Abstract
Background:
Targeting differentially activated or perturbed tumour pathways is the key idea in individualised cancer therapy, which is emerging as an important option in treating cancers with poor prognostic profiles. TP53 mutation status is known as a core determinant of survival in breast cancer. The pathways disrupted in association with TP53 mutation status in tumours are not well characterised.
Method:
In this study, we stratify breast cancers based on their TP53 mutation status and identify the set of dysregulated tumorigenic pathways and corresponding candidate driver genes using breast cancer gene expression profiles. Expressions of these genes were evaluated for their effect on patient survival first in univariate models, followed by multivariate models with TP53 status as a covariate.
Results:
The most strongly differentially enriched pathways between breast cancers stratified by TP53 mutation status include in addition to TP53 signalling, several known cancer pathways involved in renal, prostate, pancreatic, colorectal, lung and other cancers, and signalling pathways such as calcium signalling, MAPK, ERBB and vascular endothelial growth factor (VEGF) signalling pathways. We found that mutant TP53 in conjunction with active estrogen receptor (ER) signalling significantly influence survival. We also found that upregulation of VEGFA mRNA levels in association with active ER signalling is a significant marker for poor survival, even in the presence of wild-type TP53.
Conclusion:
Mutation status of TP53 in breast cancer involves wide ranging derangement of several pathways. Among the candidate genes of the significantly deranged pathways, we identified VEGFA expression as an important marker of survival even when controlled by TP53 mutation status. Interestingly, independent of the TP53 mutation status, the survival effect of VEGFA was found significant in patients with active ER signalling (ER/PgR+), but not in those with ER/PgR− status. Therefore, we propose more studies to focus on the role of complex interplay between TP53, ER and VEGF signalling from therapeutic and prognostic context in breast cancer.
Keywords: breast cancer, TP53 mutation status, estrogen receptor signalling, vascular endothelial growth factor signalling, dysregulated pathways, survival
The fact that nearly 30% of early-diagnosed breast cancer cases might eventually develop recurrent or metastatic disease (O’Shaughnessy, 2005) – underscores the priority to explore the mechanisms of advanced disease. The TP53 protein is an important clinical biomarker of breast cancer because of its association with tumour progression (Norberg et al, 2001), metastatic potential (D’Assoro et al, 2010), early relapse (Aas et al, 1996), response to chemotherapy (Aas et al, 1996; Kandioler-Eckersberger et al, 2000; Bertheau et al, 2007), and ultimately, to prognosis and survival (Børresen et al, 1995; Berns et al, 2000; Olivier et al, 2006). It is also of relevance to molecular subtypes of breast cancer (Miller et al, 2005; Langerød et al, 2007). Whereas ∼70% of breast cancers with wild-type TP53 are mostly of the Luminal A subtype, mutant TP53 is common in the remaining 30%, which have a poorer prognosis and are classified as triple negative or luminal B. The focus of this work is to identify diagnostic, prognostic and therapeutic biomarkers associated with pathways perturbed by TP53 mutations and understand their relationship to patient survival in breast cancer, under current therapeutic protocols.
TP53 is a key regulator of programmed cell death, cell cycle, DNA repair and genomic stability. In response to stimulus-specific post-transcriptional modification, TP53 regulates genes, which activate specific cellular programs. The TP53 protein has three major functional domains: a transactivation domain at its N-terminal, a central DNA-binding domain (which includes mutation hotspots) and tetramerization and regulatory domains at the C-terminal. The location and type of TP53 mutation affect the ability of TP53 to regulate its target genes, leading to aberrant functions (Blandino et al, 1999) with clinical implications (Kim and Deppert, 2006). Characterisation of the differential activation of key pathways and candidate genes according to the TP53 mutation status may therefore identify mechanisms correlated with TP53 mutation status in breast cancer.
In this study, we stratify breast cancers based on their TP53 mutation status and identify the set of dysregulated tumorigenic pathways and their candidate driver genes by using gene expression data sets obtained from tumours. The goal is to infer the class-specific candidate gene signature by identifying weak to moderate, but coherent gene expressions that significantly influence tumorigenic pathways and survival.
Results
We first categorised breast cancer samples by their corresponding TP53 mutation status, as described in Supplementary Table 1 and performed analysis as shown in the flow-chart (Supplementary Figure 1).
Candidate driver pathways differentially perturbed by TP53 mutations
Enrichment analysis of pathways between mutation status classes was performed using globaltest (Goeman et al, 2011) and SAM-GS (Dinu et al, 2007) on the primary and combined validation data set. Globaltest, although being sensitive to genes with smaller regression coefficients, its results might be influenced by the standardisation and normalisation procedures. SAM-GS on the other hand is shown to have relatively higher power in the lower alpha-level region, thus can better focus on pathways of greatest interest (Liu et al, 2007). Therefore, we use a combination of the two approaches here. The list of differentially enriched KEGG (Kanehisa and Goto, 2000) pathways identified by each of the methods on each of the data set is shown together in Supplementary Table 2. A set of 40 pathways inferred as commonly significant by both the methods in both data sets (Table 1) – are graphically presented as an enrichment map color-coded according to globaltest FDR corrected P-values (Supplementary Figure 2). The most dysregulated pathways included a group of key signalling pathways – such as p53 signalling, calcium signalling, MAPK, ErbB, vascular endothelial growth factor (VEGF) signalling and various cancer pathways.
