Background: An interaction network involving soluble factors and nuclear pore proteins underlies nucleocytoplasmic transport.
Results: Nup50 interacts with two regions of Nup153, one of which is bridged by importin α.
Conclusion: Efficient import is dependent on the interaction between Nup153 and Nup50.
Significance: Nup153 provides a scaffold to facilitate interactions that contribute to trafficking through the nuclear pore.
Keywords: Cell Biology, Nuclear Membrane, Nuclear Pore, Nuclear Transport, Nucleus, Npap60, Nup153, Nup50, Nuclear Pore Complex, Nucleocytoplasmic Transport
Abstract
Interactions between Nup50 and soluble transport factors underlie the efficiency of certain nucleocytoplasmic transport pathways. The platform on which these interactions take place is important to building a complete understanding of nucleocytoplasmic trafficking. Nup153 is the nucleoporin that provides this scaffold for Nup50. Here, we have delineated requirements for the interaction between Nup153 and Nup50, revealing a dual interface. An interaction between Nup50 and a region in the unique N-terminal region of Nup153 is critical for the nuclear pore localization of Nup50. A second site of interaction is at the distal tail of Nup153 and is dependent on importin α. Both of these interactions involve the N-terminal domain of Nup50. The configuration of the Nup153-Nup50 partnership suggests that the Nup153 scaffold provides not just a means of pore targeting for Nup50 but also serves to provide a local environment that facilitates bringing Nup50 and importin α together, as well as other soluble factors involved in transport. Consistent with this, disruption of the Nup153-Nup50 interface decreases efficiency of nuclear import.
Introduction
The basic cycle of nucleocytoplasmic trafficking involves transport receptors along with the cofactor Ran and regulators of its nucleotide status (1). However, a broader repertoire of proteins guides the efficiency of transport and provides additional layers of regulation and fidelity to this process. One such protein is Nup50/Npap60 (referred to here as Nup50), which was discovered as a nuclear pore-associated protein (2) and first examined in the context of binding and regulating the cell cycle factor p27Kip1 (3, 4). The pore association of Nup50 was further characterized by immunoelectron microscopy, which mapped Nup50 to the basket element of the nuclear pore complex (NPC),3 in close proximity to Nup153 (5). Indeed, Nup50 and Nup153 were found to associate (4), and since then, Nup153 was determined to be essential for the nuclear pore localization of Nup50 (6). Interestingly, both of these nucleoporins are highly dynamic components of the NPC (7).
Nup50 is a regulator of nuclear transport (5, 8, 9), with most studies to date focused on a role in facilitating nuclear import. Although there is not complete agreement on the precise mechanism by which Nup50 contributes to import, its interactions with soluble transport factors are well characterized. In this regard, Nup50 has three main functional domains: an N-terminal region that interacts with importin α (N); a central region, with phenylalanine-glycine (FG) motifs shared by many nucleoporins, that provides a docking site for importin β (F); and a C-terminal region that confers binding to the transport regulator Ran (R) (see Fig. 1A) (10). Yoneda and co-workers (11) recently extended this groundwork by comparing the two main isoforms of Nup50, which arise from alternative splicing and differ in the extreme N-terminal sequences (see Fig. 1A). This divergence lies at a critical region, where Nup50 binds to importin α. Thus, although the distinction between the isoforms is quite discrete, it results in significantly different behavior with respect to the ability of Nup50 to displace the cognate nuclear localization signal (NLS) cargo of importin α.
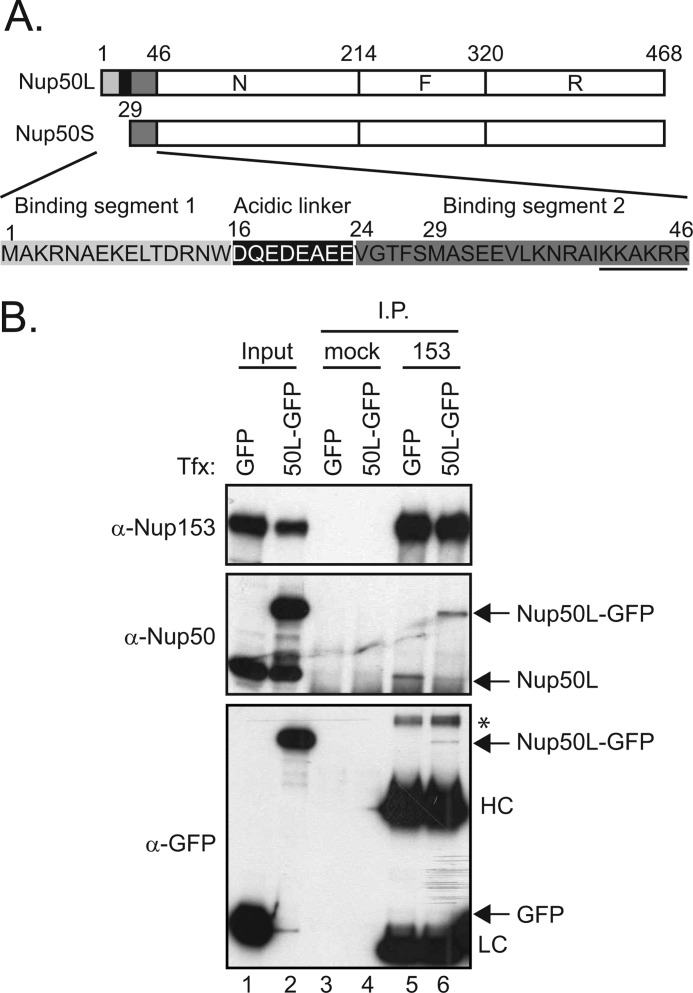
FIGURE 1.
Limited levels of Nup50 associate with Nup153. A, schematic depicting the three regions of Nup50, designated N, F, and R (9), as well as the sequence differences between the long and short isoforms of Nup50, as annotated previously (11). B, Nup153 was immunoprecipitated (I.P.) from lysates of HeLa cells expressing GFP or Nup50L (50L)-GFP (lanes 5 and 6); material isolated with equivalent levels of protein A-Sepharose beads alone was run alongside (lanes 3 and 4). The precipitated material was immunoblotted for the presence of Nup153, Nup50 (both recombinant and endogenous), and GFP. For comparison, samples of the input material are shown (7.5% input for Nup153 and Nup50 detection and 10% input for GFP detection) (lanes 1 and 2). The immunoglobulin heavy chain (HC) and light chain (LC) and an unknown background band (*) are indicated. Tfx, transfection.
