Background: The RNA polymerase II C-terminal domain (CTD) recruits RNA processing complexes spatio-temporally through its phosphorylation patterns.
Results: The CTD kinases, CDK7, BRD4, and CDK9, interact and phosphorylate each other and TAF7.
Conclusion: CTD kinases regulate each other both directly and indirectly through TAF7.
Significance: CTD kinase cross-talk indicates a novel mechanism for ensuring orderly, sequential phosphorylation of Pol II CTD.
Keywords: CDK (Cyclin-dependent Kinase), Phosphorylation Enzymes, RNA Polymerase II, Transcription Elongation Factors, Transcription Initiation Factors, BRD4, CDK7, CDK9, Cross-talk, TAF7
Abstract
The RNA polymerase II (Pol II) C-terminal domain (CTD) serves as a docking site for numerous proteins, bridging various nuclear processes to transcription. The recruitment of these proteins is mediated by CTD phospho-epitopes generated during transcription. The mechanisms regulating the kinases that establish these phosphorylation patterns on the CTD are not known. We report that three CTD kinases, CDK7, CDK9, and BRD4, engage in cross-talk, modulating their subsequent CTD phosphorylation. BRD4 phosphorylates PTEFb/CDK9 at either Thr-29 or Thr-186, depending on its relative abundance, which represses or activates CDK9 CTD kinase activity, respectively. Conversely, CDK9 phosphorylates BRD4 enhancing its CTD kinase activity. The CTD Ser-5 kinase CDK7 also interacts with and phosphorylates BRD4, potently inhibiting BRD4 kinase activity. Additionally, the three kinases regulate each other indirectly through the general transcription factor TAF7. An inhibitor of CDK9 and CDK7 CTD kinase activities, TAF7 also binds to BRD4 and inhibits its kinase activity. Each of these kinases phosphorylates TAF7, affecting its subsequent ability to inhibit the other two. Thus, a complex regulatory network governs Pol II CTD kinases.
Introduction
The RNA polymerase II (Pol II)2 C-terminal domain (CTD) consists of 52 tandem repeats of the amino acid heptad Y1S2P3T4S5P6S7. CTD phosphorylation dictates many Pol II interactions and is a critical step during eukaryotic transcription (1, 2). It not only triggers co-transcriptional RNA capping and splicing, but also couples transcription with DNA repair, chromatin remodeling, and RNA transport (3). The pattern of phosphorylation on each heptad repeat and across the entire CTD is dynamic during transcription, resulting in transient and functionally distinct phospho-epitopes (3). These transient marks signal the spatio-temporal recruitment of over 100 different proteins with diverse roles (4).
The mechanisms that establish the timing and patterns of CTD phosphorylation are unknown. Although interplay between the CTD kinases has been proposed as a possibility (5, 6), no direct evidence for its existence has been found thus far. The CTD kinases CDK7 and CDK9 phosphorylate CTD Ser-5/7 and Ser-2/5, respectively (7). CDK7 phosphorylation of Ser-5 is associated with transcription initiation, RNA capping, and histone modifications, whereas CDK9 phosphorylation of Ser-2/5 is associated with elongation, splicing, and DNA repair (8). CDK7, the kinase component of TFIIH, resides within a CDK activating kinase (CAK) subcomplex consisting of CDK7, Cyclin H, and MAT1. CDK9 is the kinase component of the PTEFb complex (7). We recently identified a third CTD kinase, the bromodomain protein BRD4. BRD4, a candidate therapeutic target for multiple cancers, uniquely phosphorylates CTD Ser-2 sites (9). How the activities of these kinases are coordinated to ensure a proper temporal and spatial sequence of CTD phosphorylation remains unknown. Therefore, we investigated the structural and functional interactions between BRD4, CDK9, and CDK7 and whether such interactions modulate Pol II CTD phosphorylation.
We report here that the three CTD kinases, CDK7, CDK9, and BRD4, engage in direct and indirect cross-talk to modulate each others ability to phosphorylate the Pol II CTD. BRD4 and CDK9 phosphorylate each other at multiple sites, affecting their respective CTD kinase activities: CDK9 phosphorylates BRD4, enhancing its CTD kinase activity. Concomitantly, BRD4 directly phosphorylates the Thr-186 activation site on CDK9, identifying it as the hitherto unknown kinase responsible for activating PTEFb. At high molar ratios, BRD4 also phosphorylates CDK9 at the Thr-29 inactivation site. Furthermore, CDK7 interacts with and phosphorylates BRD4, inhibiting its CTD kinase activity. In addition to these direct interactions that modulate the CTD kinases, TAF7, a general transcription factor, regulates them indirectly. We show that, TAF7, which inhibits CDK9 and CDK7 kinase activities (10), also binds BRD4 and inhibits its kinase activity. Phosphorylation of TAF7 by these kinases in turn modulates its inhibitory effects. These findings thus reveal the existence of a complex network of Pol II CTD kinase cross-talk and regulation, suggesting a potential regulatory mechanism for CTD phosphorylation.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture
HeLa cells were grown as described previously (10). Drosophila SF9 cells were grown in TNM-FH insect medium (BD Biosciences Pharmingen) and maintained at 27 °C.
Reagents
Antibodies used for either immunoprecipitation or immunoblotting were: anti-RNA Pol II, mAb 8WG16 (Santa Cruz Biotechnology); anti-BRD4, H-250 (Santa Cruz Biotechnology); anti-Pol II phospho-Ser-2, mAb 3E10 (Millipore); anti-Pol II phospho-Ser-5, mAb 3E8 (Millipore); anti-CDK9, mAb D-7 (Santa Cruz Biotechnology); anti-phospho-CDK9, Thr-29, anti-phospho-CDK9, Thr-187 (Cell Signaling); anti-cyclin T1 and anti-cyclin H (Santa Cruz Biotechnology); anti-tubulin (Abcam), anti-phosphoserine clone 4A4 (Millipore), anti-TAF7 (Abcam), and anti-CDK7 (Santa Cruz Biotechnology). Kinase inhibitors flavopiridol, roscovitine, and apigenin (Sigma) were dissolved in dimethyl sulfoxide to 10 mm. The BRD4 inhibitor (+)-JQ1 (BPS Bioscience) was dissolved in ethanol to 2 mm. Stock solutions of all inhibitors were stored at −80 °C.
Plasmid Constructs
Murine FLAG-BRD4 WT and ΔB1, ΔB2, ΔB1B2, ΔN, and ΔC mutants are as described before (9) and were a kind gift from K. Ozato (NICHD, NIH, Bethesda, MD). The human FLAG-tagged BRD4 WT and FAA-AAA mutant in the pCMV2 mammalian vector were a gift from E. Verdin (Gladstone Institute of Virology, University of California, San Francisco, CA). The human GST-CTD construct in pGEX vector, the CDK9 WT, and CDK9 D167N mutant in pCDNA vectors were a gift from J. N. Brady (NCI, NIH, Bethesda, MD). The TAF7 in pcDNA3 vector, and the GST- and FLAG-tagged full-length and fragments of TAF7 in pGEX and pET vectors for protein expression have been described previously (10, 11).
Protein Purification
Recombinant FLAG-tagged BRD4 and associated mutant proteins were purified from Sf9 cells as described earlier (9). The FLAG peptide was eliminated on a microcon column (Millipore) and proteins were recovered in HKEG buffer (20 mm Hepes, pH 7.9, 100 mm KCl, 0.2 mm EDTA, 20% (v/v) glycerol). The GST-CTD and GST-TAF7 full-length and TAF7 fragments was purified from a bacterial expression system (BL21 Escherichia coli cells) using GST-Sepharose beads (Amersham Biosciences). HA-tagged CDK9 and CDK9 D167N were purified from transiently transfected HeLa cells. The proteins were immunoprecipitated with anti-HA affinity matrix (Roche Applied Science), eluted with HA peptide (1 mg/ml), and recovered in HKEG buffer after removing the HA peptide on a microcon column (Millipore). Purified native TFIIH was obtained from Protein One. Purified, enzymatically active PTEFb and CAK complexes were procured from Millipore.
In Vitro Kinase Assays
In vitro kinase assays with BRD4, PTEFb, and CAK were performed in 20 μl of 50 mm Tris (pH 7.5), 5 mm DTT, 5 mm MnCl2, 5 mm MgCl2 with 10 μCi of [γ-32P]ATP (6000 Ci/mm) and/or 40 μm ATP where indicated in the figure legends. The kinase reactions for incubated for 1 h at 30 °C, following which the proteins were resolved by SDS-PAGE and the extent of phosphorylation was quantitated by a PhosphorImager. When CTD phosphorylation was determined by immunoblotting with specific antibodies as indicated in the figures, kinase assays were performed with cold ATP. When kinase inhibitors were used, appropriate dilutions of the inhibitor were added at the start of the kinase reaction. Mock treated kinase reactions were treated with equivalent volumes of dimethyl sulfoxide.
