Background: SR141716A binds the CB1 receptor selectively and exerts inverse agonist activity.
Results: We identify a key role of the minor binding pocket for SR141716A binding.
Conclusion: SR141716A exerts inverse agonist activity by securing the Trp rotameric switch, restraining CB1 from activation.
Significance: This is the first reported molecular description of the superimposition of SR141716A- and CP55940-binding sites.
Keywords: Cannabinoid Receptors, Cell Signaling, Cholesterol, G Protein-coupled Receptors (GPCR), Molecular Docking, Inverse Agonism, Toggle Switch, Water-mediated H-bonding
Abstract
SR141716A binds selectively to the brain cannabinoid (CB1) receptor and exhibits a potent inverse agonist/antagonist activity. Although SR141716A, also known as rimonabant, has been withdrawn from the market due to severe side effects, there remains interest in some of its many potential medical applications. Consequently, it is imperative to understand the mechanism by which SR141716A exerts its inverse agonist activity. As a result of using an approach combining mutagenesis and molecular dynamics simulations, we determined the binding mode of SR141716A. We found from the simulation of the CB1-SR141716A complex that SR141716A projects toward TM5 to interact tightly with the major binding pocket, replacing the coordinated water molecules, and secures the Trp-3566.48 rotameric switch in the inactive state to promote the formation of an extensive water-mediated H-bonding network to the highly conserved SLAXAD and NPXXY motifs in TM2/TM7. We identify for the first time the involvement of the minor binding pocket formed by TM2/TM3/TM7 for SR141716A binding, which complements the major binding pocket formed by TM3/TM5/TM6. Simulation of the F1742.61A mutant CB1-SR141716A complex demonstrates the perturbation of TM2 that attenuates SR141716A binding indirectly. These results suggest SR141716A exerts inverse agonist activity through the stabilization of both TM2 and TM5, securing the Trp-3566.48 rotameric switch and restraining it from activation.
Introduction
The cannabinoid receptor 1 (CB1)2 (Fig. 1A) is a G protein-coupled receptor (GPCR) found predominantly in the central nervous system (1). CB1 agonists such as (−)-Δ9-tetrahydrocannabinol (Fig. 1B), the main psychoactive component of cannabis sativa, elicit a stimulating effect on appetite as well as other physiological responses such as anxiety and hypertension (2) prompting research examining the therapeutic potential of inhibiting CB1 activation. SR141716A, also known as rimonabant, was developed by Sanofi-Aventis and is the first CB1 inverse agonist to go into clinical trials (3). It showed great potential as an anti-obesity agent, although psychiatric side effects prevented it from being approved for use by the Food and Drug Administration (4). Various other CB1 inverse agonists have since been developed as therapeutics, such as ibipinabant by Solvay/Bristol-Myers Squibb, surinabant by Sanofi-Aventis, otenabant by Pfizer, and taranabant by Merck, although all exhibited at least some psychiatric side effects in clinical trials (4).
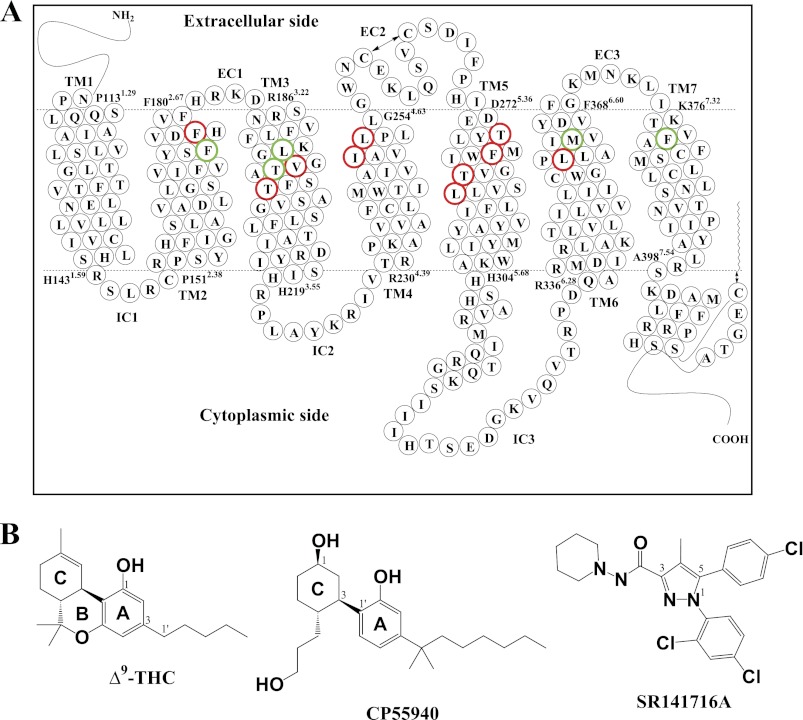
FIGURE 1.
CB1 receptor. A, human CB1 receptor in two-dimensional representation. Seven TM helices (TM1 to TM7), three extracellular (EC1, EC2, and EC3), and three intracellular (IC1, IC2, and IC3) regions are presented. Putative TM helical boundaries (TM1, Pro-1131.29–His-1431.59; TM2, Pro-1512.38–Phe-1802.67; TM3, Arg-1863.22–His-2193.55; TM4, Arg-2304.39–Gly-2544.63; TM5, Asp-2725.36–His-3045.68; TM6, Arg-3366.28–Phe-3686.60; and TM7, Lys-3767.32–Ala-3987.54) are represented by dotted lines. The EC2 intraloop disulfide linkage between Cys-257EC2 and Cys-264EC2 and the palmitoylation of the C-terminal Cys-415 are represented by a two-sided arrow. Amino acid residues examined in the present mutational studies are depicted in boldface type; residues having a role in SR141716A binding (Phe-1742.61, Leu-1933.29, Thr-1973.33, Met-3636.55, and Phe-3797.35) are shown in green, and noncontacting residues (Phe-1772.64, Val-1963.32, Thr-2013.37, Ile-2474.56, Leu-2504.59, Thr-2745.38, Phe-2785.42, Thr-2835.47, Leu-2875.51, and Leu-3596.51) are shown in red. The details of the binding pocket are represented in Fig. 4. B, molecular structures of the classical (ABC-tricyclic) cannabinoids (−)-Δ9-tetrahydrocannabinol (Δ9-THC), the nonclassical (AC-bicyclic) cannabinoid CP55940, and SR141716A.
By definition, inverse agonists bind the basal active form (R′) of the receptor and promote the inactive form (R) of the receptor, whereas agonists promote the active form (R*) of the receptor. Thus, binding of an inverse agonist would shift the equilibrium toward the inactive state, whereas binding of an agonist would shift the equilibrium toward the active state (5). In the absence of a CB1 x-ray structure, details of the mechanism by which SR141716A binds to and promotes the inactive state of the receptor are probed using mutagenesis and ligand binding analysis coupled with molecular dynamics (MD) simulations. The aromatic residues Phe-2003.36, Trp-2795.43, and Trp-3566.48 in transmembrane domains three (TM3), five (TM5), and six (TM6), respectively, have been shown to be involved in high affinity inverse agonist binding (6). (The Ballesteros-Weinstein numbering (7) is used (in superscript) to indicate the relative position of amino acid residues within the TM helical bundle; for loop residues, only the loop positions are indicated.) Alanine substitutions of Phe-2003.36 or Trp-3566.48 resulted in 3- and 7-fold reductions in SR141716A binding affinity, respectively, compared with the wild-type (WT) CB1.3 A more substantial change in SR141716A binding affinity was observed following the replacement of Trp-2795.43 with alanine, as SR141716A exhibited a greater than 1,000-fold decrease in binding affinity for the CB1 W2795.43A receptor. In addition, a lysine to alanine substitution of Lys-1923.28 in TM3 resulted in a 17-fold reduction in SR141716A binding affinity relative to CB1 WT in HEK293 cells indicating Lys-1923.28 also mediates SR141716A binding (8). Interestingly, Hurst et al. (8) demonstrated treatment of CB1 K1923.28A receptors transiently expressed in a neuronal cell line with 1 μm SR141716A resulted in no detectable SR141716A-mediated inverse agonism implicating Lys-1923.28 in the efficacy of SR141716A as well as the affinity. However, because the SR141716A concentration was just 25-fold greater than the Kd value of this mutant, this aspect requires further investigation.
CB1 has been shown to be capable of activating multiple isoforms of G protein, and recent studies suggest certain GPCRs can signal in a G protein-independent manner (9). A better understanding of the molecular mechanism by which inverse agonists such as SR141716A bind to and promote the inactive state of the receptor could lead to the development of a novel class of CB1 inverse agonists. These compounds could potentially selectively deactivate specific signaling pathways and/or stabilize a receptor conformation in a partially deactivated state for therapeutic purposes. Consequently, it is important that we delineate the binding site for SR141716A and the contact residues involved.
In this study, we have utilized an approach combining mutagenesis and MD simulations to determine the binding mode of SR141716A to gain insight into ligand interactions in the binding pocket and its inverse agonist activity. We identified residues Phe-1742.61, Leu-1933.29, and Phe-3797.35 as critical residues for SR141716A by mutational analysis and determined essential SR141716A-binding domains by MD simulations. We identified for the first time the involvement of the minor binding pocket for SR141716A binding, which complements the major binding pocket.
The simulation of the CB1-SR141716A complex revealed that SR141716A projects toward TM5 to interact tightly with the major binding pocket, replacing the coordinated water molecules, and secures the Trp-3566.48 rotameric switch in the inactive state to promote the formation of an extensive water-mediated H-bonding network to the highly conserved SLAXAD and NPXXY motifs in TM2/TM7. These results suggest SR141716A exerts inverse agonist activity through the stabilization of both TM2 and TM5, securing the Trp-3566.48 rotameric switch and restraining it from activation.
EXPERIMENTAL PROCEDURES
Plasmid Preparation
Human CB1 cDNA cloned into pcDNA3.1 was used as a template for site-directed mutagenesis (QuikChange, Stratagene, La Jolla, CA). DNA sequencing verified the presence of the mutations.
CB1 Expression and Membrane Preparation
CB1 receptor-containing membrane preparations were obtained as described previously (10).
