Abstract
Background
Congenital adrenal hyperplasia (CAH) due to 21-hydroxylase deficiency (21-OHD) is an autosomal recessive disorder of cortisol biosynthesis caused by CYP21A2 mutations. An increase in gene copy number variation (CNV) exists at the CYP21A2 locus. CNV of C4, a neighboring gene that encodes complement component 4, is associated with autoimmune disease susceptibility.
Methods
Comprehensive genetic analysis of the RCCX (RP-C4-CYP21-TNX) region was conducted in 127 unrelated 21-OHD patients (100 classic, 27 nonclassic). C4 copy number was determined by Southern blot. C4 CNV and serum C4 levels were evaluated in relation to CYP21A2 mutations and relevant phenotypes.
Results
The most common CYP21A2 mutation associated with the nonclassic form of CAH, V281L, was associated with high C4 copy number (p=7.13×10−16). Large CYP21A2 deletion was associated with low C4 copy number (p=1.61×10−14). Monomodular RCCX with a short C4 gene, a risk factor for autoimmune disease, was significantly less frequent in CAH patients compared to population estimates (2.8 vs. 10.6%; p=1.08×10−4).
Conclusions
CAH patients have increased C4 CNV, with mutation-specific associations that may be protective for autoimmune disease. The study of CYP21A2 in relation to neighboring genes provides insight into the genetics of CNV hotspots, an important determinant of human health.
Keywords: Congenital adrenal hyperplasia, 21-hydroxylase, complement component 4, copy number variation
INTRODUCTION
Congenital adrenal hyperplasia (CAH, OMIM 201910) due to 21-hydroxylase deficiency (21-OHD) is an autosomal recessive disorder of the adrenal cortex characterized by impaired cortisol biosynthesis with or without aldosterone deficiency. The defect in cortisol biosynthesis results in a compensatory increase in corticotrophin and stimulation of the adrenal cortex, leading to androgen excess.[1] CAH manifests a broad spectrum of phenotypes and is classified according to clinical severity: the classic or severe form occurs in approximately one case per 15,000 live births worldwide, [1] while a more common mild or nonclassic (NC) form has a prevalence of one in 1,000 in Caucasians.[2]
The CYP21A2 (OMIM 201910) gene encoding 21-hydroxylase is mapped at the central region of the human major histocompatibility complex on chromosome 6, and is in tandem with three neighboring genes (RP1, C4 and TNXB) from the telomeric to centromeric ends to constitute a genetic module termed RCCX (RP-C4-CYP21-TNX). RP1 encodes a serine/threonine nuclear protein kinase, C4 encodes for the immune effector protein complement component, and TNX (tenascin) is a member of the extracellular matrix protein family. With the exception of C4, each of the other functional genes (RP1, CYP21A2 and TNXB) has a corresponding highly homologous pseudogene (RP2, CYP21A1P and TNXA). Due to the highly homologous sequences between functional and pseudogenes, the RCCX modules are characterized by modular duplication or deletion events in which each duplicated or deleted module usually covers a CYP21A1P-TNXA-RP2-C4 unit. Although bimodular RCCX is the most common haplotype, great variation of RCCX length has been observed to generate misalignment between homologous genes during meiosis, leading to unequal crossovers.[3] Consequently, copy number variation (CNV) of C4, CYP21, and TNX is widely found in humans and this complexity has challenged the genetic analysis of relevant human conditions.
One of the major clinical aspects of the genetics of RCCX is C4 CNV. C4 is an important effector protein that plays a crucial role in the classical and the mannose-binding lectin activation pathways of the immune system.[4–6] In addition to two isotopes, the human C4 gene can be either 21kb (C4 long, C4L) or 14.6kb (C4 short, C4S) in length, depending on the integration of the endogenous retrovirus HERV-K (C4) into the ninth intron. While complete C4 deficiency is extremely rare, susceptibility to autoimmune diseases such as systemic lupus erythematosus has been associated with lower C4 copy number and lower serum C4 protein levels.[7]
The present study is among our series of comprehensive genetic investigations of RCCX in a large cohort of patients with CAH.[8–10] In CAH, the majority of CYP21A2 mutations originate from the CYP21A1P pseudogene and are transferred by gene conversion during meiosis. [11] This high frequency of RCCX crossover events led us to hypothesize that CAH patients may have C4 CNV associated with specific CYP21A2 genotypes that may modulate autoimmune disease risk.
