Abstract
Dietary factors and the associated lifestyle play a major role in the pathophysiology of many diseases. Several diets, especially a Western lifestyle with a high consumption of meat and carbohydrates and a low consumption of vegetables, have been linked to common diseases, such as metabolic syndrome, atherosclerosis, inflammatory bowel diseases, and colon cancer. The gastrointestinal tract harbors a complex and yet mainly molecularly defined microbiota, which contains an enormous number of different species. Recent advances in sequencing technologies have allowed the characterization of the human microbiome and opened the possibility to study the effect of "environmental" factors on this microbiome. The most important environmental factor is probably "what we eat," and the initial studies have revealed fascinating results on the interaction of nutrients with our microbiota. Whereas short-term changes in dietary patterns may not have major influences, long-term diets can affect the microbiota in a substantial manner. This issue may potentially have major relevance for human gastrointestinal health and disease because our microbiota has features to regulate many immune and metabolic functions. Increasing our knowledge on the interaction between nutrients and microbiota may have tremendous consequences and result in a better understanding of diseases, even beyond the gastrointestinal tract, and finally lead to better preventive and therapeutic strategies.
Keywords: Nutrition, Inflammation, Intestinal immunity, Microflora
INTRODUCTION
The human gastrointestinal tract harbors more than 100 trillion bacteria defining the gut microbiota.1 A huge number of other organisms such as archaea, viruses, parasites, or fungi are also part of the gut microflora. The microbiota consists of ten times more bacteria than human cells in the body, including up to more than 1,000 species dominated by anaerobic bacteria and encode for 100- to 200-fold more genes than our own genome.2 It is well known that our microbiota controls the development of the immune system, regeneration of the epithelium and recruitment of various leukocytes into the epithelium. Importantly, evolutionary conserved mechanisms allow these microorganisms not only to live in peace with the host but also to exert complementary especially metabolic functions, which cannot be performed by the host itself. Tolerance of the microbiota is only possible by an efficient physical barrier which is exerted by the mucus and in addition by reduction of antigenic moieties of the microbiota and active immune processes achieving this state of tolerance. Moreover, recent studies have demonstrated that the gut microbiota with its products interacts with host pathways (e.g., epithelial cells) and thereby controls host energy expenditure and storage. Abnormal and impaired microbiota has been identified recently in many diverse diseases such as inflammatory bowel diseases, colorectal cancer, irritable bowel syndrome, metabolic syndrome, or non-alcoholic fatty liver disease.3-6
The composition of microbial communities is generally considered stable within each individual.7 In this study, the authors confirmed such a stability, however, also showed that not only antibiotic therapy but also other features such as overseas travelling or temporary illness affected the microbiota. A human core microbiome has been suggested and may include a common group of organisms, gene/protein families and/or metabolic functions.8 Also elderly persons demonstrate a remarkably stable microbiota although the core microbiota of elderly subjects seems to differ from younger people with greater numbers of Bacteroides spp. and different abundance patterns of certain Clostridium groups.9 Recent studies have demonstrated that there might exist a huge impact of environmental factors such as diet on the gut's microbiome.10 Genetic host factors may be less important as dissimilarity in gut bacterial communities is huge in identical twins.8 In this article we will discuss the role of dietary factors modulating the gut's microbiota.
METAGENOMIC INSIGHTS INTO OUR MICROBIOTA
Metagenomics is defined as recovery of genetic material directly from environmental surfaces, e.g., the gut and therefore includes analysis of the entire DNA in an organism. Even in the first sequence-based characterizations of the human microbiome it became evident that there exists a significant enrichment in metabolic pathways favoring energy harvest from diet.11,12 The development of functional metagenomics allowed to identify new functions of the microbiota especially in the metabolism of dietary fibers by carbohydrate active enzymes to degrade them into stable monosaccharides and disaccharides.13 A landmark publication has recently presented for the first time a human gut microbial catalogue,1 describing more than 3 million non-redundant microbial genes in our microbiota suggesting that our microbiota contains 150 times more genes than it's host. Furthermore, over 99% of genes are bacterial and each individual might contain more than 150 different species. Importantly, they further observed that around 40% of one's individual bacterial genes are shared by at least 50% of subjects highlighting the concept of a core microbiome and high level of functional similarities between individuals. Recently, Arumugam et al.14 suggested the presence of certain enterotypes in humans based on functional metagenomic analysis of three different patient cohorts from different areas in the world. It is still unclear what "enterotype" means with respect to functional consequences but further studies should enable us to prove this very interesting concept. Furthermore, studies will demonstrate whether certain enteroytpes are associated with diseases respectively disease patterns. The evolution of metagenomic analysis already had a major impact on the understanding of our microbiota and opened a fascinating rapidly evolving field in human science.
