Background: Little is known about BACE1 functions.
Results: BACE1−/− mice have axon guidance defects similar to those of CHL1−/− mice; CHL1 undergoes BACE1-dependent processing in vivo; CHL1 and BACE1 co-localize in terminals and growth cones.
Conclusion: BACE1 deficiency produces a CHL1 loss-of-function phenotype.
Significance: BACE1 inhibition may cause axon guidance defects, adding a note of caution to BACE1 inhibitor drug development.
Keywords: Alzheimer Disease, Amyloid, Axon, Drug Development, Gene Knock-out, Secretases, BACE1, CHL1, Axon Guidance, β-Secretase
Abstract
BACE1 is the β-secretase enzyme that initiates production of the β-amyloid peptide involved in Alzheimer disease. However, little is known about the functions of BACE1. BACE1-deficient mice exhibit mild but complex neurological phenotypes suggesting therapeutic BACE1 inhibition may not be completely free of mechanism-based side effects. Recently, we have reported that BACE1 null mice have axon guidance defects in olfactory sensory neuron projections to glomeruli in the olfactory bulb. Here, we show that BACE1 deficiency also causes an axon guidance defect in the hippocampus, a shortened and disorganized infrapyramidal bundle of the mossy fiber projection from the dentate gyrus to CA3. Although we observed that a classical axon guidance molecule, EphA4, was cleaved by BACE1 when co-expressed with BACE1 in HEK293 cells, we could find no evidence of BACE1 processing of EphA4 in the brain. Remarkably, we discovered that the axon guidance defects of BACE1−/− mice were strikingly similar to those of mice deficient in a recently identified BACE1 substrate, the neural cell adhesion molecule close homolog of L1 (CHL1) that is involved in neurite outgrowth. CHL1 undergoes BACE1-dependent processing in BACE1+/+, but not BACE1−/−, hippocampus, and olfactory bulb, indicating that CHL1 is a BACE1 substrate in vivo. Finally, BACE1 and CHL1 co-localize in the terminals of hippocampal mossy fibers, olfactory sensory neuron axons, and growth cones of primary hippocampal neurons. We conclude that BACE1−/− axon guidance defects are likely the result of abrogated BACE1 processing of CHL1 and that BACE1 deficiency produces a CHL1 loss-of-function phenotype. Our results imply the possibility that axon mis-targeting may occur in adult neurogenic and/or regenerating neurons as a result of chronic BACE1 inhibition and add a note of caution to BACE1 inhibitor development.
Introduction
Alzheimer disease (AD)3 is a devastating neurodegenerative disorder that is the leading cause of dementia in the elderly. Two hallmark histopathological lesions are found in the AD brain, amyloid plaques composed of the β-amyloid (Aβ) peptide and neurofibrillary tangles made of hyperphosphorylated Tau protein (2). A wide range of studies suggests that cerebral accumulation of the Aβ peptide plays a critical early role in AD pathogenesis. Most compellingly, autosomal dominant mutations in the amyloid precursor protein (APP) and the presenilins 1 and 2 cause early onset familial AD by increasing the production of the neurotoxic 42-amino acid isoform of Aβ, Aβ42 (2, 3).
The production of Aβ is initiated through the cleavage of APP by the protease β-secretase or β-site APP-cleaving enzyme 1 (BACE1) (4–8). The Swedish double mutation of APP (K670N,M671L) lies immediately N-terminal to the β-secretase site of APP and increases BACE1 cleavage and Aβ production severalfold (9). Another mutation, A673V, also increases BACE1 cleavage of APP and causes familial AD (10, 11). Conversely, a mutation was recently discovered at the same site in APP, A673T, that decreases BACE1 cleavage, reduces Aβ production, and protects against AD (12). Taken together, these results strongly suggest the following: 1) excessive cerebral Aβ levels can cause AD; 2) reducing cerebral Aβ levels should be beneficial for AD, and 3) therapeutic strategies that decrease BACE1 cleavage of APP should prevent, if not treat, AD. Indeed, the latter point has been intensely pursued since the discovery of BACE1, and several pharmaceutical companies have recently entered clinical trials with small molecule BACE1 inhibitor drug candidates.
To determine whether therapeutic BACE1 inhibition would cause mechanism-based toxicity, several lines of BACE1 null mice were generated by gene targeting (13–15). Initial reports indicated that BACE1−/− mice were viable, fertile, and devoid of abnormalities. However, upon closer examination, BACE1−/− mice were found to have several subtle phenotypes that involved memory deficits (16–18), hypomyelination (19, 20), reduced survival and growth retardation (21), irregularities in neuronal excitability and electrophysiology (17, 21, 22), seizure (22, 23), reduced dendritic spine density (24), schizophrenia endophenotypes (24), and axon guidance defects (1, 25). Although these BACE1 null abnormalities are relatively mild, they are complex neurological phenotypes that raise a concern that BACE1 inhibitors may not be completely free of mechanism-based side effects.
The phenotypes of BACE1−/− mice are the result of deficient processing of BACE1 substrates. For example, hypomyelination is caused by reduced BACE1 cleavage of type III neuregulin-1 (NRG1) that results in decreased release of the NRG1 EGF-like domain that signals to glia to myelinate axons (19, 20). Initially, a small number of BACE1 substrates were discovered through individual studies (26). However, the list of potential BACE1 substrates has recently grown considerably because of the use of traditional and novel proteomics screening methods (27–29). Recent reports suggest that molecules involved in axon guidance, neurite outgrowth, and synapse formation are BACE1 substrates. For example, members of the ephrin and Eph receptor families of axon guidance molecules were identified as BACE1 substrates in a proteomics screen using non-neuronal cell lines (27). More recently, the neural cell adhesion molecules L1 and close homolog of L1 (CHL1) were shown to be BACE1 substrates in primary neuron cultures and in the brain (28, 29). The roles of L1 and CHL1 in neurite outgrowth and the ephrins and Eph receptors in axon guidance, together with the fact that BACE1 expression is elevated during the early postnatal period (19), a time of substantial axon outgrowth, guidance, and synapse formation, suggest that BACE1−/− mice may have phenotypes related to axon outgrowth and guidance caused by deficient processing of these substrates. Association of specific substrates with given BACE1 null phenotypes is critical for molecular understanding of BACE1 functions and for devising future strategies to avoid potential mechanism-based side effects of therapeutic BACE1 inhibition.
Recently, we (1) and other investigators (25) reported that BACE1−/− mice exhibit defects in the guidance of OSN axons to odorant receptor-specific glomeruli in the olfactory bulb. Because OSNs regenerate throughout life and require on-going axon guidance in the adult, we reasoned that BACE1 deficiency might cause axon mis-targeting in other adult axon regenerating and neurogenic systems in BACE1−/− mice. Therefore, we sought to determine whether axon guidance errors occur in other neuron populations in BACE1−/− mice and, if so, to identify the responsible BACE1 substrate. We chose to investigate axon guidance in a population of neurons that undergo adult neurogenesis and that are critical for hippocampus-dependent memory function, i.e. dentate gyrus granule cells, which project axons (mossy fibers) to CA3 pyramidal neurons. Importantly, mossy fiber terminals exhibit robust expression of BACE1 (17, 30).
We report here that BACE1 null mice exhibit mossy fiber axon guidance abnormalities consisting of a shortened IPB and IPB axons that traverse the CA3 pyramidal cell layer prematurely. The hippocampal mossy fiber and OSN axon guidance defects of BACE1−/− mice strikingly resemble those of CHL1 null mice (31, 32). BACE1−/− primary hippocampal neurons in culture had decreased axon length, in accord with the role of CHL1 in axon outgrowth. Although EphA4, an axon guidance molecule involved in mossy fiber topographic mapping, was cleaved by BACE1 in vitro, it did not appear to be a BACE1 substrate in vivo. In addition, BACE1−/− hippocampal neurons had deficient, rather than enhanced, growth cone collapse induced by the EphA4 ligand ephrinB3. In contrast, CHL1 exhibited robust processing by BACE1 in the hippocampus and olfactory bulb, brain regions that exhibit axon-targeting errors in BACE1−/− mice. CHL1 co-localized with BACE1 in presynaptic terminals of the hippocampus and olfactory bulb and in growth cones of primary hippocampal neurons in culture. Moreover, CHL1 levels were higher in BACE1−/− hippocampus and olfactory bulb than in wild type, as expected from abrogated BACE1 cleavage and subsequent elevation of CHL1 steady-state levels. Our results confirm that CHL1 is a BACE1 substrate in vivo. Furthermore, we conclude that deficient BACE1 processing of CHL1 produces a CHL1 loss-of-function phenotype that involves axon guidance defects in the brain.
EXPERIMENTAL PROCEDURES
Mice
BACE1−/− mice were purchased from The Jackson Laboratory (Bar Harbor, ME) (15). BACE1+/− mice were used for breeding, and all wild-type controls used were BACE1+/+ littermates. OMP-GFP is a gene-targeted mouse strain in which GFP is expressed from the locus encoding OMP (33). BACE1−/− mice were bred to OMP-GFP mice to produce heterozygous OMP-GFP mice that were either BACE1+/+ or BACE1−/−. All mice were maintained in microisolator cages in the Barrier Facilities of Northwestern University Center for Comparative Medicine. All animal procedures were in strict accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Northwestern University Animal Care and Use Committee.
Tissue Preparation
BACE1−/− and littermate BACE1+/+ mice were euthanized at the ages of 7 days, 3 weeks, 6 weeks, and 3 months of age, and brain tissue was harvested for biochemical and histological analyses. Mice older than 8 days were deeply anesthetized with intraperitoneal injection of ketamine (200 mg/kg)/xylazine (25 mg/kg) and then transcardially perfused with HEPES buffer containing protease inhibitors (20 μg/ml phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, 20 μm sodium orthovanadate, and 100 μm dithiothreitol (DTT)) before brain harvest. One hemibrain per mouse was fixed at 4 °C in 4% paraformaldehyde in PBS for 24 h and then cryopreserved in 30% (w/v) sucrose for >24 h. The other hemibrain was dissected to isolate the hippocampus and olfactory bulb, and these tissue portions were flash-frozen in liquid nitrogen for biochemical analysis. Hippocampi and olfactory bulbs were homogenized in PBS 1% Triton X-100 with 1× Calbiochem protease inhibitor mixture set I (EMD Biosciences, La Jolla, CA) using a tube and pestle single use Dounce homogenizer. Total protein concentrations of hippocampal homogenates were determined by the BCA method (Pierce). Hemibrains and olfactory bulbs used for immunofluorescence tissue section staining were drop-fixed in 10% formalin/PBS for 16 h at 4 °C, then cryopreserved in 20% (w/v) sucrose/PBS for 24 h, and then maintained in 30% (w/v) sucrose at 4 °C until sectioning. 20–40-μm coronal sections were cut from formalin-fixed brains on a freezing sliding microtome and collected in 0.1 m PBS with 0.01% sodium azide.
Immunofluorescence Microscopy of Tissue Sections
Coronal sections of either 20 μm (for synaptoporin/MAP2 co-labeling of mossy fiber terminals) or 40 μm (for all other experiments) with equivalent rostral-caudal locations were selected using anatomical landmarks and the size and shape of the hippocampus and ventricles. Free-floating sections were washed in TBS + 0.25% Triton X-100 (TBS+T) and blocked for 90 min in 5% goat serum before being incubated in primary antibodies (synaptoporin: rabbit pAb, Synaptic Systems, 102 002, 1:250; MAP2: mouse mAb, gift from Lester Binder, 1:250; BACE1: rabbit mAb D10E5, Cell Signaling, 5606, 1:250 or rabbit mAb, Epitomics, 2882-1, 1:250; CHL1: goat pAb, R&D Systems, AF2147, 1:250; NCAM1: rat mAb, Genetex H28-123, 1:200; synaptophysin, mouse mAb, Millipore, MAB5258-20UG, 1:500) in TBS+T, 1% bovine serum albumin (BSA) at 4 °C overnight on an orbital shaker. In some cases, sections were incubated in primary antibody at 37 °C for 2 h before being moved to 4 °C overnight. For synaptoporin/MAP2 co-labeling of mossy fiber terminals, primary antibody incubation was at 4 °C for 48 h. Sections were then washed in TBS+T, 1% BSA and incubated in the dark for 90 min in Alexa-Fluor donkey anti-goat 488 (A11055), anti-mouse 594 (A21203), and anti-rabbit 647 (A31573) secondary antibodies (1:500) (Invitrogen) along with DAPI (300 nm concentration). Secondary antibody incubation was at 4 °C for 48 h for synaptoporin/MAP2 co-labeling of mossy fiber terminals. The extended incubation times for primary and secondary antibodies and the thinner sections were necessary in the experiments examining mossy fiber terminals to permit full tissue penetration of the synaptoporin antibody, allowing three-dimensional Z-stack imaging. Sections were washed in the dark in TBS before mounting with ProLong gold antifade reagent (Invitrogen P36934) and coverslipping 1.5 (VWR).
Imaging was performed using a 40× (NA 1.3) or 60× (NA 1.4) oil-immersion objective lens on Nikon A1R (olfactory bulb) or a 10× air objective (hippocampus), 20× air objective (hippocampus), or a 40× oil objective (SPB/IPB and stratum lucidum) on a Nikon C1Si laser scanning confocal microscope (Tokyo, Japan). Laser power percentage, gain, and offset settings were held constant for all images acquired, and saturation was never reached. OMP-GFP olfactory bulb sections were imaged for GFP and DAPI fluorescence followed by acquisition of Z-stack images, which were collapsed to the maximum intensity projection using NIS-Elements Advanced Research Version 4.0 software. Any further image analysis was performed using ImageJ (National Institutes of Health).
