Background: The measles virus hemagglutinin stalk transmits the signal that triggers membrane fusion.
Results: Functional analyses based on covalent tetramerization followed by disulfide bond reduction identify three modules in the upper half of the stalk.
Conclusion: The modules have following functions (top-to-bottom): linker, spacer/signal conduction, contact with the F-trimer.
Significance: Modules with similar structure and function may exist in attachment protein stalks of other paramyxoviruses.
Keywords: Animal Viruses, Membrane Fusion, Negative-strand RNA Viruses, Virology, Virus Entry, Hemagglutinin, Measles Virus
Abstract
The measles virus (MV) fusion apparatus consists of a fusion protein and an attachment protein named hemagglutinin (H). After receptor-binding through its cuboidal head, the H-protein transmits the fusion-triggering signal through its stalk to the fusion protein. However, the structural basis of signal transmission is unclear because only structures of H-heads without their stalk have been solved. On the other hand, the entire ectodomain structure of the hemagglutinin-neuraminidase protein of another Paramyxovirus revealed a four-helix bundle stalk. To probe the structure of the 95-residue MV H-stalk we individually substituted head-proximal residues (positions 103–153) with cysteine, and biochemically and functionally characterized the resultant proteins. Our results indicate that most residues in the central segment (positions 103–117) can be cross-linked by engineered disulfide bonds, and thus may be engaged in a tetrameric structure. While covalent tetramerization disrupts fusion triggering function, disulfide bond reduction restores it in most positions except Asp-113. The next stalk segment (residues 123–138) also has high propensity to form covalent tetramers, but since these cross-links have little or no effect on function, it can conduct the fusion-triggering signal while remaining in a stabilized tetrameric configuration. This segment may act as a spacer, maintaining H-heads at an optimal height. Finally, the head-proximal segment (residues 139–154) has very limited propensity to trap tetramers, suggesting bifurcation into two flexible linkers clamped by inter-subunit covalent links formed by natural Cys-139 and Cys-154. We discuss the modular structure of the MV H-stalk in the context of membrane fusion triggering and cell entry by Paramyxoviruses.
Introduction
Measles virus (MV)2 is an enveloped, negative-strand RNA virus of the family Paramyxoviridae (1). This family includes deadly emerging viruses like Hendra and Nipah, and prevalent human pathogens like mumps, parainfluenza, and respiratory syncytial viruses that still cause significant morbidity and mortality (2). MV, while long targeted for eradication (3), still affects 10 million people annually and killed 139,000 worldwide in 2010 (4).
While many enveloped viruses take advantage of low pH (5) or proteases (6) in the endosomal compartment to trigger membrane fusion, most Paramyxoviruses including MV fuse directly with the plasma membrane (2, 7). The membrane fusion apparatus of the Paramyxoviruses is composed of two glycoproteins, the attachment and fusion (F) proteins. The attachment protein is named H (hemagglutinin) for MV and other members of the genus Morbilliviruses (8), G for the Henipaviruses (9), and HN (hemagglutinin-neuraminidase) for most other Paramyxoviruses (2). The attachment proteins are responsible for receptor binding and subsequent F-activation. While HN binds sialic acid at the cell surface to effect cell entry, H and G-proteins bind proteinaceous receptors (10–12). Nevertheless, certain G-HN chimeric proteins are functional (13, 14) suggesting a modular architecture of the fusion apparatus of Paramyxoviruses, and conserved fundamental characteristics of their fusion triggering mechanism (15).
MV H is a 617-amino acid type II glycoprotein comprised of a 34-residue cytoplasmic tail, a membrane spanning segment, and an ectodomain with a 95-residue stalk and a six-bladed β-propeller head domain that contacts the receptors (16–18). The presumably tetrameric stalk holds together a pair of H-head dimers, which are stabilized by a disulfide bond between Cys-154 at their base (16) and a second inter-subunit disulfide bond at Cys-139 in the stalk (19). While both H-head dimer structures reported to date exhibit an identical dimer interface (16, 20), it is unclear how the dimers may interact to form tetramers: only incomplete H-heads missing their stalks and stalk-proximal residues crystallized with two alternative tetrameric interfaces (21). Intriguingly, one of these H-proteins crystallized in a hexameric space group, and this structure could only be solved at lower resolution (21).
On the other hand, recently a partial ectodomain structure of the HN-protein of Newcastle disease virus (NDV) (22), and a stalk structure of the human parainfluenza virus 5 (PIV5) (23) were solved, revealing four-helix bundle (4HB) stalk structures. Remarkably, in the NDV structure the head-dimers interact with one flank of the stalks, rather than forming tetrameric contacts above it (22). This stalk-adherent conformation of the heads has been named “heads-down”, to differentiate it from the “heads-up” conformation with H-heads forming tetramers above the stalk. It is possible that the pre-triggering MV H-tetramer conformation is “heads-down.”
Another critical insight gained from the HN-stalk structural analyses is segmentation: part of the lower half is a supercoiled 4HB with 3-to-4 heptad repeats; the next 25 residues are in a straight 11-mer repeat conformation; and since no density was observed for the 7–9 head-proximal residues a flexible linker was postulated (22, 23). These and other observations suggested the NDV HN stalk-extension model of fusion triggering: receptor pulling could cause the lower half of the stalk to uncoil and extend, destroying the pre-formed interactions of the central segment with the F-trimer and triggering its refolding resulting in membrane fusion (22).
The MV H-stalk starts with a membrane-proximal charged residue (Arg-59) (24) and ends with Cys-154 covalently linking the base of the head-dimer (16). Interestingly, this stalk is predicted to be 15–20 residues longer than the HN-stalks. In analogy with the NDV HN-stalk, it may be comprised of a membrane proximal module that acts as a spring, a central module that interacts with the F-trimer and primes it for refolding, and an upper segment that connects with the dimeric H-heads. Indeed, functional analyses of the central segment of the MV and canine distemper virus (CDV) H-stalks are consistent with F-triggering function of this segment (25, 26). Here we focused our analyses on the structure and function of the upper half of the stalk.
To probe the structure of the H-stalk and to identify segments with different function we sought to covalently cross-link residues of different subunits. Toward tetramer trapping, after aligning the HN- and H-stalks and obtaining a structure-based model, we performed comprehensive Cys mutagenesis from the predicted supercoiled-to-straight transition point (residue 103) to the base of the head (residue 153).
