Background: Oocydin A is an anticancer haterumalide with strong antimicrobial activity against agriculturally important plant pathogenic fungi and oomycetes.
Results: The oocydin A gene cluster has been identified and characterized in four plant-associated enterobacteria.
Conclusion: The ooc gene cluster is organized in three transcriptional units encoding enzymes that belong to a growing class of trans-acyltransferase polyketide synthases.
Significance: Oocydin A has potential agricultural, pharmacological, and chemotherapeutic applications.
Keywords: Bacterial Metabolism; Microbiology; Multifunctional Enzymes; Natural Product Biosynthesis; Polyketides; Haterumalide; Oocydin A; Polyketide Synthase; Anti-cancer, Antifungal and Anti-oomycete Compound; trans-Acyltransferase
Abstract
Haterumalides are halogenated macrolides with strong antitumor properties, making them attractive targets for chemical synthesis. Unfortunately, current synthetic routes to these molecules are inefficient. The potent haterumalide, oocydin A, was previously identified from two plant-associated bacteria through its high bioactivity against plant pathogenic fungi and oomycetes. In this study, we describe oocydin A (ooc) biosynthetic gene clusters identified by genome sequencing, comparative genomics, and chemical analysis in four plant-associated enterobacteria of the Serratia and Dickeya genera. Disruption of the ooc gene cluster abolished oocydin A production and bioactivity against fungi and oomycetes. The ooc gene clusters span between 77 and 80 kb and encode five multimodular polyketide synthase (PKS) proteins, a hydroxymethylglutaryl-CoA synthase cassette and three flavin-dependent tailoring enzymes. The presence of two free-standing acyltransferase proteins classifies the oocydin A gene cluster within the growing family of trans-AT PKSs. The amino acid sequences and organization of the PKS domains are consistent with the chemical predictions and functional peculiarities associated with trans-acyltransferase PKS. Based on extensive in silico analysis of the gene cluster, we propose a biosynthetic model for the production of oocydin A and, by extension, for other members of the haterumalide family of halogenated macrolides exhibiting anti-cancer, anti-fungal, and other interesting biological properties.
Introduction
Plants have to cope with potentially devastating biotic and abiotic environmental stresses during their life cycle. Among these stresses, fungal and oomycete plant pathogens are responsible for many of the most agriculturally important diseases worldwide (1). Effective chemical control of these infections is extremely difficult, and biopesticides (pest management agents based on living microorganisms or natural products) are considered to be one of the most promising methods for rational crop management (2). Many bacteria can synthesize bioactive secondary metabolites, some of which can combat key fungal and oomycete plant pathogens, and thus these metabolites may have utility in biocontrol systems (3, 4).
Oocydin A is a chlorinated macrolide that was isolated in 1999 from the plant epiphytic bacterial strain Serratia marcescens MSU97 because of its strong bioactivity against plant pathogenic oomycetes (5). The same macrolide, named haterumalide NA, was also isolated from the sponge Ircinia sp. (6) and subsequently from the rhizosphere bacterium, Serratia plymuthica A153, where it showed the ability to inhibit the hyphal growth of the plant pathogenic fungus, Sclerotinia sclerotiorum (7). Furthermore, in 2005, it was also isolated from Serratia liquefaciens number 1821 on the basis of its anti-hyperlipidemic activity and given the code FR177391 (8). Spectroscopic data for haterumalide NA, oocydin A, and FR177391 are identical, although there are differences in their reported optical rotations, so it remains formally possible that the bioactive metabolite from Serratia species is the enantiomer of that derived from the sponge.
Various haterumalides (6, 9) and their oxygenated analogs, the biselides (10, 11), also show powerful antitumor activity against human lung, breast, leukemia, and colon cancer cells. Interestingly, haterumalide NA/oocydin A exhibited cytotoxicity against P388 leukemia (6) and breast (5) cancer cells. These broad biological impacts made the haterumalides/oocydins very attractive targets for total chemical synthesis, which has been achieved by several research groups (9, 12, 13).
Although there are no reports to date on the biosynthetic route to oocydin A, its structure suggests a polyketide origin. During the biosynthesis of polyketides, the polyketide chain is assembled and elongated while covalently attached to acyl carrier protein (ACP)3 domains. The elongation is performed by the C–C bond-forming ketosynthase (KS) domains, and acyltransferase (AT) domains are responsible for introducing (mostly) malonyl- or methylmalonyl-building units to the ACP during each cycle of elongation. Type I polyketide synthases (PKSs) are usually multidomain proteins in which the domains form an assembly line, consisting of multiple modules, each of which is responsible for one round of chain elongation. The minimal KS-AT-ACP module can be supplemented with one or more of a ketoreductase (KR) domain (converting the β-keto group to a β-hydroxy group), a dehydratase (DH) domain (eliminating water to generate a C C double bond), and an enoylreductase (ER) domain, which reduces double bonds to saturated intermediates (14). According to the textbook model, the number of modules in a polyketide gene cluster is correlated with the number of extension cycles executed by the PKS and therefore with the structure of the eventual secondary metabolite. After the synthesis of the polyketide backbone, the chain is released from the PKS, usually by hydrolysis or cyclization catalyzed by a thioesterase (TE) domain, and in many cases, the polyketide is then modified by a range of tailoring enzymes. These modifications include glycosylations, hydroxylations, acyl transfers, epoxidations, and halogenations (14–16). In bacteria, polyketide biosynthesis genes are normally organized in gene clusters with the PKS genes being part of a single operon, which reflects both the coordinate regulation required for the activation of the biosynthetic pathway and the evolutionary origin of the cluster, in most cases, by horizontal gene transfer between microbial genomes (17).
C double bond), and an enoylreductase (ER) domain, which reduces double bonds to saturated intermediates (14). According to the textbook model, the number of modules in a polyketide gene cluster is correlated with the number of extension cycles executed by the PKS and therefore with the structure of the eventual secondary metabolite. After the synthesis of the polyketide backbone, the chain is released from the PKS, usually by hydrolysis or cyclization catalyzed by a thioesterase (TE) domain, and in many cases, the polyketide is then modified by a range of tailoring enzymes. These modifications include glycosylations, hydroxylations, acyl transfers, epoxidations, and halogenations (14–16). In bacteria, polyketide biosynthesis genes are normally organized in gene clusters with the PKS genes being part of a single operon, which reflects both the coordinate regulation required for the activation of the biosynthetic pathway and the evolutionary origin of the cluster, in most cases, by horizontal gene transfer between microbial genomes (17).
In this study, we employed genome sequencing, comparative genomics, mutagenesis, and chemical analysis to identify the PKS gene cluster responsible for the biosynthesis of oocydin A. The oocydin A gene cluster is organized in three different transcriptional units, and it is present in four different plant-associated enterobacteria, three of them belonging to the genus Serratia. Based on the structure of the gene cluster and analysis of its conserved motifs and catalytic residues, we propose a model for the biosynthesis of oocydin A.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, Phages, Culture Media, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Serratia, Dickeya, and their derivative strains were routinely grown at 30 °C, unless otherwise indicated, in Luria Broth (LB: 5 g of yeast extract liter−1,10 g of bactotryptone liter−1, and 5 g of NaCl liter−1), potato dextrose (24 g of potato dextrose broth liter−1), enriched potato dextrose medium (24 g of potato dextrose broth liter−1, 6 g of bactopeptone liter−1, 4 g of yeast extract liter−1, and 100 mg of NaCl liter−1), half-strength tryptic soy medium (15 g of tryptic soy broth liter−1), or minimal medium (0.1%, w/v, (NH4)2SO4, 0.41 mm MgSO4, 0.2% (w/v) glucose, 40 mm K2HPO4, 14.7 mm KH2PO4, pH 6.9–7.1). Escherichia coli strains were grown at 37 °C in LB. E. coli DH5α was used for gene cloning. Media for propagation of E. coli β2163 were supplemented with 300 μm 2,6-diaminopimelic acid. When appropriate, antibiotics were used at the following final concentrations (in μg ml−1): ampicillin, 100; kanamycin, 25 (E. coli strains) and 75 (Serratia strains); streptomycin, 50; erythromycin, 200. Sucrose was added to a final concentration of 10% (w/v) when required to select derivatives that had undergone a second crossover event during marker exchange mutagenesis.
TABLE 1.
Bacteria, oomycete, fungi, plasmids and phages used in this study
| Bacteria/fungi/oomycete/ phage/plasmid(s) | Genotype or relevant characteristica | Reference or source |
|---|---|---|
| E. coli DH5α | supE44 lacU169 (φ80lacZΔ M15) hsdR17 (rK−mK−) recA1 endA1 gyrA96 thi-1 relA1 | 18 |
| E. coli CC118λpir | araD, Δ(ara, leu), ΔlacZ74, phoA20, galK, thi-1, rspE, rpoB, argE, recA1, λpir | 19 |
| E. coli HH26 | Mobilizing strain for conjugal transfer | 20 |
| E. coli EPI100-T1R | F− mcrAΔ (mrr-hsdRMS-mcrBC) (φ80lacZΔ M15) ΔlacX74 recA1 | Epicentre |
| E. coli β2163 | F− RP4-2-Tc::Mu DdapA:: (erm-pir), KmrEmr | 21 |
| E. coli ESS | β-Lactam super-sensitive indicator strain | 22 |
| B. subtilis JH642 | pheA1 trpC2 | J.A. Hoch |
| S. plymuthica A153 | Wild type, rhizosphere isolate | 23 |
| MMnO2 | A153 transposon mutant oocS::Tn-KRCPN1; Kmr | This study |
| MMnO4 | A153 transposon mutant oocQ::Tn-KRCPN1; Kmr | This study |
| MMnO9 | A153 transposon mutant oocN::Tn-KRCPN1; Kmr | This study |
| MMnO13 | A153 transposon mutant oocJ::Tn-KRCPN1; Kmr | This study |
| MMnO14 | A153 transposon mutant oocC::Tn-KRCPN1; Kmr | This study |
| MMnO15 | A153 transposon mutant oocU::Tn-KRCPN1; Kmr | This study |
| MMnO18 | A153 transposon mutant oocQ::Tn-KRCPN1; Kmr | This study |
| OocK | A153 oocK::Km, constructed by marker exchange, Kmr | This study |
| OocM | A153 oocM:: Km, constructed by marker exchange, Kmr | This study |
| S. marcescens MSU97 | Wild type, plant epiphyte, pigmented | 5 |
| S. odorifera 4Rx13 | Wild type, rhizosphere isolate | 24 |
| D. dadantii Ech703 | Wild type, plant pathogen | CP001654 |
| Fungi/oocymete strains | ||
| P. ultimum | Wild type, plant pathogen | R. Cooper |
| V. dahliae 5368 | Wild type, plant pathogen | R. Cooper |
| T. cucumeris | Wild type, plant pathogen | STCC 2813 |
| A. solani | Wild type, plant pathogen | STCC 2997 |
| F. oxysporum | Wild type, plant pathogen | STCC 2715 |
| S. cerevisiae | Wild type | S.G. Oliver |
| S. pombe | Wild type | J. Mata |
| Phages/plasmids | ||
| φMAM1 | Generalized transducing phage for S. plymuthica A153 | Footnote 4 |
| pWEB-TNC | Apr, Cmr; Cosmid cloning vector | Epicentre |
| pKNG101 | Smr; oriR6K mob sacBR, | 20 |
| pUC18Not | Apr; identical to pUC18 but with two NotI sites flanking pUC18 polylinker | 19 |
| p34S-Km3 | Kmr, Apr; Km3 antibiotic cassette | 25 |
| pKCPRN1 | Kmr, Tcr; Derivative of pDS1028uidA with the uidA and cat genes replaced with lacZ and km genes. | 26 |
| pMAMV52 | Apr; 2.2-kb EcoRI/PstI PCR product containing oocK was inserted into the EcoRI/PstI sites of pUC18Not | This study |
| pMAMV53 | Apr, Kmr; 0.95-kb SmaI fragment containing km3 cassette of p34S-Km3 was inserted into HincII site of oocK in pMAMV52 | This study |
| pMAMV54 | Smr, Kmr; 3.2 kb NotI fragment of pMAMV53 was cloned at the same site in pKNG101 | This study |
| pMAMV60 | Apr; 2.0-kb EcoRI/PstI PCR product containing oocM was inserted into the EcoRI/PstI sites of pUC18Not | This study |
| pMAMV61 | Apr, Kmr; replacement of 0.26-kb BamHI fragment internal to oocM of pMAMV60 for a 0.96-kb BamHI km3 cassette of p34S-Km3 | This study |
| pMAMV62 | Smr, Kmr; 2.7 kb NotI fragment of pMAMV61 was cloned at the same site in pKNG101 | This study |
| pNJ5000 | Tcr; Mobilizing plasmid used in marker exchange | 27 |
a The following abbreviations are used: Ap, ampicillin; Km, kanamycin; Cm, chloramphenicol; Rif, rifampin; Sm, streptomycin; Tc, tetracycline; Em, erythromycin; STCC, Spanish Type Culture Collection.
