Background: NAIP5 and NLRC4 induce an innate immune response to intracellular flagellin.
Results: Flagellin fragments were identified that induce signaling-competent NAIP5-NLRC4 inflammasomes with 11- and 12-fold symmetry.
Conclusion: Conserved flagellin terminal regions induce an inflammasome in which NAIP5 and NLRC4 appear to occupy equivalent positions.
Significance: We provide fundamental insights into the formation and structure of hetero-oligomeric inflammasomes.
Keywords: Electron Microscopy (EM), Immunology, Ligand-binding Protein, Nod-like Receptors (NLR), Protein Complexes, Protein Purification, Protein-Protein Interactions, Receptor Structure-Function, Structural Biology, Inflammasomes
Abstract
The NOD-like receptors NAIP5 and NLRC4 play an essential role in the innate immune response to the bacterial tail protein flagellin. Upon flagellin detection, NAIP5 and NLRC4 form a hetero-oligomeric inflammasome that induces caspase-1-dependent cell death. So far, both the mechanism of formation of the NAIP5-NLRC4 inflammasome and its structure are poorly understood. In this study we combine inflammasome reconstitution in HEK293 cells, purification of inflammasome components, and negative stain electron microscopy to address these issues. We find that a Salmonella typhimurium flagellin fragment comprising the D0 domain and the neighboring spoke region is able to co-precipitate NAIP5 and induce formation of the NAIP5-NLRC4 inflammasome. Comparison with smaller fragments indicates that flagellin recognition is mediated by its C-terminal residues as well as the spoke region. We reconstitute the inflammasome from purified flagellin, NAIP5, and NLRC4, thus proving that no other cellular components are required for its formation. Electron micrographs of the purified inflammasome provide unprecedented insight into its architecture, revealing disk-like complexes consisting of 11 or 12 protomers in which NAIP5 and NLRC4 appear to occupy equivalent positions. On the basis of our data, we propose a model for inflammasome formation wherein direct interaction of flagellin with a single NAIP5 induces the recruitment and progressive incorporation of NLRC4, resulting in the formation of a hetero-oligomeric inflammasome.
Introduction
The NOD-like receptor (NLR)2 family comprises cytosolic receptors of the innate immune system that respond to a wide variety of pathogen and danger associated molecular patterns. The NLR family members NLRC4 (also known as IPAF) and NAIP5 play important roles in the control of Legionella and Salmonella infections in mice by sensing the bacterial tail protein flagellin (1–6).
NLRs are characterized by a highly conserved nucleotide binding NACHT domain. The NLRs share a C-terminal leucine-rich repeat domain, which is thought to be involved in ligand recognition and retaining the NLR in an inactive state. At their N terminus, NLRs possess one or more copies of an effector domain, the identity of which varies between family members; NLRC4 contains an N-terminal caspase activating and recruitment domain (CARD) that interacts with the CARD domain of pro-caspase-1 (7–9), whereas NAIP5 contains three N-terminal baculovirus inhibitor of apoptosis protein repeat (BIR) domains.
Upon ligand recognition, several NLRs form high molecular weight complexes, known as inflammasomes, that activate pro-caspase-1 (10–12). Activated caspase-1 mediates the proteolytic processing of pro-interleukins pro-IL-1β and pro-IL-18 (10, 13) and induces an inflammatory cell death known as pyroptosis (14). Several purified NLR NACHT domains were shown to bind nucleotides (15–19), but the exact role of ATP binding and whether hydrolysis takes place is unclear. In some studies, mutation of the conserved Walker A lysine that coordinates the nucleotide γ-phosphate reduces spontaneous and ligand-induced self-association and downstream signaling (6, 16–18, 20), whereas others reported no significant effects (6).
Information on inflammasome structure so far derives from negative stain electron microscopy on NLRP1, which revealed pentameric and heptameric ring-like structures (21). Likely, the architecture of inflammasomes resembles that of apoptosomes formed by Apaf-1, CED4, and Dark, which, like NLRs, contain a NACHT domain essential for oligomerization. For apoptosomes, higher resolution data are available that show heptameric or octameric rings that are stabilized via homotypic interactions between the NACHT domains (22–26).
Recently, it has been shown that ligand specificity of the NLRC4 inflammasome is determined by members of the NAIP family (5, 6). In mice, NLRC4 and NAIP2 form an inflammasome in response to PrgJ, a component of the bacterial type III secretion system, whereas NLRC4 together with either NAIP5 or NAIP6 recognizes flagellin. In humans only one NAIP variant exists, which forms a complex with NLRC4 upon detection of CprI, a PrgJ homologue. NAIP5-NLRC4-dependent cell death can be induced by a 35-residue C-terminal region of flagellin and was suggested to depend on three conserved leucine residues near the C terminus (4, 5).
In this study we show that in addition to the flagellin C-terminal 35 residues, other elements in its N- and C-terminal regions contribute to inflammasome formation. We reconstitute the inflammasome from its purified components and find by electron microscopy that NAIP5-NLRC4 complexes contain 11 or 12 subunits and therefore are considerably larger than was expected based on homologous structures. We propose a model for the formation of the NAIP5-NLRC4 inflammasome in which flagellin binding to NAIP5 leads to the recruitment of multiple copies of NLRC4, resulting in the formation of a disk-shaped hetero-oligomeric complex in which both NLRs occupy equivalent positions.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293E cells were cultured in Freestyle medium (Invitrogen), containing 0.2% FCS, 50 μg/ml G418 disulfate at 37 °C in a 5% CO2 humidified atmosphere. The cells were grown in suspension at 120 rpm. Small scale (4 ml) and large scale (1L) transfections were performed according to Durocher et al. (27). When appropriate, cell viabilities were measured 48 h post-transfection using a Casy Model TTC cell counter (Schärfe Systems, Reutlingen, Germany).
Plasmids
cDNA was purchased from Invivogen (human Nlrc4), Imagenes (murine Naip5 and murine Casp1), and OriGene (human Naip). Murine spleen cDNA, used for the isolation of Nlrc4, was a kind gift from G. Folkerts (Department of Pharmacology and Pathophysiology, Utrecht University, Utrecht, The Netherlands). DNA encoding Salmonella typhimurium and Salmonella enteritidis flagellin (FliC) were a kind gift from J. P. M. van Putten (Faculty of Veterinary Medicine, Department of Infection and Immunity, Utrecht University).
Coding sequences were amplified by PCR using forward primers that introduce a BamHI restriction site while omitting the start codon and reverse primers that introduce a NotI restriction site and omit the stop codon, unless indicated otherwise. At the protein level this procedure results in the introduction of a Gly-Ser sequence at the N terminus and three Ala residues at the C terminus. Domain boundaries of S. typhimurium flagellin constructs are indicated in Fig. 2A. The F41 fragment of S. enteritidis comprises amino acids Arg-53 to Arg-461. For the FliC-C construct a reverse primer was used that preserves the stop codon, and therefore no triple Ala sequence is present at its C terminus. For the FliC-D0L and FliC-D0S fragments, synthetic DNA (DNA2.0) was used that also included a stop codon preceding the NotI site. PCR products were subcloned into pCR-TOPO vector (Invitrogen).
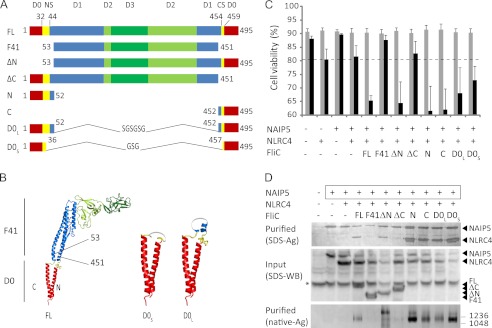
FIGURE 2.
Flagellin fragments contribute differently to caspase-1-dependent cell death and inflammasome formation. A, schematic representation of the flagellin constructs used in this study. The numbers indicate the inclusive amino acid boundaries of the various constructs or of the N-terminal spoke (NS) and C-terminal spoke (CS). For the F41 fragment, domain boundaries were taken from Samatey et al. (34); boundaries for D0 and the spokes were taken from Yonekura et al. (33). B, cartoon representation of the flagellin EM structure (Protein Data Bank code 1UCU). The numbers indicate the boundaries of the F41 fragment. The domains are colored as in A. The Gly-Ser repeat loops that connect the N- and C-terminal halves of D0L and D0S are depicted in gray. C, viabilities of HEK293E cells 48 h after transfection with the indicated proteins. For further explanation, see Fig. 1A. D, analysis of inflammasome formation in HEK293E cells transfected with the proteins indicated. The boxed proteins carry the His6-StrepII3 purification handle. Other proteins were expressed with a His6-FLAG3-TEV tag. Purified protein was analyzed by SDS-PAGE (top panel) or native PAGE (3–12%; bottom panel) followed by silver staining. For the native gel, the numbers on the right indicate the molecular masses of the markers in kDa. The cleared cell lysate was analyzed on Western blot probed with anti-His tag antibody (middle panel). The asterisk denotes a protein that interacts nonspecifically with the antibody.
Point mutations were created using the QuikChange method (Stratagene). The FliC 3LA mutants (L491A, L493A, and L494A) contain a stop codon preceding the NotI site.
The BamHI/NotI DNA fragments were subcloned into pUPE vectors (U-Protein Express BV, Utrecht, The Netherlands) that contain an expression cassette under control of a CMV promoter. Expression vectors encoded a C-terminal TEV-StrepII3-His6, FLAG3-His6, or StrepII3-His6 tag or an N-terminal His6-FLAG3 or His6-BAP-TEV tag. The biotin acceptor peptide (BAP) tag comprises amino acids GLNDIFEAQKIEWHE. A vector encoding the Escherichia coli BirA enzyme was a kind gift from H. T. M. Timmers (Faculty of Medicine, Utrecht University). Murine Casp1 was expressed without tags.