Indeed, we observed that certain Cys substitutions near the center of the stalk (residues 103–117) supported covalent tetramerization that interfered with function. Importantly, fusion function was partially restored by reduction of the disulfide bonds, consistent with similar results from the anaylsis of the CDV H-stalk (26). On the other hand, cross-linking the segment between residues 123 and 138 occurred efficiently with little or no impact on function. Finally, Cys substitutions between the natively occurring Cys-139 and Cys-154, clamping the top of the stalk, exhibited limited propensity to trap covalent tetramers. These data identify three functional modules in the upper half of the MV H-stalk.
EXPERIMENTAL PROCEDURES
Cells, Plasmids, and Mutagenesis
Vero (African green monkey kidney) cells were maintained in Dulbecco modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). All H-protein mutants were generated in the pCG-H plasmid by the QuikChange site-directed mutagenesis (Stratagene) protocol according to the manufacturer's instructions. All Cys substitution mutants were introduced in a vaccine-lineage H-protein backbone (H-NSe) (27, 28). The integrity of the clones was confirmed by sequencing the H-protein gene in the vicinity of the mutation.
Mammalian Cell Transfection and Determination of the H-protein Oligomeric State
Lipofectamine 2000 (Invitrogen) was used for Vero cell transfections according to the manufacturer's instructions. The standard pCG-H (27) or mutated plasmids (2 μg) were transfected in to Vero cells seeded in 12-well plates (1.5 × 105 cells). Twenty-four hours post-transfection cells were washed three times with phosphate buffered saline (PBS) and lysed using RIPA buffer (50 mm Tris, pH 8.0, 62.5 mm EDTA, 0.4% deoxycholate, 1% Igepal, and 1 mm phenylmethylsulfonyl fluoride) supplemented with 50 mm iodoacetamide and Complete protease inhibitor mixture (Roche, Basel, Switzerland). Samples were clarified, boiled in Laemmli buffer, separated on 4–15% gradient SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (PVDF) (Millipore, Billerica, MA). The membranes were probed with a polyclonal antibody specific for the H-cytoplasmic tail (29) and visualized by an ECL Plex Cy3-conjugated secondary antibody on the Typhoon FLA 7000 imager (GE healthcare, Piscataway, NJ). Image analysis software ImageQuant TL was used for quantification of protein band intensity.
Fusion Assays and the Partial Reduction of Disulfide Bonds to Rescue Fusion Function
The semi-quantitative fusion assay was done as described previously (17). Briefly, 0.8 μg each of pCG-H, the fusion protein expression plasmid, pCG-F (27) and peGFP (enhanced GFP expression plasmid) were transfected into 1.5 × 105 Vero cells. Fusion scores were determined 24 h after transfection. A fusion score of 3 denotes wild-type fusion levels (in average more than 15 nuclei per syncytium), 2 denotes 5–15 nuclei per syncytium, 1 denotes 3–5 nuclei per syncytium, and 0 denotes less than 3 nuclei per syncytium. To rescue the fusion function of Cys substitution mutants, the Vero cells were transfected as described and then incubated with 200 nm FIP (fusion inhibitory peptide) (30, 31). Twenty-four hours after transfection Vero cells were washed with PBS and treated with 15 mm dithiothreitol (DTT) or PBS for 30 min. Fusion was recorded 3 h after DTT treatment.
Sequence Alignment and H-stalk Homology Model
The three-dimensional model of the MV H protein stalk from residues 75 to 127 was built based on the recently published crystal structures of PIV5 (23) and NDV (22). A sequence alignment with default parameters by ClustalW (32) between NDV, PIV5, and MV H sequences produced a gap of 4 residues in the NDV and PIV5 sequences corresponding to the residues IIGD in MV H stalk. This gap was reduced to a one residue gap at residue Asp-101 in MV H by a comparison of the buried residues in the crystal structures of the NDV and PIV5 stalk regions to hydrophobic residues in the MV and CDV H-stalk sequences (Fig. 1B). In this alignment the position of Pro-108 is absolutely conserved among all Morbilliviruses and the residue Asp-101 in MV H corresponds to the kink in the helical regions of the template structures. The Getarea program (33) was used to calculate the positions of inside-outside residues in the structural alignment between NDV and PIV5 stalk region. The homology model package MPACK was used to build the model structure of MV H stalk monomer from residue Ser-75 to Pro-127. The model structure obtained from MPACK was further energy minimized using the FANTOM program. Each monomer model structure of the MV H-stalk was then fitted into the PIV5 stalk-tetramer crystal structure to obtain the model structure of MV H tetramer-stalk. NAMD (34) software tool was used to energy minimize the MV H tetramer-stalk model using TIP3P water molecules for 10,000 steps. We note that both our model and an independently generated model of the CDV stalk (26) predicted the same alignment upstream of residue 91 and downstream of residue 101. Residues 91–100 are in a different helical phase due to the insertion of a one-residue gap at different positions. Nevertheless, the supercoiled-to-straight transition was consistently predicted at Val-103.
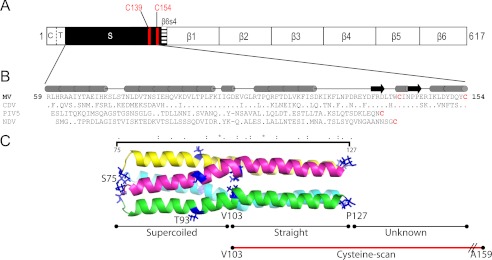
FIGURE 1.
