Background: How are mitochondrial functions regulated in response to mitochondrial defects?
Results: Acute OXPHOS defects lead rapidly to a glucose-dependent repression of complex I activity.
Conclusion: We discovered an “emergency shutdown” system adapting respiratory chain function to mitochondrial defects.
Significance: This study reveals a novel type of metabolic regulation that could open the way to medically useful alterations of OXPHOS.
Keywords: Glucose, Mitochondrial Diseases, Respiratory Chain, Signaling, Bioenergetics/Electron Transfer Complex, Cell Metabolism
Abstract
Proper coordination between glycolysis and respiration is essential, yet the regulatory mechanisms involved in sensing respiratory chain defects and modifying mitochondrial functions accordingly are unclear. To investigate the nature of this regulation, we introduced respiratory bypass enzymes into cultured human (HEK293T) cells and studied mitochondrial responses to respiratory chain inhibition. In the absence of respiratory chain inhibitors, the expression of alternative respiratory enzymes did not detectably alter cell physiology or mitochondrial function. However, in permeabilized cells NDI1 (alternative NADH dehydrogenase) bypassed complex I inhibition, whereas alternative oxidase (AOX) bypassed complex III or IV inhibition. In contrast, in intact cells the effects of the AOX bypass were suppressed by growth on glucose, whereas those produced by NDI1 were unaffected. Moreover, NDI1 abolished the glucose suppression of AOX-driven respiration, implicating complex I as the target of this regulation. Rapid Complex I down-regulation was partly released upon prolonged respiratory inhibition, suggesting that it provides an “emergency shutdown” system to regulate metabolism in response to dysfunctions of the oxidative phosphorylation. This system was independent of HIF1, mitochondrial superoxide, or ATP synthase regulation. Our findings reveal a novel pathway for adaptation to mitochondrial dysfunction and could provide new opportunities for combatting diseases.
Introduction
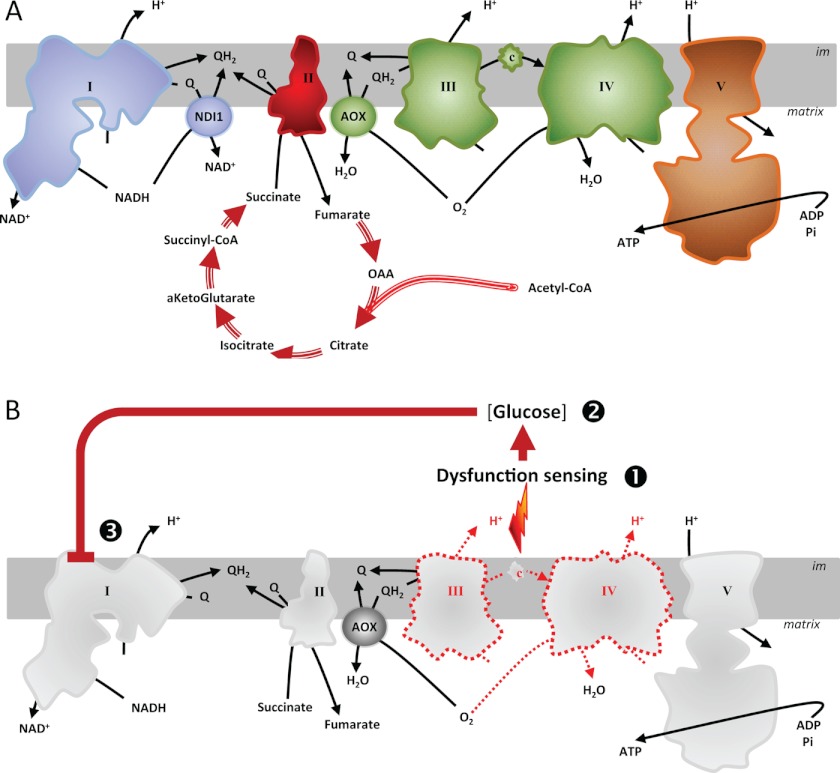
Mitochondrial diseases encompass a large group of pathologies, each presenting a dysfunctional oxidative phosphorylation (OXPHOS)4 system (1, 2). The five complexes of the OXPHOS system participate in a single orchestrated process (see Fig. 1A): the production of cellular energy via the coupling of respiratory electron transport to proton translocation across the inner mitochondrial membrane (complexes I–IV) combined with an energy transfer system that converts the resulting proton gradient into ATP (complex V). The interdependence of the respiratory chain complexes suggests that the consequences of any dysfunction should depend on which complexes are blocked in an oxidized and which ones end up in a reduced state. However, dysfunctional subunits of the same complex, expected to lead to similar biochemical consequences, can be associated with different pathologies, sometimes even presenting tissue specificity, despite a widespread or even ubiquitous respiratory dysfunction (3).
FIGURE 1.
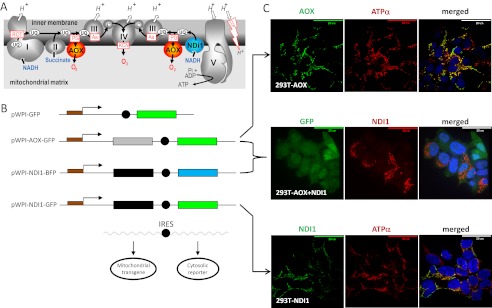
AOX and NDI1 provide alternative respiratory pathways in HEK293T cells. A, schematic diagram of the respiratory chain including AOX and NDI1. The mitochondrial respiratory chain conducts the flow of electrons (black arrows) from NADH to oxygen through four respiratory complexes (I–IV, respiratory chain complexes I–IV). Electron transfer between complexes is permitted by specific transporters: ubiquinones (UQ) and cytochrome c (c). Energy produced by the flow of electrons allows pumping of protons (H+) across the inner mitochondrial membrane. Proton re-entry through ATP synthase (V) drives the generation of ATP. CCP, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazones (e.g., FCCP) are chemical protonophores allowing H+ to cross the otherwise proton-impermeable inner mitochondrial membrane. NDI1 allows electron flow in presence of complex I inhibitors (e.g., rotenone (ROT)), whereas AOX allows electron flow in the presence of inhibitors of complex III (e.g., antimycin (Aa)) or IV (cyanide, KCN). AOX activity is specifically inhibited by n-propyl gallate (PG). B, lentiviral vectors. The pWPI-AOX-GFP lentivector allows co-expression of a cytosolic GFP (under the control of an internal ribosome entry site, black circle) together with C. intestinalis AOX; both are under the control of the EF1α promoter. The pWPI-NDI-GFP is a similar lentiviral vector allowing co-expression of a cytosolic GFP and S. cerevisiae NDI1. The pWPI-NDI-BFP is a modified pWPI-NDI-GFP where the coding sequence of GFP has been replaced by that blue fluorescent protein. C, AOX and NDI1 proteins are localized in mitochondria. Immunofluorescent staining of fixed HEK293T cells transduced as follows. Top panel, cells transduced with pWPI-AOX-GFP. ATPα, subunit α of the mitochondrial ATPase. Bottom panel, HEK293T cells transduced with pWPI-NDI1-GFP. Middle panel, cells transduced with pWPI-AOX-GFP (see top panel) and reinfected with viral particles generated using pWPI-NDI1-BFP. GFP indicates green fluorescent protein (a marker for the pWPI-AOX-GFP transduced cells) (see also supplemental Fig. S1A, top panel). The merged image also shows DAPI nuclear counterstaining. The images are representative pictures (n = 10).