Immunoprecipitation and Co-immunoprecipitation
To co-immunoprecipitate CDK7 or TAF7 with BRD4 from HeLa nuclear extracts, magnetic beads (Dynabeads Protein A) were coated with 5 μg of anti-BRD4 antibody and incubated with 25 μg of HeLa nuclear extract (Promega) for 3 h at 4 °C. The beads were then washed three times with 50 mm Tris (pH 8.0), 200 mm NaCl, and 0.2% Nonidet P-40. Bound proteins were separated on SDS-PAGE gels and immunoblotted with antibodies against BRD4, CDK7, or TAF7. To test for BRD4 binding to CDK7 or TAF7, purified His-CDK7 or GST-TAF7 was immobilized on His/GST-agarose beads and incubated with purified BRD4 for 3 h at 4 °C. Beads were washed twice with 50 mm Tris (pH 8.0), 150 mm NaCl, and 0.2% Nonidet P-40 and bound proteins were immunoblotted. CTD binding to BRD4 mutants was detected by immobilizing equimolar amounts of BRD4 proteins on anti-FLAG M2-agarose beads and incubating with 200 ng of GST-CTD for 2 h at 4 °C. The beads were washed twice with 50 mm Tris (pH 8.0), 150 mm NaCl, and 0.2% Nonidet P-40 and bound GST-CTD was immunoblotted with anti-GST antibody. All immunoblot analyses were performed using the Odyssey infrared scanner and secondary antibodies from Li-Cor.
Transient Transfections
Transient transfections were done in HeLa cells using Lipofectamine (Invitrogen). Where indicated, cells were treated with 5 μm (+)-JQ1 for 48 h. Whole cell extracts were made from the cells with Tissue extraction reagent I (Invitrogen) and analyzed through immunoprecipitation and immunoblotting as described above. BRD4 knockdown was done by Lipofectamine transfection of 80 nm siRNA (ON-TARGETplus smartpool human BRD4 siRNA; Dharmacon) into HeLa cells. Whole cell extracts of HEK293T cells with or without transient transfection of CDK7 were procured from OriGene Technologies.
RESULTS
BRD4 and CDK9 Phosphorylate Each Other
PTEFb/CDK9 is recruited to transcription sites through its interaction with BRD4 (12, 13). Our recent discovery that BRD4 is also a kinase led us to ask whether BRD4 and CDK9 phosphorylate each other. To this end, we first asked whether the PTEFb/CDK9 could phosphorylate purified recombinant BRD4. As shown in Fig. 1A (upper panel), BRD4 is robustly phosphorylated by PTEFb in an in vitro kinase assay to a level that significantly exceeds that of BRD4 autophosphorylation. To map the regions of BRD4 targeted by PTEFb, we compared the relative phosphorylation by PTEFb/CDK9 of the full-length BRD4 (WT) with a series of BRD4 mutants (ΔB1, ΔB2, ΔB1B2, ΔN, ΔC; Fig. 1A). Mutants deleted of either or both of the BRD4 bromodomains (ΔB1, ΔB2, ΔB1B2) or either the entire amino terminus (ΔN) or carboxyl terminus (ΔC) were all efficiently phosphorylated by PTEFb/CDK9. Significantly, the ΔN BRD4 mutant, which lacks intrinsic kinase activity was highly phosphorylated in the presence of PTEFb/CDK9, demonstrating that CDK9 directly phosphorylates BRD4 (Fig. 1A). Interestingly, both the N-terminal (ΔC) and C-terminal (ΔN) sections of BRD4 were phosphorylated by PTEFb/CDK9 independently, indicating a minimum of two distinct CDK9 phosphorylation sites on BRD4. Taken together, these results demonstrate that PTEFb/CDK9 not only phosphorylates BRD4 but does so at multiple sites across the extended protein.
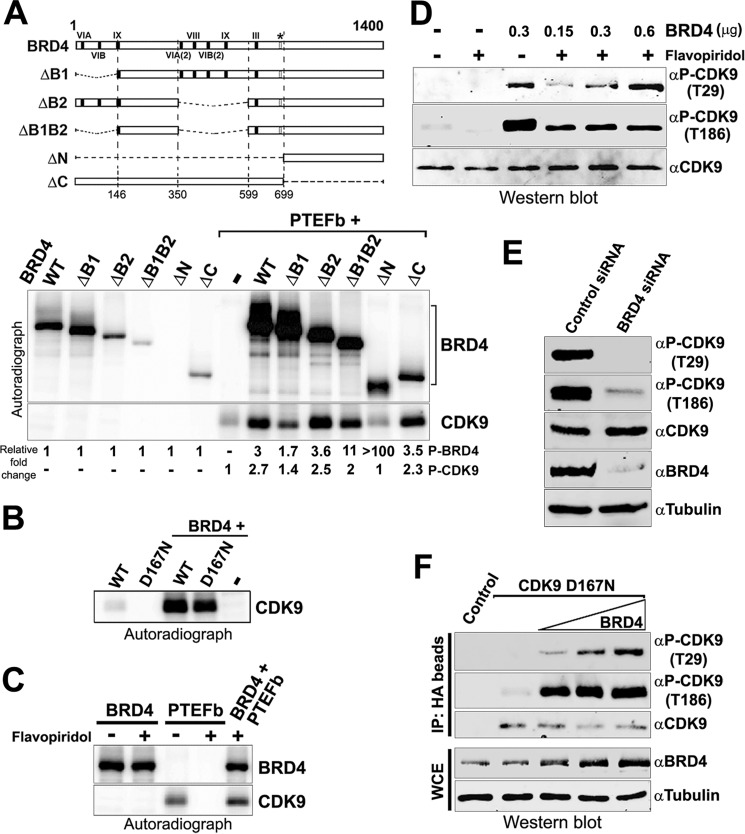
FIGURE 1.
BRD4 and CDK9 engage in cross-talk. A, BRD4 and CDK9 phosphorylate each other. BRD4 (0.3 μg), or equimolar amounts of BRD4 mutants were used in an in vitro kinase assay alone or with 0.15 μg of PTEFb. Maps of the BRD4 proteins used are shown (upper) with an autoradiograph of the phosphorylated proteins (lower). Numbers below each lane show the quantitation of total phosphorylation relative to autophosphorylation of each protein. B, BRD4 phosphorylates the CDK9 D167N kinase-dead mutant. Autoradiograph showing the results of a kinase reaction with 50 ng of wild type CDK9 or D167N mutant protein incubated with or without 0.2 μg of BRD4. C, BRD4 phosphorylates CDK9 in the presence of flavopiridol. Autoradiograph showing phosphorylation of BRD4 (0.2 μg) or PTEFb (0.1 μg) used in a kinase assay individually or together, with or without 1 μm flavopiridol. D, BRD4 phosphorylates CDK9 at both Thr-29 and Thr-186 sites in vivo. Kinase reactions were done in the presence or absence of flavopiridol (1 μm) with 0.15 μg of PTEFb alone or with 0.3 μg of BRD4 (first to third lanes). Reactions were also done with 0.15 μg of PTEFb and 0.15, 0.3, and 0.6 μg of BRD4 in the presence of flavopiridol (fourth to sixth lanes). The samples were immunoblotted with α-CDK9, phospho-specific α-CDK9 Thr(P)-186 or α-CDK9 Thr(P)-29. E, BRD4 is required for phosphorylation of CDK9 Thr-29 and Thr-186 sites in vivo. Whole cell extracts of HeLa cells transfected with control siRNA or BRD4-specific siRNA were immunoblotted with α-BRD4, α-CDK9, phospho-specific α-CDK9 Thr(P)-186, or α-CDK9 Thr(P)-29. F, BRD4 phosphorylates CDK9 at both Thr-29 and Thr-186 sites in vivo. Immunoprecipitates (IP) from HeLa cells untransfected or transfected with 2.5 μg of HA-D167N CDK9 alone or co-transfected with 1.0, 2.5, or 5.0 μg of BRD4 immunoblotted with α-CDK9, Thr(P)-29, or Thr(P)-186 α-phospho-CDK9 (upper). Immunoblots of whole cell extracts (WCE) show BRD4 levels relative to tubulin control (lower).
Conversely, BRD4 also phosphorylated CDK9. A significant increase in CDK9 phosphorylation relative to its autophosphorylation was observed in the presence of all the BRD4 proteins except the ΔN BRD4 kinase-dead mutant (Fig. 1A, lower panel). To confirm the direct phosphorylation of CDK9 by BRD4, we tested its ability to phosphorylate D167N, a point mutant of CDK9, which lacks kinase activity (14). Purified D167N CDK9 mutant, which is incapable of autophosphorylation, was highly phosphorylated by BRD4 in an in vitro kinase assay (Fig. 1B). The extent of D167N CDK9 phosphorylation was similar to that of the wild type CDK9, confirming that BRD4 directly phosphorylates CDK9 (Fig. 1B). The direct phosphorylation of CDK9 by BRD4 was further demonstrated by using flavopiridol, a potent and specific inhibitor of CDK9 kinase activity that does not affect BRD4 (9). Whereas the presence of flavopiridol completely abrogated CDK9 autophosphorylation, it did not inhibit the phosphorylation of CDK9 by BRD4 (Fig. 1C). Together, these results show that BRD4 and CDK9 directly phosphorylate each other.