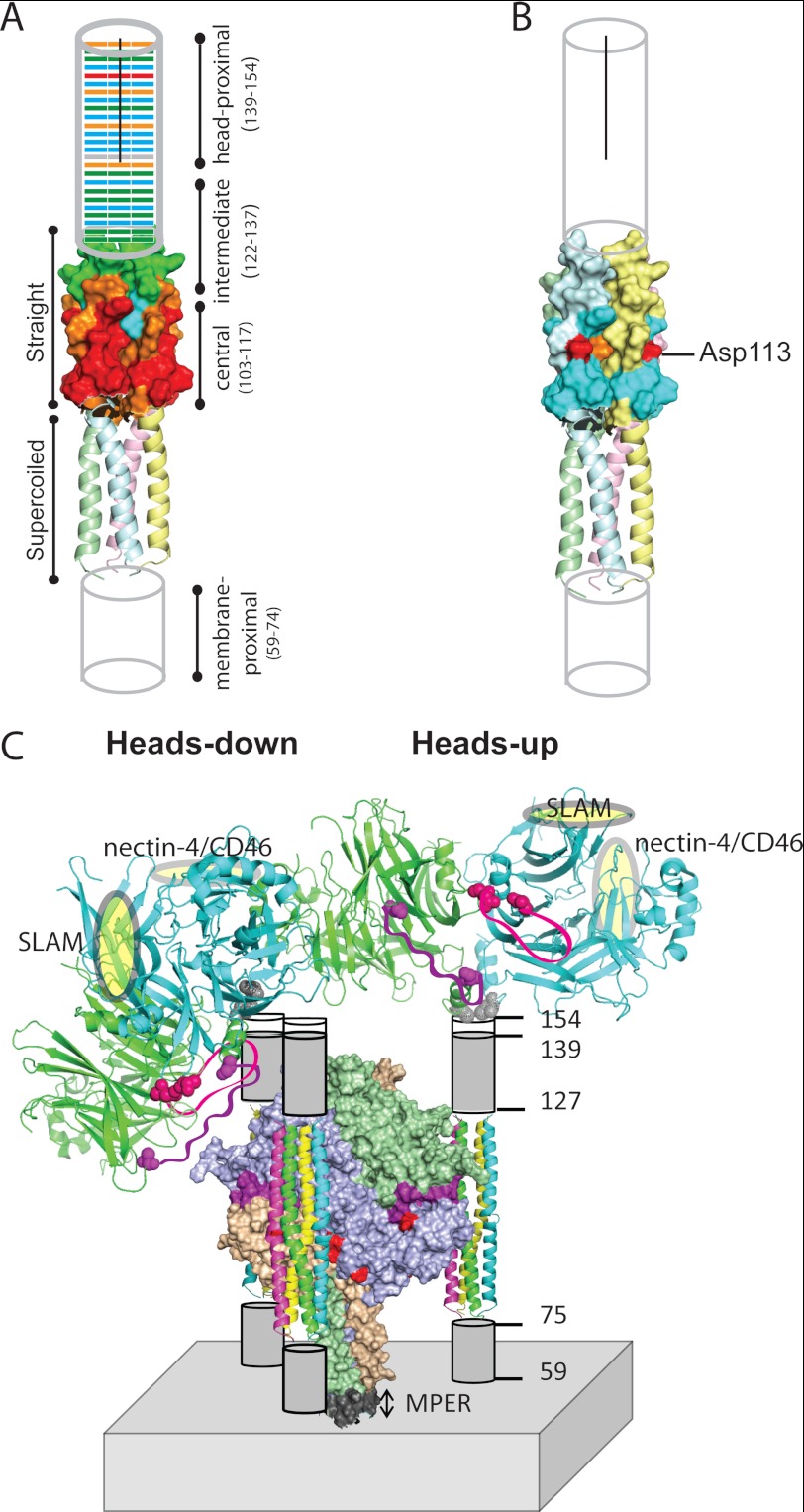
Stalk sequence alignment and homology model of MV H-stalk. A, H linear structure. From left to right: C, cytoplasmic tail; T, transmembrane region; S, stalk; β1–6, head beta propeller blades 1–6. The two stalk Cys are indicated as vertical red lines. B, predicted secondary structure of the MV stalk (top) and its alignment (bottom) with the sequences of the CDV, PIV5, and NDV stalks. Only residues differing from MV are shown. All Cys are shown in red. The horizontal black line indicates residues that comprise the MV H-stalk homology model. C, structure-based model of the MV H-stalk residues 75–127. Each monomer is shown with a different color. The N- and C-terminal residues are indicated in blue. Val-103, which approximately delineates the supercoiled segment from the relatively straight segment is also indicated. The region subjected to cysteine-scanning mutagenesis is indicated by a horizontal red line.
RESULTS
Sequence Alignment and Homology Modeling of the MV H-stalk
A schematic of the entire MV H-protein is presented in Fig. 1A. The 95-residue stalk is preceded by a cytoplasmic tail (residues 1–34) and a single transmembrane region (residues 35–58); and followed by the large cuboidal head with six propeller blades (residues 154–617). Fig. 1B shows the H-stalk sequences of the two morbilliviruses MV and CDV aligned with those of the HN-stalks which structure has been solved (PIV5 and NDV). Fig. 1C presents the 4HB MV H-stalk model; residues 75–127 (MV numbering) exhibit the highest sequence identity with the structure template (23). The lower half of the model (residues 75–102) is predicted to be a supercoiled heptad repeat, while residues 103–127 are predicted to form a straight helix with an 11-residue repeat. The membrane-proximal stalk residues 59–74 and the head-proximal residues 128–153 were not modeled due to lack of a structure template.
Similar to the PIV5 HN-stalk, most of the hydrophobic residues in the MV H tetrameric stalk are predicted to be involved in the formation of a hydrophobic core known to stabilize the 4HB. On the other hand, compared with this segment of the PIV5 or NDV stalk, MV H has one additional residue, Asp-101. This residue is located equidistant between two prolines, Pro-94 and Pro-108. This arrangement may be crucial for introducing the kink in the transition region from supercoiled to straight.
Many Cys Substitutions in the MV H-stalk Trap Covalently Linked Tetramers
We first asked whether and which Cys substitutions in the upper half of the MV H-stalk allow for cross-linking of the two dimers in a tetramer. Toward this Vero cells were transfected with the H-stalk mutants and cytoplasmic extracts were loaded onto Blue Native PAGE (BN-PAGE) gels under native conditions (18).
Fig. 2A shows two control analyses: standard H-protein, which forms both dimers and tetramers (left lane), and a mutant in which both Cys-forming disulfide bonds in the stalk have been mutated to Ala (Fig. 2A, 139A_154A, middle lane). As expected, mutation of both Cys greatly compromised the stability of the dimer, resulting in increased intensity of the H-monomer band. On the other hand, only dimers of the standard H-protein were observed in non-reducing SDS-PAGE gels (35) because denaturation disrupts non-covalent tetramer interactions (Fig. 2B, H lane). As expected, in non-reducing SDS-PAGE gels only monomers of the 139A_154A mutant were observed (Fig. 2B, 139A_154A lane).