Protein Purification
Native flagellin of S. enteritidis was purified as described previously (28). For NLR purification from small scale (4 ml) HEK293E cultures, cells were harvested 48 h post-transfection by centrifugation at 600 × g for 5 min. Cell pellets were stored for 1 h up to 1 week at −20 °C. For purification, pellets were thawed at room temperature. Cytoplasmic extracts were obtained using a method described by Tsai and Carstens (29), with omission of the PBS washing step. Cell pellets were resuspended in 320 μl of cold small scale lysis buffer containing 10 mm HEPES, pH 7.6, 5 mm MgCl2, 10 mm KCl, 5 mm DTT, 1 μg/ml DNase, and 1 tablet Complete mini, EDTA-free tablets (Roche Applied Science)/20 ml of buffer. StrepII-tagged proteins were purified from the cleared cytoplasmic extracts using StrepTactin-Sepharose beads (GE Healthcare). After incubation for 1–2 h at 4 °C, the beads were washed in StrepTactin wash buffer (100 mm NaCl, 25 mm HEPES, pH 7.5, 5 mm benzamidin, 5% glycerol, 2 mm DTT), and subsequently protein was eluted in wash buffer supplemented with 5 mm d-desthiobiotin (Sigma). In the case of biotinylated proteins, washed beads were boiled for 10 min in SDS sample buffer supplemented with 3 mm d-desthiobiotin.
For large scale purification of NAIP5-TEV-StrepII3-His6 and NLRC4-StrepII3-His6, cells were harvested 72–96 h post-transfection by centrifugation for 20 min at 500 × g. Cell pellets were resuspended in 1/10 culture volume of cold large scale lysis buffer (100 mm NaCl, 50 mm HEPES, pH 7.5, 5 mm benzamidin, 5% glycerol, 1 tablet Complete mini, EDTA-free tablets (Roche Applied Science)/20 ml of lysis buffer, 5 mm DTT, 5 mm MgCl2, 1 μg/ml DNase, 0.3% Nonidet P-40) and subjected to two freeze/thaw cycles in liquid nitrogen before storage at −80 °C until further purification. Frozen lysates were thawed at room temperature, and cell debris was removed by centrifugation at 60,000 × g for 20 min. Concentrated NaCl and imidazole solutions were added to the cleared supernatant to final concentrations of 300 and 10 mm, respectively. Nickel-Sepharose 6 Fast Flow beads (GE Healthcare) were added to the cleared lysate, incubated for 1 h at 4 °C, and poured into a Tricorn column (GE Healthcare). The column was washed with immobilized metal ion affinity chromatography wash buffer (300 mm NaCl, 50 mm HEPES, pH 7.5, 5 mm benzamidin, 5% glycerol, 5 mm DTT) containing 20 mm imidazole. Protein was eluted in wash buffer containing 250 mm imidazole. Pooled fractions mixed with an equal volume of 50 mm HEPES, pH 7.6, 5% glycerol, 5 mm benzamidin, and 2 mm DTT were incubated with StrepTactin-Sepharose beads (GE Healthcare) for 1 h. After extensive washing of the beads with StrepTactin wash buffer as described for small scale purifications, protein was eluted in StrepTactin wash buffer supplemented with 5 mm d-desthiobiotin and stored in 25–50-μl portions at −80 °C.
Gel Electrophoresis and Protein Detection
Reduced protein samples were separated on standard Laemmli 10% SDS-PAGE gels. Samples for native gel electrophoresis were run on 3–12% native PAGE Novex Bis-Tris gels (Invitrogen) according to the manufacturers protocol, using NativeMark Unstained Protein Standard (Invitrogen) as marker. Purified hNLRC4 was separated on 4–15% PhastGel with native buffer strips (GE Healthcare) according to the manufacturers protocol. The gels were silver-stained or transferred to PVDF membrane (Bio-Rad). Proteins were detected on Western blot using either a mixture of mouse anti-polyhistidine (Sigma) with mouse anti-pentahistidine (Qiagen) or mouse anti-FLAG tag (Sigma) as primary antibodies and rabbit anti-mouse-HRP (Dako) as the secondary antibody. The signal was detected using ECL (GE Healthcare).
In Vitro Inflammasome Reconstitution
Microtubes were precoated with 1 mg/ml BSA for 30 min at room temperature, washed with incubation buffer (150 mm NaCl, 50 mm HEPES, pH 7.6, 5% glycerol, 5 mm benzamidin, 2 mm DTT), and dried. Next, 100 ng of mNAIP5, 100 ng of hNLRC4, and 400 ng of S. typhimurium FliC were mixed in a total volume of 4 μl of incubation buffer supplemented with either 0.5 mm ATP and 4 mm MgCl2 or 4 mm EDTA. The mixtures were incubated overnight at 4 °C and then separated on 3–12% native gel (Invitrogen) according to the manufacturer's protocol.
Protein Purification and Sample Preparation for Electron Microscopy
For purification of the inflammasome, HEK293E cells were transfected with Naip5-TEV-StrepII3-His6, Nlrc4-FLAG3-His6, and tagless FliC-D0L. The cells were harvested 72–96 h post-transfection by spinning at 700 × g for 10 min. The pellets were stored at −80 °C until further purification. Purification was performed as described for small scale purifications, scaled up according to the culture volume.
For EM grid preparation NLRC4, NAIP5, and freshly purified inflammasome diluted to concentrations of 5–25 μg/ml were applied on 400 mesh copper grids with continuous carbon support film (Aurion, Wageningen, The Netherlands) and stained with freshly prepared 0.75% uranyl formate solution. For the polymerized NLRC4 sample, 100 mesh copper grids with home-made Formvar/carbon support film and 2% uranyl acetate stain were used.
EM Data Collection and Processing
Human NLRC4, murine NAIP5, and polymerized NLRC4 were imaged using a Tecnai BioTWIN12 TEM with a LaB6 electron source operated at 120 kV and an Eagle 4k CCD camera (FEI Company, The Netherlands). Imaging of the freshly purified inflammasome was performed at 200 kV on a Tecnai F20 TEM, equipped with a field emission gun and a USC4000 CCD camera (Gatan, München, Germany). Binned two-dimensional images were acquired at −1.5-μm defocus and 50,000× magnification resulting in a pixel size of 0.32 nm at specimen level.
Tomograms of polymerized human NLRC4 and freshly purified inflammasome were taken in low dose mode at −4-μm or −2-μm defocus and magnifications of 30,000× (binned by 2) or 50,000× (unbinned), resulting in pixel sizes of 0.77 and 0.22 nm at specimen level. For both specimens, the tilt schemes with a constant increment of 2° in ranges of +−60° and +−50°, respectively, were used.
Tomograms were reconstructed in IMOD version 4.2.15 (30) using patch tracking for image alignment and the SIRT reconstruction algorithm. In the case of polymerized NLRC4, the contrast transfer function was corrected by phase flipping after defocus estimation using TOMOCTF (31).
Symmetry analysis was performed on tomographic subvolumes that showed inflammasomes close to top view. To this end, ∼10-nm-thick virtual sections of the volumes were reprojected along the putative symmetry axis, and rotational autocorrelation was performed after circular masking. Subsequent iterative refinement of particle center and symmetry axis orientation was guided by the maximization of peaks in the autocorrelation function. Finally, averaged reprojections of individual particles were calculated according to symmetry classification.
RESULTS
Full-length Flagellin, but Not Its F41 Fragment, Induces Inflammasome Formation
To study flagellin-induced NAIP5-NLRC4 inflammasome formation, we reconstituted the inflammasome by transient transfection of its components in HEK293E cells, which do not endogenously express Naip5, Nlrc4, and caspase-1. Expression of caspase-1 alone does not lead to a significant reduction in cell viability, showing that caspase-1 autoactivation is negligible (Fig. 1A). Likewise, in the absence of caspase-1, expression of Naip5 or Nlrc4 does not cause a decrease in cell viability. As described previously (6), we noted some loss of cell viability upon co-expression of Nlrc4 with caspase-1. This effect depends on the Nlrc4 expression level and is diminished when less Nlrc4 DNA is transfected (data not shown). Co-expression with flagellin likewise reduces Nlrc4 expression to a level at which no effect on cell viability is detected (Fig. 1A and supplemental Fig. S1), and therefore NLRC4 autoactivation does not affect the measurements of flagellin-induced activation in our reconstituted system.
FIGURE 1.
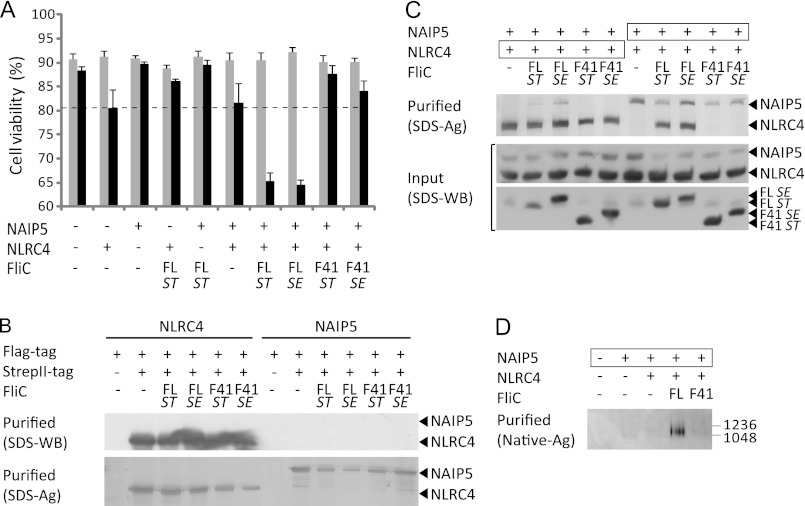
NLRC4 and NAIP5 form a caspase-1-activating complex in the presence of full-length but not F41 Salmonella flagellin. A, viabilities of HEK293E cells 48 h after transfection with the indicated proteins. Flagellin (FL) or its F41 fragment originate from the flagellin gene (FliC) of S. typhimurium (ST) or S. enteritidis (SE). The gray and black bars represent transfections without and with caspase-1, respectively. The average values and standard deviations were derived from at least three independent transfections. Expression of all proteins was verified by Western blot (not shown). The dashed line represents nonspecific cell death during NLRC4-caspase-1 co-expression in the absence of ligand as explained in the text. B, analysis of NAIP5 and NLRC4 homo-oligomerization in HEK293E cells transfected with flagellin and differentially tagged mNAIP5 or mNLRC4 as indicated. Proteins isolated by StrepII tag purification were analyzed on Western blot probed with anti-FLAG tag antibody (top panel) or analyzed by SDS-PAGE followed by silver staining (bottom panel). C, analysis of inflammasome formation in HEK293E cells transfected with the proteins indicated. The boxed proteins carry the His6-StrepII3 purification handle. Other proteins were expressed with a His6-FLAG3-TEV tag. Purified protein was analyzed by SDS-PAGE followed by silver staining (top panel). The cleared cell lysate was analyzed on Western blot probed with anti-His tag antibody (middle and bottom panel). D, native PAGE analysis of proteins purified as described for C on silver-stained native PAGE (3–12%). The numbers on the right indicate the molecular masses of the markers in kDa.