CB1 Ligand Binding
Saturation binding assays were performed by incubating 10–25 μg of CB1 receptor containing membrane preparation with at least nine concentrations of [3H]SR141716A (56 Ci/mmol; PerkinElmer Life Sciences) between 0.24 and 68 nm in Tris/Mg2+/EDTA (TME) buffer (25 mm Tris-HCl, 5 mm MgCl2, and 1 mm EDTA, pH 7.4) containing 0.1% fatty acid-free bovine serum albumin (BSA) (w/v) in a final volume of 200 μl at 30 °C. Nonspecific binding was determined with 1 μm unlabeled ligand. Following a 1-h incubation period, 250 μl of chilled TME + 5% BSA (w/v) was added to each tube to terminate the reaction. A 24-manifold Brandel cell harvester was used to separate bound from unbound ligand with four washes of the filters with cold TME buffer. Bound radioactivity was determined by scintillation counting. Data analysis was performed using GraphPad Prism (GraphPad Software Inc., San Diego).
SR141716A Docking
Our previously reported ligand-unbound CB1 receptor model (10), which was homology-built using the inactive state crystal structure of β2AR (Protein Data Bank code 2RH1) (11) as the template, was used to determine the SR141716A binding mode. When this receptor was embedded in the hydrated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine bilayer, the hydrocarbon chain of a lipid molecule was deeply inserted into the CB1 core region through the opening created by the loosely packed TM4 to TM5 domains, potentially contributing to the stabilization of the ligand-unbound form of the receptor. After removal from the lipid bilayer, the stripped protein was used to explore SR141716A docking poses by employing the genetic algorithm-based flexible docking program GOLD (12, 13).
For the ligand, an initial structure of SR141716A was built by the Build Fragment tool in Discovery Studio (Accelrys, San Diego). The lowest energy conformation of SR141716A obtained by the Generate Conformations tool, using the BEST conformation algorithm as implemented in Discovery Studio, was used for docking. Because Phe-3797.35 was identified as a residue critical for SR141716A binding in this study (Table 1), all the receptor residues within 20 Å of Phe-3797.35, whose aromatic ring was positioned toward the TM core, were defined as the binding site. This region adequately covered all the reported SR141716A contact residues (6, 14, 15), including Phe-2003.36, Trp-2795.43, and Trp-3566.48, as well as the TM4-EC2-TM5 segment important for SR141716A binding (16, 17). For the GOLD docking experiment, we used the pre-defined default GOLD generic algorithm settings. GOLDScore was used for evaluating SR141716A docking modes. The GOLD cavity detection algorithm was used to locate the ligand within the TM binding core of the protein. No ligand bumping to any part of the protein was allowed. We performed a docking experiment and retained 50 docking poses from 10,000 docking runs and repeated the same docking experiment several times to obtain convergent binding poses. Among a diverse set of SR141716A docking poses, we selected high score and highly diverse docking poses. Using the key selection rule that the N1- or C5-aromatic ring moiety should be placed near the proposed aromatic microdomain, TM3/TM5/TM6 (15), we determined four distinct poses of SR141716A within the receptor.
TABLE 1.
SR141716A binding affinities for CB1 receptors with substitutions in residues potentially involved in SR141716A binding
| Receptor | SR141716A |
|
|---|---|---|
| Kd | Bmax | |
| nm | fmol/mg | |
| WT | 2.3 ± 0.4 | 4600 ± 200 |
| F1742.61A | 11.0 ± 1.8a | 6400 ± 400 |
| L1933.29A | 7.2 ± 1.3a | 6000 ± 400 |
| T1973.33A | 6.5 ± 0.8 | 7700 ± 300 |
| M3636.55A | 6.8 ± 0.9 | 5900 ± 300 |
| F3797.35A | 8.4 ± 1.3a | 4200 ± 200 |
a Statistically significant differences were from wild type (p < 0.05) using analysis of variance followed by Dunn's post hoc test (statistical analysis conducted on Kd data of CB1 receptors determined from three SR141716A saturation binding assays).
Each of these selected SR141716A poses was inserted into the ligand-unbound CB1 receptor model, and any water within 1.0 Å of the newly introduced ligand was removed. The resulting SR141716A-bound CB1 receptor was embedded in the hydrated lipid bilayer containing ∼68,000 atoms in a box of ∼75 × ∼80 × ∼110 Å3. To relax the protein in the presence of the ligand tightly fit in the cavity, the system was initially subjected to an energy minimization of 2,500 iterations. Then the whole system was simulated at 310 K in the NPT ensemble until the root-mean-square deviation (r.m.s.d.) values indicated structural convergence (50 ns). Repeating these procedures, we obtained four distinct poses of SR141716A within the CB1 receptor simulated in the fully hydrated lipid bilayer (CB1-SR141716A_a, CB1-SR141716A_b, CB1-SR141716A_c, and CB1-SR141716A_d) (Fig. 2). We performed ligand contact analyses for these poses to select the pose that was best fit to the SR141716A contact residues reported by others (6, 14, 15) as well as in this study (Table 1). Among these distinct poses, we selected the pose (CB1-SR141716A_c) and continued the simulation for the duration of ∼200 ns to obtain CB1-SR141716A (Table 2).
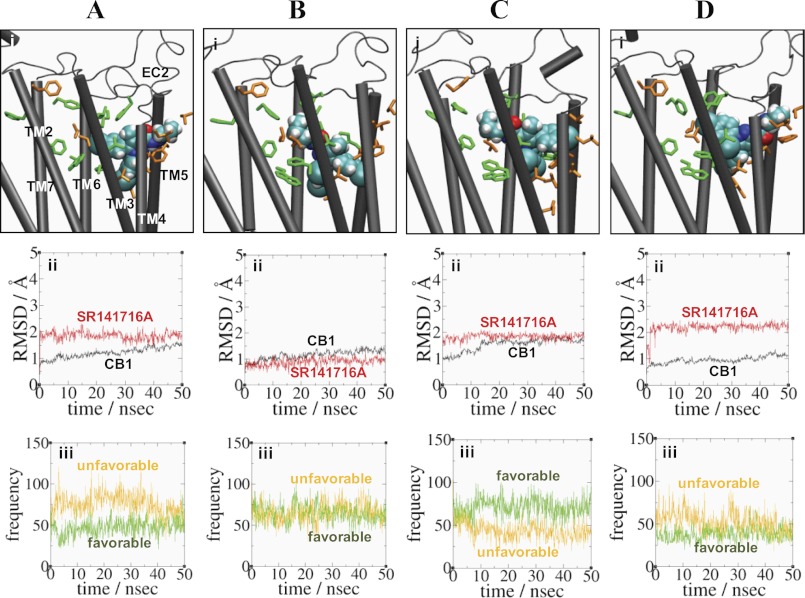
FIGURE 2.
Four binding poses CB1-SR141716A_a (A), CB1-SR141716A_b (B), CB1-SR141716A_c (C), and CB1-SR141716A_d (D) of SR141716A. Panel i, ligand contact and noncontact residue interactions with SR141716A in the four binding poses. The ligand is represented by space-filling (in atom type: C, cyan; O, red; and H, white). Ligand contact residues Phe-1702.57, Leu-1933.29, Thr-1973.33, Phe-2003.36, Phe-268EC2, Trp-2795.43, Trp-3566.48, Met-3636.55, and Phe-3797.35 selected based upon the results from this study (Table 1) as well as from the previous studies (6, 14, 15, 17, 31) that interact with the ligand are represented in stick form (green), whereas ligand noncontacting residues Phe-1772.64, Val-1963.32, Thr-2013.37, Ile-2474.56, Leu-2504.59, Thr-2745.38, Phe-2785.42, Thr-2835.47, Leu-2875.51, and Leu-3596.51, which interacted little with the ligand based upon the results from this study (Table 1), are represented in stick form (orange). EC, extracellular loop. Panel ii, r.m.s.d. for the protein (black) and the ligand (red). Panel iii, estimated (favorable) contact number between the ligand and the ligand contact residues (green) and estimated (unfavorable) contact number between the ligand and the ligand noncontact residues (orange). A criterion of 3.5 Å was used between nonbonded atoms. The contact numbers of the ligand with the contact and noncontact residues, with the standard deviation in parentheses, averaged over the last 10.0 ns of the simulation are as follows: 51(11) and 66(11) for CB1-SR141716A_a; 62(9) and 68(11) for CB1-SR141716A_b; 69(11) and 40(9) for CB1-SR141716A_c; and 39(8) and 44(11) for CB1-SR141716A_d.
TABLE 2.
MD simulations performed in this study
| Systema | Trp-3566.48 rotamer changeb | TM3/TM5 coordinated waterc | MD simulation |
|---|---|---|---|
| ns | |||
| CB1-SR141716A_a | 50 | ||
| CB1-SR141716A_b | 50 | ||
| CB1-SR141716A_c | 50 | ||
| CB1-SR141716A_d | 50 | ||
| CB1-SR141716Ad | No | No | 164 |
| F1742.61A-SR141716A | No | No | 106 |
| CB1-CP55940 | Yes | Yes | 131 |
| CB1(apo) | No | Yes | 44 |
a All simulations were performed in a fully hydrated lipid bilayer using 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (see “Experimental Procedures”). The CB1-CP55940 system was obtained by performing an additional simulation of the CP55940-CB1y system, and the CB1(apo) system was obtained by performing an additional simulation of the CB1n system in a previous study (10).
b Judged by the Trp-3566.48 χ1 angle: if trans (i.e. +120° < χ1 < +240°), yes; and otherwise, no. In this study, the Trp-3566.48 rotameric change was used to monitor the destabilization of the inactive state.
c A cluster of water molecules was coordinated to the polar residues, including Thr-1973.33, Thr-2013.37, Tyr-2755.39, and Thr-2835.47, in the region between TM3 and TM5 of the major binding pocket in the CB1 receptor.
d Simulated was continued from CB1-SR141716A_c.
