Background: SALL4 is a key stem cell transcription factor that transactivates OCT4.
Results: SALL4B is modified by sumoylation.
Conclusion: Sumoylation plays an important role in regulating SALL4B stability, subcellular localization, and transcriptional activities.
Significance: Sumoylation functions a major post-translational mechanism for regulating stem cell transcription factors.
Keywords: Post-translational Modification, Stem Cells, Sumoylation, Transcription Factors, Ubiquitination, OCT4, SALL4
Abstract
SALL4 is a transcription factor that plays a key role in the maintenance and self-renewal of embryonic stem cells and hematopoietic stem cells. Given that little is known about regulation of SALL4, we studied biochemical modifications of SALL4B, a major splicing variant of SALL4, and elucidated their biological function. SALL4B was primarily modified by ubiquitination when it was expressed in both Sf9 and HEK293T cells. A significant fraction of SALL4B was further modified by sumoylation when it was expressed in HEK293T cells. Constitutive SUMO-modification of SALL4B was also detected in Tera-1, a cell line of the teratocarcinoma origin. SALL4B sumoylation was independent of ubiquitination and lysine residues 156, 316, 374, and 401 were essential for sumoylation. Chromatin fraction contained more SUMO-deficient SALL4B. Despite a shorter half-life than the wild-type counterpart, SUMO-deficient SALL4B interacted with OCT4 more efficiently than the wild-type SALL4B. RNAi-mediated silencing of SALL4 expression caused significant down-regulation of both OCT4 and SOX2, which was rescued by ectopic expression of SALL4B but not by SUMO-deficient mutant. Significantly, compared with the wild-type SALL4B, SUMO-deficient mutant exhibited compromised trans-activation or trans-repression activities in reporter gene assays. Combined, our studies reveal sumoylation as a novel form of post-translational modification for regulating the stability, subcellular localization, and transcriptional activity of SALL4.
Introduction
Transcription factors are crucial for maintaining and self-renewing the pluripotency of embryonic stem cells. Recent studies have shown that merely manipulating expression of a set of transcription factors including OCT4, SOX2, and Nanog can reprogram differentiated cells into pluripotent stem cells (1–3). SALL4 is also an essential transcription factor that functions to maintain pluripotency of embryonic stem cells and hematopoietic stem cells (4, 5), in part through physically and functionally interacting with OCT4, SOX2, and Nanog (6–8). SALL4 also functions to increase the efficiency of reprogramming of induced pluripotent stem (iPS)2 cells (9). Extensive studies in the past have focused on the transcriptional regulation of stem cell transcription factors. SALL4 positively regulates expression of OCT4 through binding to the conserved regulatory region of the OCT4 promoter (10). However, SALL4 negatively regulates its own gene expression through a feedback loop whereas SALL4 and OCT4 work in concert to balance the expression of genes of the SALL family (10). Given the critical role of SALL4 in stem cell maintenance and self-renewal, deregulated expression of SALL4 or its structural abnormalities frequently leads to developmental abnormalities or malignant transformation (11–14).
Post-translational modifications play an essential role in the regulation of the activities of stem cell factors including OCT4, SOX2, and Nanog. Transcription factor OCT4 is the master regulator for the maintenance of pluripotency and self-renewal (15). A recent study reveals that OCT4 is phosphorylated on multiple sites and that phosphorylation in its homeobox domain reduces its transactivation activity through interfering with the DNA binding (16). OCT4 is also a target for modification by SUMO (17), a small ubiquitin-related modifier that post-translationally regulates protein molecules that are involved in many cellular processes, including gene transcription (18). Sumoylation of OCT4 enhances it stability, as well as its DNA binding and transactivation (17). Transcription factor SOX2 is essential for maintaining the pluripotency of embryonic stem cells (19). SOX2 is modified by several post-translational mechanisms including phosphorylation, acetylation, methylation, and ubiquitination (20–22). For example, SOX2 is associated with CARM1, an arginine methyltransferase, and is methylated by the enzyme; the methylation enhances its self-association (21). SOX2 is also SUMO-modified at K247 and sumoylation appears to negatively regulate its transcriptional activity (23).
Given that SALL4 physically and/or functionally interacts with OCT4, SOX2, and Nanog (7, 10) and that the transcription factor is crucial in the regulation of stem proliferation and differentiation (5, 9, 11), we focused on characterization of post-translational modifications of SALL4B, a major splicing variant. We observed that SALL4B existed primarily as a ubiquitinated form and that a fraction of SALL4B was modified by sumoylation. Mass spectrometry analysis revealed that SALL4B was also phosphorylated. Our detailed biochemical and molecular studies reveal that several lysine residues were essential for SALL4B sumoylation, which plays an important role in its stability and subcellular localization. Moreover, SALL4B sumoylation also affects its trans-activation/trans-repression activities.
EXPERIMENTAL PROCEDURES
Cell Culture
Tera-1, HEK293T, HeLa, Jurkat, and Sf9 cell lines were obtained from the American Type Culture Collection (ATCC). Cells were cultured under conditions as described in the manual provided by the supplier.
Antibodies
Antibodies to SALL4 and ubiquitin were purchased from Abcam (Boston). Antibodies to HA, FLAG, and β-actin were purchased from Cell Signaling Technology Inc. Antibodies to OCT4 and Nanog were purchased from Santa Cruz Biotechnology. Mouse anti-SUMO-1 and mouse anti-SUMO-2/3 antibodies were kindly provided by Dr. Michael Matunis (Johns Hopkins University) and Dr. Xiang-dong Zhang (Wayne State University).
Plasmids, Mutagenesis, and siRNAs
The original SALL4B expression plasmid was described previously (11). SALL4B cDNA was subcloned into pcDNA3 plasmid with the in-frame addition of 3-tandem HA tags and the His6 tag in the C-terminal. SALL4B mutants with lysine 156 (K156), K316, K374, and/or K401 residues replaced with arginines (R) were generated using the QuickChange Lightning Multi Site-directed Mutagenesis Kit (Strategene). Individual mutations were confirmed by DNA sequencing (Seqwright). Synthetic siRNA specific to SALL4A mRNA (5′-GCA UCG AUG UAG AGG AAG-3′) and to the SALL4 gene 3′-untranslated region (5′-CAA UGC AGA CAC AGU GAA A-3′), as well as the control siRNA, were purchased from Dharmacon RNAi Technology. Transfection of plasmids or siRNAs was carried out using Lipofectamine 2000 according to the protocol provided by the supplier (Invitrogen).
Western Blot
SDS-PAGE was carried out using the mini-gel system purchased from Bio-Rad. Fractionated proteins were transferred to PVDF membranes. After blocking in TBS/T containing 5% nonfat dry milk for 1 h, the membranes were incubated with primary antibodies overnight followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology) for 1 h at room temperature. After thorough washing with TBS/T buffer, specific signals on the membranes were developed with an enhanced chemiluminescent system (Pierce).