To date, the focus for Nup50 analysis has been on its interfaces with soluble transport factors, and this has been very informative with respect to building functional models (8, 12) as well as gaining insight into how these interactions are modulated (13). However, consideration of Nup50 function needs to further incorporate the fact that, in the context of the nuclear pore, Nup153 provides not just a targeting site but also a broader scaffold that likely influences the role of Nup50. Here, we have characterized the Nup153-Nup50 partnership to provide critical information about the context of Nup50 function.
EXPERIMENTAL PROCEDURES
DNA Constructs
Mammalian plasmids that express an enhanced GFP (EGFP) fusion of Nup50 and its domains were generated by cloning their PCR fragments into the pEGFP-N2 vector (Clontech). A plasmid encoding a HA-tagged version of the Nup50 N terminus was established by replacing the EGFP gene with synthetic oligonucleotides encoding 3×HA. To make integration plasmids for T-REx HeLa cells, genes cloned into the pEGFP-N2 vector described above were digested with BglII and NotI, and the resultant fragments were ligated into the BamHI and NotI sites of pcDNA5/FRT/TO (Invitrogen). A bacterial expression plasmid that expresses GST fused to Nup153(401–609) was generated by cloning this region as a PCR fragment into pGEX4T-1 (GE Healthcare). Escherichia coli expression plasmids for T7-tagged versions of Nup153 and its N-terminal deletion mutant were described by Dimaano et al. (14).
Cell Culture and Transfection Conditions
Plasmids were introduced into HeLa cells with Lipofectamine LTX reagent (Invitrogen) according to the manufacturer's instructions. T-REx HeLa cells containing genes encoding the Tet repressor and Flp recombination target site were a kind gift from Drs. Dobrikova and Gromeier (Duke University) (15). To establish stable cell lines, the parental cells were cotransfected with pcDNA5/FRT/TO containing genes of interest and pOG44 (Invitrogen), which expresses Flp recombinase, and selected in medium containing 100 μg/ml hygromycin B and 2.5 μg/ml blasticidin. Protein expression in the established cell lines was stimulated with 0.3 μg/ml doxycycline in medium without selection drugs for 24–48 h.
Preparation of Mammalian Cell Lysate
HeLa cells were harvested after trypsinization and two washes with ice-cold PBS. For immunoprecipitation and GST pulldown assays, cells were resuspended in Nonidet P-40 lysis buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 5 mm EDTA, 15 mm magnesium chloride, 1% Nonidet P-40, 60 mm β-glycerophosphate, 1 mm DTT, 0.1 mm sodium vanadate, 100 μm PMSF, and 0.1 mm sodium fluoride) or in PBS lysis buffer (PBS containing 0.2% Triton X-100) with cOmplete protease inhibitor (Roche Applied Science) and then incubated on ice for 5 min. For Western blotting, cells were resuspended in radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 7.4). 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.05% SDS) and then incubated on ice for 5 min.
Preparation of E. coli Cell Lysate
Bacteria harvested by centrifugation were frozen once, resuspended in E. coli lysis buffer (50 mm Tris-HCl (pH 8.0), 5 mm EDTA, 200 mm KCl, 1 mm DTT, and 0.05% Triton X-100) containing 400 μg/ml lysozyme, and then incubated on ice for 30 min with gentle shaking. After lysis, the supernatant was collected by centrifugation. The levels of recombinant protein were normalized by mixing with bacterial lysate without expressed protein to achieve approximately similar levels (determined by Western analysis) of the T7-tagged fusion protein in each input.
Purification of GST Fusion Proteins
Bacterial lysates containing GST, GST-Nup50, and GST-Nup153(401–609) were incubated overnight with glutathione-Sepharose 4B beads (GE Healthcare) at 4 °C with rotation. Proteins were eluted from the beads with E. coli lysis buffer containing 10% glycerol and 20 mm reduced glutathione. Eluates were dialyzed overnight against lysis buffer without Triton X-100 at 4 °C.
GST Pulldown Assay
500 μg of mammalian cell lysate (1 mg/ml) was incubated with GST and GST fusion proteins immobilized on glutathione-Sepharose 4B beads and incubated at 4 °C for 1 h. After the incubation, beads were washed three times with ice-cold lysis buffer containing protease inhibitors, and proteins bound to the beads were eluted with SDS sample buffer.
GFP-Trap® Assay
GFP fusion proteins were retrieved from mammalian cell lysates using GFP-Trap A beads (ChromoTek). 500 μg of mammalian cell lysate (1 mg/ml) containing the appropriate EGFP fusion protein was incubated with the beads at 4 °C for 15 min with rotation. After the reaction, the beads were washed three times with mammalian lysis buffer at 4 °C and incubated with SDS sample buffer to elute proteins.
Immunofluorescence and Live Imaging Import Assay
Immunofluorescence analysis was performed as described previously (16). For live import assay, HeLa cells were grown in CO2-independent medium supplemented with 10% charcoal-stripped FBS (Invitrogen). Following cotransfection with Rev-GFP-glucocorticoid receptor (RGG) (17) and constructs encoding either mCherry fused to Nup153(401–609) or mCherry alone, cells were seeded in 4-well Lab-Tek II chambers (Thermo Scientific) coated with fibronectin. After 24 h, dexamethasone was added to a final concentration of 25 nm. Images were acquired with a DeltaVision system every 3 min for 40 min at 37 °C using a 20× objective. Image analysis and intensity measurements were performed on background subtracted images of specific time points in ImageJ. Thresholds were applied to GFP montages to measure total raw integrated intensity. Freehand selections were used to measure raw integrated intensity of nuclei.
RESULTS
A Limited Pool of Nup50 Interacts with Nup153
Nup50 is an established protein partner of Nup153 (4), and the two proteins share specific attributes. Both associate with protein phosphatase complexes (18, 19), and both are highly mobile nuclear pore proteins (7). They are also both recruited relatively early during the process of post-mitotic NPC formation (20). Some of the assays underlying these observations also point to distinctions between the two nucleoporins, as their off-rates from the pore are different, and their recruitment to newly forming pores is not in precise synchrony (7, 20). Thus, although Nup50 and Nup153 overlap spatially and participate in similar processes (likely in concert), a fundamental question that has not been addressed is whether this partnership is limited to a subset of Nup50 found in the cell.