The aim of our study was to extend the genetic analysis of the RCCX module to evaluate C4 CNV in our large CAH cohort. We determined copy number of total C4, C4L and C4S and evaluated C4 CNV in relation to CYP21A2 genotype, expected population C4 CNV, [12] autoimmune disease, and serum C4 protein levels.
METHODS
Patients
We evaluated 127 unrelated patients with CAH due to 21-OHD (100 classic, 27 NC; 63 males, 64 females; mean age±SD: 18.5±13.1 years), a subgroup of our previously reported cohort enrolled in a Natural History Study at the NIH Clinical Center in Bethesda, Maryland, USA (Clinical Trial No. NCT00250159).[10] Only patients with complete genetic data, including Southern blot, were included. Autoimmune disease status was determined per clinical history and medical records. The study was approved by The Eunice Kennedy Shriver National Institute of Child Health and Human Development Institutional Review Board. Informed consent and assent for minors ≥ 8 years old were obtained.
Genotyping of the RCCX module
Defective CYP21A2 alleles of each patient were confirmed in a comprehensive approach, including sequencing, Southern blot, multiplex ligation-dependent probe amplification (MLPA), and other PCR-based methods.[10]
Per established protocol, [13] TaqI and PshAI Southern blot was performed to characterize the RCCX modules.[10] The probe A used in the TaqI Southern blots provided genotyping of four possible combinations between the RP (RP1 and RP2) and C4 (C4L and C4S) genes. The PshAI digestion was employed to confirm copy number ratio between RP1 and RP2 genes, which confirmed the total number of RCCX modules, hence C4 gene dosages seen in TaqI digestion.
C4 serum protein concentrations
Serum protein concentrations of C4 were determined by a Vista analyzer (NIH Clinical Center Laboratory, Bethesda, Maryland, USA).
Statistical analysis
Gene copy number of total C4, C4L, and C4S were compared amongst groups using Chi-square test, Fisher’s exact test, as appropriate. Group comparisons of C4 serum concentrations were carried out using the independent t test or one-way analysis of variance, as appropriate. Average values were expressed as mean±SD. A two-tailed p value of <0.05 was considered significant. Analyses were performed using SPSS statistical software V19.0.0 (SPSS Inc, Chicago, Illinois, USA).
RESULTS
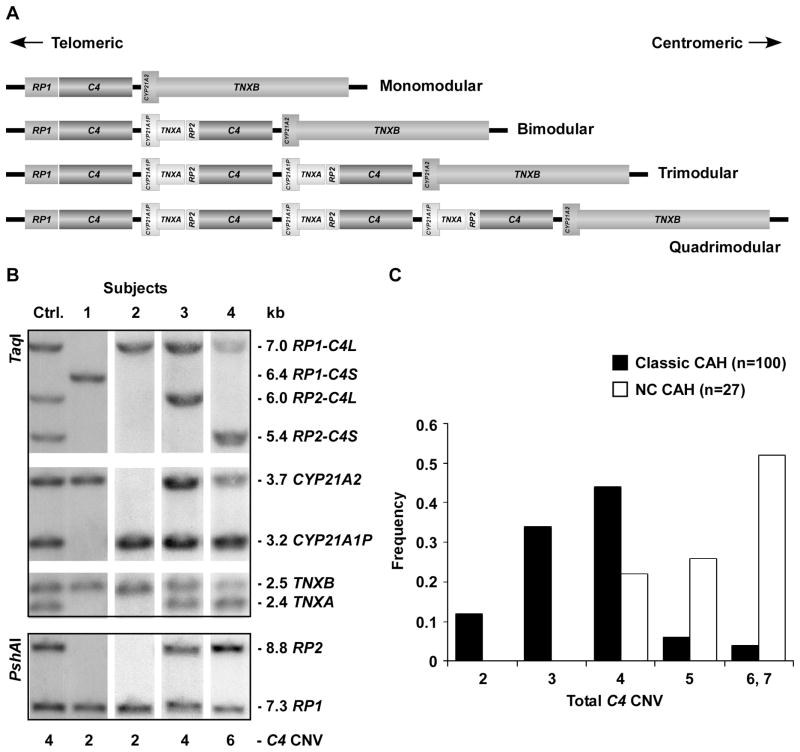
Great RCCX length variation was observed and the CNV of each gene corresponded to the number of RCCX modules (figure 1A–B). Overall, only 39% of our CAH population had the most commonly reported four copies of C4, as compared to 61% of population estimates.[12] C4 copy number was significantly higher in the nonclassic than the classic group (5.30±0.82 vs. 3.57±0.96; p=3.52×1014) (figure 1C). Correspondingly, the serum level of total C4 was greater in NC than classic patients (33.0±6.3 vs. 21.2±6.6; p=4.14×10−5).