Because Phe-1742.61 was suggested to be important for SR141716A binding (Table 1), insight on the possible role of Phe-1742.61 in SR141716A binding was gained by performing simulations of the F1742.61A mutant CB1-SR141716A complex (F1742.61A-SR141716A). We modified the CB1-SR141716A complex 40 ns into the simulation, at which point it showed structural convergence, by replacing Phe-1742.61 with an Ala residue and continued the simulation for ∼100 ns.
Simulation in a 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine Bilayer
Simulations were performed, as described in our previous study (10), by the NAMD simulation package (version 2.7 Linux-x86_64) (18), using CHARMM22 force field parameters with the φ/ψ angle cross-term map (CMAP) correction (19, 20) for the protein and the TIP3 water model (21, 22), and CHARMM27 force field parameters for the lipids (23). The topology definitions and the parameters for the palmitoylated Cys residue (CYP), including the parameters around the bond connecting Cys-415 of the CB1 receptor and the carbonyl carbon of the palmitoyl moiety, as used in the literature (24), were found on the NAMD Parameter Topology Repository site. The temperature was maintained at 310 K through the use of Langevin dynamics (25) with a damping coefficient of 1/ps. The pressure was maintained at 1 atm by using the Nosé-Hoover method (26) with the modifications as described in the NAMD Users Guide. The van der Waals interactions were switched at 10 Å and zero smoothly at 12 Å. Electrostatic interactions were treated using the Particle Mesh Ewald method (27). A pair list for calculating the van der Waals and electrostatic interactions was set to 13.5 Å and updated every 10 steps. A multiple time-stepping integration scheme, the impulse-based Verlet-I reversible reference system propagation algorithm method (28), was used to efficiently compute full electrostatics. The time step size for integration of each step of the simulation was 1 fs. The structures taken every 100 ps of these simulations were used for the analysis. All the simulations performed are summarized in Table 2.
CHARMM Parameterization
To describe SR141716A in the MD simulations using the CHARMM force field, we determined missing parameters. To determine the atomic charges of each ligand atom, charges from electrostatic potentials using a grid-based method (29) at the ab initio RHF/6–31G* level were computed using the Gaussian program (30). The charge from electrostatic potentials using a grid-based method at every atom was averaged over five representative conformations from a short (500 ps) MD simulation in a water box (40 × 40 × 40 Å3). To minimize any inconsistency with the existing CHARMM parameters, most of the missing parameters were borrowed, if possible, from the parameter values of chemically relevant structures. If necessary, employing ab initio RHF/6–31G* and MP2/6–31G* level calculations, we adjusted the values of bonds, angles, and torsion angles preferentially over adjustments to the values of force constants. We checked the validity of the newly determined CHARMM parameters for describing SR141716A by comparing molecular geometries from the MD simulations and key torsional energy barriers by CHARMM with those obtained by ab initio RHF/6–31G* and MP2/6–31G* level calculations (supplemental Table 1 and supplemental Fig. 1).
RESULTS
Potential SR141716A CB1 contact residues were substituted with alanine and the resulting receptors transiently expressed in an HEK293T cell line. Saturation binding analysis with [3H]SR141716A indicated the Bmax values for all CB1 variants tested were comparable with that of the wild-type CB1 receptor (wild-type CB1 Bmax = 4600 fmol/mg) suggesting the mutations did not impair the assembly or trafficking of the receptors. Ligand binding analysis further indicated that residues Phe-1742.61, Leu-1933.29, and Phe-3797.35 are critical for SR141716A binding affinity as the F1742.61A, L1933.29A, and F3797.35A receptors bound SR141716A with Kd values of 11, 7.2, and 8.4 nm, respectively, and were statistically different from that of the wild-type receptor (wild-type SR141716A Kd = 2.3 nm) (Table 1). Although the SR141716A binding affinity to T1973.33A and M3636.55A was reduced nearly 3-fold, this was not found to be statistically different from the wild-type receptor affinity.
Four Binding Poses of SR141716A
Using the key selection rule that the N1- or C5-aromatic ring moiety should be placed near the proposed aromatic microdomain TM3/TM5/TM6 (15), we significantly reduced the number of plausible binding poses of SR141716A to four. We initially performed four MD simulations of the CB1-SR141716A complex for four distinct poses of SR141716A within the CB1 receptor (CB1-SR141716A_a, CB1-SR141716A_b, CB1-SR141716A_c, and CB1-SR141716A_d) (see “Experimental Procedures”) (Table 2). Briefly, in CB1-SR141716A_a, the C5-aromatic rings interacted with the minor binding pocket, whereas the N1- and the C3-piperidinyl rings loosely interacted with the bottom and top regions of the major binding pocket, respectively. In CB1-SR141716A_b, the ligand solely occupied the major binding pocket, and both the N1- and the C5-aromatic rings were deeply engaged in the bottom of the major binding pocket, whereas the C3-piperidinyl rings closely interacted with the top of the major binding pocket. In CB1-SR141716A_c, the C3-piperidinyl ring interacted with the minor binding pocket, whereas the N1- and the C5-aromatic rings closely interacted with the major binding pocket bottom and top regions, respectively. Although symmetrical to the ligand in CB1-SR141716A_a with respect to the membrane normal, the ligand in CB1-SR141716A_c showed a closer interaction with the minor binding pocket than in CB1-SR141716A_a. Finally, in CB1-SR141716A_d, the position of the C3-piperidinyl ring was similar as in CB1-SR141716A_a, but the positions of the N1- and the C5-piperidinyl rings were swapped (Fig. 2A).
We estimated the contact numbers between SR141716A and the residues potentially involved in SR141716A binding based upon the results from this study (Table 1) as well as from the previous studies (6, 14, 15, 17, 31). These residues were as follows: Phe-1702.57, Leu-1933.29, Thr-1973.33, Phe-2003.36, Trp-2795.43, Trp-3566.48, Met-3636.55, and Phe-3797.35. Phe-1742.61 was excluded from the list of the ligand contact residues, for it was not directly involved in SR141716A binding (see below). Similarly, we estimated the contact numbers between SR141716A and the residues putatively selected as the ligand noncontact residues, including Phe-1772.64, Val-1963.32, Thr-2013.37, Ile-2474.56, Leu-2504.59, Thr-2745.38, Phe-2785.42, Thr-2835.47, Leu-2875.51, and Leu-3596.51, which were selected based upon the results from this study. Thus, the best SR141716A pose was determined such that the number of interactions between SR141716A and the ligand contact residues was high (favorable), whereas the number of interactions between SR141716A and the ligand noncontact residues was low (disfavorable). The contact numbers of the ligand with the contact and noncontact residues, with the standard deviation in parentheses, averaged over the last 10 ns of the simulation were as follows: 51(11) and 66(11) for CB1-SR141716A_a; 62(9) and 68(11) for CB1-SR141716A_b; 69(11) and 40(9) for CB1-SR141716A_c; and 39(8) and 44(11) for CB1-SR141716A_d. It was shown that the unfavorable noncontacts were apparently higher than the favorable contacts in CB1-SR141716A_a, although the unfavorable noncontacts were similar to the favorable contacts in CB1-SR141716A_b and CB1-SR141716A_d (Fig. 2). In contrast, it was in SR141716A_c that the favorable contacts were higher than the unfavorable noncontacts. Thus, among four distinct poses, we selected SR141716A_c as the best binding pose of SR141716A. Performing an additional simulation (>150 ns) of SR141716A_c, we obtained CB1-SR141716A (Table 2) for further analysis of the SR141716A binding mode as described below.
MD Simulations of CB1-SR141716A
As shown in Fig. 3A, panel i, the protein quickly converged at 20 ns into the simulation. The overall r.m.s.d. values were small in CB1-SR141716A throughout the simulation. Both an increase in the r.m.s.d. values for the protein 20 ns into the simulation and a decrease in the r.m.s.d. values after 100 ns were due to alterations mainly in TM4, as revealed in the r.m.s.d. value for the individual TM helices. As shown in Fig. 3A, panel ii, a noticeable structural change in TM4 was the breakage and the recovery of an H-bond between the side chain O of Ser-1582.45 and the side chain N of Trp-2414.50, as demonstrated by the increase in distance between the O and N from ∼3.0 Å to >5.0 Å at 100 ns followed by the subsequent decrease in distance between these atoms from >5.0 Å to ∼3.6 Å (Fig. 3A, panel ii). The side chain torsion angle of Ser-1582.45 remained unchanged, but the side chain indole ring of Trp-2414.50 was flipped (i.e. its χ1 angle changed from ∼90° to −30°) initially at 50 ns of the simulation for a short period of time and flipped again after 150 ns of the simulation. Interestingly, the ring flipping of Trp-2414.50 on TM4 near the end of the simulation was preceded by the ring flipping of Trp-2795.43 on TM5, as shown in Fig. 3A, panel iv. The ligand in CB1-SR141716A showed a low degree of fluctuation throughout the simulation, and the ligand binding pose remained unchanged (Fig. 3A, panel i).
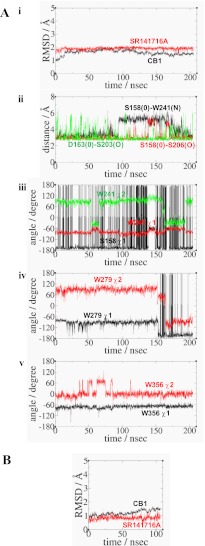
FIGURE 3.
A, simulation results of CB1-SR141716A. Panel i, r.m.s.d. for the protein (black) and the ligand (red). Panel ii, distance between the side chain O atom in Ser-1582.45 and the side chain N atom in Trp-2414.50 (black); the distance between the side chain O atom in Ser-1582.45 and the side chain O atom in Ser-2063.42 (red); and the distance between the side chain O atom in Ser-1632.50 and the side chain O atom in Ser-2033.39 (green). Panel iii, rotameric angles, the χ1 angle (black) of Ser-1582.45, and the χ1 angle (red) and the χ2 angle (green) of Trp-2414.50. Panel iv, rotameric angles, the χ1 angle (black) and the χ2 angle (red), of Trp-2795.43. Panel v, rotameric angles, the χ1 angle (black) and the χ2 angle (red), of Trp-3566.48. B, simulation results of F1742.61A-SR141716A. r.m.s.d. for the protein (black) and the ligand (red).