Protein Co-immunoprecipitation
For immunoprecipitation, cells were lysed in a lysis buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton, 2 mm sodium pyrophosphate, and 1 mm EDTA, 1 mm NaF, 1 mm sodium orthovanadate, 500 μm PMSF, 2 μm pepstatin A, 10 units/ml aprotinin, 20 mm NEM). The antibody (1 μg) and protein-G-agarose slurry (40 μl) (50/50, Millipore) were added to cell lysates and incubated at 4 °C overnight followed by thorough washing with the lysis buffer. Proteins specifically bound to resin were eluted in the SDS-PAGE sample buffer. After boiling for 5 min, protein samples in the supernatant were analyzed by SDS-PAGE followed by Western blotting.
Nickel-affinity Pull-down
For pull-down assays under the denaturing condition, HEK293T cells transfected with various expression plasmids were lysed in the lysis buffer (8 m urea, 50 mm Na2HPO4/NaH2PO4 (pH 7.4), 300 mm NaCl, 0.1% Triton X-100) containing 20 mm imidazole. Ni Superflow Resin (Clontech) was added to the cell lysates and incubated with gentle agitation at the room temperature for 2 h. The resin was then washed five times at room temperature with the lysis buffer containing 40 mm imidazole. After last wash, His6-tagged proteins were eluted in the lysis buffer containing 300 mm imidazole and blotted for SALL4, the HA tag, or the FLAG tag. For pull-down assays under the native condition, HEK293T cells transfected with various expression plasmids were lysed in the lysis buffer (50 mm Na2HPO4/NaH2PO4 ( pH 7.4), 300 mm NaCl, 20 mm NEM, 0.2% Triton X-100) containing 20 mm imidazole. Ni-IDA Superflow Resin (Clontech) was the added to the cell lysates and incubated with gentle agitation overnight at 4 °C. The resin was then washed five times at 4 °C with the lysis buffer containing 40 mm imidazole. After last wash, His6-tagged proteins were eluted in the lysis buffer containing 300 mm imidazole and blotted for SALL4, the HA tag, and/or OCT4.
Luciferase Reporter Gene Assays
Expression constructs for the firefly (Photinus pyralis) luciferase gene driven by the SALL4 or OCT4 promoter were described previously (10, 24). Tera-1 cells seeded in 12-well plate for 16 h were co-transfected with the firefly reporter plasmid (0.5 μg/well), pRL Renilla luciferase reporter plasmid (50 ng) (for monitoring transfection efficiency), and a SALL4B expression plasmid (0.5 μg) (or control pcDNA3 vector). To reduce the effect of endogenous SALL4, these cells were also transfected with 40 nm siRNAs specific to the 3′-untranslated region of SALL4 gene. 48 h after transfection, cells were lysed, and luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega). Aliquots of the cell lysates were also blotted with antibodies to SALL4 and HA, respectively.
Protein Fractionation
Cells washed twice with cold PBS were lysed in buffer #1 (10 mm Tris-HCl (pH7.9), 10 mm KCl, 1.5 mm MgCl2, 1 mm DTT) and left on ice for 10 min. After centrifugation (2000 × g × 10 min), the supernatant (the cytoplasmic extract) were collected, The pellet fraction was re-suspended in buffer #2 (20 mm Tris-HCl (pH 7.9), 1.2 m KCl, 1.5 mm MgCl2, 25% (w/v) glycerol, 0.2 mm EDTA) and left on ice for 30 min. The supernatant (the nucleoplasmic fraction) were harvested after centrifugation (16,000 × g × 10 min). The pellets were re-suspended in buffer #3 (10 mm Tris-HCl (pH 7.9), 10 mm NaCl, 3 mm MgCl2, 1.5 mm CaCl2) containing Benzonase (Novagen). The suspension was then mixed at room temperature for 5 min followed by incubation on ice for 30 min, after which the supernatant (S1) was collected (2000 × g × 10 min). The pellets were again re-suspended into buffer #3 containing 0.25 m EDTA and incubated on ice for 30 min, after which the supernatant (S2) was collected after centrifugation (16,000 × g × 15 min). Fractions S1 and S2 were designated as the chromatin-associated protein extract.
Half-life Assays
HEK293T cells were transfected with either His-HA-SALL4B-WT or His-HA-SALL4B-4R plasmids for 24 h. Transfected cells were then collected by trypsinization and reseeded into new dishes. After 16 h of incubation, cycloheximide (CHX, Sigma) was added to the culture to a final concentration of 10 μg/ml. At various times of CHX treatment, cells were harvested and lysed. Equal amounts of total proteins were blotted with antibodies to HA and β-actin.
Statistical Analysis
The Student's t test was used to evaluate the difference between two groups. A value of p < 0.05 was considered to be statistically significant.
RESULTS
SALL4B Is Post-translationally Modified
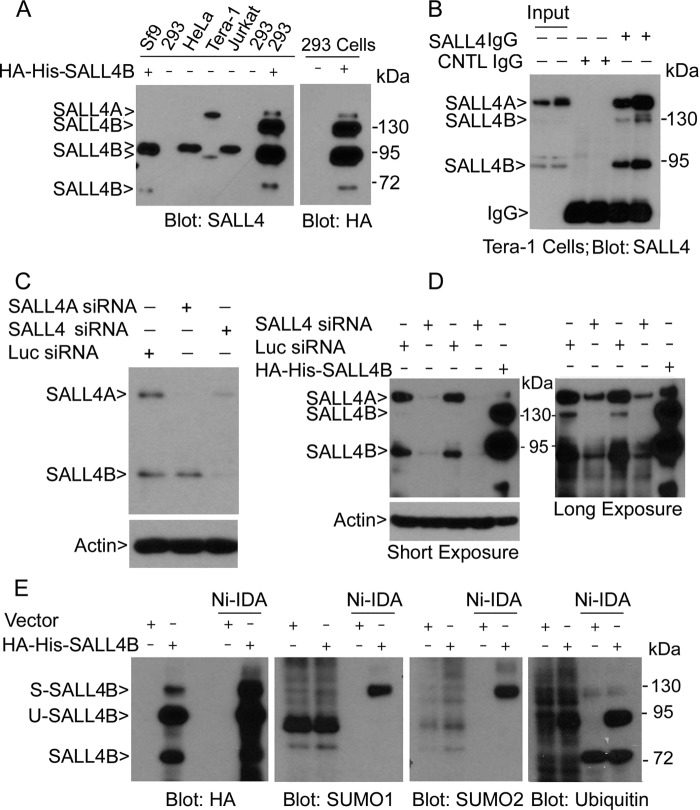
SALL4 is primarily expressed in embryonic stem cells and lineage-specific stem cells (8). Deregulated expression of SALL4 is also observed in transformed cells (11–13). As the first step to study post-translational regulation of SALL4, we examined several common cell lines for SALL4 expression. We noticed that HeLa and Jurkat cells, but not HEK293T cells, expressed a major form of SALL4 that migrated at about 95 kDa (Fig. 1A). Tera-1, a cell line of the teratocarcinoma origin, expressed two major forms of SALL4 with estimated molecular weights of about 160 and 90 kDa, respectively. Both HEK293T cells and insect Sf9 cells transfected with a SALL4B plasmid expressed SALL4B with a slower mobility than endogenous SALL4B in Tera-1 cells due to fusion with both His6 and HA tags. Of note, ectopically expressed SALL4B apparently co-migrated with SALL4 detected in HeLa and Jurkat cells. Intriguingly, HEK293T cells transfected with His6-tagged SALL4B plasmid also contained several additional bands with an apparent molecular mass of 165, 130, and 70 kDa, respectively. These bands were SALL4B-specific as they were all positive to the HA tag antibody and no corresponding bands were present in vehicle-transfected control cells (Fig. 1A). Moreover, these SALL4B-related bands were also detected in an independent study (25). Given that the predicted molecular weight of SALL4B is about 70 kDa, our results strongly suggest that ectopically expressed SALL4B with a slow mobility was modified by unknown post-translational mechanisms.