Figure 1.
Copy number variation (CNV) of RCCX (RP-C4-CYP21-TNX) modules and C4 genes. (A) Schematic representation of four types of RCCX haplotypes resulting from modular duplication or deletion events at the central region of the human major histocompatibility complex (MHC) on chromosome 6. (B) CNV of the RCCX constituting genes was found by Southern blotting with TaqI and PshAI restriction fragment length polymorphism (RFLP). The control shows equal intensity of RP1 and RP2, indicating 4 copies of C4: one is short and three are long. Patient 1 is homozygous for a monomodular RCCX with a C4S gene and lacks RP2, CYP21A1P and TNXA. Similarly, Patient 2 is homozygous for a monomodular RCCX with a C4L gene. Patient 3 has 4 copies of C4, which are all long. Patient 4 has 6 copies of C4: four are short and the other two are long because the 5.4 kb RP2-C4S TaqI band is twice as intense as the 7.0 kb RP1-C4L TaqI band (confirmed by a PshAI Southern blot), and the same intensity ratio of CYP21A1P:CYP21A2 and TNXA:TNXB. (C) Phenotypic variation was observed for total C4 copy number.
Similar numbers of CAH patients had extremely low (9.4% with 2) and high (14.2% with 6–7) copies of C4. When compared with a previously published group of healthy European Americans, [12] our CAH patients had a different C4 copy number distribution (p=5.24×10−12), which was greatest for C4S (p=7.96×10−12). The copy numbers of RP1-C4L, RP1-C4S, RP2-C4L and RP2-C4S in our CAH population were 247, 7, 110, and 136, respectively. There was a significant association between RP and long or short C4 genes (p=1.31×10−38) regarding relative location. This suggests that the RP1 gene is overwhelmingly in tandem with the C4L gene in RCCX modules. Three of the 7 RP1-C4S alleles were determined to be a monomodular RCCX module with a C4S gene (mono-S). Although we were not able to determine the phases of RCCX modules where the remaining 4 RP1-C4S alleles were located, the allele frequency of mono-S in our CAH patients was significantly lower than published norms (p=1.08×10−4). None of our NC CAH patients had the mono-S haplotype, a risk factor for autoimmune disease; 1.5% of the classic CAH patients had this haplotype.
V281L, the most common nonclassic CYP21A2 mutation, was associated with high C4 copy number (table 1). Additionally, higher V281L mutation copy number was associated with increased copies of C4 (table 1). No association was found between other less frequent nonclassic CYP21A2 mutations and C4 copy number.
Table 1.