Measurement of IPB
40-μm coronal sections from seven BACE1+/+ and seven BACE1−/− brains were selected. Three sections/brain from equivalent rostral-caudal locations were selected using anatomical landmarks and the size and shape of the hippocampus and ventricles. Sections were immunofluorescently labeled for synaptoporin (SPO) as described. Confocal images of all sections were acquired using a Nikon C1Si confocal microscope (Tokyo, Japan) with a 10× air objective. Images were inspected for morphological characteristics of mossy fiber projections and analyzed using ImageJ (National Institutes of Health). The following measurements were made: IPB length, free-hand line drawn through middle of the IPB (SPO-positive axon bundle inferior to the pyramidal cell layer) from the end of the pyramidal cell layer to the most distal point of uninterrupted SPO labeling beneath the pyramidal cell layer; SPB length, free-hand line drawn through the middle of the SPB (SPO-positive axon bundle superior to the pyramidal cell layer) from the interblade line (line drawn between the tips of the superior and inferior blades of the dentate gyrus) to the IPB/slu junction (line drawn perpendicular to SPB at the point where SPO labeling begins to taper outward); slu length, free-hand line drawn through the middle of the slu from the SPB/slu junction to the distal end of the slu. For each section, IPB length was normalized to total length of SPB + slu (IPB/slu ratio). These ratios were averaged over the three sections from each brain. Statistical significance was determined by a two-tailed, unpaired Student's t test.
Primary Hippocampal Neuron Culture and Microscopy
Glass coverslips were washed in nitric acid for 48 h, washed in distilled water three times for 1 h, baked at 180 °C overnight, and then placed in 60-mm culture dishes and coated with 1 mg/ml poly-l-lysine (Sigma) overnight at room temperature. Immediately before dissociation of neurons, dishes were washed three times with sterile water; neuron plating medium (Neurobasal A media (Invitrogen), 10% horse serum, 1× B-27 supplement, 1% penicillin/streptomycin, 0.5 mm glutamine, 2.5 μm glutamate) was added, and dishes were placed in a 37 °C, 5% CO2 incubator. Brains were removed from BACE1+/+ and BACE1−/− P0 pups, and hippocampi were dissected and placed in tubes (1 tube/brain) with balanced salt solution, 0.25% trypsin and incubated at 37 °C for 15 min. Hippocampi were washed three times with balanced salt solution and dissociated via trituration with a sterile glass Pasteur pipette followed by a fire-polished glass Pasteur pipette. The concentration of viable cells in suspension from each brain was determined using a Countess cell counter (Invitrogen); cell suspensions were diluted and added to culture dishes at a density of 10,000 cells/cm2. After 2 h, neuron plating medium was carefully replaced with maintenance media (Neurobasal A, 1× B-27 supplement, 0.5 mm glutamine).
For BACE1/EphA4/phalloidin co-labeling, coverslips of neurons were fixed 48 h after plating. For growth cone collapse assay, neurons were treated and then fixed beginning 18 h after plating. Media were aspirated from dishes; coverslips were briefly rinsed with PBS and then fixed for 20 min at room temperature with 4% paraformaldehyde, 0.12 m sucrose in PBS. Coverslips were again washed briefly with PBS and then permeabilized in 0.3% Triton X-100 for 5 min followed by three PBS washes. Fixed neurons were blocked in 10% BSA in PBS for 1 h at room temperature. Coverslips were rinsed briefly with PBS and then placed on a sheet of dental wax. Primary antibodies were added in a 75-μl meniscus over each coverslip in 1% BSA in PBS (BACE1: rabbit mAb D10E5, Cell Signaling, 5606, 1:250, or rabbit mAb, Epitomics, 2882-1, 1:250; CHL1: goat pAb, R&D Systems, AF2147, 1:250; EphA4 N terminus: goat pAb, R&D Systems, AF641, 1:200; tubulin: mouse mAb Tuj1, gift from Lester Binder, 1:20,000) and incubated at 4 °C overnight. Coverslips were washed three times in PBS and incubated in secondary antibodies in 1% BSA in PBS in a 75-μl meniscus over each coverslip for 1 h at room temperature, protected from light (1:500 goat anti-rabbit Alexa-Fluor 488 (Invitrogen); 1:500 donkey anti-goat Alexa-Fluor 488 (Invitrogen); 1:500 goat anti-mouse Alexa-Fluor 488 (Invitrogen)). 2 μl/ml rhodamine-phalloidin (Invitrogen) and 300 nm DAPI were also added with the secondary antibodies. Coverslips were rinsed three times with PBS and mounted on slides with Prolong Gold antifade reagent (Invitrogen). Imaging of neurons for growth cone collapse and axon length measurement was performed on Keyence integrated fluorescence microscope at 40× objective lens (NA 0.95). Any further image analysis was performed using ImageJ (National Institutes of Health).
Growth Cone Collapse Assay
Human ephrin-B3-Fc chimera (Fc-EB3) (R&D Systems) and Fc (Jackson ImmunoResearch) alone were preclustered by incubating with an anti-Fc IgG (Jackson ImmunoResearch) at a molar ratio of 5:1 (five Fc-EB3 or Fc:1 IgG) for 1 h at room temperature. 18 h after plating, primary hippocampal neuron cultures were treated with 1 μg/ml clustered Fc-EB3 or the molar equivalent of clustered Fc for 1 h at 37 °C. The neurons were then fixed and immunofluorescently labeled as described above. One coverslip/dish was labeled for EphA4 to verify clustering of receptors with clustered Fc-EB3 treatment. Four coverslips/dish were labeled with rhodamine-phalloidin to visualize actin and immunofluorescently labeled for tubulin. Neurons from these coverslips were imaged via fluorescence microscopy by an experimenter blind to genotype and treatment group, looking only at the green (tubulin) channel, and 25 neurons/coverslip were selected for assessment of growth cone collapse. Selected cells met the following criteria. The neuron must have more than one visible neurite, with one neurite at least twice as long as all other neurites (the axon). The ends of all neurites must be visible and must not cross or contact any part of any other cell. Images of these neurons were taken on the green (tubulin) and red (phalloidin) channels. These images were analyzed by an experimenter blind to genotype and treatment group using ImageJ (National Institutes of Health). Growth cone area was measured by drawing a free-hand selection around all dense phalloidin staining at the end of each axon. In addition, each axon was rated as having an un-collapsed growth cone (dense phalloidin staining that spreads out to form filopodia/lamellipodia) or collapsed growth cone (axon ends with no phalloidin-dense extensions). The mean growth cone area and percentage of growth cone collapse were calculated for each group (total of 800 cells/treatment condition from eight individual P0 brains). Mean and S.E. values were determined for each genotype/treatment combination. Statistical significance between treated and untreated dishes of a given genotype and between genotypes with the same treatment was determined using a two-tailed unpaired Student's t test.
Axon Length Measurements
Primary hippocampal neurons from P0 BACE1+/+ and BACE1−/− pups were cultured, labeled, and imaged as described above. Images of tubulin labeling in these neurons were analyzed in ImageJ by an experimenter blind to genotype. Axon length was measured by drawing a free-hand line along the axon from the axon hillock to the end of tubulin labeling. Mean axon length and S.E. were calculated for BACE1+/+ and BACE1−/− neurons. Statistical significance was determined by a two-way unpaired Student's t test.
Cell Line Culture and Transfections
HEK293 cells were maintained in 1× Dulbecco's modified Eagle's medium (DMEM)/high glucose (Invitrogen), 10% FBS, 1% penicillin/streptomycin. 24 h before transfection, cells were trypsinized and plated in antibiotic-free media at a concentration of 300,000 cells/ml media in poly-l-lysine-treated 12-well cell culture plates (Nunc) in a volume of 1 ml. Cells were transfected for 6 h with Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's recommendations. A mixture of 2.2 μl of Lipofectamine 2000 per 0.88 μg of DNA was combined in 175 μl of OptiMEM media (Invitrogen) and added to each well. The following DNA constructs were used: WT hBACE1 in CMV1 plasmid vector (4); hBACE1-D93A (inactive) in CMV1 plasmid vector (34); WT hEphA4-FLAG in LZRS plasmid vector (gift from Spiro Getsios and Kazushige Sakaguchi); and empty CMV1 vector (4). After 6 h, cells were washed with Dulbecco's PBS, and media were replaced with 500 μl of OptiMEM + 1% penicillin/streptomycin and various concentrations of BACE1 inhibitor IV (C3) (Millipore) in DMSO or DMSO alone. After conditioning cells for 24 h, cells were placed on ice, and 500 μl of conditioned media were collected from each well. Cells were then washed with Dulbecco's PBS and lysed in 120 μl of lysis buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm EDTA, 1 mm EGTA) + 1× Calbiochem Protease Inhibitor Mixture Set I (EMD). Lysates were briefly sonicated on ice and cleared at 20,000 × g at 4 °C for 10 min and transferred to a fresh tube. Total protein concentrations of lysates were determined by the BCA method (Pierce).
Immunoblotting
10–30 μg of protein from hippocampal and olfactory bulb homogenates or entire eluates from immunoprecipitations were boiled in SDS sample boiling buffer before being separated on 4–12% NuPAGE BisTris gels in 1× MES or MOPS running buffer (Invitrogen) or on 8% Tris-glycine gels (CHL1 blot) in 1× Tris-glycine running buffer and transferred to Millipore Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA). Total protein was stained with Ponceau S, and images of the stained blots were obtained with a scanner. Blots were blocked in 5% nonfat dry milk in Tris-buffered saline (TBS), 0.1% Tween 20 (TBST; Sigma) for 1 h at room temperature or overnight at 4 °C and then incubated in primary antibody (EphA4 N-terminal: goat pAb, R&D Systems, AF641, 1:1000; EphA4 C-terminal: mouse mAb 6H7, Millipore, AP1173, 1:1000; CHL1: goat pAb, R&D Systems, AF2147, 1:1000; β-tubulin, mouse mAb (gift from Lester Binder) 1:5000, Millipore, MAB5564, 1:25,000) for 1 h at RT or overnight at 4 °C. Blots were washed in TBST and incubated for 1 h in horseradish peroxidase (HRP)-conjugated goat anti-rabbit (Jackson ImmunoResearch Laboratories, West Grove, PA) or horse anti-mouse (Vector Laboratories) secondary antibodies diluted 1:10,000 in 5% milk in TBST. Immunosignals were detected using enhanced chemiluminescence (EMD Millipore Luminata Classico, Crescendo, or Forte) and quantified using a Kodak Image Station 400R imager (Rochester, NY). Densitometric analyses of immunoblots and images of Ponceau S-stained blots were performed using Kodak Molecular Imaging Software SE. Immunosignals were normalized to whole-lane Ponceau S signal or β-tubulin immunosignal. Values were expressed as percentages of the mean of the control.
Immunoprecipitation
EphA4 was immunoprecipitated from hippocampal homogenates from 8-day-old and 4-month-old BACE1+/+ and BACE1−/− mice using N-terminal EphA4 antibodies bound to protein G Dynabeads (Invitrogen). 1–3 μg of antibody (EphA4 N terminus: Gt pAb, R&D Systems) was combined with 0.75 mg of protein G Dynabeads in 200 μl of PBS, 0.02% Tween 20 (PBS-T) in a microcentrifuge tube and rotated for 1 h at room temperature. Beads were collected via magnetic rack and washed once with PBS-T, and 500 μg of protein from hippocampal homogenates was added to each tube. Tubes were rotated for 1 h at room temperature. Beads were washed three times in PBS and resuspended in PBS in a fresh tube. PBS was removed, and beads were resuspended in 1× Laemmli sample buffer. Tubes were heated to 95 °C for 5 min to elute/heat denatured proteins, and sample was removed from the beads via magnetic rack.