FIGURE 1.
SALL4B is post-translationally modified. A, equal amounts of protein lysates from HEK293T, HeLa, Tera-1, and Jurkat cells, as well as from Sf9 and 293 cells transfected with HA-His6-SALL4B, were blotted for SALL4. HEK293T cells transfected with or without SALL4B were also blotted for the HA tag. B, Tera-1 cell lysates in duplicates were immunoprecipiated with either control IgG or SALL4 IgG. Immunoprecipitates, along with lysate controls, were blotted with the anti-SALL4 antibody. C, Tera-1 cells transfected with SALL4A-specific siRNA or SALL4 siRNA (corresponding to the 3′-untranslated region) or luciferase siRNA for 48 h, after which equal amounts of cell lysates were blotted with antibodies to SALL4 and β-actin. D, Tera-1 cells transfected with SALL4 siRNA or luciferase siRNA for 48 h, after which equal amounts of cell lysates, along with the lysates from HEK293T cells transfected with SALL4B expression construct, were blotted with antibodies to SALL4 and β-actin. Blots of short and long exposures are shown. E, HEK293T cells transfected with SALL4B expression plasmid (or vector control) for 48 h were incubated with Ni-IDA resin. Proteins specifically bound to the resin, along with the lysate inputs, were blotted with antibodies to HA, SUMO-1, SUMO-2, and ubiquitin. S-SALL4B and U-SALL4B denote sumoylated and ubiquinated SALL4B, respectively.
Co-immunoprecipitation analysis coupled with immunoblotting revealed that SALL4 bands detected in Tera-1 cells were specific (Fig. 1B). Moreover, endogenous SALL4B migrated at about 130 kDa was also enriched by immunoprecipitation. As the large form (∼160 kDa) of the endogenous SALL4 in Tera-1 cells migrated at the similar position as the minor form of ectopically expressed SALL4B, we transfected these cells with siRNAs specific to SALL4A, as well as siRNAs corresponding to the 3′-untranslated region (3′-UTR) of SALL4 mRNA. Immunoblotting revealed that transfection with SALL4A siRNAs specifically silenced the 160 kDa form, but not the 130 kDa form (Fig. 1C), indicating that the large form is SALL4A. Consistent with this observation, transfection with 3′-UTR siRNAs (SALL4 siRNAs) knocked down both SALL4A and SALL4B (Fig. 1, C and D). Moreover, SALL4 siRNAs specifically down-regulated the minor form of SALL4B that co-migrated with ectopically expressed 130 kDa SALL4B (Fig. 1D, long exposure).
SALL4B Is SUMO-modified
Given the significant increase in the size of 95 and 130 kDa SALL4B bands, we suspected that these bands could be the result of post-translational modification by ubiquitination and/or sumoylation. SALL4B-transfected HEK293T cells lysates before and after Ni-IDA pull-down were blotted with antibodies to SUMO-1, SUMO-2, or ubiquitin, respectively. SALL4B band migrated at 130 kDa was positive to both SUMO-1 and SUMO-2 antibodies (Fig. 1E), indicating that it is modified by both SUMO-1 and SUMO-2. On the other hand, SALL4B migrated at 95 kDa was only positive to ubiquitin, indicating that it was modified by ubiquitination. Our further analysis confirmed that the 95 kDa band was a ubiquitinated one (supplemental Fig. S1). SALL4B with a molecular weight at about 70 kDa was positive to neither SUMO nor ubiquitin antibody, consistent with the prediction that this is an unmodified form. Additional signals (above 72 kDa marker) that occurred in pull-down samples blotted with the anti-ubiquitin antibody were nonspecific as they were also present in vehicle-transfected cells.
As the first step in determining whether SALL4B was sumoylated in vivo, we transfected Tera-1 cells with a plasmid construct expressing SENP1, an isopeptidase specifically removing the SUMO moiety (26, 27). Ectopic expression of SENP1, but not the enzymatically inactive SENP1 (SENP1-Mut) significantly diminished 130 kDa SALL4B but left other SALL4 signals intact (Fig. 2A), suggesting that SUMO-modified SALL4 was constitutively present in Tera-1 cells. To confirm sumoylation of SALL4, HEK293 cells were co-transfected with SALL4B expression plasmid and the plasmid expressing SENP1 (or SENP2). Ectopic expression of SENP1 or SENP2, but not SENP1 mutant, greatly reduced 130 kDa SALL4B signal. In contrast, SENP1 or SENP2 expression did not reduce 95 or 70 kDa signals at all (Fig. 2B). These observations are thus consistent with the sumoylated nature of 130 kDa band. We next co-transfected HEK293T cells with FLAG-tagged SUMO-1 and His-HA-tagged SALL4B expression constructs, along with plasmids expressing SENP1, SENP2, and/or FLAG-USP28. Ni-IDA pull-down samples, along with the cell lysate input, were blotted with antibodies to the FLAG and HA tags, respectively. Ectopic expression of FLAG-SUMO-1 resulted in the appearance of numerous FLAG-tagged proteins and co-transfection with either SENP1 or SENP2, significantly reduced the incorporation of FLAG signals, thus confirming their biochemical activities after expression (Fig. 2C, upper panel). Pull-down analysis revealed that ectopically expressed SALL4B was heavily modified by FLAG-SUMO-1, which was significantly reduced by co-transfection of either SENP1 or SENP2. An additional SALL4B band that migrated more slowly than the 130 kDa band was also detected. Again, this signal was greatly reduced or abolished upon transfection with SENP1 or SENP2, indicating that it is also a super-sumoylated form of SALL4B (arrow SS-SALL4B).
FIGURE 2.
