Abstract
Objective. The growth of the biomedical literature presents special challenges for both human readers and automatic algorithms. One such challenge derives from the common and uncontrolled use of abbreviations in the literature. Each additional abbreviation increases the effective size of the vocabulary for a field. Therefore, to create an automatically generated and maintained lexicon of abbreviations, we have developed an algorithm to match abbreviations in text with their expansions.
Design. Our method uses a statistical learning algorithm, logistic regression, to score abbreviation expansions based on their resemblance to a training set of human-annotated abbreviations. We applied it to Medstract, a corpus of MEDLINE abstracts in which abbreviations and their expansions have been manually annotated. We then ran the algorithm on all abstracts in MEDLINE, creating a dictionary of biomedical abbreviations. To test the coverage of the database, we used an independently created list of abbreviations from the China Medical Tribune.
Measurements. We measured the recall and precision of the algorithm in identifying abbreviations from the Medstract corpus. We also measured the recall when searching for abbreviations from the China Medical Tribune against the database.
Results. On the Medstract corpus, our algorithm achieves up to 83% recall at 80% precision. Applying the algorithm to all of MEDLINE yielded a database of 781,632 high-scoring abbreviations. Of all the abbreviations in the list from the China Medical Tribune, 88% were in the database.
Conclusion. We have developed an algorithm to identify abbreviations from text. We are making this available as a public abbreviation server at \url{http://abbreviation.stanford.edu/}.
The amount of literature in biomedicine is exploding as MEDLINE grows by 400,000 citations each year. With biomedical knowledge expanding so quickly, professionals must acquire new strategies to cope with it. To alleviate this problem, the biomedical informatics community is investigating methods to organize,1 summarize,2 and mine3 the literature.
Understanding biomedical literature is particularly challenging because of its expanding vocabulary, including the unfettered introduction of new abbreviations. An automatic method to define abbreviations would help researchers by providing a self-updating abbreviation dictionary and also facilitate computer analysis of text.
This article defines abbreviation broadly to include all strings that are shortened forms of sequences of words (its long form). Although the term acronym appears more commonly in literature, it is typically defined more strictly as a conjunction of the initial letter of words; some authors also require them to be pronounceable. Using such a strict definition excludes many types of abbreviations that appear in biomedical literature. Authors create abbreviations in many different ways, as summarized in Table 1▶.
Table 1 .
| Abb. | Definition | Description |
|---|---|---|
| VDR | vitamin D receptor | The letters align to the beginnings of the words. |
| PTU | propylthiouracil | The letters align to a subset of syllable boundaries. |
| JNK | c-Jun N-terminal kinase | The letters align to punctuation boundaries. |
| IFN | interferon | The letters align to some other place. |
| SULT | sulfotransferase | The abbreviation contains contiguous characters from a word. |
| ATL | adult T-cell leukemia | The long form contains words not in the abbreviation. |
| CREB-1 | CRE binding protein | The abbreviation contains letters not in the long form. |
| beta-EP | beta-endorphin | The abbreviation contains complete words. |
Nevertheless, the numerous lists of abbreviations covering many domains attest to broad interest in identifying them. Opaui, a web portal for abbreviations, contains links to 152 lists.4 Some are compiled by individuals or groups.5,6 Others accept submissions from users over the internet.7,8 For the medical domain, a manually collected published dictionary contains over 10,000 entries.9
Because of the size and growth of the biomedical literature, manual compilations of abbreviations suffer from problems of completeness and timeliness. Automated methods for finding abbreviations are therefore of great potential value. In general, these methods scan text for candidate abbreviations and then apply an algorithm to match them with the surrounding text. Most abbreviation finders fall into one of three types.
The simplest type of algorithm matches an abbreviation’s letters to the initial letters of the words around it. The algorithm for recognizing this type is relatively straightforward, although it must perform some special processing to ignore common words. Taghva gives as an example the Office of Nuclear Waste Isolation (ONWR), where the O can be matched with the initial letter of either “Office” or “of.”10
More complex methods relax the first letter requirement and allow matches to other characters. These typically use heuristics to favor matches on the first letter or syllable boundaries, upper case letters, length of acronym, and other characteristics.11 However, Yeates notes the challenge in finding optimal weights for each heuristic and further posits that machine learning approaches may help.12
Another approach recognizes that the alignment between an abbreviation and its long form often follows a set of patterns.13,14,15 Thus, a set of carefully and manually crafted rules governing allowed patterns can recognize abbreviations. Furthermore, one can control the performance of the system by adjusting the set of rules, trading off between the leniency in which a rule allows matches and the number of errors that it introduces.