DIETARY FACTORS AFFECTING THE HUMAN MICROBIOME
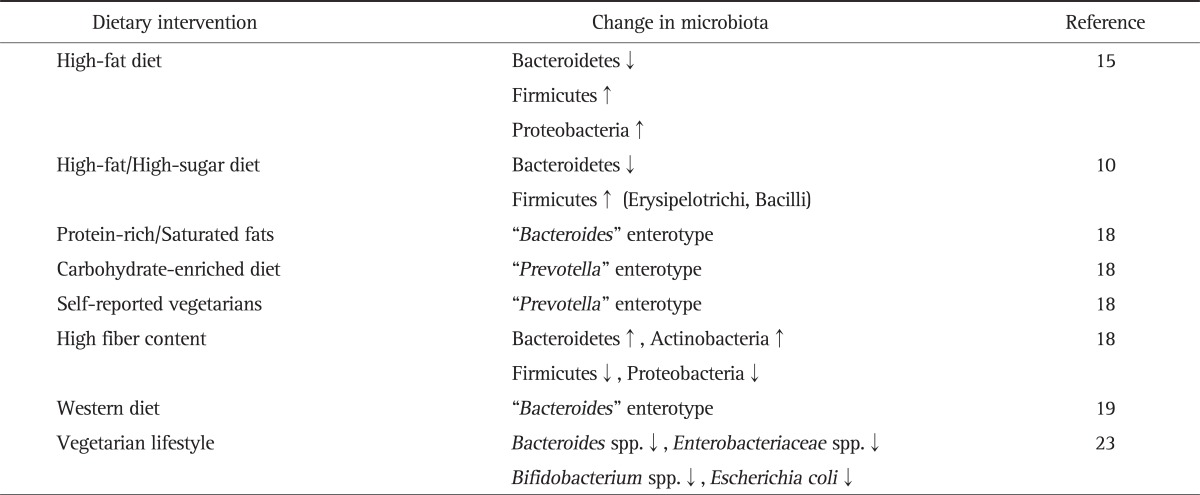
As sequencing techniques have only evolved recently it is not surprising that there is still only moderate evidence available how certain dietary factors affect the gut's microbiota/microbiome. One of the key and central questions is the fact whether and how diet might affect the composition of the gut microbiome. This question is essential to address as otherwise recently generated microbiome data might become irrelevant or limited in their interpretation. Hildebrandt et al.15 recently presented data how a high-fat diet might affect the composition of the murine gut microbiome even independently of obesity. In their study, the investigators compared wild type and resistin-like molecule beta/FIZZ2-deficient mice and assessed the influence of diet, genotype and obesity on the microbiome composition. Importantly, the authors found substantial changes in the gut microbiome when switching to a high-fat diet with a decrease in Bacteroidetes and an increase in Firmicutes and Proteobacteria and observed changes were independent of obesity. Turnbaugh et al.10 recently presented a further study into this direction. They transplanted fresh or frozen adult human fecal microbial communities into germ-free C57BL/6J mice. Interestingly, these humanized mice were stably and heritably colonized and reproduced much of the bacterial diversity of the donor's microbiota. A change in the diet (i.e., from a low-fat, plant polysaccharide to a high-fat, high-sugar diet) shifted the structure of the microbiota even in a single day, changed respective metabolic pathways in the microbiome, and affected microbiome gene expression. These humanized mice showed increased adiposity and this trait was also transmissible via microbiota transplantation. Therefore, both studies clearly show that diet critically affects the gut microbiome, changes occur rapidly even within a single day and adiposity is transmissible by stool transplantation.
