) approved research use of their DNA. Samples from subjects (P52-P61) with benign DNA copy number variations within 9q34.3 were anonymized. Initial G-banding chromosome analyses showed apparently normal karyotypes in 17 children, one male patient showed an XYY sex chromosome complement, and in 2 patients additional chromatin material at the distal 9q was detected by routine chromosome analysis. We also performed FISH analyses on anonymized samples from 6 patients (S1–S6) with constitutional triplications involving various subtelomeric regions (Table 3) under an IRB approved protocol to further study genomic rearrangements using additional molecular technologies.
Table 1.
Summary of array CGH and FISH results in patients with 9q34.3 duplications and triplications.
| Case | Size, Mba | Rearrangement | Region | Localization, Orientation |
|---|---|---|---|---|
| P6 | 5.160 | Complex tri-dup-tri-dup-nl-del | terminal | 9q34.2-q34.3, direct-inverted-direct |
| P20 | 0.085 | Insertion | interstitial | Xq28, ND |
| P25 | 2.385 | dup | interstitial | 9q34.3, direct |
| P33 | 0.258 | dup | interstitial | 9q34.3, direct |
| P33.1 | 0.258 | dup | interstitial | 9q34.3, direct |
| P46 | 0. 913 | dup | interstitial | 9q34.3, direct |
| P46.1 | 0. 913 | dup | interstitial | 9q34.3, direct |
| P47 | 0.710 | Complex dup-tri | terminal | 9q34.3, direct-inverted-direct |
| P48 | 0.728 | Complex dup-tri-dup | interstitial | 9q34.3, direct-inverted-direct |
| P50 | 7.025 | Complex del-nl-dup | terminal | 9q34.2-q34.3, inverted |
| P51 | 0.180 | dup | interstitial | 9q34.3, direct |
| P52 | 0.440 | dup | terminal | 9q34.3, direct |
| P53 | 0.440 | dup | terminal | 9q34.3, direct |
| P54 | 0.420 | dup | terminal | 9q34.3, direct |
| P55 | 0.420 | dup | terminal | 9q34.3, direct |
| P56 | 0.420 | dup | terminal | 9q34.3, direct |
| P57 | 0.397 | dup | terminal | 9q34.3, direct |
| P58 | 0.397 | dup | terminal | 9q34.3, direct |
| P59 | 0.399 | dup | terminal | 9q34.3, direct |
| P60 | 0.191 | dup | terminal | 9q34.3, direct |
| P61 | 0.440 | dup | terminal | 9q34.3, direct |
| P64 | 0.592 | Complex del-nl-dup | terminal | 9q34.3 |
CNV-copy number variant, del- deletion, dup- duplication, ND-not determined, nl- no alteration in DNA copy number, tri- triplication
In patients with terminal rearrangements, duplication-triplication size is calculated from the breakpoint to the end of the chromosome 9 assembly (UCSC, hg18).
Table 3.
Summary of array CGH and FISH results in subjects with triplication of the subtelomeric regions.
| Case | Subtelomeric region | Size, Mb | Rearrangement | Orientation of triplicated segment | Reference |
|---|---|---|---|---|---|
| 1p36.3 | 2.2 | terminal complex del-trip-dup | Not resolved | D’Angelo et al. 2009 | |
| S1 | 1p36.3 | 2.0 | terminal triplication | direct-inverted-direct | |
| S2 | 1p36.3 | 2.1 | interstitial complex trip-dup | direct-inverted-direct | |
| 2q37 | ND | terminal triplication | Not resolved | Rauch et al. 1996 | |
| S3 | 3q25-q26 | 11.7 | interstitial complex trip-dup | direct-inverted-direct | |
| 5p14-p15.3 | ~ 25* | terminal triplication | direct-inverted-direct | Harrison et al. 199 | |
| S4 | 6p25.3 | 2.5 | terminal triplication | direct-inverted-direct | |
| S5 | 6p25.3 | 1.6 | interstitial triplication | direct-inverted-direct | |
| 7p21.3-p22 | ~ 30* | complex trip-dup | direct-inverted-direct | Rivera et al. 1998 | |
| 9q34 | 2.93 | interstitial complex dup-tri-dup | direct-inverted-direct | Gijsbers et al. 2008 | |
| 10q26 | ~ 6* | terminal triplication | direct-inverted-direct | Devriendt et al. 1999 | |
| S6 | 17p13.3 | 0.62 | interstitial complex dup-tri-dup | direct-inverted-direct |
ND-not determined;
a minimal size of the rearrangement estimated based on FISH analysis.
Array-CGH studies
Patients’ genomic DNA was extracted from peripheral blood samples using the Puregene kit (Gentra Systems, Minnesota, USA) according to the manufacturer’s protocol. High resolution array CGH studies were performed using a custom 9q34 oligonucleotide microarray as previously described (Yatsenko et al. 2009a). The size, extent, and genomic content of each rearrangement were determined to 0.5–3 kb resolution. Gender-matched reference DNAs were obtained from either a normal male or female individual (Yatsenko et al. 2009b). The same reference DNA has been used for all patients studied by whole genome as well as custom 9q34 microarray. The procedures for DNA digestion, labeling, and hybridization were performed according to the manufacturer’s instructions (Agilent Technologies, Inc, CA, USA) with some modifications.
FISH Analysis
We performed a series of FISH experiments in order to independently confirm array CGH genome-dosage results, determine the location and orientation of duplicated-triplicated segments in each patient, and further characterize the structure of complex chromosomal rearrangements. Metaphase chromosome spreads and interphase nuclei were obtained for all patients and their parents (when available) from PHA-stimulated blood lymphocyte cultures. BAC (bacterial artificial chromosomes) and fosmid clones were selected from the Genome Browser database (http://genome.ucsc.edu/) based on their map location within the 9q34 region or the appropriate other subtelomeric regions. FISH analyses with locus-specific probes labeled directly with Spectrum Orange-dUTP or Spectrum Green-dUTP (Abbot Molecular/Vysis) were implemented on interphase nuclei and/or metaphase chromosomes as described elsewhere (Yatsenko et al. 2005). At least 50 interphase and/or 10 metaphase cells were scored for each hybridization. Parental samples were studied to verify if the rearrangement represents a de novo or inherited event. The Xq (305J–T7) subtelomere specific probes (Abbot Molecular/Vysis) was used to study the integrity of the Xq subtelomeric region due to ins(X;9)(q28;q34.3q34.3)mat in patient P20.
Breakpoint junction
Rearranged 9q34 genomic DNA sequences were identified in each patient from array CGH and FISH analyses. The genomic coordinates for rearrangement breakpoints were estimated from CGH arrays and surrounding DNA sequences were downloaded from the UCSC Genome browser (http://genome.ucsc.edu, hg18 assembly). Several “nested” PCR oligonucleotide primers were designed from both the proximal and the distal breakpoint segments to amplify unique junction fragments. Primers were used in different combinations under optimized conditions to obtain patient rearrangement specific junctions in comparison to parental controls. Genomic DNA sequences were amplified with Qiagen HotStarTaq or TaKaRa LA Taq (TaKaRa) kits according to the manufacturers’ protocols using conditions as previously described (Yatsenko et al. 2009a). Control samples from two normal male individuals and both parents (if available) were included in the PCR experiments. PCR products were visualized and their specificity was assessed by 1.2 % TBE agarose gel electrophoresis. DNA sequencing was performed using either the ABI Prism BigDye v3.1 terminator kit or BigDye terminator chemistry (Applied Biosystems, Foster City, CA) and an ABI 377 DNA sequencer or a 3730 DNA sequencer (Applied Biosystems) by conventional Sanger dideoxy sequencing. The chromatograms were analyzed using Sequencher 4.2 (Gene Codes).