In this study, we used the Trp-3566.48 rotameric change to indicate the stabilization or the destabilization of the inactive state. Thus, the retention of the Trp-3566.48 rotameric toggle switch (i.e. the χ1 angle of Trp-3566.48 in gauche) indicated that the receptor in the inactive state was stabilized, whereas the activation of the Trp-3566.48 rotameric toggle switch (i.e. the χ1 angle of Trp-3566.48 in trans) indicated that the receptor in the inactive state was destabilized. By definition, the χ1 angles were divided into three angle categories as follows: g−, 0–120°; trans, 120–270°, and g+, 240–360° (32). In CB1-SR141716A, no activation of the Trp-3566.48 toggle switch (i.e. the χ1 angle of W356 in +g) was observed, as shown in Fig. 3A, panel v of the χ1 and χ2 angles of Trp-3566.48. This result indicated that the receptor remained stable in the inactive state.
MD Simulation of F1742.61A-SR141716A
Because the present mutational studies showed SR141716A binding affinity was significantly attenuated by the F1742.61A mutation (Table 1), we explored the F1742.61A-SR141716A system through an MD simulation. The overall r.m.s.d. values were low due to the structurally converged CB1-SR141716A_c system at 40 ns of the simulation from which the F1742.61A mutant CB1-SR141716A system was initially generated (Fig. 3B). The r.m.s.d. values of the receptor were low (<1 Å) at the early stage of the simulation but increased near the end of the simulation (∼1.5 Å) mainly due to TM2 outward movement, as revealed in the r.m.s.d. values for the individual TM helices. However, fluctuations in the ligand throughout the simulation were low (<1 Å), suggesting no large movement of the ligand within the binding pocket. Overall, our analyses of the r.m.s.d. values for the F1742.61A-SR141716A system indicated the F1742.61A mutation altered the TM2 portion of the minor binding pocket.
SR141716A Binding Mode in the CB1 Receptor
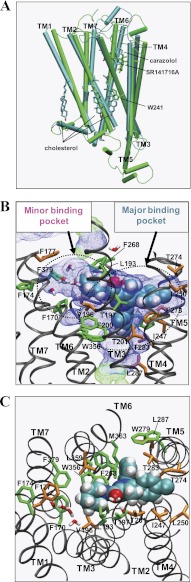
As shown in Fig. 4A, comparison of the present model of CB1-SR141716A with the inactive state x-ray structure of the β2AR (Protein Data Bank code 2RH1) (11) showed that the overall structures, including the TM topology and the location of the bound inverse agonists, look quite similar.
FIGURE 4.
Binding mode of SR141716A in CB1-SR141716A at the end of the simulation. A, superimposition of CB1-SR141716A (green) with the x-ray structures of β2AR (11) (cyan) with respect to the Cα atoms of the TM helical domain. The inverse agonists carazolol (in atom type: C, cyan; O, red) bound to β2AR and SR141716A (green) are represented in stick form. Three cholesterol molecules (in atom type) bound to β2AR and Trp-2414.50 (green) of the CB1 receptor are also represented in stick form. B, membrane-side view. C, extracellular top view. The ligand is represented by space-filling (in atom type). Ligand contact residues are represented in stick form (green), and ligand noncontacting residues are represented by stick form (orange). For the amino acids, only the side chains of the amino acids are represented. Hydrogen atoms for amino acids are omitted for clarity. Water molecules within 6 Å of the ligand are also represented. The solvent-accessible pore (blue dots for low radius surface and green dots for mid-radius surface) was created by using HOLE (44) at the ligand binding core region at the end of the MD simulations. The major binding pocket formed by TM3/TM5/TM6 and the minor binding pocket formed by TM2/TM3/TM7 are represented by dotted circles.
According to the present SR141716A binding mode, the ligand interacted with both the major binding pocket and the minor binding pocket at the central TM core region (Fig. 4, B and C). Specifically, the rigid N1- and C5-aromatic rings tightly interacted with the major binding pocket formed by TM3/TM5/TM6, although the flexible C3-piperidinyl ring moiety loosely interacted with the minor binding pocket formed by TM2/TM3/TM7. In support, replacement of the N1-dichlorophenyl ring in SR141716A with a methyl group resulted in an abolition of ligand binding (33), although the replacement of the C3-piperidinyl moiety by a methyl group resulted in a 90-fold drop in SR141716A binding affinity (34).
According to the present binding mode of SR141716A, the residues within 4.0 Å of the ligand included the following: Phe-1702.57 on TM2; Lys-1923.28, Leu-1933.29, Val-1963.32, Thr-1973.33, Phe-2003.36, and Thr-2013.37 on TM3; Ile-2474.56, Leu-2504.59, Pro-2514.60 on TM4; Ile-267EC2, Phe-268EC2, Pro-269EC2, and His-270EC2 on EC2; Thr-2745.38, Tyr-2755.39, Phe-2785.42, and Trp-2795.43 on TM5; Trp-3566.48, Leu-3596.51, Leu-3606.52, and Met-3636.55 on TM6; and Phe-3797.35 on TM7. Among these residues, the Ala mutagenesis studies indicate Leu-1933.29, Thr-1973.33, Met-3636.55, and Phe-3797.35 may be ligand contact residues (Table 1), as shown in Fig. 4, B and C. The present SR141716A binding mode appeared to be in good agreement not only with the results from the present mutation studies but also the results of mutation studies previously reported by others (6, 14, 15, 17, 31). Phe-1742.61 showed an ∼5-fold decrease in SR141716A binding (Table 1). We argue that Phe-1742.61 affects ligand binding indirectly by altering the receptor structure (see “Discussion”). A group of residues, including Phe-1772.64, Val-1963.32, Thr-2013.37, and Leu-3596.51, however, are less likely to be ligand contacts.
As shown in Fig. 4, B and C, a detailed examination revealed each moiety of SR141716A interacted with the receptor as follows. (i) The N1-dichlorophenyl ring moiety, positioned deep into the center of the major binding pocket, contacted the aromatic residues, including Phe-2003.36, Trp-2795.43, and Trp-3566.48. (ii) The C3-piperidinyl moiety, positioned toward the minor binding pocket, mainly contacted with the aromatic residues Phe-1702.57, Phe-268EC2, and Phe-3797.35 and the nonaromatic residue Leu-1933.29. (iii) The C5-chlorophenyl moiety, positioned toward TM5, mainly contacted with aromatic residues, including Tyr-2755.39 and Phe-2785.42. In agreement, the structure-activity relationships and affinity studies of SR141716A and its derivatives with the CB1 receptor demonstrated the importance of the substituent on the N1-, C3-, and C5-positions (34–36). Thus, the replacement of the N1-dichlorophenyl ring in SR141716A by the methyl group resulted in the abolishment of ligand binding (33), and the replacement of the C5-chlorophenyl ring by the ethyl group caused a >10-fold drop in ligand binding affinity (37). Similarly, replacement of the C3-piperidinyl moiety by a methylamine moiety resulted in a 90-fold drop in SR141716A binding affinity (34), although substitution with a phenyl ring moiety affected ligand binding to a substantially lesser degree (37). This suggested that a bulky hydrophobic or an aromatic moiety was required for retaining SR141716A binding.
Aromatic Stacking in SR141716A Binding
Aromatic residues appeared most important for describing SR141716A binding. As shown in Fig. 4C, Phe-3797.35 at the extracellular edge region of the minor binding pocket interacted with the C3-piperidinyl ring moiety, whereas Tyr-2755.39 at the extracellular edge region of the major binding pocket interacted with the C5-aromatic ring moiety, defining the lateral boundaries for SR141716A binding. Similarly, Phe-2003.36 and Trp-3566.48 at the bottom of the major binding pocket interacted tightly with the N1-aromatic ring moiety, whereas Phe-268EC2 at the top of the SR141716A binding pocket, covering the bound ligand, determined the top and bottom boundaries of SR141716A binding. Phe-2003.36/Trp-3566.48 stacking was extended to Phe-1702.57, Phe-1742.61, Phe-1772.64, Phe-1893.25, and Phe-3797.35, and Phe-268EC2/His-270EC2 stacking was extended to Tyr-2755.39, Phe-2785.42, and Trp-2795.43.
Thus, compared with the ligand-free receptor that was limited to Phe-1702.57/Phe-2003.36/Trp-3566.48 aromatic stacking in the region formed mainly by TM2/TM3/TM6 (10), the SR141716A-bound receptor was now extensively stabilized by the ligand, including a network in the region formed by TM3/TM5/TM6. Interestingly, these two aromatic stacking networks were inter-connected by Phe-2003.36/Trp-3566.48 stacking at the bottom of the major binding pocket, forming an extensive network of aromatic stacking. Thus, it appeared that Phe-2003.36/Trp-3566.48 stacking at the center of this extensive aromatic stacking network played a key role in stabilizing the receptor. In this view, the Trp-3566.48 rotameric switch in the CB1 receptor in association with SR141716A ligand binding and receptor regulation appeared to be of particular significance, as proposed by McAllister et al. (15).
Structure-activity relationship studies supported the importance of aromatic interactions in SR141716A binding. Thus, the replacement of the N1-dichlorophenyl ring in SR141716A by the nonaromatic hydrocarbon groups resulted in a >16-fold drop in ligand binding affinity (35). Similarly, mutational studies of Phe-2003.36, Tyr-2755.39, Phe-268EC2, Trp-2795.43, and Trp-3566.48 also supported the importance of these aromatic residues in SR141716A binding (6, 14, 17).