Beyond bacteria and archaea an incredible number of viruses are part of the microflora.16 In this first report, Gordon and colleagues reported sequencing of the viromes of virus-like particles isolated from faecal samples collected from healthy adult female monozygotic twins and their mothers at three time points over a 1-year period. Co-twins and their mothers shared a significantly greater degree of similarity in their faecal bacterial communities than did unrelated individuals. Minot et al.17 recently studied the human virome and effects of certain diets. The largest source of variance among virome samples was interpersonal variation. Interestingly, dietary intervention was associated with a change in the virome community in which individuals on the same diet converged. This important study therefore suggests that dietary factors not only affect the bacteriota but also the virome, a fascinating new world.
Probably the most important clinical study investigating interaction between diet and the microbiome came from Wu et al.18 In this study, they assessed the microbiota by pyrosequencing of 16S rDNA gene segments in 98 subject undergoing different diets. Whereas short-term diets had no influence on their enteroytpes, long-term diets indeed were able to influence and affect enterotype of individuals: whereas diets enriched in protein and animal fat favoured the "Bacteroides" enterotype, a carbohydrate enriched diet supported the "Prevotella" enterotype. The enterotype clustering was driven primarily by the ratio of the two dominant genera, Prevotella to Bacteroides, which defines a gradient across the two enterotypes. The Bacteroides enterotype was highly associated with animal protein, a variety of amino acids, and saturated fats suggesting that meat consumption as in a Western diet characterized this enterotype. The Prevotella enterotype, in contrast, was associated with low values for these groups but high values of carbohydrates and simple sugars, indicating association with a carbohydrate-based diet as used mainly in agrarian societies. Self-reported vegetarians showed enrichment in the Prevotella enterotype. Whether this finding is clinically important is not yet known, as these enterotypes so far have not been associated with certain disease patterns. Phyla positively associated with fiber were Bacteroidetes and Actinobacteria, whereas Firmicutes and Proteobacteria showed the opposite association. It is important to mention though, that a short-term diet over 24 hours with either high-fat/low-fiber or low-fat/high-fiber diet affected the microbiota, although in a moderate way. Taxa affected differed among individuals. However, one cannot rule that even minor changes could have certain consequences for human health and disease. Interestingly, several other factors affected microbiome composition such as body mass index, red wine and aspartame consumption, raising other important questions. The fact that an artificial sweetener can modify substantially our microbiota is remarkable and warrants further studies. Interestingly, bacteria with presumed health benefits such as Faecalibacterium prausnitzii have not been associated in this study with a certain enterotype.19 The data of Wu et al.18 are in accordance with a recent study comparing European and African children.19 In this study, a Western diet was also associated with the dominance of Bacteroides whereas a more vegetarian diet in Africa was dominated by the Prevotella enterotype. Despite this being a first study,18 it opened a new and exciting field and hopefully we will learn rapidly whether "enterotypes" indeed exist generally in human beings and their potential associations with human disease. Many more studies assessing the role of dietary factors on our microbiota are needed, as it seems likely that our diet is the "environmental" factor regulating and modifying the microbiota. Table 1 summarizes the effect of diets on the intestinal microbiota.
Table 1.
Effect of Various Diets on the Intestinal Microbiota
ENERGY HARVESTING OF OUR MICROBIOTA AND EFFECTS OF DIET
Analysis of the composition of or microbiota has demonstrated that obese subjects harboured a variety of mainly two prevailing phyla, Firmicutes and Bacteroidetes.20 It is still a matter of debate how and why our microbiota could affect energy harvesting, thereby improving overweight and metabolic functions. Increased harvesting had been proposed to occur via major shifts in the Firmicutes/Bacterodetes ratio. Murphy et al.21 addressed this issue in a recent publication looking at the effects of various diets, especially a high-fat diet in studying ob/ob mice. Seven-week-old ob/ob mice were fed a low-fat diet, whereas wild type mice received either a low-fat or high-fat diet. In their thoroughly performed study they not only assessed the microbiota (by metagenomic pyrosequencing) but also studied fecal energy consumption by calorimetry and faecal short chain fatty acid content. As in earlier studies they also found a progressive increase in Firmicutes after high-fat diet and in ob/ob mice, as they have seen changes in Bacteroidetes. Importantly, however, these changes were not paralleled by changes in energy harvesting and proportions of the various phyla did not correlate with energy harvest markers. Similar to earlier studies,22 Murphy et al.21 observed reduced energy content and an increase in short chain fatty acid levels in the caecum of young ob/ob mice. Importantly, these changes were not observed after high-fat feeding and especially in older mice. The authors concluded that the relationship between our microbiota and energy consumption is far more complex, needs further studies and is potentially affected by microbial adaptation to a certain diet over time. Therefore, beyond various diets other factors such as aging might play a central role.