The reconstituted system was used to study inflammasome activation by flagellin from S. typhimurium and Salmonella enterica. In addition to full-length flagellin, we tested the flagellin F41 fragment that lacks the N- and C-terminal helices that are essential for flagellin filament assembly (32, 33). All transfections were performed with and without caspase-1, and the difference in cell viability, determined 48 h post-transfection, was taken as a measure for caspase-1-induced cell death (Fig. 1A). No significant increase in cell death is observed when flagellin is co-expressed with either Naip5 or Nlrc4. However, co-expression of Salmonella flagellin with Naip5 plus Nlrc4 leads to a 20–25% reduction of cell viability, indicating formation of a functional inflammasome. Notably, co-expression of the flagellin F41 fragment with Nlrc4 and Naip5 does not induce caspase-1-dependent cell death, suggesting that the F41 fragment lacks a region that is essential for NAIP5-NLRC4 inflammasome formation.
To analyze the formation and composition of flagellin-induced complexes, we purified the inflammasome from cell lysates using streptavidin affinity purification. We selected StrepII tag-based purification, which involves a mild elution procedure, to minimize inflammasome dissociation. To this end we fused either NLRC4 or NAIP5 to a C-terminal His6-StrepII3 tag, whereas the other NLR and flagellin were fused to a His6-FLAG3 tag. Self-association of NLRs was analyzed by co-transfection of two variants carrying either a His6-StrepII3 tag or a His6-FLAG3 tag. We find that NLRC4 self-associates in the absence of ligand (Fig. 1B), consistent with the mild increase in caspase-1-dependent cell death associated with Nlrc4 expression in the viability assay (Fig. 1A). For NAIP5 we do not observe self-association even in the presence of flagellin (Fig. 1B). NAIP5 and NLRC4, when co-expressed in the absence of ligand, do not co-purify (Fig. 1C). In the presence of flagellin, NAIP5 and NLRC4 do co-purify, whereas co-expression with the F41 fragment does not induce complex formation. Analysis of the purified inflammasome by silver-stained native PAGE shows that flagellin, unlike the F41 fragment, induces formation of a large complex that runs at about the same height as the 1.0- and 1.2-MDa marker proteins (i.e., IgM pentamer and hexamer respectively; Fig. 1D). NAIP5-NLRC4 complex formation is observed independent of which protein harbors the StrepII3 tag (Fig. 1C). No other proteins were seen to co-purify in amounts detectable by silver-stained SDS-PAGE, suggesting that the inflammasome complex consists of NAIP5 and NLRC4 only. Flagellin was also not detectable on silver-stained gels but could be detected on Western blot (not shown), which indicates that flagellin is not present in the inflammasome complex in stoichiometric amounts.
Thus we show that flagellin, but not its F41 fragment, induces binding between NAIP5 and NLRC4, resulting in the formation of a high molecular weight complex. The formation of this NAIP5-NLRC4 complex is correlated with the ability to activate caspase-1, resulting in cell death. We conclude that the part of flagellin that is missing in the F41 fragment contains one or more determinants that are essential for NAIP5-NLRC4 inflammasome formation.
Elements within Flagellin Contributing to Inflammasome Formation
Flagellin essentially consists of four distinct domains: D0, D1, D2, and D3 (Fig. 2A) (33, 34). The D0 and D1 domains, which form the flagellar inner and outer tube respectively, are connected by the so-called spokes. The F41 fragment lacks the complete D0 domain, the spokes, and a small part of the D1 domain. Because this region is clearly indispensible for NAIP5-NLRC4 inflammasome formation, we wondered which element(s) within this region are involved. We designed two constructs comprising the D0 domain of S. typhimurium flagellin. The longer construct, D0L, is exactly complementary to the F41 fragment. The design of the shorter construct, D0S, was based on the EM structure of flagellin (33) and comprises the D0 domain plus a small part of the spokes (Fig. 2, A and B). In both D0L and D0S, a Gly-Ser repeat loop connects the N- and C-terminal halves of the construct. In addition, we made deletion constructs of S. typhimurium flagellin lacking either the N-terminal half of D0L (FliC-ΔN) or its C-terminal half (FliC-ΔC), as well as constructs comprising the isolated N-terminal half of D0L (FliC-N) or its C-terminal half (FliC-C) (Fig. 2A). For these constructs, domain boundaries are complementary to the F41 fragment.
When co-expressed with Nlrc4 and Naip5, the D0L construct as well as the shorter D0S construct induce caspase-1-dependent cell death (Fig. 2C). The same is true for the N-terminal deletion mutant FliC-ΔN. The deletion of the C-terminal region in FliC-ΔC, however, abrogates caspase-1-induced cell death. Whereas this finding suggests that the main determinant for caspase-1 activation is located in the C terminus, co-expressions with the isolated N- and C-terminal flagellin fragments show that these induce caspase-1-dependent cell death equally effectively. The effect of the N- and C-terminal regions therefore appears to be context-dependent. All of the flagellin fragments that induced caspase-1-dependent cell death also induce NAIP5-NLRC4 complex formation, as evidenced by co-purification of NLRC4 with StrepII-tagged NAIP5 (Fig. 2D, top panel). The flagellin fragments do not co-purify with the complex in stoichiometric amounts, as we also observed for full-length flagellin, suggesting that their interaction with the complex is relatively weak. As expected, the FliC-ΔC fragment that failed to induce cell death also failed to induce complex formation. When analyzed by native PAGE, the purified complexes give rise to at least two bands of high molecular weight, indicating heterogeneity in the complexes formed. The relative abundance of these bands varies between individual purifications and appears to be independent of the flagellin fragment employed (not shown).
Taken together, we show that all elements required for NAIP5-NLRC4 inflammasome formation are located within the flagellin D0L fragment and that, although their effect appears to be context-dependent, both the N-terminal and the C-terminal half of D0L are capable of inducing inflammasome formation and the associated caspase-1-induced cell death.
Contribution of Flagellin C-terminal Leucines to Inflammasome Formation
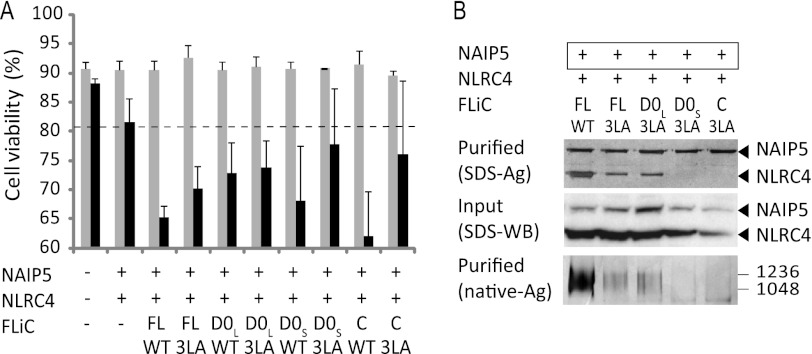
Previously, it was shown that mutation of three conserved leucines near the C terminus of flagellin severely reduces its capacity to induce NAIP5-NLRC4 inflammasome formation (4, 5). Our observation that FliC-N also induces inflammasome formation indicates, however, that other segments in addition to the C terminus are involved. To investigate the relative importance of the conserved leucine residues, we mutated them to alanine (3LA) in the various flagellin constructs.
Flagellin-3LA and D0L-3LA still induce a caspase-1-dependent decrease in cell viability, as well as formation of a NAIP5-NLRC4 complex, although the amount of complex detected was less than observed for wild type flagellin (Fig. 3). In contrast, the capacity of the shorter constructs D0S-3LA and especially FliC-C-3LA to decrease cell viability appears reduced, and these constructs are no longer able to induce formation of detectable amounts of the NAIP5-NLRC4 complex. Thus, whereas our data confirm the importance of the three leucine residues near the flagellin C terminus, their role appears less prominent for larger fragments. The different responses to D0L-3LA versus D0S-3LA suggests that the spoke region, which is mostly missing in D0S, also contributes to NAIP5-NLRC4 complex formation.
FIGURE 3.
Contribution of flagellin C-terminal leucines to inflammasome formation. A, viabilities of HEK293E cells 48 h after transfection with the indicated proteins. For further explanation, see Fig. 1A. B, analysis of inflammasome formation in HEK293E cells transfected with the proteins indicated. The boxed proteins carry the His6-StrepII3 purification handle. Other proteins were expressed with a His6-FLAG3-TEV tag. Purified protein was analyzed by SDS-PAGE (top panel) or native PAGE (3–12%; bottom panel) followed by silver staining. For the native gel, the numbers on the right indicate the molecular masses of the markers in kDa. Cleared cell lysate was analyzed on Western blot probed with anti-His tag antibody (middle panel).