DISCUSSION
C5-aromatic Ring of SR141716A Stabilizes TM5 through Aromatic Stacking
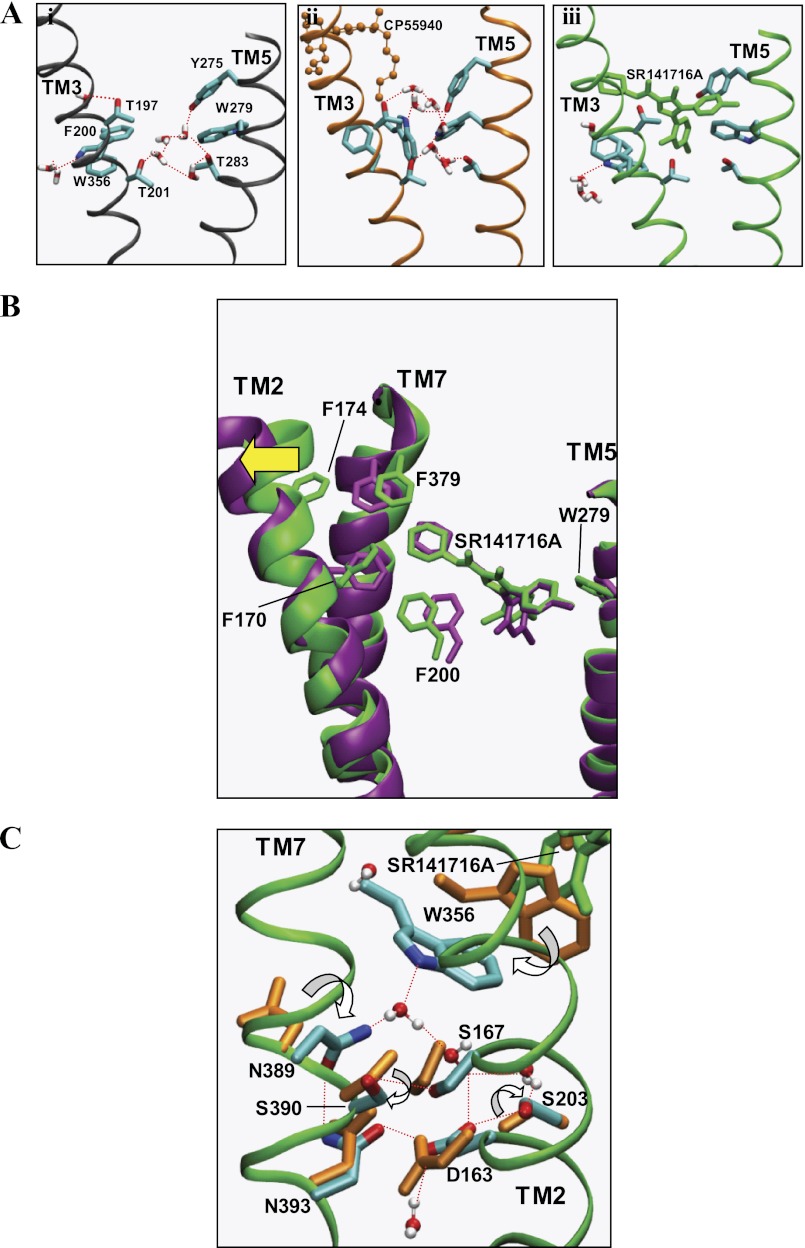
The findings that the Y2755.39A mutation abolished receptor expression (16) and that W2795.43A mutation resulted in lower receptor expression levels than in the wild type (15) suggested that TM5 was important for receptor stabilization. In our recent study examining the binding mode of the potent nonclassical cannabinoid agonist CP55940 (Fig. 1B) with the CB1 receptor (10), it was observed that a water-mediated H-bonding network was conserved in the major binding pocket near TM5 in both the ligand-unbound and the CP55940-bound receptors (Fig. 5A, panels i and ii). This water-mediated H-bonding network was coordinated by a group of polar residues from TM3 and TM5, including Thr-1973.33, Thr-2013.37, Tyr-2755.39, and Trp-2795.43, contributing to receptor stabilization. Assuming that an inward movement in the middle of TM5 is a characteristic of the ligand-induced receptor activation among rhodopsin class GPCRs (38), CB1 receptor activation would involve a similar TM5 inward movement. According to the SR141716A binding mode revealed here, the C5-aromatic ring of SR141716A replaced the water-mediated H-bond network and formed a tight aromatic stacking interaction with Tyr-2755.39 and Trp-2795.43, suggesting SR141716A stabilized TM5 (Fig. 5A, panel iii).
FIGURE 5.
A, water-mediated H-bond network coordinated to the polar residues (in stick form), including Thr-1973. 33, Thr-2013.37, Tyr-2755.39, and Thr-2835.47, and Trp-3566.48 in the region between TM3 and TM5 of the major binding pocket in the ligand-free form of the CB1 receptor (panel i), in CB1-CP55940 (panel ii), and in CB1-SR141716A (panel iii). Only water molecules within 6 Å of the side chains of Thr-2013.37, Thr-2835.47, and Trp-3566.48 are represented. H-bonds are represented by red dotted lines. Hydrogen atoms of amino acid residues are omitted for clarity. Color coding for ligands is as follows: CP55940, orange; and SR141716A, green. Color coding for amino acid residues and water: C, cyan; O, red; N, blue; and H, white. B, superposition of the ligand and the protein in CB1-SR141716A (green) and in F1742.61A-SR141716A (purple) at the end of the simulations, with respect to the backbone atoms of TM helices, viewed from the membrane side. Only aromatic residues that showed noticeable displacement are represented. Only TM2, TM5, and TM7 are represented for clarity. An outward movement of TM2 from the TM core in F1742.61A-SR141716A is represented by a yellow arrow. C, water-mediated H-bonding network proximal to the SLAXAD and NPXXY motifs is promoted by residue Trp-3566.48 in CB1-SR141716A. Prominent rotameric changes in the amino acids (stick form) in CB1-SR141716A (in atom type), compared with CB1-CP55940 (orange), are indicated by white arrows. Only water molecules (ball-and-stick) within 6 Å of Asp-1632.50, Ser-1672.54, and Asn-3937.49 are represented. H-bonds are represented by red dotted lines. Hydrogen atoms of amino acid residues are omitted for clarity.
Considering the findings that SR141716A exhibited only partial displacement of the tracer, [3H]CP55940, at a 5 μm concentration from the W2795.43A CB1 receptor, compared with 3- and 9-fold decreases in SR141716A binding affinity to the F2003.36A and W3566.48A receptors, respectively (6), the binding interaction of SR141716A with Trp-2795.43 appeared to be of particular importance. Although it was difficult to pharmacologically characterize the effect of the W2795.43A mutation on the CB1 receptor due to the drastic decrease in binding affinity, Trp-2795.43 appeared to be important not only for SR141716A binding but also for receptor function. In fact, Trp-2795.43 formed direct ligand contact in conjunction with aromatic stacking interactions with Trp-3566.48, according to the SR141716A binding mode; it helped stabilize not only the N1-aromatic ring but also aromatic stacking with Trp-3566.48. In this way, the Trp-3566.48 rotameric switch was retained in the inactive state, precluding an inward movement of TM5 (see below).
N1-aromatic Ring Moiety of SR141716A Locks the Proposed Toggle Switch Trp-3566.48 at the Bottom of the Major Binding Pocket
According to the present SR141716A binding mode, the N1-dichlorophenyl ring moiety closely interacted with Phe-2003.36/Trp-3566.48 at the major binding pocket (Fig. 4B), such that the proposed rotamer toggle switch residue Trp-3566.48 (6) was locked in the inactive state. The interaction of the C3-piperidinyl moiety of SR141716A with the minor binding pocket and the interaction of the C5-aromatic ring moiety of SR141716A with the major binding pocket possibly contributed to maintaining the N1-dichlorophenyl ring moiety in place near Phe-2003.36/Trp-3566.48 enabling it to secure the Trp-3566.48 rotameric switch in the inactive state.
SR141716A Binding Involves the Minor Binding Pocket
We identified for the first time the involvement of the minor binding pocket formed by TM2/TM3/TM7 in SR141716A binding that complements the major binding pocket formed by TM3/TM5/TM6. By mutational analysis, we identified residues Phe-1742.61, Leu-1933.29, and Phe-3797.35 as critical residues for SR141716A (Table 1). The present simulation of the F1742.61A mutant CB1-SR141716A complex revealed that some of the key ligand contacts in the minor binding pocket, Phe-2003.36 and Phe-3797.35 in particular, were displaced compared with the case of the wild-type receptor (Fig. 5B). Accordingly, the nearly 5-fold reduction in SR141716A binding affinity for F1742.61A (Table 1) was due to the altered receptor structure that attenuated SR141716A binding indirectly and was not attributed to the removal of a direct ligand contact. Although Phe-1742.61 was not directly involved in SR141716A binding, the present simulation of the F1742.61A mutant CB1-SR141716A complex demonstrated the importance of the minor ligand pocket in SR141716A binding.
SR141716A Plays an Important Role in TM2/TM3/TM7 Stabilization
In our previous study of the CB1 receptor, we observed the conserved SLAXAD motif on TM2 and the NPXXY motif on TM7 were involved in the formation of extensive water-mediated H-bond networks within the core region (10), possibly facilitating the active state of the receptor. In comparison with CB1-CP55940, where a water channel connecting the intracellular side and the highly conserved middle TM core occurred, CB1-SR141716A suggested that the bound ligand contributed to the stabilization of the receptor in the inactive state (supplemental Fig. 2).
In CB1-SR141716A, the N1-dichlorophenyl ring moiety of SR141716A interacted tightly with Trp-3566.48 so that the Trp-3566.48 rotameric toggle switch was secured from activation, promoting the formation of an extensive water-mediated H-bonding network to the conserved SLAXAD and NPXXY motifs, leading to the stabilization of TM2/TM3/TM7 (10). Thus, the side chain ring N of Trp-3566.48 formed water-mediated H-bonds to the side chain carboxylic carbonyl O of Asp-1632.50, the side chain O of Ser-2033.39, and the side chain amide N of Asn-3897.45. Furthermore, the side chain amide N of Asn-3937.49 formed an H-bond to the side chain amide O of Asn-3897.45; the side chain amide O of Asn-3937.49 formed an H-bond to the side chain carboxylic O of Asp-1632.50; and the side chain carboxylic carbonyl O of Asp-1632.50 formed an H-bond to the side chain O of Ser-2033.39 (Fig. 5C). Ser-2033.39, Asn-3897.45, and Ser-3907.46, all of which are highly conserved in family A GPCRs (39), showed prominent rotameric changes in CB1-SR141716A (supplemental Fig. 3), compared with CB1-CP55940, suggesting that the Trp-3566.48 rotameric switch in the inactive state restrained by SR141716A plays a crucial role in promoting the water-mediated H-bonding network important for the stabilization of TM2/TM3/TM7.