This is an important issue as certain studies already suggested that manipulation of our microbiota, e.g., via fecal transplants could an attractive new weight loss strategy. Before such strategies should be initiated, it is mandatory to improve our understanding of the complexity of the relationship between the gut microbiota and energy harvesting. International ongoing sequencing projects at the moment generate enormous amounts of information about our microbiota and potential metabolic functions. These data alone, however, will not allow to address functional aspects which on the one hand are mandatory to understand the complex interaction between microbiota and metabolic host functions. New tools are needed, e.g., colonization of gnotobiotic mice with selective human flora and the effects of various diets. Such models have recently been introduced to study the role of Firmicutes and Bacteroidetes in carbohydrate metabolism.10
EFFECT OF A VEGETARIAN LIFESTYLE?
Another approach to better define the role of certain dietary factors on the gut's microbiota could be investigating people with a well-defined diet such as vegans or vegetarians. Zimmer et al.23 examined faecal samples of vegetarians, vegans and a similar number of control people using ordinary omnivorous diet. Total counts of Bacteroides spp., Bifidobacterium spp., Escherichia coli and Enterobacteriaceae spp. were significantly lower in vegan samples than in controls, whereas others (E. coli biovars, Klebsiella spp., Enterobacter spp., other Enterobacteriaceae, Enterococcus spp., Lactobacillus spp., Citrobacter spp., and Clostridium spp.) were not affected. Interestingly, subjects on a vegetarian diet showed results in between vegans and controls. The authors concluded, that a strict vegan or vegetarian diet results in a significant shift in the microbiota supporting above discussed findings by Wu et al.,18 that long-term diets might indeed affect the microbiota.23
Another study compared the fecal microbiota of vegetarian and omnivorous young women in southern India. Fecal samples were collected from 32 lacto-vegetarian and 24 omnivorous young adult women. Fecal microbiota of was quantified by real-time PCR with SYBR Green using primers targeting 16S rRNA genes of groups, including: Clostridium coccoides group (Clostridium cluster XIVa), Roseburia spp.-Eubacterium rectale, Bacteroides-Prevotella group, Bifidobacterium genus, Lactobacillus group, Clostridium leptum group (Clostridium cluster IV), F. prausnitzii, Ruminococcus productus-C. coccoides, Butyrivibrio, Enterococcus species, and Enterobacteriaceae. Importantly, the fecal microbiota of the omnivorous group was enriched with Clostridium cluster XIVa bacteria, specifically Roseburia-E. rectale. Omnivores showed an increase of Clostridium cluster XIVa bacteria and butyryl-CoA CoA-transferase gene compared with vegetarians, but the authors failed to identify the components of the diet responsible for this difference.24 Evidence therefore is increasing that long-term diet is relevant in influencing the microbiota.
ENDOTOXIN: ANOTHER DIET-REGULATED PLAYER IN METABOLIC INFLAMMATION?