Table 1. Consensus list of differentially enriched pathways between two TP53 mutation status classes (wild-type TP53 profiles compared with the mutant TP53 profiles), based on pathway analysis performed by using two approaches – globaltest and SAM-GS on primary (n=111 samples) and validation data sets (a combined cross-platform data set with n=327).
| Primary data set | Validation data set | ||||
|---|---|---|---|---|---|
| Asymptotic global test | SAM-GS | Asymptotic global test | SAM-GS | ||
| KEGGID | KEGG pathway name | BH corrected P-value | FDR adj P-value | BH corrected P-value | FDR adj P-value |
| hsa:00230 | Purine metabolism | 1.8E−09 | <10e−6 | 2.37E−36 | <10e−6 |
| hsa:04115 | p53-signalling pathway | 1.8E−09 | <10e−6 | 2.43E−34 | <10e−6 |
| hsa:05211 | Renal cell carcinoma | 3.72E−09 | <10e−6 | 1.32E−17 | <10e−6 |
| hsa:05200 | Pathways in cancer | 1.1E−08 | <10e−6 | 6.86E−29 | <10e−6 |
| hsa:05215 | Prostate cancer | 1.1E−08 | <10e−6 | 1.32E−29 | <10e−6 |
| hsa:04020 | Calcium-signalling pathway | 4.12E−08 | <10e−6 | 4.81E−27 | <10e−6 |
| hsa:00260 | Glycine, serine and threonine metabolism | 4.73E−08 | <10e−6 | 1.44E−25 | <10e−6 |
| hsa:05212 | Pancreatic cancer | 5.65E−08 | <10e−6 | 1.15E−39 | <10e−6 |
| hsa:04340 | Hedgehog-signalling pathway | 6.02E−08 | <10e−6 | 5.76E−21 | <10e−6 |
| hsa:05222 | Small-cell lung cancer | 7.93E−08 | <10e−6 | 6.75E−40 | <10e−6 |
| hsa:04120 | Ubiquitin-mediated proteolysis | 0.00000012 | <10e−6 | 3.26E−40 | <10e−6 |
| hsa:04910 | Insulin signalling pathway | 0.00000012 | <10e−6 | 5.83E−27 | <10e−6 |
| hsa:00051 | Fructose and mannose metabolism | 1.28E−07 | <10e−6 | 2.51E−30 | <10e−6 |
| hsa:05218 | Melanoma | 0.00000014 | <10e−6 | 1.7E−17 | <10e−6 |
| hsa:04150 | mTOR-signalling pathway | 1.68E−07 | <10e−6 | 9.66E−26 | <10e−6 |
| hsa:00380 | Tryptophan metabolism | 1.96E−07 | <10e−6 | 1.48E−08 | <10e−6 |
| hsa:04144 | Endocytosis | 2.39E−07 | <10e−6 | 4.96E−24 | <10e−6 |
| hsa:00330 | Arginine and proline metabolism | 0.00000025 | <10e−6 | 1.29E−18 | <10e−6 |
| hsa:05214 | Glioma | 0.00000025 | <10e−6 | 1.47E−14 | <10e−6 |
| hsa:04010 | MAPK-signalling pathway | 0.00000031 | <10e−6 | 2.44E−34 | <10e−6 |
| hsa:04012 | ErbB-signalling pathway | 3.65E−07 | <10e−6 | 2.68E−17 | <10e−6 |
| hsa:04520 | Adherens junction | 4.03E−07 | <10e−6 | 9.78E−13 | <10e−6 |
| hsa:05217 | Basal cell carcinoma | 0.00000048 | <10e−6 | 6.47E−11 | <10e−6 |
| hsa:00600 | Sphingolipid metabolism | 4.94E−07 | <10e−6 | 4.67E−14 | <10e−6 |
| hsa:05120 | Epithelial cell signalling in Helicobacter pylori infection | 5.79E−07 | <10e−6 | 1.45E−11 | <10e−6 |
| hsa:04722 | Neurotrophin-signalling pathway | 6.72E−07 | <10e−6 | 1.09E−21 | <10e−6 |
| hsa:04912 | GnRH-signalling pathway | 8.22E−07 | <10e−6 | 6E−18 | <10e−6 |
| hsa:05219 | Bladder cancer | 8.23E−07 | <10e−6 | 1.61E−17 | <10e−6 |
| hsa:05210 | Colorectal cancer | 0.00000116 | <10e−6 | 3.9E−11 | <10e−6 |
| hsa:04070 | Phosphatidylinositol-signalling system | 0.00000117 | <10e−6 | 2.16E−12 | <10e−6 |
| hsa:04110 | Cell cycle | 0.00000125 | <10e−6 | 3.7E−27 | <10e−6 |
| hsa:04370 | VEGF-signalling pathway | 0.00000153 | <10e−6 | 1.01E−07 | <10e−6 |
| hsa:05221 | Acute myeloid leukaemia | 0.00000205 | <10e−6 | 6.36E−12 | <10e−6 |
| hsa:00270 | Cysteine and methionine metabolism | 0.0000036 | <10e−6 | 1.24E−25 | <10e−6 |
| hsa:04530 | Tight junction | 0.00000531 | <10e−6 | 6.85E−18 | <10e−6 |
| hsa:04350 | TGF-β-signalling pathway | 0.00000725 | <10e−6 | 5.93E−14 | <10e−6 |
| hsa:04310 | Wnt-signalling pathway | 0.0000103 | <10e−6 | 8.24E−19 | <10e−6 |
| hsa:00590 | Arachidonic acid metabolism | 0.0000146 | <10e−6 | 1.16E−11 | <10e−6 |
| hsa:05213 | Endometrial cancer | 0.000018 | <10e−6 | 0.00000131 | <10e−6 |
| hsa:04142 | Lysosome | 0.0000489 | <10e−6 | 6.09E−18 | <10e−6 |
Abbreviations: BH, Benjamini-Hochberg; FDR, false discovery rate; MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin; SAM-GS, significance analysis of microarrays for genesets; TGF, tumour growth factor; VEGF, vascular endothelial growth factor.