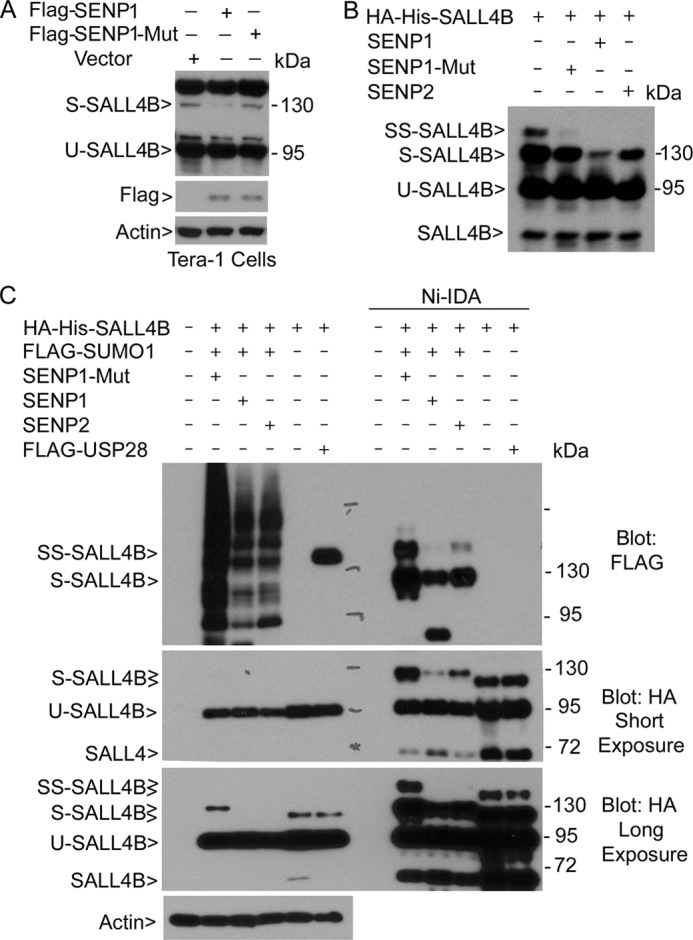
SALL4B is SUMO-modified. A, Tera-1 cells were transfected with a plasmid expressing SENP1, SENP1-Mut (enzymatically inactive), or vehicle for 48 h after which equal amounts of cell lysates were blotted with antibodies to SALL4, SENP1 and β-actin. B, HEK293T cells co-transfectd with various expression plasmids as indicated for 48 h, after which equal amounts of cell lysates were blotted with the anti-HA antibody. Arrow SS-SALL4B denotes super-sumoylated SALL4B. C, HEK293T cells were transfected with various expression plasmids as indicated for 48 h. Cell lysates of various treatments were incubated with Ni-IDA resin. Proteins specifically bound to the resin, along with the lysate inputs, were blotted with antibodies to FLAG, HA, and β-actin. Blots of both short and long exposure are shown. SS-SALL4B denotes super-sumoylated form of SALL4B.
Blotting with the HA antibody also confirmed the expression of both ubiquitinated and sumoylated SALL4B, signals of which were significantly enriched after Ni-IDA pull-down (Fig. 2C, middle panels). Again, ectopic expression of SENP1 or SENP2 but not USP28 greatly reduced sumoylated SALL4B. Moreover, expression of neither SENP1 nor SENP2 affected the level of ubiquitinated SALL4B. Of note, SUMO-modified SALL4B with a fast mobility was also detected in cells without transfection of FLAG-SUMO-1. This was likely due to the modification by the endogenous SUMO molecule as transfected SUMO-1 was conjugated with three-tandem FLAG tags. Ectopic expression of USP28 did not modulate the level of ubiquitinated SALL4B either (Fig. 2C), which is not surprising as most de-ubiquitinating enzymes including USP28 exhibit substrate specificity (28, 29).
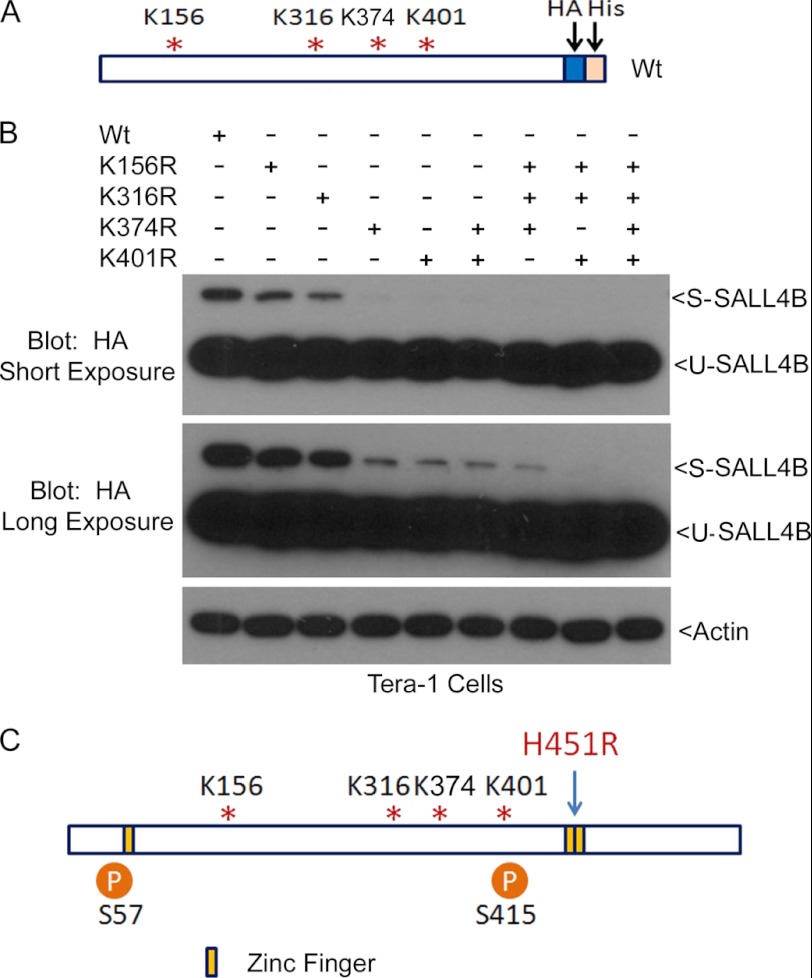
SALL4B Is SUMO-modified on Multiple Lysine Residues
To identify the potential lysine residues of SALL4B that were sumoylated, we analyzed SALL4B amino acid sequences for optimal sumoylation sites using the criteria available at Abgent Inc. (www.abgent.com). Four lysines sites (K156, K316, K374, and K401) with the highest scores were subjected to mutagenic analysis. The relative position of these sites to other domains is shown in Fig. 3A. We first made plasmid constructs expressing an HA- and His-tagged SALL4B (Wt), and its corresponding mutants with different lysine residues replaced with arginines. Immunoblotting with antibodies to SALL4 confirmed that both Wt and mutant SALL4B were efficiently expressed (Fig. 3B). However, SALL4B mutants all exhibited compromised sumoylation and K401 was more critical for sumoylation than K156, K316, or K347 (Fig. 3B, long exposure). Mutations of K156, K316, and K401 or mutations of all four residues essentially wiped out SALL4B sumoylation.
FIGURE 3.
SALL4B is sumoylated on multiple sites. A, schematic presentations of wild-type SALL4B, as well as its various mutants. Lysine residues subjected to mutation into arginines are shown. HA and His6 tags are fused in-frame at the C terminus. B, HEK293T cells were transfected with HA-tagged wild-type SALL4B (Wt) and various mutants with single or multiple lysine residues replaced with arginines for 48 h, after which equal amounts of cell lysates were blotted with antibodies to HA and β-actin. Blots of both short and long exposure are shown. C, schematic presentation of important residues of SALL4B for sumoylation and phosphorylation, as well as a histidine residue in the zinc finger domain mutation of which causes mild Okihiro syndrome.