Earlier studies in experimental animals have convincingly demonstrated that high-fat diets result in endotoxemia with evidence of systemic inflammation suggesting that dietary-modification of the gut's microbiota may be involved.25,26 A similar mechanism might be effective in humans. Pendyala et al.27 recently presented data where they treated eight healthy subjects with a Western-style diet for 1 month inducing a 71% increase in plasma levels of endotoxin activity, whereas a moderate and balanced diet reduced levels by 31%. The Western-style diet might, therefore, contribute to endotoxemia by causing changes in gastrointestinal barrier function or the composition of the microbiota. Endotoxemia might also develop in individuals with gastrointestinal barrier impairment. Therapeutic reagents that reduce endotoxemia might reduce systemic inflammation in patients with gastrointestinal diseases or metabolic syndrome. These data are in favour of the view that certain diets affect the microbiota thereby generating pro-inflammatory, detrimental pathways for the host. Another piece of evidence into this direction has been recently reported by Wang et al.28 Research by this group suggests that the link to cardiovascular disease could be through the gut. Phospatidylcholine is a fatty substance found commonly in certain types of food. Three metabolites of the dietary lipid phosphatidylcholine-choline, trimethylamine N-oxide (TMAO), and betaine-exist. Wang et al.28 identified after screening for small-molecule metabolites circulating in people's plasma and observed the presence of byproducts generated after gut bacteria break down of phospatidylcholine. Presence and levels of such byproducts importantly correlated with later development of cardiovascular disease. Experiments in mice then showed that the microbiota has a central role in setting off a metabolic chain reaction that leads to this dietary lipid getting converted into TMAO, which boosts the formation of artery-forming plaques. These findings add to an increasing evidence showing that the microbiota can cause or worsen certain conditions, including obesity and immune disorders. The work also suggests that drugs might be able to target TMAO to prevent atherosclerosis and ischemic heart disease. This study is another beautiful link between dietary factors, the microbiota and systemic inflammation/disease.
LESSONS FROM OTHER MAMMALIANS
This important study addressed whether dietary factors might affect the composition of the microbiota in 33 different mammalians.29 They observed that the adaptation to diet is similar in different mammalian lineages and importantly found that the relationship among mammalian gut microbiomes is that they share a large core repertoire of functions. Studies also included functional aspects where they could demonstrate that carnivorous microbiomes have specialized to degrade proteins as an energy source, whereas herbivorous communities have specialized to synthesize amino acids. Herbi- and carbivorous not only differed in their metabolic potential to affect amino acid metabolism, such a difference was also observed in glucose metabolism. Carbivorous and herbivorous microbiomes showed opposing directionality at the central phosphoenolpyruvate (PEP)-pyruvate-oxaloacetate (OAA) node. When gluconeogenesis is needed, OAA can be converted to PEP and pyruvate. All of the genes encoding enzymes catalyzing OAA production from pyruvate to PEP are significantly increased in the carnivore microbiomes, whereas the reverse reactions are catalyzed by enzymes whose representation is increased in herbivore microbiomes. In the human part of the study, 18 lean individuals adherent to a strict diet (i.e., members of the Calorie Restriction Society) were included with detailed assessment of their dietary behavior. Both structure and function of their gut microbiome were significantly associated with dietary intake. Overall, this fascinating story tells us that dietary factors might be highly associated with consecutive functional properties of our microbiome such as metabolism of amino acids and glucose.
HOW TO PROCEED IN THE FUTURE? SMART EXPERIMENTAL APPROACHES
Faith et al.30 recently presented an attractive animal model to study effects of various diets on human microbial communities. In their studies, they used gnotobiotic animals (germ-free mice) and transferred 10 sequenced human gut bacteria containing the most common four bacterial phyla into these animals. Shotgun sequencing of fecal DNA in these animals was performed on days 1, 2, 4, 7, and 14 of a given diet period. The total DNA yield per stool pellet increased as the amount of casein (i.e., reflects protein consumption) in the host diet increased. Interestingly, changes in species abundance as a function of changes in the concentration of casein in the host diet were also apparent for all 10 species: seven species such as Bacteroides caccae were positively correlated with amount of casein consumption whereas others, e.g., E. rectale were negatively correlated. These data not only show that such a model might be perfectly suitable to study effects of certain diets on microbial communities but also support the evidence that protein rich diets result in an increase of Bacteroides as reported in the study of Wu et al.18 In their model, Faith et al.30 could also investigate the effects of complex diets on bacterial community members. For these studies they created 48 meals consisting of random combinations and concentrations of four ingredients selected from a set of eight pureed human baby foods. Importantly, and this again reflects the strength of this model, the authors were able to explain more than half of the variation in species abundance only knowing the concentrations of the pureed foods present in each meal.