Statistical Analysis
Data are presented as means ± S.E., represented by error bars in histograms. Statistical significance between means of experimental and control groups was determined using the two-tailed Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
RESULTS
BACE1−/− Hippocampal Mossy Fibers Have Reduced Infrapyramidal Bundle Length
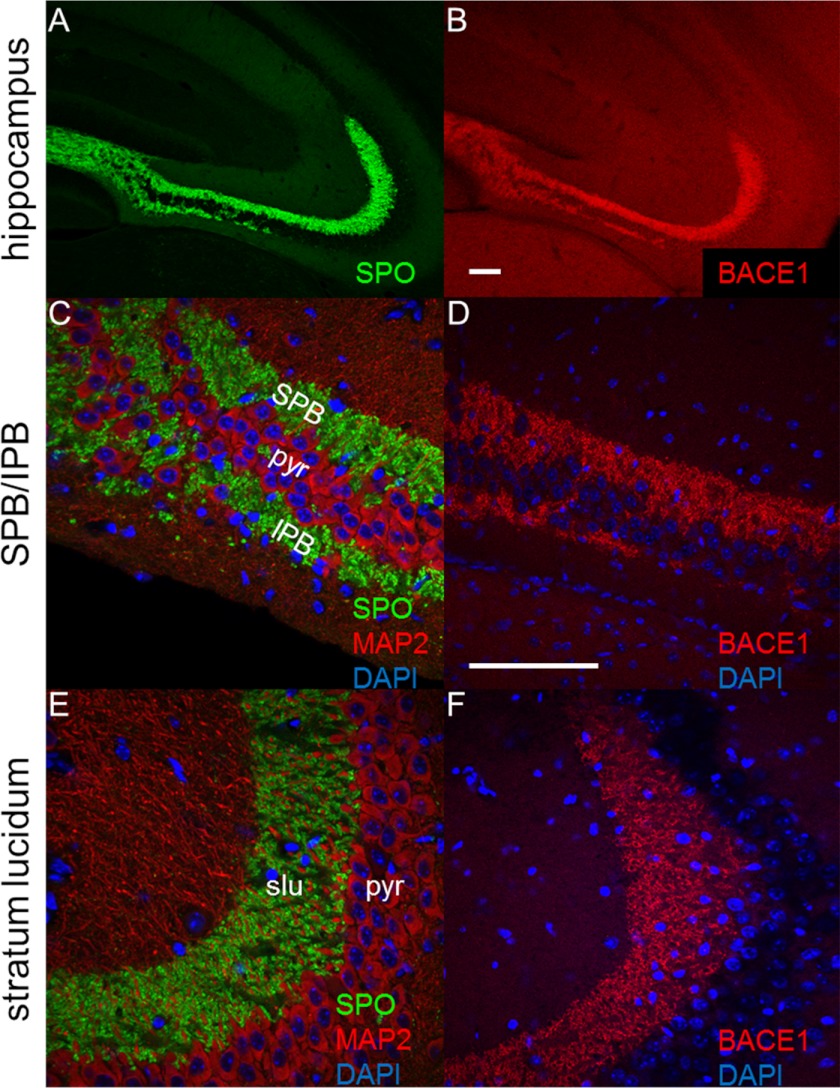
We sought to determine whether axon guidance defects were more widespread, beyond OSN axon targeting errors in the olfactory bulb (1, 25), in the brains of BACE1−/− mice. We chose to investigate the hippocampal mossy fiber axons that project from dentate gyrus granule cells to CA3 pyramidal neurons, because they are highly plastic and crucial for memory formation. Moreover, dentate gyrus granule cells undergo adult neurogenesis and therefore require ongoing axon guidance throughout life. BACE1 is highly concentrated in mossy fiber terminals and exhibits an immunostaining pattern that coincides with that of the presynaptic markers synaptophysin (17, 30) and synaptoporin, but not with the somatodendritic marker MAP2 (Fig. 1). Proximal to the dentate gyrus, mossy fibers are divided into the SPB and IPB bundles that course on the dorsal and ventral sides of the pyramidal cell layer in CA3, respectively (Fig. 1C). Mossy fibers of the IPB originate from the ventral blade of the dentate gyrus and pass through the pyramidal cell layer at a relatively defined position to join SPB mossy fibers in the stratum lucidum, at which point the IPB ends (Fig. 1A). We found robust BACE1 immunostaining in SPB and IPB, as well as in stratum lucidum (Fig. 1, D and F).
FIGURE 1.
BACE1 is concentrated in hippocampal mossy fiber terminals. Coronal sections (20 μm) from a 3-month-old wild-type mouse brain were immunostained for BACE1, synaptoporin, or MAP2 and imaged with confocal microscopy. A, hippocampal mossy fibers exhibit robust staining for the presynaptic marker SPO (green). B, adjacent section immunostained with anti-BACE1 antibody shows robust BACE1 labeling (red) in mossy fibers. C, higher magnification of synaptoporin immunostaining (green) in the region of the hippocampus proximal to the dentate gyrus reveals the SPB and IPB of the mossy fibers, whereas immunostaining for the somatodendritic marker MAP2 (red) shows the intervening pyramidal cell layer (pyr). D, higher magnification of BACE1 immunostaining of a section adjacent to that in C demonstrates BACE1 labeling (red) in the IPB and SPB of the mossy fibers. E, in the slu region of the mossy fibers that is distal to the dentate gyrus, synaptoporin (green) immunostaining reveals large mossy fiber terminals that surround MAP2-labeled (red) dendrites of pyramidal cells in CA3. F, BACE1 immunostaining (red) corresponds to that of synaptoporin (E) and indicates that BACE1 is highly concentrated in large mossy fiber terminals. DAPI staining for nuclei is shown in blue. Scale bar in B, 250 μm for A and B; scale bar in D, 250 μm for C–F.
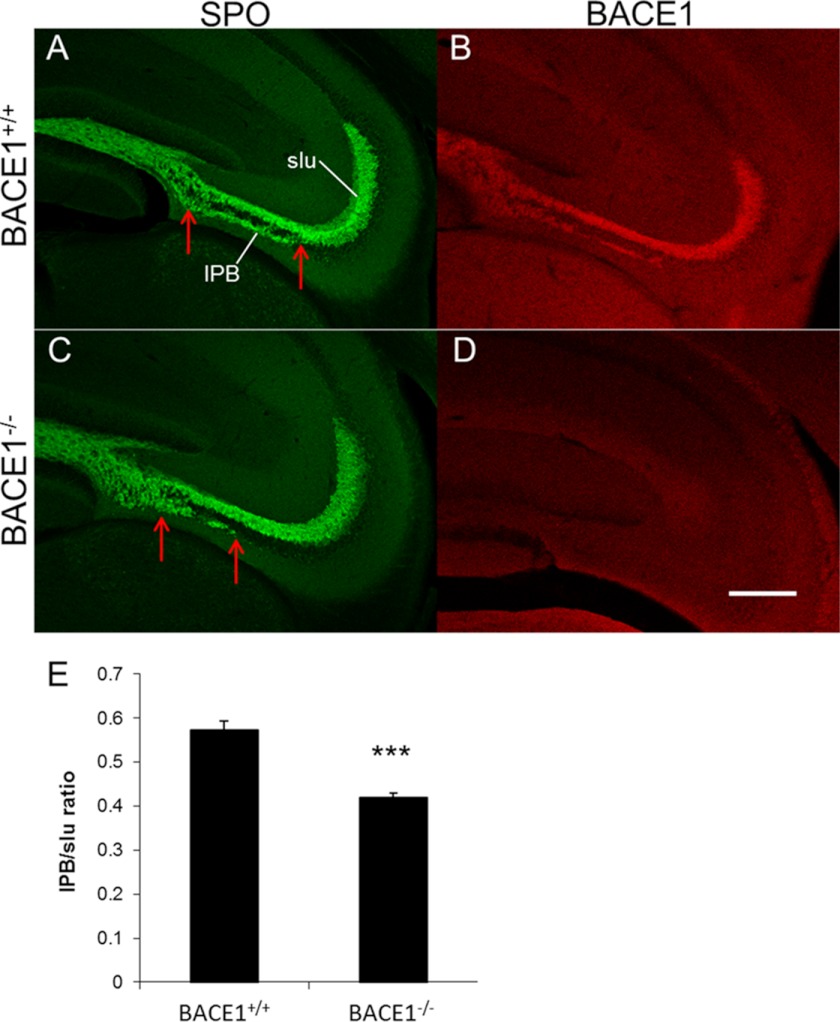
Previous studies have shown that the length of the IPB varies in different mouse strains and that longer IPB lengths correlate with improved memory performance (35). Because BACE1−/− mice exhibit memory deficits (16–18), we measured the IPB length of BACE1 null and wild-type mice. Although immunostaining of BACE1−/− adult hippocampal sections for the presynaptic terminal marker synaptoporin showed that the structure of mossy fibers appeared relatively normal overall, close examination revealed that the BACE1−/− IPB was only ∼70% of the length of the wild-type IPB (compare Fig. 2, C to A and E). Our BACE1−/− colony has been backcrossed with the C57BL/6 strain for over seven generations; therefore, it is unlikely that the short BACE1 null IPB length was the result of strain differences. These results raise the intriguing possibility that impaired BACE1 null memory performance may relate to the short IPB length of BACE1−/− mice, at least in part. Moreover, they suggest that mossy fiber axon outgrowth is perturbed by BACE1 deficiency.
FIGURE 2.
Mossy fiber infrapyramidal bundle is significantly shorter in BACE1−/− than wild-type mice. Coronal sections (40 μm) from brains of 3-month-old BACE1+/+ and BACE1−/− mice were immunostained for SPO (green; A and C) or BACE1 (red; B and D) and imaged with confocal microscopy. Representative sections of BACE1+/+ (A and B) and BACE1−/− (C and D) are shown. Note that the length of the BACE1−/− IPB is markedly shorter than that of the BACE1+/+ IPB (distances between the red arrows). Scale bar in D, 250 μm for A and D. E, lengths of the IPBs were determined using a free-form line measurement in ImageJ beginning at the point where the pyramidal cell layer ends and ending with the most distal point of continuous synaptoporin labeling inferior to the pyramidal cell layer. IPB lengths were normalized using a measurement of the distance from the beginning of the suprapyramidal bundle to the distal end of the slu, and the ratios were averaged across three sections per brain (E). The IPBs of BACE1−/− mice were significantly shorter than those of BACE1+/+ mice: 0.42 ± 0.020 versus 0.57 ± 0.011 normalized length (error bars, S.E.; n = 7; ***, p < 0.0001).
EphA4 Does Not Appear to be a BACE1 Substrate in the Hippocampus
To initially test the molecular basis of the BACE1 null IPB abnormality, we investigated members of the ephrin and Eph receptor families because they have well established roles in axon guidance and topographic mapping in the nervous system (36). In the hippocampus, a gradient of EphA4 expression in the dentate gyrus is involved in establishing a topographic map of mossy fiber terminal arborizations in CA3 (37). In addition, ephrinB3 plays a role in determining IPB length, such that ephrinB3 deficiency results in a long IPB (38). Recently, EphA4 was shown to be a potential BACE1 substrate in vitro (27), but in vivo validation was not obtained in that study.
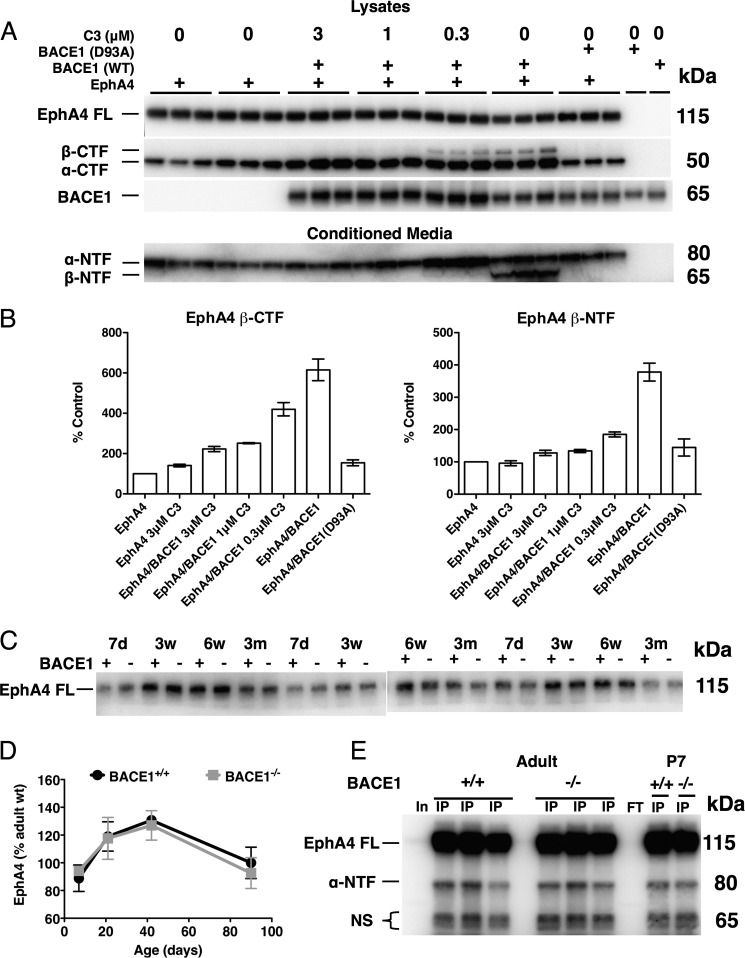
We sought to confirm that EphA4 is a BACE1 substrate in vitro and in vivo. For in vitro experiments, we co-transfected BACE1 together with EphA4 into HEK293 cells and assayed conditioned media and cell lysates for BACE1-cleaved EphA4 fragments by immunoblot (Fig. 3). BACE1 processing of EphA4, an ∼130-kDa type I transmembrane protein, is predicted to occur at a luminal juxtamembrane site to produce a secreted N-terminal fragment (NTF) and a membrane bound C-terminal fragment (CTF). In cells transfected with EphA4 alone, we observed EphA4 ∼130-kDa full-length and ∼55-kDa CTF bands in lysates and an EphA4 ∼75-kDa NTF band in conditioned media (Fig. 3A). The broad spectrum matrix metalloprotease inhibitor GM6001 blocks ectodomain shedding of EphA4 (39), and ADAM8 and ADAM9 metalloproteases have been shown to process another member of the Eph receptor family, EphB4, to produce an NTF and a CTF (40, 41). Therefore, we reasoned that the fragments we observed with EphA4 transfection alone were likely derived from ADAM (A Disintegrin and Metalloprotease) processing, in analogy to the α-secretase shedding of APP (42). Following co-transfection of EphA4 and BACE1, in addition to full-length EphA4 and ADAM-derived EphA4 fragments, immunoblot analysis revealed new bands at ∼65 kDa representing BACE1-cleaved EphA4 NTF and CTF in conditioned media and cell lysates, respectively (Fig. 3A). These BACE1-associated EphA4 fragments were produced at equal (NTF) or lower (CTF) amounts compared with those that were ADAM-derived (Fig. 3, A and B), analogous to levels of BACE1-processed APP NTF and CTF (4). Importantly, their production was prevented in a dose-dependent manner by BACE1 inhibitor treatment or by transfection with active site mutant BACE1(D93A), proving that these fragments were BACE1-dependent. Taken together, our results suggest that EphA4 is a BACE1 substrate, at least in vitro.
FIGURE 3.