The full pathway lists that show significance of differential enrichment in each individual data set are shown with their respective P-values of significance in Supplementary Table 2.
Candidate genes deregulated according to the TP53 mutation status
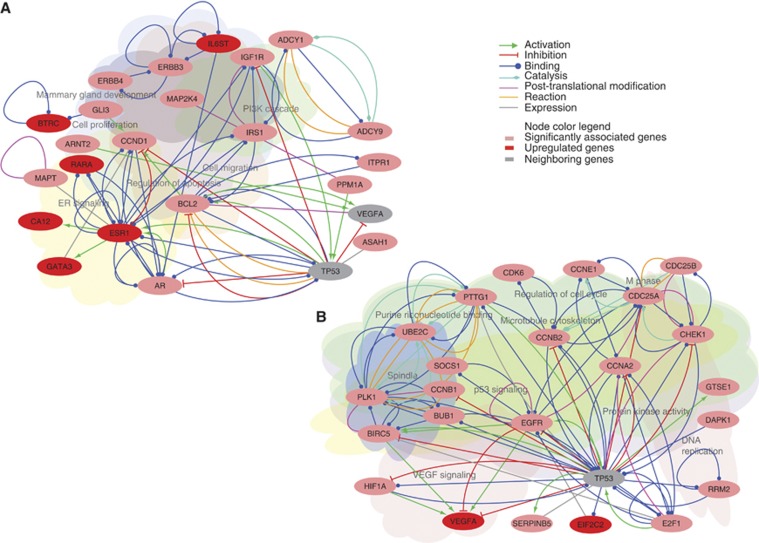
Candidate genes were identified by applying a combination of two mutually complementary approaches: pathway-based gene-search that infers class-specific association (globaltest) and pathway-independent search that identifies individual genes with class-specific upregulation (modified Kolmogorov-Smirnov approach) on both primary and validation data sets (Supplementary Tables 3 and 4). Combining genesets inferred by these two approaches would help to account for the genes with smaller as well as larger effects on the overall biological condition. A consensus genelist (Supplementary Table 5) of 112 genes consists of genes inferred as significant at least by either of the two statistical tests (but not necessarily by the same test) in both primary and validation data sets, as shown in the Venn diagram (Supplementary Figure 3). Class-specific predicted functional networks based on these genesets are plotted in Figures 1A and B for wild-type and mutant TP53 samples, respectively. These networks reflect the key genes and corresponding processes that have potential functional implication in association with the one of the TP53 mutation status class. Wild-type TP53 samples showed significance of genes involved in estrogen receptor (ER) signalling, whereas mutant TP53 samples in proliferative processes. Besides, GO terms–response to insulin stimulus and mammary gland development in wild-type and protein kinase activity, mitotic cell cycle, microtubule cytoskeleton in mutant TP53 class were over-represented (Supplementary Figure 4).
Figure 1.
TP53 mutation status-specific network of potential candidate driver genes shown based on their known and predicted functional interactions. (A) Network for wild-type TP53 breast cancer profiles. (B) Network for mutant TP53 breast cancer profiles. Significant association of gene means significant non-zero regression coefficient of a gene in a significantly differentially enriched KEGG pathway. Gene upregulation means its class-specific upward biased expression pattern, inferred by the rank-sum statistic of the modified Kolmogorov–Smirnov test. Relevant biological processes represented by these genes are also highlighted in background.
Association of EMT and stemness to TP53 mutation status
Aberrant TP53 function is shown to induce epithelial-mesenchymal transition (EMT) and thereby confers stemness properties to the cancer cells (Dhar et al, 2008). Therefore, we compared our inferred TP53 status-specific candidate genesets with the published EMT and stemness marker sets. We found that mutant TP53-marker geneset was significantly associated with embryonic stem cell (ESC) and its TP53 targets (p53ESC) genesets (P-value<0.05). Whereas wild-type TP53 signature was found significantly associated with PRC2 targets (P-value: 0.003) (Table 2). Top 1000 upregulated genes (according the signal-to-noise ratio) in mutant TP53 class were significantly associated with EMT, ESC and induced pluripotent stem cell marker genesets. Moreover, KEGG pathways involved in stemness and EMT properties such as TGFβ, wnt signalling were found differentially enriched (Supplementary Table 6b).