Mass spectrometry analysis of SALL4B that were expressed and purified from Sf9 cells revealed two serine residues (Ser-57 and Ser-415) were phosphorylated (Fig. 3C; supplemental Figs. S3 and S4). Intriguingly, both phosphorylation sites precede zinc finger domains. The zinc finger domains are essential for SALL4 function since deletions or non-sense mutations lead to developmental disorders (14, 30–32). In fact, a mutation with histidine (His-888 in SALL4A corresponding to His-451 in SALL4B) changed into arginine within the double zinc finger domain is associated with Okihiro syndrome in human (33). Lys-401, a crucial residue for sumoylation, also lies in close proximity to the second phosphorylation site, suggesting a possible regulatory interaction between these residues. Supporting this notion, it has been shown that the removal of sumoylated Gcn4, a transcription factor, from the target gene promoter requires the protein kinase activity (34).
Sumoylation Is Important for SALL4 Stability and Its Localization
To understand how sumoylation regulates SALL4 function, we first determined whether sumoylation would affect its stability. Cells transfected with wild-type and mutant SALL4B (HA-His-SALL4B-4R) for 24 h were treated with cycloheximide for various times. Immunoblotting analysis revealed that SUMO-resistant SALL4B exhibited a much shorter half-life (>2.0 h) than the wild-type counterpart (>6 h) (Fig. 4A and supplemental Fig. S4), indicating that sumoylation positively regulating its stability.
FIGURE 4.
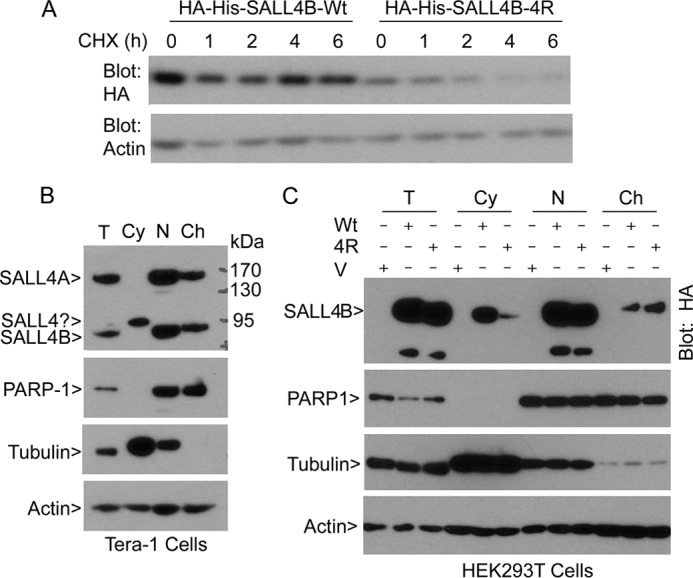
Sumoylation is important for the stability and subcellular localization of SALL4B. A, HEK293T cells transfected with a plasmid expressing HA-His-SALL4B (Wt) or HA-His-SALL4B mutant (4R) for 24 h were treated with cycloheximide (CHX) for various times as indicated. At the end of the treatment, cells were collected and lysed and approximately equal amounts of cell lysates were blotted with the antibody to HA and β-actin, respectively. B, cytoplasmic, nucleoplasmic, and chromatin fractions of Tera-1 cells were blotted with antibodies to SALL4, PARP-1, α-tubulin, and β-actin. Total cell lysates were also analyzed as control. C, HEK293T cells were transfected with the wild-type SALL4B (Wt) expression plasmid, its 4R mutant (4R), or vehicle (V) for 48 h, after which equal amounts of protein lysates of cytoplasmic, nucleoplasmic, and chromatin fractions were blotted with antibodies to HA, PARP-1, α-tubulin, and β-actin.
Constitutively expressed SALL4A and SALL4B were primarily localized to the nucleus of Tera-1 cells with a significant fraction associated with chromatin (Fig. 4B). The SALL4-specific antigen with a molecular mass of 95 kDa was enriched in the cytoplasm, but not in the nucleus. We then transfected HEK293T cells with HA- and His-tagged wild-type SALL4B and SALL4B-4R. Ectopically expressed SALL4B and its mutant proteins were blotted with the antibody to HA after fractionation. Both wild-type and mutant proteins were enriched in the nucleoplasm (Fig. 4C). A relatively small amount of wild-type of SALL4B but not 4R mutant protein was also detected in the cytoplasm. On the other hand, 4R mutant protein was enriched in the chromatin fraction despite its overall reduced level in the cell, suggesting that sumoylation may negatively regulate its chromatin association.
Sumoylation Regulates SALL4B Transcriptional Activity
As the first step to examining whether SALL4B sumoylation may affect its transactivation, we examined the physical and functional interaction between SALL4B and OCT4. OCT4 antibody, but not the control IgG, not only precipitated OCT4 but also SALL4B (Fig. 5A). We then determined the physical interaction between ectopically expressed tagged SALL4B (or its sumoylation-deficient mutants) and endogenous OCT4. SALL4B precipitates enriched after Ni-IDA pull-down were blotted for both HA and OCT4. Compared with the wild-type SALL4B, 3R (K156R/K316R/K401R), and 4R (K156R/K316R/K374R/K401R) mutants was more efficient in precipitating a small fraction of OCT4 (Fig. 5B), suggesting that sumoylation may negatively regulate the physical interaction between SALL4B and OCT4.
FIGURE 5.
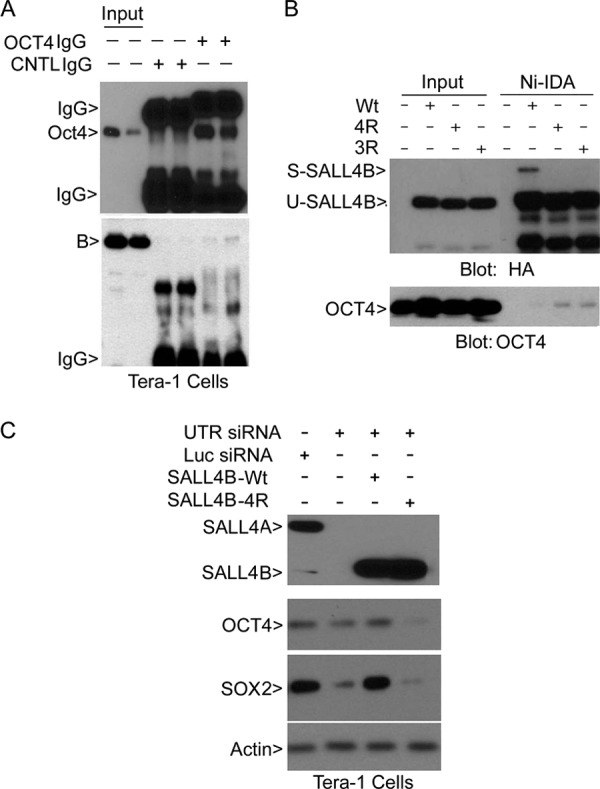
SALL4B sumoylation regulates its physical interaction with OCT4, as well as expression of Oct4 and SOX2. A, Tera-1 cell lysates in duplicate were immunoprecipitated with OCT4 IgG or control IgG. OCT4 immunoprecipitates, along with the lysate inputs, were blotted with antibodies to OCT4 and SALL4. B, HEK293T cells were transfected with wild-type SALL4B or its mutants for 48 h. 3R and 4R stand for SALL4B mutants with three (K156, K316, and K401) and four (K156, K316, K374, and K401) lysine residues replaces with arginines, respectively. Ectopically expressed SALL4B or its mutant proteins were enriched by incubation with Ni-IDA resin. Proteins bound to the resin, along with the lysate input, were blotted with antibodies to HA and OCT4. Tera-1 cells were co-transfected with SALL4 siRNA (or control siRNA) and SALL4B (or SALL4B-4R mutant) for 48 h, after which equal amounts of cell lysates were blotted with antibodies to SALL4 and HA. C, Tera-1 cells were co-transfected with SALL4 siRNA (or luciferase siRNA) and a plasmid expressing wild-type, or sumoylation-resistant, SALL4B for 48 h. Cells were then lysed, and equal amounts of cell lysates were blotted with antibodies to SALL4, OCT4, SOX2, and β-actin.