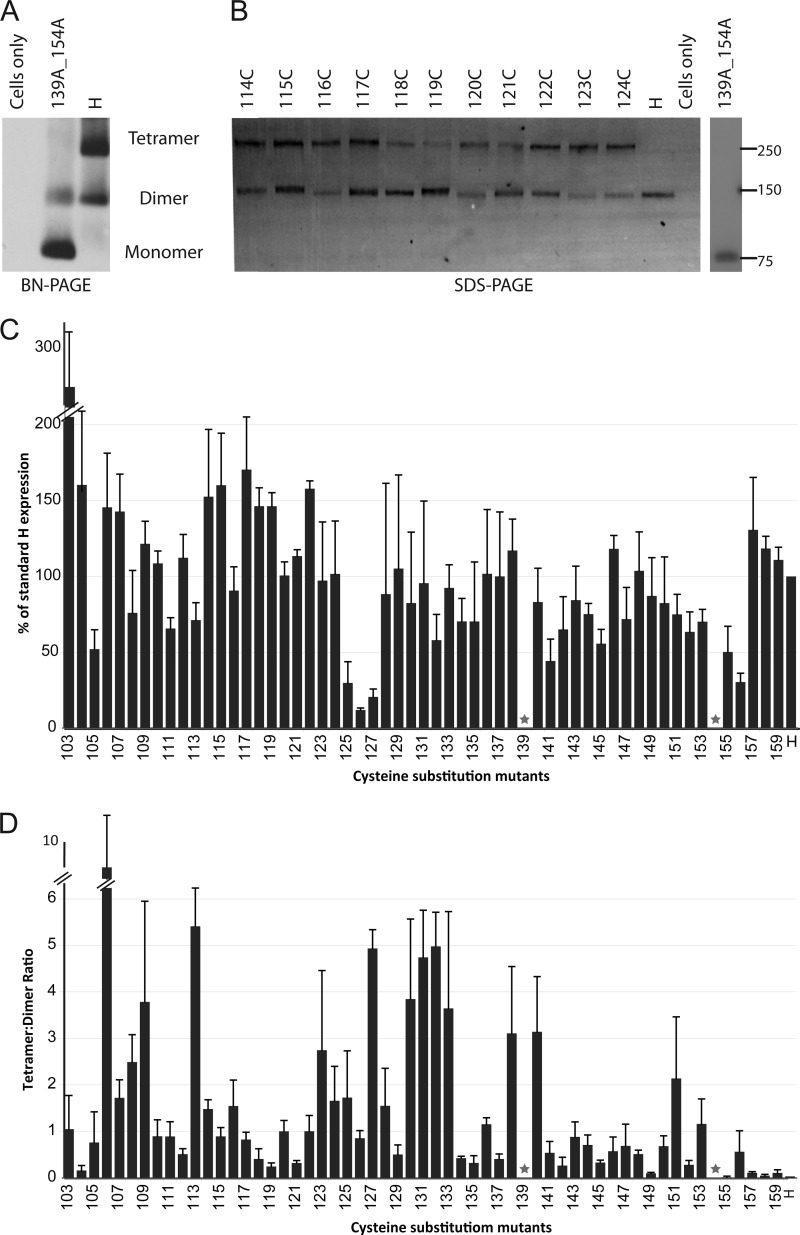
FIGURE 2.
Expression and tetramer forming propensity of Cys substitution mutants. A, H-mutant lacking both stalk Cys and standard H-protein separated on a Blue Native PAGE gel and detected by immunoblot with an H-cytoplasmic tail specific antibody. B, mutants (114C-124C) and standard H-protein extracted from cells in the presence of 50 mm iodoacetamide, separated on a non-reducing SDS-PAGE gel and detected by immunoblot analysis. Molecular weight markers are indicated on the right in kDa. C, mutant H-protein expression levels as percentage of standard H expression level. Total protein expression levels were determined by quantifying H-dimer and H-tetramer protein band intensities using a Typhoon FLA laser scanner and the Image Quant TL software. The height of the columns represents the mean of three experiments; standard deviation is indicated with error bars. Native Cys at positions 139 and 154 are indicated by stars. D, tetramer forming propensity of each mutant. The tetramer:dimer ratio was calculated by dividing the tetramer protein band intensity by that of the dimer protein band. Mean and standard deviation of three experiments are indicated.
Based on non-reducing SDS-PAGE we then assessed whether the engineered Cys in the stalk may cross-link two dimers, thus generating a covalent tetramer. Fig. 2B, lanes 114–124 shows the results for 11 of the 55 mutants. For each mutant a band corresponding to the dimer was documented, and certain mutants also produced a tetramer band. This suggested that, at least in part, the H-stalk is organized as a tetramer. However, levels of expression varied: 49 of 55 mutants had levels of expression ranging between 50 and 170% of the standard, but four mutants were expressed at low levels (Fig. 2C). Remarkably, fusion efficiency was maintained in three of these mutants, which reflects the high sensitivity of the fusion assay system (35). The variation in protein expression levels, with several mutants expressed at slightly higher levels as standard H, may reflect the inherent metastable nature of this protein.
We also compared the tetramer-to-dimer ratio observed for each of the mutants (Fig. 2D). These ratios were higher at certain positions, but there was only one instance of 3-to-4 helical periodicity in the pattern of tetramer-supporting positions: three residues near the center of the stalk (106, 109, and 113) had high tetrameric ratios while the neighboring residues had low ratios. On the other hand, several residues in the 123–133 segment, and in particular residues 130–133, had high tetramerization propensity, with tetramer-to-dimer ratios close to 4:1. In contrast, substitutions in the head-proximal segment of the stalk did not promote efficient tetramer formation: all tetramer-to-dimer ratios were below 1 with four exceptions, all located near Cys-139 (residues 138 and 140) or Cys-154 (residue 151 and 153). While our experimental approach cannot detect additional intra-dimer disulfide bonds, our analysis documents a remarkably high propensity of most residues between positions 103 and 137 to trap covalent tetramers, implying rotational flexibility of this segment of the stalk.