We observed that relatively little Nup50 co-precipitated in an immunoprecipitation of endogenous Nup153 (supplemental Fig. S1). Because the absolute level of co-immunoprecipitation is influenced by the arbitrary conditions of the experiment, we tested whether a limited pool of Nup50 associates with Nup153 by challenging the system with exogenous GFP-Nup50. If the lack of efficient co-precipitation were due simply to a failure to maintain the association during the procedure, GFP-Nup50 would be predicted to be recovered in the same proportion relative to its input and independent of endogenous Nup50 recovery. In contrast, if the population of Nup153 available for interaction with Nup50 is limited, then the total levels of Nup50 recovered would be restricted, and GFP-Nup50 would displace comparable levels of endogenous Nup50. For this experiment, we expressed the Nup50L isoform, as this is the isoform we confirmed was expressed in the line of HeLa cells used here (supplemental Fig. S1A). As shown in Fig. 1B, despite robust levels of GFP-Nup50 expression (lane 2), its recovery (lane 6) did not exceed the recovery of endogenous Nup50 in the presence of control GFP protein expression (lane 5), and in fact, endogenous Nup50 recovery was reduced in the presence of GFP-Nup50 (lane 6 versus lane 5). This could be due to limiting levels of Nup153 or to other factors that affect accessibility of the two partners. In either case, models that integrate the context of Nup153 in understanding the role of Nup50 must account for a limited pool of Nup50 in association with Nup153 at any one time (see “Discussion”).
The N-terminal Domain of Nup50 Mediates Interaction with Two Regions within Nup153
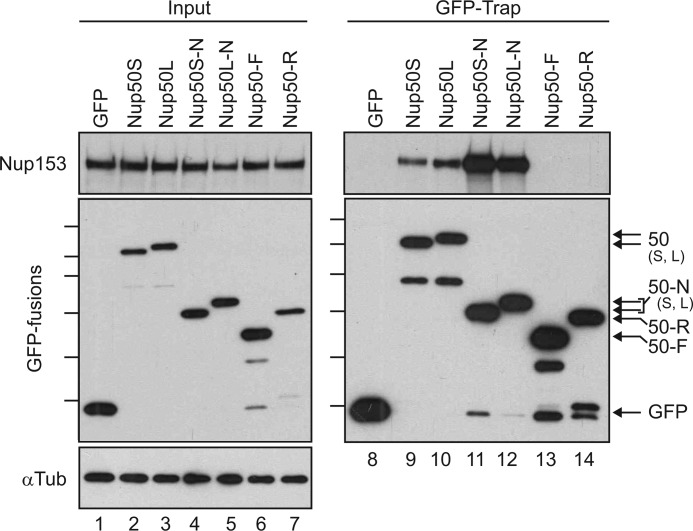
To better understand the molecular determinants of the Nup50-Nup153 partnership, expression constructs encoding either full-length Nup50 or domain fragments were expressed as GFP fusions in HeLa cells. Proteins with the GFP moiety were then recovered from cell lysates using a previously employed affinity-trap method (21) and probed for the co-precipitation of Nup153. The N-terminal domain of Nup50 robustly isolated Nup153, whereas the central FG domain region (F) or the Ran-binding region (R) had no detectable interaction despite equivalent expression and recovery. Notably, Nup153 associated to a similar degree with both short and long isoforms of Nup50, whether in the context of the full-length protein or the N-terminal domain fragment (Fig. 2).
FIGURE 2.
Nup153 interacts with the N-terminal domains of both Nup50 isoforms. A panel of Nup50-derived and control GFP fusion proteins were expressed in HeLa cells and recovered on an affinity matrix as indicated. Immunoblotting was performed to track the co-recovery of Nup153 (upper panels) along with the recovery of GFP fusion protein (middle left panel and lower right panel). The input levels of these proteins were assessed in parallel (5% loaded for Nup153 and 10% loaded for GFP) (lanes 1–7). The levels of α-tubulin (αTub) were probed (lower left panel) to confirm that equivalent levels of lysate were used for each condition. Molecular mass markers indicated are 130, 100, 70, 55, 40, and 35 kDa. S, short; L, long.
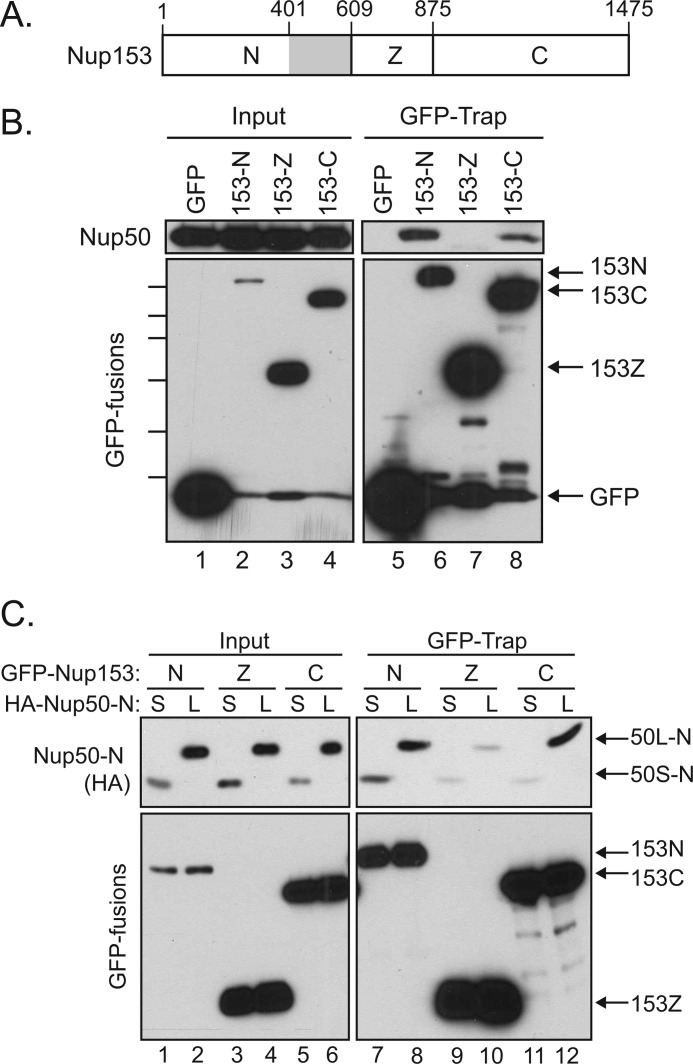
To next determine the interface on Nup153 that mediates interaction with Nup50, fragments of Nup153 corresponding to its unique N-terminal domain (N), tandem zinc finger domain (Z), or FG-rich C-terminal domain (C) were expressed in HeLa cells (Fig. 3A). These GFP fusions proteins were then recovered and probed for the co-isolation of endogenous Nup50. Both the N- and C-terminal domains were found to recover Nup50, although interaction with the C-terminal domain of Nup153 was somewhat less robust (Fig. 3B).
FIGURE 3.