EphA4 is a BACE1 substrate in vitro, but not in vivo. A and B, HEK293 cells were transiently co-transfected as indicated with EphA4 or empty vector control plus either wild-type (WT) BACE1, active site D93A mutant BACE1 (BACE1(D93A)), or empty vector control and treated with the indicated concentrations of the BACE1 inhibitor C3. Conditioned media and cell lysates were harvested after 24 h, and 10 μg of protein was resolved via SDS-PAGE and immunoblotted with an antibody against the EphA4 N terminus for conditioned media or C terminus for lysates. A, full-length EphA4 (EphA4 FL) runs at ∼130 kDa. The EphA4 N-terminal ADAM-cleaved fragment (α-NTF) is detected at ∼75 kDa, whereas the C-terminal ADAM-cleaved fragment (α-CTF) is detected at ∼55 kDa. Additional bands representing EphA4 β-NTF and β-CTF in conditioned media and lysates, respectively, are resolved at ∼65 kDa only in cells co-transfected with EphA4 and wild-type BACE1. These BACE1-dependent fragments show a dose-dependent reduction with BACE1 inhibitor C3 treatment (concentrations indicated). B, normalized quantifications (percentage of EphA4 transfection alone) of BACE1-dependent EphA4 β-NTF and β-CTFs. (Error bars, S.E.) C, BACE1+/+ (+) and BACE1−/− (−) brain homogenates from mice of the indicated ages were subjected to EphA4 immunoblot analysis using an anti-EphA4 N-terminal antibody. EphA4 FL signal is shown. (d = days; w = weeks; m = months). D, normalized quantifications (percentage of adult wild type) of BACE1+/+ and BACE1−/− EphA4 FL show no difference between wild-type and BACE1 null EphA4 FL at any age. E, BACE1+/+ and BACE1−/− hippocampal homogenates were immunoprecipitated (IP) with anti-N-terminal EphA4 antibody, resolved by SDS-PAGE, immunoblotted, and detected with the same antibody. Input (In) and IP flow-through (FT) representative lanes are also shown. The BACE1-dependent cleavage product is predicted at ∼65 kDa but is absent from IP eluates of 4-month-old and 7-day-old hippocampi. (NS, nonspecific bands.)
Overexpression of a protease and its substrate in cell culture could artifactually force cleavage that would not normally occur at endogenous enzyme and substrate levels found in tissues. To determine whether EphA4 is a BACE1 substrate in vivo, we performed EphA4 immunoblot analysis of hippocampal homogenates from postnatal and adult BACE1+/+ and BACE1−/− mice. Although we observed ADAM-processed ∼75- and ∼55-kDa EphA4 fragments by immunoblot, we found no evidence of BACE1-dependent ∼65-kDa EphA4 NTF and CTF in wild type compared with BACE1 null samples (data not shown). If EphA4 was a BACE1 substrate, then we predict that full-length EphA4 steady-state levels should become elevated in BACE1−/− samples. However, EphA4 immunoblot analysis of hippocampal homogenates from different aged postnatal to adult BACE1−/− mice showed no differences in full-length EphA4 levels compared with wild type (Fig. 3, C and D). Because BACE1-derived fragments could be produced at very low levels in vivo and therefore might be below the level of detection in immunoblots, we concentrated EphA4 fragments from hippocampal homogenates by immunoprecipitation (IP) followed by immunoblot analysis. Again, we were able to detect full-length EphA4 and the ADAM-cleaved EphA4 CTF (data not shown) and NTF (Fig. 3E) in IP immunoblots of postnatal and adult samples. Although a doublet was observed just below ∼65 kDa in BACE1+/+ samples where the BACE1-cleaved EphA4 NTF should migrate, the doublet was also present in BACE1−/− samples demonstrating that it was not BACE1-dependent (Fig. 3E). Our results indicate that either BACE1-cleaved EphA4 NTF and CTF levels are below the detection limit of IP immunoblot analysis or EphA4 is not a physiological BACE1 substrate in the hippocampus.
BACE1−/− Primary Hippocampal Neurons Have Impaired Growth Cone Collapse
Despite the fact that we could not detect the presence of BACE1-cleaved EphA4 fragments in vivo by IP immunoblot, we explored the possibility that low levels of BACE1 processing of EphA4 could modulate EphA4 signaling and thus regulate axon outgrowth and guidance. To test this, we used a sensitive functional assay of EphA4 signaling, growth cone (GC) collapse of primary hippocampal neurons in response to clustered ephrinB3. EphrinB3 is a known EphA4 ligand (43), and it plays a role in pruning of infrapyramidal mossy fiber axons (38). Therefore, it seemed reasonable that deficient BACE1 cleavage of EphA4 might elevate EphA4 steady-state levels, increase ephrinB3-EphA4 signaling, and promote mossy fiber GC collapse and axon retraction, resulting in the short BACE1−/− IPB that we had observed.
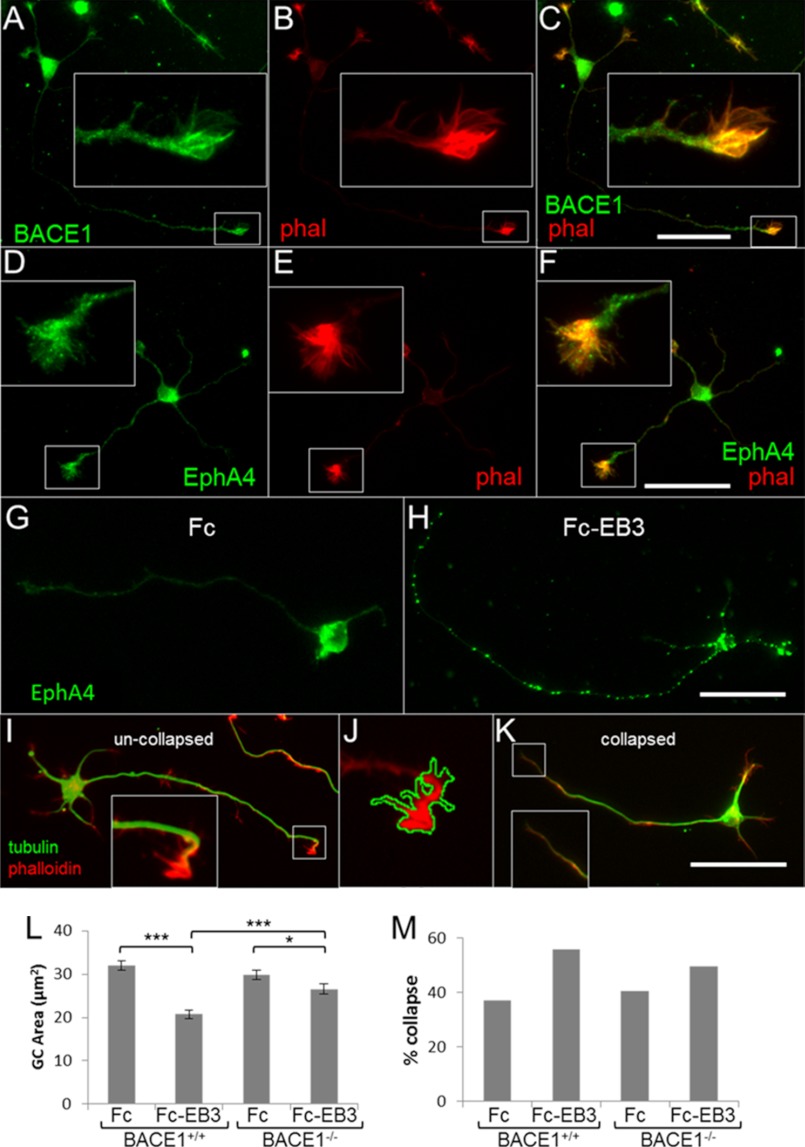
To test this hypothesis, we performed GC collapse assays on primary hippocampal neurons derived from BACE1 null and wild-type mice. Immunofluorescence microscopy showed that robust levels of both BACE1 (Fig. 4, A and C) and EphA4 (Fig. 4, D and F) were present in GCs. Primary neuron cultures were treated with either clustered ephrinB3-Fc fusion protein or Fc control (Fig. 4, G and H), and GC area and collapsed GC percentage were quantified (Fig. 4, L and M). EphA4 immunofluorescence verified that clustered ephrinB3 ligand treatment induced receptor clustering (Fig. 4H), which is necessary for full Eph signaling (44, 45). Following clustered Fc control treatment, BACE1−/− and BACE1+/+ neurons exhibited similar GC areas and percentages of GC collapse (Fig. 4, L and M). In response to clustered ephrinB3-Fc treatment, both BACE1−/− and BACE1+/+ neurons underwent GC collapse. However, contrary to our hypothesis, BACE1 null neurons showed reduced ephrinB3-induced GC area decreases and percent GC collapse increases compared with wild-type neurons (Fig. 4, L and M). In other words, BACE1−/− neurons exhibited impaired, rather than enhanced, GC collapse in response to ephrinB3. Therefore, our data did not support the hypothesis that EphA4 signaling is elevated in BACE1 null neurons, and hence it was not consistent with the notion that EphA4 is a physiological BACE1 substrate.
FIGURE 4.
BACE1−/− primary hippocampal neurons show impaired and not enhanced ephrin-B3-induced growth cone collapse. A–F, both BACE1 (green, A) and EphA4 (green, D) exhibit robust expression in wild-type primary hippocampal neuron growth cones as demonstrated by co-localization with phalloidin (phal) (red; B, C, E, and F). Insets show higher magnification of growth cones in boxed regions. Scale bar in C, 50 μm for A–C; scale bar in F, 50 μm for D–F. G and H, primary hippocampal neurons from P0 BACE1+/+ and BACE1−/− littermates were grown in low density cultures on glass coverslips for 18 h and then treated with preclustered Fc-ephrin-B3 (Fc-EB3; 1 μg/ml) or clustered Fc alone (Fc). Control coverslips (G and H) stained with anti-EphA4 antibody verify that treatment with clustered ephrin-B3 ligand induces clustering of the receptor EphA4 (green). Scale bar in H, 50 μm for G and H. I–K, fixed coverslips were stained with an anti-tubulin antibody (green) and rhodamine-phalloidin to reveal F-actin (red). Uncollapsed growth cones appear as dense phalloidin staining spreading out to form filopodia and lamellipodia (I and J), although axons with collapsed growth cones do not have dense phalloidin staining (K). Neurons from all groups were imaged, and the area of dense phalloidin staining at the end of the primary neurite (axon) was calculated by a free-form outline in ImageJ (J). Scale bar in K, 50 μm for I–K. L, histogram of mean growth cone areas. BACE1+/+ neurons showed a significant decrease in growth cone area with clustered Fc-EB3 treatment. The growth cone area of Fc-EB3-treated BACE1−/− neurons is greater and shows a smaller ephrin-B3 response than Fc-EB3-treated BACE1+/+ neurons. M, histogram of percentages of growth cone collapse shows a similar pattern. n = 800 neurons. *, p < 0.05; ***, p < 0.001; error bars, S.E.
BACE1−/− Mice Exhibit Axon Guidance Defects That Are Very Similar, If Not Identical, to Those of CHL1−/− Mice
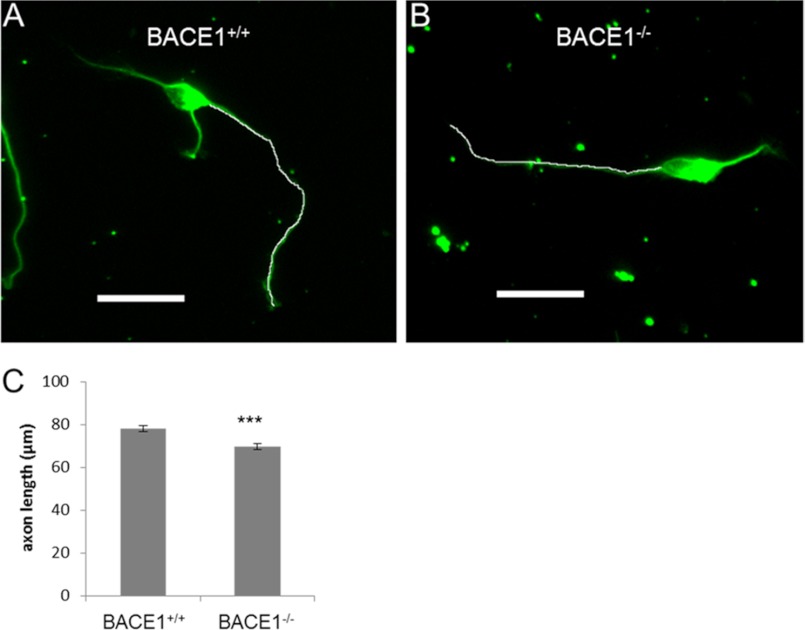
Despite the lack of evidence that EphA4 is a physiological BACE1 substrate, GC collapse was still impaired in BACE1−/− neurons, suggesting that a more general problem with growth cone or axon outgrowth and retraction might be caused by BACE1 deficiency. During our study, two groups reported that the neural cell adhesion molecules L1 and CHL1 are novel BACE1 substrates (28, 29). Because L1 and CHL1 play important roles in neurite outgrowth and axon guidance (46), we reasoned that these molecules might be involved in the axon targeting defects we have observed in BACE1−/− mice, at least in part. In support of this notion, we measured the axon lengths of BACE1−/− primary hippocampal neurons from our GC collapse experiments and discovered that they were modestly but significantly shorter than those of BACE1+/+ neurons (Fig. 5), suggesting that BACE1 deficiency mildly impairs axon outgrowth in vitro.
FIGURE 5.
BACE1−/− primary hippocampal neurons have impaired axonal outgrowth. Primary hippocampal neurons from P0 BACE1+/+ and BACE1−/− littermates were grown in low density cultures on glass coverslips for 18 h, fixed, incubated with anti-tubulin antibody, and imaged with immunofluorescence microscopy. A and B, lengths of axons were determined via free-form line measurement in ImageJ (white lines). Scale bars, 50 μm. C, BACE1−/− axons showed a modest but significant reduction in axon length by 11% compared with those of BACE1+/+ neurons ((69.6 ± 1.42 μm versus 77.9 ± 1.44 μm; n = 800 neurons). ***, p < 0.001; error bars, S.E.