Association of Flagellin Fragments with the NAIP5-NLRC4 Inflammasome and Its Components
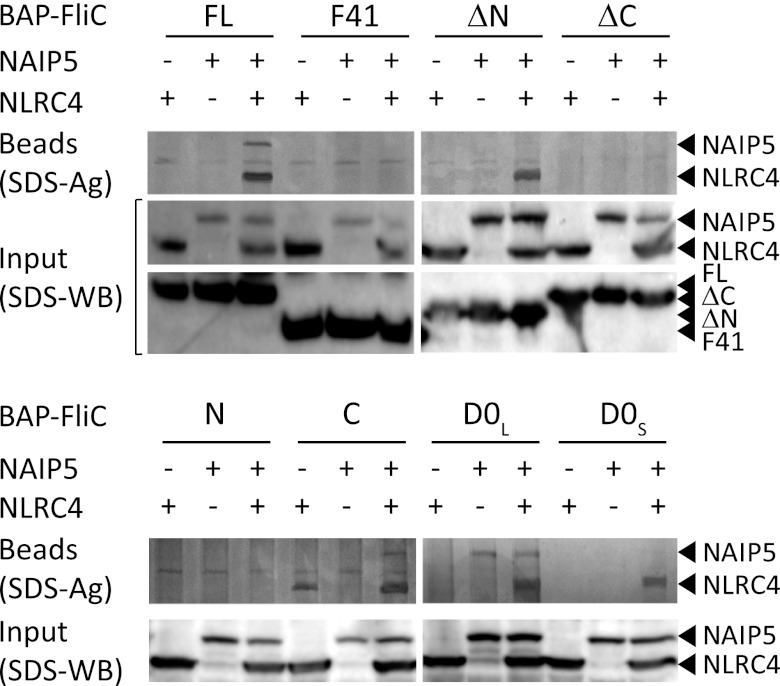
Flagellin induces formation of a high molecular weight NAIP5-NLRC4 complex. However, our inability to detect flagellin in purified samples of the complex on silver-stained SDS-PAGE gels may indicate that the flagellin-inflammasome interaction is rather weak. Because interactions between full-length flagellin and the inflammasome components have been observed (5, 6), we decided to use an alternative approach in which flagellin and its fragments were fused to a 15-residue BAP that is biotinylated in vivo by co-expression of the E. coli biotin ligase BirA (35). The BAP-tagged flagellin constructs were co-expressed with His6-FLAG3-tagged Naip5 and/or Nlrc4 and precipitated using StrepTactin-Sepharose beads. In this setup, biotinylated flagellin precipitated a NAIP5-NLRC4 complex that was clearly visible on silver-stained SDS-PAGE gels (Fig. 4), showing that direct binding of flagellin to the NAIP5-NLRC4 complex does occur.
FIGURE 4.
Association of NLRC4 and NAIP5 with biotinylated flagellin fragments. HEK293E cells were transfected with His6-FLAG3-TEV tagged NAIP5 and NLRC4 and BAP-tagged flagellin fragments as indicated. The BAP-tagged flagellin fragments were biotinylated in vivo by co-expressing biotin ligase from E. coli. StrepTactin beads were used for pulldown. Proteins bound to the beads were visualized by SDS-PAGE followed by silver staining (top panel). Cleared cell lysate was analyzed on Western blot probed with anti-His tag antibody (middle and bottom panel).
Biotinylated versions of the flagellin fragments that induced inflammasome formation and caspase-1-dependent cell death also co-precipitate the NAIP5-NLRC4 complex, except for biotinylated FliC-N. Possibly the interaction between the N terminus and the complex, even in this setup, is too weak to withstand the isolation procedure.
Binding of biotinylated flagellin to the individual NLRs was not detected (Fig. 4). Remarkably, however, the D0L fragment interacted with NAIP5 even in the absence of NLRC4 as evidenced by the co-precipitation of significant quantities of NAIP5. None of the other flagellin fragments was able to precipitate NAIP5 in detectable amounts. The C-terminal fragment FliC-C was unique in its ability to precipitate NLRC4 albeit in relatively small amounts and not consistently so in all experiments.
Thus, D0L shows relatively strong binding to NAIP5, whereas the C terminus of flagellin shows a weaker interaction with NLRC4. Most other inflammasome-inducing fragments did not precipitate the individual NLRs; they were, however, able to precipitate the NAIP5-NLRC4 complex. Higher affinity for the complex possibly results from a conformational change in one or both of the NLRs upon inflammasome formation. Alternatively, flagellin could be stabilized in the complex by binding at the interface of NAIP5 and NLRC4.
In Vitro Reconstitution of the Inflammasome
To show unequivocally that apart from NAIP5 and NLRC4 no other cellular factors are required for flagellin-induced inflammasome formation, we set out to reconstitute the inflammasome in vitro from its purified components. Flagellin was purified from S. enteritidis according to established protocols (28) (Fig. 5A). NLR purification proved challenging because insoluble aggregates tend to form upon overexpression. Production of soluble NAIP5 and NLRC4 was tested in several expression hosts using different combinations of N- and C-terminal purification tags. We succeeded in purifying small quantities of murine NAIP5 (mNAIP5) from transiently transfected HEK293E cells using tandem affinity purification on nickel-Sepharose and StrepTactin-Sepharose columns. One liter of HEK293E culture yielded 5–10 μg of pure mNAIP5 (Fig. 5A). Unfortunately, we did not succeed in purifying murine NLRC4 (mNLRC4); we did, however, obtain relatively large quantities (up to 150 μg/liter of culture) of human NLRC4 (hNLRC4) using the same expression and purification strategy as used for mNAIP5. Purified hNLRC4 is monomeric at low concentrations, but when concentrated above ∼1 mg/ml the protein forms oligomers as indicated by the formation of a ladder pattern on native gel (Fig. 5B).
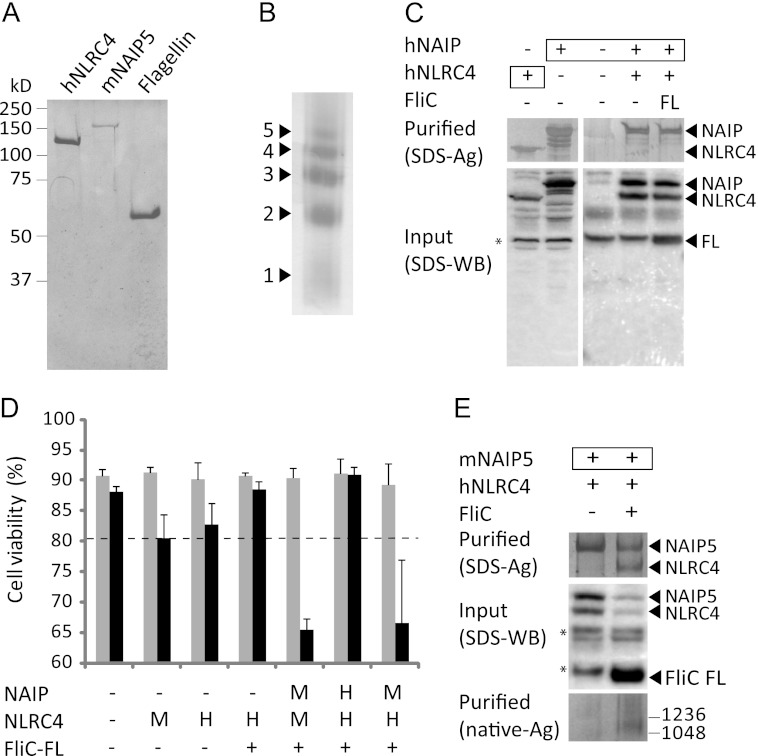
FIGURE 5.
Murine NAIP5 and human NLRC4 form an inflammasome in response to flagellin. A, SDS-PAGE analysis of purified proteins visualized by Coomassie staining. B, analysis of the spontaneous multimerization of concentrated (>1 mg/ml) solutions of purified hNLRC4 on native PAGE (4–15%). The protein was visualized by silver staining. The numbers indicate the probable number of monomers present in each multimer. C, inability of hNAIP and hNLRC4 to form a flagellin-induced inflammasome in HEK293E cells. Boxed proteins carry the His6-StrepII3 purification handle; other proteins were expressed with a His6-FLAG3-TEV tag. Purified fractions were analyzed by SDS-PAGE followed by silver staining (top panel). The cleared cell lysate was analyzed on Western blot probed with His tag antibody (bottom panel). The asterisk denotes a protein that interacts nonspecifically with the antibody. D, viabilities of HEK293E cells 48 h after transfection with the indicated proteins. The letters H and M indicate whether the human or mouse protein was expressed. For further explanation, see Fig. 1A. E, analysis of inflammasome formation in HEK293E cells transfected with mNaip5 and hNlrc4 in the presence and absence of flagellin. mNAIP5 carried the His6-StrepII3 purification handle. Other proteins were expressed with a His6-FLAG3-TEV tag. Purified protein was analyzed by SDS-PAGE (top panel) or native PAGE (3–12%; bottom panel) followed by silver staining. For the native gel, the numbers on the right indicate the molecular masses of the markers in kDa. The cleared cell lysate was analyzed on Western blot probed with anti-His tag antibody (middle panel). The asterisks denote proteins that interact nonspecifically with the antibody.
Our ability to purify hNLRC4 but not mNLRC4 posed a problem for reconstitution of a flagellin-responsive inflammasome. hNLRC4 and the sole human NAIP (hNAIP) do not form a complex in the presence of flagellin, nor do they cause a flagellin induced caspase-1-dependent decrease in cell-viability (Fig. 5, C and D). Instead, a hNAIP-hNLRC4 inflammasome is formed in response to the Type III secretion system needle protein CprI (5). Thus, a flagellin-responsive inflammasome cannot be reconstituted from hNAIP and hNLRC4. We therefore assessed whether hNLRC4, which shares 75% sequence identity with mNLRC4, can substitute its murine orthologue in flagellin-induced inflammasome formation. Overexpression of hNlrc4, like mNlrc4, causes a mild caspase-1-dependent decrease of cell viability, which is reduced when flagellin is co-expressed (Fig. 5D). Upon co-expression of hNlrc4 and mNaip5 with flagellin, we observed a clear decrease in cell viability similar to what was observed for mNLRC4 plus mNAIP5 and flagellin. Furthermore, hNLRC4 co-purifies with mNAIP5 in the presence of flagellin, and this mNAIP5-hNLRC4 complex runs in native PAGE at a molecular weight comparable with the mNAIP5-mNLRC4 complex (Fig. 5E). Thus, hNLRC4 and mNAIP5 are capable of forming a fully active inflammasome in response to flagellin.