In their rule-based system, Pustejovsky et al. introduced an interesting innovation by including lexical information.14 Their insight is that abbreviations are often composed from noun phrases and that constraining the search to definitions in the noun phrases closest to the abbreviation will improve precision. With the search constrained, they found that they could further tune their rules to also improve recall.
Finally, there is one completely different approach to abbreviation search based on compression.16 The idea here is that a correct abbreviation gives better clues to the best compression model for the surrounding text than an incorrect one. Thus, a normalized compression ratio built from the abbreviation gives a score capable of distinguishing abbreviations.
This article presents three contributions: a novel algorithm for identifying abbreviations, a set of features descriptive of various types of abbreviations, and a publically accessible abbreviation server containing all abbreviation definitions found in MEDLINE.
Methods
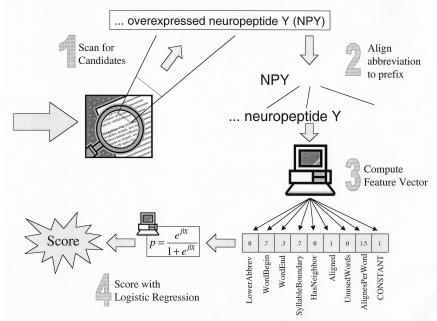
We decompose the abbreviation-finding problem into four components: (1) scanning text for occurrences of possible abbreviations, (2) aligning the candidates to the preceding text, (3) converting the abbreviations and alignments into a feature vector, and 4) scoring the feature vector using a statistical machine learning algorithm (Figure 1▶).
Figure 1 .
System architecture. We used a machine-learning approach to find and score abbreviations. First, we scan text to find possible abbreviations, align them with their prefix strings, and then collect a feature vector based on eight characteristics of the abbreviation and alignment. Finally, we apply binary logistic regression to generate a score from the feature vector.
Finding Abbreviation Candidates
We searched for possible abbreviations inside parentheses, assuming that they followed the pattern: long form (abbreviation).
Within each pair of parentheses, we retrieved the words up to a comma or semicolon. We rejected candidates longer than two words, candidates without any letters, and candidates that exactly matched the words in the preceding text.
For each abbreviation candidate, we saved the words before the open parenthesis (the prefix) so that we could search them for the abbreviation’s long form. Although we could have included every word from the beginning of the sentence, as a computational optimization, we only used 3N words, where N was the number of letters in the abbreviation. We chose this limit conservatively based on an informal observation that we always found long forms well within 3N words.
Aligning Abbreviations with Their Prefixes
For each pair of abbreviation candidate and prefix, we found the alignment of the letters in the abbreviation with those in the prefix. This is a case of the longest common substring (LCS) problem studied in computer science and adapted for biological sequence alignment in bioinformatics.17
We found the optimal alignments, those that maximize the number of matched letters, between two strings X and Y using dynamic programming in O(NM) time, where N and M were the lengths of the strings. This algorithm is expressed as a recurrence relation:
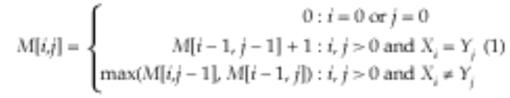 |
(1) |
M is a score matrix, and M[i,j] contains the total number of characters aligned between the substrings X1 . . . i and Y1 . . . j. To recover the aligned characters, we created a traceback parallel to the score matrix. This matrix stored pointers to the indexes preceding M[i,j]. After generating these two matrices, we recovered the alignment by following the pointers in the traceback matrix.
Computing Features from Alignments
Next we calculated feature vectors that quantitatively described each candidate abbreviation and the alignment to its prefix. For the abbreviation recognition task, we used 9 features described in Table 2▶. We chose that features we believed would be informative based on a manual examination of abbreviations found in arbitrarily chosen MEDLINE abstracts. Each feature constituted one dimension of a 9-dimension feature vector.
Table 2 .