Table 2. Association between the inferred TP53 mutation status-specific signatures with previously reported EMT and stemness markers. Statistical significance of differential expressed geneset overlapping the stemness and epithelial-mesenchymal transition (EMT) marker genelistsa. Statistical significance was computed by applying hypergeometric testb.
| wtTP53 signature | Mutant TP53 signature | Top 1000 genes ranked acc to absolute SNR (wt vs mtTP53 BC) | Top 1000 mt TP53 -upregulated genes ranked acc to SNR | |||||
|---|---|---|---|---|---|---|---|---|
| EMT and stemness geneset and its transcript size | Number of overlapping genes | P -value | Number of overlapping genes | P -value | Number of overlapping genes | P -value | Number of overlapping genes | P -value |
| EMT (n=497) | 0 | NS | 1 | NS | 11 | NS | 15 | 0.031 |
| ESC (n=553) | 0 | NS | 14 | 2.65E−13 | 22 | 2.60E−04 | 35 | 4.34E−11 |
| PRC2 (n=1016) | 7 | 3.25E−03 | 0 | NS | 25 | NS | 19 | NS |
| iPSC (n=597) | 1 | NS | 3 | NS | 17 | 4.50E−02 | 22 | 1.50E−03 |
| p53esc (n=912) | 2 | NS | 5 | 2.66E−02 | 12 | NS | 15 | NS |
Abbreviations: BC, breast cancer; ESC, embryonic stem cell; iPSC, induced pluripotent stem cell; NS, not significant; p53esc, p53 targets identified in murine embryonic stem cells; PRC2, polycomb repressive complex 2; SNR, signal-to-noise ratio.
Sources of the genelists are described in the Supplementary Table 6A.
Statistical significance was evaluated by Fisher’s exact test, in instances where number of overlapping genes ⩽5.
Vascular endothelial growth factor A upregulation with wild-type TP53 associates with activation of pro-angiogenic and pro-metastatic biological processes
Among the inferred candidate genes that were found upregulated and/or significantly associated to one of the TP53 mutation status class, 47 genes showed univariate significance to overall patient survival. Vascular endothelial growth factor A (VEGFA) maintained significance in multivariate model (Supplementary Table 7), even after adjusting for TP53 mutation status. VEGFA might be induced by estrogen receptor in breast cancer cells (Buteau-Lozano et al, 2002; Applanat et al, 2008). Besides, wild-type TP53 could block VEGFA function induced by active estrogen receptor signalling (Liang et al, 2005). However, implications of VEGFA in wild-type TP53/ER+ patients are less understood. We therefore analysed this subgroup separately by using the globaltest and moderated t-test (Smyth, 2004).
Using moderated t-test of differential expression on a cross-platform compiled data set, we found 516 gene features (Supplementary Table 8a) differentially expressed between VEGFA upregulation (VEGFA+) vs VEGFA normal/− samples (VEGFA−/N). IGF1 and PPARG were found differentially downregulated in samples with VEGFA upregulation. A GO analysis identified pathways associated with blood vessel morphogenesis, cell migration and regulation of VEGF signalling pathway. The complete list of over-represented GO terms and predicted functional interactions are shown in Supplementary Table 9 and Supplementary Figure 5, respectively. Notably, VEGFA+ vs VEGFA−/N comparison for mutant TP53 subgroup does not show any remarkable difference apart from differential expression of VEGFA itself and pH regulator CA9 (Supplementary Table 8b).
Tumours overexpressing VEGFA (both ER+ wild-type TP53 and mutant TP53 irrespective of ER status) show a differential enrichment of the mTOR-signalling pathway compared with normal/downregulated VEGFA samples. VEGFA+/ER+ wild-type TP53 samples showed significant association of EIF4EBP1, MAPK1 (P-value<0.05) and weak association of MTOR, ULK3 and RPTOR. Conversely, PIK3CA and IGF1 were significantly associated with VEGFA N/− tumours (Supplementary Figure 5 and Supplementary Table 10a). Interestingly, different sets of genes, although involved in the same pathways were found associated with VEGFA status in the mutant TP53 subgroup (Supplementary Table 10b).
TP53 mutation, ER status and VEGFA upregulation influence survival
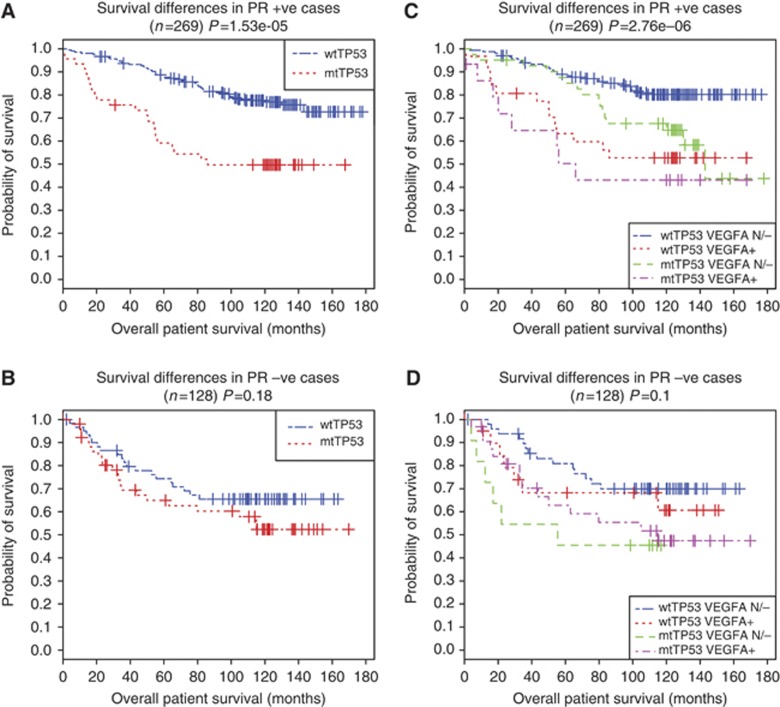
Samples were substratified according to the ER status in each TP53 mutation class. While comparing the ER+/mutant TP53 to the ER+/wild-type TP53 samples, we noted a death hazard ratio (HR) of 2.15 (95% CI: 1.25–3.70) and likelihood P-value <0.01. On the other hand, ER− samples showed weaker significance (P=0.2; HR: 2.6; 95% CI: 1.14–5.91). As progesterone receptor (PgR) positivity is a better marker of active ER signalling (Bardou et al, 2003), we also used PgR status as an indicator of active ER signalling. PgR+ samples showed a significant survival difference between mutant and wild-type tumours (P=1.53e−05, HR: 7.2, 95% CI: 3.03–17.1). However, PgR-tumours do not show significant survival differences (P>0.1) (Figure 2A and B). On the basis of these findings, we propose that active ER signalling can influence the effect of mutant TP53 on survival.