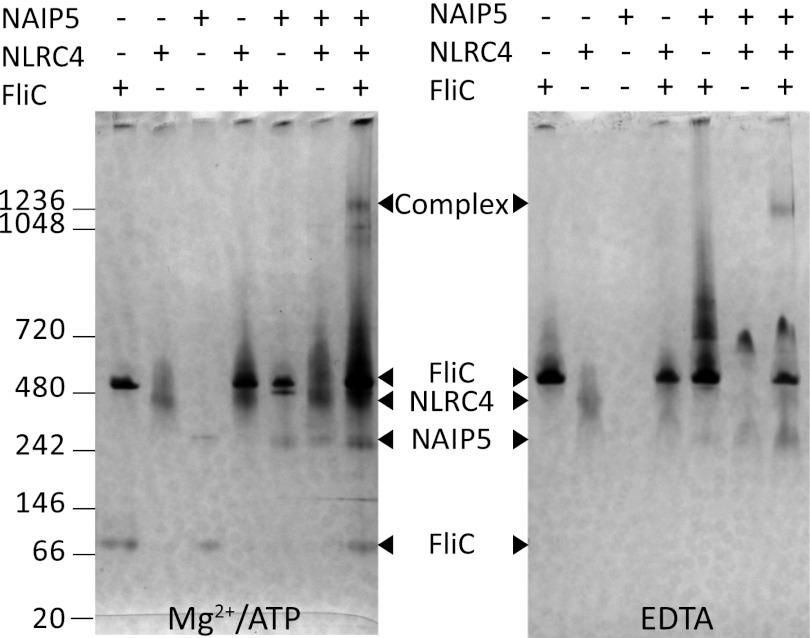
For in vitro reconstitution the purified proteins were mixed in the presence of either Mg2+-ATP or EDTA; complex formation was analyzed by native PAGE (Fig. 6). Incubation of flagellin with either mNAIP5 or hNLRC4 causes no band shift, confirming our previous finding that flagellin does not form a stable complex with NLRC4 or mNAIP5 in vivo (Fig. 4). When all three proteins are present, a high molecular weight complex is formed that runs at a similar apparent molecular weight as the NAIP5-NLRC4 complexes purified from HEK293E cell lysates. Complex formation occurs in the presence of Mg2+-ATP, as well as EDTA. In both cases, in vitro formation of the NAIP5-NLRC4 complex appears rather inefficient because only a small fraction of the proteins is incorporated. Nevertheless, our observations prove that the flagellin-induced inflammasome can be formed by direct interaction of flagellin, NAIP5, and NLRC4 only and that no additional cellular factors are required.
FIGURE 6.
In vitro reconstitution of a flagellin-induced NAIP5-NLRC4 inflammasome. Inflammasome components were mixed and incubated in the presence of Mg2+-ATP (left panel) or EDTA (right panel). The samples were analyzed after 3 and 24 h of incubation by native PAGE (3–12%) followed by silver staining. Because the results at both time points are indistinguishable, only the 24-h samples are shown. The numbers on the left indicate the molecular masses of the markers in kDa.
The Role of ATP Binding in Inflammasome Formation
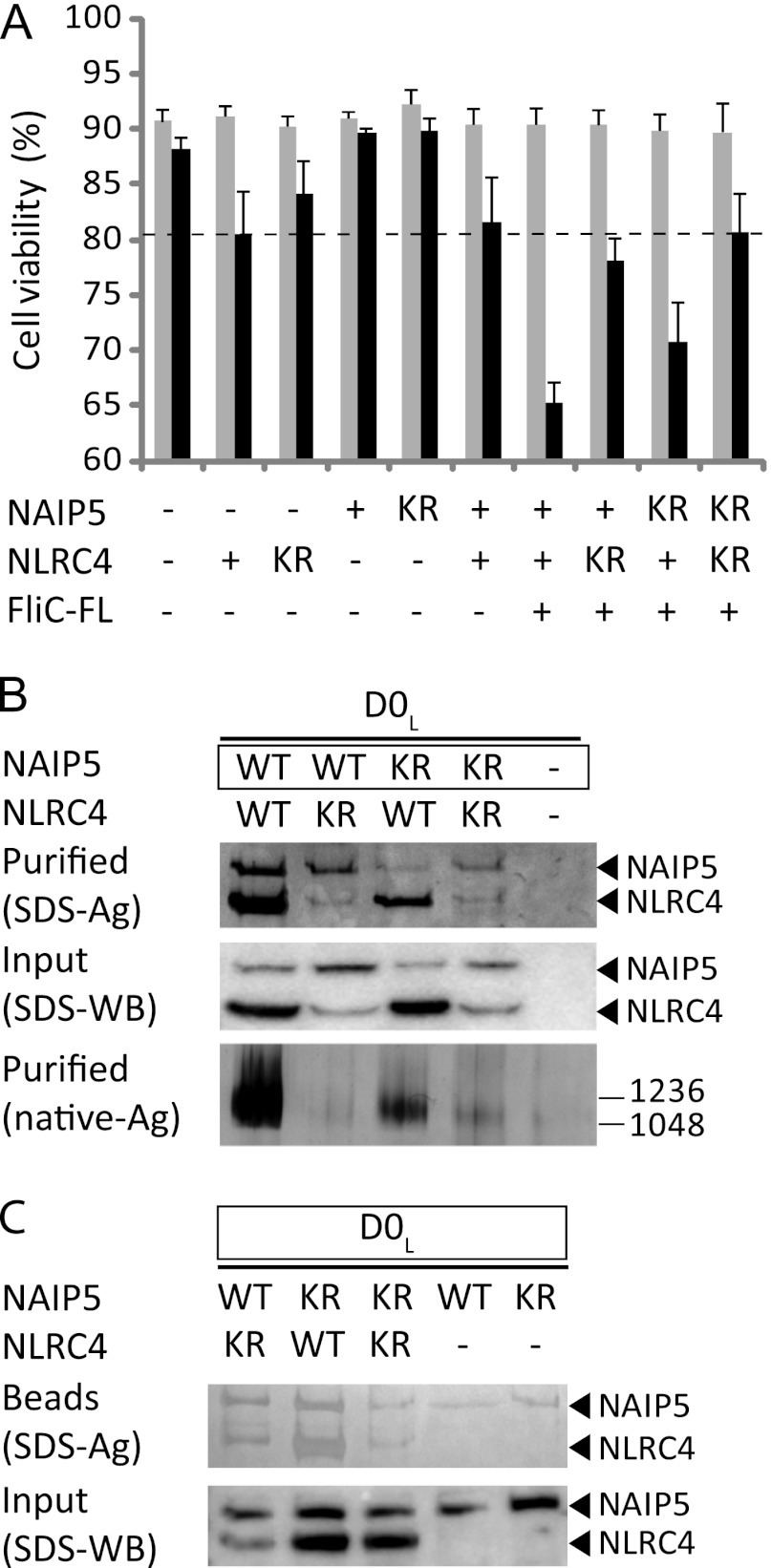
It has been suggested that ATP binding by NLRs is essential for their oligomerization (16–18), as well as for responsiveness to their specific ligand (20). The reconstitution of a NAIP5-NLRC4 inflammasome in the presence of EDTA (Fig. 6, right panel) suggests, however, that ATP binding might not be required for these NLRs. To explore the role of ATP binding further, we mutated the conserved lysine residue in the Walker A motif of both mNAIP5 (Lys-475) and mNLRC4 (Lys-175) to arginine. This mutation has been shown to strongly reduce ATP binding and downstream signaling in NLRs (36, 37) and homologous proteins containing NACHT domains (38–42). The mutants will be referred to as NAIP5-KR and NLRC4-KR, respectively. The ability of the NAIP5 and NLRC4 mutants to assemble into the inflammasome and to induce caspase-1-dependent cell death was analyzed (Fig. 7, A–C). The NAIP5-KR mutant causes a slight, but not significant, reduction in caspase-1-dependent cell death, and the amount of complex formed, albeit reduced, is still substantial (Fig. 7, A and B). In addition, NAIP5-KR, like NAIP5, co-precipitates with biotinylated FliC-D0L (Fig. 7C). Therefore, NAIP5-KR appears fully functional in inflammasome formation and induction of cell death. Analysis of the NLRC4-KR mutant was complicated by the fact that the amount of soluble NLRC4-KR in cell lysates is markedly reduced compared with the wild type protein (Fig. 7B, middle panel), indicating that NLRC4 stability is compromised by the mutation. NLRC4-KR does not induce significant caspase-1-dependent cell death, neither in ligand-independent autoactivation when expressed alone nor when co-expressed with Naip5 and flagellin (Fig. 7A). Whereas these observations suggest that ATP binding contributes to inflammasome activity, the lack of activity could also be caused by the strongly reduced expression level of the NLRC4-KR mutant. Complex formation is not abolished completely: small amounts of NLRC4-KR do co-purify with wild type and mutant NAIP5 in the presence of FliC-D0L (Fig. 7B, top panel), and a high molecular weight complex is just visible on native PAGE (Fig. 7C, bottom panel). In addition, biotinylated FliC-D0L precipitates a NLRC4-KR-NAIP5 complex and even a NLRC4-KR-NAIP5-KR double mutant complex (Fig. 7C). The observed complex formation suggests that nucleotide binding by NLRC4 may not be essential for inflammasome assembly. However, as a consequence of NLRC4-KR instability, only small amounts of complexes are formed, and we cannot determine whether these are signaling-competent. In conclusion, our data strongly indicate that ATP-binding by NAIP5 is not essential for inflammasome formation. For NLRC4 our data are not conclusive.