Features Used for Scoring Abbreviations*
| Feature | Description | β |
|---|---|---|
| Describes the abbreviation | ||
| Lower case vs. upper case | Percent of letters in abbreviation in lower case. | –1.21 |
| Describes where the letters are aligned | ||
| Beginning of word | Percent of letters aligned at the beginning of a word. | 5.54 |
| End of word | Percent of letters aligned at the end of a word. | –1.40 |
| Syllable boundary | Percent of letters aligned on a syllable boundary. | 2.08 |
| After aligned letter | Percent of letters aligned immediately after another letter. | 1.50 |
| Describes the alignment | ||
| Letters aligned | Percent of letters in the abbreviation that are aligned. | 3.67 |
| Words skipped | Number of words in the prefix not aligned to the abbreviation. | –5.82 |
| Aligned letters per word | Average number of aligned letters per word. | 0.70 |
| Miscellaneous | ||
| CONSTANT | Normalization constant for logistic regression. | –9.70 |
*These features are used to calculate the score of an alignment using Equation 3. We identified syllable boundaries using the algorithm used in TEX [22]. The β column indicates the weight given to each feature. The sign of the weight indicates whether that feature is favorably associated with real abbreviations.
Scoring Alignments with Logistic Regression
Finally, we used a supervised machine-learning algorithm to recognize abbreviations. To train this algorithm, we created a training set of 1000 randomly-chosen candidates identified from a set of MEDLINE abstracts pertaining to human genes, which we had compiled for another purpose. From these abstracts, we identified 93 abbreviations and hand-annotated the alignment between the abbreviation and prefix.
Next we generated all possible alignments between the abbreviations and prefixes in our set of 1000. This yielded our complete training set, which consisted of (1) alignments of incorrect abbreviations, (2) correct alignments of correct abbreviations, and (3) incorrect alignments of correct abbreviations. We converted these alignments into feature vectors.
Using these feature vectors, we trained a binary logistic regression classifier.18 We chose this classifier based on its lack of assumptions on the data model, ability to handle continuous data, speed in classification, and probabilistically interpretable scores.
Binary logistic regression fits the feature vectors into a log odds (logit) function:
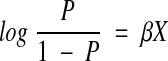 |
(2) |
with some manipulation:
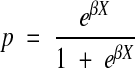 |
(3) |
where p is the probability of seeing an abbreviation, X is the feature vector, and β is the vector of weights. Thus, training this model consists of finding the β vector that maximizes the difference between the positive and negative examples.
We found the optimal β by maximizing the likelihood over all the training examples using:
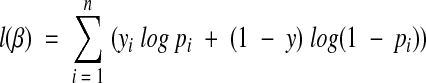 |
(4) |
where yi is 1 if training example i was a real abbreviation and 0 otherwise.
Since there is no known closed form solution to this equation, we used Newton’s method to optimize this equation to a global maximum. We initialize β to zeros, and although this is not guaranteed to converge, it usually does in practice.18 To alleviate singularity problems, we removed all the duplicate vectors from the training set.
Finally, an alignment’s score is the probability calculated from equation 3 using the optimal β vector. An abbreviation’s score is the maximum score of all the alignments to its prefix.
Evaluation
We evaluated our algorithm against the Medstract acronym gold standard,14 which contains MEDLINE abstracts with expert-annotated abbreviations and forms the basis of the evaluation of Acromed. The gold standard is publically available as an XML file at <http://www.medstract.org/gold-standards.html>.
We ran our algorithm against the Medstract gold standard (after correcting 6 typographical errors in the XML file) and generated a list of the predicted abbreviations, definitions, and their scores. With these predictions, we calculated the recall and precision at every possible score cutoff generating a recall/precision curve. Recall is defined as:
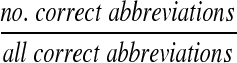 |
(5) |
and measures how thoroughly the method finds all the abbreviations. Precision is defined as:
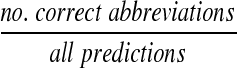 |
(6) |
and indicates the number of errors produced.
We counted an abbreviation/long form pair correct if it matched the gold standard exactly, considering only the highest scoring pair for each abbreviation. To be consistent with Acromed’s evaluation on Medstract, we allowed mismatches in 10 cases where the long form contained words not indicated in the abbreviation. For example, we accepted protein kinase A for PKA and did not require the full cAMP-dependent protein kinase A indicated in the gold standard.