Figure 2.
Overall patient survival differs significantly according to the TP53 mutation status and VEGFA expression status in PgR+ and PgR− subgroups of patients. Survival differences between wild-type TP53 and mutant TP53 in each of the subgroups are shown in Kaplan–Meier plots shown in A and B. Survival differences of four classes: (1) wild-type TP53 and VEGFA normal/downregulation (wtTP53 VEGFA N/−); (2) wild-type TP53 and VEGFA upregulation (wtTP53 VEGFA+); (3) mutant TP53 and VEGFA normal/downregulation (mtTP53 VEGFA N/−); and (4) mutant TP53 and VEGFA upregulation (mtTP53 VEGFA+) – in PgR+ and PgR− subgroups are shown in C and D. Significance of overall model is based on the likelihood ratio test P-value.
As VEGFA expression is observed here as a significant influencer on survival even after controlling for TP53 status, we reanalyzed the above effects by adding VEGFA expression status as a covariate. Among ER+ group, the overall patient survival was significantly influenced by TP53 mutation status and VEGFA (model significance=0.0005) with their corresponding HR=2.02 and 2.08, compared with baseline risk for wild-type TP53 and VEGFA normal/downregulation. Even stronger effect was observed after excluding samples with non-missense mutant TP53 (P-value=0.0001, HR=2.38 and 2.11, respectively). Survival effect of TP53 mutation status and VEGFA was stronger in PgR+ cases (HR=2.35, 95% CI: 1.17–4.74 for VEGFA upregulation, HR=5.2, 95% CI: 2.43–11.1 for mutant TP53 status, and overall likelihood ratio test P=2.76e−6), but non-significant effect in PgR–cases (Figure 2C and D). Although active ER signalling in general is known to predict better prognosis, these findings show that irrespective of the TP53 mutation status, ER+ cases with high mRNA levels of VEGFA indicates poor prognosis. Interestingly, despite of the lowest occurrence of cases with upregulated VEGFA in ER+/wtTP53 subgroup (Supplementary Figure 6), its prognostic significance underscores further exploration.
Discussion
Our findings show predominance of ER signalling in breast cancers with wild-type TP53, marked by the upregulation of ESR1, GATA-binding protein 3, retinoic acid receptor alpha (RARα) and CA12. Estrogen receptor α, a direct transcriptional activator of RARα (Han et al, 1997), mediates anti-proliferative response by vitamin A metabolite (all-trans-retinoic acid ) in breast cancer cells (Dawson et al, 1995). Retinoic acid receptor α is a rate-limiting factor for ER transcriptional activity (Ross-Innes et al, 2010). Co-expression of BCL2, ERBB4, IGF1R, IRS1 was also found in this group. Our observation of consistent upregulation of CA12, AGR3, IL6ST and STC2 genes is in agreement with their previously reported association with ER+ breast cancers. Our findings also showed upregulation of SIRT3, a mitochondrial p53 activity regulator, necessary for averting TP53-mediated growth arrest (Li et al, 2010). Predicted functional network (Figure 1A) provides a hint that genes involved in ER signalling form a core group of interactions in TP53 wild-type tumours. A strong relationship between ER signalling and TP53 can be observed in our results. This relationship also has got implications on proliferation and treatment responsiveness. The presence of wild-type TP53 improves sensitivity to Tamoxifen (Berns et al, 2000) and inhibits ER cross-talk with the EGFR/HER2 pathways (Fernandez-Cuesta et al, 2010). Experimental observations have provided evidence about potential direct ER–TP53 interactions (Liu et al, 2006). However, these complex interactions and their effects on transactivation activity of TP53 and ERα in ER+ breast cancer remains to be understood. Given that TP53 status is an important predictor of response in patients receiving therapy targeting the ER pathway (SERM), we expect that TP53 retains a subset of functions necessary for the response to such therapy.
Genes in pathways related to cell cycle, angiogenesis, chromosomal instability and metastasis were significantly affected in mutant TP53 tumours. We found the gene BUB1 and spindle-checkpoint associated kinases were significantly associated with TP53 mutant tumours. In the presence of dysfunctional TP53, their aberrant expression can cause genomic instability, leading to aneuploidy and malignant transformation (Gjoerup et al, 2007). Other genes associated with mutant TP53 included ones involved in proliferation, angiogenesis and metastasis-VEGFA, HIF1α, E2F1, CDK6 and EGFR.