Recent studies suggest that cells adapt to mitochondrial dysfunction by switching to glycolysis, despite aerobic conditions (4): this is reminiscent of the long known Warburg and Crabtree effects. The latter describes the ability of rapidly proliferating cells to favor glycolysis, depending on substrate availability, despite normoxic conditions. Despite its discovery in the 1920s, the precise mechanisms involved in the Crabtree effect are multiple and still debated (5–7). The Warburg effect describes the regulation of mitochondrial respiration in response to hypoxia. It is now known to involve the stabilization of HIF1, leading to the nuclear translocation of the transcription factor hypoxia-inducible factor 1 (HIF1). Interestingly, HIF1 stabilization can also be induced even in normoxic conditions by cytosolic accumulation of succinate and/or reactive oxygen species (8, 9), which are common although not universal consequences of mitochondrial OXPHOS dysfunction. One recently identified mechanism of metabolic regulation of mitochondrial activity involves the protein deacetylase SIRT3, which can deacetylate the NDUFS9 subunit of complex I leading to its activation (10). Acetylation of other respiratory chain subunits has also been observed (11), but the physiological roles of these modifications, as well as how they are controlled, require further investigation. Another type of regulation involves the transcription factor STAT3, which has been shown to translocate to mitochondria in response to IFN-β, down-regulating complex I and II activities (12). Metabolic adaptation in cases of mitochondrial disease have only recently begun to be investigated (13), and the effectors for such regulation are still unknown. To understand how cells adapt to mitochondrial defects before transcriptional changes can occur, we took advantage of the fact that alterative respiratory enzymes from lower eukaryotes can be expressed in mammalian cells.
Alternative oxidases (AOX) can bypass mitochondrial complex IV deficiencies in human cells, improving the viability of cells exposed to ROS or to low glucose culture conditions (14). Similarly, alternative NADH dehydrogenases (e.g., NDI1) can complement complex I defects in mice and in cultured cells (15). With the aim of understanding how mammalian mitochondrial function is regulated after a severe perturbation of the respiratory chain, we tested whether the activity of these respiratory bypass enzymes in HEK293T cells was suppressed by acute mitochondrial dysfunction (see Figs. 1A and 6A). Because the ability to produce energy via fermentation should be a critical factor for cellular adaptation to respiratory dysfunction, the cells to be tested were grown either on an efficiently fermented substrate (glucose) or on one that is poorly fermentable (galactose) (16).
FIGURE 6.
A, the alternative enzymes are respiratory bypass enzymes. Blue, respiratory complex (complex I) bypassed by NDI1; green, respiratory complexes (complexes III and IV) bypassed by AOX; brown, respiratory complex (complex V) bypassed by the simultaneous activity of AOX and NDI1; red, activities that cannot be bypassed by these alternative enzymes in any combination. Inhibition or down-regulation of a complex will be suppressed in presence of the correct bypass enzyme. B, cells can adapt to dysfunction of complexes III and/or IV by modulating complex I activity according to their metabolic fuel. In response to complex III and IV dysfunction, cells can decrease electron transfer through the respiratory chain by down-regulation of complex I activity. The extent of such inhibition is proportional to the glucose availability, but glucose itself is not the regulatory molecule.
This strategy allowed us to discover a glucose-dependent down-regulation of mitochondrial complex I activity in response to the inhibition of the downstream respiratory chain complexes III or IV. Our data indicate the critical involvement of a small diffusible molecule, while excluding some of the most likely candidates.
EXPERIMENTAL PROCEDURES
Cell Culture and Proliferation Assays
The cells were maintained in high glucose DMEM plus glutamine, pyruvate, penicillin/streptomycin, and 10% FCS in a 5% CO2, 37 °C incubator. 24 h before each experiment cells were transferred to the relevant assay medium: minimal essential medium supplemented with glutamine, pyruvate, penicillin/streptomycin, and 10% FCS, plus 25 mm glucose, 5 mm glucose, or 25 mm galactose, hereafter described as high glucose, low glucose, and galactose assay media, respectively. Assay media were renewed once, 30 min before the experimental measurements. When cells producing decreased levels of mitochondrial ROS were required, 10 mm N-acetylcysteine was added to the assay medium.
To estimate cell doubling time, the cells were seeded in assay media in the presence (1 × 105 cells/cm2) or in absence (5 × 104 cells/cm2) of respiratory chain inhibitors. Assay media were renewed after 48 h, and cells were counted after 72 h using a Burker hemocytometer. Doubling time was calculated as timehours × log(2)/log (cellscounted/cellsseeded). Each measure is the average of 3 counting of the same sample.
Lentivector Generation and Infection
Ciona intestinalis AOX cDNA was cloned into pWPI, creating pWPI-AOX as reported earlier (14). The full-length NDI1 coding sequence was amplified from Saccharomyces cerevisiae and cloned into pWPI, creating the pWPI-NDI1 plasmid. pWPI-NDI-BFP was generated by replacing GFP from pWPI by BFP from pTag-BFP-C (Evrogen). Restriction digestions were carried out under the manufacturers' recommended conditions (New England Biolabs and Fermentas). Constructs were verified by sequencing.
Lentivector production used standard procedures and the second generation packaging system, which incorporates inbuilt safety features (17). All lentivectors were generated and tested by the Tampere Virus Facility.
Immunoblots
Post-nuclear extracts and mitochondrial fractions (18) were prepared from cells washed in PBS, pelleted, and suspended for 10 min in hypotonic buffer (10 mm NaCl, 1.5 mm MgCl2, 10 mm Tris-HCl, pH 7.5). After Dounce homogenization, the buffer was adjusted to 210 mm mannitol, 70 mm sucrose, 1 mm EDTA, 5 mm Tris-HCl, pH 7.5. The nuclei were pelleted by centrifugation for 5 min at 1,300 × gmax. The supernatant was recentrifuged for 15 min at 17,000 × gmax to recover crude mitochondrial and cytosolic fractions. Protein concentration was determined using a Nanodrop2000 spectrophotometer (Thermo Scientific). Immunoblots were performed as described previously (19) using antibodies against AOX (21st Century Biochemicals), NDI1 (gift of T. Yagi), Actin (Novus Biologicals), Lamin A + C (AbCam), NDUFS3, COX2, and ATP synthase subunit (Mitoscience). Anti-HIF1 ab463 antibody from AbCam was used according to the manufacturer's protocol. Western blots were quantified densitometrically, using ImageJ software.
Oxygen Consumption
Oxygen consumption was measured with a Clark-type electrode (Hansatech Oxytherm system). Intact cell respiration was recorded from 5 × 106 cells suspended in 500 μl of assay medium at 37 °C. Antimycin A (30 ng/ml) or rotenone (150 nm) concentrations were set to twice the optimal concentration for full respiratory inhibition (supplemental Fig. S3, C and D). Cyanide and n-propyl gallate were used at 100 μm. Oxygen consumption from 5 × 106 cells permeabilized by 80 μg/ml digitonin was recorded in respiratory buffer A (20) at 37 °C. The substrate concentrations were: ADP, 10 mm; for complex I: pyruvate + malate, 5 mm each; for complex II: succinate, 10 mm; for complex IV: TMPD + ascorbate, 50 μm and 1 mm, respectively; glucose and galactose: 25 mm. All measures were corrected by subtracting the residual oxygen consumption present after full inhibition of the respiratory chain (supplemental Fig. S4).
Fluorescent Biomarkers
After overnight culture in assay medium, 3 × 105 cells were treated with cell-permeant fluorescent indicators of cell death (2 μg/ml propidium iodide for 10 min on ice), mitochondrial mass (200 nm 10-nonyl acridine orange (NAO) for 30 min at 37 °C), mitochondrial superoxide (MitoSox, 2.5 μm for 45 min at 37 °C), or mitochondrial membrane potential (200 nm tetramethylrhodamine methyl ester (TMRM) for 30 min at 37 °C). When needed, inhibitors were added 5 min before the fluorescent indicator. Medium was replaced with PBS, and cells were kept on ice (MitoSox, propidium iodide) or at 37 °C (TMRM or NAO) before flow cytometry analysis. Negative controls for mitochondrial membrane potential were obtained by adding a mitochondrial uncoupler (FCCP, 1 μm) 1 min before flow cytometry analysis.