Nup50 interacts with two distinct sites within Nup153. A, schematic of Nup153 with the unique N-terminal region (N), the zinc finger domain (Z), and the FG-rich C-terminal region (C) indicated. The gray box indicates an interface between Nup153 and Nup50 identified in Fig. 4. B, GFP proteins were recovered from lysates of cells expressing GFP alone or GFP fusions of the N-terminal, zinc finger, or C-terminal domain of Nup153. Recovery of Nup50 (upper panels) along with each GFP protein (lower panels) was tracked by immunoblotting. Molecular mass markers indicated are 130, 100, 70, 55, 40, and 35 kDa. C, lysates of cells expressing GFP fusions of Nup153-N, Nup153-Z, or Nup153-C in conjunction with either the HA-tagged Nup50S-N (S) or Nup50L-N (L) were used for a GFP-Trap assay. Immunoblotting was performed to track the co-recovery of the Nup50 fragments (anti-HA; upper panels) along with the recovery of GFP fusion protein (lower panels).
As the N-terminal region of Nup50 accounted for interaction with Nup153 (Fig. 2), we next specifically tested if the Nup50 N-terminal domain binds to these two distinct regions within Nup153. We further wanted to probe both Nup50 isoforms for the two contacts with Nup153. HA-tagged Nup50 N-terminal domain constructs representing both the short and long isoforms were therefore expressed along with the same panel of Nup153-derived GFP fusion proteins. The N-terminal domain from Nup50L indeed interacted with both Nup153-N and Nup153-C (Fig. 3C, lanes 8 and 12). In contrast, whereas the Nup50S N-terminal domain was recovered proportionately to input with Nup153-N, little was recovered with Nup153-C (Fig. 3C, lanes 7 and 11), suggesting that the sequence differences between these isoforms selectively impact one interface with Nup153.
Docking of Nup50 at the Nuclear Pore Is Dependent on Its Interaction with the N-terminal Interface on Nup153
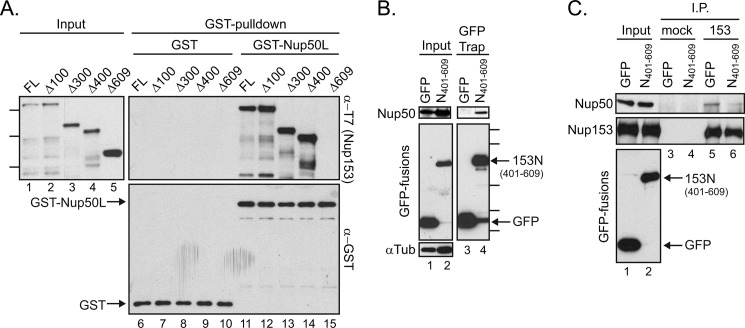
To test for direct binding as well as to map the determinants of Nup50 binding to Nup153 more finely, we produced a series of N-terminal truncations of Nup153 in E. coli. Bacterial lysates were then incubated with GST-Nup50L or GST alone. Full-length Nup153 and several constructs with residues truncated from the N-terminal end of this nucleoporin were recovered specifically with GST-Nup50L (Fig. 4A, lanes 11–14), strongly suggesting a direct interaction between Nup50 and Nup153. When the full-length N-terminal domain of Nup153 was deleted (Δ609), the Nup50-binding activity lost (Fig. 4A, lane 15). In contrast, truncation of the first 400 amino acids (Δ400) retained full binding capacity (Fig. 4A, lane 14). This result identifies residues between positions 401 and 609 (Fig. 3A, gray box) as necessary for this interaction. To determine whether these residues are sufficient for interaction with Nup50, this small region was expressed as a fusion protein with GFP and recovered as before. The co-isolation observed for endogenous Nup50L with Nup153(401–609) (Fig. 4B, lane 4) establishes this distal part of the Nup153 N-terminal domain as sufficient for contact with Nup50. Expressing a GFP fusion of this fragment was also found to reduce recovery of Nup50 in a Nup153 immunoprecipitation (Fig. 4C). Although contact between Nup153-N and Nup50 may not be restricted to this region, this experiment confirms the necessity of this interface for Nup50-Nup153 interaction.
FIGURE 4.
Amino acids 401–609 within Nup153 are necessary and sufficient for contact with Nup50. A, bacterial lysates containing a panel of N-terminally truncated, recombinantly expressed Nup153 constructs were incubated with GST (lanes 6–10) or GST-Nup50L (lanes 11–15), followed by affinity purification on glutathione-Sepharose beads. Co-purifying Nup153-derived proteins were tracked by immunoblotting with the anti-T7 antibody (left panel and upper right panel), and recovery of GST proteins themselves was monitored by immunoblotting with anti-GST antibody (lower right panel). Molecular mass markers indicated are 170, 130, and 70 kDa. FL, full-length. B, GFP was recovered from lysates of cells expressing GFP alone or a GFP fusion with amino acids 401–609 of Nup153. Recovery of any co-isolated Nup50 (upper panels) along with each GFP protein (middle left panel and lower right panel) was tracked by immunoblotting. The levels of α-tubulin (αTub) were tracked to ensure equivalent loading of samples (lower left panel). Molecular mass markers indicated are 100, 70, 40, 35, and 25 kDa. C, Nup153 was immunoprecipitated (I.P.) from lysates of cells expressing GFP or GFP-Nup153(401–609) (lanes 5 and 6); material isolated with equivalent levels of protein A-Sepharose beads alone was run alongside (lanes 3 and 4). The precipitated material was immunoblotted for the presence of Nup153 and Nup50. For comparison, samples of the input material are shown (lanes 1 and 2).