VEGFA upregulation is an important indicator of pro-angiogenic and pro-metastatic activity. Dysregulation of TP53-VEGF signalling may potentially be a key event in breast cancers with mutant TP53. Mutant TP53 may facilitate this tumorigenic programme by: passing the direct survival advantage to malignant cells, by facilitating the VEGF-mediated enhanced cell migration, angiogenesis and metastasis or by overcoming the regulation by ETS1 (Dittmer, 2003). Active ER signalling and mutant TP53 are also reported to activate VEGF and mark poor prognosis (Berns et al, 2003). In our data, we see that mutant TP53 and VEGF upregulation significantly affects patient survival in ER+/PgR+ samples, but not in ER−/PgR− samples. Activation of VEGFA may also be attributed to the expression of EGFR (Maity et al, 2000) or CDK6, which can correlate with the expression of mutant TP53 (Wyllie et al, 2003) and potentially delay cell senescence. Thus, besides the direct effects of lost TP53 function, other related opportunistic mechanisms, such as dysregulated proliferative effects of VEGFA may contribute the overall manifestation.
ER+/wild-type TP53 samples showed relatively low occurrence of VEGFA upregulation but poor survival profile. ER-mediated induction of VEGF (Berns et al, 2003; Applanat et al, 2008) and VEGF regulation by TP53 (Liang et al, 2005) suggests a complex interplay between these three signalling mechanisms. This group also showed the differential enrichment of mTOR signalling. Co-activation of VEGF and mTOR pathway components has been previously reported (Trinh et al, 2009). Thus, VEGFA may represent a biomarker of interest to identify the target subset of ER+ breast cancer patients who might benefit from early administration of VEGFA or mTOR-targeted therapy.
Materials and Methods
Agilent chip based gene expression data for a subset of 111 breast cancer cases from (Enerly et al, 2011) GEO (accession number GSE19783) was used as the primary data set. TP53 mutations for the primary data in coding regions of exons 2–11 and clinical data were obtained from (Naume et al, 2007). Expression data used for validation was obtained from GEO (accession number GSE3494) and from Stanford Microarray Database. Clinical and TP53 data for these data sets were obtained from (Miller et al, 2005; Langerød et al, 2007).
Methods used to merge data sets to form a validation data set
Two expression data sets (Miller et al, 2005; Langerød et al, 2007) from independent studies and different technology platform were preprocessed, quantile normalised and combined based on UniGene identifiers. Batch effects were corrected by applying parametric empirical Bayes method (Johnson et al, 2007).
Differential enrichment of pathways and candidate genes
The globaltest (Goeman et al, 2011) uses a regression model where genes are covariates and sample classes are response variables. Significant association of gene means significant non-zero regression coefficient of a gene in a geneset (here a particular KEGG pathway). SAM-GS is another geneset enrichment analysis method based on the t-like statistic for assessing the permutation-based significance of association between an individual pathway and a phenotype of interest. KEGG pathways inferred as significant by globaltest at FDR corrected P-value of 10e−5 and validated by SAM-GS (Dinu et al, 2007) at FDR corrected P-value cutoff=10e−6 on both primary and validation data sets were analysed by post-hoc covariate test to identify significant genes. Gene upregulation means its class-specific upward biased expression pattern, inferred by the rank-sum statistic of the modified Kolmogorov–Smirnov test (Yang et al, 2010).
Class-specific predicted functional interactions between genes in the genesets were obtained from STRING database (Jensen et al, 2009).
Pathways enrichment and GO analysis
Gene Ontology (GO) analysis was performed for each TP53 mutation status-specific genesets using DAVID (Huang et al, 2009) by Fisher's exact test with human whole genome as a background. Differentially enriched pathways and GO terms were graphically presented as Enrichment map (Merico, 2009), with nodes color-coded by FDR-adjusted P-value of significance and node-size proportionate to number of genes in the pathway. Fraction of overlapping genes between any two pathways is represented by the edge thickness, with cutoff overlap coefficient of 0.1.
Association of TP53 biology with EMT and stemness marker signatures
Inferred class-specific genesets were tested by hypergeometric test for their association to the published EMT and stemness marker genesets shown in Supplementary Table 6a. A larger genelist inferred by using signal-to-noise ratio between TP53 mutation status classes was also tested for its association to these published genesets.
Survival analysis
A combined cohort of 438 cases obtained by merging clinical data from three individual clinical data sets (Supplementary Table 1) was used. Kaplan–Meier estimation of survival and computation of Cox proportional hazards frailty model for the death event was performed by using R package survival (Therneau and Lumley, 2009). Inferred candidate genes were assessed for their uni-/multivariate effect on survival. The effect of TP53 mutation status together with genes that maintain significance in a multivariate model (VEGFA expression status) and predicted subtype (Parker et al, 2009)- was computed with and without stratification by ER/PgR status.
Discretisation of gene expression
The mRNA expression levels of candidate genes were discretised into two levels using mean (μ)+0.5*standard deviation (s.d.) as a cutoff in each data set.
Analysis was performed by using R (R Development Core Team, 2011).
Acknowledgments
This paper is a part of the doctoral thesis work of HJ. His work was supported by grant number 2789119 from Helse Sør-Øst and internal grants from Akershus University Hospital (number: 2679030 and 2699015 to VNK).