Two phosphorylation sites on CDK9 are critical for its role during transcription: Thr-29 and Thr-186. Phosphorylation of Thr-29 inactivates its kinase activity, whereas phosphorylation of CDK9 at Thr-186 is crucial for its activity (15, 16). The kinase(s) directly responsible for these phosphorylation events has not been identified although the presence of BRD4 is known to be a requirement (16, 17). We therefore asked whether either of these sites is phosphorylated directly by BRD4. Following an in vitro kinase assay, CDK9 phosphorylated by BRD4 was immunoblotted with phosphospecific antibodies for CDK9 Thr(P)-29 and Thr(P)-186 (Fig. 1D). In the absence of BRD4, there was no detectable autophosphorylation at Thr-29, whereas autophosphorylation of Thr-186 occurs only at barely detectable levels. Surprisingly, BRD4 efficiently phosphorylated CDK9 at both Thr-29 and Thr-186 sites. Part of the observed phosphorylation reflects the activation of CDK9 autophosphorylation by BRD4, as previously reported (16) because the CDK9 inhibitor flavopiridol markedly reduces the levels of phosphorylation (Fig. 1D, compare third and fifth lanes). However, even in the presence of flavopiridol, both Thr-29 and Thr-186 are significantly phosphorylated, demonstrating that BRD4 directly phosphorylates CDK9 at both sites, although the patterns of phosphorylation differ. Thus, whereas the extent of Thr-29 phosphorylation increases with increasing BRD4 concentration, Thr-186 phosphorylation is constant. Therefore, BRD4 phosphorylation of CDK9 at the CDK9 Thr-186 site is efficient and preferred, whereas phosphorylation at Thr-29 is less efficient and requires an excess of BRD4 relative to CDK9.
BRD4 phosphorylated CDK9 Thr-29 and Thr-186 sites in vivo, as shown in two ways. First, siRNA-mediated knockdown of endogenous BRD4 resulted in abrogation of CDK9 Thr-29 and Thr-186 phosphorylation in vivo (Fig. 1E). Second, overexpression of BRD4 increased CDK9 phosphorylation, as follows. The HA-tagged D167N CDK9 kinase-dead mutant was transfected into HeLa cells, either alone or co-transfected with increasing amounts of Brd4. The HA-tagged CDK9 D167N was recovered from whole cell extracts of the transfected cells with HA-agarose beads and subjected to immunoblotting with Thr(P)-29 and Thr(P)-186 antibodies. As shown in Fig. 1F, increasing concentrations of BRD4 result in increasing phosphorylation of Thr-29 on the D167N mutant in vivo. Consistent with the in vitro results, the Thr-186 site of the D167N CDK9 mutant also was efficiently phosphorylated by BRD4. Together, these results demonstrate that BRD4 directly phosphorylates CDK9 at both its Thr-29 inactivation site and Thr-186 activation site, in vitro and in vivo.
CDK7 Interacts with and Phosphorylates BRD4
We next asked whether BRD4 also interacts with the other major CTD kinase, CDK7. Such an association in vivo was indicated by the finding that BRD4 co-immunoprecipitated CDK7 from HeLa nuclear extracts (Fig. 2A). To further investigate the interaction between BRD4 and CDK7, an in vitro pulldown assay was done using a recombinant CAK complex in which the CDK7 component was His-tagged. BRD4 was efficiently bound by the recombinant CAK immobilized on nickel-nitrilotriacetic acid-agarose beads (Fig. 2B). Thus, BRD4 and CDK7/CAK directly interact, both in vitro and in vivo.
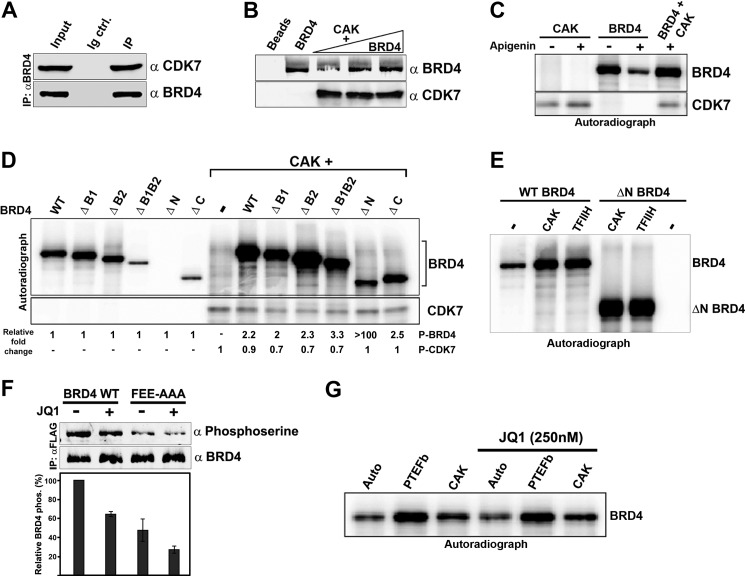
FIGURE 2.
CDK7 interacts with and phosphorylates BRD4. A, BRD4 associates with CDK7 in vivo. BRD4 was immunoprecipitated from HeLa nuclear extract using α-BRD4 and immunoblotted with α-CDK7. B, BRD4 interacts directly with CDK7. BRD4 (0.1, 0.4, and 0.6 μg) was pulled down with 0.2 μg of His-CDK7 (CAK) immobilized on Ni-NT beads. Beads alone or 0.6 μg of BRD4, loaded directly on the gel, are shown as negative and positive controls. C, CDK7 phosphorylates BRD4 in the presence of apigenin. Autoradiograph showing phosphorylation of BRD4 (0.3 μg) or CDK7/CAK (0.15 μg) in kinase assays individually or in combination, with or without 100 μm apigenin. D, CDK7 phosphorylates BRD4 at multiple sites. Autoradiograph of in vitro kinase assays of BRD4 (0.3 μg) or equimolar amounts of BRD4 mutants individually or with 0.12 μg of CAK. Numbers below each lane shows the quantitation of total phosphorylation relative to autophosphorylation of each protein. E, BRD4 is phosphorylated equally by the CAK complex and the holo-TFIIH complex. Autoradiograph showing wild type BRD4 (0.3 μg) and the BRD4 ΔN kinase-dead mutant (0.15 μg) phosphorylated by equimolar amounts of purified recombinant CAK (0.1 μg) or purified native TFIIH (0.26 μg). F, CDK7 phosphorylates BRD4 in vivo. HeLa cells were transfected with 3 μg of human BRD4 or a BRD4 mutant incapable of binding PTEFb (FEE-AAA) and were treated or not with (+)-JQ1. Equal amounts of BRD4 immunoprecipitated from these cells were immunoblotted with α-phosphoserine or α-BRD4 (upper). Quantitation of averages ± S.E. from two independent experiments (lower). G, auto- or transphosphorylation of BRD4 is not affected by the JQ1 inhibitor. Autoradiograph showing BRD4 (0.3 μg) either autophosphorylated, or transphosphorylated by PTEFb (0.1 μg) and CAK (0.1 μg), in the presence or absence of 250 nm JQ1.
CDK7/CAK is known to activate several kinases by phosphorylating them (18). To test if CDK7/CAK phosphorylates BRD4, in vitro kinase reactions were done with or without apigenin, which inhibits BRD4 kinase activity but has no effect on CDK7 kinase activity (Fig. 2C). BRD4 was efficiently phosphorylated by CDK7/CAK, even in the presence of apigenin, demonstrating that CDK7 directly phosphorylates BRD4. To localize the CDK7 phosphorylation target site(s) on BRD4, the BRD4 WT and mutants described in Fig. 1A were used as substrates in a kinase reaction with or without purified CDK7/CAK. In the presence of CDK7/CAK, all the BRD4 proteins were phosphorylated to an extent at least 2-fold greater than that of autophosphorylation alone (Fig. 2D). Notably, the ΔN BRD4 mutant, which lacks intrinsic kinase activity, was efficiently phosphorylated in the presence of CDK7/CAK. Because both the N-terminal (ΔC) and C-terminal (ΔN) fragments of BRD4 were phosphorylated by CDK7, CDK7 has at least two distinct phosphorylation sites on BRD4. In contrast to the reciprocity of phosphorylation observed with PTEFb, BRD4 does not phosphorylate CDK7 (Fig. 2D). Thus, CDK7/CAK interacts directly with BRD4, resulting in the unidirectional phosphorylation of BRD4 in vitro. The ability of the native TFIIH holo-complex, containing the CDK7/CAK with the TFIIH core, to phosphorylate BRD4 was also examined. In an in vitro kinase assay, purified native TFIIH and CDK7/CAK phosphorylated both WT BRD4 and the kinase-dead ΔN BRD4 mutant with equal efficiency (Fig. 2E). Thus, BRD4 is phosphorylated by CDK7 equally well in the context of CAK or TFIIH.
Whether CDK7 phosphorylates BRD4 in vivo was addressed as follows. As a CTD kinase, TFIIH is most likely to interact with chromatin-bound BRD4, possibly during preinitiation complex assembly. Consequently, release of BRD4 from chromatin would be predicted to abrogate TFIIH-mediated BRD4 phosphorylation. Accordingly, because the small molecule inhibitor JQ1 specifically inhibits binding of BRD4 to acetylated chromatin/transcription (19), it should also disrupt TFIIH phosphorylation of BRD4. To test this prediction, HeLa cells were transfected with FLAG-tagged WT BRD4 and treated with or without JQ1. To eliminate the effect of CDK9/PTEFb phosphorylation of BRD4, FLAG-BRD4 FEE-AAA, a BRD4 mutant incapable of binding PTEFb/CDK9 (12) was transfected in parallel. BRD4 was immunoprecipitated from JQ1-treated or untreated cells and immunoblotted with anti-phosphoserine antibody. JQ1 treatment resulted in a 40% decrease in WT BRD4 phosphorylation (Fig. 2F). This result was consistent with the interpretation that preventing BRD4 recruitment to the chromatin, and thereby its interaction with TFIIH/CDK7, causes a decrease in BRD4 phosphorylation. It is important to note that JQ1 does not inhibit BRD4 kinase activity (9) or directly influence CDK7 and CDK9 phosphorylation of BRD4 (Fig. 2G). The BRD4 FEE-AAA mutant, which is unable to interact with PTEFb, was 45% less phosphorylated than WT BRD4, even in the absence of JQ1. In the presence of JQ1, this level of BRD4 phosphorylation was reduced further by another 30% (Fig. 2F). Taken together, these results suggest that chromatin-bound BRD4 is phosphorylated by CDK7/TFIIH in vivo, apart from its phosphorylation by CDK9/PTEFb.