Copy number variation of the C4 gene and the CYP21A2 V281L mutation or chimera gene
| Copy number of CYP21A2 V281L mutation |
C4 Copy Number Variation (CNV)
|
Total | C4 Gene Copy Index1 | P value of t test | ||||
|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6, 7 | ||||
| 0 | 12 | 33 | 43 | 5 | 1 | 94 | 3.47±0.83 | 7.00×10−11 (0 vs. 1) |
| 1 | 0 | 1 | 6 | 8 | 6 | 21 | 4.95±0.97 | 7.71×10−3 (1 vs. 2) |
| 2 | 0 | 0 | 1 | 0 | 11 | 12 | 5.83±0.58 | 3.49×10−10 (0 vs. 2) |
|
| ||||||||
| Fisher’s exact test | p=7.13×10−16 | |||||||
|
| ||||||||
| Copy number of chimera gene |
C4 Copy Number Variation (CNV)
|
Total | C4 Gene Copy Index1 | P value of t test | ||||
| 2 | 3 | 4 | 5 | 6, 7 | ||||
|
| ||||||||
| 0 | 1 | 3 | 35 | 8 | 15 | 62 | 4.53±0.97 | 3.78×10−6 (0 vs. 1) |
| 1 | 2 | 27 | 14 | 5 | 3 | 51 | 3.63±1.00 | 7.47×10−5 (1 vs. 2) |
| 2 | 9 | 4 | 1 | 0 | 0 | 14 | 2.43±0.65 | 4.50×10−11 (0 vs. 2) |
|
| ||||||||
| Fisher’s exact test | p=1.61×10−14 | |||||||
Gene Copy Index (GCI) indicates that mean±SD of C4 copy number within a selected population.[7]
C4 copy number was also significantly different based on the copy number of chimera genes at the RCCX locus (table 1); having a chimera gene at the RCCX locus was associated with a decrease in C4 copy number. The overall distribution difference of C4 copy number was significant when deletion copy number varied (table 1). In particular, 75% (9 out of 12) of patients who carried two copies of C4 had two copies of chimera genes, accounting for approximately two-thirds of the patients who carried two chimera genes.
In our cohort of 127 CAH patients, four had history of autoimmune disease: two with Hashimoto thyroiditis, one with inflammatory bowel disease, and one with seronegative juvenile rheumatoid arthritis. No association was found between history of autoimmunity and C4 CNV or C4 serum levels.
DISCUSSION
In this study, we report a comprehensive investigation of the complement component 4 gene in a large cohort of North American patients with CAH due to 21-OHD. In genetic studies of the RCCX module, the C4, CYP21A2, and TNXB genes have been studied extensively in autoimmune disease, CAH, and connective tissue disorders, respectively. Extending genetic analysis from a causative single gene perspective to the whole RCCX module in relation to phenotype is of great importance, as exemplified by cases of TNX-deficiency found in CAH patients.[8, 14] We found that CAH patients have greater C4 CNV, with mutation-specific associations that may be protective for autoimmune disease.
Our patients with CAH demonstrated a significantly different distribution of C4 copy number than expected, [12, 15] with an increase in extremely low and high C4 copy number. Interestingly, the approximate 20% decrease in the expected four copies of C4 in our cohort was almost equally distributed between the groups with either 2 or ≥ 6 C4 copies. This suggests that unequal crossover events in the population of defective CYP21A2 carriers resulted in an increase of monomodular and longer (trimodular or quadrimodular) RCCX haplotypes, and that these haplotypes were transmitted to and concentrated in the CAH population. Therefore, having CAH appears to increase the probability of having either very low or very high C4 copy number.
Nonclassic CAH is among the most common autosomal recessive diseases, with Ashkenazi Jews having the highest prevalence.[16] We report the novel finding of an association between the V281L mutation, the most frequent CYP21A2 mutation found in NC CAH patients, [10, 16] and an increase in C4 copy number, indicating that the V281L mutation may be much more frequent in longer RCCX modules at a haplotype level. Since higher C4 copies are protective against autoimmune disease, our finding leads to an intriguing hypothesis that carrier status for V281L may present an evolutionary advantage. Under the alignment model proposed by Blanchong et al. to explain possible pairings of heterozygous RCCX length variants during meiosis, [3] the CYP21A2 gene carrying a V281L mutation may discourage alignment to the homologous sequence on the paired chromosome. Consequently, the V281L mutation is enriched in longer RCCX modules with higher C4 copy number. Furthermore, higher C4S copy number and greater serum level of total C4 in the NC patients support the role of C4S in increasing C4 protein concentration.[12]
The RCCX module is characterized by modular duplication or deletion events. In particular, large gene deletions result in chimera genes at the centromeric tail of the RCCX module, which account for approximately 30% of CYP21A2 mutations.[10, 17] These chimera genes are mostly present in a form of CYP21A1P/CYP21A2, [9] but also TNXA/TNXB in subset of patients in which the functional CYP21A2 gene is completely deleted and the deletion extends into the TNXB gene.[8, 14] As expected, we found that having a chimera gene at the RCCX locus was associated with a decrease in total C4 copy number, but this was not associated with autoimmune disease.