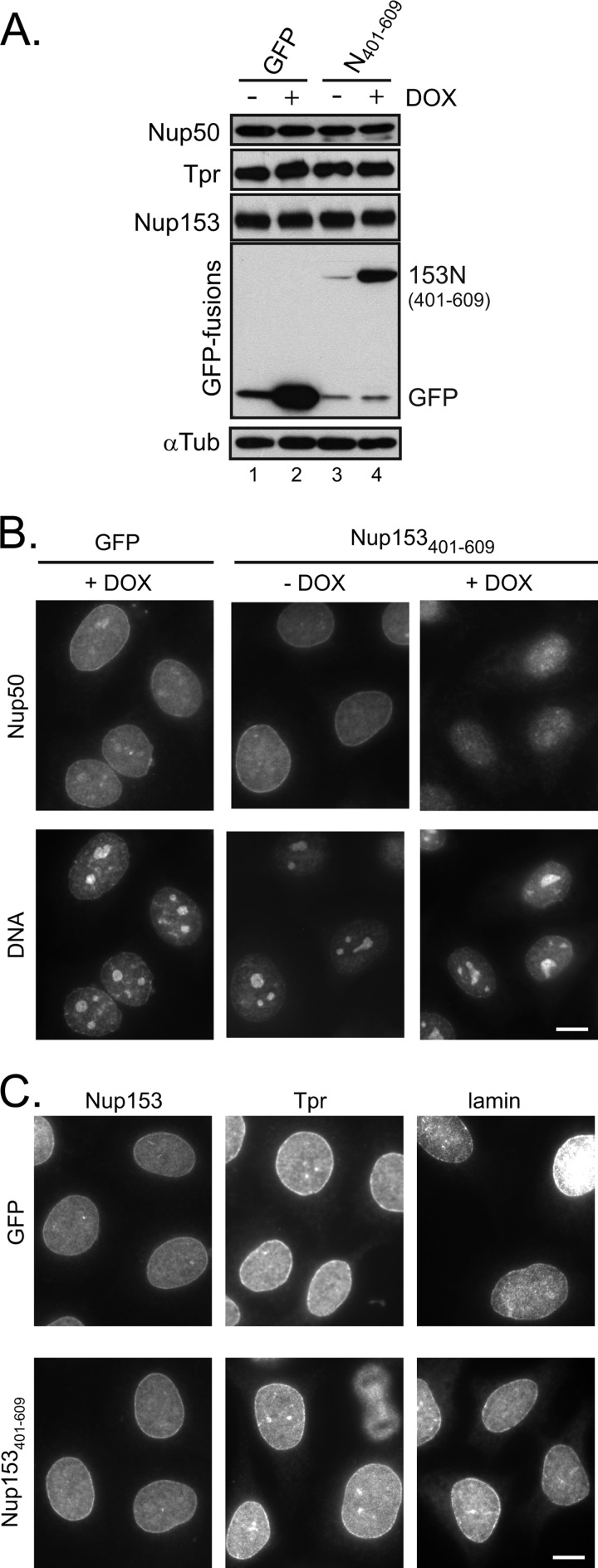
Next, to investigate the contribution of this interface to the localization of Nup50, we engineered HeLa cells to inducibly express the minimal binding fragment (Nup153(401–609)). Induction of this fragment did not alter the levels of Nup50 or other basket nucleoporins (Fig. 5A). Under non-induced conditions, Nup50 was detected throughout the nucleus and at the nuclear rim (Fig. 5B), consistent with its dynamic localization at the NPC (7) and its additional nucleoplasmic roles (22). The specificity of this pattern was confirmed by pretreating cells with Nup50-specific siRNA and similarly performing immunolocalization of Nup50 (supplemental Fig. S2). A reduction in signal at both the rim and nucleoplasm was seen using two independent siRNAs. In the presence of doxycycline-induced GFP-Nup153(401–609), the population of Nup50 at the rim of the nucleus was notably reduced (Fig. 5B) compared with either the non-induced cells or cells induced to express GFP alone. This implicates the Nup153 N-terminal site of interaction as a key determinant of Nup50 NPC targeting. Importantly, expression of this fragment did not alter the localization of Nup153 itself, and it did not interfere with the localization of the nuclear pore basket component Tpr (Fig. 5C), which binds an adjacent region of Nup153 (6).
FIGURE 5.
Disruption of Nup50-Nup153 interaction prevents nuclear rim localization of Nup50. A, HeLa cells were engineered to express either GFP or GFP-Nup153(401–609) in response to doxycycline (DOX). Cells that were either untreated or incubated with doxycycline for 24 h were harvested, and the cell lysates were probed for levels of Nup50, Tpr, and Nup153 as indicated. The induction was confirmed by tracking GFP fusion proteins, and α-tubulin (αTub) was tracked to ensure equivalent loading of samples. B, following doxycycline-induced expression of GFP (left panels) or before (middle panels) and after (right panels) similarly induced GFP-Nup153(401–609) expression, the localization of Nup50 was tracked by indirect immunofluorescence, with DNA detection by DAPI staining shown in the accompanying panels. C, the nucleoporins Nup153 and Tpr, as well as lamin, were detected by indirect immunofluorescence under similar conditions in which either GFP or GFP-Nup153(401–609) had been induced by doxycycline. Scale bars = 10 μm.
A Second Site of Interaction between Nup153 and Nup50 Is Bridged by Importin α
The lack of detectable binding upon deletion of the Nup153 N-terminal domain in the context of recombinant proteins (Fig. 4A) suggested that interaction between Nup50 and the Nup153 C-terminal domain requires an additional factor(s). Moreover, any bridging factors are likely to interact differentially with the short and long isoforms of the Nup50, as the isoforms differ in their interaction with Nup153-C (Fig. 3C). These data pointed to importin α as a good candidate for modulating the interaction of Nup50 with Nup153-C. Both binding segments BS1 and BS2 make contact with importin α (8, 12), but BS1 is not present in the short form of Nup50 (Fig. 1A). The resulting absence of contact sites for Nup50S in the minor NLS-binding site of importin α results in lower affinity between the short Nup50 isoform and importin α compared with the long isoform (Fig. 6B, lane 7 versus lane 9).
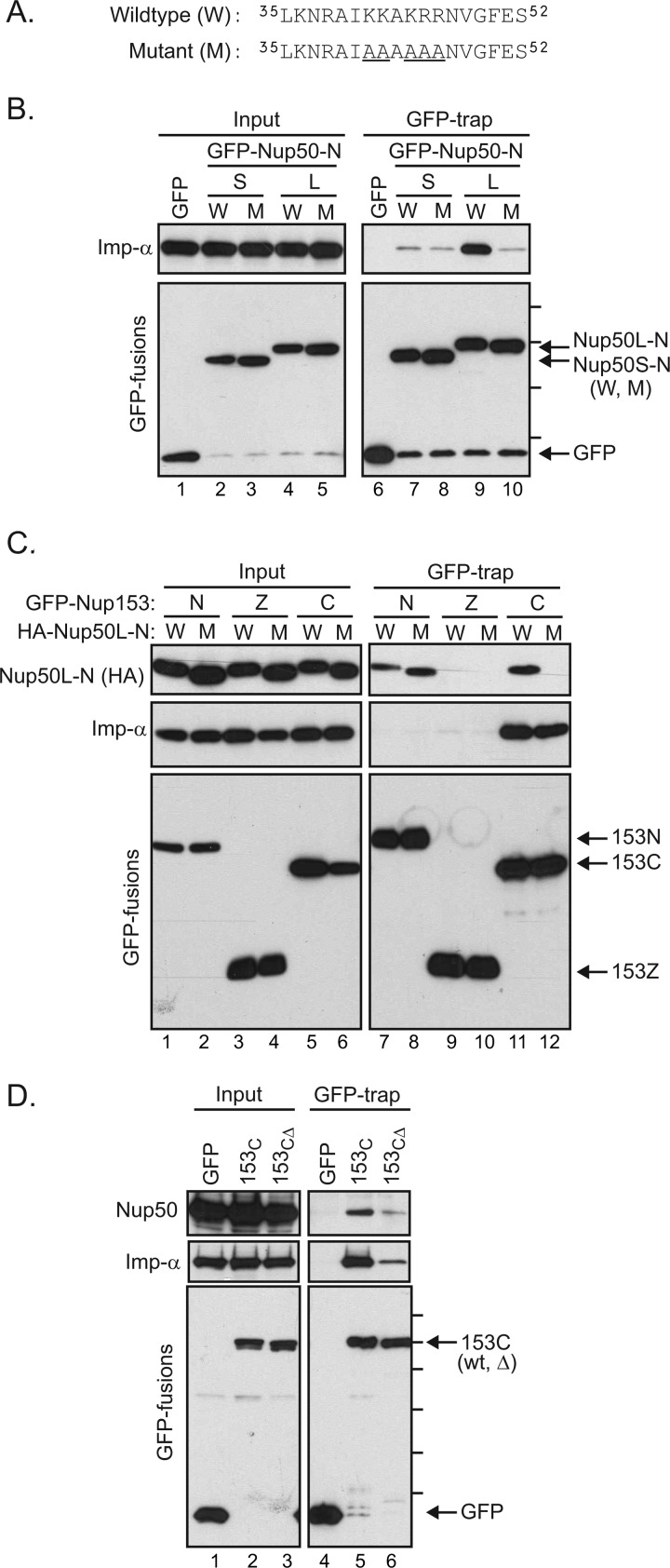
FIGURE 6.