In Vitro Nucleic Acid Techniques
A S. marcescens MSU97 cosmid library was constructed from high molecular weight genomic DNA (35–45 kb) using the pWEB-TNC Cosmid cloning kit following the manufacturer's instructions (Epicenter Biotechnologies). Plasmid DNA was isolated using the Anachem Keyprep plasmid kit. For DNA digestion, the manufacturer's instructions were followed (New England Biolabs and Fermentas). Separated DNA fragments were recovered from agarose using the Anachem gel recovery kit. Ligation reactions, total DNA extraction, and Southern blots were performed by standard protocols (28). DNA digoxigenin-dUTP probes were obtained via PCR following the instructions of the manufacturer (Roche Applied Science). Competent cells were prepared using calcium chloride, and transformations were performed by standard protocols (28). Phusion® high fidelity DNA polymerase (New England Biolabs) was used in the amplification of PCR fragments for cloning. Sequences of these PCR fragments were verified to discard amplicons containing mutations. Routine DNA sequencing was carried out at the University of Cambridge DNA Sequencing Facility on an Applied Biosystems 3730xl DNA analyzer.
Genome Sequencing and Bioinformatics Analyses
Genomic DNA sequencing was performed at the DNA Sequencing Facility, Department of Biochemistry (University of Cambridge), using 454 DNA pyrosequencing technology on a Pico Titer Plate for a Roche Applied Science Genome Sequencer FLX system. The shotgun assemblies were carried out using 454 GS de novo assembler software (Newbler version 2.6). For the S. marcescens MSU97 genome sequence, the assembly used 521,156 reads or 204 MB of raw data to give 38× coverage of the genome and resulted in 81 contigs, 68 of which were larger than 500 bp. The average contig size was 77,365 bp, and the largest contig was 418,649 bp. The Serratia plymuthica A153 assembly used 308,585 reads or 129 MB of raw data to give a 22× coverage of the estimated genome size and resulted in a total of 36 contigs, 24 of them larger than 500 bp. The average contig size was 230,980, and the largest contig was 1,516,666 bp.
Automated annotation of the bacterial sequences was done using the BASys web server (29). Anti-SMASH was used to analyze the potential secondary metabolite biosynthesis gene clusters present in the genomes (30). Genome comparison analyses were performed employing wgVISTA on-line tool (31). Open reading frames (ORFs) in the oocydin A gene cluster were automatically predicted using Glimmer 3.0 (32). Blast analyses were made for the functional gene assignment. Protein domain organization was identified using the NCBI conserved domains database (33) and the Pfam database (34). Multiple sequence alignments were carried out with ClustalW2 (European Bioinformatics Institute). Artemis software (Wellcome Trust Sanger Institute) was used to visualize genomic sequences.
Transposon Mutagenesis
Random transposon mutagenesis of S. plymuthica A153 using Tn-KRCPN1 was performed as described below. In a biparental conjugal mating, 500 μl of overnight cultures of E. coli β2163 (pKRCPN1) and S. plymuthica A153 were mixed, collected by centrifugation, resuspended in 30 μl of fresh LB, and spotted on an LB agar plate supplemented with 300 μm 2,6-diaminopimelic acid. After overnight incubation at 30 °C, cells were scraped off the plate and resuspended in 1 ml of LB. Serial dilutions were plated on LB agar medium containing 75 μg ml−1 kanamycin. 2,6-Diaminopimelic acid was not added to the LB agar medium, to allow counterselection of the E. coli donor. In total, 5000 kanamycin-resistant insertion mutants were screened for their inability to inhibit Pythium ultimum growth. The insertion site of transposon Tn-KRCPN1 in mutants of interest was determined using random primed PCR following the method described previously (35) and using primers described in supplemental Table S5. From the 22 isolated mutants exhibiting no, or reduced, bioactivity, all the insertions were located in the oocydin A gene cluster. Within the isolated mutants, some of the insertions sites were identical, suggesting that the strains were most likely clonal isolates.
Marker Exchange Mutagenesis
Specific site-directed mutants defective in oocK and oocM were constructed by homologous recombination using derivative plasmids of the suicide vector pKNG101 (20). These plasmids, which are listed in Table 1, were confirmed by DNA sequencing, and they carried mutant alleles for the replacement of wild type genes in the chromosome. In all cases, plasmids were transferred to S. plymuthica A153 by triparental conjugation using E. coli CC118λpir and E. coli HH26 (pNJ500) as helper. Mutants defective in oocK and oocM were generated using plasmids pMAMV54 and pMAMV62, respectively (Table 1). All relevant mutations were confirmed by PCR, sequencing, and Southern blot analysis. Primers used in this marker exchange mutagenesis are listed in Table S5.
Generalized Transduction
The newly isolated generalized transducing phage, φMAM1,4 was used for transduction of chromosomal mutations using a method similar to that described previously (36). Transductants were selected on plates containing kanamycin, and retention of phage sensitivity was confirmed in the transductants.
RNA Extraction, cDNA Synthesis, and Reverse Transcription-PCR (RT-PCR) Analyses
RNA was extracted from late exponential (12 h) cultures grown in enriched potato dextrose medium using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. RNA concentration was determined spectrophotometrically, and RNA integrity was assessed by agarose gel electrophoresis. Genomic DNA contamination was eliminated by treating total RNA with Turbo DNA-free (Ambion). The synthesis of cDNA was performed using random hexamers (GE Healthcare) and SuperScript II reverse transcriptase (Invitrogen) in a 20-μl reaction with 2.5 μg of total RNA and incubation at 42 °C for 2 h. A negative control reaction was also performed, omitting the reverse transcriptase enzyme. Then the equivalent of 50 ng of total RNA was subjected to PCR amplification using primers to amplify across the junctions (supplemental Table S5). Positive and negative control PCRs were performed using genomic DNA and no-RT cDNA samples, respectively, as templates. PCR conditions consisted of 30 cycles of denaturation for 1 min at 94 °C, annealing for 1 min at 62 °C, and extension for 40 s at 72 °C.
Antibacterial, Antifungal, and Anti-oomycete Activity in Vitro
Production of antibiotic compounds was tested against E. coli ESS and Bacillus subtilis JH642 using lawn assays as described previously (37). Antagonistic activities of bacterial strains against the fast growing plant pathogenic oomycete P. ultimum were assayed by spotting 5 μl of overnight cultures of the selected strains on a PDA plate. Following incubation for 16 h at 25 °C, the plates were inoculated with 5-mm diameter mycelial plugs taken from a culture of P. ultimum grown on PDA. Plates were incubated at 25 °C for 3–5 days. For the fungicide assays, indicator top agars of Verticillium dahliae, Thanatephorus cucumeris, Alternaria solani, and Fusarium oxysporum f. sp. lycopersici were prepared by vortexing a 5-mm fungal plug in 10 ml of sterile distilled water. Then 15 ml of PDA was added and mixed, and 5 ml of top lawns were poured into PDA plates. Five microliters of overnight cultures of the selected strains were spotted on the surface of the fungal agar lawn and incubated for 7–10 days at 25 °C. To determine fungicide/anti-oomycete levels in bacterial supernatants, culture samples were taken, and cells were pelleted by centrifugation (14,000 × g, 10 min), and the supernatant was filtered (0.2 μm). Three hundred microliters of the filter-sterilized supernatant were added to wells cut into the PDA plate and incubated at 25 °C for 3–5 days. All the experiments were repeated at least five times.
Caenorhabditis elegans Virulence Assays
The C. elegans virulence assays were adapted from those carried out for Kurz et al. (38). Briefly, NGM plates were inoculated with 50 μl of an overnight culture of the oocydin A-producing wild type strains (S. marcescens MSU97, S. plymuthica A153, S. odorifera 4Rx13, and Dickeya dandatii Ech703) or the oocydin A-deficient S. plymuthica A153 mutants. Plates inoculated with E. coli OP50 were also included as a negative control. The plates were incubated for 16 h at 25 °C. For each assay, worms previously fed on E. coli OP50 were transferred to the plates inoculated with the strains to test. Fifty L4 stage hermaphrodite DH26 worms (genotype fer-15(b26)II, sterile at 25 °C) were used for each strain tested (10 worms per plate). Plates were incubated at 25 °C, and the number of live worms was scored every 24 h. Worms were considered dead when they failed to respond to touch. Survival curves were analyzed using GraphPad Prism software. p values <0.05 were considered statistically significant.
Motility and Biofilm Formation Assays
Motility and biofilm assays were performed at 25 °C in enriched potato dextrose medium, conditions where we observed enhanced oocydin A production. Swimming assays were performed on 0.3% agar plates. Biofilm formation assays were carried out in 96-well microtiter plates with shaking at 75 rpm. Crystal violet staining and quantification method were performed as described previously (39).
LC-MS Studies
For the analysis of the bacterial supernatants, 25 ml of enriched potato dextrose broth was inoculated with the strains to analyze and incubated at 25 °C. After 48 h of incubation, cells were pelleted by centrifugation and filtered. Before the extraction, the pH of the culture supernatant was adjusted to 3.8 with citric acid. Six milliliters of the pH-adjusted supernatant were extracted twice with dichloromethane (6 ml). The organic layers were combined, dried over sodium sulfate, and evaporated under reduced pressure. The residue was resuspended in 1 ml of H2O/acetonitrile (1:1, v/v), and 10 μl was analyzed by LC-MS on a Finnigan MAT LCQ instrument using a Phenomenex Kinetex 2.6 μm XB-C18 100A column of size 100 × 2.1 mm eluted at 0.3 ml min−1 with a linear gradient over 15 min from 95:5 to 5:95 of water to acetonitrile, each containing 0.1% formic acid.
RESULTS AND DISCUSSION
Identification of the Oocydin A Gene Cluster
To identify the oocydin A gene cluster, the genome sequence of S. marcescens MSU97 was obtained using 454 pyrosequencing technology, automatically annotated, and analyzed. Based on the presence of NRPS and PKS encoding genes, at least five candidate biosynthetic gene clusters were identified in the 5.3-Mb MSU97 draft genome. Among them are gene clusters involved in the biosynthesis secondary metabolites such as siderophores and the red pigmented linear tripyrrole, prodigiosin.