Gene-targeted null mice for L1 and CHL1 have been generated and characterized previously. L1−/− mice have hydrocephalus, enlarged ventricles, and severe hypoplasia of certain CNS structures (e.g. hippocampus, corpus callosum, corticospinal tract, and cerebellar vermis) (47–49), whereas CHL1−/− mice exhibit relatively mild axon mis-targeting in the hippocampus and olfactory bulb (31, 32). Because L1−/− abnormalities are markedly more serious than those of BACE1 null, it appeared less likely that abrogation of BACE1 cleavage of L1 could be responsible for the axon guidance defects of the BACE1−/− mice. Therefore, we compared the BACE1 null phenotype in detail with that of CHL−/− to determine their degree of similarity, and hence we evaluated whether deficient BACE1 processing of CHL1 might play a role in the axon outgrowth and guidance phenotypes of the BACE1−/− mice.
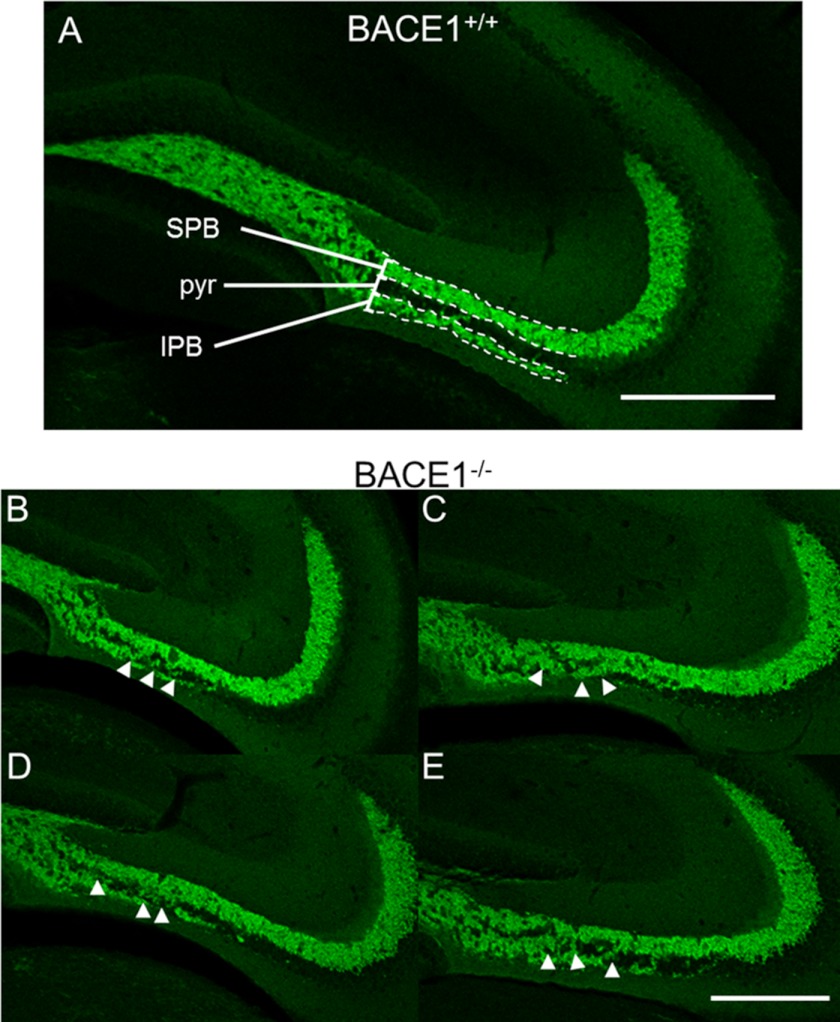
Close examination of BACE1−/− hippocampus and olfactory bulb revealed axon guidance mistakes that are strikingly similar to those previously published for CHL1−/− mice (31, 32). In the wild-type hippocampus, we noted that the IPB was relatively uniform in length and thickness from animal to animal, and mossy fibers rarely crossed into the CA3 pyramidal cell layer (Fig. 6A). In contrast, BACE1−/− hippocampi often exhibited IPBs that appeared disorganized or disrupted and had multiple groups of mossy fiber axons that invaded and/or crossed the pyramidal cell layer prior to the termination of the IPB, often obscuring the pyramidal cell layer (Fig. 6, B–E). These BACE1−/− IPB axon guidance defects appeared nearly indistinguishable from those previously reported for CHL1−/− mice (compare Fig. 6, A–E with Fig. 3, A and B, in Montag-Sallaz et al. (31) and Fig. 2, A–D, in Heyden et al. (32)).
FIGURE 6.
Mossy fibers exhibit abnormal invasion and crossing of the CA3 pyramidal cell layer in BACE1−/− mice. 40-μm coronal sections from BACE1+/+ (A) and BACE1−/− (B–E) mice were immunostained for synaptoporin (green) and examined for abnormalities in morphology by confocal microscopy. Mossy fibers travel in two bundles, the SPB and IPB, above and below the pyramidal cell layer (pyr), respectively, before the IPB crosses the pyramidal cell layer to terminate in the stratum lucidum. In wild-type mice, the pyramidal cell layer is typically clear of mossy fiber axons along the majority of the region bordered by the SPB and IPB (dotted lines in A). In contrast, BACE1−/− mice exhibit large groups of mossy fibers from the IPB that invade and/or cross the pyramidal cell layer prior to the termination of the IPB, often obscuring the pyramidal cell layer (arrowheads in B–E). These BACE1 null IPB defects are similar to those exhibited by CHL1−/− mice (compare with Fig. 3, A and B, in Montag-Sallaz et al. (31) and Fig. 2, A–D, in Heyden et al. (32)). Scale bar in A = 250 μm; scale bar in E, 250 μm for B–E.
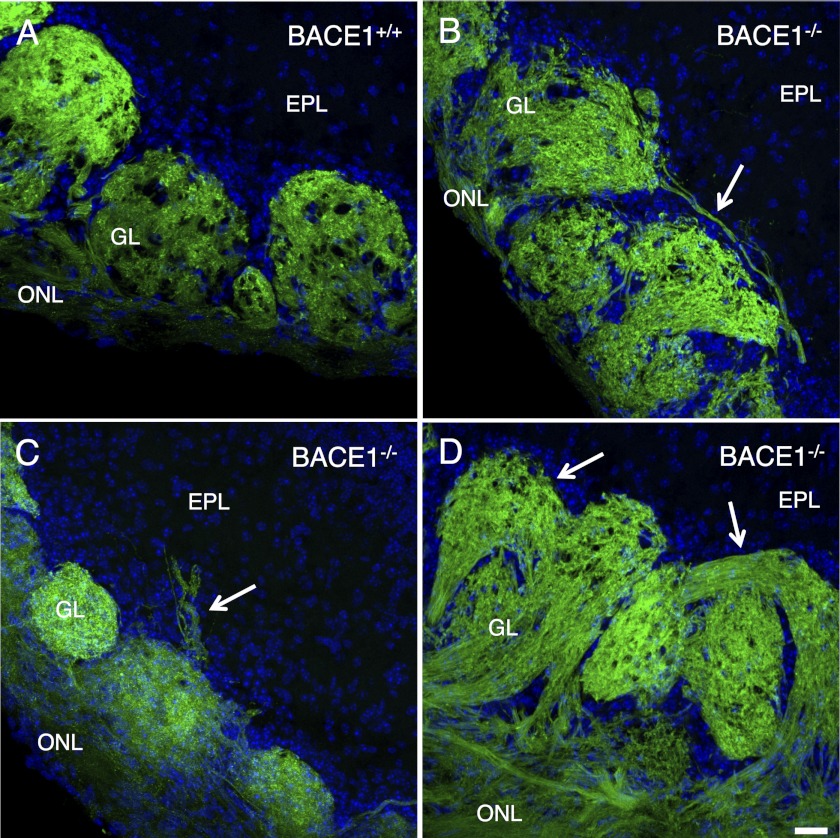
We previously reported that BACE1−/− mice have olfactory bulbs with malformed glomeruli and OSN axons that aberrantly target ectopic glomeruli (1). To determine whether the OSN axon guidance defects of BACE1−/− mice were similar to those of CHL1−/−, we bred BACE1−/− mice with gene-targeted mice in which the gene encoding GFP was inserted into the OMP gene locus (OMP-GFP mice) (33). OMP-GFP mice express GFP in all OSNs, and all OSN axons projecting to the olfactory bulb exhibit green fluorescence, so that we could sensitively probe axon guidance in the BACE-deficient olfactory system. We observed that OMP-GFP;BACE1−/− olfactory bulb sections displayed grossly malformed glomeruli and mis-targeted OSN axons that extended beyond the boundaries of the glomerulus as defined by periglomerular cells, including axons that connected more than one glomerulus or passed through the glomerular layer to terminate in the external plexiform layer (Fig. 7). These BACE1−/− OSN axon guidance errors appeared highly similar to those exhibited in CHL1−/− mice (compare Fig. 7, A–D with Fig. 4, A–F, in Montag-Sallaz et al. (31) and Fig. 3, A–D, in Heyden et al. (32)). Taken together, our results demonstrate that BACE1−/− axon outgrowth and guidance defects are very similar, if not identical, to those previously reported for hippocampal mossy fibers and OSN axons of CHL1−/− mice, suggesting that they arise from abnormalities in a common underlying pathway.
FIGURE 7.
Olfactory sensory neuron axons exhibit axon-targeting errors in the olfactory bulbs of BACE1−/− mice. 30-μm coronal sections of olfactory bulbs from adult OMP-GFP, BACE1+/+ (A) and OMP-GFP, and BACE1−/− (B–D) mice were imaged by laser scanning confocal microscopy for GFP (green) and DAPI (blue) to identify periglomerular cells and cells in the external plexiform layer (EPL). The olfactory nerve layer (ONL) and glomerular layer (GL) are labeled. In BACE1+/+ olfactory bulbs (A), axons of a specific odorant receptor normally innervate a single glomerulus. However, in BACE1−/− olfactory bulbs (B–D), some olfactory axons terminate in two glomeruli (arrow, B) or pass through the glomerular layer and terminate in the external plexiform layer (arrow, C). BACE1−/− olfactory bulbs also have grossly malformed glomeruli, such as torturously curved glomeruli (arrows in D), in contrast to typical spherical BACE1+/+ glomeruli (A). Scale bar in D, 25 μm for A–D.
CHL1 Is a BACE1 Substrate in the Hippocampus and Olfactory Bulb
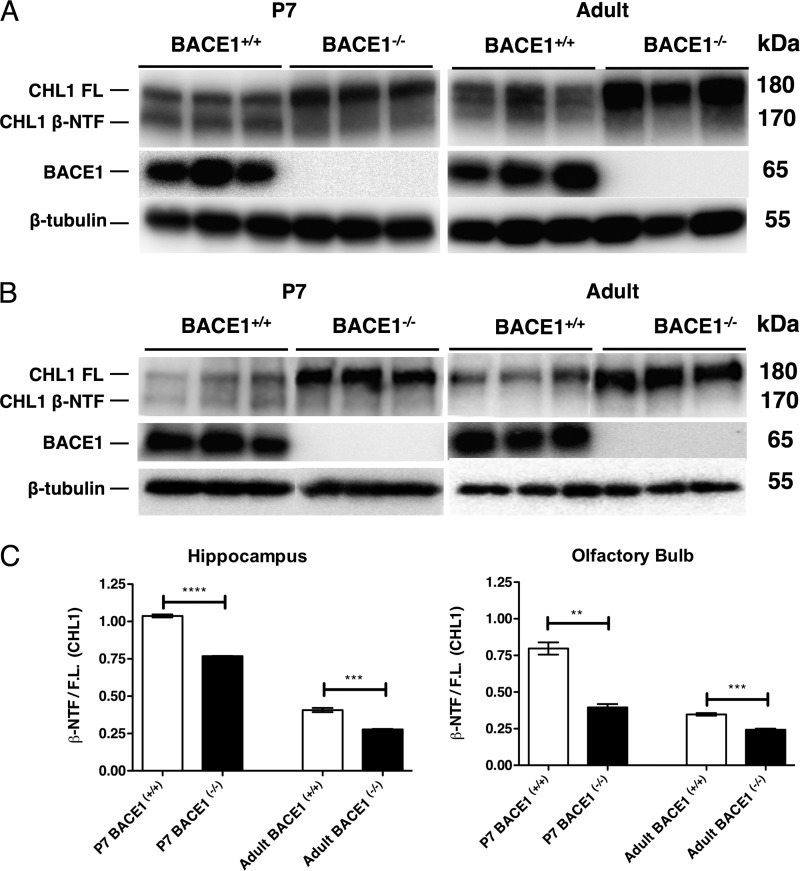
Given the similarity between the axon guidance defects in BACE1−/− and CHL1−/− mice, we next sought to determine whether CHL1 is a BACE1 substrate in the hippocampus and olfactory bulb. CHL1 is an ∼180-kDa type I transmembrane protein that is cleaved by BACE1 to produce an ∼170-kDa secreted NTF and an ∼10-kDa membrane-bound CTF (28, 29). To investigate BACE1 processing of CHL1 in vivo, we analyzed hippocampal and olfactory bulb homogenates from P7 and adult BACE1+/+ and BACE1−/− mice by immunoblot using an anti-N-terminal CHL1 antibody (Fig. 8). In wild-type P7 hippocampal and olfactory bulb homogenates, we detected two bands of approximately equal intensity representing ∼180-kDa full-length CHL1 and ∼170-kDa CHL1 NTF (Fig. 8, A and B, left panels), consistent with previous findings (28, 29). Importantly, P7 BACE1 null homogenates exhibited significantly increased intensity of the upper full-length CHL1 band, although the level of the lower CHL1 NTF band was diminished compared with wild type, suggesting that the ∼170-kDa band did indeed represent BACE1-cleaved CHL1 NTF. In the P7 BACE1−/− hippocampal homogenate, we observed an additional CHL1 band that migrated slightly faster than the CHL1 NTF in BACE1+/+ homogenates (Fig. 8A, left panel). CHL1 is also a substrate of ADAM8 (50), and it is possible that the faster migrating BACE1 null CHL1 NTF is derived from increased ADAM8 cleavage of CHL1 as a compensatory mechanism in response to BACE1 deficiency.