Importin α mediates an interaction between the C-terminal tail of Nup153 and Nup50. A, the sequence context of mutations (underlined) used to test the role of the N-terminal region of Nup50 (9). Mutations were made in HA-tagged Nup50 N-terminal domain constructs. B, GFP fusions with the Nup50 N-terminal domain from the short (S) and long (L) isoforms in either the wild-type (W) or mutant (M) form were expressed. Material recovered by GFP-Trap was then immunoblotted to detect association of importin α (Imp-α). The molecular mass markers indicated are 70, 55, 40, and 30 kDa. C, following coexpression of Nup153 domain constructs (N, Z, and C) fused to GFP along with HA-tagged Nup50L-N in either the wild-type or mutant form, GFP proteins were recovered from cell lysates. GFP proteins, as well as co-isolating Nup50-N and endogenous importin α, were tracked by immunoblotting as indicated (inputs are 2% for HA-Nup50-N, 4% for importin α, and 8% for GFP). D, GFP fusion proteins were recovered from lysates of cells expressing GFP alone, a GFP fusion with the Nup153 C-terminal region, or a GFP fusion with a truncated version of the Nup153 C-terminal domain lacking the terminal 18 amino acids, previously defined as an importin α-binding motif. Recovery of endogenous Nup50 and importin α, along with each GFP protein, was tracked by immunoblotting as indicated (inputs are 6% for Nup50, 16% importin α, and 8% for GFP). Molecular mass markers indicated are 130, 100, 70, 55, 40, and 35 kDa.
To test for a role of importin α in the Nup50-Nup153 interaction, residues within the BS2 region of Nup50 critical to the Nup50-importin α interface (8) were mutated. This five-amino acid substitution (Fig. 6A) was previously found to disrupt association with importin α (Fig. 6B, lane 9 versus lane 10) (9, 11). Epitope (HA)-tagged versions of the Nup50L N-terminal domain in either wild-type or mutant form were coexpressed with GFP fusion proteins of Nup153 domain fragments (Fig. 6C). Recovery of these GFP fusion proteins again revealed an interaction with both the N- and C-terminal regions of Nup153 (Fig. 6C). Interestingly, whereas both wild-type and mutant Nup50 associated equivalently with Nup153-N (Fig. 6C, lanes 7 and 8), mutation of Nup50 abrogated its association with Nup153-C (lane 12). Thus, binding of importin α by Nup50 appears to aide specifically in mediating the association of the Nup153 C-terminal domain and Nup50.
The C-terminal region of Nup153 is FG-rich. Nup domains rich in this motif share the ability to interact with import and export transport receptors (23, 24), suggesting that this general property may confer a Nup50/importin α/β-docking site. In addition to the general affinity for karyopherin family members via its FG repeats, however, Nup153 has been noted to have a specific binding site for importin α (25, 26). This independent motif is found in the terminal residues of Nup153 (amino acids 1458–1475). To determine whether Nup50 docking at the C-terminal region of Nup153 relies on this specific binding site for importin α or whether the general properties of the FG-rich terminal domain confer this indirect interaction with Nup50, we compared the recovery of endogenous Nup50L with GFP fusions of the Nup153 C-terminal domain in its wild-type form or with a 18-amino acid truncation of the terminal residues (153CΔ). Recovery of Nup50 was markedly decreased by truncation of Nup153-C (Fig. 6D). As expected, recovery of importin α was decreased by this mutation. Of note, this was the case for a range of importin α isoforms (supplemental Fig. S3). Probing for importin β revealed that it too relies on these residues in Nup153 for robust interaction (supplemental Fig. S3); presumably, the more general interaction with FG repeat domains is below the threshold of detection in this pulldown assay. Together, this mutational analysis supports the notion that importin α bridges Nup50 to Nup153 specifically at a discrete region at the C terminus of Nup153.
Efficient Nuclear Import Relies on the Context of Nup153 for Nup50 Function
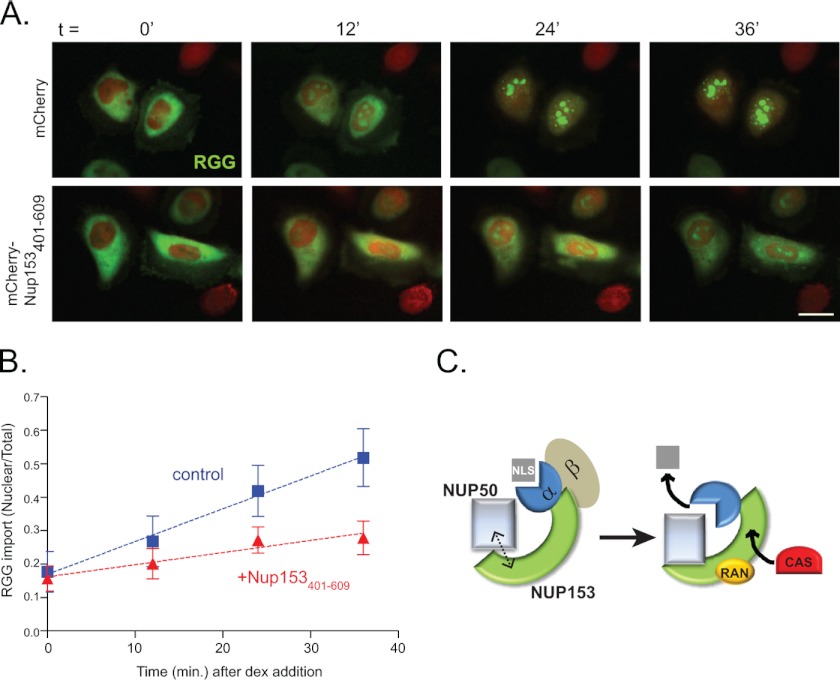
To test the importance of contact between Nup153 and Nup50, HeLa cells were transfected with the Nup153(401–609) fragment to interfere with this partnership (Fig. 4C). Nuclear import was then assessed by tracking a cotransfected glucocorticoid-responsive import cargo, RGG (17). Using low levels of dexamethasone and tracking import in real time (Fig. 7, A and B), we observed that nuclear accumulation of the RGG cargo was notably slower when the Nup50-Nup153 interaction was disrupted by Nup153(401–609). This was also the case in an independent experiment tracked at intervals with fixed samples (data not shown). These results suggest that the contribution of Nup50 to transport efficiency relies on its interaction with Nup153.