A number of studies have reported that genes encoding enzymes responsible for the halogenation of secondary metabolites are closely associated with particular NRPS and PKS gene clusters (40–44). Therefore, we hypothesized that the gene cluster involved in the biosynthesis of oocydin A was likely to encode a halogenase enzyme. During the in silico screening of the MSU97 genome, we identified a non-heme chloroperoxidase and a peroxidase that were not associated with any of the candidate biosynthetic gene clusters. However, two flavin-dependent monooxygenases were identified in a 77.5-kb gene cluster involved in the biosynthesis of an unknown polyketide. Flavin-dependent halogenases are the main class of enzyme involved in the halogenation of aliphatic compounds (15, 45), so it seemed possible that this was the oocydin A gene cluster, although its complexity made it difficult to predict the structure of the polyketide synthesized.
In a first direct attempt to isolate the oocydin A gene cluster, we constructed a cosmid library of S. marcescens MSU97. Specific primers for five of the MSU97 biosynthetic gene clusters were designed, and screening of 2000 clones yielded 90 cosmid clones that hybridized positively to the probes. However, none of these cosmids could induce the production of oocydin A in E. coli, a host generally appropriate for the expression of cloned enterobacterial genes (46–48). Random mutagenesis was then employed as a second strategy to isolate S. marcescens MSU97 mutants with reduced antimicrobial activity toward the oomycete P. ultimum. However, the MSU97 strain proved to be recalcitrant to various genetic tools that we have previously used successfully for the genetic analysis of secondary metabolite production in Serratia and Erwinia (36, 47, 49). This genetic intractability, coupled with high intrinsic multidrug resistance, made a classical mutagenesis approach unfeasible.
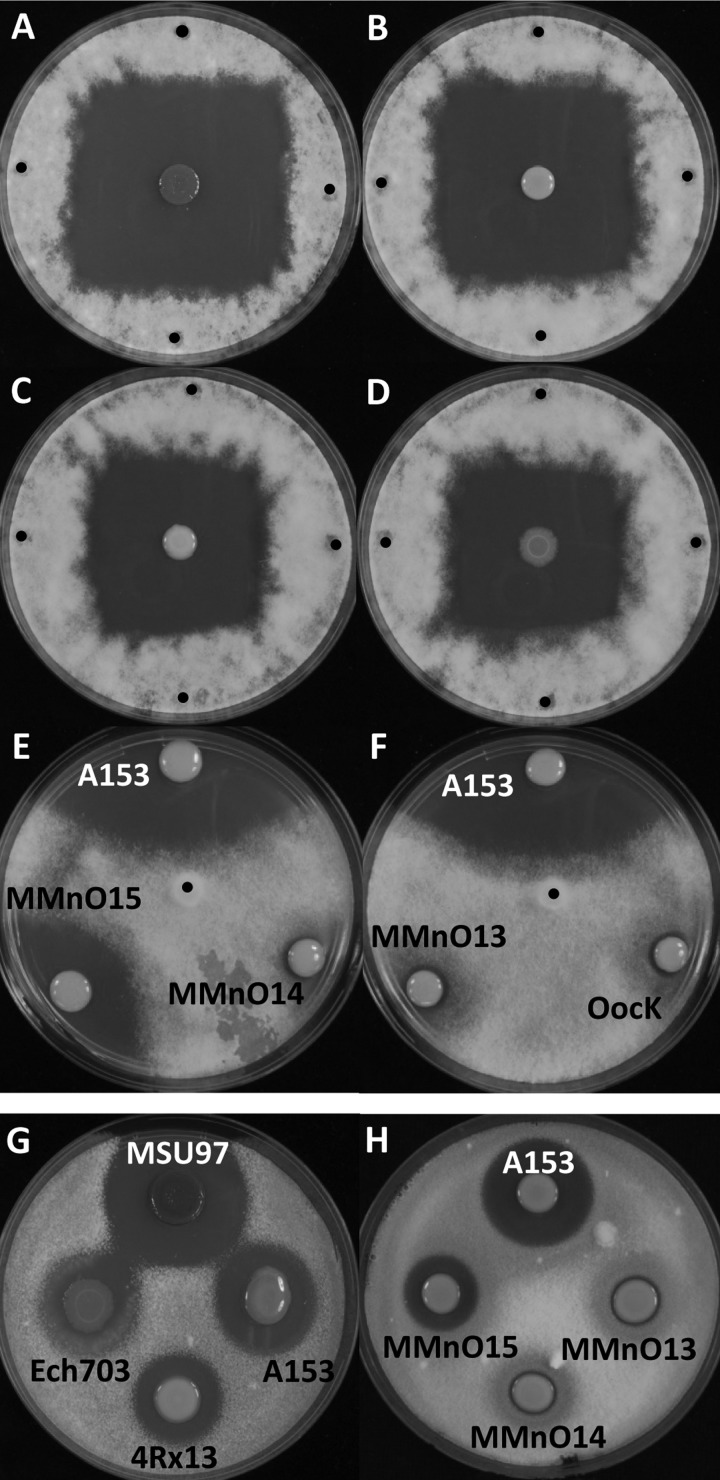
Besides S. marcescens MSU97, we had access to another oocydin A-producing strain, S. plymuthica A153 (7). We postulated that both strains should carry the same gene cluster for oocydin A biosynthesis. S. plymuthica A153 is a genetically tractable strain and therefore susceptible to efficient transposon mutagenesis. With the aim of isolating oocydin A-defective mutants, a transposon mutant library was screened in dual culture plate bioassays for mutants defective in antimicrobial activity toward the oomycete P. ultimum. We identified six independent transposon insertion mutants showing complete loss of the bioactive properties against the oomycete (Fig. 1, E and F, and supplemental Fig. S1A). We also isolated a mutant (MMnO15) that showed reduced anti-Pythium activity (Fig. 1E). All the insertion mutations were transduced back into a S. plymuthica A153 wild type genetic background using a new phage (φMAM1) that we have recently isolated4 to ensure that the phenotypes were indeed associated with single insertions and to confirm association between mutation and phenotype. The resulting transductants displayed the same phenotype as the original random mutants confirming genetically that loss of bioactivity was caused by the transposon insertion. Random primed PCR and genome sequence analysis of S. plymuthica A153 confirmed that all the transposon insertions mapped to a gene cluster homologous to the 77.5-kb MSU97 gene cluster (supplemental Table S2) that we had proposed as the main candidate for the biosynthesis of oocydin A (Fig. 2A). This corroborated the hypothesis that this particular polyketide gene cluster is also carried by S. plymuthica A153 (Table 2). We then successfully inactivated, in A153, the two candidate halogenase-encoding genes present in the gene cluster (oocK and oocM) using allelic replacement through the insertion of a kanamycin cassette. The resulting mutants were incapable of inhibiting the growth of P. ultimum (Fig. 1F and supplemental Fig. S1A) as expected.
FIGURE 1.
Antifungal and anti-oomycete activities of the oocydin A producing strains. Bioactivities against P. ultimum (A–F) and V. dahliae (G and H) are shown. P. ultimum bioassays were performed by inoculating 5-mm diameter mycelial plugs in the center (E and F) or at four different positions at the edge (A–D) of the bacterium-inoculated plates, as described under “Experimental Procedures.” P. ultimum inoculation points are indicated with a black dot. For the fungicide assays, an indicator V. dahliae top agar was prepared as described under “Experimental Procedures,” and 5 μl of overnight cultures of the selected strains were spotted on the surface of the fungal indicator agar lawn. The bioassays were repeated at least five times, and representative results are shown. P. ultimum and V. dahliae pictures were taken after 72 and 96 h of incubation at 25 °C, respectively.
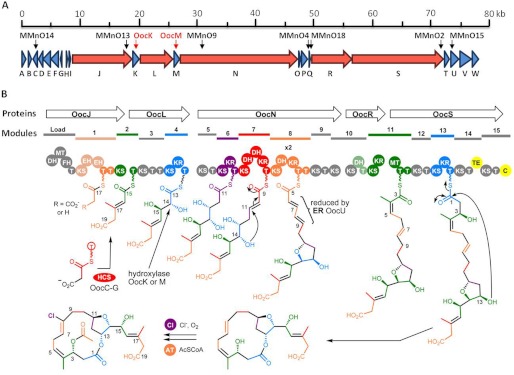
FIGURE 2.
Proposed biosynthetic pathway for oocydin A. A, genetic organization of the ooc biosynthetic gene cluster in S. plymuthica A153. The same genetic organization was found in S. marcescens MSU97, S. odorifera 4Rx13, and D. dadantii Ech703. Location of the Tn-KRCPN1 transposon insertions and marker exchange mutants are indicated by black and red arrows, respectively. Modular polyketide synthase genes are shown in red. B, model for the biosynthesis of oocydin A showing the structures of some of the proposed intermediates. T, acyl carrier protein; AT, acyltransferase; C, NRPS condensation domain; Cl, chlorinase; DH, dehydratase; EH, enoyl-CoA hydratase; FH, FkbH-like domain; HCS, 3-hydroxy-3-methylglutaryl-CoA synthase cassette; KR, ketoreductase; KS, ketosynthetase; MT, methyltransferase; TE, thioesterase. Domains in gray are proposed to be inactive or not involved in chain extension. Domains in grayish colors may be active.
TABLE 2.