In CB1-CP55940, however, the Trp-3566.48 rotameric switch in the active state participated in the water-mediated H-bonding network conserved in the major binding pocket near TM5 (Fig. 5A, panel ii), without contributing to the stabilization of the highly conserved SLAXAD and NPXXY motifs (supplemental Fig. 2, panel ii). As a result, the ligand induced the activation of the Trp-3566.48 rotameric switch (i.e. the χ1 angle of Trp-3566.48 in trans), which facilitated the formation of a water channel that gave access to the highly conserved SLAXAD and NPXXY motifs, leading to the destabilization of TM2/TM3/TM7.
Overall, these results suggested that SR141716A plays an important role in TM2/TM3/TM7 stabilization. In support, it was well established that TM2/TM3/TM7 stabilization was important for the inactive state of the family A GPCRs (40, 41).
Comparison of the Present SR141716A Binding Model with the Previously Published SR141716A Binding Models
McAllister et al. (6) reported a model of SR141716A binding that was determined in consideration of the region formed by TM3/TM4/TM5/TM6 as well as TM4/EC2/TM5 as the primary binding pocket region. This model was determined by imposing an H-bond between the C3-amide O atom and the side chain N atom of Lys-1923.28, under the assumption that Lys-1923.28 was a key contact for SR141716A (8). Similarly, Salo et al. (42) and Silvestri et al. (43) reported models of SR141716A binding that were determined according to the postulate that Lys-1923.28 directly interacts with SR141716A. However, the role of Lys-1923.28 has not been fully established. It would be desirable not to be biased by limiting the SR141716A binding mode by Lys-1923.28. In support, it was shown that Lys-1923.28 was not crucial for the binding of taranabant, whose binding pocket is believed to be shared with SR141716A based upon mutagenesis data (31).
Accordingly, this study of the SR141716A binding mode was determined by exhaustively sampling the available space allowed for SR141716A binding within the TM core region, without imposing any constraint. We employed the MD simulation approach carefully guided by the results of the mutational studies reported previously (6, 14, 15, 17, 31) and presented in this study. Thus, our SR141716A-binding model was a dynamic model, where the binding mode of SR141716A was dynamically identified as the ligand moved within the binding pocket as a result of a long MD simulation performed in a lipid bilayer. In contrast, the SR141716A mode by McAllister et al. (6) was a static model obtained by a simple energy minimization, and the SR141716A modes by Salo et al. (42) and Silvestri et al. (43) were dynamic models obtained by a short MD simulation performed in the gas or aqueous phase. It should be noted that the template used for the construction of the present model of the CB1 receptor was the x-ray structure of β2AR (11), although the template used in the CB1 receptor models by McAllister et al. (6), Salo et al. (42), and Silvestri et al. (43) were the x-ray structures of rhodopsin. Despite the similarity in overall structural topology, homology receptor models derived from different x-ray structures as templates may result in structures that are locally quite different.
The present SR141716A binding model is similar to the model proposed by McAllister et al. (6) with respect to the positioning of the three substituents within the TM-binding core (i.e. the N1-aromatic ring moiety oriented toward the bottom of the major binding pocket, the C3-piperidinyl ring moiety toward the minor binding pocket, and the C5-aromatic ring toward TM5). Interestingly, in the models by Salo et al. (42) and Silvestri et al. (43), the positions of the N1-aromatic ring moiety and the C5-aromatic ring were switched, compared with the present SR141716A binding mode. The detailed receptor-ligand interactions in the present SR141716A binding mode were significantly different from those in the SR141716A binding modes by McAllister et al. (6), Salo et al. (42), and Silvestri et al. (43). Because of the use of Lys-1923.28 as a key contact for SR141716A (8), SR141716A in these binding models was shifted more toward TM3 and the minor binding pocket. Thus, the N1-aromatic ring failed to form aromatic stacking with Trp-3566.48 and the C5-aromatic ring loosely interacted with TM5. In contrast, the engagement of the N1-aromatic ring moiety in an interaction with Trp-3566.48 appears to be essential for not only ligand binding but also for receptor regulation according to the present SR141716A binding model. As we demonstrated in the present simulation of the CB1-SR141716A model, SR141716A was able to stabilize the receptor, securing the Trp-3566.48 toggle switch in the inactive state.
Comparison of the Binding Mode of SR141716A with the Binding Mode of CP55940
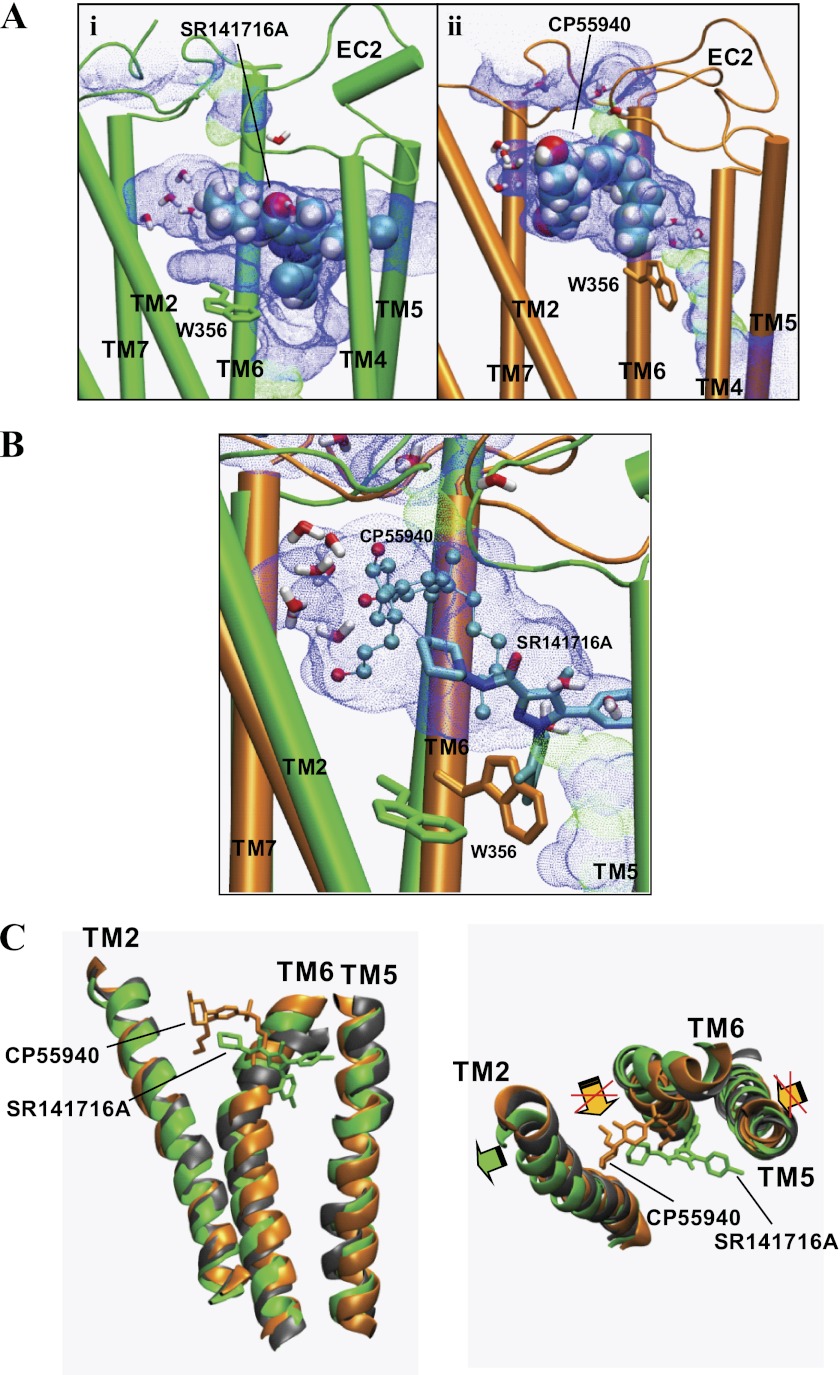
Comparison of the previously published model of the CB1-CP55940 complex (10) and the CB1-SR141716A complex revealed that SR141716A commonly shares the minor binding pocket with CP55940 but is the only ligand that occupies the major binding pocket toward TM5 (Fig. 6A). Significant displacements in the side chain orientation of the residues consisting of the major binding pocket suggested that the major binding pocket in the central core of the TM helical bundle of the CB1 receptor was highly flexible and could easily accommodate different classes of chemical structures. The rearrangement of the Trp-3566.48 side chain was of particular interest. Thus, as shown in Fig. 6A, the major binding pocket in CP55940 binding, which was limited by Trp-3566.48 in the trans form, expanded as Trp-3566.48 was converted to the g+ form in SR141716A binding. Our results of ligand-specific receptor interactions were in line with recent findings that different β2AR ligands induced distinctive receptor conformations (45) and that essentially none of the compounds identified in a structure-based screen against the antagonist-bound dopamine D3 receptor demonstrated agonist activity (46).
FIGURE 6.
Comparison of the SR141716A binding model with the CP55940-binding model. A, comparison of the major binding pocket and the minor binding pocket in CB1-SR141716A (green) (panel i) and in CB1-CP55940 (orange) (panel ii). Each of the solvent-accessible pores in the ligand pocket of the system were created using HOLE (44) at the end of the simulation. The ligand is represented in a space-filling model (in atom type). The ligand is represented by space-filling (in atom type). The toggle switch Trp-3566.48 is represented in stick form. Water molecules within 6 Å of the ligand are also represented. EC, extracellular loop. B, overlap between SR141716A (stick form) to CP55940 (ball-and-stick) within the solvent-accessible pore created by using HOLE (44) at the receptor binding pocket of CB1-CP55940. The overlap between SR141716A and CP55940 was obtained by superimposing the systems of CB1-SR141716A (green) with CB1-CP55940 (orange) at the end of the simulations, with respect to the receptor TM backbone atoms. Water molecules within 6 Å of CP55940 are also represented. Trp-3566.48 residues in CB1-CP55940 (orange) and in CB1-SR141716A (green) are represented in stick form. It should be noted that the receptor in CB1-CP55940 was considered to be in a state not fully activated (10). C, superposition of CB1-SR141716A (green) and CB1-CP55940 (orange) with respect to the ligand-unbound CB1 receptor (gray). SR141716A (green) and CP55940 (orange) are represented in stick form. Hydrogen atoms are omitted for clarity. Only TM2, TM5, and TM6 are represented for clarity. An outward movement of TM2 promoted in CB1-SR141716A is indicated by a green arrow, and an inward movement of TM5 and a rigid-body rotation of TM6 promoted in CB1-CP55940 are indicated by orange arrows. Suppression of these TM5 and TM6 movements by CB1-SR141716A are marked by red Xs.