FIGURE 7.
Nup153-Nup50 interaction is required for efficient nuclear import. A, frames from a live imaging movie showing import kinetics of RGG (green) in cells expressing mCherry (upper panels) or mCherry-Nup153(401–609) (lower panels) at 0, 12, 24, and 36 min after addition of dexamethasone (dex). Scale bar = 10 μm. B, quantification of raw fluorescence signal graphed as nuclear to total ratios (the mean for 25 control (blue squares) and 31 Nup153(401–609)-expressing (red triangles) cells), with error bars representing S.D. C, a working model is depicted in which two interfaces between the N-terminal region of Nup50 and Nup153, one mediated by importin α, aide in releasing the NLS cargo. Interaction between Nup153 and soluble factors CAS and Ran may create local concentrations that facilitate rapid capture of importin α for export.
DISCUSSION
Taken together, these results suggest a model in which Nup153 works as a scaffold for Nup50 by means of an anchoring interaction at the Nup153 N-terminal domain in conjunction with an indirect interaction with the distal C terminus of Nup153 via importin α (Fig. 7C). The role of Nup153 as a scaffold likely extends to its ability to bind other participants in the nuclear import cycle as well, such as Ran and CAS (cellular apoptosis susceptibility protein) (27, 28). Thus, at any one time, a limited population of Nup50 (which includes and may be restricted to the population dynamically localized to the nuclear pore) is in a local environment where factors necessary for delivery of the NLS cargo and recycling of importins α and β are readily accessible to facilitate this process efficiently. Consistently, importin α interaction with the Nup153 C terminus was found to be important for efficient import (26). The results reported here also underscore a distinction between Nup50S and Nup50L that may underlie certain functional differences (11). Specifically, Nup50S lacks determinants that contribute to a second site of interaction with Nup153.
The C-terminal importin α-docking site on Nup153 is highly conserved across species (29) and has affinity for a range of importin α isoforms (supplemental Fig. S3) (29). In the context of the Saccharomyces cerevisiae basket nucleoporin Nup1p, this tail region has been found to be important for the efficiency of import (30). In mammalian cells, this motif has been proposed to contribute to export of importin α (25). In both cases, association was shown to take place (even enhanced) between the Nup protein and importin α in the presence of the NLS peptide cargo (25, 26). In the context of Nup50L docked at the N-terminal domain of Nup153, the NLS cargo would be subject to displacement by the high affinity binding of the Nup50L N-terminal region (BS1 and BS2) as described (12). Although previous studies have indicated that Nup153 is not required for import to take place, subtle effects on import were noted in mammalian cells (16, 31) and characterized in Drosophila cells (32) as well in nuclei reconstituted in Xenopus egg extracts (33). The results reported here are consistent with a role for Nup153 in the efficiency of import, perhaps by promoting the Nup50-importin α interaction in a spatially restricted manner.
Intriguingly, the more recent analysis of importin α binding at the Nup153 C terminus suggested that there could be a non-canonical role for importin α at this site (26). The model proposed, in which importin α can occupy this site and contribute to import without binding the NLS cargo itself, could further influence the interpretation of the Nup153-Nup50 interaction. For instance, two molecules of Nup50 could be in contact with Nup153 (one at region 401–609 and one bridged by importin α at the C terminus), playing separate roles in transport. It is also important to keep in mind that RGG is a complex cargo, with multiple sequences that can direct transport, including those that bind importins α and 7 (34). For all of these reasons, to continue moving beyond the core machinery of transport and to understand regulatory layers that ensure efficiency, further study will be required.
In Nup124p (Schizosaccharomyces pombe), this C-terminal motif was found to be critical retrotransposon activity, specifically for Tf1-Gag import (29). Interestingly, the other region of Nup124p critical to retrotransposon function shares homology with Nup153 as well, in this case, at the site within the N-terminal domain of Nup153 (residues 448–634) that we found here is important for the interface with Nup50 (35). This dual mode of interaction with Nup153 (or similar Nup proteins in other species) may be a paradigm used more generally, as two distinct types of Nup153-binding proteins, lamins (36) and NPC-associated SUMO (small ubiquitin-like modifier) proteases (37), have each recently been found to interact with sites in both the N- and C-terminal regions of Nup153. The characterization here of interaction regions within both the N- and C-terminal domains of Nup153 with respect to Nup50 provides an important framework for understanding the cooperative roles of these dynamic nucleoporins.
Acknowledgments
The SA1 (anti-Nup153) hybridoma was generously provided by Dr. Brian Burke (Institute of Medical Biology, Singapore). We gratefully acknowledge Dr. Akihiko Kuniyasu for generous support.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM61275 (to K. S. U.). Shared resources used in this work are supported in part by National Institutes of Health Grant P30 CA042014 awarded to the Huntsman Cancer Institute.

This article contains supplemental Figs. S1–S3.
- NPC
- nuclear pore complex
- NLS
- nuclear localization signal
- EGFP
- enhanced GFP
- RGG
- Rev-GFP-glucocorticoid receptor
- BS1/2
- binding segment 1/2.