FIGURE 7.
Effect of Walker A mutations on inflammasome formation. A, viabilities of HEK293E cells 48 h after transfection with the indicated wild type (+) or mutant (KR) proteins. For further explanation, see Fig. 1A. B, analysis of inflammasome formation in HEK293E cells transfected with D0L and WT or mutant (KR) NLRs as indicated. For further explanation, see Fig. 2D. C, analysis of the binding of WT or mutant (KR) inflammasome components to biotinylated D0L in HEK293E cells. For further explanation, see Fig. 4.
Analysis of the NAIP5-NLRC4 Inflammasome by Electron Microscopy
To gain insight in the macromolecular organization of the NAIP5-NLRC4 inflammasome, we visualized the complex as well as the individual NLRs by negative stain EM. Electron micrographs of the mNAIP5 and hNLRC4 preparations that were used for the in vitro reconstitution experiments show particles with a globular shape and a diameter of about 8 nm, which presumably represent monomers or at most dimers (Fig. 8, A and B). Micrographs of a hNLRC4 sample that had been concentrated to ∼8 mg/ml before it was diluted for EM analysis show large rod-shaped particles that are about 25 ± 2 nm in width and vary in length from about 30 nm to over 200 nm (Fig. 8C). Studied in closer detail by electron tomography (Fig. 8D), these rods appear to have a layered structure composed of stacked rings or, alternatively, a helical arrangement. The limited quality of the data did not allow us to discriminate between both arrangements, because Fourier analysis and subvolume averaging were unsuccessful. Because these rod-shaped particles are only observed at very high protein concentrations, their physiological relevance is debatable. Nevertheless, their formation may be related to the previously observed induction of caspase-1-dependent cell death upon overexpression of Nlrc4, as well as its ligand-independent self-association (Fig. 1, A and B).
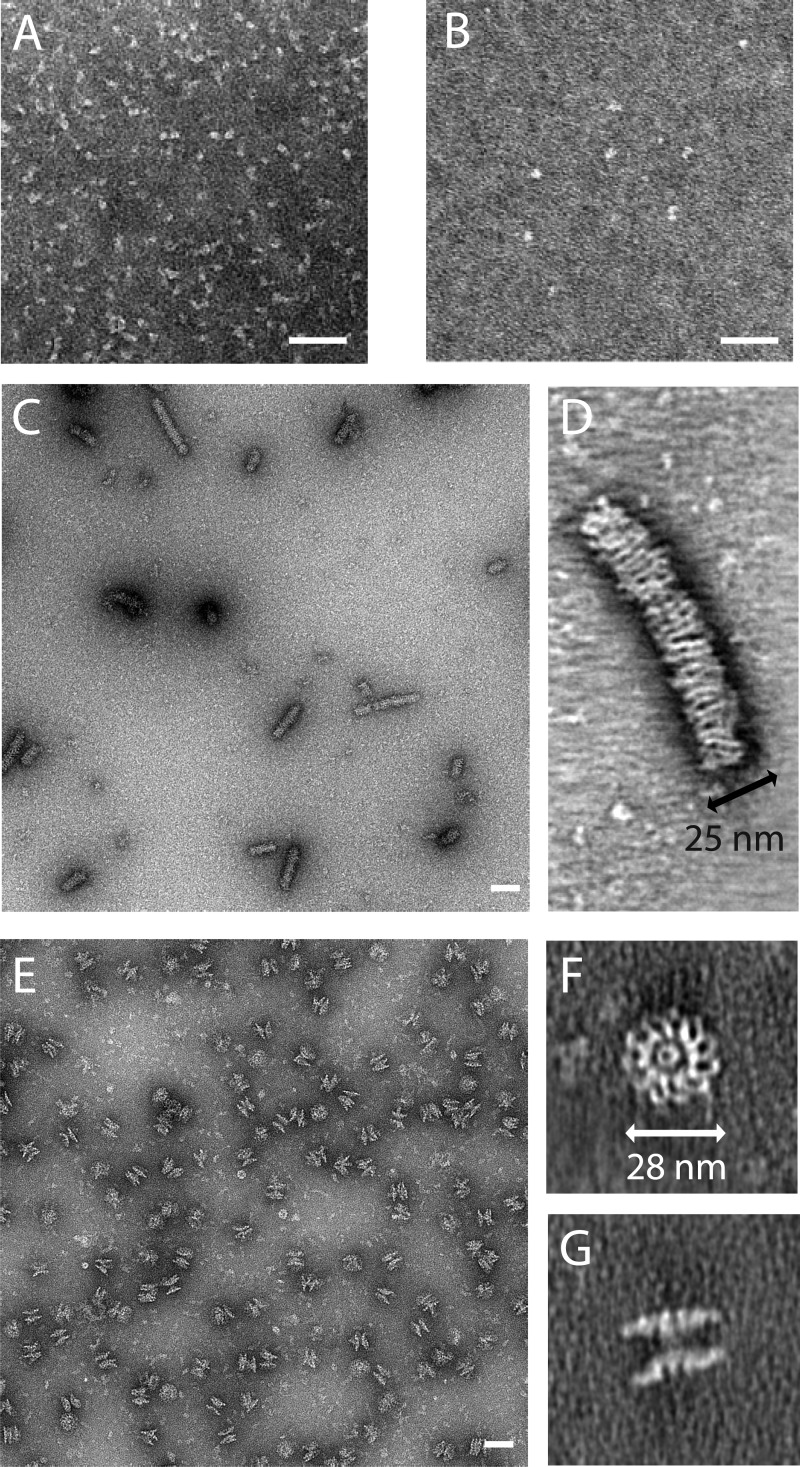
FIGURE 8.
Electron micrographs of the NAIP5-NLRC4 inflammasome. A–C and E, EM projection images of negatively stained samples of purified hNLRC4 (A), mNAIP5 (B), hNLRC4 stored at high concentration (C), and D0L-induced mNAIP5-mNLRC4 inflammasome freshly purified from HEK293E cells (E). D, F, and G, virtual sections through electron tomograms of the concentrated hNLRC4 sample (D) and the freshly purified inflammasome sample in top view (F) and side view (G). The scale bars represent 50 nm. Quantitative analysis of inflammasome symmetry (supplemental Fig. S3) shows that the particle depicted in F has 11-fold symmetry.
Because the in vitro reconstitution of the NAIP5-NLRC4 complex was rather inefficient, we purified the inflammasome directly from cells co-transfected with murine Naip5, Nlrc4, and FliC-D0L. Electron micrographs reveal top and side views of disk-shaped particles with a radius of 28 ± 2 nm (Fig. 8, E–G). A top view tomogram of a particularly well stained NAIP5-NLRC4 inflammasome (Fig. 8F) reveals considerable detail. The flagellin D0L fragment would be too small to be resolved, but the curved leucine-rich repeat domains of the individual NLR monomers can clearly be distinguished at the perimeter of the disk. From the number of leucine-rich repeat domains, it is evident that this particular particle consists of 11 protomers. Rotational autocorrelation analysis of 15 putative top view inflammasomes reveals that among six particles that could be classified unambiguously, four display 11-fold symmetry, whereas two particles display 12-fold symmetry (supplemental Fig. S3). Because of the limited number of particles analyzed, we cannot exclude that other symmetries occur as well. The number of protomers is remarkable because EM structures of the NLRP1 inflammasome, as well as the Apaf-1, CED4, and Dark apoptosomes, revealed similarly shaped, but considerably smaller disks consisting of 5–8 protomers only (21, 23, 25, 26). Although at this resolution no distinction can be made between NAIP5 and NLRC4, the absence of nonsymmetric density features that are sufficiently large to represent an NLR monomer strongly suggests that the two proteins are incorporated into the disks at equivalent positions. In analogy with the EM structures of homologous inflammasomes and apoptosomes, the central region, where the individual monomers interact, is expected to be formed by the NACHT domains of NAIP5 and NLRC4. The N-terminal effector-binding domains likely are centrally located on one side of the disk, forming a platform for procaspase-1 to bind. In the side view, tomograms of the NAIP5-NLRC4 complex (Fig. 8G) disks are seen to stack in pairs. Similar pairwise stacking was observed in EM studies of the Dark and Apaf-1 apoptosomes and was attributed to nonphysiological CARD-CARD domain interactions occurring at high protein concentrations (24, 26, 43). Assuming that a similar mechanism is responsible for the formation of the stacked NAIP5-NLRC4 disks, the density connecting two disks most likely represents the NLR effector-binding domains. In this case, the physiologically active unit of the NAIP5-NLRC4 inflammasome is the single disk.
In summary, we show that flagellin induces formation of disk-shaped inflammasomes, consisting of 11 or 12 protomers, in which NAIP5 and NLRC4, to a first approximation, occupy equivalent positions. Inflammasomes of this size have not been previously observed and may represent a unique feature of NAIP-NLRC4 complexes.
DISCUSSION
The flagellin D0 domain and the neighboring spoke region are essential for formation and stabilization of the flagellar filament and therefore indispensible for bacterial motility (32, 33). The structural importance of this region is reflected by high sequence conservation between bacterial species (supplemental Fig. S2). From an evolutionary viewpoint, this makes the extended D0 domain an appropriate target for recognition by the innate immune system. Previously, inflammasome activation was attributed to the C-terminal 35 amino acids of flagellin (4). We now show that the N-terminal 52 amino acids of flagellin likewise contain an element that induces the association of NAIP5 with NLRC4 and concomitant caspase-1-dependent cell death. The difference in response to D0L and D0S suggests that this element resides in the spoke region; D0L precipitates NAIP5, whereas D0S, which lacks this region (supplemental Fig. S2), does not (Fig. 4). Moreover, the role of the three conserved leucine residues located near the C terminus of flagellin is more prominent in fragments that lack the spoke region (Fig. 3). This shows that, in addition to the C terminus, the spoke region contributes significantly to inflammasome activation.