Results
High resolution array CGH and FISH analyses of 9q34 duplications
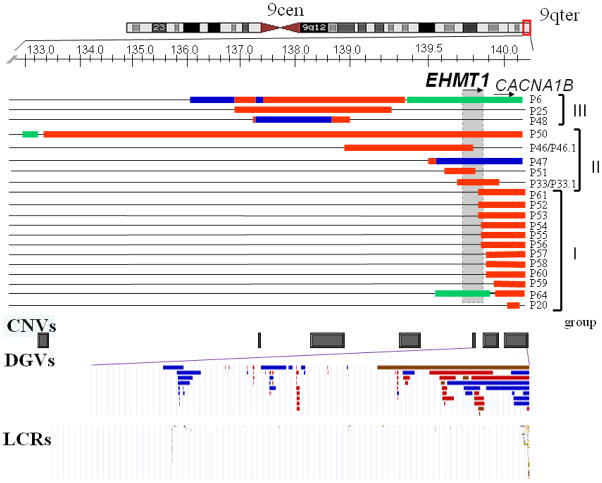
We assessed the genomic content, size, and extent of the imbalances in 20 children with segmental aneusomy due to gains of the 9q34 region using a high resolution custom designed 9q34 oligonucleotide microarray. In our subjects, the 9q34.3 rearrangements range from 85 kb to 7 Mb in size with variant breakpoint locations (Figure 1, Table 1). To classify these nonrecurrent rearrangements as either potentially pathogenic imbalances or benign copy number variations (CNVs) subjects were divided into three groups based on the extent of the duplicated genomic interval. Group I comprises twelve patients (P20, P52-P61, P64) with duplications involving the most distal part of the 9q34.3 subtelomere region. This region encompasses the CACNA1B gene and the 3′ end (exons 24–26) of the EHMT1 gene. P64 also has a deletion of the EHMT1 gene resulting in Kleefstra syndrome [MIM#610253].
Figure 1.
Genomic intervals of the 9q34 rearrangements identified by array-CGH in 20 patients. Top, an ideogram of chromosome 9. Below, a magnified view of the 9q34 genomic region. The locations of the EHMT1 and CACNA1B genes are indicated by horizontal arrows showing the direction of transcription. The genomic region encompassing the EHMT1 gene is shown as a gray box. Unchanged chromosome regions are represented by black horizontal lines. A loss in copy number (deletion) is shown by a horizontal green bar. A gain, duplications and triplications are represented as red and blue bars, respectively. Within this 9q34.3 interval, copy number variants CNVs (below) have been detected in normal individuals and are shown here as gray boxes. Below is a magnified view of CNVs from Database of Genomic Variants DGVs within the EHMT1 and CACNA1B genes. Blue, red and brown DGVs indicate regions with a gain, loss and both gain and loss, respectively observed in healthy individuals.
High-resolution analysis of at least 1,000 individuals from various population groups revealed multiple copy number variations in the 9q34.3 region (Figure 1). Within this duplicated interval, CNVs have been reported in normal individuals for either part of, or the entire CACNA1B and the 3′ end of EHMT1 gene but not for either the 5′ end or the entire EHMT1 gene (Figure 1) (Database of Human Genomic Variants, http://projects.tcag.ca/variation, Mar 25, 2010 update). Thus, duplications of the 9q34.3 genomic region in patients P52-P61, P64 are likely benign CNVs if copy number gains are present in the distal 9q34.3 and do not include the entirety of the EHMT1 gene. Potentially, the same DNA segment can be inserted into a different chromosome locus (like observed for P20) resulting in gene disruption or alteration and therefore leading to clinical manifestations.
Within Group I, ten patients out of twelve had a tandem duplication of the 9q34.3 (Figure 1), although two of the 10 patients, P53 and P56, had additional chromosomal abnormalities (Table 2). Parental FISH analyses in five families demonstrated that these 9q34.3 duplications are inherited from healthy parents in three patients and represent de novo abnormalities in two subjects (Table 2). Parents were unavailable for testing in other families. Duplications of this genomic interval have been reported previously in unaffected controls (Redon et al. 2006), and found in normal parents in our study, thus we conclude that duplications encompassing ~600 kb of the distal 9q34.3 most likely represent benign CNVs. In the remaining two patients, P20 and P64, the interpretation of the rearrangement gain and gene dosage is more complex. A microarray detected gain encompassing 85 kb of the CACNA1B gene in a male patient P20; FISH studies showed that the duplicated 9q34.3 sequence was inserted into the distal long arm of the X chromosome, near Xq subtelomere. FISH analysis using an Xqter specific probe revealed a normal hybridization pattern, consistent with an insertion of 9q34.3 segment into Xq. Maternal FISH studies revealed the same abnormal chromosome X with an additional 9q sequence, thus P20 has apparently inherited the derivative X chromosome from a healthy mother who has the unbalanced X;9 rearrangement. Whole genome array CGH analysis (CMA version 8) in P20 did not identify any clinical relevant copy number changes in other interrogated loci. aCGH in P64 showed a complex deletion-duplication rearrangement on 9q34.3.
Table 2.
Clinical presentation in patients with a gain of the 9q34 region.
| Case | Age, years | Sex | Genes involved | Inheritance | Phenotype | Additional chromosome abnormalities | Interpretation of the 9q34 gain |
|---|---|---|---|---|---|---|---|
| P6 | 12 | M | multiple | de novo | 9q34.3 microdeletion/Kleefstra syndrome | none | Pathogenic |
| P20 | 10 | M | CACNA1B | maternal | Asperger syndrome | der(X)ins(X;9) | Potentially Pathogenic |
| P25 | 11 | M | multiple | de novo | Speech delay, aggressive behavior | XYY | Pathogenic |
| P33 | 6 | M | EHMT1, CACNA1B | Maternal (P33.1) | Speech delay, nonsyndromic mild ID | none | Potentially Pathogenic familial |
| P33.1 | 32 | F | EHMT1, CACNA1B | ND | Speech delay, learning disability | none | Potentially Pathogenic |
| P46b | 7 | F | EHMT1 (exons 1-15), multiple | Maternal (P46.1) | Speech delay, congenital cataract, brain tumor, nonsyndromic ID | none | Potentially Pathogenic familial |
| P46.1 | 35 | F | EHMT1 (exons 1-15), multiple | ND | Speech delay, nonsyndromic ID | none | Potentially Pathogenic |
| P47 | 11 | M | Multiple, EHMT1 | de novo | Speech delay, learning disability, ASD | none | Pathogenic |
| P48 | 8 | F | multiple | de novo | Speech delay, mild ID | none | Pathogenic |
| P50 | 6 | F | Multiple, EHMT1 | de novo | ID, speech delay | none | Pathogenic |
| P51 | 8 | M | EHMT1 (exons 1-16) | de novo | ASD (high-functioning) | none | Potentially Pathogenic |
| P52 | 5 | M | EHMT1 (exons 24-26), CACNA1B | Phenotypically normal mother | Speech delay, learning disability | none | Benign CNV |
| P53 | 1 | F | EHMT1 (exons 24-26), CACNA1B | ND | MCA | der(X)t(X;16) | Benign CNV |
| P54 | 4 | F | EHMT1 (exons 25-26), CACNA1B | Phenotypically normal mother | MCA | none | Benign CNV |
| P55 | 5 | M | EHMT1 (exons 25-26), CACNA1B | de novo | Expressive language disorder | none | Benign CNV |
| P56 | 2 | M | EHMT1 (exons 25-26), CACNA1B | ND | Dysmorphic features | rec(6) (dup6p/del6q) | Benign CNV |
| P57 | 6 | F | CACNA1B | ND | ASD | none | Benign CNV |
| P58 | 9 | M | CACNA1B | de novo | Nonsyndromic ID | none | Benign CNV |
| P59 | 10 | M | CACNA1B | Phenotypically normal father | DD, ASD, obesity | none | Benign CNV |
| P60 | 15 | M | CACNA1B | ND | Dysmorphic features | none | Benign CNV |
| P61 | 3 | F | EHMT1 (exons 24-26), CACNA1B | ND | MCA | none | Benign CNV |
| P64 | 16 | F | multiple | de novo | 9q34.3 microdeletion/Kleefstra syndrome | none | Benign CNV |
ASD-autistic spectrum disorder, CNV-copy number variant; F-female, ID-intellectual disability, M-male, MCA-multiple congenital anomalies, ND-not determined;
results of molecular analysis on this patient is reported by Cheung et al., 2011.