Deduced functions of ORFs in S. plymuthica A153 ooc biosynthetic gene cluster
| Protein | Size (amino acid) | Proposed functiona | Sequence similarity (protein, origin) | Identity/similarity | GenBankTM accession no. |
|---|---|---|---|---|---|
| % | |||||
| OocA | 295 | α/β-Hydrolase | BYI23_C008640, Burkholderia sp. YI23 | 28/48 | AET93010 |
| OocB | 201 | Efflux protein | RSMK01349, R. solanacearum MolK2 | 40/60 | YP_002254246 |
| OocC | 249 | Enoyl-CoA hydratase | BatE, P. fluorescens | 62/75 | ADD82946 |
| OocD | 258 | Enoyl-CoA hydratase | PksH, Burkholderia pseudomallei 406e | 50/69 | ZP_04967830 |
| OocE | 420 | HMG-CoA synthase | PksG, Paenibacillus polymyxa E681 | 66/82 | YP_003871376 |
| OocF | 417 | Ketosynthase | KAOT1_04360, Kordia algicida OT-1 | 43/61 | ZP_02163873 |
| OocG | 83 | Acyl carrier protein | AcpK, Streptomyces griseoflavus Tu4000 | 43/69 | ZP_07315130 |
| OocH | 44 | Hypothetical protein | |||
| OocI | 53 | Hypothetical protein | |||
| OocJ | 3657 | PKS (DH-MT-FkbH-T-KS-EH-EH-T-T-KS) | |||
| - DH | BSU6633_07526, B. subtilis ATCC 6633 | 24/50 | ZP_06873407 | ||
| - MT-FkbH-T | OciA, Planktothrix agardhii NIES-205 | 39/57 | ABW84363 | ||
| - Module1 (KS-EH-EH-T-T) | OnnB, symbiont bacterium of Theonella swinhoei | 38/54 | AAV97870 | ||
| - KS (module 2) | PksM, P. polymyxa SC2 | 69/82 | ADO57347 | ||
| OocK | 434 | Flavin-dependent monooxygenase | PedG, symbiont bacterium of Paederus fuscipes | 61/78 | AAS47561 |
| OocL | 2403 | PKS (T-KS-T-T-KS-KR-T) | |||
| -T (module 2) | BaeL, Bacillus amyloliquefaciens FZB42 | 46/66 | YP_001421293 | ||
| - Module3 (KS-T-T) | SorA, Sorangium cellulosum | 42/57 | ADN68476 | ||
| - Module4 (KS-KR-T) | KAOT1_04320, K. algicida OT-1 | 38/54 | ZP_02163865 | ||
| OocM | 377 | Flavin-dependent monooxygenase | ORF2, Nostoc sp. “Peltigera membranacea” | 54/69 | ADA69238 |
| OocN | 6614 | PKS (KS-T-KS-KR-T-KS-DH-DH-KR-T-KS-DH-KR-T-T-KS-T-KS) | |||
| - Module5 (KS-T) | Saccy_3500, Saccharomonospora cyanea NA-134 | 34/51 | ZP_09747982 | ||
| - Module6 (KS-KR-T) | SorD, S. cellulosum | 39/54 | ADN68479 | ||
| - Module7 (KS-DH-DH-KR-T) | SorE, S. cellulosum | 39/55 | ADN68480 | ||
| - Module8 (KS-DH-KR-T-T) | Gura_3095, Geobacter uraniireducens Rf4 | 44/60 | YP_001231833 | ||
| - Module9 (KS-T) | DfnH, Paenibacillus elgii B69 | 51/68 | ZP_09073715 | ||
| - KS (module 10) | BaeR, Bacillus sp. 5B6 | 37/54 | IF13282 | ||
| OocO | 89 | Acyl carrier protein | BSn5_20710, B. subtilis BSn5 | 32/60 | ZP_07900867 |
| OocP | 389 | Hypothetical protein | BuboB_26000, Burkholderia ubonensis Bu | 25/40 | ZP_02381200 |
| OocQ | 119 | Hypothetical protein | SpiBuddy_0991, Sphaerochaeta globus st. Buddy | 25/42 | YP_004247012 |
| OocR | 2264 | PKS (KS-DH-T-KS-KR) | |||
| - KS-DH-T (module 10) | BBR47_39880, Brevibacillus brevis 100599 | 33/51 | YP_002773469 | ||
| - KS-KR (module 11) | RhiC, Burkholderia rhizoxinica | 42/58 | CAL69890 | ||
| OocS | 5341 | PKS (MT-T-T-KS-T-KS-KR-T-KS-T-TE-KS-T-C) | |||
| - MT-T-T (module 11) | RhiC, R. solanacearum CFBP2957 | 44/62 | YP_003748157 | ||
| - Module 12 (KS-T) | PedF, symbiont bacterium of Paederus fuscipes | 39/55 | AAS47564 | ||
| - Module 13 (KS-KR-T) | BaeM, Bacillus amyloliquefaciens IT-45 | 42/59 | EHM05017 | ||
| - KS-T (module 14) | BATR1942_06405, Bacillus atrophaeus 1942 | 49/63 | YP_003973166 | ||
| - TE (module 14) | BryX, Candidatus Endobugula sertula | 45/67 | ABM63529 | ||
| - Module 15 (KS-T-C) | Fr9C, Pseudomonas sp. 2663 | 35/49 | ADH01484 | ||
| OocT | 276 | Hypothetical protein | Krac_1408, Ktedonobacter racemifer DSM 44963 | 26/46 | ZP_06972719 |
| OocU | 482 | Flavin-dependent nitroreductase | BatK, P. fluorescens | 53/73 | ADD82952 |
| OocV | 624 | Acyl transferase (AT1-AT2) | |||
| - AT-1 | BryP, Candidatus Endobugula sertula | 59/75 | ABK51299 | ||
| - AT-2 (candidate acyl hydrolase) | BryP, Candidatus Endobugula sertula | 41/63 | ABK51299 | ||
| OocW | 372 | Acyl transferase | PedD, B. pseudomallei 1710b | 49/65 | YP_337765 |
a C, NRPS condensation domain; T, acyl carrier protein.
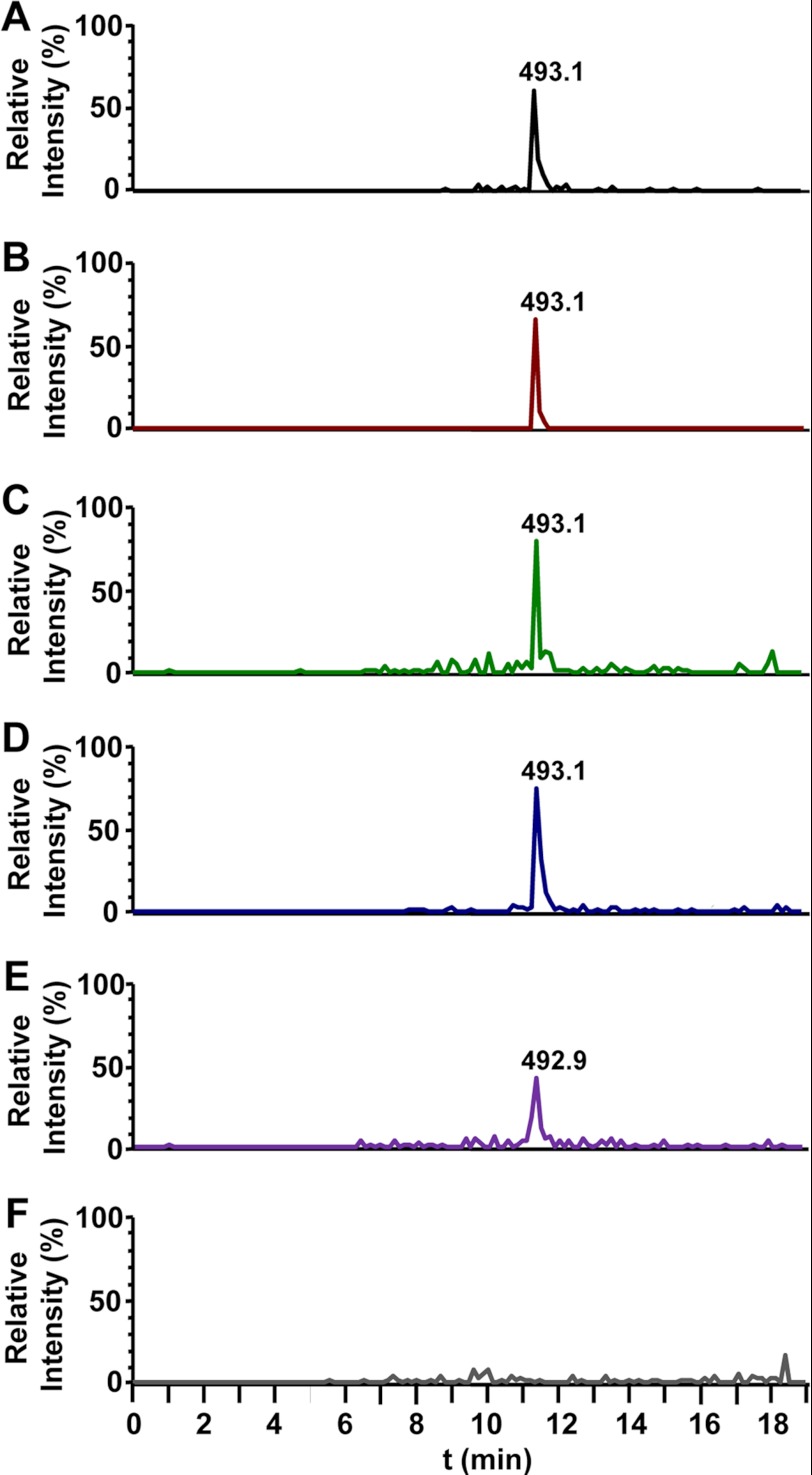
To confirm the identity of the oocydin A gene cluster and to corroborate its role in the biosynthesis of the macrolide, we analyzed by LC-MS the culture supernatants of S. marcescens MSU97 and S. plymuthica A153 wild type strains and identified a metabolite with the same molecular weight as oocydin A (Fig. 3, A and B). This metabolite showed the characteristic isotope pattern of a monochlorinated compound, with peaks at m/z 493 and 495 [M + Na]+ in a 3:1 ratio as a result of the two isotopes 35Cl and 37Cl (supplemental Fig. S2A). The MS/MS fragmentation of [M + Na]+ ion m/z 493 yielded two major ions, at m/z 433 [M + Na-AcOH]+ and 449 [M + Na-CO2]+ (supplemental Fig. S2, B and C). The same analysis showed that all the nonbioactive mutants displayed a complete loss of oocydin A production (Fig. 3F and supplemental Fig. S3, B–D), confirming the role of the gene cluster in the biosynthesis of the halogenated macrolide. There were no dechloro analogs of oocydin A detected in the extracts of oocK or oocM mutants that formally could be due to potential polar effects of the kanamycin cassette insertion in the respective genes. The analysis of the MMnO15 extracts showed that oocydin A was produced at significantly low levels (3–5%) as compared with the wild type strain A153 (Fig. 3E). This result suggests that other trans-acting ATs and ERs encoded in the genome of A153 could be catalyzing the required reactions, although less efficiently than OocU, -V, and -W. Taking all these data into account, we have named the genes of this cluster ooc, for oocydin A biosynthetic genes.
FIGURE 3.
LC-MS analysis of culture supernatant extracts of oocydin A producing strains and mutants in the ooc gene cluster. A, S. marcescens MSU97 wild type strain; B, S. plymuthica A153 wild type strain; C, S. odorifera 4Rx13 wild type strain; D, D. dadantii Ech703 wild type strain; E, S. plymuthica A153 MMnO15 mutant strain; F, S. plymuthica A153 MMnO13 mutant strain. The chromatograms show selective ion monitoring at m/z 493 ± 0.5. The peaks at 11.4 min correspond to oocydin A (m/z 493.1 [M + Na]+). The chromatograms of the MSU97 (A) wild type A153 (B) and Ech703 (D) strains were scaled down 4, 20, and 2 times, respectively, as compared with the ooc mutants MMnO15 (E) and MMnO13 (F). Fragmentation patterns of oocydin A are shown in supplemental Fig. S2. Chemical analyses of other mutants in the ooc gene cluster are shown in supplemental Fig. S3.
Oocydin A Gene Cluster Is Present in Several Plant-associated Enterobacteria
In addition to S. marcescens MSU97 and S. plymuthica A153, genome comparison analysis revealed that the ooc gene cluster is also present in the rhizobacterium, S. odorifera 4Rx13 (24) (GenBankTM accession no. ADBX01000002.1; genomic coordinates 102562-180808), and in the phytopathogenic bacterium D. dadantii Ech703 (GenBankTM accession NC_012880; genomic coordinates 1624970–1705054). Chemical analysis of 4Rx13 and Ech703 by LC-MS showed that they also produced oocydin A (Fig. 3, C and D, and supplemental Fig. S2, D and E), and, as expected, they show very clear anti-Pythium properties (Fig. 1, C and D). Interestingly, the genomic context of the ooc gene clusters in S. marcescens MSU97, S. plymuthica A153, and D. dadantii Ech703 is completely different. Therefore, the upstream and downstream ends of the different loci were assigned based on the homologies between the three biosynthetic clusters. The ooc gene clusters of S. plymuthica A153 and S. odorifera 4Rx13 show the highest homology (94.8% at DNA level; supplemental Table S1), and they also share the same genomic context. Therefore, these data suggest that the biosynthetic cluster could have been transferred horizontally between different genera and species of bacteria. In fact, although the G + C content of the ooc gene clusters of MSU97 (57.8%), A153 (56.6%), 4Rx13 (56.0%), and Ech703 (52.3%) are similar to the genomic G + C content in the chromosome, sequences reminiscent of mobile genetic elements, such as a bacteriophage P4 integrase, border the ooc gene cluster in D. dadantii Ech703.