FIGURE 8.
CHL1 undergoes BACE1-dependent cleavage in hippocampus and olfactory bulb. A and B, hippocampi and olfactory bulbs were harvested from P7 and 3-month-old (Adult) BACE1+/+ and BACE1−/− mice, homogenized, and subjected to immunoblot analysis using an anti-N-terminal CHL1 antibody. A significant increase in ∼180-kDa full-length CHL1 (CHL1 FL) can be seen in BACE1−/− P7 and adult hippocampus (A) and the olfactory bulb (B). Additionally, an ∼160-kDa BACE1-specific CHL1 fragment (CHL1 β-NTF) is present just below the CHL1 full-length band in BACE1+/+ samples but is diminished or absent in BACE1−/− homogenates (A and B). Note the presence of a third band between CHL1 FL and CHL1 β-NTF bands in adult BACE1+/+ hippocampus that may represent an alternative CHL1 splice variant or glycosylated isoform (A, right panel). C, ratio of CHL1 β-NTF to full-length CHL1 is significantly increased in BACE1+/+ hippocampus and olfactory bulb homogenates compared with those of BACE1−/−, indicating that CHL1 is a substrate of BACE1 in both the postnatal and adult hippocampus and olfactory bulb. **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
We observed that the relative levels of CHL1 full-length and NTF in BACE1−/− versus BACE1+/+ P7 hippocampal and olfactory bulb homogenates followed similar patterns in the different brain regions as follows: full-length CHL1 was greater than CHL1 NTF in BACE1−/− samples, whereas full-length CHL1 and CHL1 NTF were closer to equal in BACE1+/+ samples. In agreement, the ratios of the immunoblot signal intensities of CHL1 NTF/full-length CHL1 were significantly higher in BACE1+/+ versus BACE1−/− homogenates (Fig. 8C) and is further evidence that the ∼170-kDa band represents BACE1-cleaved CHL1 NTF.
Finally, immunoblot analysis of adult hippocampal and olfactory bulb homogenates produced findings that were similar to those of the P7 samples, except that BACE1+/+ CHL1 NTF levels appeared lower in adult compared with P7 samples (Fig. 8, A and B, right panels). The lower CHL1 NTF levels in adult BACE1+/+ homogenates may reflect both the reduced BACE1 levels in adult brain (19) and the reduced neurite outgrowth in mature compared with prenatal brain, a process in which CHL1 is involved. In addition, we observed that the pattern of CHL1 fragments in the adult was different from that in P7, in that there was the presence of an additional band between CHL1 FL and CHL1 β-NTF bands in adult BACE1+/+ hippocampus that may represent an alternative CHL1 splice variant or glycosylated isoform. In any case, our results are consistent with those previously reported for whole brain (28, 29) and indicate that deficient BACE1 processing of CHL1 in BACE1−/− hippocampus and olfactory bulb results in increased and decreased steady-state levels of full-length CHL1 and cleaved CHL1 NTF, respectively, supporting the hypothesis that CHL1 is a BACE1 substrate in vivo.
CHL1 and BACE1 Localization Overlaps in Terminals of Mossy Fibers and OSN Axons and Growth Cones of Primary Hippocampal Neurons
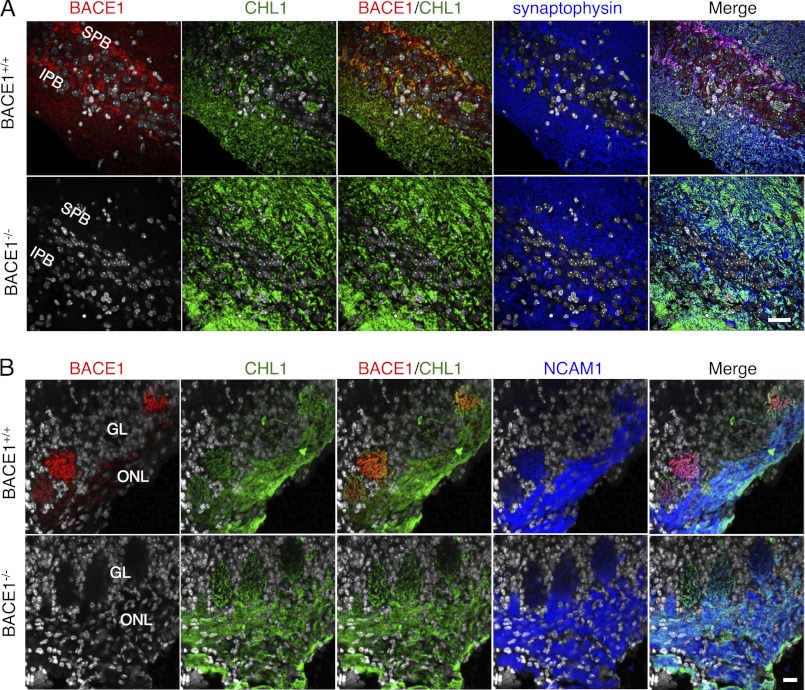
If CHL1 is the BACE1 substrate that accounts for axon guidance defects in BACE1−/− hippocampus and olfactory bulb, then the two proteins should co-localize, at least partially, in these brain regions in BACE1+/+ mice. To test this, we co-immunostained P7 BACE1+/+ and BACE1−/− hippocampus and olfactory bulb sections with anti-N-terminal CHL1 and anti-BACE1 antibodies followed by confocal microscopy (Fig. 9). P7 mice were chosen for analysis because we reasoned that the higher levels of BACE1 and neurite outgrowth in P7 brain would provide more robust co-localization than in adult brain. In BACE1+/+ hippocampus, we observed that CHL1 and BACE1 immunostaining overlapped significantly in the IPB and SPB/stratum lucidum that contains mossy fiber axons and terminals (Fig. 9A). Similar to hippocampus, immunostaining of P7 BACE1+/+ olfactory bulb glomeruli showed significant co-localization of CHL1 and BACE1 in the terminals of OSN axons (Fig. 9B). CHL1 and BACE1 immunostaining appeared to co-localize within the presynaptic synaptophysin-positive mossy fiber and NCAM1-positive OSN axon terminals, both of which exhibit a high degree of synaptic plasticity and require ongoing axon guidance in the adult. CHL1 localization in BACE1−/− hippocampus or olfactory bulb did not appear significantly different from that in BACE1+/+.
FIGURE 9.
CHL1 and BACE1 co-localize in terminals of mossy fibers and olfactory sensory neuron axons. 30–40-μm coronal sections from BACE1+/+ and BACE1−/− P7 hemibrains (A) and olfactory bulbs (B) were immunostained with anti-BACE1, CHL1, synaptophysin (hemibrains), and NCAM1 (olfactory bulbs) antibodies and imaged by confocal microscopy. A, top row, BACE1 (red) co-localizes with CHL1 (green) in mossy fiber terminals of the wild-type IPB and SPB of the hippocampus (yellow/orange in BACE1/CHL1); there is significant overlap of CHL1 and BACE1 with synaptophysin (blue), confirming presynaptic localization (Merge). A, bottom row, CHL1 localization is not significantly altered in BACE1−/− hippocampus. B, top row, BACE1 (red) co-localizes with CHL1 (green) in olfactory sensory neuron axon terminals in glomeruli (GL) of the olfactory bulb (yellow/orange in BACE1/CHL1); there is significant overlap of CHL1 and BACE1 with NCAM1 (blue) in glomeruli, confirming presynaptic localization (Merge), although most of the CHL1 and NCAM1 appear in olfactory sensory neuron axons of the olfactory nerve layer (ONL). B, bottom row, CHL1 localization is not significantly altered in BACE1−/− olfactory bulb. Nuclear staining with DAPI (gray)/ Scale bar in lower right frames in A and B, 25 μm for hippocampal and olfactory bulb images, respectively.
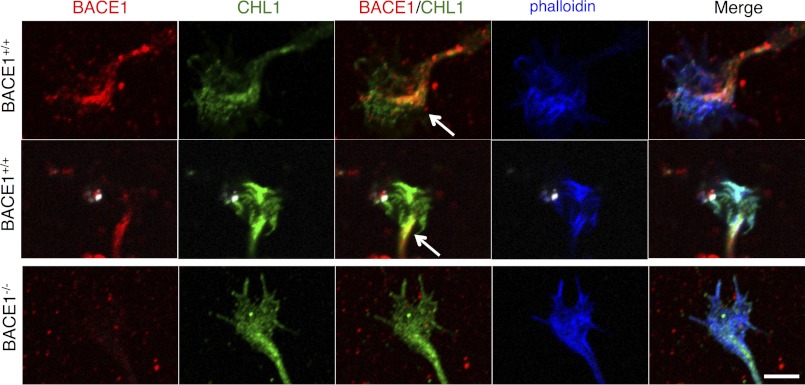
In order for BACE1 processing of CHL1 to regulate axon outgrowth, we reasoned that BACE1 and CHL1 should co-localize, at least partially, within growth cones of primary hippocampal neurons in culture. Therefore, we triple stained wild-type and BACE1 null hippocampal neuron cultures with anti-BACE1 and anti-CHL1 antibodies and fluorescently tagged phalloidin to label F-actin in growth cones. In wild-type growth cones, BACE1 immunostaining was primarily punctate, probably reflecting BACE1 localization in intracellular vesicles, with most of BACE1 puncta concentrated in the axon and central region of the growth cone (Fig. 10, top two rows). Relatively little BACE1 immunostaining appeared in the region of phalloidin staining in the growth cone. Immunostaining of CHL1 was more distributed throughout the growth cone than that of BACE1 and overlapped considerably with phalloidin staining. Importantly, the highest concentration of CHL1 coincided with the majority of BACE1 in the growth cone central region. This significant overlap of BACE1 and CHL1 is consistent with the hypothesis that CHL1 is a BACE1 substrate in the growth cone during axon outgrowth. The localization of CHL1 in BACE1−/− growth cones and their morphology appeared unaltered by BACE1 deficiency (Fig. 10, bottom row).
FIGURE 10.
CHL1 and BACE1 co-localize in growth cones of primary hippocampal neurons. Primary hippocampal neurons from P0 BACE1+/+ (top two rows) and BACE1−/− (bottom row) littermates were grown in low density cultures on glass coverslips for 18 h, fixed, incubated with antibodies against BACE1 (red) and CHL1 (green), stained with rhodamine-labeled phalloidin (blue) to visualize F-actin in growth cones, and imaged with confocal microscopy. In wild-type neurons, punctate BACE1 immunostaining is concentrated in the central region of the growth cone that appears continuous with the axon (BACE1, top two rows). Although CHL1 immunostaining is distributed throughout the growth cone (CHL1, top two rows) and overlaps with phalloidin (Merge), CHL1 is also concentrated in the central region of the growth cone. Note the significant overlap of BACE1 and CHL1 in the growth cone central region (arrows, yellow/orange in BACE1/CHL1, top two rows). Phalloidin-stained F-actin in the expanding growth cone co-localizes with CHL1 only (cyan) as well as CHL1 and BACE1 (white) in Merge. BACE1−/− growth cones do not appear to have abnormal morphology or CHL and F-actin localization (bottom row). Scale bar, 5 μm for all frames.
Taken together, our results demonstrate that CHL1 and BACE1 overlap to a significant extent in mossy fiber terminals in the hippocampus, OSN axon terminals in the olfactory bulb, and in growth cones of primary hippocampal neurons, obviously a necessary prerequisite for cleavage of CHL1 by BACE1. Together, these results strongly support the conclusion that CHL1 is a BACE1 substrate in the brain, and that the axon guidance defects of BACE1−/− mossy fibers and OSN axons are likely the result of deficient BACE1 processing of CHL1. Interestingly, the nearly indistinguishable axon guidance phenotypes of BACE1−/− and CHL1−/− mice suggest that BACE1 deficiency and the consequent decrease in BACE1 processing of CHL1 results in CHL1 loss-of-function.
DISCUSSION
Correspondence between BACE1 Deficiency and CHL1 Loss-of-Function
Previously, we demonstrated that BACE1 is expressed in the terminals of OSN axons in the olfactory bulb and that BACE1−/− mice exhibit malformed glomeruli and have mis-targeted OSN axons in the bulb (1). These phenotypes indicated that BACE1 has a function in axon guidance, at least in the initial segment of the olfactory system, and they suggested BACE1 might play a role in axon guidance in other parts of the nervous system. To investigate this question, we examined the hippocampus of BACE1−/− mice, a region of the brain essential for learning and memory. Indeed, BACE1 null mice exhibit memory impairment (16–18) and seizures (22, 23), suggesting hippocampal dysfunction. We discovered that the BACE1−/− mouse has a shortened mossy fiber IPB (Fig. 2), a part of the hippocampus that closely correlates with memory performance in mouse strains. The IPB consists of mossy fiber axons that originate from granule cells located in the ventral blade of the dentate gyrus. IPB mossy fibers traverse the ventral side of the CA3 pyramidal cell layer before they pass through the cell layer to join other mossy fibers on the dorsal side of CA3 in the stratum lucidum. Mouse strains exhibit strain-specific IPB sizes, and strains that have smaller IPBs do not perform as well on memory tests as do strains that have larger IPBs (35). Although the physiological basis of the memory performance-IPB length correlation is unclear, we speculate that the relative memory impairment of BACE1−/− mice may be a consequence of short IPB length, at least in part.