In addition, we evaluated the coverage of the database against a list of abbreviations from the China Medical Tribune, a weekly Chinese language newspaper covering medical news from Chinese journals.17 The website includes a dictionary of 452 commonly used English medical abbreviations with their long forms. We searched the database for these abbreviations (after correcting 21 spelling errors) and calculated the recall as
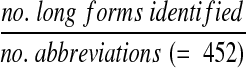 |
(7) |
Scanning MEDLINE
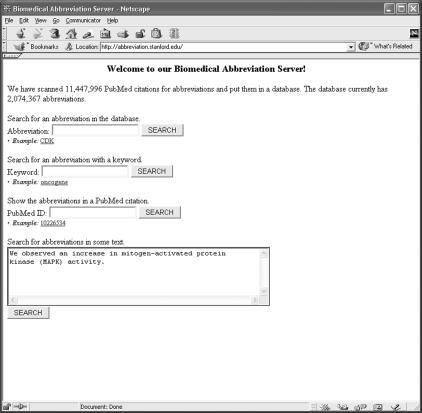
We searched MEDLINE abstracts to the end of the year 2001 for abbreviations and kept all predictions that scored at least 0.001. We then put those predictions into a relational database and built an abbreviation server, a web server that, given queries by abbreviation or word, returns abbreviations and their definitions. The server can also search for abbreviations in text provided by the user (Figure 2▶).
Figure 2 .
Abbreviation server. Our abbreviation server supports queries by abbreviation or keyword.
We implemented the code in Python 2.220 and C with the Biopython 1.00a4 and mxTextTools 2.0.3 libraries. The website was built with RedHat Linux 7.2, MySQL 3.23.46, and Zope 2.5.0 on a Dell workstation with a 1.5GHz Pentium IV and 512Mb of RAM.
Results
We ran our algorithm against the Medstract gold standard and calculated the recall and precision at various score cutoffs (Figure 3▶). Identifying 140 out of 168 correctly, it obtained a maximum recall of 83% at 80% precision (Table 3▶). The recall/precision curve plateaued at two levels of precision, 97% at 22% recall (score = 0.88) and 95% at 75% recall (score = 0.14).
Figure 3 .
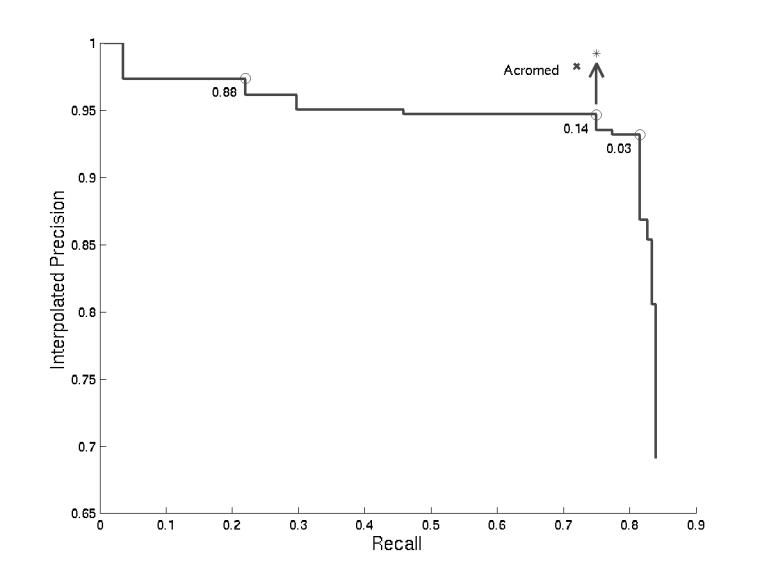
Abbreviations Predicted in Medstract Gold Standard. We calculated the recall and precision of the abbreviations found with every possible score cutoff. Some scores are labelled on the curve. When the score cutoff is 0.14, seven of the abbreviations the algorithm found were not identified in the gold standard but nevertheless looked correct (primary ethylene response element (PERE), basic helix-loop-helix (bHLH), intermediate neuroblasts defective (ind), Ca2+-sensing receptor (CaSR), GABA(B) receptor (GABA(B)R1), Polymerase II (Pol II), GABAB receptor (GABA(B)R2)). The arrow points to the adjusted performance if these abbreviations had been included in Medstract. The performance of the Acromed system on this gold standard, as reported in Pustejovsky et al.,14 is shown for comparison.
Table 3 .