Group II includes five patients (P33, P46, P47, P50, and P51), in which aCGH analysis revealed a gain in copy number encompassing either the entire or the 5′ end of EHMT1 (Figure 1, Table 1). FISH studies demonstrated tandem duplications in three patients (P33, P46 and P51), whereas subjects P47 and P50 had complex rearrangements. In P47, the derivative chromosome 9 consist of a triplication of approximately 682 kb in size, interrupted by a small duplicated segment 7.9 kb, and a duplication of the more proximal region 20.3 kb (Figure 2A). There are no low copy repeats (LCRs) or other repetitive sequences that may be attributed to the difference in DNA copy number detection. In P50, both deletion and duplication were found on a derivative chromosome 9 (data not shown). FISH analyses demonstrated an inverted orientation of the apparent duplicated region (Table 1). P33 had intellectual and learning disability, speech and language delay. His mother (P33.1) required speech and language therapy, and attended special education classes. P46 and P46.1 presented with nonsyndromic intellectual and learning disabilities, and speech delay (Cheung et al. 2011). P51 has been diagnosed with autism spectrum disorder, behavioral problems and anxiety.
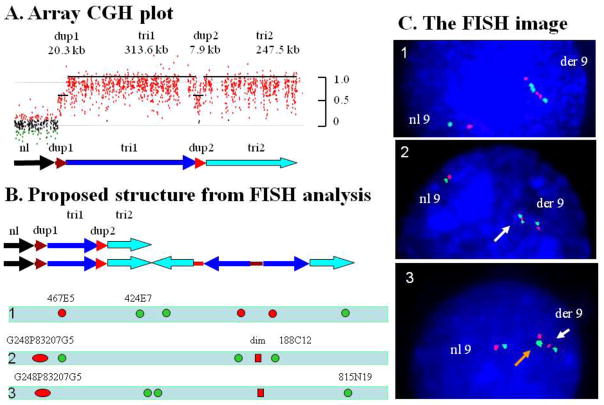
Figure 2. Complex rearrangement in patient P47.
(A) Array-CGH profile in patient P47 showing a complex duplication-triplication-duplication-triplication structure. (B) Proposed structure of the normal and derivative chromosome 9. Bars with arrowheads indicate the orientation of the DNA segments. Bars without arrowheads indicate the location of DNA segments however the orientation of the structure is uncertain. The approximate location of FISH probes labeled in green and red are shown. A combination of probes is given for each FISH experiment [1], [2], and [3] (C) The results of FISH analyses in each of FISH experiments. [1] Hybridization with probes RP11-467E5 in red) and RP11-424E7 in green) depicting the derivative chromosome 9 has a triplication with an inverted middle segment. [2] A diminished signal produced by fosmid G248P83207G5 in red is indicated by a white arrow. [3] RP11-815N19 (AL954642) probe produced two signals in green close proximity to each other seen as a signal with double intensity (orange arrow). The pattern red-green-green-red indicates an inverted orientation of the middle copy of the trip1-trip2 region.
In Group II, patients presented with autistic behavior, cognitive impairment, intellectual disability, learning difficulties, speech and developmental delay. Parental analyses demonstrated de novo rearrangements in three subjects, whereas two patients, P33 and P46, inherited the duplication from their similarly affected mothers. Comparison of the molecular findings (Table 2) in conjunction with clinical features, i.e. genomotype-phenotype correlations, indicate that increased dosage of EHMT1 may be responsible for the neurodevelopmental impairment, speech delay, and autism spectrum disorders in these patients. We were not able to correlate the specific differences in clinical severity between patients with duplications and triplications leading to further increase in dosage of the EHMT1 gene due to variability in size and gene content in each patient, the variability of the duplication phenotype, and the limited number of triplication patients available for study.
Group III includes three patients (P6, P25, and P48, Figure 1) with a gain in copy number involving a genomic region proximal to EHMT1, but within 9q34.3. One patient (P6) had a complex chromosome rearrangement (CCR) (Figure 3A) resulting in 9q34.3 deletion syndrome (Yatsenko et al. 2009a). Two subjects, P25 with a de novo duplication and P48 with complex duplication-triplication rearrangements (Figure 4A), respectively, presented with speech and developmental delay suggesting that other dosage sensitive genes, which may be involved in CNS development or function, are located within this genomic interval. One alternative possibility is that amplification of non-coding regions may result in abnormal expression of flanking genes (Ricard et al. 2010; Zhang et al. 2010). Duplications of 9q34.3 that are similar to those observed in patients from Group II and III are not seen amongst healthy individuals.
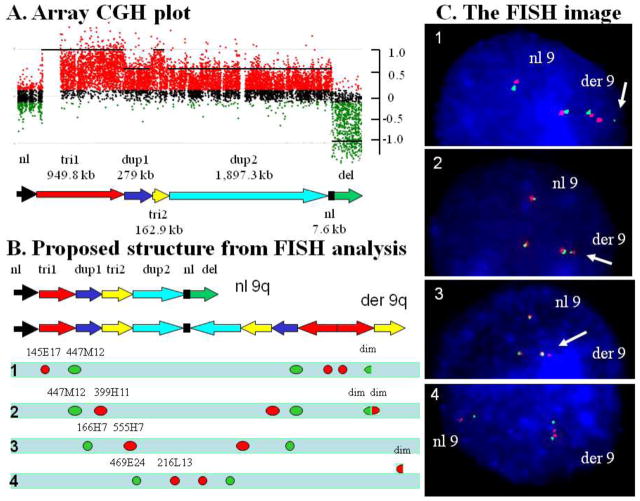
Figure 3. Complex rearrangement in patient P6.
(A) A magnified view of array-CGH profile in patient P6 showing a complex interrupted deletion-duplication-triplication rearrangement. (B) Schematic view of the normal and proposed derivative chromosome 9 structure. Bars with arrowheads indicate the orientation of the DNA segments. Bars without arrowheads indicate the location of DNA segments however the orientation of the structure is uncertain. The approximate location and the relative position of the FISH probes labeled in green and red are shown. A combination of probes in four FISH experiments (1, 2, 3 and 4) is given to demonstrate an orientation and structure of the duplicated and triplicated segments in this patient. (C) The results of FISH analyses in each of four experiments. [1] Hybridization with probes RP11-145E17 (AL354796, red) and RP11-447M12 (AL353611, green) showing the orientation and relative position of the “tri1” and “dup1” segments. For probe RP11-447M12 (green), the telomeric signal was of diminished intensity (arrow) as compared to the proximal signals. [2] Hybridization with probes RP11-399H11 (AL390778, red) and RP11-447M12 (green) showing two red and green diminished signals (arrow), both co-localized to each other. [3] Hybridization with probes RP11-166H7 (AL159992, green) and RP11-555H7 (AL353615, red) showing the orientation of the “dup1” and “tri2” segments. [4] Inverted orientation of the two “dup2” segments.