CONCLUSIONS AND OUTLOOK
The excitement of metagenomics has just started allowing a whole-genome approach to our microbiota, which so far could not have been assessed properly using conventional methodology as most gut bacteria cannot be cultured. Our microbial community may profoundly affect the development of fat mass development, glucose intolerance, diabetes, and low-grade systemic inflammation. It evolved as a fascinating insight in the last years that the microbiota in itself exemplifies many important biological functions regulating important metabolic functions of the host. This insight has boosted the interest of many various disciplines in this topic. Assuming that the microbiota plays a fundamental role in directing metabolic and immune functions, to identify and understand the so-called "environmental" factors controlling the microbiota are of even greater interest. Dietary factors are very likely to be on the "very top" of this list. First studies have highlighted that rather long-term dietary strategies might impact composition of our microbiota. Much more information and studies are needed into this direction.
The notion that the 'obese microbiota' might harvest more energy from the diet, and that the intestinal microbiota might at the same time direct the host response to energy intake, could offer new therapeutic approaches to obesity.31 What are the logical next steps to achieve? We should approach and search for nutritional interventions to manipulate specific gut microbial species. Both prebiotics and probiotics could have the potential to affect gut microbiota/microbiome modifying such "an obese microbiome." The germ-free mouse system as recently reported by Jeff Gordon's group could be an ideal model to study new pre-/probiotics into this direction. Furthermore, certain antibiotics could also be developed which might selectively modulate an "obese microbiome." It is fascinating to recognize that indeed the gut microbiome might reflect this critical "intestinal trigger" linking environment and host in obesity. This "wonderful box" has just been opened and a new area of clinical science has been started. Further insights might not only improve our understanding of gut's biology but also redefine our current view of many diseases far beyond the gastrointestinal tract.
ACKNOWLEDGEMENTS
This study was supported by the Christian Doppler Research Society.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashida H, Ogawa M, Kim M, Mimuro H, Sasakawa C. Bacteria and host interactions in the gut epithelial barrier. Nat Chem Biol. 2011;8:36–45. doi: 10.1038/nchembio.741. [DOI] [PubMed] [Google Scholar]
- 3.DuPont AW, DuPont HL. The intestinal microbiota and chronic disorders of the gut. Nat Rev Gastroenterol Hepatol. 2011;8:523–531. doi: 10.1038/nrgastro.2011.133. [DOI] [PubMed] [Google Scholar]
- 4.Lepage P, Häsler R, Spehlmann ME, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Thomas LV, Ockhuizen T. New insights into the impact of the intestinal microbiota on health and disease: a symposium report. Br J Nutr. 2012;107(Suppl 1):S1–S13. doi: 10.1017/S0007114511006970. [DOI] [PubMed] [Google Scholar]
- 6.Tilg H, Kaser A. Gut microbiome, obesity, and metabolic dysfunction. J Clin Invest. 2011;121:2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jalanka-Tuovinen J, Salonen A, Nikkilä J, et al. Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS One. 2011;6:e23035. doi: 10.1371/journal.pone.0023035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claesson MJ, Cusack S, O'Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurokawa K, Itoh T, Kuwahara T, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tasse L, Bercovici J, Pizzut-Serin S, et al. Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res. 2010;20:1605–1612. doi: 10.1101/gr.108332.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyes A, Haynes M, Hanson N, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minot S, Sinha R, Chen J, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy EF, Cotter PD, Healy S, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- 22.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 23.Zimmer J, Lange B, Frick JS, et al. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur J Clin Nutr. 2012;66:53–60. doi: 10.1038/ejcn.2011.141. [DOI] [PubMed] [Google Scholar]
- 24.Kabeerdoss J, Shobana Devi R, Regina Mary R, Ramakrishna BS. Faecal microbiota composition in vegetarians: comparison with omnivores in a cohort of young women in southern India. Br J Nutr. 2011 Dec 20; doi: 10.1017/S0007114511006362. Epub. DOI: http://dx.doi.org/10.1017/S0007114511006362. [DOI] [PubMed] [Google Scholar]
- 25.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 26.Cani PD, Neyrinck AM, Fava F, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 27.Pendyala S, Walker JM, Holt PR. A high-fat diet is associated with endotoxemia that originates from the gut. Gastroenterology. 2012;142:1100–1101. doi: 10.1053/j.gastro.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muegge BD, Kuczynski J, Knights D, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]