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Aas T, Børresen AL, Geisler S, Smith-Sørensen B, Johnsen H, Varhaug JE, Akslen LA, Lønning PE (1996) Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med 2: 811–814 [DOI] [PubMed] [Google Scholar]
- Applanat MP, Buteau-Lozano H, Herve MA, Corpet A (2008) Vascular endothelial growth factor is a target gene for estrogen receptor and contributes to breast cancer progression. Adv Exp Med Biol 617: 437–444 [DOI] [PubMed] [Google Scholar]
- Bardou V-J, Arpino G, Elledge RM, Osborne CK, Clark GM (2003) Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases. J Clin Oncol 21: 1973–1979 [DOI] [PubMed] [Google Scholar]
- Berns EM, Foekens JA, Vossen R, Look MP, Devilee P, Henzen-Logmans SC, Van Staveren IL, Van Putten WL, Inganas M, Meijer-Van Gelder ME, Cornelisse C, Claassen CJ, Portengen H, Bakker B, Klijn JG (2000) Complete sequencing of TP53 predicts poor response to systemic therapy of advanced breast cancer. Cancer Res 60: 2155–2162 [PubMed] [Google Scholar]
- Berns EMJJ, Klijn JGM, Look MP, Grebenchtchikov N, Vossen R, Peters H, Geurts-Moespot A, Portengen H, Van Staveren IL, Meijer-Van Gelder ME, Bakker B, Sweep FCGJ, Foekens JA (2003) Combined vascular endothelial growth factor and TP53 status predicts poor response to tamoxifen therapy in estrogen receptor-positive advanced breast cancer. Clin Cancer Res 9: 1253–1258 [PubMed] [Google Scholar]
- Bertheau P, Turpin E, Rickman DS, Espié M, De Reyniès A, Feugeas J-P, Plassa L-F, Soliman H, Varna M, De Roquancourt A, Lehmann-Che J, Beuzard Y, Marty M, Misset J-L, Janin A, De Thé H (2007) Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin−cyclophosphamide regimen. PLoS Med 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandino G, Levine AJ, Oren M (1999) Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene 18(2): 477–485 [DOI] [PubMed] [Google Scholar]
- Børresen AL, Andersen TI, Eyfjörd JE, Cornelis RS, Thorlacius S, Borg A, Johansson U, Theillet C, Scherneck S, Hartman S (1995) TP53 mutations and breast cancer prognosis: particularly poor survival rates for cases with mutations in the zinc-binding domains. Genes Chromosomes Cancer 14: 71–75 [DOI] [PubMed] [Google Scholar]
- Buteau-Lozano H, Ancelin M, Lardeux B, Milanini J, Perrot-Applanat M (2002) Transcriptional regulation of vascular endothelial growth factor by estradiol and tamoxifen in breast cancer cells: a complex interplay between estrogen receptors alpha and beta. Cancer Res 62(17): 4977–4984 [PubMed] [Google Scholar]
- D’Assoro AB, Leontovich A, Amato A, Ayers-Ringler JR, Quatraro C, Hafner K, Jenkins RB, Libra M, Ingle J, Stivala F, Galanis E, Salisbury JL (2010) Abrogation of p53 function leads to metastatic transcriptome networks that typify tumor progression in human breast cancer xenografts. Int J Oncol 37: 1167–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MI, Chao WR, Pine P, Jong L, Hobbs PD, Rudd CK, Quick TC, Niles RM, Zhang XK, Lombardo A (1995) Correlation of retinoid binding affinity to retinoic acid receptor alpha with retinoid inhibition of growth of estrogen receptor-positive MCF-7 mammary carcinoma cells. Cancer Res 55(19): 4446–4451 [PubMed] [Google Scholar]
- Dhar G, Banerjee S, Dhar K, Tawfik O, Mayo MS, Vanveldhuizen PJ, Banerjee SK (2008) Gain of oncogenic function of p53 mutants induces invasive phenotypes in human breast cancer cells by silencing CCN5/WISP-2. Cancer Res 68(12): 4580–4587 [DOI] [PubMed] [Google Scholar]
- Dinu I, Potter JD, Mueller T, Liu Q, Adewale AJ, Jhangri GS, Einecke G, Famulski KS, Halloran P, Yasui Y (2007) Improving gene set analysis of microarray data by SAM-GS. BMC Bioinformatics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer J (2003) The Biology of the Ets1 Proto-Oncogene. Mol Cancer 2: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerly E, Steinfeld I, Kleivi K, Leivonen S-K, Aure MR, Russnes HG, Rønneberg JA, Johnsen H, Navon R, Rødland E, Mäkelä R, Naume B, Perälä M, Kallioniemi O, Kristensen VN, Yakhini Z, Børresen-Dale A-L (2011) miRNA-mRNA Integrated Analysis Reveals Roles for miRNAs in Primary Breast Tumors. PLoS One 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cuesta L, Anaganti S, Hainaut P, Olivier M (2010) p53 status influences response to tamoxifen but not to fulvestrant in breast cancer cell lines. Int J Cancer 128(8): 1813–1821 [DOI] [PubMed] [Google Scholar]
- Gjoerup OV, Wu J, Chandler-Militello D, Williams GL, Zhao J, Schaffhausen B, Jat PS, Roberts TM (2007) Surveillance mechanism linking Bub1 loss to the p53 pathway. Proc Natl Acad Sci USA 104: 8334–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ, Van Houwelingen HC, Finos L (2011) Testing against a high-dimensional alternative in the generalized linear model: asymptotic type I error control. Biometrika 98: 381–390 [Google Scholar]