CDK9, BRD4, and CDK7 Cross-phosphorylation Modulates Their CTD Kinase Activities
The finding that the three CTD kinases cross-phosphorylate raised the question of whether this influenced their ability to phosphorylate the CTD. To address this question, we first asked whether BRD4 and CDK9 synergized or antagonized in their phosphorylation of the Pol II CTD because they both phosphorylate CTD Ser-2. To this end, various ratios of PTEFb to BRD4 were titrated into in vitro kinase reactions in the presence of an excess of CTD substrate (Fig. 3A). Interestingly, at equimolar ratios, the combination of PTEFb and BRD4 resulted in an apparent synergistic increase in their CTD kinase activity (Fig. 3A, compare lane 7 to lane 1 and 4). However, as the concentration of BRD4 increased relative to PTEFb, not only was the synergy progressively lost, phosphorylation was no longer even additive (compare lane 9 with lane 1 plus 6). Thus, cross-talk between BRD4 and CDK9 resulted in an activation of the activity of one or both kinases at equimolar concentrations but inhibition at higher BRD4 concentrations.
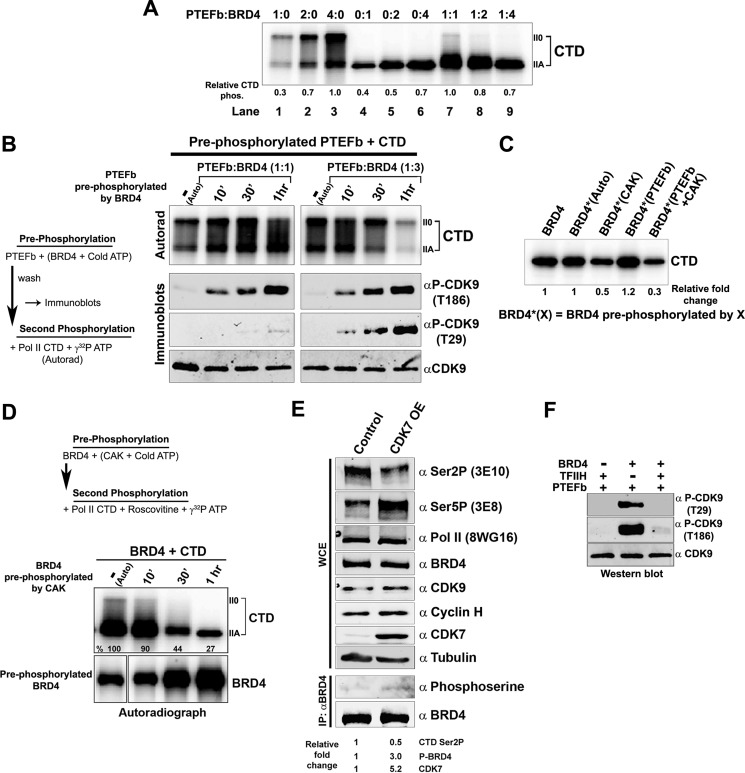
FIGURE 3.
Cross-phosphorylation between CDK9, CDK9, and BRD4 affects their Pol II CTD kinase activities. A, the kinase activities of BRD4 and PTEFb exhibit dose-dependent synergy in phosphorylating the Pol II CTD. Autoradiograph of an in vitro kinase assay where GST-CTD (0.1 μg) was phosphorylated with either PTEFb alone (50, 100, or 200 ng; lanes 1–3), BRD4 alone (100, 200, or 400 ng; lanes 4–6), or 50 ng of PTEFb with BRD4 (100, 200, or 400 ng; lanes 7–9). Numbers below the lanes indicate quantitation of relative Pol II CTD phosphorylation. B, BRD4 activates or represses CDK9 phosphorylation of the Pol II CTD depending on the relative BRD4 concentration. Left, experimental design. Right, upper panel, autoradiograph of GST-CTD (0.1 μg) phosphorylated by 0.1 μg of PTEFb pre-phosphorylated by either 0.2 (left panel) or 0.6 μg (right panel) of BRD4. Right, lower panels, immunoblots with α-CDK9, α-Thr(P)-29, or α-Thr(P)-186 phospho-CDK9 antibodies, showing patterns of PTEFb phosphorylation following: 1 h of autophosphorylation in the absence of BRD4 or pre-phosphorylation by BRD4 for 10 min, 30 min, or 1 h. (Note that PTEFb alone generates both IIA and IIO forms of phosphorylated CTD, whereas BRD4 primarily generates the IIA form.) C, BRD4 CTD kinase activity is affected when it is phosphorylated by other kinases. BRD4 (0.3 μg) that was unphosphorylated, autophosphorylated, or pre-phosphorylated with CAK (0.1 μg), PTEFb (0.1 μg), or both was used to phosphorylate 0.1 μg of GST-CTD in the presence of 100 μm roscovitine and 1 μm flavopiridol. Numbers below the lanes indicate quantification of relative phosphorylation of the CTD. D, phosphorylation of BRD4 by CDK7 suppresses the CTD kinase activity of BRD4 in vitro. Top, experimental design; BRD4 (0.3 μg) was either autophosphorylated for 1 h or phosphorylated by CDK7/CAK (0.1 μg) for 10 min, 30 min, and 1 h in a kinase reaction with unlabeled ATP; control reactions were run in parallel with [γ-32P]ATP (lower panel). The BRD4 pre-phosphorylated with unlabeled ATP was used in secondary kinase reactions with 0.1 μg of GST-CTD, [γ-32P]ATP, and 100 μm roscovitine (upper panel). E, in vivo phosphorylation of BRD4 by CDK7 suppresses the Pol II CTD kinase activity of BRD4. Whole cell extracts (WCE) of HEK293 cells untransfected (control) or transfected with 5 μg of CDK7 (CDK7 OE) were immunoblotted with α-CTD Ser-2P, α-CTD Ser-5P, α-Pol II, α-BRD4, α-CDK9, α-CDK7, or α-tubulin (upper). Equal amounts of BRD4 immunoprecipitated (IP) from untransfected or CDK7-transfected cells were immunoblotted with α-phosphoserine or α-BRD4 (lower). Numbers below the lanes indicate quantitation of immunoblots. F, BRD4 phosphorylation of CDK9 is inhibited by TFIIH. In vitro kinase reactions were done with PTEFb (0.1 μg) in the presence of either TFIIH (0.4 μg), BRD4 (0.2 μg), or both and immunoblotted with α-CDK9, Thr(P)-29, and Thr(P)-186 α-phospho-CDK9.
To investigate the basis for the differential effects on CTD phosphorylation of differing ratios of BRD4 and PTEFb, we examined the effect of pre-phosphorylation by one on the subsequent kinase activity of the other. As BRD4-phosphorylated CDK9 at both its activation (Thr-186) and inactivation (Thr-29) sites (Fig. 1, D and E), we first tested how these phosphorylations affected CDK9 CTD kinase activity. His-tagged PTEFb/CDK9 immobilized on beads was pre-phosphorylated for increasing periods by BRD4 with cold ATP at molar ratios of either 1:1 or 3:1 BRD4:PTEFb. An identical amount of PTEFb was autophosphorylated for 1 h in parallel. The immobilized pre-phosphorylated PTEFb, free of detectable BRD4, was then used in kinase assays with CTD as a substrate and [γ-32P]ATP. At a ratio of 1:1, BRD4 phosphorylated the Thr-186 activation site of PTEFb almost exclusively, as shown in immunoblots (Fig. 3B). In contrast, at a 3:1 ratio of BRD4:PTEFb, phosphorylation of the PTEFb Thr-29 inactivation site was clearly evident and increased over time (Fig. 3B). Interestingly, the CTD kinase activity of PTEFb/CDK9 increased in tandem with pre-phosphorylation at Thr-186 alone. However, PTEFb/CDK9 CTD kinase activity decreased as the extent of Thr-29 pre-phosphorylation increased. These results show that equimolar BRD4 activates the PTEFb CTD kinase activity by phosphorylating the Thr-186 site. However, at higher BRD4 concentrations, the CDK9 Thr-29 site is also phosphorylated, resulting in a net inhibition of CTD kinase activity at high BRD4/CDK9 ratios (Fig. 3, A and B). Thus, BRD4 is capable of alternatively activating or inactivating PTEFb/CDK9 CTD kinase activity. In contrast, PTEFb pre-phosphorylation of BRD4 resulted in only a small, albeit reproducible, increase in the CTD kinase activity of BRD4 (Fig. 3C). Thus, changes in CTD phosphorylation in the presence of both BRD4 and CDK9 (Fig. 3A) were primarily the result of altered CDK9 CTD kinase activity.