Flow Cytometry
Cell fluorescence was counted from 40,000 objects. The region of interest was defined using forward scatter/side scatter values: excluding debris and dead cells for mitochondrial mass (NAO), membrane potential (TMRM), and superoxide (Mitosox) measurements, but selecting all objects for life/death assessments (propidium iodide). MitoSox and TMRM staining were measured with a Coulter® EPICS®-XL-MCL instrument using 488-nm excitation and 620 ± 15-nm (FL3). GFP intensity was recorded at 520 ± 10-nm emission (FL1). Cell fluorescence was measured with an accuri® C6 flow cytometer (488-nm excitation) for propidium iodide (FL3: >670-nm emission), and NAO staining (FL2: 585 ± 40-nm emission). GFP intensity was recorded at 533 ± 30-nm emission (FL1) with this instrument. Compensations were estimated independently for each flow cytometer and each series of experiments.
Blue Native PAGE and In-gel Activity
400 μg of isolated mitochondria were solubilized in 80 μl of 1.5 m aminocaproic acid, 50 mm Bis-Tris and 10 μl of 10% dodecylmaltoside for 30 min on ice. The proteins were purified by centrifugation at 16,000 × gmax for 30 min at 4 °C and quantified using Nanodrop2000 spectrophotometer (Thermo Scientific). 2.5 μl of 5% (w/v) Coomassie Blue in aminocaproic acid was added for each 100 μg of protein. Electrophoresis and in gel activity assays for complex I and IV were conducted as previously described (21).
Immunofluorescence Staining and Confocal Imaging
Immunofluorescence protocols followed standard procedures (22). Alexa Fluor 568 and Alexa Fluor 488 (Invitrogen) were used as secondary antibodies. The nuclei were stained with DAPI during mounting (Pro Long Gold + DAPI; Molecular Probes). Images were captured using a Nikon Eclipse Ti inverted Microscope, Andor camera, spinning disk confocal system, and Andor IQ2 software. Image editing was limited to resizing and addition of scale bars.
Statistical Analyses
Comparison between populations was performed after testing for equality of the variances (F-test) followed by unpaired two-tailed Student's t tests corrected or not for variance inequality. Analyses of variance were used when more than two samples were compared. When analyses of variance indicated significant differences, post-hoc multiple unpaired two-tailed Student's t test Bonferroni corrected for multiple comparisons were performed. *, **, and *** indicate statistical significance and correspond to Bonferroni corrected thresholds of 5, 1, and 0.1%, respectively. All values are presented as the means ± S.E.
RESULTS
AOX and NDI1 Can Be Expressed in HEK293T Cells
Internal NADH dehydrogenases (e.g., NDI1) and AOX constitute alternative respiratory pathways in many lower eukaryotes, providing resistance to respiratory chain inhibitors such as rotenone and antimycin, acting as inhibitors of complex I and III, respectively (Fig. 1A). HEK293T cells expressing AOX or NDI1 were generated by lentivector-mediated transduction (23) using the pWPI expression vector (24), in which the transgene is co-transcribed with a cytosolic GFP reporter (Fig. 1B). Translation of the GFP is supported by an internal ribosome entry site. More than 75% of cells were reproducibly transduced using a multiplicity of infection of three infective units/cell. Co-immunostaining of transduced cells with anti-GFP and either anti-AOX or anti-NDI1 antibodies demonstrated concomitant expression of the transgenes with the fluorescent marker (supplemental Fig. S1A). Localization of AOX and NDI1 in mitochondria was confirmed by co-immunostaining with a mitochondrial marker (Fig. 1C) and by cell fractionation followed by immunoblotting (supplemental Fig. S1C). The stability of transgene expression was verified by tracking GFP expression using flow cytometry (supplemental Fig. S1B).
AOX and NDI1 Are Functional in HEK293T Cells
Two type of control cells were used throughout the study, namely untransduced and pWPI-GFP transduced HEK293T cells. For simplicity, a single “controls” group including both is shown whenever the properties of these cell populations were indistinguishable from each other.
When alternative pathways are functional, oxygen consumption should become resistant to specific respiratory chain inhibitors (Fig. 1A). To confirm AOX and NDI1 functionality, we therefore analyzed oxygen consumption, initially in digitonin-permeabilized cells (Fig. 2A). Digitonin permeabilization permits access of the respiratory substrates to mitochondria without damaging mitochondria but also disrupts cytosolic processes. Cells, ADP, and digitonin were added to the respiration buffer at the beginning of each experiment. Substrates and inhibitors were successively added in the order displayed in supplemental Fig. S2, C and D, for example. In the absence of respiratory chain inhibitors, neither AOX nor NDI1 altered the rate of oxygen consumption induced by substrate addition (pyruvate/malate, succinate, or TMPD + ascorbate), indicating that the expression of the alternative enzymes do not significantly interfere with mitochondrial respiration via the OXPHOS system.
FIGURE 2.
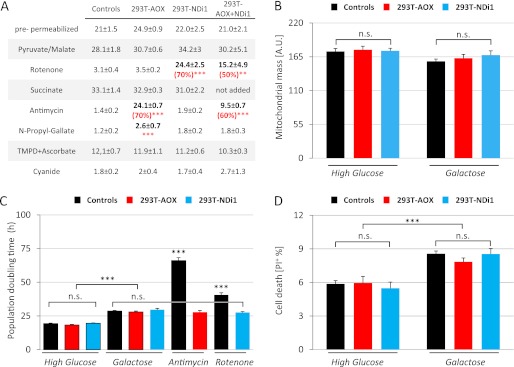
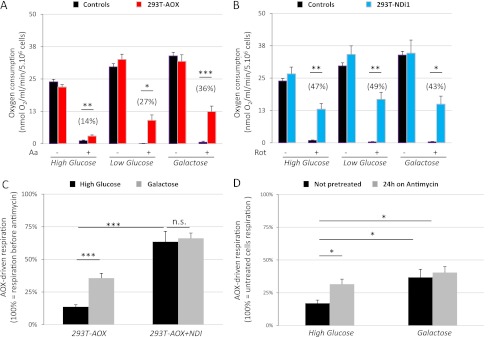
Expression of the alternative pathways does not alter cell physiology. A, alternative respiratory enzymes do not alter substrate-driven oxygen consumption but provide resistance to specific inhibitors. Oxygen consumption, in the presence of ADP, of 5 × 106 digitonin-permeabilized (80 μg/ml) cells after the addition of various respiratory substrates (pyruvate/malate, succinate, and TMPD + ascorbate) and inhibitors (rotenone, antimycin, n-propyl gallate, and cyanide). Measures were performed using a Clark-type electrode, and cells were grown for 24 h in high glucose medium before oxygen consumption analysis. All of the measurements were independently corrected for nonrespiratory oxygen consumption. The data are expressed as the means ± S.E. (error bars). From left to right, n = 8 (4 + 4), 6, 6, and 4. **, p < 0.01; ***, p < 0.001. B, mitochondrial mass is similar in transduced and untransduced cells. Flow cytometric analysis of 10-nonyl acridine orange fluorescence. The cells were grown for 24 h on high glucose or galactose medium before staining. The data are expressed as the means ± S.E. (error bars): controls, n = 24 (12 + 12); 293T-AOX, n = 12; and 293T-NDI1, n = 12. C, alternative respiratory enzymes protect cells exposed to specific respiratory chain inhibitors but do not alter cellular proliferation. For population doubling time, measures were made using a Bürker hemocytometer 72 h after plating. Untreated cells were plated in high glucose (5 × 104/cm2; 293T and 293T-AOX, n = 18; and 293T-NDI1, n = 9) or galactose medium (1 × 105/cm2; 293T, 293T-AOX, and 293T-NDI1, n = 18). Antimycin-treated (30 ng/ml) and rotenone-treated (150 nm) cells were plated in high glucose medium (1 × 105/cm2; 293T, 293T-AOX, and 293T-NDI1 n = 9). Each data point is the average of three independent measurements. The data are expressed as the means ± S.E. (error bars). Important similarities (n.s.) are emphasized. D, alternative respiratory enzymes do not alter cell viability. Flow cytometry of propidium iodide (PI; 2 μg/ml) stained cells. 3 × 105 cells were grown for 24 h in the indicated medium before staining. For controls, n = 21 (293T, 12; and 293T-GFP, 9); and for 293T-AOX and 293T-NDI1, n = 9. The data are expressed as the means ± S.E. (error bars).