As SALL4 regulates OCT4 and SOX2 expression (10), we next studied whether SALL4B sumoylation played a role in controlling expression of these stem cell transcription factors. Specific silencing of SALL4 via RNAi resulted in reduction of OCT4 and SOX2 (Fig. 5C). Ectopic expression of wild-type SALL4B, but not its sumoylation-resistant mutant, was capable of rescuing down-regulation of OCT4 and SOX2. These results strongly suggest that sumoylation is essential for the maintenance of transcriptional activity of SALL4B.
To functionally study the effect of SALL4 sumoylation in the transaction of its target gene expression, we first established the system in which endogenous SALL4 expression was silenced and transfected SALL4B could be expressed. Immunoblotting with antibodies to both SALL4 and the HA tag showed that endogenous SALL4A and SALL4B were efficiently down-regulated after transfection with 3′-UTR siRNAs whereas wild-type SALL4B or its 4R mutant plasmid was efficiently expressed after transfection (Fig. 6A). SALL4 binds to the OCT4 promoter and transactivates its gene expression (10). Therefore, we next co-transfected Tera-1 cells with the plasmid expressing firefly luciferase driven by the OCT4 promoter and the plasmid expressing wild-type SALL4B or 4R mutant. To minimize the influence of endogenous SALL4 proteins, these cells were co-transfected with SALL4 3′-UTR siRNAs. Reporter gene assays revealed that transfection with the wild-type SALL4B expression plasmid significantly activated the reporter gene activity as compared with the vector-transfected control and that 4R mutant was much less efficient in activating the reporter gene (Fig. 6B). Given that SALL4 is capable of trans-repressing its own gene expression (10), we next analyzed the reporter gene activity in Tera-1 cells that were co-transfected with a plasmid expressing firefly luciferase driven by the SALL4 promoter and the plasmid expressing wild-type SALL4B or 4R mutant. Expression of wild-type SALL4B greatly reduced the activity of the reporter gene as compared with the vehicle-transfected control. Although expression of 4R mutant also reduced the reporter gene activity, the magnitude of reduction was less pronounced compared with that of the wild-type SALL4B (Fig. 6C), implicating a negative effect of sumoylation on trans-repression of its own gene expression.
FIGURE 6.
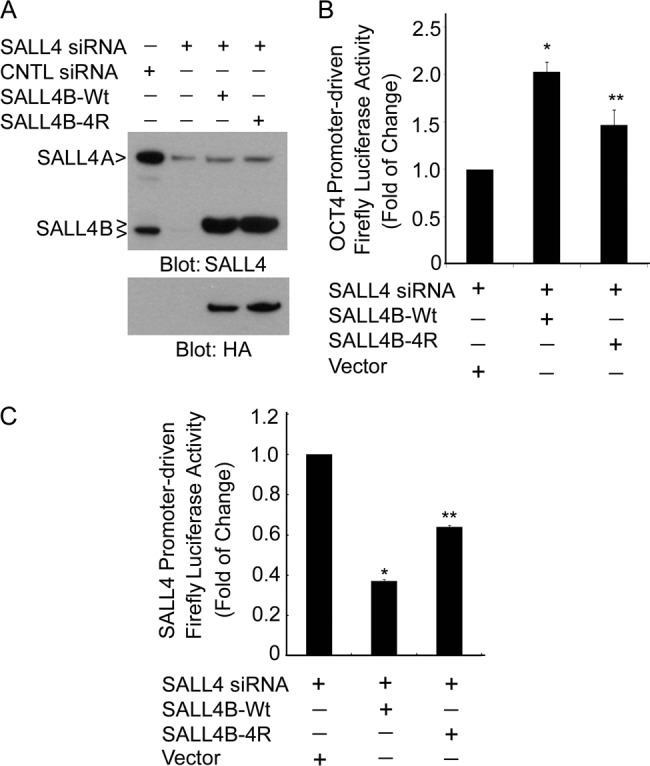
Sumoylation regulates transcriptional activity of SALL4B. A, Tera-1 cells were co-transfected with siRNAs to SALL4 (or to luciferase as control) and a plasmid expressing wild-type either wild-type or sumoylation-resistant SALL4B for 48 h. At the end of treatment, cells were collected and lysed, and equal amounts of cell lysates were blotted with antibodies to SALL4 and HA, respectively. B, Tera-1 cells were seeded in triplicate and co-transfected with the firefly luciferase reporter construct driven by the OCT4 gene promoter and various SALL4B expression plasmids as indicated for 48 h. Endogenous SALL4 proteins were down-regulated by co-transfection with SALL4 siRNA. At the end of transfection, cell lysates were prepared and assayed for firefly luciferase activity. Data are expressed as the fold-change of firefly luciferase activity after normalization. Each experiment was repeated for at least three times. C, Tera-1 cells were co-transfected with the firefly luciferase reporter construct driven by the SALL4 gene promoter and various SALL4B expression plasmids as indicated for 48 h. Endogenous SALL4 proteins were down-regulated by co-transfection with SALL4 siRNA. At the end of transfection, cell lysates were prepared and assayed for firefly luciferase activity. Data are expressed as the fold-change of firefly luciferase activity after normalization. Each experiment was repeated for at least three times.