Because CDK7/CAK also directly phosphorylates BRD4 (Fig. 2), we next tested the effect of CDK7/CAK pre-phosphorylation of BRD4 on its CTD kinase activity. BRD4 was pre-phosphorylated in the presence of CDK7/CAK with cold ATP for 10 min, 30 min, or 1 h. Increasing incubation times resulted in increased CDK7 pre-phosphorylation of BRD4, as determined in parallel reactions with [γ-32P]ATP (Fig. 3D, lower autoradiogram). The pre-phosphorylated BRD4 was then used in a kinase reaction with CTD as substrate in the presence of [γ-32P]ATP and roscovitine, which inactivates CDK7. As shown in Fig. 3D (upper autoradiogram), pre-phosphorylation of BRD4 dramatically decreased its CTD kinase activity. The extent of inhibition increased in tandem with increased pre-phosphorylation. The suppression of BRD4 CTD kinase activity by CDK7 phosphorylation was not overcome by simultaneous PTEFb pre-phosphorylation (Fig. 3C). Thus, phosphorylation by CDK7 suppresses BRD4 CTD kinase activity in vitro.
To confirm these findings in vivo, the effect of CDK7 overexpression on BRD4 phosphorylation and CTD kinase activity was examined in HEK293 cells transfected with exogenous CDK7. The transfected, exogenous CDK7 was enzymatically active in vivo, as evidenced by the fact that CTD Ser-5 phosphorylation was increased in CDK7 overexpressing cells (Fig. 3E). Whole cell extracts from these cells were then probed for levels of phosphorylated BRD4 and CTD Ser-2 phosphorylation, as an in vivo measure of BRD4 CTD kinase activity. To measure phosphorylated BRD4 levels, equal amounts of immunoprecipitated BRD4 from nontransfected control cells and CDK7 overexpressing cells were immunoblotted with anti-phosphoserine antibody. As predicted, phosphorylation of BRD4 was 3-fold higher in CDK7 overexpressing cells than in control cells (Fig. 3E). Importantly, and consistent with the in vivo results, CTD Ser-2 phosphorylation, the target of BRD4, was decreased by 2-fold relative to controls and correlated with increased BRD4 phosphorylation. The decrease in CTD Ser-2 phosphorylation in CDK7 overexpressing cells can be directly correlated with CDK7 inhibition of BRD4 kinase activity for the following reasons. 1) It is known that CDK7 does not interact with or influence the kinase activity of CDK9/PTEFb (20). 2) The 3E10 Ser-2P antibody used here does not recognize phosphorylation by the only other CTD Ser-2 kinase, CDK12 (21). Interestingly, whereas the total amounts of BRD4, CDK9, and Pol II did not differ between CDK7-overexpressing cells and control cells, the regulatory component of CAK, cyclin H, is slightly induced in CDK7-overexpressing cells, consistent with CDK7 activation. Taken together, these results indicate that CDK7/CAK phosphorylates BRD4 in vitro and in vivo, resulting in decreased BRD4 CTD Ser-2 kinase activity. CDK7/TFIIH inhibits BRD4 phosphorylation of CDK9 as well, demonstrating that TFIIH is a potent inhibitor of BRD4 kinase activity in general (Fig. 3F). Thus, the above results demonstrate that cross-talk between BRD4 and CDK9 results in the reciprocal modulation of their kinase activities, whereas the interaction of CDK7 and BRD4 leads to the suppression of BRD4 kinase activity.
TAF7 Interacts with BRD4 and Represses Its CTD Kinase Activity
Originally identified as a component of the TFIID general transcription factor, TAF7 functions as a regulator of transcription initiation through its interactions with TAF1, TFIIH, and PTEFb (10). It inhibits the acetyltransferase activity of TAF1 and the kinase activities of CDK7 and CDK9 (10, 11). In turn, TAF7 is phosphorylated by each of the transcription factors. Because TAF7 regulates the kinase activity of these kinases, we asked if it also associates with BRD4 and affects its CTD kinase activity. As shown in Fig. 4A, endogenous BRD4 co-immunoprecipitated TAF7 from HeLa nuclear extracts, demonstrating their interaction in vivo. A reciprocal in vitro pulldown of recombinant BRD4 by GST-TAF7 bound to GST beads confirmed a direct interaction between the two (Fig. 4B). The interacting domains were mapped in reciprocal pulldown assays using the BRD4 mutants described in Fig. 1A and TAF7 mutants (Fig. 4, C and D). The efficient binding of TAF7 to BRD4 requires the BRD4 bromodomains (Fig. 4C). Conversely, BRD4 interacts primarily with the serine-rich central segment of TAF7, which is also the PTEFb and TAF1 interaction domain (Fig. 4D).
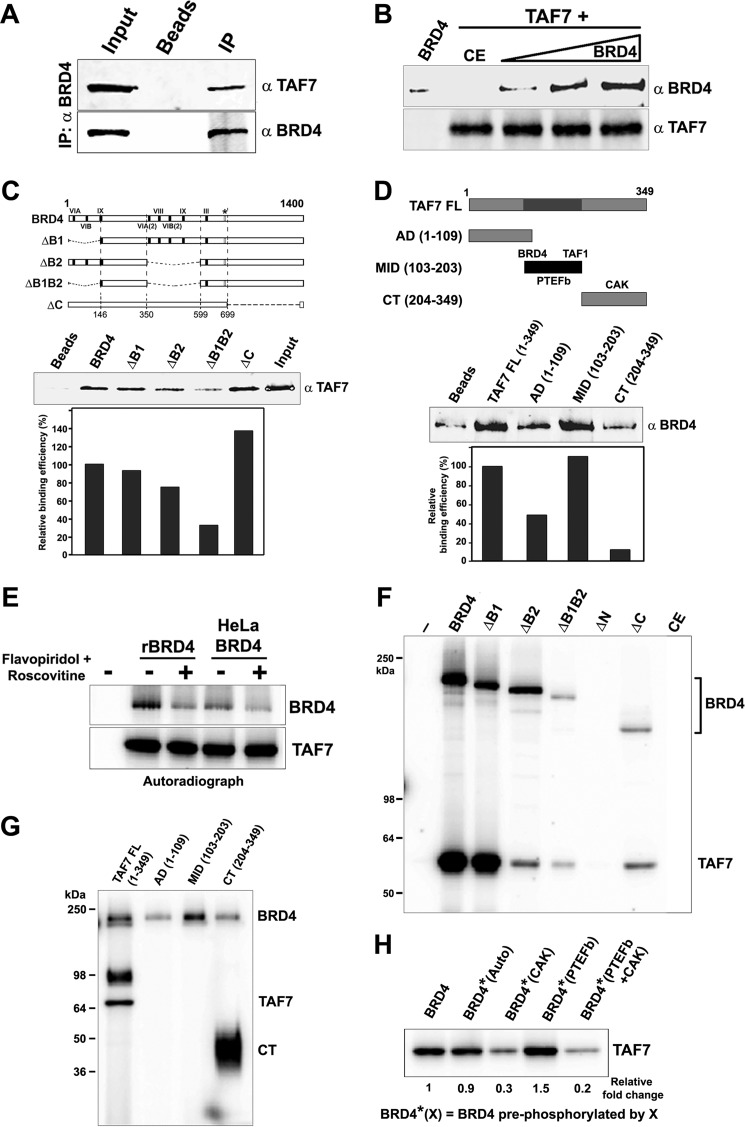
FIGURE 4.
TAF7 binds and is phosphorylated by BRD4. A, TAF7 associates with BRD4 in vivo. BRD4 was immunoprecipitated (IP) from HeLa nuclear extract using α-BRD4 and immunoblotted with α-TAF7. B, TAF7 interacts directly with BRD4. BRD4 (0.5, 1.0, and 1.5 μg) was pulled down with 0.5 μg of GST-TAF7 immobilized on GST beads and immunoblotted with α-BRD4. BRD4 (1.5 μg) with GST beads alone, or insect cell control extract (CE) made in parallel with recombinant BRD4 were used as negative controls. C, TAF7 binds BRD4 kinase subdomains. Upper, map of BRD4 proteins used. Middle, α-TAF7 immunoblot of 0.3 μg of GST-TAF7 (input) recovered by pull-down with 1.0 μg of wild type FLAG-BRD4 or equimolar amounts of FLAG-BRD4 mutants. Lower, quantitation of recovered GST-TAF7. D, BRD4 interacts with the central serine-rich region of TAF7. Upper, α-BRD4 immunoblot of 0.6 μg of BRD4 recovered by pull-down with 1 μg of full-length GST-TAF7 or equimolar amounts of GST-TAF7 sections. Middle, quantitation of recovered BRD4. Lower, map of TAF7 proteins used showing interaction sites of kinases. E, TAF7 is phosphorylated by BRD4. Autoradiograph of a kinase assay with TAF7 (0.15 μg) alone (first lane) or with recombinant murine BRD4 (0.3 μg) or human BRD4 immunoprecipitated from HeLa nuclear extract, with or without 1 μm flavopiridol and 100 μm roscovitine. F, TAF7 phosphorylation is dependent on BRD4 kinase domains. Autoradiograph showing 0.1 μg of TAF7 phosphorylated by 0.3 μg of BRD4 WT or equimolar amounts of BRD4 mutants. G, BRD4 phosphorylates the C-terminal end of TAF7. Autoradiograph showing 0.75 μg of full-length GST-TAF7 or equimolar amounts of GST-TAF7 sections phosphorylated by 0.6 μg of BRD4 (additional band migrating at ∼98 kDa is most likely hyper-phosphorylated TAF7 accentuated in the 15% gel used here). H, phosphorylation of BRD4 modulates its ability to phosphorylate TAF7. BRD4 (0.3 μg) that was unphosphorylated, autophosphorylated, or pre-phosphorylated with CAK (0.1 μg), PTEFb (0.1 μg), or both was used to phosphorylate 0.1 μg of TAF7 in the presence of 100 μm roscovitine and 1 μm flavopiridol.