It has been observed that polymerized flagellin is less potent in activating NAIP5 (44), which is consistent with the role of D0L in inflammasome induction, because D0 is not exposed in the flagellar filament (33). Polymerization of overexpressed flagellin in our reconstituted system in HEK293E cells could explain why we observe no co-precipitation of NAIP5 with biotinylated flagellin, whereas biotinylated D0L, which we presume does not polymerize, does precipitate NAIP5 (Fig. 4). Likewise, polymerization of purified flagellin may have contributed to the low efficiency of complex formation in the in vitro reconstitution of the NAIP5-NLRC4 inflammasome (Fig. 6A).
Electron microscopy reveals that the NAIP5-NLRC4 inflammasome is far larger than previously observed for complexes of other NACHT domain-containing proteins and contains 11 or 12 protomers arranged in a disk-shaped complex. Because of the limited number of particles analyzed, we cannot exclude that other sizes occur as well. Although the resolution of the EM micrographs is insufficient to distinguish NAIP5 from NLRC4, the highly symmetrical structure and the absence of obvious protrusions or deviations (Fig. 8F) indicates that both NLRs occupy equivalent positions within the inflammasome and thus likely exhibit equivalent NACHT-NACHT interactions. As explained in the results section, the observed dimerization of the inflammasome disks (Fig. 8G) is presumably nonphysiological. The functional unit is expected to be a single disk that exposes the effector binding domains on one side, thereby forming a platform for procaspase-1 recruitment and activation. Low level caspase-1 activation also occurs independent of flagellin as a consequence of overexpression-induced homo-oligomerization of NLRC4 (Fig. 1, A and B). Similar overexpression-induced homo- and hetero-oligomerization of NLR NACHT domains has been reported more frequently (8, 45–47). Although NLRC4 homo-oligomers are unlikely to play a role at physiological expression levels, electron micrographs (Fig. 8, C and D) show that NLRC4 assembles into a rod-like structure with a diameter that is very similar to that of the NAIP5-NLRC4 disk. We are unable to determine whether these rods consist of stacked NLRC4 disks or an NLRC4 helix; nevertheless the similarity in diameters suggests that the arrangement of monomers in the NLRC4 rod and the NAIP5-NLRC4 disk is similar.
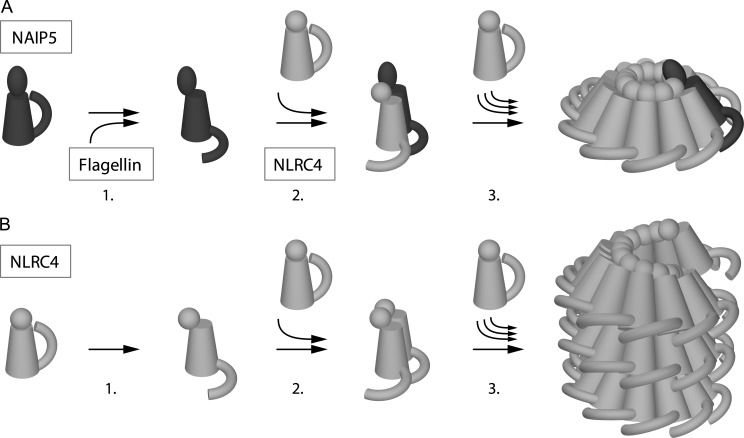
Our data lead us to propose a model for NAIP5-NLRC4 inflammasome formation by the following sequence of events (Fig. 9). The initiation step involves recognition of flagellin by NAIP5. This is supported by our co-precipitation experiments, which show association of D0L with NAIP5, and is consistent with a role for NAIP proteins in providing specificity to NLRC4 containing inflammasomes (5, 6). Because NAIP5 does not oligomerize in the presence of flagellin (Fig. 1B), the next step in inflammasome formation must involve binding of NLRC4 to the NAIP5-flagellin complex. Finally, because NLRC4 is able to self-associate, we speculate that a conformational change in complex-incorporated NLRC4 can be recognized by the free protein and induces the progressive incorporation of NLRC4 monomers into the complex through homotypic interactions until a disk-shaped inflammasome is formed.
FIGURE 9.
Model of NAIP5-NLRC4 inflammasome formation versus NLRC4-multimerization. A, steps involved in inflammasome formation. Resting NAIP5 undergoes a conformational change upon interaction with flagellin (1). Activated NAIP5 subsequently recruits resting NLRC4, which in turn undergoes a conformational change (2). This activated form of NLRC4 is recognized by resting NLRC4, which results in the progressive incorporation of NLRC4 (3) into a disk-like complex that exposes the NLRC4 CARD domains at one face, creating a platform suitable for procaspase-1 recruitment and activation. B, steps involved in the nonphysiological formation of NLRC4 multimers; NLRC4 is spontaneously activated at low frequency (1). As with the formation of the hetero-oligomeric complex, activated NLRC4 is recognized by resting NLRC4 (2), which, in overexpression conditions, results in the progressive incorporation of additional NLRC4 monomers and the formation of rod-like structures (3). In these rods, consisting either of stacked disks or, as depicted here, a helical arrangement, the CARD domains are possibly only partially exposed, resulting in limited procaspase-1 activation. Leucine-rich repeat domains are depicted as arches; NACHT domains are tubes; CARD domains are spheres; and BIR domains are ellipsoids.
The sequence of events described above would produce disks that contain a single NAIP5 molecule. Although at this point we can only speculate about the exact ratio between NAIP5 and NLRC4 in the complex, our data strongly indicate that the major part of the inflammasome is formed by NLRC4. In inflammasome preparations we generally observe a clear excess of NLRC4, even though the isolation procedure results in co-purification of any residual monomeric NAIP5. Likewise, an excess of NLRC4 is also observed in co-precipitation of the inflammasome with biotinylated flagellin (Fig. 4), which does not co-precipitate the monomeric NLRs. Thus, we can exclude a 1:1 composition and therefore models with disks composed of alternating NAIP5 and NLRC4 monomers, as well as a structure consisting of two stacked disks each consisting of only one of the NLRs. Our data, however, does not exclude the presence of more than one NAIP5 molecule per disk.
The question arises of whether flagellin also interacts with NLRC4. We find no solid evidence for a direct interaction other than the precipitation of NLRC4 by the flagellin C-terminal fragment in some, but not all experiments (Fig. 4). Also, NLRC4 self-association and related caspase-1 activation are not enhanced in the presence of flagellin (Fig. 1, A and B). NLRC4 does, however, appear to increase flagellin binding affinity because many flagellin fragments are not capable of precipitating NAIP5 but do precipitate the complex (Fig. 4). Although this observed stabilization may arise from NLRC4-induced conformational changes in NAIP5, it could also result from flagellin binding at the NAIP5-NLRC4 interface. Flagellin binding at the NAIP5-NLRC4 interface, or to NAIP5 only, would both be consistent with our finding that relatively small amounts of flagellin co-purify with the inflammasome, but we cannot exclude that this finding results from flagellin dissociation after inflammasome formation. The C-terminal residues of flagellin show a striking similarity to the C-terminal residues of the ligands of other NLRC4-containing inflammasomes (5, 6, 48). This region includes the three leucines that proved essential for induction of inflammasome formation by FliC-C and D0S (Fig. 3). If NLRC4 has a direct interaction with the different NAIP-inflammasome ligands, the conserved C-terminal region may well constitute a common NLRC4-binding motif.
The role of ATP binding and/or hydrolysis in inflammasome formation remains unclear. Our mutagenesis data suggest that at least for NAIP5, an intact ATP-binding site is not a prerequisite for inflammasome formation or signaling. For NLRC4, data analysis is complicated by the instability of the NLRC4-KR mutant. The observation of in vitro inflammasome formation in the presence of EDTA (Fig. 6) does, by itself, not rule out a possible role for ATP binding; for instance, nucleotide-dependent apoptosome formation by Apaf-1 and Dark is enhanced by EDTA (24, 26). Thus far, various studies on the effect of ATP binding by NAIP5 and NLRC4, including mutations and deletions of the ATP-binding site, yielded inconsistent outcomes on NLR association and inflammasome signaling (6, 17, 46). Further biochemical studies using purified NAIP5, NLRC4, and their ATP-binding mutants may resolve this issue.
In conclusion, using a number of experimental approaches, we show that (i) a flagellin-responsive NAIP5-NLRC4 inflammasome can be reconstituted from the purified components, and thus no additional cellular factors are required for its formation; (ii) flagellin recognition is determined by its conserved N- and C-terminal regions; (iii) NLRC4 is the main inflammasome constituent; and (iv) the inflammasome is a disk-like complex consisting of 11 or 12 NLR molecules. We propose a model of the inflammasome in which NAIP5 functions as a flagellin sensor and NLRC4 provides a platform for caspase-1 recruitment. Although the exact ratio of NAIP5 and NLRC4 in the inflammasome remains to be determined, we speculate that it contains a single NAIP5 molecule. Our data provide a solid basis for further investigations into the formation of NAIP-NLRC4 inflammasomes, and we anticipate that the ability to purify the individual NLRs, as well as the complete inflammasome, will boost studies aimed at the elucidation of their three-dimensional structures.
This work was supported by the Dutch Top Institute Pharma Project D1-101.

This article contains supplemental Figs. S1–S3.
- NLR
- NOD-like receptor
- BAP
- biotin acceptor peptide
- BIR
- baculovirus inhibitor of apoptosis protein repeat
- CARD
- caspase activating and recruitment domain
- TEV
- tobacco etch virus
- h
- human
- m
- murine.