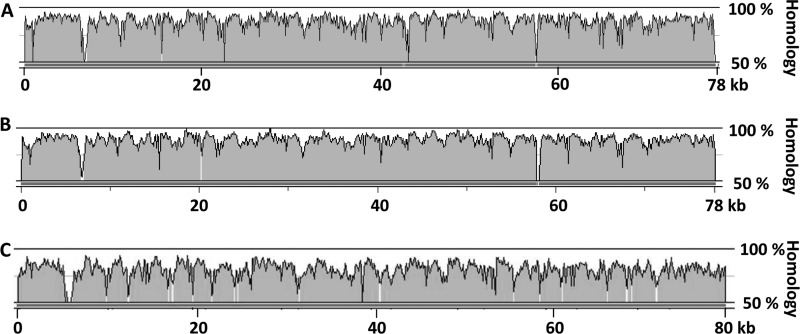
The oocydin A gene clusters of A153, 4Rx13, and Ech703 are 78.2, 78.3, and 80.1 kb, respectively, and they are 81.7, 81.8, and 70.9% identical, respectively, at the DNA level to the 77.5-kb cluster of MSU97 (Fig. 4 supplemental Table S1). Unsurprisingly from the high nucleotide homology, the module organization within the PKS proteins encoded by the four ooc gene clusters revealed that all share the same domains and domain organization. Furthermore, on average, the proteins encoded by the S. marcescens MSU97 ooc gene cluster are 87.1, 87.3, and 78.6% identical to those of A153, 4Rx13, and Ech703, respectively. However, there are some interesting differences between them. First, the initial gene of the cluster in MSU97, A153, and 4Rx13 (encoding a putative alpha/beta hydrolase) is not present in Ech703 (supplemental Table S4). Second, OocN and OocS of the MSU97, A153, and Ech703 ooc gene clusters are divided into three and two genes, respectively, in S. odorifera 4Rx13 (supplemental Table S3), although it is formally possible that the difference might be due to an error in genome sequencing. In accordance with this notion, domains that are encoded by two different proteins in 4Rx13 were found contiguous within the same PKS protein in other bacteria (supplemental Table S3). Finally, the region containing the putative small genes, oocH and oocI, of MSU97 is poorly conserved between the strains showing a 49.4, 46.5, and 41.7% identity at the DNA level, compared with A153, 4Rx13, and Ech703, respectively. Because this is a region of divergence of two transcriptional units (Fig. 2A), it is reasonable to hypothesize that this region has an important regulatory role(s) in the expression of this complex gene cluster. Interestingly, the G + C content of this intergenic region in MSU97, A153, 4Rx13, and Ech703, which is 42.6, 41.7, 42.1, and 37.1%, respectively, is significantly lower than the mean G + C content of the gene clusters. OocB, a putative transporter present in the gene cluster of MSU97, A153, and Ech703 (supplemental Table S3), was not present in the GenBankTM draft annotation of the 4Rx13 cluster, but the reannotation of the cluster showed that it is, in fact, present.
FIGURE 4.
DNA homology between the oocydin A gene cluster of S. marcescens MSU97 and the oocydin A gene cluster of the other producing strains. The alignments represent the percentage of homology between the ooc gene cluster of S. marcescens MSU97 and the ooc gene cluster of S. plymuthica A153 (A), S. odorifera 4Rx13 (B), and D. dadantii Ech703 (C). Alignments were performed using wgVISTA (31).
Bioinformatic Analysis of the ooc Biosynthetic Gene Clusters
As described above, the biosynthetic gene cluster of oocydin A spans between 77 and 80 kb and consists of 23 ORFs. The likely function for each gene product was deduced by sequence comparison with proteins of known function, and 16 of the proteins can be assigned possible roles in oocydin A biosynthesis (Table 2 and supplemental Tables S2–S4).
The oocydin A biosynthetic cluster encodes five multifunctional PKS enzymes (OocJ, OocL, OocN, OocR, and OocS), which include a total of 16 β-KS, 7 KR, 20 ACP, 5 DH, 2 enoyl-CoA hydratases (EH), 2 methyltransferase (MT) domains, a TE, and an NRPS condensation (C) domain (Fig. 2B, Table 2, and supplemental Tables S2–S4). One of the first observations derived from the analysis was the absence of integrated AT domains in all the biosynthetic modules of the five PKS proteins encoded by the ooc gene cluster. This places the oocydin A PKS in the growing class of PKSs that use trans-acting AT domains. In these enzymes, each module receives its acyl building block from one or more discrete ATs encoded by separate genes (50). The ooc gene cluster encodes two such proteins, OocV and OocW, containing two and one AT domains, respectively, with all three of the domains containing the catalytic Ser–His dyad and the highly conserved N-terminal GQGSQ loop (supplemental Fig. S4) (51, 52). In accordance with the trans-AT nature of the ooc gene cluster, analysis of its interdomain sequences identified regions with homology to truncated AT domains following most of the KS domains. Recently, it has been suggested that these regions are the binding sites for the separate AT domains in trans-AT PKSs (53). Two or more free-standing ATs are infrequently found in trans-AT PKSs, and to our knowledge, only nine examples have been described as follows: bacillaene (54); elansolid (55); etnangien (56); kirromycin (57); mupirocin (58); pederin (59); rhizopodin (60), rhizoxin (61), and sorangicin (62). Multiple sequence alignments of OocV and OocW against known trans-ATs revealed that OocV-AT1 and OocW share all the residues characteristic of a malonyl-CoA-specific AT, whereas OocV-AT2 possesses more active site residue diversity (supplemental Fig. S4) (51, 52, 62). It has been suggested that this residue diversity may be associated with a broader range of substrate specificity or specialized functions (52, 62–64). BLAST analyses and multiple sequence alignments also revealed that OocV-AT1 and OocV-AT2 are most similar to BryP-AT1 and BryP-AT2, respectively (Table 2 and supplemental Tables S2–S4; supplemental Fig. S4), which have been grouped in two different classes of trans-ATs (64). Lopanik et al. (64) showed that both BryP-AT1 and BryP-AT2 prefer to transfer malonyl-CoA, although BryP-AT1 is more active than BryP-AT2. Multiple alignment analyses (supplemental Fig. S4) also grouped OocV-AT2 with the stand-alone acyltransferase PedC. A recent study showed that PedC does not exhibit AT activity but possesses acyl hydrolase activity and cleaves acyl groups bound to ACP, suggesting a role in PKS biosynthetic proofreading (65).
The number of domains in each of the five Ooc PKS proteins varies between 5 (OocR) and 18 (OocN). Interestingly, the number of KS (16 as domains within the modular proteins and one as a separate ORF) and ACP (22 domains, two of them present in separate ORFs) domains exceeds the number needed for the biosynthesis of oocydin A, which would be 8 and 9 domains, respectively. Presumably, some of the KS domains are either inactive and skipped or are simply used for transferring the acyl intermediate to the next ACP domain without extending it. In accordance with this observation, the catalytic CHH triad present in active KS domains is absent in KS1 (SHH), KS5 (CQH), KS10a (CEH), and OocF (SHH) (supplemental Figs. S5–S8, the numbering of domains refers to the module number as shown in Fig. 2B) (66). Furthermore the characteristic HGTGT catalytic motif, which is essential for the decarboxylative condensation, is poorly conserved in KS5 (QGMGL), KS10a (EANGS), KS10b (HGASS), KS12 (HATGG), KS14 (HSTGS), and KS15 (HAA/TGM) which may indicate that these domains are not extending the polyketide chain (supplemental Fig. S5–S8) (62). All ACP domains show the highly conserved signature motif GXDS except for ACP7 (GFNS), ACP14 (GLNS), OocG (GANS), and OocO (GFES) (supplemental Fig. S5–S8) (67).
All seven potential KR domains identified showed the characteristic Rossmann fold for cofactor binding. The SYN conserved triad located in the active site (68) was modified in KR4 (SYA) and KR11 (S(Y/S)(A/I)) (supplemental Fig. S5–S8). Module 3 contains a region of 55 amino acids, which shows homology with the Rossmann fold of NAD(P)-binding proteins, but it lacks the SYN conserved triad, which indicates that it does not function as a ketoreductase. Of the five predicted DH domains found in the PKS, three of them (DH7a, DH8, and DH10) contain the HXXXGXXXXP motif characteristic of DH domains (supplemental Fig. S5–S8) (69). Module 1 contains two EH domains showing altered conserved motifs essential for the oxyanion hole that stabilizes the enolate anions (supplemental Fig. S9) (70). However, the “oxyanion hole” consensus motifs are present in OocC and OocD (supplemental Fig. S9). A TE and a C domain containing the GXSXG (71) and the HHXXXDG (72) conserved motifs, respectively, were identified in the last multimodular PKS of the cluster.
Finally, at least three putative tailoring enzymes are encoded by genes in the cluster, and they probably catalyze oxidative/reductive transformations (OocK, OocM, and OocU). The cluster also encodes a protein (OocB) 40% identical (60% similar) to the putative efflux protein RRSL_03865 present in Ralstonia solanacearum UW551 and in other genera such as Enterobacter, Photorhabdus, and Burkholderia. This protein might be involved in the efflux of the bioactive molecule to the environment. Perhaps surprisingly, no genes encoding obvious candidate regulatory proteins were found in the ooc gene cluster.
Model for the Biosynthesis of Oocydin A
It has been observed that trans-AT systems possess numerous functional peculiarities as compared with cis-AT PKSs, in which the AT domains are located within the multidomain PKS. These peculiarities are often translated into unusual chemistry and include the loss of co-linearity found in canonical PKSs, lack of expected domains, unusual domain orders and domain sets, and the splitting of modules between two PKS proteins. Additionally, PKS modules can be “skipped” or used more than once during the biosynthetic pathway (16, 50). These unusual characteristics make it difficult to establish an obvious predictive correlation between the polyketide structure and the architecture of the trans-AT PKS modules. However, the substrate specificity of KS domains of trans-AT PKSs has been correlated with their sequence, and they have been found to be most similar to other KS domains that process similar substrates (50, 55). Thus, based on an extensive in silico analysis, we propose a mechanism for the biosynthesis of oocydin A as shown in Fig. 2B. In this model, functional KS domains have been compared with the nearest characterized homologs to determine what acyl groups could be accepted by the respective domains. As has been observed in other trans-AT PKSs, the oocydin A PKS deviates from the textbook PKS model architecture in several features.
The structure of oocydin A shows carboxyl groups at both ends of the linear chain of the polyketide, so its biosynthesis could, in principle, be started at either end. However, the organization of the domains makes it much more likely that the starter unit is C-17 to C-19. The predicted DH-MT-FkbH domains in the Load module are uncommon but have some similarity to the DH-KR-FkbH Load module of BryA, involved in the biosynthesis of bryostatin, which functions to load a lactyl starter unit onto an ACP domain (73). However, the structure of oocydin A does not require any such starter unit; the expected starter unit is a malonyl unit instead of the glyceric acid unit normally loaded by the FkbH domain (74). Therefore, we propose that the first three domains (DH, MT, and FkbH) might be nonfunctional relics in the process of evolutionary degeneration. In accordance with this, the DH and MT domains present in this unusual DH-MT-FkbH module lack the characteristic HXXXGXXXXP and LEXGXGXG conserved motifs, respectively. Furthermore KS1 has a serine in place of the active site cysteine and so is probably incapable of chain extension. Thus, the proposed biosynthesis would begin with the transfer of an activated malonyl starter unit from malonyl-CoA to the ACP1 domain of OocJ by one of the proposed trans-ATs. It is possible that KS1 catalyzes the decarboxylation of this malonyl group to an acetyl group, but it is equally likely that this carboxyl is retained, and it is the acetic acid unit introduced by the 3-hydroxy-3-methylglutaryl-CoA synthase (HCS) cassette (see below) that is decarboxylated. KS2 would then catalyze the first round of chain extension to give a β-keto-thioester attached to ACP2.