We then tested if BRD4, like the other three kinases, phosphorylates TAF7. Both recombinant murine BRD4 and human BRD4 immunoprecipitated from HeLa nuclear extracts efficiently phosphorylated recombinant TAF7 (Fig. 4E). BRD4 phosphorylation of TAF7 was unaffected by the combined effect of flavopiridol and roscovitine, neither of which inhibits BRD4 kinase activity, demonstrating that the phosphorylation was not a result of contaminating CDK kinases. The phosphorylation of TAF7 by BRD4 depends on the kinase activity of BRD4, because TAF7 is not phosphorylated by the kinase-dead ΔN mutant (Fig. 4F). Interestingly, whereas BRD4 binds primarily to the central serine-rich region of TAF7 (Fig. 4D), it phosphorylates TAF7 exclusively on its C-terminal end (Fig. 4G). BRD4 phosphorylation of TAF7 is either substantially inhibited or slightly activated when BRD4 is pre-phosphorylated by CDK7 or CDK9, respectively (Fig. 4H). This is consistent with the effect of BRD4 pre-phosphorylation on its ability to phosphorylate the Pol II CTD (Fig. 3C).
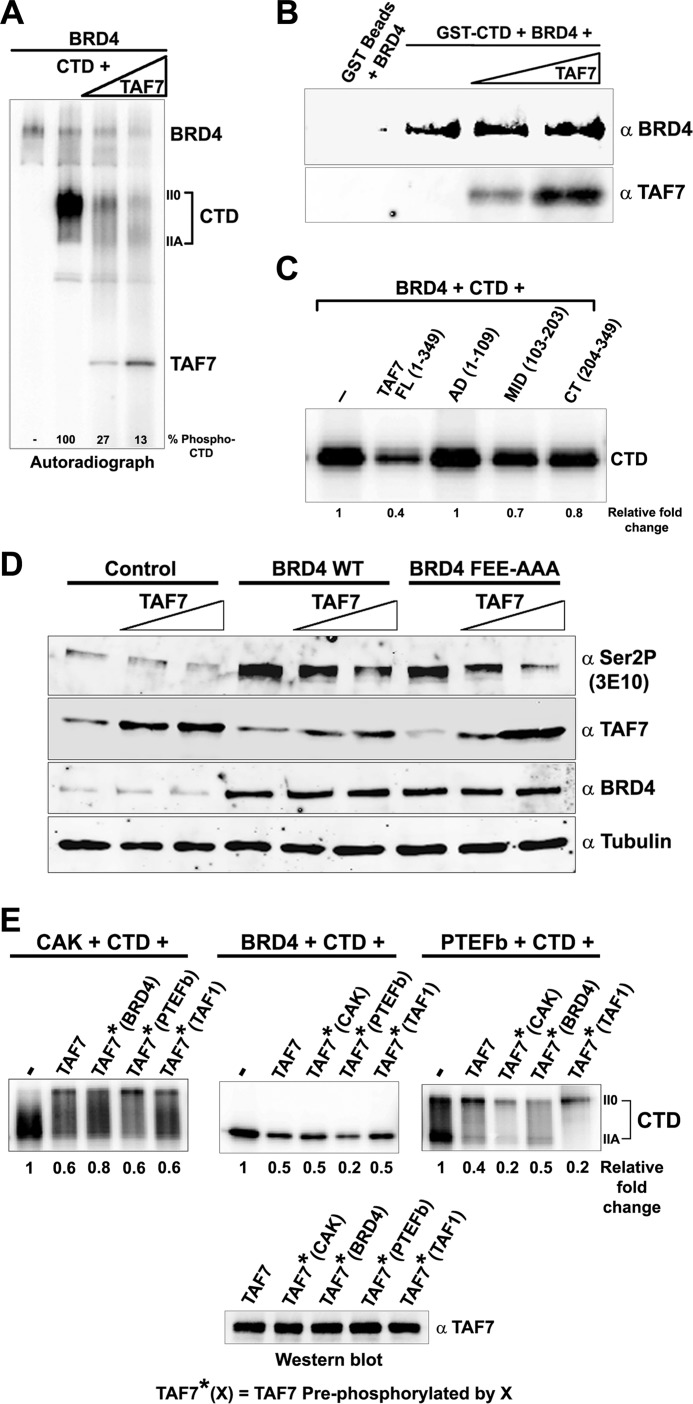
Because TAF7 directly interacts with BRD4, we next tested if BRD4 kinase activity, like CDK9 and CDK7, is regulated by TAF7. In an in vitro kinase assay, increasing amounts of TAF7, increasingly inhibited Pol II CTD phosphorylation by BRD4 (Fig. 5A). The suppression was not due to competitive binding of TAF7 on BRD4. In a pulldown assay, increasing TAF7 did not compete with binding of BRD4 to immobilized GST CTD, indicating that both TAF7 and CTD can bind BRD4 simultaneously (Fig. 5B). TAF7 does not bind the CTD directly (10). BRD4 CTD kinase activity is most efficiently inhibited by the full-length TAF7, although both the mid- and C-terminal domains are weakly inhibitory (Fig. 5C). TAF7 suppression of BRD4 kinase activity was confirmed in vivo: overexpression of TAF7 in HeLa cells resulted in a concomitant decrease in CTD Ser-2 phosphorylation mediated by both endogenous and exogenous BRD4. A similar decrease in CTD Ser-2 phosphorylation was observed when the BRD4 mutant incapable of recruiting PTEFb to the transcription site, BRD4 FEE-AAA, was co-transfected with TAF7 (Fig. 5D). Together, these results confirm that TAF7 suppresses BRD4-mediated CTD Ser-2 phosphorylation both in vitro and in vivo.
FIGURE 5.
TAF7 inhibits BRD4 phosphorylation of Pol II CTD. A, TAF7 inhibits BRD4 phosphorylation of Pol II CTD in vitro. Autoradiograph of a kinase assay with 0.1 μg of GST-CTD phosphorylated by 0.2 μg of BRD4 in the absence or presence of increasing amounts of TAF7 (0.1 and 0.2 μg). B, TAF7 and Pol II CTD do not compete in binding to BRD4. BRD4 (0.5 μg) was pulled down with 0.2 μg of GST-CTD immobilized on GST beads and 65 μm ATP in the absence or presence of increasing amounts of TAF7 (0.2 and 0.4 μg; ∼1:1 and 1:3 CTD:TAF7) and immunoblotted with α-BRD4. TAF7 bound to BRD4 was detected with α-TAF7. C, specific domains on TAF7 influence its inhibition of BRD4 CTD kinase activity. Autoradiograph showing 0.1 μg of GST-CTD phosphorylated by 0.2 μg of BRD4 in the absence or presence of 0.1 μg of full-length TAF7 or equimolar amounts of TAF7 AD, MID, and C-terminal sections. Numbers below the lanes indicate quantification of relative phosphorylation. D, TAF7 inhibits Pol II CTD Ser-2 phosphorylation by BRD4 in vivo. Five micrograms of human BRD4, BRD4 FEE-AAA mutant, or vector alone (control) were transfected into HeLa cells with or without co-transfection with increasing amounts of TAF7 (2.5 and 5.0 μg). Whole cell extracts of these cells were immunoblotted with α-CTD Ser-2P, α-TAF7, α-BRD4, and α-Tubulin control. E, phosphorylation of TAF7 by Pol II CTD kinases selectively affects its inhibitory potential. TAF7 (0.5 μg) was pre-phosphorylated or not by TAF1 (0.5 μg), BRD4 (0.5 μg), CAK (0.25 μg), or PTEFb (0.25 μg) for 1 h with unlabeled ATP. Nonphosphorylated or pre-phosphorylated TAF7, heat treated for 15 min at 70 °C, was added to kinase reactions with [γ-32P]ATP, 0.1 μg of GST-CTD and CAK (0.1 μg; in the presence of 100 μm Apigenin), BRD4 (0.2 μg: in the presence of 100 μm roscovitine and 1 μm flavopiridol), or PTEFb (0.1 μg) (upper panels). Numbers below the lanes indicate quantitation of relative Pol II CTD phosphorylation. The α-TAF7 immunoblot (lower panel) documented equal amounts of unphosphorylated and pre-phosphorylated TAF7 in the reactions.