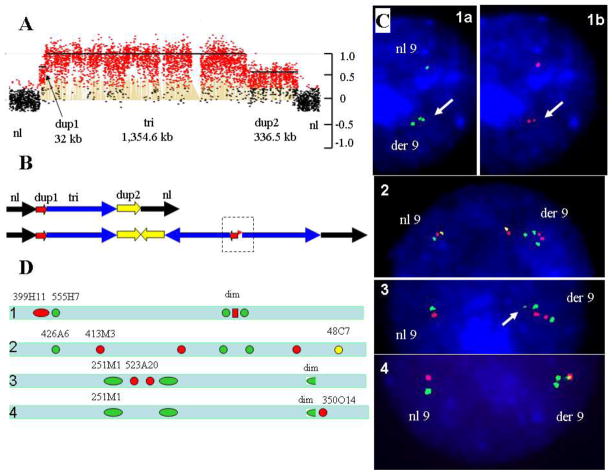
Figure 4. Complex rearrangement in patient P48.
(A) Array-CGH profile in patient P48 showing a complex deletion-duplication rearrangement. (B) Proposed structure of the normal and derivative chromosome 9. A combination of probes in four FISH experiments (1, 2, 3 and 4) is given to demonstrate an orientation and structure of the duplicated and triplicated segments in this patient. (C) The results of FISH analyses in each of FISH experiments. [1] Hybridization with probes RP11-399H11 (AL390778, red) and RP11-555H7 (AL353615, green) showing the relative position of the “tri” and “dup1” segments. Images of the same cell were taken with a selective green (1a) and red filter (1b) respectively. [1a] Hybridization with RP11-555H7 (green) demonstrates a triplication. A double signal is indicated by an arrow. [1b] Probe RP11-399H11 (red) showed a diminished intensity signal (arrow) co-localized with a green double signal. [2] Hybridization with probes RP11-413M3 (AL592301, red), RP11-426A6 (AL161452, green) and RP11-48C7 (AL365502, yellow) produced a hybridization pattern consistent with normal- inverted- normal position of the triplicated segment. [3] Hybridization with probes RP11-251M1 (AL590226, green) and RP11-523A20 (AL449425, red) showing the inverted orientation of the “dup2” and “tri” segments. Diminished green signal (arrow) indicates skipping of the “dup2” segment at the DNA junction. [4] Co-hybridization of RP11-251M1 (green) and RP11-350O14 (AL929554, red) indicates that abnormal DNA junction localized in the vicinity of the 9q telomere.
FISH analyses in patients with complex 9q34 rearrangements
Five out of twenty (25%) patients among our cohort of subtelomeric 9q34.3 gains had a complex rearrangement of the 9q34.2-q34.3 region. In three cases (P47, P6, and P48), high resolution array CGH analysis revealed evidence for triplicated genomic segments interspersed with the duplicated regions. In two cases (P50 and P64), a deletion of the proximal 9q34.3 region and a duplication of the distal region were also identified. Array CGH provides neither genome positional nor orientation information. We sought to further characterize these complex rearrangements using independent molecular approaches to refine structure and potentially provide insights into molecular mechanism for formation.
To determine the genomic position, orientation and structure of the rearranged genomic interval within 9q34 we performed two - and three - color FISH analyses using BAC and fosmid clones in each patient with complex 9q34 triplications. FISH probes from the regions of interest were hybridized together in different combinations. In subject P47, segments of duplications and triplications extending to the 9qter were identified (Figure 2A,B). FISH analysis using BAC clones RP11-467E5 (AL590627) and RP11-424E7 (AL591424) as probes revealed a triplication with an apparently inverted orientation of the middle segment in comparison to the two flanking segments that are directly orientated (Figure 2B,C). Localization of the duplicated segment was determined based on FISH analysis using fosmid G248P83207G5 (Figure 2B[2], 2B[3]). This probe spans the entire “dup1” region as well as about 17 kb of flanking unaltered genomic region. FISH analysis showed one red signal of normal size and one diminished signal (Figure 2C[2], 2C[3], white arrow) on a derivative chromosome 9. This duplicated segment is located in proximity to the triplicated “tri1” region as shown in Figure 2B[2] and 2B[3]. FISH analysis with probe RP11-815N19 (AL954642, green) revealed one enlarged signal (Figure 2C[3], orange arrow) and a normal size signal consistent with an inverted orientation for the middle segment of the triplicated interval as well as showing the relative position of the triplication and duplication segments. Because of the limited genomic resolution of FISH studies the structure of the abnormal 9q can not be determined definitively and may be more complex; however one proposed structure based upon the interpretation of available data is presented in Figure 2B.
Based on results from both G-banding and FISH studies of derivative chromosome 9 in patient P6, the rearrangement was originally interpreted as a 9q34.3 terminal deletion stabilized by a duplication of the more proximal 9q34.2-q34.3 segment (Yatsenko et al. 2009a). Using high resolution custom 9q34 microarray we determined multiple segments with alterations in DNA copy number: “tri1”, “dup1”, “tri2”, “dup2” and “del” (Figure 3A,B). Further FISH analyses enabled us to propose a structure for this complex rearrangement (Figure 3B,C); note that for the triplicated segment the middle copy appears to be inverted in orientation with respect to the other flanking direct copies: a DUP-TRP/INV-DUP structure (Carvalho et al. 2011).
P48 had a complex aberration consisting of a triplication embedded within a duplication segment. In addition, the structure of the triplication in derivative chromosome 9 was similar in arrangement to that of P47 with an apparent inverted orientation of the middle segment of the triplication; i.e. DUP-TRP/INV-DUP (Figure 4).
Breakpoint junction analyses
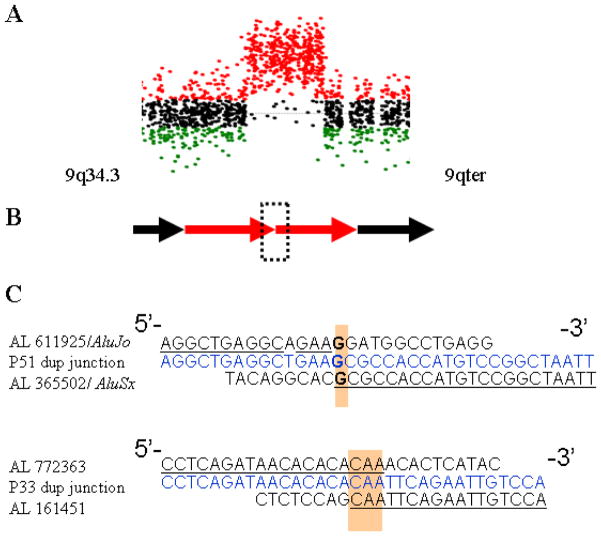
We obtained junction sequences for the rearrangements in two subjects (P33 and P51) with interstitial duplications of the 9q34.3 region (Figure 5A). These patients were found to have a tandem duplication of 276,825 bp, and 202,911 bp in size, respectively. Amplification was achieved across the junction between the proximal and distal breakpoints using outwardly oriented PCR primers (Figure 5B). For subject P33, the duplication breakpoints were located within unique sequences; no nearby repetitive elements were identified in the draft reference haploid human genome. Microhomology, 3 bp (CAA), was identified at the breakpoint junction (Figure 5C). The same junction fragment was amplified using genomic DNA of the patient’s mother (P33.1), but neither from the patient’s father nor control DNAs. The breakpoints in subject P51 were located within two Alu elements, AluSx and AluJo family members, both oriented in the same direction, with limited similarity between each other. The proximal breakpoint was identified within the AluSx, and the distal breakpoint mapped in AluJo element (Figure 5C). As expected, junction fragments were amplified from the patient P51 genomic DNA, but not from the parental or control DNAs, indicating de novo duplication in a child.
Figure 5. Rearrangement mechanism revealed by high-resolution array CGH and analyses of patient-specific junction sequences from patients P33 and P51.
(A) Array-CGH profile in patient P33 showing a duplication of the 9q34.3 region. (B) A schematic presentation of simple tandem duplication in patients P33 and P51. Junction fragment is indicated by a dashed rectangle. (C) Sequence analysis of the duplication junctions. Top, normal distal flanking sequence; bottom, normal proximal flanking sequence; middle (blue), duplication junction sequence.