Rotenone or antimycin treatment, respectively inhibiting complex I and complex III of the respiratory chain, blocked oxygen consumption in control cells. In contrast, oxygen consumption of permeabilized AOX-transduced cells was resistant to antimycin. This antimycin resistance was abolished by n-propyl gallate, a specific inhibitor of AOX. Similarly, oxygen consumption of NDI1-transduced cells was resistant to rotenone. These results demonstrate that both AOX and NDI1 proteins are enzymatically functional in the mitochondria of lentivector-transduced HEK293T cells, when the corresponding segments of the OXPHOS system are inhibited.
AOX or NDI1 Expression Preserves Cell Physiology
To estimate the potential alterations caused by the introduction of alternative respiratory enzymes into HEK293T cells, growth rate (Fig. 2C) and mortality (Fig. 2D) were quantified. Whether cells were grown in high glucose or galactose assay medium, the alternative respiratory enzymes did not modify cell doubling time in the absence of inhibitors. In contrast, the growth of 293T-AOX cells was significantly improved when exposed to antimycin, whereas 293T-NDI1 cells were similarly protected from the effects of rotenone. Cell survival, as assessed by flow cytometry of propidium iodide-stained cells, was not modified by the alternative respiratory enzymes; in the absence of OXPHOS inhibitors, analysis of cell viability by trypan blue exclusion confirmed this observation (data not shown).
Potential changes in mitochondrial mass were evaluated by flow cytometry, measuring intracellular accumulation of NAO (a cell-permeant fluorescent dye accumulating in mitochondria because of its affinity to cardiolipin). Whether transduced or not, all cell populations accumulated NAO in similar amounts (Fig. 2B). Because NAO accumulation in mitochondria is sensitive to mitochondrial membrane potential, signal intensity was remeasured after mitochondrial uncoupling with FCCP, but once again, no differences could be observed (data not shown).
Oxygen consumption was also assessed in intact cells, with the expectation that we would observe the same effects of AOX or NDI1 expression as in permeabilized cells. After 24 h of culture in high glucose, low glucose, or galactose assay media, respiration was measured; in each case the same assay medium was used for growth of the cells, using a Clark-type oxygraph. In the absence of respiratory chain inhibitors, AOX- and NDI1-expressing cells exhibited the same oxygen consumption as control cells (Fig. 3, A and B). Note that, independently of the transgene expressed, respiration of cells grown in high glucose medium was ∼25% less than in galactose-grown cells, as is typical of rapidly dividing cells, which need to increase respiration rates in galactose medium to maintain ATP production (25). In summary, expression of the alternative respiratory enzymes NDI1 or AOX preserved HEK293T cell physiology and respiration.
FIGURE 3.
In intact cells, AOX-driven respiration is controlled by prior growth in glucose, whereas NDI1-driven respiration is insensitive to glucose. The cells were cultured for 24 h in different media. Respiration was measured with a Clark-type electrode using 5 × 106 intact cells resuspended in the growth medium. The data are expressed as the means ± S.E. (error bars). All of the measurements were independently corrected for nonrespiratory oxygen consumption. *, p < 0.05; **, p < 0.01; ***, p < 0.001. A, in intact cells AOX-driven respiration is repressed by prior growth in glucose. Cellular respiration rates before (Aa −) and after (Aa +) antimycin injection into the oxygraph chamber are shown. The values in parentheses represent antimycin-resistant respiration relative to the respiration observed before the addition of antimycin (see also Fig. 3C). For controls, n = 20 in high glucose (10 + 10), 8 in low glucose (4 + 4), and 9 in galactose (293T, 5; and 293T-GFP, 4). For 293T-AOX, n = 22 in high glucose, 9 in low glucose, and 18 in galactose. B, in intact cells NDI1-driven respiration is insensitive to glucose in the growth medium. Respiration rates before (Rot −) and after (Rot +) rotenone treatment are shown. The values in parentheses represent rotenone-resistant respiration relative to the respiration observed before the addition of rotenone. For each of the tested growth media, n = 7 for controls (293T, 4; and 293T-GFP, 3), and n = 7 for 293T-NDI1. C, in intact cells, when respiration is driven by NDI1 instead of complex I, AOX-driven respiration becomes insensitive to prior growth on glucose. Oxygen consumption by intact cells expressing AOX or AOX + NDI1. The values represent antimycin-resistant respiration rates relative to the respiration observed before the addition of antimycin. For 293T-AOX, n = 22 in high glucose and 18 in galactose; for 293T-AOX + NDI, n = 3 in both media. D, glucose repression of respiration is alleviated after long term exposure to antimycin. Oxygen consumption in intact 293T-AOX cells (5 × 106) grown for 24 h in different media, with or without the addition of 30 ng/ml antimycin. The values represent antimycin-resistant respiration rates relative to the respiration observed before the addition of antimycin. For each cell type and growth medium, n = 7.
Growth in Glucose Suppresses the Ability of AOX but Not NDI1 to Preserve Whole Cell Respiration after RC Inhibition
We next tested the effects of growth in different media on the response to respiratory chain inhibition of intact cells via the successive injection of inhibitors into the oxygraph chamber (see supplemental Fig. S4 for examples). In control cells, antimycin or rotenone fully inhibited respiration regardless of the growth medium (Fig. 3, A and B, black bars). As expected, respiration of 293T-AOX cells exposed to rotenone and of 293T-NDI1 cells exposed to antimycin was also fully inhibited, regardless of the growth medium (not shown).
In contrast, after complex III inhibition with antimycin, 293T-AOX cells that had been grown in galactose medium showed high residual respiration (Fig. 3A). This antimycin-resistant respiration was lower in 293T-AOX cells grown on low glucose medium and lower still in the same cells grown on high glucose medium. Intact cell respiration via the AOX bypass is thus suppressed by growth in glucose. This inhibitory effect of glucose was not limited to complex III inhibition: although in isolated mitochondria AOX provides a bypass for cyanide inhibition of complex IV (26), this bypass was lost in intact cyanide-treated cells grown in high glucose (supplemental Fig. S2, A and B).
Surprisingly, when the corresponding experiments were performed on intact 293T-NDI1 cells, growth on different media had no effect on rotenone-resistant respiration, which was robust in all cases (Fig. 3B). To exclude any artifact arising from the fact that the respiration rates of cells varied according to the growth medium used, we reanalyzed the data, expressing the drug-resistant respiration as a proportion of that before inhibition (Fig. 3, A and B, values in parentheses; see also supplemental Fig. S4 for examples of calculations). After this correction, AOX-driven respiration in the presence of antimycin was still sensitive to the level of glucose in the growth medium, whereas NDI1-driven respiration in the presence of rotenone remained insensitive to glucose.
Analysis of Mitochondrial Membrane Potential and Superoxide Production Confirms That AOX-driven Respiration Is Inhibited by Growth on Glucose, whereas NDI1-driven Respiration Is Not
Because AOX and NDI1 both transfer electrons without pumping protons across the mitochondrial inner membrane (Fig. 1A), the active participation of either enzyme in respiratory electron flow should decrease mitochondrial membrane potential (m). Moreover, complex I inhibition by rotenone and complex III inhibition by antimycin enhance superoxide production, whereas the corresponding alternative respiratory enzymes should suppress this, in line with their ability to restore respiration (27).