The TAF7 regulation of the kinases involved in every step of transcription, TAF1, CDK7, BRD4, and CDK9, led us to investigate if pre-phosphorylation of TAF7 by one of the kinases influenced its subsequent ability to suppress the other kinases. The effect of pre-phosphorylating TAF7 by TAF1, CDK7/CAK, PTEFb/CDK9, or BRD4, was assessed in kinase reactions with Pol II CTD as the substrate (Fig. 5E). TAF7 pre-phosphorylated by either CDK7/CAK or TAF1, inhibited CTD phosphorylation by PTEFb significantly better than unphosphorylated TAF7; pre-phosphorylation of TAF7 by BRD4 had no effect (Fig. 5E, right-hand panel). TAF7 inhibition of BRD4 kinase activity was only affected by PTEFb pre-phosphorylation, which enhanced TAF7 inhibition (Fig. 5E, center panel). Conversely, BRD4 pre-phosphorylation modestly, but reproducibly, decreased TAF7 inhibition of CDK7/CAK kinase activity (Fig. 5E, left panel). Interestingly, whereas all forms of TAF7 inhibited the CDK7/CAK kinase activity, the proportion of IIA to II0 forms of phosphorylated CTD was visibly altered in the presence of each form of TAF7 (Fig. 5E, left panel). These results suggest that phosphorylation of TAF7 by each of the kinases modulates the ability of TAF7 to inhibit other CTD kinases.
DISCUSSION
RNA Pol II CTD phosphorylation has multiple roles in eukaryotic transcription. It was originally found to be a regulatory mechanism for transcription initiation and elongation. More recently, it has been shown to function simultaneously as a signal for the sequential recruitment of numerous factors that coordinate an array of nuclear processes with transcription (8). Among those are proteins responsible for RNA maturation and processing, DNA repair, and chromatin remodeling that are recruited during transcription initiation and elongation in response to differing patterns of Ser-5 and Ser-2 CTD phosphorylation (3). CTD phosphorylation patterns change throughout each transcription cycle, creating a series of distinct platforms necessary for the binding of factors mediating different nuclear functions. The current model for the role of changing RNA Pol II CTD phosphorylation, the “phospho-CTD cycle,” describes phosphorylation and dephosphorylation of the CTD at Ser-5, Ser-2, and Ser-7 occurring dynamically and in waves (3). Characterization of the CTD phosphorylation patterns has been focused primarily on the sequential timing of CTD phosphorylation patterns created by respective CTD kinases thus far. However, little is known about the mechanisms responsible for the regulation of CTD kinases that result in the shifting patterns of CTD phosphorylations. In the present study, we have reported that three primary CTD kinases interact directly, as well as indirectly through TAF7, to actively modulate their ability to phosphorylate the Pol II CTD. These findings lead to a speculative model in which CTD kinase cross-talk serves to ensure that CTD phosphorylation events occur in an orderly and sequential fashion.
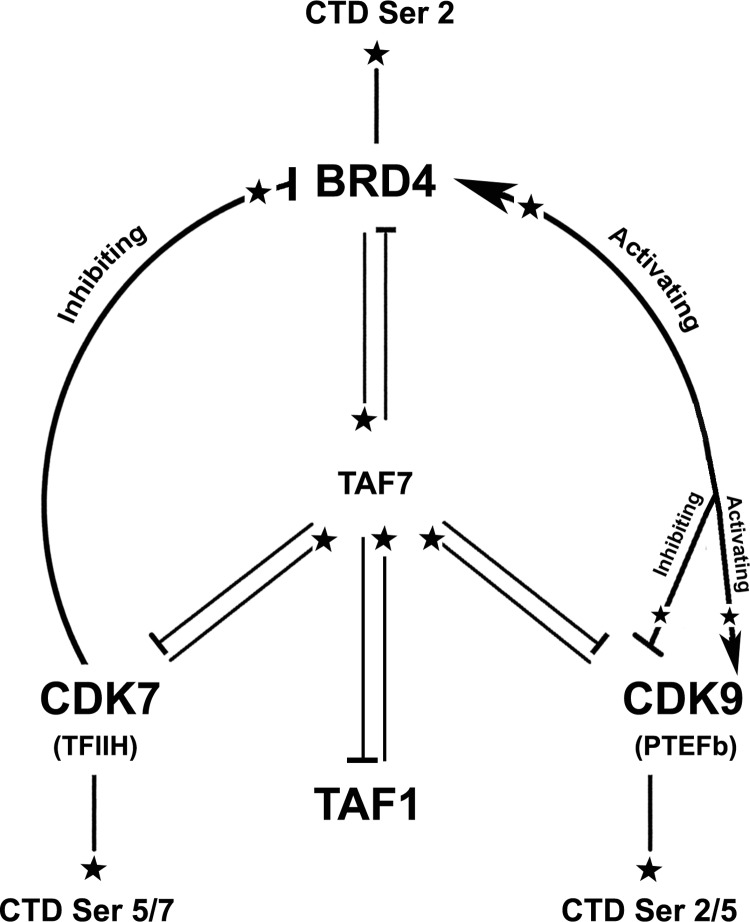
We have identified two distinct layers that contribute to the modulation of CTD kinase activity: 1) direct regulatory mechanisms mediated by reciprocal phosphorylations of BRD4, PTEFb, and TFIIH kinases and 2) indirect mechanisms mediated by TAF7 interactions. Until recently, the role of BRD4 in transcription had been presumed to be passive as either a bookmark of actively transcribed genes or as a scaffold that recruits PTEFb to chromatin (22). Our recent discovery that BRD4 is a Pol II CTD kinase that phosphorylates Ser-2 suggested that it is an active participant in transcription. In the present study, we have provided further evidence that BRD4 is an integral component in regulating CTD phosphorylation. We report that BRD4 interacts with both PTEFb and TFIIH, in a complex interplay in which their respective kinase activities are regulated. We also identify another layer of regulation mediated by TAF7, which functions to further regulate the CTD activities of these kinases. Although originally characterized as a component of TFIID, TAF7 has been shown to function independently of TFIID as a regulator of transcription initiation through its modulation of the activities of TAF1, PTEFb, and TFIIH (10, 11, 23). The present studies extend the scope of function of TAF7 to BRD4 whose CTD kinase activity it inhibits. In aggregate, these studies reveal an intricate network of kinase interactions regulating CTD phosphorylation and thereby transcription (Fig. 6).
FIGURE 6.
Summary of Pol II CTD kinase cross-talk pathways. CDK7, a CTD Ser-5/7 kinase, phosphorylates BRD4 and inhibits its Pol II CTD kinase activity. BRD4, a CTD Ser-2 kinase, phosphorylates CDK9 either activating or inactivating its Pol II CTD kinase activity depending on their relative concentrations. CDK9, a CTD Ser-2/5 kinase, phosphorylates BRD4 modestly activating its kinase activity. All three kinases autophosphorylate as well as phosphorylate TAF7. Phosphorylation of TAF7 by each of the kinases affects its subsequent effect on the others. Phosphorylation, activation, and repression are indicated by stars ( ), arrowheads (
), arrowheads ( ), and cross-bars (⊣), respectively.
), and cross-bars (⊣), respectively.
The sequential ordering of the interactions among the CTD kinases and TAF7 during transcription remains to be determined. We speculate that the regulatory events occur dynamically, with an initial inhibition of PTEFb kinase activity by BRD4 during assembly of the transcription preinitiation complex. Multiple BRD4 molecules are recruited to the transcription site and are known to transiently dimerize on chromatin (22, 24). Because BRD4 at higher molar ratios relative to PTEFb phosphorylates the Thr-29 site on CDK9, which inactivates PTEFb (16), these BRD4 dimers may inactivate CDK9 kinase activity during early transcription and prevent premature elongation. All chromatin-bound PTEFb is known to be phosphorylated at CDK9 Thr-186 (25), presumably by BRD4 during their initial interaction prior to chromatin binding. However, CDK9 Thr-29 phosphorylation by BRD4 dominates over the positive Thr-186 phosphorylation, enabling BRD4 to control PTEFb kinase activity. The interaction between BRD4 and PTEFb also results in the phosphorylation of BRD4, which activates its Pol II CTD kinase activity. Thus, prior to productive elongation, the CTD kinase activity of PTEFb is likely to be inhibited, whereas that of BRD4 is activated.
Upon completion of preinitiation complex assembly, TFIIH phosphorylates CTD Ser-5 coincident with recruitment of the 5′ RNA capping complex (7). We speculate that TFIIH regulates subsequent CTD Ser-2 phosphorylation through its phosphorylation of BRD4. The novel interaction between BRD4 and TFIIH results in a unidirectional phosphorylation of BRD4 by CAK/CDK7. Phosphorylation by CAK/CDK7, until now known only to be an activating event (18), inhibits BRD4 CTD kinase activity both in vitro and in vivo. These results suggest a transcriptional mechanism where CDK7 phosphorylates CTD Ser-5 sites while transiently suppressing BRD4 CTD kinase activity. We speculate that once the CTD is phosphorylated at Ser-5, the inhibition of BRD4 kinase is released and the CTD is then phosphorylated at Ser-2. In support of this model, we have shown previously that BRD4 preferentially phosphorylates Ser-2 of Pol II CTD pre-phosphorylated at Ser-5 by CDK7 (9). Similarly, CTD Ser-5 phosphorylation of Pol II enhances CTD Ser-2 phosphorylation in vivo in both Schizosaccharomyces pombe and Saccharomyces cerevisiae (8). In addition, PTEFb, which has been described as the kinase that phosphorylates CTD Ser-2 after Ser-5 phosphorylation by CDK7, is unable to phosphorylate CTD pre-phosphorylated at Ser-5 (26). These findings are thus consistent with the interpretation that BRD4 is the initial CTD Ser-2 kinase whose activity is regulated by TFIIH.