Functional Analysis of Cys Substitution Mutants Define Distinct H-stalk Segments
We then assessed the functional consequences of the introduction of Cys mutations in the upper half of the H-stalk. For this the H-mutants were co-transfected with the standard F-protein expression plasmid, and the amount of cell fusion measured with a semi-quantitative assay. Co-transfection of standard H and F expression plasmids into Vero cells, which express the vaccine strain receptor CD46, resulted in extensive syncytia formation (Fig. 3A, H+F panel). Mutating either one of the native stalk Cys to Ala results in a decrease in the average number of nuclei per syncytium (Fig. 3A, two intermediate levels of fusion, as exemplified by mutants 139A or 154A, respectively).
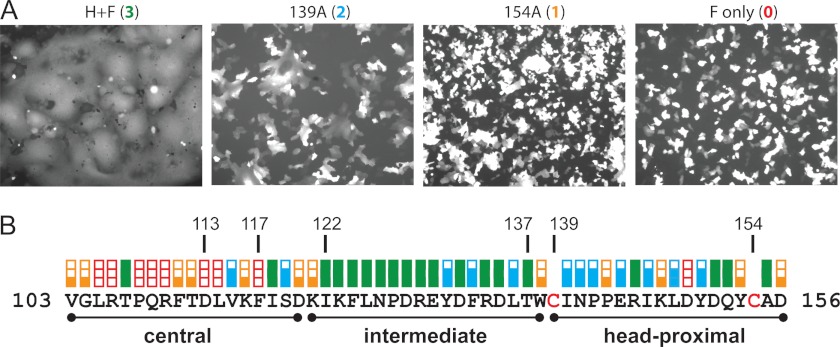
FIGURE 3.
Fusion-support function of H-stalk mutants. A, examples of visual assessment of syncytium formation. Vero cells were co-transfected with an F-expression plasmid, a standard or mutated H-expression plasmid and a GFP-expression plasmid. The fusion score was determined 24-hours post transfection. Fusion score of 3 (left panel) was observed after expression of wild-type proteins. Fusion scores of 2 (second panel) and 1 (third panel) were observed after co-expression of the mutant H-proteins indicated with wild-type F-protein. A 0 fusion score was documented in a control transfected without the H-expression plasmid (fourth panel). The parameters (number of nuclei per syncytium) used to assess the level of fusion are indicated in the experimental procedures. B, fusion scores for all the Cys substitution mutants, in linear sequence. Red empty box, fusion score of zero; one-third filled orange box, fusion score of 1; two-thirds filled blue box, fusion score of 2; full green box, fusion score of 3. The central, intermediate and head-proximal segments are indicated by black lines.
Fig. 3B documents the fusion score, on a scale of 0 (no fusion) to 3 (wild type levels of fusion), of each of the stalk mutants. Eight Cys substitutions in the center of the stalk (residues 103–117) completely abolished the fusion triggering function of the H-protein, and 5 other substitutions significantly decreased it. In stark contrast, 13 Cys substitutions in the intermediate stalk segment (residues 122–137) had no effect on fusion function and the other 3 substitutions had a small effect. Thus, while both the central and intermediate stalk segments have tetrameric structure, they must be subject to different functional constraints. Since mutations in the head-proximal segment (residues 139–153) rarely resulted in efficient tetramer formation, their effects on function were less informative.
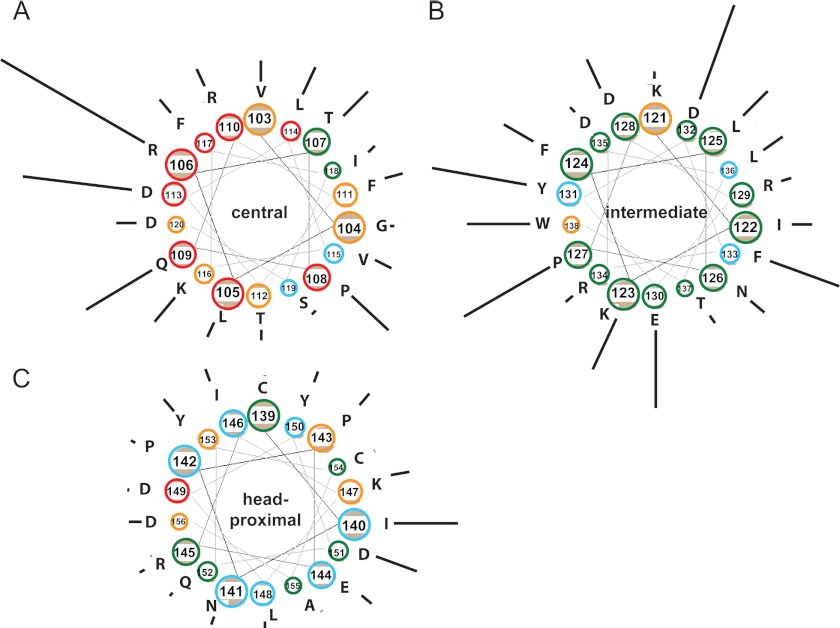
We then examined in more detail helical periodicity in the pattern of fusion interference. Fig. 4 presents the stalk in a hypothetical helical wheel secondary structure. For each residue the fusion function is color-coded and the tetramer formation propensity is indicated by the length of a radial black line. While confirming that residues 103–117 (Fig. 4A) are highly susceptible to disruption of fusion function by cross-linking, this representation also clearly shows that residues on one face of the helix (residues 106, 109, 113, and 116) are significantly more efficient at tetramerization. Indeed, helical periodicity was previously detected in this segment: carbohydrate insertions in residues 111, 114, and 118 were shown to interfere with fusion function (25). In contrast, residues in the next segment (123–138) (Fig. 4B) can efficiently support tetramer formation irrespective of their predicted location in the helix. Finally, most head-proximal residues (Fig. 4C) are unable to support tetramerization. Altogether, these data indicate that the head-proximal half of the stalk is divided into three distinct segments with different functions.
FIGURE 4.
Tetramer-to-dimer ratios in three stalk segments shown as helical wheels. Residue number and identity are indicated. The color of each circle denotes the fusion score; red (0), orange (1), blue (2), and green (3). The length of the line next to the circle is proportional to the tetramer:dimer ratio. A, central segment. B, intermediate segment. C, head-proximal segment.
Inhibition of Fusion Function by Stabilization of the Central Segment Is Reversible
We then asked whether those disulfide bonds in the central segment of the stalk that completely abolished function act by constraining its conformation. Toward this we assessed whether fusion-triggering function could be restored by exposing Vero cells that had been transfected with the relevant H and standard F expression plasmids to mild reducing conditions.