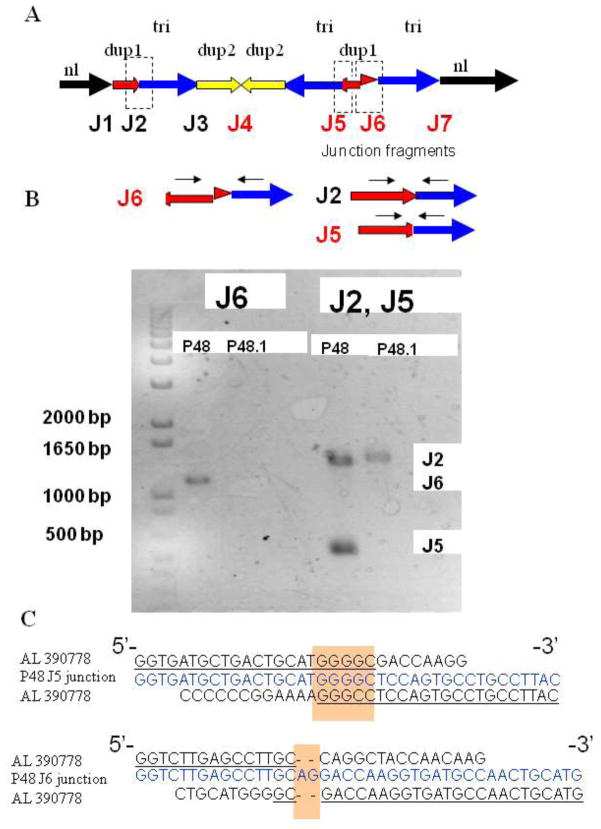
Five subjects (P6, P48, P50, P47, and P64) had complex 9q34.3 rearrangements. Based on results of array CGH analysis and FISH studies we determined the structure of the derivative chromosome 9 in each subject with complex alterations and attempted to amplify patient-specific breakpoint junctions as well as wild type junction sequences that are expected to be present on both normal and derivative chromosome 9. Two patient-specific junction fragments were obtained in P48 (Figure 6). For subject P48 junction fragments J1–J3 are expected to be present on both normal chromosome 9 and der(9) (Figure 6A), while J4–J7 are rearrangement-specific junctions. Using primers from the proximal regions of the “dup1” and “tri” segments we obtained PCR products (J6, Figure 6A,B) of ~ 1200 bp in size, which is ~ 900 bp longer than predicted based on primer positions in comparison to the haploid human reference genome sequence. Sequence analysis of this product reveals an insertion of 905 bp that apparently originated from the distal part of the “dup 1” segment (Figure 6A,B). Thus, the breakpoint junction between the “dup1” and “tri” segments was actually located about 900 bp proximal to the one identified by array CGH analysis. Interestingly, using primers from the distal region of the “dup1” and proximal “tri” segments (J2, Figure 6A) in P48 we obtained two PCR products of ~ 1400 bp, and ~ 470 bp in size. Only one PCR product was detected after amplification of parental (P48.1) and control genomic DNAs with the same primers (Figure 6B). Sequence analysis demonstrates two junction fragments J2 and J5. In J5 fragment, a 947 bp sequence was missing when compared to the J2 sequence. Part of this sequence, a 905 bp segment was present at the J6 sequence as described above, and 42 bp were deleted. Sequence analyses in J5 or J6 showed no large homology between the breakpoint-flanking regions. The junction fragment in J5 had a microhomology of five bases (GGGGC) and there was an insertion of two bases (AG) at the J6 junction (Figure 6C). Attempted amplifications across the J4 and J7 in P48 did not recover any PCR products. Possible breakpoints may occur at locations distinct from the CGH determined CNV segments as shown for J6 fragment in patient P48. In addition, other genome variations such as inversions, insertions, or deletions may be present in the vicinity of the rearrangement-specific junctions in this individual subject’s personal genome, thus complicating amplifications using strategies that depend on preconceived notions of genome structure based upon the current haploid reference human genome sequence.
Figure 6. Complex rearrangement revealed by analysis of junction sequences in patient P48.
(A) Proposed structure of the derivative chromosome 9 based on FISH study and analysis of the patient-specific junction fragments. Junction sequences are designated by dashed rectangles. Junction sequences J1–J3 are expected to be present on a normal chromosome 9. Junction fragments J4–J7 are patient-specific sequences proposed to exist on P48 derivative chromosome 9. (B) Two patient-specific junction fragments J6 and J5 were obtained in P48. Black arrows show locations of PCR primers for cloning the breakpoint junctions. Neither junction was observed using the same PCR primers to amplify the genomic DNA of the parents. The J2 junction fragment was obtained for patient P48 as well as for the patient’s mother (P48.1). (C) Direct sequencing of the amplified fragment in patient P48 revealed the breakpoint junction. The breakpoint junction sequence (middle, blue) is aligned to the 9q reference sequence. The homologous nucleotides at the breakpoints are shown in yellow shaded boxes.
Subtelomeric triplication with an inverted middle copy
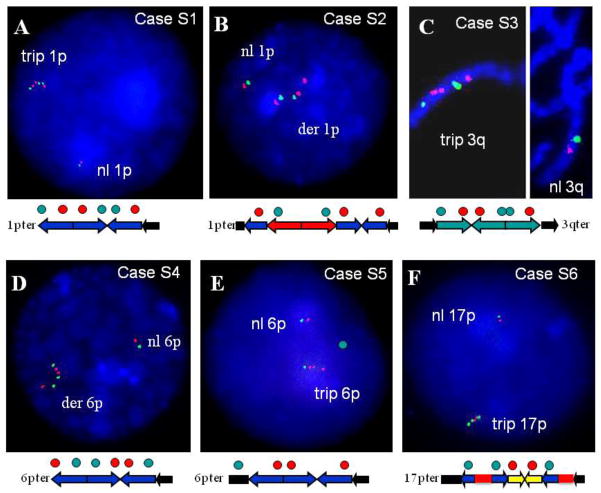
Constitutional triplications involving subtelomeric regions appear to be relatively rarely identified. Only 6 patients have been described with triplications involving 1p, 2q, 5p, 7p, 9q, and 10q distal chromosome regions (Table 3). The orientation of the triplicated segments has been determined only in a few patients with large, cytogenetically visible aberrations (Harrison et al. 1998; Rivera et al. 1998; Devriendt et al. 1999; Gijsbers et al. 2008). Molecular characterization by FISH analyses demonstrated an unusual pattern of the triplication with inversion of the middle copy, however the extent, order and orientation of the rearranged segments were not resolved in the majority of patients with triplications and complex aberrations.
Interestingly, we identified three subjects with triplications involving the 9q34.3 region. Moreover, we show that in each patient the middle copy of the triplicated segment appears to be inverted in orientation: a DUP-TRP/INV-DUP structure. To determine if the observed pattern, with an inversion of the middle copy flanked by directly oriented copies, represents a general phenomenon we performed FISH analyses in six patients S1–S6 with triplications within the subtelomeric regions (Table 3).
In patients S1–S6, a gain in DNA copy number consistent with triplications of either 1p, 3p 6p, or 17p was identified by array CGH analysis at Baylor College of Medicine. Dual-color FISH demonstrates that in each case the middle copy of the triplication is inverted in comparison to the two flanking segments that are directly orientated (Figure 7). Similarly, in patients with complex duplication-triplication aberrations, the middle copy of the triplicated segments embedded within a duplicated region also has an inverted orientation. Thus, the observed triplication pattern appears to represent a general rearrangement product pattern that may occur in complex triplication containing rearrangements at several and potentially all human subtelomeric regions (Carvalho et al 2011).
Figure 7. FISH analyses demonstrating an inverted orientation of the middle copy in subjects S1–S6 with triplications of the subtelomere regions.