DISCUSSION
In this study, we report for the first time that SALL4B is SUMO-modified and that SALL4B sumoylation plays an important role in regulating its subcellular localization, as well as its transcriptional activities. SALL4B appears to be modified by both SUMO-1 and SUMO-2 as the sumoylated form of SALL4B is positive to both SUMO-1 and SUMO-2 antibodies. Supporting this notion, the major SUMO-modified band is about 60 kDa larger than the predicted, unmodified form and thus large enough for both SUMO-1 and SUMO-2 modifications. The observation that mutation of several lysine residues is required for the complete elimination of SALL4B sumoylation strongly suggests that sumoylation occurs on multiple sites. Furthermore, super-sumoylated forms of SALL4B are also detected in cells transfected with SALL4B expression plasmid, particularly after enrichment with Ni-IDA (Fig. 2C). We have demonstrated that SALL4B is modified by ubiquitination and phosphorylation as well although the exact mechanism by which these modifications in regulating SALL4 remains to be elucidated. It appears that sumoylation and ubiquitination are mutually exclusive as the sumoylated SALL4B is not immunoreactive to the ubiquitin antibody or vice versa. One caveat for this explanation is that the antigen epitope for each type of modification could be masked by other type of modification.
SALL4B exists primarily as the ubiquitinated form and the modification appears to be conserved, as SALL4B expressed in Sf9 cells also co-migrates with the ubiquitinated form (Fig. 1A). On the other hand, no SALL4B that co-migrates with the sumoylated form is detected on denaturing gels after expression in Sf9 cells, suggesting that sumoylation is evolved as a regulatory mechanism in higher animal cells. It is noted that SALL4A detected in Tera-1 cells migrates at the position similar to the super sumoylated SALL4B (Figs. 1A and 2D). However, SALL4A is not a SUMO-modified form of SALL4B as SENP1 expression does not reduce the intensity of this band (Fig. 2A). More importantly, SALL4A is specifically silenced after transfection with SALL4A-specific siRNAs (Fig. 1C). HeLa and Jurkat cells express a protein that is immunoreactive to SALL4 antibody and migrates at about 95 kDa (Fig. 1A), although the nature of this band remains unknown. Intriguingly, Tera-1 cells also contain a small amount of the 95 kDa antigen, which is primarily localized to the cytoplasm (Fig. 4B). It is tempting to speculate that it may represent a new spliced form of SALL4. Ectopically expressed SALL4B has an apparent molecular mass of 95 kDa (co-migrating with the SALL4B band form in HeLa and Jurkat cells) and is larger than endogenous SALL4B in Tera-1. The increased mobility is caused by the addition of both His tag and 3 tandem FLAG tags.
Increasing evidence indicates that the sumoylation pathway plays an essential role in embryogenesis and development (35–37). Sumoylation modulates the affinity with DNA binding and the activity of stem cell transcription factors (23), as well as the subcellular localization of general transcription factors (38, 39). Our current study indicates that SALL4B sumoylation regulates its transcriptional activity. SUMO-resistant mutant exhibits an enhanced binding to chromatin as compared with the wild-type control, suggesting that sumoylation may directly impact on its transcriptional activity. However, reporter gene assays indicate that SUMO-resistant mutant SALL4B has a reduced transcription activity. One possibility is that the mutant protein may be associated with a different subdomain of chromatin through interacting with a different set of protein partners. Moreover, given that SUMO-resistant mutant binds more efficient with OCT4 (Fig. 5B), it is possible that the mutant SALL4B may sequestrate OCT4 and reduce its transcriptional activity because OCT4 also auto-regulates its expression (40). Similar to our finding, SOX2 is modified by SUMO-1 at lysine 247 and sumoylated SOX2 exhibits reduced DNA binding to the enhancer region of its target gene (23).
Although embryonic stem cells express both SALL4A and SALL4B, both forms appear to have a function in the maintenance and self-renewal of stem cells (5, 10, 25). SALL4A and SALL4B bind to overlapping but not identical sites within the genome; when it is expressed alone, SALL4B can maintain the pluripotent state of mouse stem cells (25). A recent study reveals that either SALL4A or SALL4B can stimulate a robust ex vivo expansion of CD34+ hematopoietic stem cells when it is ectopically expressed via the lentiviral approach (41). More importantly, SALL4B-mediated stem cell expansion is associated with their engraftment and long-term repopulation in vivo (41). Our current study reveals a novel regulatory mechanism by which SALL4B functions to control the transactivation of OCT4, the master gene for maintaining stem cell properties and their renewal. Sumoylation-deficient SALL4B is less efficient in enhancing the transactivation activity for the OCT4 promoter, resulting in a compromised activity of the reporter gene product (Fig. 5C). This is consistent with the early observation that SALL4 positively regulates OCT4 gene expression (10). Further supporting the positive regulation of OCT4 by SALL4, we have observed that down-regulation of SALL4 expression in Tera-1 cells via transfection of SALL4-specific siRNA leads to decreased levels of SOX2, as well as OCT4 (Fig. 5C).
Acknowledgments
We thank coworkers in the laboratory for valuable discussions and suggestions. We thank Dr. Jingke Chen for providing us with SENP1 expression constructs and Michael Matunis for SUMO-1 and SUMO-2/3 antibodies, as well as SENP2 expression construct.
This study was supported, in whole or in part, by US Public Service Awards (to W. D.) (CA090658 and ES019929), NIEHS Center Grant ES000260, and a key State project of China on iPS and stem cell research (2011ZX09102-010-04).

This article contains supplemental Figs. S1–S4.
- iPS
- induced pluripotent stem
- CHX
- cycloheximide.