The methyl group at C-17 is attached to a carbon that would initially have been a carbonyl group. Such methyl groups are usually added by HCS cassettes that consist of stand-alone HCS, ketosynthase, and ACP proteins (63, 73, 75). Importantly, these three proteins are contiguous in the cluster (OocE–G). In this mechanism for generating a methyl group, two dehydratase (DH)-like EH domains are also required. Two genes for stand-alone EH proteins (OocC and OocD) that complete the operon encoding the HCS cassette are present in the ooc gene cluster. The similarity between the EH domains of OocD and OocC is only 19.1% (supplemental Fig. S9), and it has been hypothesized that such EH domains have different functional roles catalyzing, respectively, the dehydration and decarboxylation required to produce the methyl group (70). Two side-by-side EH domains are also present in module 1 of OocJ. The integration of EHs within a PKS module is unusual, although it has been described in the gene clusters of the related PKSs that make pederin (59) and onnamide A (76) as examples of the biosynthetic versatility of PKSs. The fact that the motifs essential for the oxyanion hole are poorly conserved in EH1a and EH1b and that a mutant defective in the EH OocC (MMnO14) does not produce oocydin A suggest that OocC and OocD are responsible for the required dehydration and decarboxylation. Thus, the HCS cassette (or alternatively the DH-like domains in module 1) would function at module 2 by adding a methyl group at C-17 (or adding –CH2CO2H if decarboxylation of the starter unit does occur). The domain structures of the other four PKSs (OocL, OocN, OocR, and OocS) do not show clearly the order in which they act. However, the structure of oocydin A suggests that they act in the same order as the genetic organization of their cognate genes, which is normally the case. Thus, the subsequent biosynthetic steps can be largely rationalized by the co-linearity rule with several deviations because some domains have mutant catalytic domains and several modules lack domains that would be predicted from the oocydin A structure. Module 3 appears to contain a fully active KS domain but only has a fragment of a KR domain, which lacks the active site groups and cannot be functional. As oocydin A has no keto groups that have not been reduced, this suggests that this module may be skipped. Module skipping has been described in other trans-AT PKS such as difficidin (54), myxovirescin A (77), leinamycin (78), and chivosazol (79). Following the assembly line, KS5 at the N-terminal end of OocN has a mutated CHH motif, so if probably does not elongate the chain. However, it may be involved in transferring the chain to the next module without elongating it.
Further along OocN, KS7 is predicted, by homology with other PKS domains, to accept the (S)-β-hydroxy-acyl group that is produced by module 6. Domain 7 has two DH-like domains. In other PKSs, such as the sorangicin one (62), the extra DH domain is proposed to catalyze a cyclization in which a hydroxyl group attacks the β-position of an α,β-unsaturated thioester in a Michael addition mechanism (interestingly, many domains in the ooc PKS show high similarity to equivalent domains in the sor PKS). In accordance with this model, KS8 has homology to KSs that accepts acyl groups where a cyclization has occurred. In the case of oocydin A, this cyclization needs to be from a hydroxyl at C-14, as shown in Fig. 2B. It appears therefore that hydroxylation at C-14 occurs, whereas the acyl chain is incomplete and still attached to the PKS (most likely when it was attached to module 4 before the reduction of the keto group at C-15 occurs so that deprotonation of C-14 is facilitated). The enzyme responsible for hydroxylation of C-14 is probably one of the two flavin-dependent monooxygenases OocK and OocM, the genes for which are located between PKS-encoding genes, immediately before and after oocL. Because PKS genes are very often contiguous in a cluster, this location of oocK and oocM may suggest that the encoded enzymes act on substrates that are still bound to one of the PKS proteins.
Given that the fragment of a KR domain in module 3 lacks the catalytic triad, the number of active KR domains identified in the ooc biosynthetic cluster is insufficient for the biosynthesis of the macrolide. Therefore, one of the modules is probably used twice, catalyzing two rounds of elongation. This unusual feature has mainly been reported in trans-AT systems, such as rhizopodin (60), lankacidin (53), and oxazolomycin (50, 80). We propose that module 8 would be used twice, but another possibility is that KS9 is in fact active in chain elongation but then passes the β-keto acyl group back to module 8 for processing by its KR and DH domains (which would be an example of nonlinearity often found in trans-AT PKSs).
At this point in assembly, an ER domain would be needed but OocN (like all of the other four PKS proteins) lacks this type of domain. However, OocU has high similarity to proteins found in PKS clusters that have been shown to be trans-acting flavin-dependent ERs such as PksE from B. subtilis (81) or BatK from Pseudomonas fluorescens (66). Therefore, we postulate that this ER domain might reduce the α,β,γ,δ-conjugated diene attached to module 8, as shown in Fig. 2B, leaving the remaining double bond in the β,γ-position.
Further along the PKS production line, module 11, which spans OocR and OocS, contains an MT domain that we suggest adds the methyl group present at C-4. The structure of oocydin A also requires a DH to act at this stage to generate the C-4/C-5 double bond. There is no DH in module 11, but there is one in module 10, so perhaps the β-hydroxyacyl group from module 11 gets passed back for dehydration by that domain. Alternatively a trans-acting DH may effect the dehydration.
The TE and the NRPS C domains of OocS show high similarity to the domains present in the PKS proteins BryX and BryD, respectively, involved in the biosynthesis of bryostatin (73). Interestingly, the last PKS proteins involved in the chain extension of oocydin A and bryostatin both terminate with these C domains. It is not known whether it is the TE or the C domain that catalyzes the macrolactonization in bryostatin biosynthesis. By analogy, we propose that either the TE or the NRPS C domain in OocS is the likely candidate for the macrolactonization that releases the acyl group from the PKS. After the release of the polyketide from the PKS, the final acetylation (shown in Fig. 2B) can be catalyzed by a separate acyltransferase. However, there is a possibility that one of the KS14, KS15, TE, or C domains of OocS catalyze this acetylation. KSs, TEs, and C domains all catalyze acyl transfers of some complexion, so in principle, O-acylation would be only a small change of function.
Finally, the chlorine atom at C-8 should be introduced by a chlorinase. Chlorinating enzymes have been classified into highly specific halogenases requiring dioxygen and either a reduced flavin or α-ketoglutarate as co-substrates and less specific haloperoxidases that use hydrogen peroxide, often P450 enzymes (15, 45). Because there are no predicted P450 enzymes in the ooc gene cluster, the main candidates for the halogenase are the flavin-dependent monooxygenases OocK and OocM. In agreement with this, the mechanism of action of flavin-containing halogenases is just a slight modification of that of monooxygenases (82), and crystallization of some of these halogenases has revealed a flavin monooxygenase domain in most of them (83). The chlorination could be a post-PKS biosynthesis step (Fig. 2B) or it could happen on a PKS-bound intermediate, perhaps in module 7 at the β-keto-thioester stage, when C-8 is easy to deprotonate.
Oocydin A Cluster Is Formed by Three Transcriptional Units
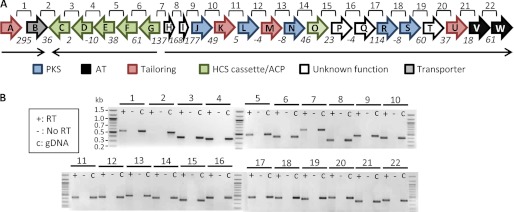
The genetic organization of the oocydin A cluster suggests the presence of at least three transcriptional units. To further investigate this hypothesis, transcript analysis by RT-PCR was performed on cultures of S. marcescens MSU97. To select the conditions where the oocydin A genes were being expressed, MSU97 culture samples were taken at different points along the growth curve. Filter-sterilized supernatants from these samples were added to holes punched in P. ultimum bioassay plates. Oomycete growth inhibition was observed in the supernatants after 12 h of growth in enriched potato dextrose medium (5), and these growth conditions were therefore used for the RT-PCR studies. For this analysis, primers were designed to cover the region between the 3′ end of the upstream gene and the 5′ end of the contiguous downstream gene (Fig. 5A). Two smaller transcriptional units, one consisting of oocA and oocB and a second consisting of oocG, oocF, oocE, oocD, and oocC, were identified. In addition, RT-PCR products were detected across all the intergenic regions containing oocJ–W, indicating the existence of a large polycistronic transcript (Fig. 5B). Unexpectedly, prominent PCR products were also obtained for the region covering the genes oocG–I (Fig. 5B). This might indicate the presence of divergent overlapping messenger RNAs in this region, consistent with the proposed regulatory role of this locus in the ooc cluster. Our analysis confirmed the presence of the three predicted transcriptional units in the oocydin A cluster, although the absence of additional internal promoters cannot be assumed.
FIGURE 5.
Analysis of the transcriptional units in the oocydin A gene cluster by RT-PCR. A, schematic representation of the ooc gene cluster in S. marcescens MSU97. Lines labeled 1–22 above the gene cluster represent the regions amplified in the RT-PCR shown in B. Numbers below the arrows represent the intergenic distance between contiguous genes, although negative numbers indicate overlapping genes. Black arrows represent the three transcriptional units found in the ooc gene cluster. B, transcript analysis by RT-PCR using primers designed to span the intergenic region between two adjacent genes. For each region, three PCR analyses were carried out: +, RT-PCR on cDNA; −, negative control with no reverse transcriptase; gDNA, positive control with genomic DNA as template.
Biological Properties of Oocydin A
Oocydin A has been shown to be very active against plant pathogenic oomycetes belonging to Pythium and Phytophthora genera (5). However, the activity of oocydin A against plant pathogenic fungi such as S. sclerotiorum was controversial because two different studies observed contradictory results (5, 7). These apparently contradictory findings perhaps could be explained by the concentration of bioactive molecules present in each bioassay because oocydin A had fungistatic or fungicidal effects, depending on the concentration (7). With the aim of determining the biological roles of oocydin A, we characterized phenotypically the strain S. plymuthica A153 and its non-oocydin A-producing mutants. The other oocydin A producers, S. marcescens MSU97, S. odorifera 4Rx13, and D. dadantii Ech703 were also analyzed. Perhaps surprisingly, given the number of enzymes that it encodes, inactivation of the large ooc gene cluster had no detectable effect on the growth rate. Neither was there any detectable impact on motility or biofilm formation of S. plymuthica A153 (data not shown). Besides the very high bioactivity that oocydin A shows against the oomycete, P. ultimum, we also tested its bioactive properties against fungal plant pathogens. In contrast to the report by Strobel et al. (5), we observed that the halogenated molecule was also active against the fungus, V. dahliae, because all the producing strains show bioactivity against the fungus, whereas the A153 mutants defective in the ooc gene cluster had lost the anti-Verticillium activity (Fig. 1H and supplemental Fig. S1B). Interestingly, as we had observed previously in the P. ultimum inhibition assays, MMnO15 also showed reduced bioactivity against V. dahliae (Fig. 1H). In our study we have also verified that oocydin A inhibits the growth of other fungal plant pathogens, including A. solani, F. oxysporum, and to a lesser degree, T. cucumeris (supplemental Fig. S1, C–E). However, oocydin A was not active against the ascomycetes Saccharomyces cerevisiae and Schizosaccharomyces pombe (data not shown).