- Han QX, Allegretto EA, Shao ZM, Kute TE, Ordonez J, Aisner SC, Rishi AK, Fontana JA (1997) Elevated expression of retinoic acid receptor-alpha (RAR alpha) in estrogen-receptor-positive breast carcinomas as detected by immunohistochemistry. Am J Surg Pathol Part B 6(1): 42–48 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1): 44–57 [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, Von Mering C (2009) STRING 8 – a global view on proteins and their functional interactions in 630 organisms. Nucl Acids Res 37: D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8: 118–127 [DOI] [PubMed] [Google Scholar]
- Kandioler-Eckersberger D, Ludwig C, Rudas M, Kappel S, Janschek E, Wenzel C, Schlagbauer-Wadl H, Mittlböck M, Gnant M, Steger G, Jakesz R (2000) TP53 mutation and p53 overexpression for prediction of response to neoadjuvant treatment in breast cancer patients. Clin Cancer Res 6: 50–56 [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1): 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Deppert W (2006) The versatile interactions of p53 with DNA: when flexibility serves specificity. Cell Death Differ 13(6): 885–889 [DOI] [PubMed] [Google Scholar]
- Langerød A, Zhao H, Borgan Ø, Nesland JM, Bukholm IR, Ikdahl T, Kåresen R, Børresen-Dale A-L, Jeffrey SS (2007) TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res 9(3): R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Banck M, Mujtaba S, Zhou M-M, Sugrue MM, Walsh MJ (2010) p53-Induced Growth Arrest Is Regulated by the Mitochondrial SirT3 Deacetylase. PLoS One 5: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Wu J, Stancel GM, Hyder SM (2005) p53-dependent inhibition of progestin-induced VEGF expression in human breast cancer cells. J Steroid Biochem Mol Biol 93(2-5): 173–182 [DOI] [PubMed] [Google Scholar]
- Liu Q, Dinu I, Adewale AJ, Potter JD, Yasui Y (2007) Comparative evaluation of gene-set analysis methods. BMC Bioinformatics 8: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Konduri SD, Bansal S, Nayak BK, Rajasekaran SA, Karuppayil SM, Rajasekaran AK, Das GM (2006) Estrogen receptor-alpha binds p53 tumor suppressor protein directly and represses its function. J Biol Chem 281(15): 9837–9840 [DOI] [PubMed] [Google Scholar]
- Maity A, Pore N, Lee J, Solomon D, O’Rourke DM (2000) Epidermal growth factor receptor transcriptionally up-regulates vascular endothelial growth factor expression in human glioblastoma cells via a pathway involving phosphatidylinositol 3′-kinase and distinct from that induced by hypoxia. Cancer Res 60(20): 5879–5886 [PubMed] [Google Scholar]
- Merico D (2009) Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5: e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, Bergh J (2005) An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA 102(38): 13550–13555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naume B, Zhao X, Synnestvedt M, Borgen E, Russnes HG, Lingjaerde OC, Strømberg M, Wiedswang G, Kvalheim G, Kåresen R, Nesland JM, Børresen-Dale A-L, Sørlie T (2007) Presence of bone marrow micrometastasis is associated with different recurrence risk within molecular subtypes of breast cancer. Mol Oncol 1(2): 160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg T, Klaar S, Kärf G, Nordgren H, Holmberg L, Bergh J (2001) Increased p53 mutation frequency during tumor progression – results from a breast cancer cohort. Cancer Res 61: 8317–8321 [PubMed] [Google Scholar]
- O’Shaughnessy J (2005) Extending survival with chemotherapy in metastatic breast cancer. Oncologist 10(Suppl 3): 20–29 [DOI] [PubMed] [Google Scholar]
- Olivier M, Langerød A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bièche I, Varley J, Bignon Y, Uhrhammer N, Winqvist R, Jukkola-Vuorinen A, Niederacher D, Kato S, Ishioka C, Hainaut P, Børresen-Dale A-L (2006) The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12: 1157–1167 [DOI] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J clin oncol 27(8): 1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2011) R: A Language and Environment for Statistical Computing. Vienna Austria R Foundation for Statistical Computing 1, ISBN 3-900051-07-0 [Google Scholar]
- Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M, Carroll JS (2010) Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev 24: 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3. [DOI] [PubMed] [Google Scholar]
- Therneau T, Lumley T (2009) Survival: Survival Analysis Including Penalised Likelihood. R package Version 2.35-8. R package version 2.35-8http://CRAN.R-project.org/package=survival [Google Scholar]
- Trinh XB, Tjalma WA, Vermeulen PB, Van den Eynden G, Van der Auwera I, Van Laere SJ, Helleman J, Berns EM, Dirix LY, van Dam PA (2009) The VEGF pathway and the AKT/mTOR/p70S6K1 signalling pathway in human epithelial ovarian cancer. Br J Cancer 100(6): 971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie F, Haughton M, Bartek J, Rowson J, Wynford-Thomas D (2003) Mutant p53 can delay growth arrest and loss of CDK2 activity in senescing human fibroblasts without reducing p21(WAF1) expression. Exp Cell Res 285(2): 236–242 [DOI] [PubMed] [Google Scholar]
- Yang Y, Kort EJ, Ebrahimi N, Zhang Z, Teh BT (2010) Dual KS: defining gene sets with tissue set enrichment analysis. Cancer Inform 9: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.