As measured by TMRM (a fluorescent dye accumulating in mitochondria in proportion with mitochondrial membrane potential) fluorescence (28, 29), m was highest in cells grown on high glucose and lowest in cells grown on galactose medium (Fig. 4, A–C). AOX, but not NDI1 expression further decreased m in uninhibited cells. Although this alteration did not otherwise perturb cell physiology, it suggests that AOX subtly altered the properties of the OXPHOS system, most likely by a low constitutive activity diverting a proportion of the electron flow. Because this decrease in m was similar in all growth media, it also confirms that any such AOX activity is independent of the growth substrate.
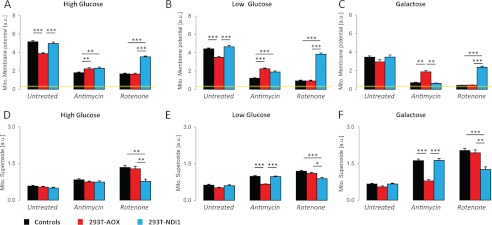
FIGURE 4.
Prior growth in glucose influences mitochondrial membrane potential and superoxide production in 293T-AOX cells treated with antimycin but not in 293T-NDI1 cells treated with rotenone. 3 × 105 cells cultured for 24 h in a given medium were treated with antimycin (30 ng/ml) or rotenone (150 nm) or mock-treated for 5 min prior to and during the incubation with the fluorescent indicator (MitoSox, 2.5 μm for 45 min at 37 °C; TMRM, 200 nm for 30 min at 37 °C). Fluorescence was analyzed by flow cytometry. A–C, mitochondrial membrane potential. Flow cytometry analysis of TMRM-stained (emission, 620 ± 15 nm) cells. The data are expressed as the means ± S.E. (error bars). For each cell population and treatment: A, n = 14; B, n = 10; and C, n = 6. Yellow line, TMRM intensity after FCCP (1 μm) uncoupling. D–F, superoxide production. Flow cytometry analysis of MitoSox-stained (emission, 620 ± 15 nm) cells. The data are expressed as the means ± S.E. (error bars). For each cell population and treatment: D, n = 13; E, n = 19; and F, n = 9.
Treatment with inhibitors decreased m in control cells, and the extent of this decrease was again dependent on the growth medium, suggesting that m was maintained under glycolytic conditions by mitochondrial consumption of cytosolic ATP (30). Following complex III inhibition by antimycin (but not complex I inhibition by rotenone), AOX expression restored m most effectively in galactose-grown cells, to a lesser extent in cells grown on low glucose medium, and least of all in cells grown on high glucose medium. In contrast, NDI1 expression restored m in rotenone-inhibited cells regardless of the growth medium, while having no effect on cells treated with antimycin.
Mitochondrial production of reactive oxygen species (mtROS) was measured by flow cytometric analysis of MitoSox (a dye accumulating in mitochondria in proportion with superoxide generated) fluorescence (Fig. 4, D–F). Cells grown in any of the media tested and not treated with any inhibitor showed very low levels of mtROS production, irrespective of whether they carried an AOX or NDI1 transgene. Control cells treated with respiratory chain inhibitors showed elevated mtROS production, which was greatest in cells grown on galactose medium and least in cells grown on high glucose medium. Regardless of the growth medium, NDI1 expression attenuated mtROS production in cells treated with rotenone, but not antimycin. In contrast, the ability of AOX to attenuate mtROS production resulting from antimycin treatment was dependent on the growth medium, being highest in galactose-grown cells, less in cells grown on low glucose, and not significant in cells grown on high glucose. AOX had no effect on mtROS production after rotenone treatment. Thus, measurements of both m and mtROS production were consistent with the inference that AOX-driven respiration was negatively regulated by growth on glucose, whereas NDI1-driven respiration was unaffected.
Glucose Suppression of Respiration Occurs by Down-regulation of Complex I
To identify the site of this glucose-dependent regulation, we analyzed the antimycin-resistant respiration of HEK293T cells simultaneously expressing AOX and NDI1. Our reasoning was that the absence of glucose regulation would suggest a control at the level of complex I (or possibly the ATP synthase, complex V), whereas the preservation of glucose regulation would indicate a control at another site upstream of complex III or possibly AOX itself (see Fig. 6A).
To distinguish AOX-expressing and NDI1-expressing cells, the coding sequence of GFP within the pWPI-NDI1 vector was replaced by that of BFP (Fig. 1B). AOX-expressing cells showed green cytosolic fluorescence, whereas cells transduced with this new NDI1 expression vector had a blue cytosolic fluorescence (not shown). 293T-AOX cells were then secondarily transduced with pWPI-NDI1-BFP. Fluorescence microscopy and co-immunofluorescence revealed that ∼85% of the cells expressed the cytosolic GFP (a marker for AOX expression), ∼55% expressed NDI1, and ∼45% expressed both transgenes (Fig. 1C).
After rotenone treatment and digitonin permeabilization, these cells maintained a strong pyruvate- and malate-driven oxygen consumption (Fig. 2A), indicating that NDI1 was active in a substantial fraction of the cells. This rotenone-resistant respiration was highly resistant to antimycin but sensitive to the further addition of n-propyl gallate (Fig. 2A), demonstrating that most of the NDI1-expressing cells also had a functional AOX.
Intact AOX- and NDI1-expressing cells were then cultured in the different growth media and treated with rotenone to direct electron flow through NDI1 instead of complex I. This rotenone-resistant respiration was set as the basal respiration rate (100%). The subsequent inactivation of complex III by antimycin revealed a strong antimycin-resistant respiration irrespective of the growth medium (Fig. 3C).
The negative effect on antimycin-resistant respiration produced by growth in high glucose was thus inferred to be exerted at the level of complex I (or complex V). In the latter case, however, complex III inhibition would be expected to increase m in control cells because of ATP synthase inhibition. In contrast, m was decreased by antimycin treatment (Fig. 4, A–C), thus eliminating complex V as a plausible site of the regulatory effect and identifying complex I itself as the molecular target thereof.
Complex I Inhibition Is a Fast, but Short Term Regulation
In intact cells, glucose down-regulation of complex I occurred less than 5 min after respiratory chain poisoning (supplemental Figs. S2A and S4) and could be maintained throughout the period of oxygraphic measurement (up to 45 min). To test how this regulation developed after extended inhibition, 293T-AOX cells were exposed to antimycin for 24 h (Fig. 3D). In high glucose medium, this treatment strongly alleviated the inhibition of complex I, whereas in galactose-grown cells it had no such effect. Furthermore, when cultured in the presence of antimycin for >24 h, AOX-expressing cells grown in high glucose exhibited an increased growth rate compared with antimycin-treated control cells (Fig. 2C). Glucose-dependent inhibition of complex I under conditions of complex III or IV inhibition is thus rapidly established but is only transient.
We next measured the antimycin-resistant respiration rate of intact 293T-AOX cells grown in a given medium immediately after transfer to other media. The cells were maintained in the new medium for ∼3 min before addition of antimycin, allowing only a very short period of adaptation to the change of carbon source. We observed (Fig. 5A) an increase in the antimycin-resistant respiration of glucose-grown cells when transferred to galactose medium but did not observe any inhibition by transfer of galactose-grown cells to high glucose. Thus, although it is rapidly detectable after treatment with a complex III inhibitor, the glucose-dependent inhibition of complex I can only be established by prolonged culture in the presence of glucose, whereas it is partially and rapidly relieved after the removal of glucose.
FIGURE 5.