Abbreviations in Medstract Gold Standard Missed*
| No. | Description | Example |
|---|---|---|
| 13 | “Abbreviation” and long form are synonyms. | apoptosis → programmed cell death |
| 7 | Abbreviation is outside parentheses. | |
| 3 | Best alignment score yields incorrect long form. | FasL → Fas and Fas ligand |
| 2 | Letters in abbreviation are out of order. | TH → helper T |
| 25 | TOTAL |
*Our algorithm failed to find 25 of the 168 abbreviations in the Medstract gold standard. This table categorizes types of abbreviations and the number of each type missed.
At a score cutoff of 0.14, the algorithm made 8 errors. 7 of those errors are abbreviations missing from the gold standard: primary ethylene response element (PERE), basic helix-loop-helix (bHLH), intermediate neuroblasts defective (ind), Ca2+-sensing receptor (CaSR), GABA(B) receptor (GABA(B)R1), polymerase II (Pol II), and GABAB receptor (GABA(B)R2). The final error occurred when an unfortunate sequence of words in the prefix yielded a higher scoring alignment than the long form: Fas and Fas ligand (FasL).
Then we scanned all MEDLINE abstracts until the end of 2001 for abbreviations. This required 70 hours of computation using five processors on a Sun Enterprise E3500 running Solaris 2.6. In all, we processed 6,426,981 MEDLINE abstracts (only about half of the 11,447,996 citations had abstracts) at an average rate of 25.5 abstracts/second.
From this scan, we identified a total of 1,948,246 abbreviations from MEDLINE, and 20.7% of them were defined in more than one abstract. Only 2.7% were found in five or more abstracts; 2,748,848 (42.8%) of the abstracts defined at least 1 abbreviation and 23.7% of them defined 2 or more.
Of the nearly two million abbreviation/definition pairs, there were only 719,813 distinct abbreviations because many of them had different definitions (e.g., AR can stand for autosomal recessive, androgen receptor, amphiregulin, aortic regurgitation, aldose reductase, among others). More than one definition, was available for 156,202 abbreviations (21.7%).
The average number of definitions for abbreviations with six characters or less was 4.61, higher than the 2.28 reported by Liu et al.21 One possible reason for this discrepancy is that Liu’s method correctly counts morphological variants of the same definition. Both methods, however, overcount definitions that have the same meaning, but different words. We found that 37.5% of the abbreviations with six characters or less had multiple definitions, which concurs with Liu’s 33.1%.
Of the abbreviations, 781,632 have a score of at least 0.14. Of those, 328,874 (42.1%) are acronyms (i.e., they are composed of the first letters of words).
The growth rate of both abstracts in MEDLINE and new abbreviation definitions is increasing (Figure 4▶). Last year 64,262 new abbreviations were introduced, and there is an average of one new abbreviation in every 5–10 abstracts.
Figure 4 .
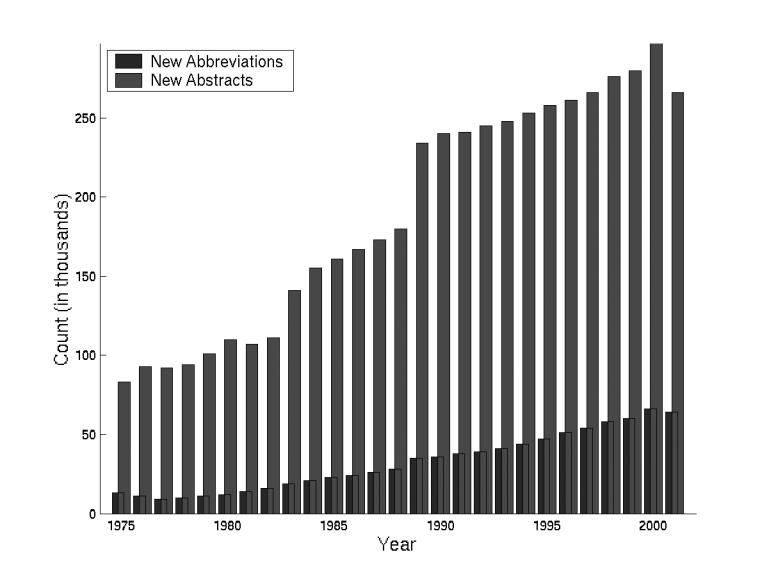
Growth of abstracts and abbreviations. The number of abstracts and abbreviations added to MEDLINE steadily increases.