Our findings support prior studies of C4 in classic CAH patients; the low frequency of mono-S allele was reported in a relatively small cohort of 22 classic CAH patients [3] and in 46 unrelated Brazilian classic CAH patients.[18] Yu et al. suggested that lower frequency of mono-S might reduce autoimmune disease risk.[3, 12] Similarly, the reduced frequency of mono-S in our CAH cohort may explain our low incidence of autoimmune disorders, however, the young age distribution of the patients may be a confounder.
The human C4 locus has been identified as a functional CNV hotspot.[19] The genome architecture at these CNV loci has a potential role in stirring up mutational events.[19] Harboring a highly homologous sequence, the RCCX module is inherently prone to gene duplication or deletion events by virtue of specific characteristics of the local DNA sequence environment.[20] The involvement of multiple genes in these events results in concurrent CNVs across the surrounding genes. Consequently, the overall clinical phenotypes of a patient go beyond the simple pattern of “single gene disorder” when the causative gene is located in a functional CNV hotspot. Our data provides insight into the genetics of copy number variation hotspots, the effect on multiple neighboring genes, and the ultimate impact on human disease.
Acknowledgments
We are grateful to the patients for their participation in this study. We are indebted to Dr. Chack-Yung Yu (Department of Pediatrics, The Ohio State University, Columbus, Ohio, USA) for providing probes for Southern blot and valuable discussion of the genetics of the RCCX module. We want to thank NIA Core Laboratory staff for DNA extraction and sample processing, and Ms. Tina Roberson for administrative assistance.
Funding: This work was supported (in part) by the Intramural Research Programs of the The Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), the National Institutes of Health Clinical Center, and the National Institute of Aging (NIA) and (in part) by The Congenital Adrenal Hyperplasia Research, Education and Support (CARES) Foundation. Dr. Merke is a Commissioned Officer in the United States Public Health Service.
Footnotes
Contributors: WC acquired the laboratory data, collected and analysed the data, and wrote the manuscript; ZX acquired the laboratory data; MN collected and analysed data; CVR collected clinical data; NBM supervised the laboratory studies and performed critical revision of the manuscript; DPM supervised the overall study design, analysis and interpretation of data and performed critical revision of the manuscript.
Competing interests: None.
Patient consent: Obtained.