SR141716A Competes with the Classical and Nonclassical Cannabinoids for the Binding of the Hydrophobic Pocket
When the present CB1 receptor-SR141716A model was superimposed on the previously published CB1 receptor-CP55940 model (10), it was shown that SR141716A, although positioned deeper in the binding pocket than CP55940, had a decent overlap with CP55940 (Fig. 6B). The C3-carbonylpiperidinyl ring moiety of SR141716A overlapped with the C3 alkyl chain of CP55940 in the major binding pocket. It should be noted that this region of the major binding pocket was equivalent to the hydrophobic pocket for the binding of the C3 alkyl chain of classical and nonclassical cannabinoids (10). Thus, it appeared that SR141716A competed with the classical and nonclassical cannabinoids for the binding of the hydrophobic pocket. It was also shown that the C3-piperidinyl moiety of SR141716A, although not exactly overlapped, was in proximity to the AC-ring moiety of CP55940 in the minor binding pocket, close enough for competing for the binding of the minor binding pocket (Fig. 6B).
Considering that both SR141716A and the classical and nonclassical cannabinoids competed for the binding of the hydrophobic pocket, ligand-specific receptor interactions at the hydrophobic pocket appeared to be important for regulating molecular switches in this region (e.g. the Trp-3566.48 rotameric toggle switch) and inducing associated receptor conformational changes required for receptor activation. In the case of SR141716A, the N1-aromatic ring of SR141716A was tightly engaged in aromatic stacking with Trp-3566.48 so that the Trp-3566.48 rotameric toggle switch was secured from activation. In contrast, for CP55940, the flexible C3 alkyl chain appeared to directly interact with Trp-3566.48 for triggering the Trp-3566.48 rotamer change and the subsequent rigid-body movement of TM6, eventually leading to receptor activation.
Is TM4 Movement due to the Stabilization by SR141716A Binding or due to the Destabilization Caused by the Lack of Cholesterol?
It was also observed in the present simulation of CB1-SR141716A that there were noticeable detachments of TM4 along the breakage of a direct H-bond between the side chain O of Ser-1582.45 and the side chain N of Trp-2414.50 (Fig. 3A, panels ii and iii). Although it was not clear whether the recovered H-bond between the side chain O of Ser-1582.45 and the side chain N of Trp-2414.50, along with the modified χ2 angles of Trp-2414.50 and Trp-2795.43 (Fig. 3A) at the end of the CB1-SR141716A simulation, represented the receptor stabilized by SR141716A, it appeared that receptor stabilization by SR141716A involves the coupling of TM4 and TM5 possibly through the bound ligand SR141716A and that the retention of the H-bond between the side chain O of Ser-1582.45 and the side chain N of Trp-2414.50 would be important for receptor stabilization.
It was also possible that TM4 was detached from the TM bundle due to the systematic limitation of the current model system. For example, cholesterol has been implicated in structural and functional roles in GPCRs (47). The cholesterol-binding site predicted to be in almost half of the rhodopsin family GPCRs (47) and confirmed in the recent x-ray structures of the β2AR (Protein Data Bank codes 2RH1 and 3D4S) (11, 48) appears to exist in the region where the H-bond is formed between Ser-1582.45 and Trp-2414.50 (Fig. 4A), or alternatively, it may be proximal to helix 8 of the CB1 receptor (49). It has been reported that cholesterol increased TM2/TM4 helical packing and that its main effect was upon TM4 not TM2 (48). Thus, it was possible that the breakage of the Ser-1582.45/Trp-2414.50 H-bond observed in the present CB1-SR141716A complex during the simulation was an artifact due to the destabilization caused by the lack of cholesterol that would otherwise stabilize the receptor structure at the TM2/TM4 junction.
Emerging Picture of the Molecular Mechanism of SR141716A Inverse Agonism in the CB1 Receptor
The results of our simulations of the CB1 receptor-SR141716A models in this study provided some insight into the molecular mechanism of the inverse agonism of SR141716A in the CB1 receptor. As shown in Fig. 6C, the superimposition of CB1-SR141716A and CB1-CP55940 with respect to the ligand-unbound CB1 receptor suggests that SR141716A promotes a small outward movement of TM2 and suppresses an inward movement of TM5 required for receptor activation (38). Here, we propose that SR141716A exerts inverse agonism through the stabilization of not only the major but the minor binding pocket as well. Thus, the binding of the C3-piperidinyl moiety of SR141716A stabilized TM2, and the binding of the C5-aromatic ring moiety of SR141716A stabilized TM5. These stabilizing interactions appeared critical for positioning the ligand's N1-aromatic ring moiety to secure the Trp-3566.48 toggle switch from activation, contributing to the TM2/TM3/TM7 stabilization important for maintaining the inactive state of the receptor (40, 41). Furthermore, these interactions prohibit a TM6 rigid-body movement required for receptor activation (50, 51), as shown in Fig. 6C, leading to stabilization of the inactive state (i.e. the inverse agonism). In support, a similar role of Trp-4286.48 of the histamine H1 receptor in securing the receptor in an inactive state has recently been observed in the x-ray structure of the human H1 receptor (52).
Acknowledgments
J.-Y. S. acknowledges the Texas Advanced Computing Center at University of Texas at Austin for providing HPC resources that have contributed to the research results. This work used the Extreme Science and Engineering Discovery Environment, which is supported by National Science Foundation Grant OCI-1053575.
This work was supported, in whole or in part, by National Institutes of Health Grant DA020663 (to J.-Y. S.) and Grant DA020763 (to D. A. K.). This work was also supported by National Science Foundation Grants ACI-0338618l, OCI-0451237, OCI-0535258, and OCI-0504075.

This article contains supplemental Tables 1, Figs. 1–3, and additional references.
Because the CB1 mutational data cross species (e.g. human and mouse), the numbering scheme for mutated residues is translated into the number that it would be in the human.
- CB1 receptor
- brain cannabinoid receptor 1
- GPCR
- G protein-coupled receptor
- SR141716A
- N-(piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-pyrazole-3-carboxamide
- CP55940
- (−)-3-[2-hydroxyl-4-(1,1-dimethylheptyl)phenyl]-4-[3-hydroxylpropyl]cyclohexan-1-ol
- TM
- transmembrane
- MD
- molecular dynamics
- r.m.s.d.
- the root-mean-square deviations
- β2AR
- β2-adrenergic receptor.