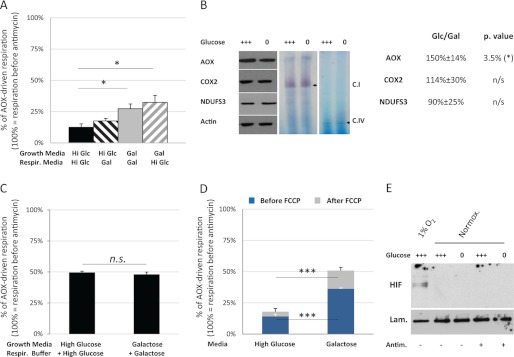
Characteristics of the glucose-dependent down-regulation of complex I induced by complex III inhibition. A, following complex III inhibition, and complex I is slowly inhibited by glucose but rapidly activated by galactose. Respiration of 5 × 106 293T-AOX cells grown in the indicated “growth media” was measured in the indicated “respiration media” using a Clark-type electrode. Hi Glc, high glucose media; Gal, galactose media. AOX-driven respiration is presented as a proportion of the respiration detected before antimycin treatment. The data are expressed as the means ± S.E. (error bars). For each cell type and assay condition, n = 10. B, growth in glucose does not alter complex I quantity nor in-gel complex I activity in extracts from antimycin-treated cells. Left panel, representative immunoblots showing AOX, complex IV (COX2), complex I (NDUFS3), and actin from post-nuclear extracts of 293T-AOX cells grown for 24 h in high glucose (+++) or galactose (0) medium and then antimycin-treated (30 ng/ml) for 10 min before protein extraction. For each cell type and assay condition, n = 4. Middle and right panels, complex I and complex IV, respectively, in-gel activity from mitochondrial extracts of 293T-AOX cells grown for 24 h in high glucose (+++) or galactose (0) medium and treated with antimycin (30 ng/ml) for 10 min before protein extraction. For each cell type and assay condition, n = 3. Table panel, estimated changes in AOX, complex I (NDUFS3), and complex IV (COX2) amounts after antimycin treatment (30 ng/ml, 10 min) of cells grown for 24 h on high glucose or galactose medium (Glc/Gal). The measurements were obtained from densitometry of nonsaturated autoradiographs and normalized to the signal for actin (e.g., Fig. 5B). For each cell type and assay condition, n = 4. C, complex I regulation by glucose depends on an indirect mechanism. Oxygen consumption by 5 × 106 digitonin-permeabilized cells previously grown for 24 h in high glucose or galactose medium was measured using a Clark-type electrode and a respiratory buffer supplemented with the sugar from the corresponding growth medium. Respiration was measured before and after the addition of antimycin (30 ng/ml) to the oxygraph chamber, as well as after full inhibition of the respiratory chain. AOX-driven respiration is presented as a proportion of the respiration prior to antimycin treatment. The measurements were independently corrected for nonrespiratory oxygen consumption. The data are expressed as the means ± S.E. (error bars). For each cell type and assay condition, n = 8. D, uncoupling does not alter glucose regulation of complex I after antimycin inhibition. Oxygen consumption by 5 × 106 intact 293T-AOX cells previously grown for 24 h in high glucose or galactose medium was measured using a Clark-type electrode, in the corresponding growth medium. Cells, antimycin (30 nm), and FCCP (1 μm) were successively added to the oxygraph chamber. AOX-driven respiration is shown as a proportion of the respiration prior to antimycin treatment. The measurements were independently corrected for nonrespiratory oxygen consumption. The data are expressed as the means ± S.E. (error bars). For each cell type and assay condition, n = 7. E, glucose regulation of mitochondrial function following complex III inhibition is HIF1 independent. Immunoblot for HIF1α in nuclear extracts of 293T-AOX cells grown for 24 h on high glucose (+++) or galactose (0) medium. Cells exposed to low (1%) or atmospheric O2 in presence or absence of antimycin (30 ng/ml, 10 min). Lam., lamin loading control; Normox., normoxic. Representative immunoblot, n = 3.
Regulation of Complex I Activity by Glucose Is Indirect and Independent of m
To test for any biochemical differences in complex I in AOX-expressing cells grown in different media and then treated (for 30 min) with antimycin, we analyzed mitochondrial protein extracts by Western blotting and blue native electrophoresis in-gel activity assays. AOX protein was at a higher level in cells grown on high glucose (Fig. 5B), as expected from the fact that the transgene promoter (EF1) is responsive to Sp1 (31, 32). In contrast, the level of the NDUFS3 subunit of complex I relative to cytosolic or mitochondrial loading controls was unaffected both by the growth medium and by the presence of antimycin (Fig. 5B and supplemental Fig. S1D). This results was as expected; because complex I down-regulation occurred so rapidly after complex III or complex IV inhibition (see supplemental Fig. S2A and S4 for example), it was unlikely to be dependent on altered rates of protein synthesis or turnover. Similarly, the growth medium had no effect on the in-gel activity of complex I (Fig. 5B). Although this assay is only semiquantitative and would not reveal small variations in enzyme activity, the >2-fold decrease in respiration rate that we measured in intact cells (Fig. 3C) should have been detected.
Cells expressing AOX and treated with antimycin showed identical complex I activity after permeabilization with digitonin, whether they were grown in glucose or galactose, and even after addition of the growth sugar to the assay buffer (Fig. 5C and supplemental Fig. S2, C and D), demonstrating that glucose itself cannot inhibit complex I. However, intact 293T-AOX cells treated with FCCP (a mitochondrial uncoupler) maintained the inhibition of antimycin-resistant respiration after growth on high glucose medium (Fig. 5D). Supplementation of the growth medium with N-acetyl cysteine (a potent antioxidant that can increase the levels of glutathione) did decrease mtROS production (supplemental Fig. S3A) but did not alleviate the glucose-dependent inhibition of antimycin-resistant respiration in intact 293T-AOX cells (supplemental Fig. S3B). The glucose-dependent down-regulation of complex I in response to respiratory chain dysfunction is thus inferred to be indirect and to require cell integrity but does not depend on m or mtROS.
Inhibition of Complex I by Glucose Is Not the Consequence of a Hypoxia-like Response
One of the best known mechanisms adapting respiration to glucose levels involves the stabilization of HIF1, either in response to hypoxia or resulting from the cytosolic accumulation of succinate (8, 9). Because the inhibition of the respiratory chain could lead to succinate accumulation, we followed HIF1 nuclear accumulation in the different culture and treatment conditions (Fig. 5E). As a control, we first verified that exposure of the 293T-AOX cells to low oxygen conditions (1%) led to the relocation of HIF1 in the nucleus. In atmospheric O2 conditions, however, complex III inhibition failed to induce nuclear translocation of HIF1, irrespective of the growth medium used. HIF1 stabilization therefore appears to play no role in the glucose-dependent inhibition of complex I in response to complex III inhibition.
DISCUSSION
The alternative respiratory enzymes are naturally absent from mammalian cells. However, by expressing them transgenically in human cells, we were able to detect and characterize a previously unknown regulatory system controlling the activity of complex I of the respiratory chain in response to inhibition of complexes III or IV. This regulation was transient, but was established rapidly, and was contingent on prior growth of the cells in a medium containing high levels of glucose.
In permeabilized cells, transgenically expressed NDI1 and AOX gave rise to functional bypasses, as expected, for respiratory complex I and complexes III and IV, respectively. The activity of these bypasses was independent of the growth medium in which the cells had previously been cultured. However, the behavior of intact cells was different. After rotenone treatment, NDI1-driven respiration was functional, irrespective of the growth medium. However, following antimycin treatment, AOX-driven respiration was fully functional only if cells had been previously grown on galactose medium, whereas prior growth on increasing concentrations of glucose suppressed respiration under these conditions. Respiratory activity was mirrored by predictable changes in m and mtROS production under all the conditions tested, consistent with unimpeded electron flow through NDI1 when complex I was inhibited, but with electron flow through AOX under tight regulation when complex III was inhibited. Our findings are consistent with a previous report of the antioxidant effects of another AOX, from Symplocarpus renifolius, being glucose-sensitive when the gene was expressed on HeLa cells (33). The antioxidant effects of C. intestinalis AOX, when expressed in a patient-derived fibroblast cell-line with COX10 deficiency (14), also appear to be glucose-sensitive. The novel, glucose-dependent regulation of respiration revealed by our findings may therefore be a general mechanism.