(A) FISH analyses in subject S1 with 1p36.3 triplication, (B) In subject S2 with complex duplication-triplication of the 1p36.3 subtelomere region, (C) in subject S3 with complex duplication-triplication of the 3q25-q26 region, (D) and (E) in subjects S4 and S5 with triplications of the 6p25 region, and (F) in subject S6 with complex duplication-triplication-duplication of the 17p13.3 subtelomere region.
Discussion
Instability of the 9q34 genomic region
We analyzed 20 patients with copy number gains involving the 9q34.3 subtelomeric region to provide experimental data regarding structural information for rearrangement products that might enable insights into potential mechanisms that generate such rearrangements. Genomic instability of the 9q34 region is revealed by the numerous reports of patients with constitutional rearrangements of the distal 9q (Yatsenko et al. 2005; Kleefstra et al. 2006; Simovich et al. 2007; Gijsbers et al. 2008; Yatsenko et al. 2009a), as well as acquired aberrations associated with malignancies (Dave et al. 2005; Andreeva et al. 2008). We show here by high resolution genomic analyses, including array CGH, FISH studies, and sequence analyses of selected breakpoint junctions, that what appears to be simple rearrangements associated with copy number gains at 9q34 are often actually complex in nature. Furthermore, we again uncovered a particular structure of DUP-TRP/INV-DUP as has been seen in multiple patients with triplications at either the MECP2 or PLP1 loci (Carvalho et al. 2011, Shimojima et al. 2012).
Gains involving the most distal 9q34.3 represent copy number variations (CNV) that are frequently observed among our patients as well as their parents and unrelated ‘healthy control’ individuals. Such common variants are considered to be benign alterations. Interestingly, duplications and triplications involving the entire EHMT1 gene, or the 5′ end of the EHMT1 gene, have not been observed among healthy populations. Whereas extensive clinical characterization has not been reported for a cohort of subjects with such duplications, patients affected with Kleefstra syndrome due to EHMT1 haploinsufficiency exhibit intellectual and learning disabilities, significant speech problems, and multiple congenital anomalies. The subjects with duplications reported herein with EHMT1 gains represent rearrangements that have not been reported in healthy control populations. We observe these patients to have associated neurodevelopmental impairment, speech delay, and autism spectrum disorders consistent with the hypothesis that increased EHMT1 dosage is associated with a neuro - behavioral phenotype. Future studies involving large numbers of subjects with duplications including EHMT1, and perhaps EHMT1 gain of function point mutations, will be required to establish the range of neurobehavioral phenotypes that can potentially be manifest in association with such rare variants.
Molecular mechanisms of complex rearrangements
Several molecular mechanisms have been proposed to explain the origin of chromosome terminal deletions and derivative chromosomes, however mechanisms giving rise to nonrecurrent, direct and inverted duplications, triplications, as well as complex rearrangements are not well understood. Previous studies have implicated BFB cycles as one mechanism (McClintock 1941, Ballif et al. 2003) involved in subtelomeric rearrangements, however, rearrangements consisting of interrupted inverted duplication-deletion are rare and the majority of those reported have been detected by conventional chromosome analyses and FISH studies. Furthermore, recent work in C. elegans telomerase deficient mutants (Lowden et al. 2011) reveals complex structures more parsimoniously explained by DNA replication models such as FoSTeS (Lee et al. 2007). The widespread implementation of array CGH in clinical diagnostics and research studies reveals a substantial number of patients with complex abnormalities present at subtelomeric regions, with complexities beyond the inverted interrupted duplication predicted by a BFB mechanism. Moreover, microarray analysis with a high density of interrogating probes at the subtelomeric region detects additional alterations in cases that appear to be inverted duplication-deletion aberrations.
We analyzed the structure of the rearranged chromosome 9 and demonstrated complex genomic alterations involving the subtelomeric 9q region (Fig. 3). Repair of a terminal deletion could be affected through a breakage-fusion-bridge cycle, as suggested by Ballif and colleagues (2003), however occurrence of multiple alterations particularly those involving higher order copy number (e.g. triplication), can be more parsimoniously explained by a replication-based repair mechanism. FISH analysis in P6 revealed a complex rearranged genomic structure formed between multiple segments of the distal 9q34. Segments that are found to be duplicated or triplicated have both directly oriented and inverted segments along the chromosome end, suggesting a potential template switching between different replication forks with inversion products reflecting template switching to a replication fork proceeding in the opposite direction as postulated by FoSTeS (Lee et al. 2007) or re-establishing of a new fork in the reverse strand as postulated by MMBIR (Hastings et al. 2009a). Alternatively, BIR mediated by inverted repeats with high nucleotide identity can also lead to inverted triplications, a mechanism that was shown to be responsible for formation of DUP-TRP/INV-DUP structures in the X-chromosome where the finding of some breakpoints without microhomology suggested both an homology/microhomology driven event like BIR/MMBIR driving the FoSTeS driven template switch occurred in combination with a non-homologous breakpoint repair event (Carvalho et al. 2011). We found similar DUP-TRP/INV-DUP structure at multiple subtelomeric gains involving chromosome ends other than 9q34.3.
The telomeric genomic sequences may be particularly susceptible to instability and rearrangements generated by DNA replication mechanisms because of the end-replication problem as well as peculiar DNA sequence and structure of the subtelomere. At the end of a linear chromosome, DNA polymerase can synthesize the leading strand, but the discontinuous lagging strand of the replication fork is challenged by the linear nature of chromosome (Verdun and Karlseder 2007). This phenomenon, known as the “end-replication problem” (Watson et al. 1972), results in a loss of a DNA sequence at the chromosome terminus during a cell division. In addition to a potential ‘end replication challenge’ arising from the 5′ to 3′ polarity of DNA polymerases, telomere replication can originate within the subtelomeric unique sequences as far as 120 kb away from telomere repeats. These subtelomere regions consist of a substantial number of repetitive elements and DNA sequences that form alternative secondary structures (Yatsenko et al. 2009a), non B DNA conformations (Mirkin and Mirkin 2007) or can become bound to protein complexes affecting DNA replication and genome integrity (Anand et al. 2011; Drosopolous et al. 2012). In P48, sequence analysis uncover multiple aberrant events at the rearrangement-specific breakpoint junctions including a 42 bp deletion, inversion and DNA misrepair by fusion with nonhomologous sequence, resulting in a complex rearrangement.
Complex nature and structure of triplications within the subtelomeric regions
It is possible that similar molecular mechanism can result in similar abnormal rearrangement products or structures that may underlie many non-recurrent rearrangements involving the subtelomeric chromosomal regions. Surprisingly, three patients (P6, P47, and P48) among our cohort had triplications of 9q34.3 region. Similar structures of complex dup- trip were found at the subtelomeric regions of chromosome 1p, 6q (Figure 7), 15q, 17p, and Xq (Carvalho et al. 2009). High resolution aCGH showed a complex rearrangement in each case with triplication.
Interestingly, each triplicated segment had flanking duplicated sequences, consistent with a replication mechanism. Comprehensive FISH studies revealed a pattern with an apparent inverted middle copy of the triplication region. The triplication integrated within flanking duplicated segments may reflect its formation due to skipping of the sequence from replication. Skipping the region due to a template switch as a part of the FoSTeS mechanism, for example, may lead to a deletion, or duplication, as seen in P47 and P48. It also can explain the presence of the region without copy number change found between deletion and duplication in P6. Among the three patients with 9q34.3 complex triplications, skipped regions were found within the most distal region of chromosome and may have resulted from template switching events as a result of a replication fork delay or replication from multiple initiation sites (Drosopolous et al. 2012). Furthermore, template switching between inverted repeats can result in a DUP-TRP/INV-DUP structure (Carvalho et al. 2011). Such complex rearrangements may be caused by certain genomic sequence features that can result in stalling of DNA replication forks such as low complexity repeats or GC-rich stretches, forming irregular DNA structures (Bacolla et al. 2004), by chromatin modifications or due to the influence of telomere assembling protein complexes. These findings, and the recent documentation of apparent FoSTeS generated complexities at chromosome ends in C elegans telomerase mutants, suggest that the most distal portion of chromosome is genomically unstable perhaps because it is prone to DNA replication errors consistent with the idea that the FoSTeS/MMBIR DNA replication-based mechanisms may play a central role in structural rearrangements within subtelomeric regions.