REFERENCES
- 1. Terry L. J., Shows E. B., Wente S. R. (2007) Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416 [DOI] [PubMed] [Google Scholar]
- 2. Fan F., Liu C. P., Korobova O., Heyting C., Offenberg H. H., Trump G., Arnheim N. (1997) cDNA cloning and characterization of Npap60: a novel rat nuclear pore-associated protein with an unusual subcellular localization during male germ cell differentiation. Genomics 40, 444–453 [DOI] [PubMed] [Google Scholar]
- 3. Müller D., Thieke K., Bürgin A., Dickmanns A., Eilers M. (2000) Cyclin E-mediated elimination of p27 requires its interaction with the nuclear pore-associated protein mNPAP60. EMBO J. 19, 2168–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smitherman M., Lee K., Swanger J., Kapur R., Clurman B. E. (2000) Characterization and targeted disruption of murine Nup50, a p27Kip1-interacting component of the nuclear pore complex. Mol. Cell. Biol. 20, 5631–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guan T., Kehlenbach R. H., Schirmer E. C., Kehlenbach A., Fan F., Clurman B. E., Arnheim N., Gerace L. (2000) Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol. 20, 5619–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hase M. E., Cordes V. C. (2003) Direct interaction with Nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell 14, 1923–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rabut G., Doye V., Ellenberg J. (2004) Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6, 1114–1121 [DOI] [PubMed] [Google Scholar]
- 8. Matsuura Y., Stewart M. (2005) Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 24, 3681–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindsay M. E., Plafker K., Smith A. E., Clurman B. E., Macara I. G. (2002) Npap60/Nup50 is a tri-stable switch that stimulates importin α/β-mediated nuclear protein import. Cell 110, 349–360 [DOI] [PubMed] [Google Scholar]
- 10. Moore M. S. (2003) Npap60: a new player in nuclear protein import. Trends Cell Biol. 13, 61–64 [DOI] [PubMed] [Google Scholar]
- 11. Ogawa Y., Miyamoto Y., Asally M., Oka M., Yasuda Y., Yoneda Y. (2010) Two isoforms of Npap60 (Nup50) differentially regulate nuclear protein import. Mol. Biol. Cell 21, 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pumroy R. A., Nardozzi J. D., Hart D. J., Root M. J., Cingolani G. (2012) Nucleoporin Nup50 stabilizes closed conformation of armadillo repeat 10 in importin α5. J. Biol. Chem. 287, 2022–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kosako H., Yamaguchi N., Aranami C., Ushiyama M., Kose S., Imamoto N., Taniguchi H., Nishida E., Hattori S. (2009) Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat. Struct. Mol. Biol. 16, 1026–1035 [DOI] [PubMed] [Google Scholar]
- 14. Dimaano C., Ball J. R., Prunuske A. J., Ullman K. S. (2001) RNA association defines a functionally conserved domain in the nuclear pore protein Nup153. J. Biol. Chem. 276, 45349–45357 [DOI] [PubMed] [Google Scholar]
- 15. Kaiser C., Dobrikova E. Y., Bradrick S. S., Shveygert M., Herbert J. T., Gromeier M. (2008) Activation of cap-independent translation by variant eukaryotic initiation factor 4G in vivo. RNA 14, 2170–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mackay D. R., Elgort S. W., Ullman K. S. (2009) The nucleoporin Nup153 has separable roles in both early mitotic progression and the resolution of mitosis. Mol. Biol. Cell 20, 1652–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Love D. C., Sweitzer T. D., Hanover J. A. (1998) Reconstitution of HIV-1 Rev nuclear export: independent requirements for nuclear import and export. Proc. Natl. Acad. Sci. U.S.A. 95, 10608–10613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moorhead G. B., Trinkle-Mulcahy L., Nimick M., De Wever V., Campbell D. G., Gourlay R., Lam Y. W., Lamond A. I. (2008) Displacement affinity chromatography of protein phosphatase 1 (PP1) complexes. BMC Biochem. 9, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vagnarelli P., Ribeiro S., Sennels L., Sanchez-Pulido L., de Lima Alves F., Verheyen T., Kelly D. A., Ponting C. P., Rappsilber J., Earnshaw W. C. (2011) Repo-Man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev. Cell 21, 328–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., Sironi L., Ellenberg J. (2008) Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M. C., Leonhardt H. (2008) A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7, 282–289 [DOI] [PubMed] [Google Scholar]
- 22. Kalverda B., Pickersgill H., Shloma V. V., Fornerod M. (2010) Nucleoporins directly stimulate expression of developmental and cell cycle genes inside the nucleoplasm. Cell 140, 360–371 [DOI] [PubMed] [Google Scholar]
- 23. Bayliss R., Littlewood T., Stewart M. (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin β in nuclear trafficking. Cell 102, 99–108 [DOI] [PubMed] [Google Scholar]
- 24. Shah S., Tugendreich S., Forbes D. (1998) Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol. 141, 31–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moroianu J., Blobel G., Radu A. (1997) RanGTP-mediated nuclear export of karyopherin α involves its interaction with the nucleoporin Nup153. Proc. Natl. Acad. Sci. U.S.A. 94, 9699–9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ogawa Y., Miyamoto Y., Oka M., Yoneda Y. (2012) The interaction between importin α and Nup153 promotes importin α/β-mediated nuclear import. Traffic 13, 934–946 [DOI] [PubMed] [Google Scholar]
- 27. Nakielny S., Shaikh S., Burke B., Dreyfuss G. (1999) Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 18, 1982–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ball J. R., Ullman K. S. (2005) Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma 114, 319–330 [DOI] [PubMed] [Google Scholar]
- 29. Sistla S., Pang J. V., Wang C. X., Balasundaram D. (2007) Multiple conserved domains of the nucleoporin Nup124p and its orthologs Nup1p and Nup153 are critical for nuclear import and activity of the fission yeast Tf1 retrotransposon. Mol. Biol. Cell 18, 3692–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pyhtila B., Rexach M. (2003) A gradient of affinity for the karyopherin Kap95p along the yeast nuclear pore complex. J. Biol. Chem. 278, 42699–42709 [DOI] [PubMed] [Google Scholar]
- 31. Zhou L., Panté N. (2010) The nucleoporin Nup153 maintains nuclear envelope architecture and is required for cell migration in tumor cells. FEBS Lett. 584, 3013–3020 [DOI] [PubMed] [Google Scholar]
- 32. Sabri N., Roth P., Xylourgidis N., Sadeghifar F., Adler J., Samakovlis C. (2007) Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J. Cell Biol. 178, 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walther T. C., Fornerod M., Pickersgill H., Goldberg M., Allen T. D., Mattaj I. W. (2001) The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring, and import of a subset of nuclear proteins. EMBO J. 20, 5703–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Freedman N. D., Yamamoto K. R. (2004) Importin 7 and importin α/importin β are nuclear import receptors for the glucocorticoid receptor. Mol. Biol. Cell 15, 2276–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Varadarajan P., Mahalingam S., Liu P., Ng S. B., Gandotra S., Dorairajoo D. S., Balasundaram D. (2005) The functionally conserved nucleoporin Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr. Mol. Biol. Cell 16, 1823–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Al-Haboubi T., Shumaker D. K., Köser J., Wehnert M., Fahrenkrog B. (2011) Distinct association of the nuclear pore protein Nup153 with A- and B-type lamins. Nucleus 2, 500–509 [DOI] [PubMed] [Google Scholar]
- 37. Chow K. H., Elgort S., Dasso M., Ullman K. S. (2012) Two distinct sites in Nup153 mediate interaction with the SUMO proteases SENP1 and SENP2. Nucleus 3, 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]