We searched for abbreviations from the China Medical Tribune against our database of all MEDLINE abbreviations. Allowing differences in capitalization and punctuation, we matched 399 of the 452 abbreviations to their correct long forms for a maximum recall of 88% (Figure 5▶). Using a score cutoff of 0.14 yields a recall of
Figure 5 .
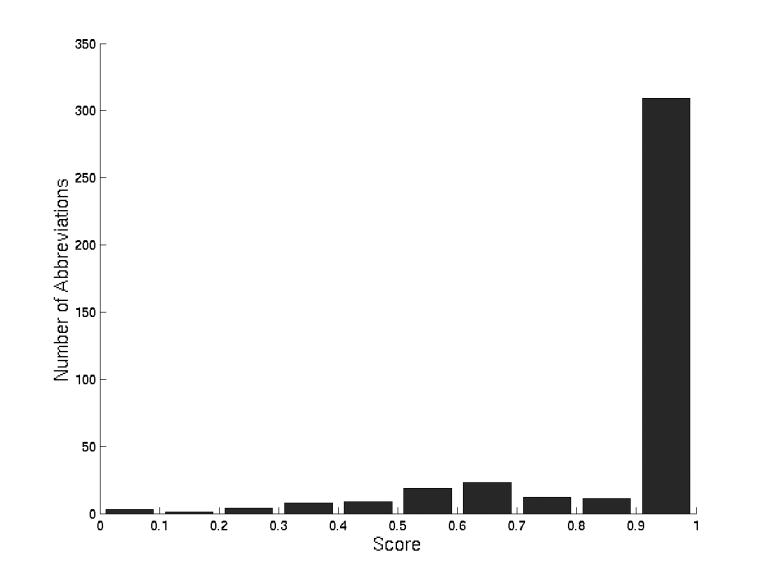
Scores of correct abbreviations from the China Medical Tribune. Using a score cutoff of 0.90 yields a recall of 68%, 0.14 87%, and 0.03 88%.
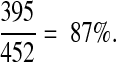 |
Of the 53 abbreviations missed, 11 were in the database as a close variation, such as elective repeat caesarean section instead of elective repeat C-section. Also, when applied on the abbreviation and long form pairs, the algorithm could identify 45 of the 53 abbreviations with a score cutoff of 0.14. However, ambiguities in free text may lead to higher error rates. Extraneous words may be included in the long form.
Discussion
With the enormous number of abbreviations currently in MEDLINE and the rate at which prolific authors define new ones, maintaining a current dictionary of abbreviation definitions clearly requires automated methods. Since nearly half of MEDLINE abstracts contain abbreviations, computer programs analyzing this text will frequently encounter them and can benefit from their identification. Since less than half of all abbreviations are formed from the initial letters of words, automated methods must handle more sophisticated and nonstandard constructs.
Thus, we created a robust method for identifying abbreviations using supervised machine learning. The method uses a set of features that describe different patterns seen commonly within abbreviations. We evaluated it against the Medstract gold standard because it was easily available, it eliminated the need to develop an alternate standard, and it provided a reference point to compare methods.
The majority of the errors on this data set (see Table 2▶) occurred because the gold standard included synonyms, words, and phrases with identical meanings. In such cases, the algorithm could not find the correspondences between letters, indicating a fundamental limitation of letter-matching techniques.
The largest remaining source of error was from our strong assumption that the abbreviation must be inside parentheses and the long form outside. The algorithm missed seven abbreviations that immediately preceded the long form, which was inside parentheses. To handle this problem, the candidate finder should also allow this pattern. However, it is unclear how adding more candidates may impact the precision.
Our precision in this evaluation was hurt by abbreviations missing from the gold standard. Our algorithm identified eight of these, and seven had scores higher than 0.14. Disregarding these cases yields a precision of 99% at 82% recall, which is comparable to Acromed at 98% and 72%.