Supplementary Material
Acknowledgments
We are thankful to the families for their cooperation. This study was supported in part by grants the IDDRC (Intellectual and Develeopmental Disabilities Research Center (P30 HD024064) and the National Institute of Neurological Disorders and Stroke (R01 NS058529) to JRL.
Footnotes
Supplemental data includes one table containing genomic coordinates of the rearrangements within the 9q34 region.
Web Resources
BLAST, http://www.ncbi.nlm.nih.gov/blast/ (for BLASTn, BLASTz, and BLAST2)
Ensembl Genome Browser, http://www.ensembl.org/
NCBI, http://www.ncbi.nih.gov/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
UCSC Genome Browser, http://genome.ucsc.edu/
Disclosure
J.R.L. is a consultant for Athena Diagnostics, owns stock in 23andMe and Ion Torrent Systems Inc., and is a co-inventor on multiple US and European patents for DNA diagnostics. Furthermore, the Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular diagnostic testing (MGL, http://www.bcm.edu/geneticlabs/).
References
- Anand RP, Shah KA, Niu H, Sung P, Mirkin SM, Freudenreich CH. Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 2012;40:1091–1105. doi: 10.1093/nar/gkr836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva SV, Drozdova VD, Ponochevnaia EV, Kavardakova NV. Rearrangements of chromosome 9 in different hematological neoplasia. Tsitol Genet. 2008;42:72–79. [PubMed] [Google Scholar]
- Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J Biol Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- Ballif BC, Wakui K, Gajecka M, Shaffer LG. Translocation breakpoint mapping and sequence analysis in three monosomy 1p36 subjects with der(1)t(1;1)(p36;q44) suggest mechanisms for telomere capture in stabilizing de novo terminal rearrangements. Hum Genet. 2004;114:198–206. doi: 10.1007/s00439-003-1029-y. [DOI] [PubMed] [Google Scholar]
- Ballif BC, Yu W, Shaw CA, Kashork CD, Shaffer LG. Monosomy 1p36 breakpoint junctions suggest pre-meiotic breakage–fusion bridge cycles are involved in generating terminal deletions. Hum Mol Genet. 2003;12:2153–2165. doi: 10.1093/hmg/ddg231. [DOI] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 Retrotransposition Activity in Human Genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Beri S, De Agostini C, Novara F, Fichera M, Grillo L, Galesi O, Vetro A, Ciccone R, Bonati MT, Giglio S, Guerrini R, Osimani S, Marelli S, Zucca C, Grasso R, Borgatti R, Mani E, Motta C, Molteni M, Romano C, Greco D, Reitano S, Baroncini A, Lapi E, Cecconi A, Arrigo G, Patricelli MG, Pantaleoni C, D’Arrigo S, Riva D, Sciacca F, Dalla Bernardina B, Zoccante L, Darra F, Termine C, Maserati E, Bigoni S, Priolo E, Bottani A, Gimelli S, Bena F, Brusco A, di Gregorio E, Bagnasco I, Giussani U, Nitsch L, Politi P, Martinez-Frias ML, Martínez-Fernández ML, Martínez Guardia N, Bremer A, Anderlid BM, Zuffardi O. Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with Phelan/McDermid syndrome. PLoS Genet. 2011;7:e1002173. doi: 10.1371/journal.pgen.1002173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Mani E, Aceti G, Anderlid BM, Baroncini A, Pramparo T, Zuffardi O. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q133 deletion syndrome. J Med Genet. 2006;43:822–828. doi: 10.1136/jmg.2005.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone PM, Liu P, Zhang F, Carvalho CM, Towne CF, Batish SD, Lupski JR. Alu-specific microhomology-mediated deletion of the final exon of SPAST in three unrelated subjects with hereditary spastic paraplegia. Genet Med. 2011;13:582–592. doi: 10.1097/GIM.0b013e3182106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, Hines-Dowell S, Seaver LH, Friehling L, Lee S, Smith R, Del Gaudio D, Withers M, Liu P, Cheung SW, Belmont JW, Zoghbi HY, Hastings PJ, Lupski JR. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. 2011;43:1074–1081. doi: 10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Zhang F, Liu P, Patel A, Sahoo T, Bacino CA, Shaw C, Peacock S, Pursley A, Tavyev YJ, Ramocki MB, Nawara M, Obersztyn E, Vianna-Morgante AM, Stankiewicz P, Zoghbi HY, Cheung SW, Lupski JR. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum Mol Genet. 2009;18:2188–2203. doi: 10.1093/hmg/ddp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HC, Yatsenko SA, Kadapakkam M, Legay H, Su J, Lupski JR, Plon SE. Constitutional tandem duplication of 9q34 that truncates EHMT1 in a child with ganglioglioma. Pediatr Blood Cancer. 2011 doi: 10.1002/pbc.23219. doi:101002/pbc23219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, Blumenthal I, Mills RE, Kirby A, Lindgren AM, Rudiger SR, McLaughlan CJ, Bawden CS, Reid SJ, Faull RL, Snell RG, Hall IM, Shen Y, Ohsumi TK, Borowsky ML, Daly MJ, Lee C, Morton CC, MacDonald ME, Gusella JF, Talkowski ME. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet. 2012;44:390–397. doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo CS, Gajecka M, Kim CA, Gentles AJ, Glotzbach CD, Shaffer LG, Koiffmann CP. Further delineation of nonhomologous-based recombination and evidence for subtelomeric segmental duplications in 1p36 rearrangements. Hum Genet. 2009;125:551–563. doi: 10.1007/s00439-009-0650-9. [DOI] [PubMed] [Google Scholar]
- Dave BJ, Wiggins M, Higgins CM, Pickering DL, Perry D, Aoun P, Abromowich M, DeVetten M, Sanger WG. 9q34 rearrangements in BCR/ABL fusion-negative acute lymphoblastic leukemia. Cancer Genet Cytogenet. 2005;162:30–37. doi: 10.1016/j.cancergencyto.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Devriendt K, Matthijs G, Holvoet M, Schoenmakers E, Fryns JP. Triplication of distal chromosome 10q. J Med Genet. 1999;36:242–245. [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos WC, Kosiyatrakul ST, Yan Z, Calderano SG, Schildkraut CL. Human telomeres replicate using chromosome-specific, rather than universal, replication programs. J Cell Biol. 2012;197:253–266. doi: 10.1083/jcb.201112083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Craddock CF, Villegas A, Bentley DP, Williams HJ, Galanello R, Cao A, Wood WG, Ayyub H, Higgs DR. Healing of broken human chromosomes by the addition of telomeric repeats. Am J Hum Genet. 1994;55:505–512. [PMC free article] [PubMed] [Google Scholar]
- Flint J, Rochette J, Craddock CF, Dode C, Vignes B, Horsley SW, Kearney L, Buckle VJ, Ayyub H, Higgs DR. Chromosomal stabilization by a subtelomeric rearrangement involving two closely related Alu elements. Hum Mol Genet. 1996;5:1163–1169. doi: 10.1093/hmg/5.8.1163. [DOI] [PubMed] [Google Scholar]