It can be interpreted as a system for limiting unnecessary damage resulting from over-reduction of the quinone pool, when complexes III and IV are inhibited or overloaded. Glucose provides an alternative substrate, metabolized fermentatively to lactate (25). In the presence of adequate levels of glucose, electron transfer via the respiratory chain is not required to produce ATP, or to regenerate NAD+, so it can be switched off entirely when the respiratory chain is dysfunctional. However, galactose cannot be metabolized fermentatively at a sufficient rate to sustain cellular ATP levels, so down-regulation of respiration under such conditions would make a bad situation worse.
Co-expression of NDI1 and AOX abolished the glucose-dependent suppression of respiration in the presence of antimycin, excluding the idea that it was exerted at a step upstream of the respiratory chain (e.g., pyruvate entry or oxidation), and thus limiting it, logically to complexes I or V of the OXPHOS system. Furthermore, the observed decrease in m following complex III inhibition excludes complex V as the site of regulation, because the resulting accumulation of protons in the intermembrane space would produce the opposite effect on m. The fact that FCCP had no effect on the glucose-dependent regulation of respiration in intact cells also rules out complex V as the target.
These interpretations are subject to the caveat that the alternative respiratory enzymes do not in themselves fundamentally alter cellular physiology. In the absence of respiratory inhibitors, substrate-driven oxygen consumption in permeabilized cells was unaffected by NDI1 or AOX. Although the culture media has a clear influence on intact cell respiration, none of the transgenes tested altered this influence. These observations are consistent with earlier findings made in AOX-expressing HEK293 cells (26), patient-derived fibroblasts (14), and HeLa cells (33), although they differ from a previous report showing increased pyruvate/malate-driven respiration in NDI1-transformed HEK293T cells (34). However, in the latter case NDI1 was under the control of the very strong CMV promoter, and cells were selected for growth in rotenone-containing medium. Our use of a weaker promoter and of a nonselected cell population infected at a moderate multiplicity of infection (∼20% of the cells remained uninfected) likely led to a lower expression level, which did not disturb cell metabolism.
We found no effects of the alternative respiratory enzymes on cell viability, cell growth, mitochondrial mass, or mtROS production. The only relevant parameter that was observed to alter in response to their expression was a small, but statistically significant decrease in m in cells transduced with AOX. Because this decrease was independent of the assay media and because the collapse of m (using FCCP) did not alter the control of the respiratory chain by glucose, it can be safely concluded that the alteration of m by AOX has no influence on this regulation.
Western blot and blue native electrophoresis in-gel histochemistry revealed no differences in the amount and activity of complex I in glucose-grown versus galactose-grown cells, excluding a direct modification (35, 36) of complex I. The addition of glucose or galactose to permeabilized cells had no effect on substrate oxidation. The glucose-dependent inhibition of complex I must therefore be indirect. Briefly exposing galactose-grown cells to glucose increased antimycin-resistant respiration rather than inhibiting it. The inhibition thus appears to depend upon a factor accumulating slowly in glucose-grown cells. Logically this factor could be a protein, as in the cytokine-dependent inhibition of complex I by STAT3 (12) or depending on enzymatically induced modifications of complex I such as acetylation/deacetylation known to be influenced by SIRT3 (11) and mitochondrial acetyl transferases (37). However, the direct mediator of inhibition must be a small molecule, because digitonin permeabilization abolished the inhibitory effect of prior growth in glucose. Digitonin creates small pores in cellular membranes, allowing free diffusion of small molecules but not the release of proteins from mitochondria, such as cytochrome c. Mitochondrial ROS are excluded from mediating the inhibition, based on three arguments. First, mtROS production in antimycin-treated cells was highest under the conditions of least inhibition of complex I, despite previous reports suggesting that complex I is inhibited by ROS (36). Second, in antimycin-treated AOX-expressing cells, mtROS production was at the same low level in all medium conditions. Third, the addition of N-acetyl cysteine decreased mtROS production by ∼20% but did not alter glucose regulation. Changes in pH are also excluded by the fact that the H+ ionophore FCCP also did not perturb the glucose regulation.
Inhibition of complex I occurred very rapidly after the addition of antimycin or cyanide (within ∼3 min) and lasted at least ∼45 min. However, it was lost after 24 h of continuous complex III inhibition. This is consistent with the idea that it constitutes an “emergency shutdown” system, limiting electron flow into a respiratory chain that the cell perceives as defective, until alterations in transcription can accommodate the stress in other ways. Such stresses that would be minimized by an emergency shutdown include the perturbation of the cataplerotic reactions of the TCA cycle, depriving cells of essential metabolites, and excess mtROS production, with extremely damaging consequences. Dysfunctional mitochondria could also undergo fission followed by ubiquitination-dependent degradation (38, 39), a highly expensive outcome for the cell. They can also release cytochrome c and other proapoptotic factors, causing cell death. All of these consequences could be prevented by the down-regulation of complex I activity (40–42), providing a rationale for a physiological mechanism to reduce complex I activity under conditions of respiratory dysfunction.
The introduction of alternative respiratory enzymes into human HEK293T cells has revealed an intrinsic regulatory pathway of mammalian mitochondria. We demonstrated that glucose rapidly and transiently down-regulates complex I activity in response to downstream inhibition of the respiratory chain (Fig. 6B). Understanding the more detailed molecular mechanism(s) involved in this regulation is now essential because this regulatory pathway could provide a novel therapeutic target for the control of cell metabolism in diverse pathological states.
Acknowledgments
We thank Dr. Alberto Sanz, Dr. Lucia Valente, and Prof. Hans Spelbrink for input and technical assistance and Dr. Jaakko Pohjoismäki for carefully reading of the manuscript.

This article contains supplemental Figs. S1–S4.
- OXPHOS
- oxidative phosphorylation
- AOX
- alternative oxidase
- HIF1
- hypoxia-inducible factor 1
- NDI1
- NADH dehydrogenase 1
- NAO
- 10-nonyl acridine orange
- TMRM
- tetramethylrhodamine methyl ester
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- mtROS
- mitochondrial production of reactive oxygen species
- TMPD
- N,N,N′, N′-tetramethyl-p-phenylenediamine.