REFERENCES
- 1. Franchi L., Amer A., Body-Malapel M., Kanneganti T. D., Ozören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., Grant E. P., Núñez G. (2006) Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in Salmonella-infected macrophages. Nat. Immunol. 7, 576–582 [DOI] [PubMed] [Google Scholar]
- 2. Miao E. A., Alpuche-Aranda C. M., Dors M., Clark A. E., Bader M. W., Miller S. I., Aderem A. (2006) Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7, 569–575 [DOI] [PubMed] [Google Scholar]
- 3. Ren T., Zamboni D. S., Roy C. R., Dietrich W. F., Vance R. E. (2006) Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog 2, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lightfield K. L., Persson J., Brubaker S. W., Witte C. E., von Moltke J., Dunipace E. A., Henry T., Sun Y. H., Cado D., Dietrich W. F., Monack D. M., Tsolis R. M., Vance R. E. (2008) Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9, 1171–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao Y., Yang J., Shi J., Gong Y. N., Lu Q., Xu H., Liu L., Shao F. (2011) The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600 [DOI] [PubMed] [Google Scholar]
- 6. Kofoed E. M., Vance R. E. (2011) Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Damiano J. S., Stehlik C., Pio F., Godzik A., Reed J. C. (2001) CLAN, a novel human CED-4-like gene. Genomics 75, 77–83 [DOI] [PubMed] [Google Scholar]
- 8. Poyet J. L., Srinivasula S. M., Tnani M., Razmara M., Fernandes-Alnemri T., Alnemri E. S. (2001) Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276, 28309–28313 [DOI] [PubMed] [Google Scholar]
- 9. Geddes B. J., Wang L., Huang W. J., Lavellee M., Manji G. A., Brown M., Jurman M., Cao J., Morgenstern J., Merriam S., Glucksmann M. A., DiStefano P. S., Bertin J. (2001) Human CARD12 is a novel CED4/Apaf-1 family member that induces apoptosis. Biochem. Biophys. Res. Commun. 284, 77–82 [DOI] [PubMed] [Google Scholar]
- 10. Martinon F., Burns K., Tschopp J. (2002) The inflammasome. A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 11. Schroder K., Tschopp J. (2010) The inflammasomes. Cell 140, 821–832 [DOI] [PubMed] [Google Scholar]
- 12. Martinon F., Tschopp J. (2007) Inflammatory caspases and inflammasomes. Master switches of inflammation. Cell Death Differ. 14, 10–22 [DOI] [PubMed] [Google Scholar]
- 13. Dinarello C. A. (1999) IL-18. A TH1-inducing, proinflammatory cytokine and new member of the IL-1 family. J. Allergy Clin. Immunol. 103, 11–24 [DOI] [PubMed] [Google Scholar]
- 14. Fink S. L., Cookson B. T. (2005) Apoptosis, pyroptosis, and necrosis. Mechanistic description of dead and dying eukaryotic cells. Infect. Immun. 73, 1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mo J., Boyle J. P., Howard C. B., Monie T. P., Davis B. K., Duncan J. A. (2012) Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J. Biol. Chem. 287, 23057–23067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duncan J. A., Bergstralh D. T., Wang Y., Willingham S. B., Ye Z., Zimmermann A. G., Ting J. P. (2007) Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl. Acad. Sci. U.S.A. 104, 8041–8046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu C., Wang A., Wang L., Dorsch M., Ocain T. D., Xu Y. (2005) Nucleotide binding to CARD12 and its role in CARD12-mediated caspase-1 activation. Biochem. Biophys. Res. Commun. 331, 1114–1119 [DOI] [PubMed] [Google Scholar]
- 18. Ye Z., Lich J. D., Moore C. B., Duncan J. A., Williams K. L., Ting J. P. (2008) ATP binding by monarch-1/NLRP12 is critical for its inhibitory function. Mol. Cell. Biol. 28, 1841–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Askari N., Correa R. G., Zhai D., Reed J. C. (2012) Expression, purification, and characterization of recombinant NOD1 (NLRC1): A NLR family member. J. Biotechnol. 157, 75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zurek B., Proell M., Wagner R. N., Schwarzenbacher R., Kufer T. A. (2012) Mutational analysis of human NOD1 and NOD2 NACHT domains reveals different modes of activation. Innate Immun. 18, 100–111 [DOI] [PubMed] [Google Scholar]
- 21. Faustin B., Lartigue L., Bruey J. M., Luciano F., Sergienko E., Bailly-Maitre B., Volkmann N., Hanein D., Rouiller I., Reed J. C. (2007) Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell 25, 713–724 [DOI] [PubMed] [Google Scholar]
- 22. Yu X., Acehan D., Ménétret J. F., Booth C. R., Ludtke S. J., Riedl S. J., Shi Y., Wang X., Akey C. W. (2005) A structure of the human apoptosome at 12.8 A resolution provides insights into this cell death platform. Structure 13, 1725–1735 [DOI] [PubMed] [Google Scholar]
- 23. Acehan D., Jiang X., Morgan D. G., Heuser J. E., Wang X., Akey C. W. (2002) Three-dimensional structure of the apoptosome. Implications for assembly, procaspase-9 binding, and activation. Mol. Cell 9, 423–432 [DOI] [PubMed] [Google Scholar]
- 24. Yuan S., Yu X., Topf M., Dorstyn L., Kumar S., Ludtke S. J., Akey C. W. (2011) Structure of the Drosophila apoptosome at 6.9 Å resolution. Structure 19, 128–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qi S., Pang Y., Hu Q., Liu Q., Li H., Zhou Y., He T., Liang Q., Liu Y., Yuan X., Luo G., Li H., Wang J., Yan N., Shi Y. (2010) Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141, 446–457 [DOI] [PubMed] [Google Scholar]
- 26. Yu X., Wang L., Acehan D., Wang X., Akey C. W. (2006) Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J. Mol. Biol. 355, 577–589 [DOI] [PubMed] [Google Scholar]
- 27. Durocher Y., Perret S., Kamen A. (2002) High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ibrahim G. F., Fleet G. H., Lyons M. J., Walker R. A. (1985) Method for the isolation of highly purified Salmonella flagellins. J. Clin. Microbiol. 22, 1040–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsai A., Carstens R. P. (2006) An optimized protocol for protein purification in cultured mammalian cells using a tandem affinity purification approach. Nat. Protoc 1, 2820–2827 [DOI] [PubMed] [Google Scholar]
- 30. Kremer J. R., Mastronarde D. N., McIntosh J. R. (1996) Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 [DOI] [PubMed] [Google Scholar]
- 31. Fernández J. J., Li S., Crowther R. A. (2006) CTF determination and correction in electron cryotomography. Ultramicroscopy 106, 587–596 [DOI] [PubMed] [Google Scholar]
- 32. Vonderviszt F., Aizawa S., Namba K. (1991) Role of the disordered terminal regions of flagellin in filament formation and stability. J. Mol. Biol. 221, 1461–1474 [DOI] [PubMed] [Google Scholar]
- 33. Yonekura K., Maki-Yonekura S., Namba K. (2003) Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424, 643–650 [DOI] [PubMed] [Google Scholar]
- 34. Samatey F. A., Imada K., Nagashima S., Vonderviszt F., Kumasaka T., Yamamoto M., Namba K. (2001) Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410, 331–337 [DOI] [PubMed] [Google Scholar]
- 35. de Boer E., Rodriguez P., Bonte E., Krijgsveld J., Katsantoni E., Heck A., Grosveld F., Strouboulis J. (2003) Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 100, 7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Inohara N., Koseki T., del Peso L., Hu Y., Yee C., Chen S., Carrio R., Merino J., Liu D., Ni J., Núñez G. (1999) Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-κB. J. Biol. Chem. 274, 14560–14567 [DOI] [PubMed] [Google Scholar]
- 37. Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. (2001) Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 276, 4812–4818 [DOI] [PubMed] [Google Scholar]
- 38. Tameling W. I., Elzinga S. D., Darmin P. S., Vossen J. H., Takken F. L., Haring M. A., Cornelissen B. J. (2002) The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14, 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tameling W. I., Vossen J. H., Albrecht M., Lengauer T., Berden J. A., Haring M. A., Cornelissen B. J., Takken F. L. (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140, 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chaudhary D., O'Rourke K., Chinnaiyan A. M., Dixit V. M. (1998) The death inhibitory molecules CED-9 and CED-4L use a common mechanism to inhibit the CED-3 death protease. J. Biol. Chem. 273, 17708–17712 [DOI] [PubMed] [Google Scholar]
- 41. Chinnaiyan A. M., Chaudhary D., O'Rourke K., Koonin E. V., Dixit V. M. (1997) Role of CED-4 in the activation of CED-3. Nature 388, 728–729 [DOI] [PubMed] [Google Scholar]
- 42. Hu Y., Ding L., Spencer D. M., Núñez G. (1998) WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. J. Biol. Chem. 273, 33489–33494 [DOI] [PubMed] [Google Scholar]
- 43. Yuan S., Yu X., Asara J. M., Heuser J. E., Ludtke S. J., Akey C. W. (2011) The holo-apoptosome. Activation of procaspase-9 and interactions with caspase-3. Structure 19, 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Molofsky A. B., Byrne B. G., Whitfield N. N., Madigan C. A., Fuse E. T., Tateda K., Swanson M. S. (2006) Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203, 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Damiano J. S., Oliveira V., Welsh K., Reed J. C. (2004) Heterotypic interactions among NACHT domains. Implications for regulation of innate immune responses. Biochem. J. 381, 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zamboni D. S., Kobayashi K. S., Kohlsdorf T., Ogura Y., Long E. M., Vance R. E., Kuida K., Mariathasan S., Dixit V. M., Flavell R. A., Dietrich W. F., Roy C. R. (2006) The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7, 318–325 [DOI] [PubMed] [Google Scholar]
- 47. Hsu L. C., Ali S. R., McGillivray S., Tseng P. H., Mariathasan S., Humke E. W., Eckmann L., Powell J. J., Nizet V., Dixit V. M., Karin M. (2008) A NOD2-NALP1 complex mediates caspase-1-dependent IL-1β secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc. Natl. Acad. Sci. U.S.A. 105, 7803–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miao E. A., Mao D. P., Yudkovsky N., Bonneau R., Lorang C. G., Warren S. E., Leaf I. A., Aderem A. (2010) Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. U.S.A. 107, 3076–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]