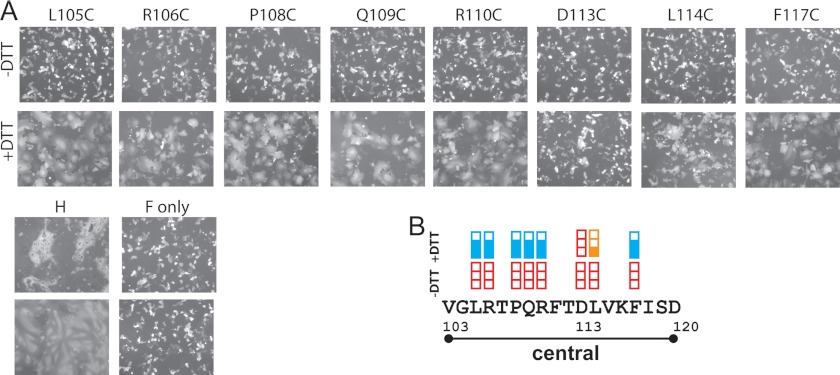
As shown in Fig. 5A, reduction of disulfide bonds allowed all but two mutants to regain at least 50% of fusion-support function (compare +DTT and −DTT fusion assays). The only position in which reduction did not restore function was Asp-113 (Fig. 5B). Consistently, it was recently observed that Cys mutation of the corresponding position (Glu-113) in the CDV H-stalk caused irreversible loss of function, whereas the loss of function in neighboring positions could be reversed by disulfide bond reduction (26). Since constraining residues in the central segment of the MV and CDV H-stalks inhibits fusion, and alleviating that constraint by disulfide reduction restores function, we conclude that a conformational change of this segment is required for transmission of the fusion triggering signal.
FIGURE 5.
Disulfide bond reduction can restore fusion support function. A, visual assessment of syncytium formation. Vero cells were co-transfected with the indicated H, standard F, and GFP expression plasmids and incubated with FIP, a fusion inhibitor. Twenty-four hours post-transfection cells were washed and treated either with PBS (−DTT, top row of panels) or with 15 mm DTT (+DTT, bottom row of panels) for 30 min. Fusion score was recorded 3 h after DTT treatment. B, fusion scores. The extent of fusion before or after DTT treatment is indicated above each residue. Red empty box, fusion score of 0; one-third filled orange box, fusion score of 1; and two-thirds filled blue box, fusion score of 2. The central segment is indicated.
DISCUSSION
The stalks of the attachment proteins of Paramyxoviruses transmit the signal that triggers membrane fusion to the homologous F-proteins, thereby controlling cell entry (36–38). While the metastable nature of these attachment proteins has been a challenge for structural studies, the recently solved structure of the HN-protein ectodomain revealed a 4HB that extends from near the membrane to within 7–9 residues below the base of the HN-head dimers (22, 23). From there two flexible linkers bifurcate to allow one head in each dimer to adhere laterally to the stalk in a tilted or “heads-down” conformation.
We asked here whether the MV H-stalk, which could be up to 15–20 residues longer than the HN-stalks, has a similar structure. We found that many residues up to Ile-140 have high tetramer formation propensity suggesting that their Cα carbon atoms are less than 7.5 Å apart. Since the 4HB of the NDV ectodomain ends with the residue collinear with MV H residue Glu130 (Fig. 1B), the tetrameric structure of MV H appears to be about 10 residues longer that of NDV HN. The following residues, 141–153 may form two flexible linkers, clamped by the intra-subunit covalent bonds of Cys-139 and Cys-154. Interestingly, even above Ile140 several residues have some tetramer-formation propensity, and head-proximal residues 151 and 153 have moderate tetramer formation propensity. This suggests that tetrameric structures may exist even above Ile140 in a fraction of the H-stalks, and that in these oligomers the four H-heads' bases may be in close proximity.
Our analyses indicate that the central segment of the MV stalk must change conformation to transmit the fusion-triggering signal, similar to residues 91–115 of the CDV H-stalk (26). In particular, Cys substitutions in 13 of the 15 positions between residues 103–117 significantly interfered with fusion-triggering function (Fig. 3B). On the other hand, in 7 of the 8 positions completely blocking function, reduction of the disulfide bonds restored it (Fig. 5B). These data are reported on the model structure of the H-stalk of Fig. 6A, showing in red the residues which mutation to Cys completely blocks fusion function. Fig. 6B illustrates that after reduction of the covalent links all positions recovered some function, with the exception of Asp-113. Interestingly, the CDV H-stalk also has a negatively charged residue (Glu) at this position, and again Cys mutation causes irreversible loss of function (26). Thus, a negative charge at position 113 of the Morbillivirus H-stalk may be necessary for function.
FIGURE 6.
Functional modules of the H-stalk, and possible arrangements of the F and H oligomers. A, fusion-support function of the Cys substitution mutants indicated on a schematic of the H-stalk. From the bottom, the membrane-proximal amino acids (59–74) are indicated as a cylinder. The homology model which covers residues 75–127 is shown next, with its supercoiled lower segment shown in the backbone representation, and the straight upper segment shown in surface representation. The head-proximal residues 128–154 are indicated by another cylinder. The black line indicates the segment of the stalk that is dimeric. Different colors denote the fusion score before reduction of disulfide bonds; red (0), orange (1), blue (2), and green (3). B, fusion-support function, after DTT treatment, of the eight Cys substitution mutants in the central stalk segment that completely blocked fusion. Color code for fusion function as in panel A. Six of the eight mutants recovered most fusion function (blue), but Asp-113 (red) did not and Leu-114 (orange) showed only modest recovery. C, alternative arrangements of the H-heads on the stalk, and proposed arrangement of F-trimers around the H-stalks. Top: two H-head dimers, one shown in the heads-down conformation (left), the other in the heads-up conformation (right). Heads are shown in backbone representation, but only one H-head dimer is shown in each tetrameric structure for clarity. One monomer comprising an H-dimer is shaded green and the other blue. The receptor-binding sites (SLAM and nectin-4/CD46) are indicated by yellow ovals. Two large gaps in the crystal structure are indicated by purple (168–187) and pink (238–248) ribbons. The residues flanking these gaps are indicated in a space-filling representation. The heads-down conformation of the H-dimer (left) would allow these loop regions to interact with H-stalk and perhaps from part of the head-stalk interface. Cys-154 at the base of the dimer is indicated in gray. Lower half: H-stalks are shown interacting with hydrophobic cavities on three sides of the F-trimer (center). The F-trimer is shown in surface representation with the three subunits indicated in different colors, and the fusion peptide in purple. Six surface residues important for fusion-triggering signal receipt (43) are shown in red. The membrane-proximal external region (MPER) is indicated in gray (44).