REFERENCES
- 1. DiMauro S. (2004) Mitochondrial diseases. Biochim. Biophys. Acta 1658, 80–88 [DOI] [PubMed] [Google Scholar]
- 2. Zeviani M., Carelli V. (2007) Mitochondrial disorders. Curr. Opin. Neurol. 20, 564–571 [DOI] [PubMed] [Google Scholar]
- 3. DiMauro S., Schon E. A. (2008) Mitochondrial disorders in the nervous system. Annu. Rev. Neurosci. 31, 91–123 [DOI] [PubMed] [Google Scholar]
- 4. Marroquin L. D., Hynes J., Dykens J. A., Jamieson J. D., Will Y. (2007) Circumventing the Crabtree effect. Replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol. Sci. 97, 539–547 [DOI] [PubMed] [Google Scholar]
- 5. Díaz-Ruiz R., Avéret N., Araiza D., Pinson B., Uribe-Carvajal S., Devin A., Rigoulet M. (2008) Mitochondrial oxidative phosphorylation is regulated by fructose 1,6-bisphosphate. A possible role in Crabtree effect induction? J. Biol. Chem. 283, 26948–26955 [DOI] [PubMed] [Google Scholar]
- 6. Rodríguez-Enríquez S., Juárez O., Rodríguez-Zavala J. S., Moreno-Sánchez R. (2001) Multisite control of the Crabtree effect in ascites hepatoma cells. Eur. J. Biochem. 268, 2512–2519 [DOI] [PubMed] [Google Scholar]
- 7. Lee Y. J., Burlet E., Galiano F., Circu M. L., Aw T. Y., Williams B. J., Witt S. N. (2011) Phosphate and succinate use different mechanisms to inhibit sugar-induced cell death in yeast. Insight into the Crabtree effect. J. Biol. Chem. 286, 20267–20274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brière J. J., Favier J., Bénit P., El Ghouzzi V., Lorenzato A., Rabier D., Di Renzo M. F., Gimenez-Roqueplo A. P., Rustin P. (2005) Mitochondrial succinate is instrumental for HIF1alpha nuclear translocation in SDHA-mutant fibroblasts under normoxic conditions. Hum. Mol. Genet. 14, 3263–3269 [DOI] [PubMed] [Google Scholar]
- 9. Selak M. A., Armour S. M., MacKenzie E. D., Boulahbel H., Watson D. G., Mansfield K. D., Pan Y., Simon M. C., Thompson C. B., Gottlieb E. (2005) Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 7, 77–85 [DOI] [PubMed] [Google Scholar]
- 10. Ahn B. H., Kim H. S., Song S., Lee I. H., Liu J., Vassilopoulos A., Deng C. X., Finkel T. (2008) A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. U.S.A. 105, 14447–14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang J. Y., Hirschey M. D., Shimazu T., Ho L., Verdin E. (2010) Mitochondrial sirtuins. Biochim. Biophys. Acta 1804, 1645–1651 [DOI] [PubMed] [Google Scholar]
- 12. Wegrzyn J., Potla R., Chwae Y. J., Sepuri N. B., Zhang Q., Koeck T., Derecka M., Szczepanek K., Szelag M., Gornicka A., Moh A., Moghaddas S., Chen Q., Bobbili S., Cichy J., Dulak J., Baker D. P., Wolfman A., Stuehr D., Hassan M. O., Fu X. Y., Avadhani N., Drake J. I., Fawcett P., Lesnefsky E. J., Larner A. C. (2009) Function of mitochondrial Stat3 in cellular respiration. Science 323, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tanaka-Esposito C., Chen Q., Lesnefsky E. J. (2012) Blockade of electron transport before ischemia protects mitochondria and decreases myocardial injury during reperfusion in aged rat hearts. Transl. Res. 160, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dassa E. P., Dufour E., Gonçalves S., Paupe V., Hakkaart G. A., Jacobs H. T., Rustin P. (2009) Expression of the alternative oxidase complements cytochrome c oxidase deficiency in human cells. EMBO Mol. Med. 1, 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marella M., Seo B. B., Nakamaru-Ogiso E., Greenamyre J. T., Matsuno-Yagi A., Yagi T. (2008) Protection by the NDI1 gene against neurodegeneration in a rotenone rat model of Parkinson's disease. PLoS One 3, e1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S. J., Capaldi R. A. (2004) Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 64, 985–993 [DOI] [PubMed] [Google Scholar]
- 17. Pellinen R., Hakkarainen T., Wahlfors T., Tulimäki K., Ketola A., Tenhunen A., Salonen T., Wahlfors J. (2004) Cancer cells as targets for lentivirus-mediated gene transfer and gene therapy. Int. J. Oncol. 25, 1753–1762 [PubMed] [Google Scholar]
- 18. Spector D. L. (1998) Isolation of mitochondria from cells and tissues, in Culture and Biochemical Analysis of Cells (Spector D. L., Goldman R. D., Leinwand L. A., eds) pp. 411–417, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 19. Cannino G., Ferruggia E., Rinaldi A. M. (2009) Proteins participating to the post-transcriptional regulation of the mitochondrial cytochrome c oxidase subunit IV via elements located in the 3'UTR. Mitochondrion 9, 471–480 [DOI] [PubMed] [Google Scholar]
- 20. Chretien D., Rustin P., Bourgeron T., Rötig A., Saudubray J. M., Munnich A. (1994) Reference charts for respiratory chain activities in human tissues. Clin. Chim. Acta 228, 53–70 [DOI] [PubMed] [Google Scholar]
- 21. Nijtmans L. G., Henderson N. S., Holt I. J. (2002) Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods 26, 327–334 [DOI] [PubMed] [Google Scholar]
- 22. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 23. Mátrai J., Chuah M. K., VandenDriessche T. (2010) Recent advances in lentiviral vector development and applications. Mol. Ther. 18, 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiznerowicz M., Trono D. (2003) Conditional suppression of cellular genes. Lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77, 8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Warburg O., Geissler A. W., Lorenz S. (1967) On growth of cancer cells in media in which glucose is replaced by galactose. Hoppe-Seyler's Z. Physiol. Chem. 348, 1686–1687 [PubMed] [Google Scholar]
- 26. Hakkaart G. A., Dassa E. P., Jacobs H. T., Rustin P. (2006) Allotopic expression of a mitochondrial alternative oxidase confers cyanide resistance to human cell respiration. EMBO Rep. 7, 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fernie A. R., Carrari F., Sweetlove L. J. (2004) Respiratory metabolism. Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7, 254–261 [DOI] [PubMed] [Google Scholar]
- 28. Maldonado E. N., Patnaik J., Mullins M. R., Lemasters J. J. (2010) Free tubulin modulates mitochondrial membrane potential in cancer cells. Cancer Res. 70, 10192–10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wong A., Cortopassi G. A. (2002) High-throughput measurement of mitochondrial membrane potential in a neural cell line using a fluorescence plate reader. Biochem. Biophys. Res. Commun. 298, 750–754 [DOI] [PubMed] [Google Scholar]
- 30. Appleby R. D., Porteous W. K., Hughes G., James A. M., Shannon D., Wei Y. H., Murphy M. P. (1999) Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur. J. Biochem. 262, 108–116 [DOI] [PubMed] [Google Scholar]
- 31. Ejiri S. (2002) Moonlighting functions of polypeptide elongation factor 1. From actin bundling to zinc finger protein R1-associated nuclear localization. Biosci. Biotechnol. Biochem. 66, 1–21 [DOI] [PubMed] [Google Scholar]
- 32. Tan N. Y., Khachigian L. M. (2009) Sp1 phosphorylation and its regulation of gene transcription. Mol. Cell Biol. 29, 2483–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsukawa K., Kamata T., Ito K. (2009) Functional expression of plant alternative oxidase decreases antimycin A-induced reactive oxygen species production in human cells. FEBS Lett. 583, 148–152 [DOI] [PubMed] [Google Scholar]
- 34. Seo B. B., Matsuno-Yagi A., Yagi T. (1999) Modulation of oxidative phosphorylation of human kidney 293 cells by transfection with the internal rotenone-insensitive NADH-quinone oxidoreductase (NDI1) gene of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1412, 56–65 [DOI] [PubMed] [Google Scholar]
- 35. Verdin E., Hirschey M. D., Finley L. W., Haigis M. C. (2010) Sirtuin regulation of mitochondria. Energy production, apoptosis, and signaling. Trends Biochem. Sci. 35, 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taylor E. R., Hurrell F., Shannon R. J., Lin T. K., Hirst J., Murphy M. P. (2003) Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J. Biol. Chem. 278, 19603–19610 [DOI] [PubMed] [Google Scholar]
- 37. Scott I., Webster B. R., Li J. H., Sack M. N. (2012) Identification of a molecular component of the mitochondrial acetyltransferase programme. A novel role for GCN5L1. Biochem. J. 443, 655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Livnat-Levanon N., Glickman M. H. (2011) Ubiquitin-proteasome system and mitochondria. Reciprocity. Biochim. Biophys. Acta 1809, 80–87 [DOI] [PubMed] [Google Scholar]
- 39. Youle R. J., Narendra D. P. (2011) Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lambert A. J., Brand M. D. (2004) Inhibitors of the quinone-binding site allow rapid superoxide production from mitochondrial NADH:ubiquinone oxidoreductase (complex I). J. Biol. Chem. 279, 39414–39420 [DOI] [PubMed] [Google Scholar]
- 41. Loschen G., Flohé L., Chance B. (1971) Respiratory chain linked H2O2 production in pigeon heart mitochondria. FEBS Lett. 18, 261–264 [DOI] [PubMed] [Google Scholar]
- 42. Liu Y., Fiskum G., Schubert D. (2002) Generation of reactive oxygen species by the mitochondrial electron transport chain. J. Neurochem. 80, 780–787 [DOI] [PubMed] [Google Scholar]