REFERENCES
- 1. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 2. Yamanaka S., Blau H. M. (2010) Nature 465, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin, Thomson J. A. (2009) Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang J., Chai L., Fowles T. C., Alipio Z., Xu D., Fink L. M., Ward D. C., Ma Y. (2008) Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 105, 19756–19761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang J., Tam W. L., Tong G. Q., Wu Q., Chan H. Y., Soh B. S., Lou Y., Yang J., Ma Y., Chai L., Ng H. H., Lufkin T., Robson P., Lim B. (2006) Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 8, 1114–1123 [DOI] [PubMed] [Google Scholar]
- 6. van den Berg D. L., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2010) Cell Stem. Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu Q., Chen X., Zhang J., Loh Y. H., Low T. Y., Zhang W., Sze S. K., Lim B., Ng H. H. (2006) Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J. Biol. Chem. 281, 24090–24094 [DOI] [PubMed] [Google Scholar]
- 8. Lim C. Y., Tam W. L., Zhang J., Ang H. S., Jia H., Lipovich L., Ng H. H., Wei C. L., Sung W. K., Robson P., Yang H., Lim B. (2008) Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem. Cell 3, 543–554 [DOI] [PubMed] [Google Scholar]
- 9. Tsubooka N., Ichisaka T., Okita K., Takahashi K., Nakagawa M., Yamanaka S. (2009) Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells 14, 683–694 [DOI] [PubMed] [Google Scholar]
- 10. Yang J., Gao C., Chai L., Ma Y. (2010) PLoS One 5, e10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ma Y., Cui W., Yang J., Qu J., Di C., Amin H. M., Lai R., Ritz J., Krause D. S., Chai L. (2006) SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood 108, 2726–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shuai X., Zhou D., Shen T., Wu Y., Zhang J., Wang X., Li Q. (2009) Overexpression of the novel oncogene SALL4 and activation of the Wnt/β-catenin pathway in myelodysplastic syndromes. Cancer Genet. Cytogenet. 194, 119–124 [DOI] [PubMed] [Google Scholar]
- 13. Cui W., Kong N. R., Ma Y., Amin H. M., Lai R., Chai L. (2006) Differential expression of the novel oncogene, SALL4, in lymphoma, plasma cell myeloma, and acute lymphoblastic leukemia. Mod. Pathol. 19, 1585–1592 [DOI] [PubMed] [Google Scholar]
- 14. Kohlhase J., Chitayat D., Kotzot D., Ceylaner S., Froster U. G., Fuchs S., Montgomery T., Rösler B. (2005) SALL4 mutations in Okihiro syndrome (Duane-radial ray syndrome), acro-renal-ocular syndrome, and related disorders. Hum. Mutat 26, 176–183 [DOI] [PubMed] [Google Scholar]
- 15. Sterneckert J., Höing S., Schöler H. R. (2012) Concise review: Oct4 and more: the reprogramming expressway. Stem Cells 30, 15–21 [DOI] [PubMed] [Google Scholar]
- 16. Brumbaugh J., Hou Z., Russell J. D., Howden S. E., Yu P., Ledvina A. R., Coon J. J., Thomson J. A. (2012) Phosphorylation regulates human OCT4. Proc. Natl. Acad. Sci. U.S.A. 109, 7162–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wei F., Schöler H. R., Atchison M. L. (2007) Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J. Biol. Chem. 282, 21551–21560 [DOI] [PubMed] [Google Scholar]
- 18. Hannoun Z., Greenhough S., Jaffray E., Hay R. T., Hay D. C. (2010) Post-translational modification by SUMO. Toxicology 3, 288–293 [DOI] [PubMed] [Google Scholar]
- 19. Mallanna S. K., Ormsbee B. D., Iacovino M., Gilmore J. M., Cox J. L., Kyba M., Washburn M. P., Rizzino A. (2010) Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells 28, 1715–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baltus G. A., Kowalski M. P., Zhai H., Tutter A. V., Quinn D., Wall D., Kadam S. (2009) Acetylation of sox2 induces its nuclear export in embryonic stem cells. Stem. Cells 27, 2175–2184 [DOI] [PubMed] [Google Scholar]
- 21. Zhao H. Y., Zhang Y. J., Dai H., Zhang Y., Shen Y. F. (2011) PLoS One 6, e27026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeong C. H., Cho Y. Y., Kim M. O., Kim S. H., Cho E. J., Lee S. Y., Jeon Y. J., Lee K. Y., Yao K., Keum Y. S., Bode A. M., Dong Z. (2010) Phosphorylation of Sox2 cooperates in reprogramming to pluripotent stem cells. Stem Cells 28, 2141–2150 [DOI] [PubMed] [Google Scholar]
- 23. Tsuruzoe S., Ishihara K., Uchimura Y., Watanabe S., Sekita Y., Aoto T., Saitoh H., Yuasa Y., Niwa H., Kawasuji M., Baba H., Nakao M. (2006) Inhibition of DNA binding of Sox2 by the SUMO conjugation. Biochem. Biophys. Res. Commun. 351, 920–926 [DOI] [PubMed] [Google Scholar]
- 24. Lu J., Jeong H. W., Kong N., Yang Y., Carroll J., Luo H. R., Silberstein L. E., Yupoma, Chai L. (2009) Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS One 4, e5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rao S., Zhen S., Roumiantsev S., McDonald L. T., Yuan G. C., Orkin S. H. (2010) Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol. Cell Biol. 30, 5364–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheng J., Kang X., Zhang S., Yeh E. T. (2007) SUMO-specific protease 1 is essential for stabilization of HIF1α during hypoxia. Cell 131, 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang F., Hu L., Chen C., Yu J., O'Connell C. B., Khodjakov A., Pagano M., Dai W. (2012) BubR1 is modified by sumoylation during mitotic progression. J. Biol. Chem. 287, 4875–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 29. Popov N., Wanzel M., Madiredjo M., Zhang D., Beijersbergen R., Bernards R., Moll R., Elledge S. J., Eilers M. (2007) The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 9, 765–774 [DOI] [PubMed] [Google Scholar]
- 30. Borozdin W., Wright M. J., Hennekam R. C., Hannibal M. C., Crow Y. J., Neumann T. E., Kohlhase J. (2004) Novel mutations in the gene SALL4 provide further evidence for acro-renal-ocular and Okihiro syndromes being allelic entities, and extend the phenotypic spectrum. J. Med. Genet. 41, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang B., Li L., Xie X., Wang J., Yan J., Mu Y., Ma X. (2010) Genetic variation of SAL-Like 4 (SALL4) in ventricular septal defect. Int. J. Cardiol 145, 224–226 [DOI] [PubMed] [Google Scholar]
- 32. Paradisi I., Arias S. (2007) IVIC syndrome is caused by a c.2607delA mutation in the SALL4 locus. Am. J. Med. Genet. A 143, 326–332 [DOI] [PubMed] [Google Scholar]
- 33. Miertus J., Borozdin W., Frecer V., Tonini G., Bertok S., Amoroso A., Miertus S., Kohlhase J. (2006) A SALL4 zinc finger missense mutation predicted to result in increased DNA binding affinity is associated with cranial midline defects and mild features of Okihiro syndrome. Hum. Genet. 119, 154–161 [DOI] [PubMed] [Google Scholar]
- 34. Rosonina E., Duncan S. M., Manley J. L. (2012) Sumoylation of transcription factor Gcn4 facilitates its Srb10-mediated clearance from promoters in yeast. Genes Dev. 26, 350–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kang X., Qi Y., Zuo Y., Wang Q., Zou Y., Schwartz R. J., Cheng J., Yeh E. T. (2010) SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell 38, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chiu S. Y., Asai N., Costantini F., Hsu W. (2008) SUMO-specific protease 2 is essential for modulating p53-Mdm2 in development of trophoblast stem cell niches and lineages. PLoS Biol. 6, e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taylor K. M., Labonne C. (2005) SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev. Cell 9, 593–603 [DOI] [PubMed] [Google Scholar]
- 38. Yang W. H., Heaton J. H., Brevig H., Mukherjee S., Iñiguez-Lluhi J. A., Hammer G. D. (2009) SUMOylation inhibits SF-1 activity by reducing CDK7-mediated serine 203 phosphorylation. Mol. Cell Biol. 29, 613–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hamard P. J., Boyer-Guittaut M., Camuzeaux B., Dujardin D., Hauss C., Oelgeschläger T., Vigneron M., Kedinger C., Chatton B. (2007) Sumoylation delays the ATF7 transcription factor subcellular localization and inhibits its transcriptional activity. Nucleic Acids Res. 35, 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okumura-Nakanishi S., Saito M., Niwa H., Ishikawa F. (2005) Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem. 280, 5307–5317 [DOI] [PubMed] [Google Scholar]
- 41. Aguila J. R., Liao W., Yang J., Avila C., Hagag N., Senzel L., Ma Y. (2011) SALL4 is a robust stimulator for the expansion of hematopoietic stem cells. Blood 118, 576–585 [DOI] [PMC free article] [PubMed] [Google Scholar]