The short BACE1 null IPB suggested a defect in axon guidance, so we initially investigated the potential role of ephrins and Eph receptors, classical axon guidance molecules. Ephrin-Eph receptor signaling causes growth cone collapse and axon repulsion (36), so a short IPB would be consistent with increased signaling. We chose to examine ephrinB3 and EphA4 because of the following: 1) ephrinB3−/− mice have abnormally long IPBs suggesting ephrinB3 decreases IPB length (38); 2) a gradient of EphA4 expression in dentate gyrus granule cells establishes a topographic map of mossy fiber projections to CA3 (37), and 3) EphA4 was recently identified as a candidate BACE1 substrate in vitro (27). Although EphA4 is cleaved by BACE1 when overexpressed in HEK293 cells, we found no evidence of physiological EphA4 processing by endogenous BACE1 in the hippocampus (Fig. 3, C–E). BACE1−/− primary hippocampal neurons exposed to ephrinB3 did exhibit growth cone collapse (Fig. 4), but it was impaired rather than the enhanced collapse that we predicted based on increased and decreased lengths of ephrinB3−/− and BACE1−/− IPBs, respectively. However, axon lengths of BACE1−/− primary hippocampal neurons were significantly reduced (Fig. 5), suggesting a potential reason for the short BACE1 null IPB. Although BACE1−/− ephrinB3-induced growth cone collapse was abnormal, the lack of evidence in support of EphA4 as a BACE1 substrate in vivo suggested that other molecular mechanisms were responsible for the short BACE1 null IPB.
During our study, we became aware of reports that L1 and CHL1 are BACE1 substrates in vivo (28, 29). Because these molecules are involved in neurite outgrowth (46), we reasoned that the short BACE1−/− IPB could be the result of L1 or CHL1 loss-of-function. Our examination of the published L1−/− and CHL1−/− phenotypes suggested that L1−/− defects are much more severe than the mild axon guidance errors of the BACE1 null. L1−/− mice exhibit severe brain deformities, including hypoplasia of the hippocampus, corticospinal tract, corpus callosum, cerebellar vermis, enlarged ventricles, and various behavioral abnormalities (47–49). However, CHL1−/− and BACE1−/− phenotypes appear nearly indistinguishable (31, 32). For example, in addition to short IPBs, CHL1−/− and BACE1−/− mice exhibit disorganized mossy fiber bundles, with an increased number of mossy fiber crossovers between the infrapyramidal and suprapyramidal bundles (Fig. 6). Additionally, both CHL1 and BACE1 null mice have OSN axons that aberrantly connect to more than one glomerulus or terminate in the external plexiform layer of the olfactory bulb (Fig. 7). Importantly, CHL1 co-localizes with BACE1 in the terminals of mossy fibers and OSN axons (Fig. 9) and in growth cones of primary hippocampal neurons (Fig. 10). Moreover, BACE1 appears to cleave endogenous CHL1 in the hippocampus and olfactory bulb (Fig. 8). Together, these data strongly suggest that CHL1 is the physiological BACE1 substrate responsible for the axon guidance defects found in BACE1 null hippocampus and olfactory bulb.
The remarkable similarity between BACE1−/− and CHL1−/− phenotypes suggests that deficient BACE1 cleavage of CHL1 results in CHL1 loss-of-function. Although we cannot exclude the possibility that partial loss of L1 function may also play a role in BACE1−/− mice, we find this less likely given the severity of the L1−/− phenotype. CHL1 is cleaved in the juxtamembrane region by the metalloprotease ADAM8, causing secretion of the ∼165-kDa CHL1 ectodomain, which is a very potent stimulator of neurite outgrowth and neuronal survival in primary neuron culture (50). We note that the ADAM8-derived CHL1 ectodomain is very similar in size to the BACE1-dependent CHL1 NTF, indicating that both ADAM8 and BACE1 cleave CHL1 at proximate sites in the CHL1 juxtamembrane region. This conclusion is supported by the identification of the in vitro BACE1 cleavage site between residues 1061 and 1062 of CHL1, a position that is 21 amino acids N-terminal to the CHL1 transmembrane region (28). It is interesting to note that L1 is processed by γ-secretase as well as by the α-secretases ADAM10 and ADAM17 (51), suggesting the possibility that CHL1 may also be a γ-secretase substrate.
We hypothesize that both ADAM8- and BACE1-derived CHL1 fragments share similar activities and functions. However, important distinctions between the two fragments could exist regarding their neuronal production and secretion sites, which in turn would determine their sites of biological activity. BACE1 is predominantly localized in endosomes of the soma and the presynaptic terminal (4, 17, 30, 52), although ADAM8 is likely found on the plasma membrane. We determined that BACE1 and CHL1 co-localize in growth cones and terminals of hippocampal mossy fibers and OSN axons (Figs. 9 and 10). Therefore, production and secretion of BACE1-dependent CHL1 NTF are likely to occur in growth cones and presynaptic terminals, precisely at the sites of CHL1 action for axon outgrowth and guidance. In support of this notion, it is interesting to note that secreted CHL1 ectodomain acts as a potent stimulator of neurite outgrowth in vitro (50). Notably, CHL1 interacts with semaphorin 3A during neurite outgrowth and growth cone collapse (53, 54). Intriguingly, soluble L1 converts Sema3A-induced axonal repulsion into attraction (55, 56). Given the close similarity between CHL1 and L1, soluble CHL1 may also regulate Sema3A-induced axonal repulsion and attraction. Taken together, these findings suggest that a major function of CHL1 is provided by the secreted CHL1 ectodomain as opposed to membrane-bound CHL1 and that BACE1 deficiency produces CHL1 loss-of-function as a result of decreased CHL1 ectodomain secretion from presynaptic terminals and growth cones.
Implications of BACE1 Inhibition for AD
The phenotypes of BACE1−/− mice, caused by deficient processing of CHL1 and other BACE1 substrates, suggest that BACE1 inhibition for AD may be associated with mechanism-based side effects. In particular, the axon guidance defects observed in BACE1−/− mice imply the possibility that similar adverse events may occur in adult neurogenic and/or regenerating neural systems as a result of chronic BACE1 inhibition and therefore add a note of caution to BACE1 inhibitor drug development. This would appear to contraindicate BACE1 as a therapeutic target. However, several arguments suggest that BACE1 inhibitor drugs may be relatively safe in humans. First, the phenotypes of BACE1 null mice, although complex, are relatively mild. In fact, years of analysis were required to uncover the BACE1 null phenotypes. On an outbred or mixed genetic background, BACE1−/− mice are viable, fertile, and have no apparent phenotype (13, 14). Only when mice are backcrossed onto an inbred genetic background (e.g. C57BL/6) do phenotypes become noticeable (21). This suggests that modifier genes in specific genetic backgrounds contribute to BACE1 null phenotypes. Second, BACE1 null phenotypes arise only in the BACE1−/− homozygotes, equivalent to 100% BACE1 inhibition, but not in BACE1+/− heterozygotes. Based on AD mouse model studies, it is hypothesized that 50% BACE1 inhibition is necessary to delay or prevent amyloid deposition (17, 57). Finally, many if not all the BACE1 null phenotypes may be related to development in the absence of BACE1, whereas the fully developed adult may not require BACE1 activity. Future experiments with conditional BACE1−/− mice will be necessary to address this question. Although the BACE1 phenotypes are a note of caution moving forward, the points discussed above offer the hope that partial BACE1 inhibition in the genetically outbred adult human population may result in relatively few mechanism-based toxicities. Given the devastating effects on individuals with AD, accepting a modicum of side effect risk may be acceptable to provide a benefit for this debilitating and ultimately fatal neurodegenerative disease.
Acknowledgments
We acknowledge the Getsios (Northwestern University) and the Sakaguchi (Wakayama Medical University) laboratories for providing the EphA4 plasmids used in this work. We thank Drs. Aryeh Routtenberg, Adriana Ferreira, and Thomas Bozza (Northwestern University) for helpful discussions. Confocal imaging work was performed at the Northwestern University Cell Imaging Facility generously supported by National Institutes of Health Grants CCSG P30 CA060553 from NCI awarded to the Robert H. Lurie Comprehensive Cancer Center. Growth cone collapse imaging and axon length measurement was accomplished through the generosity of Keyence Corp. (Japan).
This work was supported, in whole or in part, by National Institutes of Health Grant R01AG022560 (to R. V.), Northwestern University Mechanisms of Aging and Dementia Training Grant T32 AG20506 (to L. K.), Drug Discovery in Age-related Disorders Training Grant T32 AG260-12 (to S. R.), Medical Scientist Training Program Training Grant T32 GM08152-19, and Cellular and Molecular Basis of Disease Training Grant T32 GM08061 (to B. H.). This work was also supported by American Health Assistance Foundation Grant A2012063 (to R. V.).
This article was selected as a Paper of the Week.
- AD
- Alzheimer disease
- Aβ
- β-amyloid
- APP
- amyloid precursor protein
- IPB
- infrapyramidal bundle
- SPO
- synaptoporin
- OSN
- olfactory sensory neuron
- SPB
- suprapyramidal bundle
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- slu
- stratum lucidum
- OMP
- olfactory marker protein
- FL
- full-length
- IP
- immunoprecipitated
- GC
- growth cone
- NTF
- N-terminal fragment
- CTF
- C-terminal fragment.
REFERENCES
- 1. Rajapaksha T. W., Eimer W. A., Bozza T. C., Vassar R. (2011) The Alzheimer β-secretase enzyme BACE1 is required for accurate axon guidance of olfactory sensory neurons and normal glomerulus formation in the olfactory bulb. Mol. Neurodegener. 6, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tanzi R. E., Bertram L. (2005) Twenty years of the Alzheimer disease amyloid hypothesis. A genetic perspective. Cell 120, 545–555 [DOI] [PubMed] [Google Scholar]
- 3. Sisodia S. S., St George-Hyslop P. H. (2002) γ-Secretase, Notch, Aβ, and Alzheimer disease. Where do the presenilins fit in? Nat. Rev. Neurosci. 3, 281–290 [DOI] [PubMed] [Google Scholar]
- 4. Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. (1999) β-Secretase cleavage of Alzheimer amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741 [DOI] [PubMed] [Google Scholar]
- 5. Yan R., Bienkowski M. J., Shuck M. E., Miao H., Tory M. C., Pauley A. M., Brashier J. R., Stratman N. C., Mathews W. R., Buhl A. E., Carter D. B., Tomasselli A. G., Parodi L. A., Heinrikson R. L., Gurney M. E. (1999) Membrane-anchored aspartyl protease with Alzheimer disease β-secretase activity. Nature 402, 533–537 [DOI] [PubMed] [Google Scholar]
- 6. Sinha S., Anderson J. P., Barbour R., Basi G. S., Caccavello R., Davis D., Doan M., Dovey H. F., Frigon N., Hong J., Jacobson-Croak K., Jewett N., Keim P., Knops J., Lieberburg I., Power M., Tan H., Tatsuno G., Tung J., Schenk D., Seubert P., Suomensaari S. M., Wang S., Walker D., Zhao J., McConlogue L., John V. (1999) Purification and cloning of amyloid precursor protein β-secretase from human brain. Nature 402, 537–540 [DOI] [PubMed] [Google Scholar]
- 7. Hussain I., Powell D., Howlett D. R., Tew D. G., Meek T. D., Chapman C., Gloger I. S., Murphy K. E., Southan C. D., Ryan D. M., Smith T. S., Simmons D. L., Walsh F. S., Dingwall C., Christie G. (1999) Identification of a novel aspartic protease (Asp-2) as β-secretase. Mol. Cell. Neurosci. 14, 419–427 [DOI] [PubMed] [Google Scholar]
- 8. Lin X., Koelsch G., Wu S., Downs D., Dashti A., Tang J. (2000) Human aspartic protease memapsin 2 cleaves the β-secretase site of β-amyloid precursor protein. Proc. Natl. Acad. Sci. U.S.A. 97, 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Citron M., Oltersdorf T., Haass C., McConlogue L., Hung A. Y., Seubert P., Vigo-Pelfrey C., Lieberburg I., Selkoe D. J. (1992) Mutation of the β-amyloid precursor protein in familial Alzheimer disease increases β-protein production. Nature 360, 672–674 [DOI] [PubMed] [Google Scholar]
- 10. Di Fede G., Catania M., Morbin M., Rossi G., Suardi S., Mazzoleni G., Merlin M., Giovagnoli A. R., Prioni S., Erbetta A., Falcone C., Gobbi M., Colombo L., Bastone A., Beeg M., Manzoni C., Francescucci B., Spagnoli A., Cantù L., Del Favero E., Levy E., Salmona M., Tagliavini F. (2009) A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science 323, 1473–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giaccone G., Morbin M., Moda F., Botta M., Mazzoleni G., Uggetti A., Catania M., Moro M. L., Redaelli V., Spagnoli A., Rossi R. S., Salmona M., Di Fede G., Tagliavini F. (2010) Neuropathology of the recessive A673V APP mutation. Alzheimer disease with distinctive features. Acta Neuropathol. 120, 803–812 [DOI] [PubMed] [Google Scholar]