Ethics approval: The Eunice Kennedy Shriver National Institute of Child Health and Human Development Institutional Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Merke DP, Bornstein SR. Congenital adrenal hyperplasia. Lancet. 2005;365:2125–36. doi: 10.1016/S0140-6736(05)66736-0. [DOI] [PubMed] [Google Scholar]
- 2.Speiser PW, Dupont B, Rubinstein P, et al. High frequency of nonclassical steroid 21-hydroxylase deficiency. Am J Hum Genet. 1985;37:650–67. [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchong CA, Zhou B, Rupert KL, et al. Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in caucasians. The load of RCCX genetic diversity on major histocompatibility complex-associated disease. J Exp Med. 2000;191:2183–96. doi: 10.1084/jem.191.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walport MJ. Complement. Second of two parts. N Engl J Med. 2001;344:1140–4. doi: 10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- 5.Walport MJ. Complement. First of two parts. N Engl J Med. 2001;344:1058–66. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 6.Yu CY, Chung EK, Yang Y, et al. Dancing with complement C4 and the RP-C4-CYP21-TNX (RCCX) modules of the major histocompatibility complex. Prog Nucleic Acid Res Mol Biol. 2003;75:217–92. doi: 10.1016/s0079-6603(03)75007-7. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, Chung EK, Wu YL, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80:1037–54. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Kim MS, Shanbhag S, et al. The phenotypic spectrum of contiguous deletion of CYP21A2 and tenascin XB: quadricuspid aortic valve and other midline defects. Am J Med Genet A. 2009;149A:2803–8. doi: 10.1002/ajmg.a.33092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Xu Z, Sullivan A, et al. Junction site analysis of chimeric CYP21A1P/CYP21A2 genes in 21-hydroxylase deficiency. Clin Chem. 2012;58:421–30. doi: 10.1373/clinchem.2011.174037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkielstain GP, Chen W, Mehta SP, et al. Comprehensive genetic analysis of 182 unrelated families with congenital adrenal hyperplasia due to 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2011;96:E161–72. doi: 10.1210/jc.2010-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koppens PF, Hoogenboezem T, Degenhart HJ. Carriership of a defective tenascin-X gene in steroid 21-hydroxylase deficiency patients: TNXB -TNXA hybrids in apparent large-scale gene conversions. Hum Mol Genet. 2002;11:2581–90. doi: 10.1093/hmg/11.21.2581. [DOI] [PubMed] [Google Scholar]
- 12.Saxena K, Kitzmiller KJ, Wu YL, et al. Great genotypic and phenotypic diversities associated with copy-number variations of complement C4 and RP-C4-CYP21-TNX (RCCX) modules: a comparison of Asian-Indian and European American populations. Mol Immunol. 2009;46:1289–303. doi: 10.1016/j.molimm.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung EK, Wu YL, Yang Y, et al. Human complement components C4A and C4B genetic diversities: complex genotypes and phenotypes. Curr Protoc Immunol. 2005;Chapter 13(Unit 13.8) doi: 10.1002/0471142735.im1308s68. [DOI] [PubMed] [Google Scholar]
- 14.Burch GH, Gong Y, Liu W, et al. Tenascin-X deficiency is associated with Ehlers-Danlos syndrome. Nat Genet. 1997;17:104–8. doi: 10.1038/ng0997-104. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Chung EK, Zhou B, et al. Diversity in intrinsic strengths of the human complement system: serum C4 protein concentrations correlate with C4 gene size and polygenic variations, hemolytic activities, and body mass index. J Immunol. 2003;171:2734–45. doi: 10.4049/jimmunol.171.5.2734. [DOI] [PubMed] [Google Scholar]
- 16.New MI. Extensive clinical experience: nonclassical 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2006;91:4205–14. doi: 10.1210/jc.2006-1645. [DOI] [PubMed] [Google Scholar]
- 17.Stikkelbroeck NM, Hoefsloot LH, de Wijs IJ, et al. CYP21 gene mutation analysis in 198 patients with 21-hydroxylase deficiency in The Netherlands: six novel mutations and a specific cluster of four mutations. J Clin Endocrinol Metab. 2003;88:3852–9. doi: 10.1210/jc.2002-021681. [DOI] [PubMed] [Google Scholar]
- 18.Guerra-Junior G, Grumach AS, de Lemos-Marini SH, et al. Complement 4 phenotypes and genotypes in Brazilian patients with classical 21-hydroxylase deficiency. Clin Exp Immunol. 2008;155:182–8. doi: 10.1111/j.1365-2249.2008.03838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu W, Zhang F, Wang Y, et al. Identification of copy number variation hotspots in human populations. Am J Hum Genet. 2010;87:494–504. doi: 10.1016/j.ajhg.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper DN, Bacolla A, Ferec C, et al. On the sequence-directed nature of human gene mutation: the role of genomic architecture and the local DNA sequence environment in mediating gene mutations underlying human inherited disease. Hum Mutat. 2011;32:1075–99. doi: 10.1002/humu.21557. [DOI] [PMC free article] [PubMed] [Google Scholar]