The role of PTEFb in transcription elongation and its phosphorylation of Ser-2/5 on the CTD are well established. PTEFb/CDK9 kinase activity during transcription is entirely dependent on CDK9 Thr-186 phosphorylation (15). However, the kinase responsible for this phosphorylation has remained elusive despite many attempts to identify it. In the present study, we have demonstrated that BRD4 activates PTEFb/CDK9 both by directly phosphorylating at Thr-186, and by activating CDK9 autophosphorylation at this site. More importantly, BRD4 modulates PTEFb CTD kinase activity both positively by Thr-186 phosphorylation and negatively by Thr-29 phosphorylation. It has been shown previously that during early elongation, BRD4 dissociates from the elongation complex, and the protein phosphatase PP2A specifically dephosphorylates the Thr-29 sites on CDK9 (16). However, CDK9 Thr-186 phosphorylation is only dephosphorylated by PPM1A/B (17), suggesting that Thr-186 phosphorylated by BRD4 persists through elongation. Therefore, BRD4 activates PTEFb/CDK9 phosphorylation of the CTD Ser-2 during productive elongation through its prior phosphorylation of CDK9 Thr-186.
The in vivo kinase activity of each CTD kinase is likely to be a function of all their interactions with each other, enabling tight regulation of CTD phosphorylation and transcription. An important corollary to this model is that a cascade of phosphorylations regulates CTD phosphorylation, transcription, and co-transcriptional nuclear processes: CDK7 regulates BRD4, which in turn regulates PTEFb. It is noteworthy that CDK7 does not interact with CDK9 (20) although XPB, a component of TFIIH separate from CDK7/CAK, inhibits CDK9 autophosphorylation but not its CTD kinase activity (27). Layered upon the direct modifications that regulate CTD kinase activities are the interactions with TAF7 that provide indirect regulation. TAF7 binds to and directly inhibits the CTD kinase activities of BRD4, CAK/CDK7, and PTEFb. Importantly, TAF7 is also phosphorylated by each of these kinases, such that pre-phosphorylation of TAF7 by one kinase differentially affects its inhibition of another kinase. For example, pre-phosphorylation of TAF7 by CAK/CDK7 potentiates TAF7 inhibition of PTEFb CTD kinase activity, but has no effect on BRD4. In this way, PTEFb could be inhibited by CAK/CDK7 through TAF7, until transcription initiation is complete. We speculate that TAF7 could act as a messenger between the kinases involved in transcription.
In conclusion, we propose that CTD kinase cross-talk involving BRD4, TFIIH, and PTEFb governs dynamic CTD phosphorylation. Implicit in a model of regulation by dynamic CTD phosphorylation is the assumption that there must be a series of phosphatases that are similarly involved in the cross-talk. Documenting and characterizing dynamic CTD kinase cross-talk in a physiological context will thus be challenging. Nevertheless, our demonstration that such cross-phosphorylations do occur, and modulate CTD phosphorylation in vivo, supports its existence. Future studies will be directed at further characterization of the interplay among the CTD kinases and TAF7 as well as identification of cognate phosphatases. The identification of CTD kinase cross-talk and its modulation of CTD phosphorylation have broad implications in understanding how transcription is coordinated with DNA damage responses, chromatin remodeling, and other nuclear processes.
Acknowledgments
We thank members of our lab for helpful discussions. We also thank Drs. David Levens, Ranjan Sen, Brian Lewis, and Dan Larson for critical reading of the manuscript.
This work was supported, by the Intramural Research Program of the NCI, Center for Cancer Research, National Institutes of Health.
- Pol II
- polymerase II
- CTD
- C-terminal domain
- CAK
- CDK activating kinase.
REFERENCES
- 1. Phatnani H. P., Greenleaf A. L. (2006) Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20, 2922–2936 [DOI] [PubMed] [Google Scholar]
- 2. Zhang D. W., Rodríguez-Molina J. B., Tietjen J. R., Nemec C. M., Ansari A. Z. (2012) Emerging views on the CTD code. Genet. Res. Int. DOI: 10.1155/2012/347214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartkowiak B., MacKellar A. L., Greenleaf A. L. (2011) Updating the CTD story. From tail to epic. Genet. Res. Int. DOI: 10.4061/2011/623718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Phatnani H. P., Jones J. C., Greenleaf A. L. (2004) Expanding the functional repertoire of CTD kinase I and RNA polymerase II. Novel phospho-CTD-associating proteins in the yeast proteome. Biochemistry 43, 15702–15719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bataille A. R., Jeronimo C., Jacques P. É., Laramée L., Fortin M. È., Forest A., Bergeron M., Hanes S. D., Robert F. (2012) A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol. Cell 45, 158–170 [DOI] [PubMed] [Google Scholar]
- 6. Fisher R. P. (2005) Secrets of a double agent. CDK7 in cell-cycle control and transcription. J. Cell Sci. 118, 5171–5180 [DOI] [PubMed] [Google Scholar]
- 7. Prelich G. (2002) RNA polymerase II carboxyl-terminal domain kinases. Emerging clues to their function. Eukaryot. Cell 1, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Egloff S., Dienstbier M., Murphy S. (2012) Updating the RNA polymerase CTD code. Adding gene-specific layers. Trends Genet. 28, 333–341 [DOI] [PubMed] [Google Scholar]
- 9. Devaiah B. N., Lewis B. A., Cherman N., Hewitt M. C., Albrecht B. K., Robey P. G., Ozato K., Sims R. J., 3rd, Singer D. S. (2012) BRD4 is an atypical kinase that phosphorylates Serine2 of the RNA Polymerase II carboxyl-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 109, 6927–6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gegonne A., Weissman J. D., Lu H., Zhou M., Dasgupta A., Ribble R., Brady J. N., Singer D. S. (2008) TFIID component TAF7 functionally interacts with both TFIIH and P-TEFb. Proc. Natl. Acad. Sci. U.S.A. 105, 5367–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gegonne A., Weissman J. D., Zhou M., Brady J. N., Singer D. S. (2006) TAF7. A possible transcription initiation check-point regulator. Proc. Natl. Acad. Sci. U.S.A. 103, 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bisgrove D. A., Mahmoudi T., Henklein P., Verdin E. (2007) Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. U.S.A. 104, 13690–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jang M. K., Mochizuki K., Zhou M., Jeong H. S., Brady J. N., Ozato K. (2005) The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19, 523–534 [DOI] [PubMed] [Google Scholar]
- 14. Fujinaga K., Cujec T. P., Peng J., Garriga J., Price D. H., Graña X., Peterlin B. M. (1998) The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 72, 7154–7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen R., Liu M., Li H., Xue Y., Ramey W. N., He N., Ai N., Luo H., Zhu Y., Zhou N., Zhou Q. (2008) PP2B and PP1α cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 22, 1356–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou M., Huang K., Jung K. J., Cho W. K., Klase Z., Kashanchi F., Pise-Masison C.A., Brady J. N. (2009) Bromodomain protein Brd4 regulates human immunodeficiency virus transcription through phosphorylation of CDK9 at threonine 29. J. Virol. 83, 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cho S., Schroeder S., Ott M. (2010) CYCLINg through transcription. Post-translational modifications of P-TEFb regulate transcription elongation. Cell Cycle 9, 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Serizawa H., Mäkelä T. P., Conaway J. W., Conaway R. C., Weinberg R. A., Young R. A. (1995) Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature 374, 280–282 [DOI] [PubMed] [Google Scholar]
- 19. Delmore J. E., Issa G. C., Lemieux M. E., Rahl P. B., Shi J., Jacobs H. M., Kastritis E., Gilpatrick T., Paranal R. M., Qi J., Chesi M., Schinzel A.C., McKeown M. R., Heffernan T. P., Vakoc C.R., Bergsagel P. L., Ghobrial I. M., Richardson P. G., Young R. A., Hahn W. C., Anderson K. C., Kung A. L., Bradner J. E., Mitsiades C. S. (2011) BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim J. B., Sharp P. A. (2001) Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276, 12317–12323 [DOI] [PubMed] [Google Scholar]
- 21. Bartkowiak B., Liu P., Phatnani H. P., Fuda N. J., Cooper J. J., Price D. H., Adelman K., Lis J. T., Greenleaf A. L. (2010) CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 24, 2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu S. Y., Chiang C. M. (2007) The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 282, 13141–13145 [DOI] [PubMed] [Google Scholar]
- 23. Devaiah B. N., Lu H., Gegonne A., Sercan Z., Zhang H., Clifford R. J., Lee M. P., Singer D. S. (2010) Novel functions for TAF7, a regulator of TAF1-independent transcription. J. Biol. Chem. 285, 38772–38780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang R., Li Q., Helfer C. M., Jiao J., You J. (2012) The bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J. Biol. Chem. 287, 10738–10752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ai N., Hu X., Ding F., Yu B., Wang H., Lu X., Zhang K., Li Y., Han A., Lin W., Liu R., Chen R. (2011) Signal-induced Brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation. Nucleic Acids Res. 39, 9592–9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Czudnochowski N., Bösken C. A., Geyer M. (2012) Serine 7 but not serine 5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat. Commun. 3, 842. [DOI] [PubMed] [Google Scholar]
- 27. Zhou M., Nekhai S., Bharucha D. C., Kumar A., Ge H., Price D. H., Egly J. M., Brady J. N. (2001) TFIIH inhibits CDK9 phosphorylation during human immunodeficiency virus type 1 transcription. J. Biol. Chem. 276, 44633–44640 [DOI] [PubMed] [Google Scholar]