S. plymuthica A153 and S. marcescens MSU97 also show antibacterial activity against Gram-positive and Gram-negative bacteria (supplemental Fig. S10). However, the non-oocydin A-producing mutants of A153 showed the same antibacterial properties (supplemental Fig. S11), whereas these activities were not detected in the other oocydin A producers Ech703 and 4Rx13 (supplemental Fig. S10), showing that, in accordance with previous observations (8), oocydin A is not responsible for the antibacterial properties.
A similar result was observed with C. elegans. The nematode C. elegans has been used previously as a model system for the in vivo identification of bacterial virulence factors (84). A153 and MSU97 showed a dramatic virulence in C. elegans assays (supplemental Fig. S12). However, the non-oocydin A-producing mutants killed C. elegans at the same rate as the wild type strain (supplemental Fig. S13), whereas 4Rx13 and Ech703 show reduced, or no, virulence against the nematode (supplemental Fig. S12), implying that oocydin A is unlikely to be responsible for the observed virulence seen in MSU97 and A153 (supplemental Fig. S12).
Concluding Remarks
Oocydin A is an antifungal and anti-oomycete halogenated macrolide that has been shown also to possess antitumor properties. Therefore, this molecule, or analogs, may have considerable utility and promise for crop disease biocontrol and/or in medical chemotherapy (5, 6, 9, 11). In this work, we describe for the first time the identification of the ooc biosynthetic gene cluster in four different strains of plant-associated enterobacteria, including bacteria from Serratia and Dickeya genera. The ooc biosynthetic gene cluster consists of 23 genes (or 22 genes in D. dadantii Ech703) organized in three transcriptional units. The analysis of the domains and module organization in the PKS proteins has revealed that these oocydin A biosynthetic enzymes belong to the subclass of trans-AT PKSs. In accordance with this, uncommon domain orders and splits of modules between PKS proteins were found in the ooc cluster products, similar to other trans-AT PKSs involved in biosynthesis of bryostatin (73), rhizopodin (60), rhizoxin (61), and sorangicin (62). A HCS cassette, required to modify the oocydin A PKS-bound intermediate, was also identified in the ooc gene cluster.
The broad spectrum of biological activities of oocydin A has made it an attractive compound for chemical studies. Although its chemical synthesis has been accomplished, the overall yield of only 1.3% for the best current synthesis remains inefficient (9). Thus, our definition of the genes responsible for the biosynthesis of oocydin A and our proposed biosynthetic model now provide an excellent opportunity to investigate the enzymatic mechanisms involved and the nature of the biosynthetic machinery. By a combination of molecular genetics and (bio) chemistry, we can now test the model for biosynthesis and confirm the assembly pathway. Furthermore, given the distribution of the ooc cluster in different enterobacterial genera and in nonconserved genetic contexts, we are now in a position to investigate how the production of this curious bioactive molecule is regulated. Future research will provide information that enables knowledge-based strategies for enhancing productivity and for generating novel analog haterumalides by synthetic biology methods. Such novel analogs might have important agricultural, pharmacological, and chemotherapeutic applications.

This article contains supplemental Figs. S1–S13 and Tables S1–S5.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) JX315603 and JX315604.
M. A. Matilla and G. P. C. Salmond, unpublished data.
- ACP
- acyl carrier protein
- AT
- acyltransferase
- DH
- dehydratase
- EH
- enoyl-CoA hydratase
- ER
- enoylreductase
- HCS
- 3-hydroxy-3-methylglutaryl-CoA synthase
- KR
- ketoreductase
- KS
- ketosynthase
- MT
- methyltransferase
- NRPS
- nonribosomal peptide synthetase
- Ooc
- oocydin A
- PKS
- polyketide synthase
- TE
- thioesterase
- PDA
- potato dextrose agar.
REFERENCES
- 1. Fisher M. C., Henk D. A., Briggs C. J., Brownstein J. S., Madoff L. C., McCraw S. L., Gurr S. J. (2012) Emerging fungal threats to animal, plant, and ecosystem health. Nature 484, 186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chandler D., Bailey A. S., Tatchell G. M., Davidson G., Greaves J., Grant W. P. (2011) The development, regulation, and use of biopesticides for integrated pest management. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 1987–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lugtenberg B., Kamilova F. (2009) Plant growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556 [DOI] [PubMed] [Google Scholar]
- 4. Raaijmakers J. M., Paulitz T. C., Steinberg C., Alabouvette C., Moënne-Loccoz Y. (2009) The rhizosphere. A playground for soil-borne pathogen and beneficial microorganisms. Plant Soil 321, 341–361 [Google Scholar]
- 5. Srobel G., Li J. Y., Sugawara F., Koshino H., Harper J., Hess W. M. (1999) Oocydin A, a chlorinated macrocyclic lactone with potent anti-oomycete activity from Serratia marcescens. Microbiology 145, 3557–3564 [DOI] [PubMed] [Google Scholar]
- 6. Takada N., Sato H., Suenaga K., Arimoto H., Yamada K., Ueda K., Uemura D. (1999) Isolation and structures of haterumalides NA, NB, NC, ND, and NE, novel macrolides from an Okinawan sponge Ircinia sp. Tetrahedron Lett. 40, 6309–6312 [Google Scholar]
- 7. Thaning C., Welch C. J., Borowicz J. J., Hedman R., Gerhardson B. (2001) Suppression of Sclerotinia sclerotiorum apothecial formation by the soil bacterium Serratia plymuthica. Identification of a chlorinated macrolide as one of the causal agents. Soil Biol. Biochem. 33, 1817–1826 [Google Scholar]
- 8. Sato B., Nakajima H., Fujita T., Takase S., Yoshimura S., Kinoshita T., Terano H. (2005) FR177391, a new anti-hyperlipidemic agent from Serratia. I. Taxonomy, fermentation, isolation, physico-chemical properties, structure elucidation, and biological activities. J. Antibiot. 58, 634–639 [DOI] [PubMed] [Google Scholar]
- 9. Ueda M., Yamaura M., Ikeda Y., Suzuki Y., Yoshizato K., Hayakawa I., Kigoshi H. (2009) Total synthesis and cytotoxicity of haterumalides NA and B and their artificial analogs. J. Org. Chem. 74, 3370–3377 [DOI] [PubMed] [Google Scholar]
- 10. Teruya T., Shimogawa H., Suenaga K., Kigoshi H. (2004) Biselides A and B, novel macrolides from the Okinawan ascidian Didemnidae sp. Chem. Lett. 33, 1184–1185 [Google Scholar]
- 11. Teruya T., Suenaga K., Maruyama S., Kurotaki M., Kigoshi H. (2005) Biselides A–E. Novel polyketides from the Okinawan ascidian Didemnidae sp. Tetrahedron 61, 6561–6567 [Google Scholar]
- 12. Kigoshi H., Kita M., Ogawa S., Itoh M., Uemura D. (2003) Enantioselective synthesis of 15-epi-haterumalide NA methyl ester and revised structure of haterumalide NA. Org. Lett. 5, 957–960 [DOI] [PubMed] [Google Scholar]
- 13. Hoye T. R., Wang J. (2005) Alkyne haloallylation (with Pd(II)) as a core strategy for macrocycle synthesis. A total synthesis of (−)-haterumalide NA/(−)-oocydin A. J. Am. Chem. Soc. 127, 6950–6951 [DOI] [PubMed] [Google Scholar]
- 14. Sattely E. S., Fischbach M. A., Walsh C. T. (2008) Total biosynthesis. In vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat. Prod. Rep. 25, 757–793 [DOI] [PubMed] [Google Scholar]
- 15. Neumann C. S., Fujimori D. G., Walsh C. T. (2008) Halogenation strategies in natural product biosynthesis. Chem. Biol. 15, 99–109 [DOI] [PubMed] [Google Scholar]
- 16. Hertweck C. (2009) The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 48, 4688–4716 [DOI] [PubMed] [Google Scholar]
- 17. Fischbach M. A., Walsh C. T. (2006) Assembly line enzymology for polyketide and nonribosomal peptide antibiotics. Logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 [DOI] [PubMed] [Google Scholar]
- 18. Woodcock D. M., Crowther P. J., Doherty J., Jefferson S., DeCruz E., Noyer-Weidner M., Smith S. S., Michael M. Z., Graham M. W. (1989) Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17, 3469–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herrero M., de Lorenzo V., Timmis K. N. (1990) Transposon vectors containing nonantibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J. Bacteriol. 172, 6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaniga K., Delor I., Cornelis G. R. (1991) A wide-host range suicide vector for improving reverse genetics in Gram-negative bacteria. Inactivation of the blaA gene of Yersinia enterocolitica. Gene 109, 137–141 [DOI] [PubMed] [Google Scholar]
- 21. Demarre G., Guérout A. M., Matsumoto-Mashimo C., Rowe-Magnus D. A., Marlière P., Mazel D. (2005) A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 156, 245–255 [DOI] [PubMed] [Google Scholar]
- 22. Bainton N. J., Stead P., Chhabra S. R., Bycroft B. W., Salmond G. P., Stewart G. S., Williams P. (1992) N-(3-Oxohexanoyl)-l-homoserine lactone regulates carbapenem antibiotic production in Erwinia carotovora. Biochem. J. 288, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hökeberg M., Gerhardson B., Johnsson L. (1997) Biological control of cereal seed-borne diseases by seed bacterization with greenhouse-selected bacteria. Eur. J. Plant Pathol. 103, 25–33 [Google Scholar]
- 24. Berg G., Roskot N., Steidle A., Eberl L., Zock A., Smalla K. (2002) Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl. Environ. Microbiol. 68, 3328–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dennis J. J., Zylstra G. J. (1998) Plasposons. Modular self-cloning minitransposon derivatives for rapid genetic analysis of Gram-negative bacterial genomes. Appl. Environ. Microbiol. 64, 2710–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts K. J. (2010) Quorum Sensing in the Mouse Intestinal Pathogen Citrobacter rodentium. Ph.D. thesis, University of Cambridge, Cambridge, UK [Google Scholar]
- 27. Grinter N. J. (1983) A broad host range cloning vector transposable to various replicons. Gene 21, 133–143 [DOI] [PubMed] [Google Scholar]
- 28. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 29. Van Domselaar G. H., Stothard P., Shrivastava S., Cruz J. A., Guo A., Dong X., Lu P., Szafron D., Greiner R., Wishart D. S. (2005) BASys. A web server for automated bacterial genome annotation. Nucleic Acids Res. 33, W455–W459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Medema M. H., Blin K., Cimermancic P., de Jager V., Zakrzewski P., Fischbach M. A., Weber T., Takano E., Breitling R. (2011) anti-SMASH. Rapid identification, annotation, and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I. (2004) VISTA. Computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Delcher A. L., Harmon D., Kasif S., White O., Salzberg S. L. (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27, 4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz. M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Lu F., Marchler G. H., Mullokandov M., Omelchenko M. V., Robertson C. L., Song J. S., Thanki N., Yamashita R. A., Zhang D. (2011) CDD. A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A., Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fineran P. C., Everson L., Slater H., Salmond G. P. (2005) A GntR family transcriptional regulator (PigT) controls gluconate-mediated repression and defines a new, independent pathway for regulation of the tripyrrole antibiotic, prodigiosin, in Serratia. Microbiology 151, 3833–3845 [DOI] [PubMed] [Google Scholar]