We have noted that in the MV H-stalk homology model the side chains of seven of the eight residues which mutation to Cys completely blocks function point away from the hydrophobic core of the 4HB (supplemental Fig. S1; the exception is Leu-114). Clearly, if these side chains were always pointing away from the center of the 4HB, covalent tetrameric structures would not be efficiently trapped. The apparent flexibility of the central stalk segment could be accounted by either the mobility of individual side chains, or local changes in inter-peptide backbone distances (bulging), or both. We also note that Cys substitutions may cause the helices to reorient, which would be consistent with the inherent flexibility of this segment, and with previous analyses concluding that this segment of the Morbillivirus stalk is flexible (25, 39).
Our analyses defined a second functional module in the upper half of the stalk which, in contrast to the F-trimer contact module, tolerates tetrameric stabilization remarkably well. Cys at 11 out of the 16 positions between residues 123 and 138 trapped tetramers efficiently, but covalent linkage had no effect on function. Thus, this module can conduct the fusion-triggering signal while remaining in a stabilized tetrameric configuration.
Altogether, our analyses inform the model of membrane fusion triggering presented in Fig. 6C. This model considers two alternative conformations of the attachment protein ectodomain inferred from different sets of crystallographic data: either “heads-up” (Fig. 6C, right), with heads located above the stalk, making tetrameric contacts similar to those observed in crystal structures derived from incomplete H-heads (21). Or “heads-down” (Fig. 6C, left), with tilted heads in contact with the sides of the stalk, as revealed by the crystal structure of an entire HN-ectodomain (22).
Assuming that the MV H-stalk has a similar structure as the HN-stalk, with a lower supercoiled segment and an upper straight segment, our results suggest a total rise of at least 110 Å: 15 Å for presumed supercoiled 4HB residues 59–74 (1 Å per residue), 75 Å for the modeled segment (residues 75–127) and 20 Å for residues 128–140 (1.5 Å per residue). A tetrameric 141–154 segment would further extend this rise. Thus, in the “up” conformation, the H-heads would occupy a belt largely above the bulk of the F-head densities that are located 55–125 Å from the membrane (40).
On the other hand, in the “down” conformation the H-heads would occupy a lower but broader belt, implying very tight packing in the 80–120 Å region. A recently published cryo-electron tomography analysis (41) documents F/H protein densities starting about 40 Å from the membrane, peaking at 80 Å, and extending up to about 150 Å, without revealing an intermediate band of lower density that may be expected if the heads were exclusively “up” (25). While our analyses are consistent with most H-heads being “down”, stalks with an extended tetramer structure holding the heads “up” may also exist. These “zipped-up” tetrameric structures may form upon destabilization of the interactions of one head with one side of the stalk.
In conclusion, our analyses of the MV H-stalk structure and function suggest the following update of the envelope assembly and disassembly model. During intracellular transport and viral particle release H-tetramers stabilize F-trimers (42). At cell entry receptor pulling de-stabilizes the H-dimer interface (35), eliciting a conformational change in the central stalk segment (26, 39) triggering F-trimer refolding and membrane fusion. Here we observed that an intermediate segment of the upper stalk (residues 122–137) can remain in a stabilized configuration during the entire fusion-triggering process. This segment may act as a spacer allowing transmission of the fusion triggering signal while maintaining the H-heads at an optimal distance from the F-interacting stalk module.
Finally, analyses of the F-trimer reactive surface (43) suggest that the H-stalks are lodged in its sides, as shown in the bottom half of Fig. 6C. Two helices of one H-stalk would contact one side of an F-trimer (center), while the other two helices would contact the side of another trimer which is not shown. This implies that a conformational change in the stalk of one H-tetramer would be transmitted to two apposed F-trimers. When these trimers refold, each one would de-stabilize apposed H-tetramers, starting a chain reaction resulting in membrane fusion.
Acknowledgments
We thank Swapna Apte-Sengupta, Sayantan Bose, and Robert Lamb for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA090636 (to R. C.).

This article contains supplemental Fig. S1.
- MV
- measles virus
- H
- hemagglutinin
- F
- fusion
- NDV
- Newcastle disease virus
- 4HB
- four-helix bundle.