Believing the algorithm to have sufficient performance, we ran it against all of MEDLINE and put the results in a database as an abbreviation server. During validation, we found that the server contained 88% of the abbreviations from the dictionary in the China Medical Tribune. Since this list was created independently of MEDLINE, the results demonstrate that this server contains most of the common abbreviations of interest to medical professionals. To improve the recall even further, Yu has shown that linking to external dictionaries of abbreviations can augment the ability of automated methods to assign definitions that are not indicated in the text.15
We note that using the server to assign meanings to abbreviations must be done carefully. Since about one-fifth of all abbreviations were degenerate, the correct one must be disambiguated using the context of the abbreviation. Pustejovsky has done some work showing the suitability of the vector-space model for this task.14
We are making the abbreviation server available at <http://abbreviation.stanford.edu/>. This server contains all the abbreviations the algorithm found in MEDLINE and also includes an interface that will identify abbreviations from user-specified text. We will be using the algorithm in support of text mining on the biomedical literature by identifying abbreviations defined in text and by looking up abbreviations against the abbreviation server. We hope that this server will also be useful for the general biomedical community.
Acknowledgments
JTC was partially supported by a Stanford Graduate Fellowship. We thank Zhen Lin for invaluable help with translating the China Medical Tribune web site and Elmer Bernstam for thoughtful comments on earlier drafts of this paper. We thank Chris Manning for useful discussions on this project. We thank James Pustejovsky and José Castaño for helpful discussions about Acromed and the Medstract gold standard. Finally, we thank the anonymous reviewers for insightful comments that have improved this paper.
This work was supported by NIH LM 06244 and GM61374, NSF DBI-9600637, and a grant from the Burroughs-Wellcome Foundation.
References
- 1.Iliopoulos I, Enright A, Ouzounis C. Textquest: Document clustering of medline abstracts for concept discovery in molecular biology. Pac Symp Biocomput. 2001;384–95. [DOI] [PubMed]
- 2.Andrade M, Valencia A. Automatic annotation for biological sequences by extraction of keywords from medline abstracts. development of a prototype system. Proc Int Conf Intell Syst Mol Biol. 1997; 5:25–32. [PubMed] [Google Scholar]
- 3.Jenssen T, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 2001;28 (1):21–8. [DOI] [PubMed] [Google Scholar]
- 4.Opaui guide to lists of acronyms, abbreviations, and initialisms on the worldwide web: <http://www.opaui.com/acro.html>.
- 5.Acronyms and initialisms for health information resources: <http://www.geocities.com/~mlshams/acronym/acr.htm>.
- 6.Human genome acronym list: <http://www.ornl.gov/hgmis/acronym.html>.
- 7.Acronym finder: <http://www.acronymfinder.com/>.
- 8.The great three-letter abbreviation hunt: <http://www.atomiser.demon.co.uk/abbrev/>.
- 9.Jablonski S (ed). Dictionary of Medical Acronyms and Abbreviations. Philadelphia, Hanley & Belfus, 1998.
- 10.Taghva K, Gilbreth J. Recognizing Acronyms and Their Definitions. Technical Report, ISRI (Information Science Research Institute) UNLV, 1995.
- 11.Yoshida M, Fukuda K, Takagi T. Pnad-css: A workbench for constructing a protein name abbreviation dictionary. Bioinformatics. 2000;16:169–75. [DOI] [PubMed] [Google Scholar]
- 12.Yeates S. Automatic extraction of acronyms from text. In New Zealand Computer Science Research Students’ Conference. 1999, pp 117–24.
- 13.Larkey LS, Ogilvie P, Price MA, Tamilio B. Acrophile: an automated acronym extractor and server. In ACM DL. 2000, pp 205–14.
- 14.Pustejovsky J, Castaño J, Cochran B, et al. Automatic extraction of acronym-meaning pairs from medline databases. Medinfo. 2001;10 (Pt 1):371–5. [PubMed] [Google Scholar]
- 15.Yu H, Hripcsak G, Friedman C. Mapping abbreviations to full forms in biomedical articles. J Am Med Inform Assoc. 2002; 9:262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeates S, Bainbridge D, Witten IH. Using compression to identify acronyms in text. In Data Compression Conference. 2000, pp 582.
- 17.Needleman S, Wunsch C. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48 (3):443–53. [DOI] [PubMed] [Google Scholar]
- 18.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. New York, Springer-Verlag, 2001.
- 19.China Medical Tribune: <http://www.cmt.com.cn/>.
- 20.Lutz M, Ascher D, Willison F. Learning Python. Sebastopol, CA, O’Reilly, 1999.
- 21.Liu H, Lussier Y, Friedman C. A study of abbreviations in the umls. Proc AMIA Symp. 2001:393–7. [PMC free article] [PubMed]
- 22.Knuth D. The Texbook. Reading, MA, Addison-Wesley, 1986.