REFERENCES
- 1. Howlett A. C., Barth F., Bonner T. I., Cabral G., Casellas P., Devane W. A., Felder C. C., Herkenham M., Mackie K., Martin B. R., Mechoulam R., Pertwee R. G. (2002) International union of pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol. Rev. 54, 161–202 [DOI] [PubMed] [Google Scholar]
- 2. Robson P. (2001) Therapeutic aspects of cannabis and cannabinoids. Br. J. Psychiatry 178, 107–115 [DOI] [PubMed] [Google Scholar]
- 3. Colombo G., Agabio R., Diaz G., Lobina C., Reali R., Gessa G. L. (1998) Appetite suppression and weight loss after the cannabinoid antagonist SR141716. Life Sci. 63, PL113–PL117 [DOI] [PubMed] [Google Scholar]
- 4. Lazary J., Juhasz G., Hunyady L., Bagdy G. (2011) Personalized medicine can pave the way for the safe use of CB1 receptor antagonists. Trends Pharmacol. Sci. 32, 270–280 [DOI] [PubMed] [Google Scholar]
- 5. Kobilka B. K., Deupi X. (2007) Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 28, 397–406 [DOI] [PubMed] [Google Scholar]
- 6. McAllister S. D., Rizvi G., Anavi-Goffer S., Hurst D. P., Barnett-Norris J., Lynch D. L., Reggio P. H., Abood M. E. (2003) An aromatic microdomain at the cannabinoid CB(1) receptor constitutes an agonist/inverse agonist binding region. J. Med. Chem. 46, 5139–5152 [DOI] [PubMed] [Google Scholar]
- 7. Ballesteros J. A., Weinstein H. (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 [Google Scholar]
- 8. Hurst D. P., Lynch D. L., Barnett-Norris J., Hyatt S. M., Seltzman H. H., Zhong M., Song Z. H., Nie J., Lewis D., Reggio P. H. (2002) N-(Piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-pyrazole-3-carboxamide (SR141716A) interaction with Lys-3.28192 is crucial for its inverse agonism at the cannabinoid CB1 receptor. Mol. Pharmacol. 62, 1274–1287 [DOI] [PubMed] [Google Scholar]
- 9. Reiter E., Ahn S., Shukla A. K., Lefkowitz R. J. (2012) Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu. Rev. Pharmacol. Toxicol. 52, 179–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shim J. Y., Bertalovitz A. C., Kendall D. A. (2011) Identification of essential cannabinoid-binding domains. Structural insights into early dynamic events in receptor activation. J. Biol. Chem. 286, 33422–33435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) High resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones G., Willett P., Glen R. C. (1995) Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 245, 43–53 [DOI] [PubMed] [Google Scholar]
- 13. Verdonk M. L., Cole J. C., Hartshorn M. J., Murray C. W., Taylor R. D. (2003) Improved protein-ligand docking using GOLD. Proteins 52, 609–623 [DOI] [PubMed] [Google Scholar]
- 14. McAllister S. D., Tao Q., Barnett-Norris J., Buehner K., Hurst D. P., Guarnieri F., Reggio P. H., Nowell Harmon K. W., Cabral G. A., Abood M. E. (2002) A critical role for a tyrosine residue in the cannabinoid receptors for ligand recognition. Biochem. Pharmacol. 63, 2121–2136 [DOI] [PubMed] [Google Scholar]
- 15. McAllister S. D., Hurst D. P., Barnett-Norris J., Lynch D., Reggio P. H., Abood M. E. (2004) Structural mimicry in class A G protein-coupled receptor rotamer toggle switches. The importance of the F3.36(201)/W6.48(357) interaction in cannabinoid CB1 receptor activation. J. Biol. Chem. 279, 48024–48037 [DOI] [PubMed] [Google Scholar]
- 16. Shire D., Calandra B., Bouaboula M., Barth F., Rinaldi-Carmona M., Casellas P., Ferrara P. (1999) Cannabinoid receptor interactions with the antagonists SR 141716A and SR 144528. Life Sci. 65, 627–635 [DOI] [PubMed] [Google Scholar]
- 17. Ahn K. H., Bertalovitz A. C., Mierke D. F., Kendall D. A. (2009) Dual role of the second extracellular loop of the cannabinoid receptor 1. Ligand binding and receptor localization. Mol. Pharmacol. 76, 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen J., Im W., Brooks C. L., 3rd (2006) Balancing solvation and intramolecular interactions. Toward a consistent generalized Born force field. J. Am. Chem. Soc. 128, 3728–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Buck M., Bouguet-Bonnet S., Pastor R. W., MacKerell A. D., Jr. (2006) Importance of the CMAP correction to the CHARMM22 protein force field. Dynamics of hen lysozyme. Biophys. J. 90, L36–L38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. (1983) CHARMM. A program for macromolecular energy, minimization, and dynamics. J. Comp. Chem. 4, 187–217 [Google Scholar]
- 22. MacKerell A. D., Jr., Bashford D., Bellott M., Dunbrack R. L., Jr., Evanseck J., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher I. W., Roux B., Schlenkrich M., Smith J., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 23. Feller S., MacKerell A. D., Jr. (2000) An improved empirical potential energy function for molecular simulations of phospholipids. J. Phys. Chem. B 104, 7510–7515 [Google Scholar]
- 24. Saam J., Tajkhorshid E., Hayashi S., Schulten K. (2002) Molecular dynamics investigation of primary photoinduced events in the activation of rhodopsin. Biophys. J. 83, 3097–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Feller S. E., Zhang Y., Pastor R. W., Brooks B. R. (1995) Constant-pressure molecular dynamics simulation. The Langevin piston method. J. Chem. Phys. 103, 4613–4621 [Google Scholar]
- 26. Hoover W. G. (1985) Canonical dynamics. Equilibrium phase-space distributions. Phys. Rev. A 31, 1695–1697 [DOI] [PubMed] [Google Scholar]
- 27. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 28. Tuckerman M., Berne B. J., Martyna G. (1992) Reversible multiple time scale molecular dynamics. J. Chem. Phys. 97, 1990–2001 [Google Scholar]
- 29. Breneman C. M., Wiberg K. B. (1990) Determining atom centered monopoles from molecular electrostatic potentials. The need for high sampling density in formamide conformational analysis. J. Comp. Chem. 11, 361–373 [Google Scholar]
- 30. Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J. A., Jr., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam J. M., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas O., Foresman J. B., Ortiz J. V., Cioslowski J., Fox D. J. (2009) Gaussian 09, Revision A.02, Gaussian, Inc., Wallingford, CT [Google Scholar]
- 31. Lin L. S., Ha S., Ball R. G., Tsou N. N., Castonguay L. A., Doss G. A., Fong T. M., Shen C. P., Xiao J. C., Goulet M. T., Hagmann W. K. (2008) Conformational analysis and receptor docking of N-[(1S,2S)-3-(4-chlorophenyl)-2-(3-cyanophenyl)-1-methylpropyl]-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]oxy}propanamide (taranabant, MK-0364), a novel, acyclic cannabinoid-1 receptor inverse agonist. J. Med. Chem. 51, 2108–2114 [DOI] [PubMed] [Google Scholar]
- 32. Shi L., Liapakis G., Xu R., Guarnieri F., Ballesteros J. A., Javitch J. A. (2002) β2-Adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J. Biol. Chem. 277, 40989–40996 [DOI] [PubMed] [Google Scholar]
- 33. Makriyannis A., Liu Q. (2001) Preparation of pyrazole derivatives as cannabinoid receptor antagonists. PCT Int. Appl. WO 2001029007 [Google Scholar]
- 34. Francisco M. E., Seltzman H. H., Gilliam A. F., Mitchell R. A., Rider S. L., Pertwee R. G., Stevenson L. A., Thomas B. F. (2002) Synthesis and structure-activity relationships of amide and hydrazide analogues of the cannabinoid CB(1) receptor antagonist N-(piperidinyl)- 5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-pyrazole-3-carboxamide (SR141716). J. Med. Chem. 45, 2708–2719 [DOI] [PubMed] [Google Scholar]
- 35. Wiley J. L., Jefferson R. G., Grier M. C., Mahadevan A., Razdan R. K., Martin B. R. (2001) Novel pyrazole cannabinoids. Insights into CB(1) receptor recognition and activation. J. Pharmacol. Exp. Ther. 296, 1013–1022 [PubMed] [Google Scholar]
- 36. Shim J. Y., Welsh W. J., Cartier E., Edwards J. L., Howlett A. C. (2002) Molecular interaction of the antagonist N-(piperidin-1-yl)-5-(4-chlorophenyl)-1- (2,4-dichlorophenyl)-4-methyl-1H-pyrazole-3-carboxamide with the CB1 cannabinoid receptor. J. Med. Chem. 45, 1447–1459 [DOI] [PubMed] [Google Scholar]
- 37. Lan R., Liu Q., Fan P., Lin S., Fernando S. R., McCallion D., Pertwee R., Makriyannis A. (1999) Structure-activity relationships of pyrazole derivatives as cannabinoid receptor antagonists. J. Med. Chem. 42, 769–776 [DOI] [PubMed] [Google Scholar]
- 38. Rasmussen S. G., Choi H. J., Fung J. J., Pardon E., Casarosa P., Chae P. S., Devree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., Steyaert J., Weis W. I., Kobilka B. K. (2011) Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature 469, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mirzadegan T., Benkö G., Filipek S., Palczewski K. (2003) Sequence analyses of G-protein-coupled receptors. Similarities to rhodopsin. Biochemistry 42, 2759–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kleinau G., Hoyer I., Kreuchwig A., Haas A. K., Rutz C., Furkert J., Worth C. L., Krause G., Schülein R. (2011) From molecular details of the interplay between transmembrane helices of the thyrotropin receptor to general aspects of signal transduction in family a G-protein-coupled receptors (GPCRs). J. Biol. Chem. 286, 25859–25871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chugunov A. O., Simms J., Poyner D. R., Dehouck Y., Rooman M., Gilis D., Langer I. (2010) Evidence that interaction between conserved residues in transmembrane helices 2, 3, and 7 are crucial for human VPAC1 receptor activation. Mol. Pharmacol. 78, 394–401 [DOI] [PubMed] [Google Scholar]
- 42. Salo O. M., Lahtela-Kakkonen M., Gynther J., Järvinen T., Poso A. (2004) Development of a 3D model for the human cannabinoid CB1 receptor. J. Med. Chem. 47, 3048–3057 [DOI] [PubMed] [Google Scholar]
- 43. Silvestri R., Cascio M. G., La Regina G., Piscitelli F., Lavecchia A., Brizzi A., Pasquini S., Botta M., Novellino E., Di Marzo V., Corelli F. (2008) Synthesis, cannabinoid receptor affinity, and molecular modeling studies of substituted 1-aryl-5-(1H-pyrrol-1-yl)-1H-pyrazole-3-carboxamides. J. Med. Chem. 51, 1560–1576 [DOI] [PubMed] [Google Scholar]
- 44. Smart O. S., Goodfellow J. M., Wallace B. A. (1993) The pore dimensions of gramicidin A. Biophys. J. 65, 2455–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kahsai A. W., Xiao K., Rajagopal S., Ahn S., Shukla A. K., Sun J., Oas T. G., Lefkowitz R. J. (2011) Multiple ligand-specific conformations of the β2-adrenergic receptor. Nat. Chem. Biol. 7, 692–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carlsson J., Coleman R. G., Setola V., Irwin J. J., Fan H., Schlessinger A., Sali A., Roth B. L., Shoichet B. K. (2011) Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nat. Chem. Biol. 7, 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oates J., Watts A. (2011) Uncovering the intimate relationship between lipids, cholesterol, and GPCR activation. Curr. Opin. Struct. Biol. 21, 802–807 [DOI] [PubMed] [Google Scholar]
- 48. Hanson M. A., Cherezov V., Griffith M. T., Roth C. B., Jaakola V. P., Chien E. Y., Velasquez J., Kuhn P., Stevens R. C. (2008) A specific cholesterol-binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure 16, 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oddi S., Dainese E., Fezza F., Lanuti M., Barcaroli D., De Laurenzi V., Centonze D., Maccarrone M. (2011) Functional characterization of putative cholesterol-binding sequence (CRAC) in human type-1 cannabinoid receptor. J. Neurochem. 116, 858–865 [DOI] [PubMed] [Google Scholar]
- 50. Farrens D. L., Altenbach C., Yang K., Hubbell W. L., Khorana H. G. (1996) Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science 274, 768–770 [DOI] [PubMed] [Google Scholar]
- 51. Altenbach C., Kusnetzow A. K., Ernst O. P., Hofmann K. P., Hubbell W. L. (2008) High resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl. Acad. Sci. U.S.A. 105, 7439–7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shimamura T., Shiroishi M., Weyand S., Tsujimoto H., Winter G., Katritch V., Abagyan R., Cherezov V., Liu W., Han G. W., Kobayashi T., Stevens R. C., Iwata S. (2011) Structure of the human histamine H1 receptor complex with doxepin. Nature 475, 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]