REFERENCES
- 1. Griffin D. E. (2007) in Fields' Virology, Fifth Edition (Fields B., Knipe D. M., Howley P. M., eds), pp. 1551–1585, Lippincott Williams and Wilkins, Philadelphia [Google Scholar]
- 2. Lamb R. A., Parks G. D. (2007) in Fields' Virology, Fifth Edition (Fields B., Knipe D. M., Howley P. M., eds), pp. 1305–1340, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 3. Moss W. J. (2009) Measles control and the prospect of eradication. Curr. Top. Microbiol. Immunol. 330, 173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(2011) Vaccines: the case of measles. Nature 473, 434–435 [DOI] [PubMed] [Google Scholar]
- 5. Marsh M., Helenius A. (2006) Virus entry: open sesame. Cell 124, 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chandran K., Sullivan N. J., Felbor U., Whelan S. P., Cunningham J. M. (2005) Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308, 1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iorio R. M., Melanson V. R., Mahon P. J. (2009) Glycoprotein interactions in paramyxovirus fusion. Future Virol. 4, 335–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Navaratnarajah C. K., Miest T. S., Carfi A., Cattaneo R. (2012) Targeted entry of enveloped viruses: measles and herpes simplex virus I. Curr. Opin. Virol. 2, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee B., Ataman Z. A. (2011) Modes of paramyxovirus fusion: a Henipavirus perspective. Trends Microbiol. 19, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mühlebach M. D., Mateo M., Sinn P. L., Prüfer S., Uhlig K. M., Leonard V. H., Navaratnarajah C. K., Frenzke M., Wong X. X., Sawatsky B., Ramachandran S., McCray P. B., Jr., Cichutek K., von Messling V., Lopez M., Cattaneo R. (2011) Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480, 530–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tatsuo H., Ono N., Tanaka K., Yanagi Y. (2000) SLAM (CDw150) is a cellular receptor for measles virus. Nature 406, 893–897 [DOI] [PubMed] [Google Scholar]
- 12. Chang A., Dutch R. E. (2012) Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 4, 613–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mirza A. M., Aguilar H. C., Zhu Q., Mahon P. J., Rota P. A., Lee B., Iorio R. M. (2011) Triggering of the Newcastle disease virus fusion protein by a chimeric attachment protein that binds to Nipah virus receptors. J. Biol. Chem. 286, 17851–17860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Porotto M., Salah Z., DeVito I., Talekar A., Palmer S. G., Xu R., Wilson I. A., Moscona A. (2012) The second receptor binding site of the globular head of the Newcastle disease virus hemagglutinin-neuraminidase activates the stalk of multiple paramyxovirus receptor binding proteins to trigger fusion. J. Virol. 86, 5730–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bose S., Zokarkar A., Welch B. D., Leser G. P., Jardetzky T. S., Lamb R. A. (2012) Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc. Natl. Acad. Sci. U.S.A., 109, E2625–E2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hashiguchi T., Kajikawa M., Maita N., Takeda M., Kuroki K., Sasaki K., Kohda D., Yanagi Y., Maenaka K. (2007) Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. U.S.A. 104, 19535–19540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Navaratnarajah C. K., Vongpunsawad S., Oezguen N., Stehle T., Braun W., Hashiguchi T., Maenaka K., Yanagi Y., Cattaneo R. (2008) Dynamic interaction of the measles virus hemagglutinin with its receptor signaling lymphocytic activation molecule (SLAM, CD150). J. Biol. Chem. 283, 11763–11771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brindley M. A., Plemper R. K. (2010) Blue Native-PAGE and biomolecular complementation reveal tetrameric or higher order oligomer organization of the physiological measles virus attachment (H) protein. J. Virol. 84, 12174–12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Plemper R. K., Hammond A. L., Cattaneo R. (2000) Characterization of a region of the measles virus hemagglutinin sufficient for its dimerization. J. Virol. 74, 6485–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Santiago C., Celma M. L., Stehle T., Casasnovas J. M. (2010) Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat. Struct. Mol. Biol. 17, 124–129 [DOI] [PubMed] [Google Scholar]
- 21. Hashiguchi T., Ose T., Kubota M., Maita N., Kamishikiryo J., Maenaka K., Yanagi Y. (2011) Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat. Struct. Mol. Biol. 18, 135–141 [DOI] [PubMed] [Google Scholar]
- 22. Yuan P., Swanson K. A., Leser G. P., Paterson R. G., Lamb R. A., Jardetzky T. S. (2011) Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc. Natl. Acad. Sci. U.S.A. 108, 14920–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bose S., Welch B. D., Kors C. A., Yuan P., Jardetzky T. S., Lamb R. A. (2011) Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J. Virol. 85, 12855–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alkhatib G., Briedis D. J. (1986) The predicted primary structure of the measles virus hemagglutinin. Virology 150, 479–490 [DOI] [PubMed] [Google Scholar]
- 25. Paal T., Brindley M. A., St. Clair C., Prussia A., Gaus D., Krumm S. A., Snyder J. P., Plemper R. K. (2009) Probing the spatial organization of measles virus fusion complexes. J. Virol. 83, 10480–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ader N., Brindley M. A., Avila M., Origgi F. C., Langedijk J. P. M., Örvell C., Vandevelde M., Zurbriggen A., Plemper R. K., Plattet P. (2012) Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J. Biol. Chem. 287, 16324–16334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cathomen T., Buchholz C. J., Spielhofer P., Cattaneo R. (1995) Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology 214, 628–632 [DOI] [PubMed] [Google Scholar]
- 28. Radecke F., Spielhofer P., Schneider H., Kaelin K., Huber M., Dötsch C., Christiansen G., Billeter M. A. (1995) Rescue of measles viruses from cloned DNA. EMBO J. 14, 5773–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cathomen T., Naim H. Y., Cattaneo R. (1998) Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72, 1224–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Norrby E. (1971) The effect of a carbobenzoxy tripeptide on the biological activities of measles virus. Virology 44, 599–608 [DOI] [PubMed] [Google Scholar]
- 31. Richardson C. D., Choppin P. W. (1983) Oligopeptides that specifically inhibit membrane fusion by paramyxoviruses: studies on the site of action. Virology 131, 518–532 [DOI] [PubMed] [Google Scholar]
- 32. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 33. Fraczkiewicz R., Braun W. (1998) Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 19, 319–333 [Google Scholar]
- 34. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Navaratnarajah C. K., Oezguen N., Rupp L., Kay L., Leonard V. H., Braun W., Cattaneo R. (2011) The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat. Struct. Mol. Biol. 18, 128–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deng R., Wang Z., Mirza A. M., Iorio R. M. (1995) Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209, 457–469 [DOI] [PubMed] [Google Scholar]
- 37. Tanabayashi K., Compans R. W. (1996) Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70, 6112–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsurudome M., Kawano M., Yuasa T., Tabata N., Nishio M., Komada H., Ito Y. (1995) Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213, 190–203 [DOI] [PubMed] [Google Scholar]
- 39. Lee J. K., Prussia A., Paal T., White L. K., Snyder J. P., Plemper R. K. (2008) Functional interaction between paramyxovirus fusion and attachment proteins. J. Biol. Chem. 283, 16561–16572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yin H. S., Wen X., Paterson R. G., Lamb R. A., Jardetzky T. S. (2006) Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liljeroos L., Huiskonen J. T., Ora A., Susi P., Butcher S. J. (2011) Electron cryotomography of measles virus reveals how matrix protein coats the ribonucleocapsid within intact virions. Proc. Natl. Acad. Sci. U.S.A. 108, 18085–18090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Plemper R. K., Hammond A. L., Cattaneo R. (2001) Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 276, 44239–44246 [DOI] [PubMed] [Google Scholar]
- 43. Apte-Sengupta S., Negi S., Leonard V. H., Oezguen N., Navaratnarajah C. K., Braun W., Cattaneo R. (2012) Base of the measles virus fusion trimer head receives the signal that triggers membrane fusion. J. Biol. Chem. 287, 33026–33035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zokarkar A., Lamb R. A. (2012) The Paramyxovirus fusion protein C-terminal region: mutagenesis indicates an indivisible protein unit. J. Virol. 86, 2600–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]