- 12. Jonsson T., Atwal J. K., Steinberg S., Snaedal J., Jonsson P. V., Bjornsson S., Stefansson H., Sulem P., Gudbjartsson D., Maloney J., Hoyte K., Gustafson A., Liu Y., Lu Y., Bhangale T., Graham R. R., Huttenlocher J., Bjornsdottir G., Andreassen O. A., Jönsson E. G., Palotie A., Behrens T. W., Magnusson O. T., Kong A., Thorsteinsdottir U., Watts R. J., Stefansson K. (2012) A mutation in APP protects against Alzheimer disease and age-related cognitive decline. Nature 488, 96–99 [DOI] [PubMed] [Google Scholar]
- 13. Luo Y., Bolon B., Kahn S., Bennett B. D., Babu-Khan S., Denis P., Fan W., Kha H., Zhang J., Gong Y., Martin L., Louis J. C., Yan Q., Richards W. G., Citron M., Vassar R. (2001) Mice deficient in BACE1, the Alzheimer β-secretase, have normal phenotype and abolished β-amyloid generation. Nat. Neurosci. 4, 231–232 [DOI] [PubMed] [Google Scholar]
- 14. Roberds S. L., Anderson J., Basi G., Bienkowski M. J., Branstetter D. G., Chen K. S., Freedman S. B., Frigon N. L., Games D., Hu K., Johnson-Wood K., Kappenman K. E., Kawabe T. T., Kola I., Kuehn R., Lee M., Liu W., Motter R., Nichols N. F., Power M., Robertson D. W., Schenk D., Schoor M., Shopp G. M., Shuck M. E., Sinha S., Svensson K. A., Tatsuno G., Tintrup H., Wijsman J., Wright S., McConlogue L. (2001) BACE knockout mice are healthy despite lacking the primary β-secretase activity in brain. Implications for Alzheimer disease therapeutics. Hum. Mol. Genet. 10, 1317–1324 [DOI] [PubMed] [Google Scholar]
- 15. Cai H., Wang Y., McCarthy D., Wen H., Borchelt D. R., Price D. L., Wong P. C. (2001) BACE1 is the major β-secretase for generation of Aβ peptides by neurons. Nat. Neurosci. 4, 233–234 [DOI] [PubMed] [Google Scholar]
- 16. Ohno M., Sametsky E. A., Younkin L. H., Oakley H., Younkin S. G., Citron M., Vassar R., Disterhoft J. F. (2004) BACE1 deficiency rescues memory deficits and cholinergic dysfunction in a mouse model of Alzheimer disease. Neuron 41, 27–33 [DOI] [PubMed] [Google Scholar]
- 17. Laird F. M., Cai H., Savonenko A. V., Farah M. H., He K., Melnikova T., Wen H., Chiang H. C., Xu G., Koliatsos V. E., Borchelt D. R., Price D. L., Lee H. K., Wong P. C. (2005) BACE1, a major determinant of selective vulnerability of the brain to amyloid-β amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J. Neurosci. 25, 11693–11709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ohno M., Chang L., Tseng W., Oakley H., Citron M., Klein W. L., Vassar R., Disterhoft J. F. (2006) Temporal memory deficits in Alzheimer mouse models. Rescue by genetic deletion of BACE1. Eur. J. Neurosci. 23, 251–260 [DOI] [PubMed] [Google Scholar]
- 19. Willem M., Garratt A. N., Novak B., Citron M., Kaufmann S., Rittger A., DeStrooper B., Saftig P., Birchmeier C., Haass C. (2006) Control of peripheral nerve myelination by the β-secretase BACE1. Science 314, 664–666 [DOI] [PubMed] [Google Scholar]
- 20. Hu X., Hicks C. W., He W., Wong P., Macklin W. B., Trapp B. D., Yan R. (2006) Bace1 modulates myelination in the central and peripheral nervous system. Nat. Neurosci. 9, 1520–1525 [DOI] [PubMed] [Google Scholar]
- 21. Dominguez D., Tournoy J., Hartmann D., Huth T., Cryns K., Deforce S., Serneels L., Camacho I. E., Marjaux E., Craessaerts K., Roebroek A. J., Schwake M., D'Hooge R., Bach P., Kalinke U., Moechars D., Alzheimer C., Reiss K., Saftig P., De Strooper B. (2005) Phenotypic and biochemical analyses of BACE1- and BACE2-deficient mice. J. Biol. Chem. 280, 30797–30806 [DOI] [PubMed] [Google Scholar]
- 22. Hu X., Zhou X., He W., Yang J., Xiong W., Wong P., Wilson C. G., Yan R. (2010) BACE1 deficiency causes altered neuronal activity and neurodegeneration. J. Neurosci. 30, 8819–8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hitt B. D., Jaramillo T. C., Chetkovich D. M., Vassar R. (2010) BACE1−/− mice exhibit seizure activity that does not correlate with sodium channel level or axonal localization. Mol. Neurodegener. 5, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Savonenko A. V., Melnikova T., Laird F. M., Stewart K. A., Price D. L., Wong P. C. (2008) Alteration of BACE1-dependent NRG1/ErbB4 signaling and schizophrenia-like phenotypes in BACE1-null mice. Proc. Natl. Acad. Sci. U.S.A. 105, 5585–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cao L., Rickenbacher G. T., Rodriguez S., Moulia T. W., Albers M. W. (2012) The precision of axon targeting of mouse olfactory sensory neurons requires the BACE1 protease. Sci. Rep. 2, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vassar R., Kovacs D. M., Yan R., Wong P. C. (2009) The β-secretase enzyme BACE in health and Alzheimer disease. Regulation, cell biology, function, and therapeutic potential. J. Neurosci. 29, 12787–12794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hemming M. L., Elias J. E., Gygi S. P., Selkoe D. J. (2009) Identification of β-secretase (BACE1) substrates using quantitative proteomics. PLoS One 4, e8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou L., Barão S., Laga M., Bockstael K., Borgers M., Gijsen H., Annaert W., Moechars D., Mercken M., Gevaer K., De Strooper B. (2012) The neural cell adhesion molecules L1 and CHL1 are cleaved by BACE1 protease in vivo. J. Biol. Chem. 287, 25927–25940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuhn P. H., Koroniak K., Hogl S., Colombo A., Zeitschel U., Willem M., Volbracht C., Schepers U., Imhof A., Hoffmeister A., Haass C., Rossner S., Bräse S., Lichtenthaler S. F. (2012) Secretome protein enrichment identifies physiological BACE1 protease substrates in neurons. EMBO J. 31, 3157–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao J., Fu Y., Yasvoina M., Shao P., Hitt B., O'Connor T., Logan S., Maus E., Citron M., Berry R., Binder L., Vassar R. (2007) β-Site amyloid precursor protein-cleaving enzyme 1 levels become elevated in neurons around amyloid plaques. Implications for Alzheimer disease pathogenesis. J. Neurosci. 27, 3639–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Montag-Sallaz M., Schachner M., Montag D. (2002) Misguided axonal projections, neural cell adhesion molecule 180 mRNA up-regulation, and altered behavior in mice deficient for the close homolog of L1. Mol. Cell. Biol. 22, 7967–7981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heyden A., Angenstein F., Sallaz M., Seidenbecher C., Montag D. (2008) Abnormal axonal guidance and brain anatomy in mouse mutants for the cell recognition molecules close homolog of L1 and NgCAM-related cell adhesion molecule. Neuroscience 155, 221–233 [DOI] [PubMed] [Google Scholar]
- 33. Potter S. M., Zheng C., Koos D. S., Feinstein P., Fraser S. E., Mombaerts P. (2001) Structure and emergence of specific olfactory glomeruli in the mouse. J. Neurosci. 21, 9713–9723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bennett B. D., Denis P., Haniu M., Teplow D. B., Kahn S., Louis J. C., Citron M., Vassar R. (2000) A furin-like convertase mediates propeptide cleavage of BACE, the Alzheimer β-secretase. J. Biol. Chem. 275, 37712–37717 [DOI] [PubMed] [Google Scholar]
- 35. Crusio W. E., Schwegler H. (2005) Learning spatial orientation tasks in the radial maze and structural variation in the hippocampus in inbred mice. Behav. Brain Funct. 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Leary D. D., McLaughlin T. (2005) Mechanisms of retinotopic map development. Ephs, ephrins, and spontaneous correlated retinal activity. Prog. Brain Res. 147, 43–65 [DOI] [PubMed] [Google Scholar]
- 37. Galimberti I., Bednarek E., Donato F., Caroni P. (2010) EphA4 signaling in juveniles establishes topographic specificity of structural plasticity in the hippocampus. Neuron 65, 627–642 [DOI] [PubMed] [Google Scholar]
- 38. Xu N. J., Henkemeyer M. (2009) Ephrin-B3 reverse signaling through Grb4 and cytoskeletal regulators mediates axon pruning. Nat. Neurosci. 12, 268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Inoue E., Deguchi-Tawarada M., Togawa A., Matsui C., Arita K., Katahira-Tayama S., Sato T., Yamauchi E., Oda Y., Takai Y. (2009) Synaptic activity prompts γ-secretase-mediated cleavage of EphA4 and dendritic spine formation. J. Cell Biol. 185, 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guaiquil V. H., Swendeman S., Zhou W., Guaiquil P., Weskamp G., Bartsch J. W., Blobel C. P. (2010) ADAM8 is a negative regulator of retinal neovascularization and of the growth of heterotopically injected tumor cells in mice. J. Mol. Med. 88, 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guaiquil V., Swendeman S., Yoshida T., Chavala S., Campochiaro P. A., Blobel C. P. (2009) ADAM9 is involved in pathological retinal neovascularization. Mol. Cell. Biol. 29, 2694–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sisodia S. S. (1992) β-Amyloid precursor protein cleavage by a membrane-bound protease. Proc. Natl. Acad. Sci. U.S.A. 89, 6075–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pasquale E. B. (2004) Eph-ephrin promiscuity is now crystal clear. Nat. Neurosci. 7, 417–418 [DOI] [PubMed] [Google Scholar]
- 44. Davis S., Gale N. W., Aldrich T. H., Maisonpierre P. C., Lhotak V., Pawson T., Goldfarb M., Yancopoulos G. D. (1994) Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science 266, 816–819 [DOI] [PubMed] [Google Scholar]
- 45. Stein E., Lane A. A., Cerretti D. P., Schoecklmann H. O., Schroff A. D., Van Etten R. L., Daniel T. O. (1998) Eph receptors discriminate specific ligand oligomers to determine alternative signaling complexes, attachment, and assembly responses. Genes Dev. 12, 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schmid R. S., Maness P. F. (2008) L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr. Opin. Neurobiol. 18, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dahme M., Bartsch U., Martini R., Anliker B., Schachner M., Mantei N. (1997) Disruption of the mouse L1 gene leads to malformations of the nervous system. Nat. Genet. 17, 346–349 [DOI] [PubMed] [Google Scholar]
- 48. Demyanenko G. P., Tsai A. Y., Maness P. F. (1999) Abnormalities in neuronal process extension, hippocampal development, and the ventricular system of L1 knockout mice. J. Neurosci. 19, 4907–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fransen E., D'Hooge R., Van Camp G., Verhoye M., Sijbers J., Reyniers E., Soriano P., Kamiguchi H., Willemsen R., Koekkoek S. K., De Zeeuw C. I., De Deyn P. P., Van der Linden A., Lemmon V., Kooy R. F., Willems P. J. (1998) L1 knockout mice show dilated ventricles, vermis hypoplasia, and impaired exploration patterns. Hum. Mol. Genet. 7, 999–1009 [DOI] [PubMed] [Google Scholar]
- 50. Naus S., Richter M., Wildeboer D., Moss M., Schachner M., Bartsch J. W. (2004) Ectodomain shedding of the neural recognition molecule CHL1 by the metalloprotease-disintegrin ADAM8 promotes neurite outgrowth and suppresses neuronal cell death. J. Biol. Chem. 279, 16083–16090 [DOI] [PubMed] [Google Scholar]
- 51. Maretzky T., Schulte M., Ludwig A., Rose-John S., Blobel C., Hartmann D., Altevogt P., Saftig P., Reiss K. (2005) L1 is sequentially processed by two differently activated metalloproteases and presenilin/γ-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol. Cell. Biol. 25, 9040–9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yan X. X., Xiong K., Luo X. G., Struble R. G., Clough R. W. (2007) β-Secretase expression in normal and functionally deprived rat olfactory bulbs. Inverse correlation with oxidative metabolic activity. J. Comp. Neurol. 501, 52–69 [DOI] [PubMed] [Google Scholar]
- 53. Schlatter M. C., Buhusi M., Wright A. G., Maness P. F. (2008) CHL1 promotes Sema3A-induced growth cone collapse and neurite elaboration through a motif required for recruitment of ERM proteins to the plasma membrane. J. Neurochem. 104, 731–744 [DOI] [PubMed] [Google Scholar]
- 54. Wright A. G., Demyanenko G. P., Powell A., Schachner M., Enriquez-Barreto L., Tran T. S., Polleux F., Maness P. F. (2007) Close homolog of L1 and neuropilin 1 mediate guidance of thalamocortical axons at the ventral telencephalon. J. Neurosci. 27, 13667–13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Castellani V., Chédotal A., Schachner M., Faivre-Sarrailh C., Rougon G. (2000) Analysis of the L1-deficient mouse phenotype reveals cross-talk between Sema3A and L1 signaling pathways in axonal guidance. Neuron 27, 237–249 [DOI] [PubMed] [Google Scholar]
- 56. Castellani V., De Angelis E., Kenwrick S., Rougon G. (2002) Cis and trans interactions of L1 with neuropilin-1 control axonal responses to semaphorin 3A. EMBO J. 21, 6348–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McConlogue L., Buttini M., Anderson J. P., Brigham E. F., Chen K. S., Freedman S. B., Games D., Johnson-Wood K., Lee M., Zeller M., Liu W., Motter R., Sinha S. (2007) Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP transgenic mice. J. Biol. Chem. 282, 26326–26334 [DOI] [PubMed] [Google Scholar]