- Gijsbers AC, Bijlsma EK, Weiss MM, Bakker E, Breuning MH, Hoffer MJ, Ruivenkamp CA. A 400kb duplication, 24 Mb triplication and 130 kb duplication of 9q343 in a patient with severe mental retardation. Eur J Med Genet. 2008;51:479–487. doi: 10.1016/j.ejmg.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Harrison KJ, Teshima IE, Silver MM, Jay V, Unger S, Robinson WP, James A, Levin A, Chitayat D. Partial tetrasomy with triplication of chromosome (5)(p14–15.33) in a patient with severe multiple congenital anomalies. Am J Med Genet. 1998;79:103–107. [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A Microhomology-Mediated Break-Induced Replication Model for the Origin of Human Copy Number Variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CRL, Schneider AM, Lu Y, Niranjan T, Shen P, Robinson MA, Steranka JP, Valle D, Civin CI, Wang T, Wheelan SJ, Ji H, Boeke JD, Burns KH. Mobile Interspersed Repeats Are Major Structural Variants in the Human Genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural Mutagenesis of Human Genomes by Endogenous Retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Koolen DA, Nillesen WM, de Leeuw N, Hamel BC, Veltman JA, Sistermans EA, van Bokhoven H, van Ravenswaay C, de Vries BB. Interstitial 2.2 Mb deletion at 9q34 in a patient with mental retardation but without classical features of the 9q subtelomeric deletion syndrome. Am J Med Genet A. 2006;140:618–623. doi: 10.1002/ajmg.a.31123. [DOI] [PubMed] [Google Scholar]
- Lamb J, Harris PC, Wilkie AO, Wood WG, Dauwerse JG, Higgs DR. De novo truncation of chromosome 16p and healing with (TTAGGG)n in the alpha thalasaemia/mental retardation syndrome (ATP-16) Am J Hum Genet. 1993;52:668–676. [PMC free article] [PubMed] [Google Scholar]
- Ledbetter DH, Martin CL. Cryptic telomere imbalance:a 15-year update. Am J Med Genet C Semin Med Genet. 2007;145C:327–334. doi: 10.1002/ajmg.c.30149. [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kołodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, Bacino CA, Campos-Acevedo LD, Delgado MR, Freedenberg D, Garnica A, Grebe TA, Hernández-Almaguer D, Immken L, Lalani SR, McLean SD, Northrup H, Scaglia F, Strathearn L, Trapane P, Kang SH, Patel A, Cheung SW, Hastings PJ, Stankiewicz P, Lupski JR, Bi W. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowden MR, Flibotte S, Moerman DG, Ahmed S. DNA synthesis generates terminal duplications that seal end-to-end chromosome fusions. Science. 2011;332:468–471. doi: 10.1126/science.1199022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders:structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Retrotransposition and Structural Variation in the Human Genome. Cell. 2010;141:1110–1112. doi: 10.1016/j.cell.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders ten years on. Genome Med. 2009;1:42. doi: 10.1186/gm42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28:810–820. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Pfeiffer RA, Trautmann U. Deletion or triplication of the a3(VI) collagen gene in three patients with 2q37 chromosome aberrations and symptoms of collagen-related disorders. Clin Genet. 1996;49:279–285. doi: 10.1111/j.1399-0004.1996.tb03789.x. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard G, Molina J, Chrast J, Gu W, Gheldof N, Pradervand S, Schütz F, Young JI, Lupski JR, Reymond A, Walz K. Phenotypic consequences of copy number variation:insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol. 2010;8:e1000543. doi: 10.1371/journal.pbio.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera H, Bobadilla L, Rolon A, Kunz J, Crolla JA. Intrachromosomal triplication of distal 7p. J Med Genet. 1998;35:78–80. doi: 10.1136/jmg.35.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Shaw CA, Lu XY, Sahoo T, Bacino CA, Lalani SR, Stankiewicz P, Yatsenko SA, Li Y, Neill S, Pursley AN, Chinault AC, Patel A, Beaudet AL, Lupski JR, Cheung SW. Identification of chromosome abnormalities in subtelomeric regions by microarray analysis: A study of 5,380 cases. Am J Med Genet. 2008;146A:2242–2251. doi: 10.1002/ajmg.a.32399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima K, Mano T, Kashiwagi M, Tanabe T, Sugawara M, Okamoto N, Arai H, Yamamoto T. Pelizaeus-Merzbacher disease caused by a duplication-inverted triplication-duplication in chromosomal segments including the PLP1 region. Eur J Med Genet. 2012;55:400–403. doi: 10.1016/j.ejmg.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Simovich MJ, Yatsenko SA, Kang S-HL, Cheung SW, Dudek ME, Pursley A, Ward PA, Patel A, Lupski JR. Prenatal diagnosis of a 9q343 microdeletion by array-CGH in a fetus with an apparently balanced translocation. Prenat Diag. 2007;27:1112–1117. doi: 10.1002/pd.1841. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, Ernst C, Hanscom C, Rossin E, Lindgren AM, Pereira S, Ruderfer D, Kirby A, Ripke S, Harris DJ, Lee JH, Ha K, Kim HG, Solomon BD, Gropman AL, Lucente D, Sims K, Ohsumi TK, Borowsky ML, Loranger S, Quade B, Lage K, Miles J, Wu BL, Shen Y, Neale B, Shaffer LG, Daly MJ, Morton CC, Gusella JF. Sequencing Chromosomal Abnormalities Reveals Neurodevelopmental Loci that Confer Risk across Diagnostic Boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga T, Aplan PD. Chromosomal aberrations induced by double strand DNA breaks. DNA Repair (Amst) 2005;4:1038–1046. doi: 10.1016/j.dnarep.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley H, Di S, Scherer SW, Royle NJ. Characterization of terminal deletions at 7q32 and 22q133 healed by de novo telomere addition. Am J Hum Genet. 2000;67:610–622. doi: 10.1086/303050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239 (94):197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Lamb J, Harris PC, Finney RD, Higgs DR. A truncated human chromosome 16 associated with alpha thalassaemia is stabilized by the addition of telomeric repeats (TTAGGG)n. Nature. 1990;346:868–871. doi: 10.1038/346868a0. [DOI] [PubMed] [Google Scholar]
- Yatsenko SA, Brundage EK, Roney EK, Cheung SW, Chinault AC, Lupski JR. Molecular mechanisms for subtelomeric rearrangements associated with the 9q343 microdeletion syndrome. Hum Mol Genet. 2009a;18:1924–1936. doi: 10.1093/hmg/ddp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko SA, Cheung SW, Scott DA, Nowaczyk MJ, Tarnopolsky M, Naidu S, Bibat G, Patel A, Leroy JG, Scaglia F, Stankiewicz P, Lupski JR. Deletion 9q343 syndrome:genotype-phenotype correlations and an extended deletion in a patient with features of Opitz C trigonocephaly. J Med Genet. 2005;42:328–335. doi: 10.1136/jmg.2004.028258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko SA, Shaw CA, Ou Z, Pursley AN, Patel A, Bi W, Cheung SW, Lupski JR, Chinault AC, Beaudet AL. Microarray-based comparative genomic hybridization using sex-matched reference DNA provides greater sensitivity for detection of sex chromosome imbalances than array-comparative genomic hybridization with sex-mismatched reference DNA. J Mol Diagn. 2009b;11:226–237. doi: 10.2353/jmoldx.2009.080064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Seeman P, Liu P, Weterman MA, Gonzaga-Jauregui C, Towne CF, Batish SD, De Vriendt E, De Jonghe P, Rautenstrauss B, Krause KH, Khajavi M, Posadka J, Vandenberghe A, Palau F, Van Maldergem L, Baas F, Timmerman V, Lupski JR. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP:rare CNVs as a cause for missing heritability. Am J Hum Genet. 2010;86:892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuffardi O, Bonaglia M, Ciccone R, Giorda R. Inverted duplications deletions: underdiagnosed rearrangements? Clin Genet. 2009;75:505–513. doi: 10.1111/j.1399-0004.2009.01187.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.