- 36. Coulthurst S. J., Williamson N. R., Harris A. K., Spring D. R., Salmond G. P. (2006) Metabolic and regulatory engineering of Serratia marcescens: mimicking phage-mediated horizontal acquisition of antibiotic biosynthesis and quorum-sensing capacities. Microbiology 152, 1899–1911 [DOI] [PubMed] [Google Scholar]
- 37. Slater H., Crow M., Everson L., Salmond G. P. (2003) Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol. Microbiol. 47, 303–320 [DOI] [PubMed] [Google Scholar]
- 38. Kurz C. L., Chauvet S., Andrès E., Aurouze M., Vallet I., Michel G. P., Uh M., Celli J., Filloux A., De Bentzmann S., Steinmetz I., Hoffmann J. A., Finlay B. B., Gorvel J. P., Ferrandon D., Ewbank J. J. (2003) Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 22, 1451–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O'Toole G. A., Kolter R. (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways. A genetic analysis. Mol. Microbiol. 28, 449–461 [DOI] [PubMed] [Google Scholar]
- 40. Cadel-Six S., Dauga C., Castets A. M., Rippka R., Bouchier C., Tandeau de Marsac N., Welker M. (2008) Halogenase genes in nonribosomal peptide synthetase gene clusters of Microcystis (cyanobacteria). Sporadic distribution and evolution. Mol. Biol. Evol. 25, 2031–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Foulston L. C., Bibb M. J. (2010) Microbisporicin gene cluster reveals unusual features of lantibiotic biosynthesis in actinomycetes. Proc. Natl. Acad. Sci. U.S.A. 107, 13461–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fujimori D. G., Hrvatin S., Neumann C. S., Strieker M., Marahiel M. A., Walsh C. T. (2007) Cloning and characterization of the biosynthetic gene cluster for kutznerides. Proc. Natl. Acad. Sci. U.S.A. 104, 16498–16503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reeves C. D., Hu Z., Reid R., Kealey J. T. (2008) Genes for the biosynthesis of the fungal polyketides hypothemycin from Hypomyces subiculosus and radicicol from Pochonia chlamydosporia. Appl. Environ. Microbiol. 74, 5121–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiao Y., Li S., Niu S., Ma L., Zhang G., Zhang H., Zhang G., Ju J., Zhang C. (2011) Characterization of tiacumicin B biosynthetic gene cluster affording diversified tiacumicin analogs and revealing a tailoring dihalogenase. J. Am. Chem. Soc. 133, 1092–1105 [DOI] [PubMed] [Google Scholar]
- 45. Wagner C., El Omari M., König G. M. (2009) Biohalogenation. Nature's way to synthesize halogenated metabolites. J. Nat. Prod. 72, 540–553 [DOI] [PubMed] [Google Scholar]
- 46. McGowan S. J., Sebaihia M., Porter L. E., Stewart G. S., Williams P., Bycroft B. W., Salmond G. P. (1996) Analysis of bacterial carbapenem antibiotic production genes reveals a novel β-lactam biosynthesis pathway. Mol. Microbiol. 22, 415–426 [DOI] [PubMed] [Google Scholar]
- 47. Harris A. K., Williamson N. R., Slater H., Cox A., Abbasi S., Foulds I., Simonsen H. T., Leeper F. J., Salmond G. P. (2004) The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology 150, 3547–3560 [DOI] [PubMed] [Google Scholar]
- 48. Ramsay J. P., Williamson N. R., Spring D. R., Salmond G. P. (2011) A quorum-sensing molecule acts as a morphogen controlling gas vesicle organelle biogenesis and adaptive flotation in an enterobacterium. Proc. Natl. Acad. Sci. U.S.A. 108, 14932–14937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Williamson N. R., Simonsen H. T., Ahmed R. A., Goldet G., Slater H., Woodley L., Leeper F. J., Salmond G. P. (2005) Biosynthesis of the red antibiotic, prodigiosin, in Serratia. Identification of a novel 2-methyl-3-n-amyl-pyrrole (MAP) assembly pathway, definition of the terminal condensing enzyme, and implications for undecylprodigiosin biosynthesis in Streptomyces. Mol. Microbiol. 56, 971–989 [DOI] [PubMed] [Google Scholar]
- 50. Piel J. (2010) Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 27, 996–1047 [DOI] [PubMed] [Google Scholar]
- 51. Keatinge-Clay A. T., Shelat A. A., Savage D. F., Tsai S. C., Miercke L. J., O'Connell J. D., 3rd, Khosla C., Stroud R. M. (2003) Catalysis, specificity, and ACP-docking site of Streptomyces coelicolor malonyl-CoA:ACP transacylase. Structure 11, 147–154 [DOI] [PubMed] [Google Scholar]
- 52. Yadav G., Gokhale R. S., Mohanty D. (2003) Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 328, 335–363 [DOI] [PubMed] [Google Scholar]
- 53. Dickschat J. S., Vergnolle O., Hong H., Garner S., Bidgood S. R., Dooley H. C., Deng Z., Leadlay P. F., Sun Y. (2011) An additional dehydratase-like activity is required for lankacidin antibiotic biosynthesis. ChemBioChem 12, 2408–2412 [DOI] [PubMed] [Google Scholar]
- 54. Chen X. H., Vater J., Piel J., Franke P., Scholz R., Schneider K., Koumoutsi A., Hitzeroth G., Grammel N., Strittmatter A. W., Gottschalk G., Süssmuth R. D., Borriss R. (2006) Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 188, 4024–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Teta R., Gurgui M., Helfrich E. J., Künne S., Schneider A., Van Echten-Deckert G., Mangoni A., Piel J. (2010) Genome mining reveals trans-AT polyketide synthase directed antibiotic biosynthesis in the bacterial phylum bacteroidetes. ChemBioChem 11, 2506–2512 [DOI] [PubMed] [Google Scholar]
- 56. Menche D., Arikan F., Perlova O., Horstmann N., Ahlbrecht W., Wenzel S. C., Jansen R., Irschik H., Müller R. (2008) Stereochemical determination and complex biosynthetic assembly of etnangien, a highly potent RNA polymerase inhibitor from the myxobacterium Sorangium cellulosum. J. Am. Chem. Soc. 130, 14234–14243 [DOI] [PubMed] [Google Scholar]
- 57. Weber T., Laiple K. J., Pross E. K., Textor A., Grond S., Welzel K., Pelzer S., Vente A., Wohlleben W. (2008) Molecular analysis of the kirromycin biosynthetic gene cluster revealed β-alanine as precursor of the pyridone moiety. Chem. Biol. 15, 175–188 [DOI] [PubMed] [Google Scholar]
- 58. El-Sayed A. K., Hothersall J., Cooper S. M., Stephens E., Simpson T. J., Thomas C. M. (2003) Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem. Biol. 10, 419–430 [DOI] [PubMed] [Google Scholar]
- 59. Piel J. (2002) A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. U.S.A. 99, 14002–14007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pistorius D., Müller R. (2012) Discovery of the rhizopodin biosynthetic gene cluster in Stigmatella aurantiaca Sg a15 by genome mining. ChemBioChem 13, 416–426 [DOI] [PubMed] [Google Scholar]
- 61. Partida-Martinez L. P., Hertweck C. (2007) A gene cluster encoding rhizoxin biosynthesis in Burkholderia rhizoxina, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem. 8, 41–45 [DOI] [PubMed] [Google Scholar]
- 62. Irschik H., Kopp M., Weissman K. J., Buntin K., Piel J., Müller R. (2010) Analysis of the sorangicin gene cluster reinforces the utility of a combined phylogenetic/retrobiosynthetic analysis for deciphering natural product assembly by trans-AT PKS. ChemBioChem 11, 1840–1849 [DOI] [PubMed] [Google Scholar]
- 63. Gurney R., Thomas C. M. (2011) Mupirocin. Biosynthesis, special features and applications of an antibiotic from a Gram-negative bacterium. Appl. Microbiol. Biotechnol. 90, 11–21 [DOI] [PubMed] [Google Scholar]
- 64. Lopanik N. B., Shields J. A., Buchholz T. J., Rath C. M., Hothersall J., Haygood M. G., Håkansson K., Thomas C. M., Sherman D. H. (2008) In vivo and in vitro trans-acylation by BryP, the putative bryostatin pathway acyltransferase derived from an uncultured marine symbiont. Chem. Biol. 15, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jensen K., Niederkrüger H., Zimmermann K., Vagstad A. L., Moldenhauer J., Brendel N., Frank S., Pöplau P., Kohlhaas C., Townsend C. A., Oldiges M., Hertweck C., Piel J. (2012) Polyketide proofreading by an acyltransferase-like enzyme. Chem. Biol. 19, 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mattheus W., Gao L. J., Herdewijn P., Landuyt B., Verhaegen J., Masschelein J., Volckaert G., Lavigne R. (2010) Isolation and purification of a new kalimantacin/batumin-related polyketide antibiotic and elucidation of its biosynthesis gene cluster. Chem. Biol. 17, 149–159 [DOI] [PubMed] [Google Scholar]
- 67. Aparicio J. F., Molnár I., Schwecke T., König A., Haydock S. F., Khaw L. E., Staunton J., Leadlay P. F. (1996) Organization of the biosynthetic gene cluster of rapamycin in Streptomyces hygroscopicus. Analysis of the enzymatic domains in the modular polyketide synthase. Gene 169, 9–16 [DOI] [PubMed] [Google Scholar]
- 68. Reid R., Piagentini M., Rodriguez E., Ashley G., Viswanathan N., Carney J., Santi D. V., Hutchinson C. R., McDaniel R. (2003) A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 42, 72–79 [DOI] [PubMed] [Google Scholar]
- 69. Keatinge-Clay A. (2008) Crystal structure of the erythromycin polyketide synthase dehydratase. J. Mol. Biol. 384, 941–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gu L., Jia J., Liu H., Håkansson K., Gerwick W. H., Sherman D. H. (2006) Metabolic coupling of dehydration and decarboxylation in the curacin A pathway. Functional identification of a mechanistically diverse enzyme pair. J. Am. Chem. Soc. 128, 9014–9015 [DOI] [PubMed] [Google Scholar]
- 71. Konz D., Marahiel M. A. (1999) How do peptide synthetases generate structural diversity? Chem. Biol. 6, R39–R48 [DOI] [PubMed] [Google Scholar]
- 72. Marahiel M. A. (1997) Protein templates for the biosynthesis of peptide antibiotics. Chem. Biol. 4, 561–567 [DOI] [PubMed] [Google Scholar]
- 73. Sudek S., Lopanik N. B., Waggoner L. E., Hildebrand M., Anderson C., Liu H., Patel A., Sherman D. H., Haygood M. G. (2007) Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula,” the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 70, 67–74 [DOI] [PubMed] [Google Scholar]
- 74. Sun Y., Hong H., Gillies F., Spencer J. B., Leadlay P. F. (2008) Glyceryl-S-acyl carrier protein as an intermediate in the biosynthesis of tetronate antibiotics. ChemBioChem. 9, 150–156 [DOI] [PubMed] [Google Scholar]
- 75. Geders T. W., Gu L., Mowers J. C., Liu H., Gerwick W. H., Håkansson K., Sherman D. H., Smith J. L. (2007) Crystal structure of the ECH2 catalytic domain of CurF from Lyngbya majuscula. Insights into a decarboxylase involved in polyketide chain β-branching. J. Biol. Chem. 282, 35954–35963 [DOI] [PubMed] [Google Scholar]
- 76. Piel J., Hui D., Wen G., Butzke D., Platzer M., Fusetani N., Matsunaga S. (2004) Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. U.S.A. 101, 16222–16227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Simunovic V., Zapp J., Rachid S., Krug D., Meiser P., Müller R. (2006) Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthases, and trans-acting acyltransferases. ChemBioChem. 7, 1206–1220 [DOI] [PubMed] [Google Scholar]
- 78. Tang G. L., Cheng Y. Q., Shen B. (2006) Polyketide chain skipping mechanism in the biosynthesis of the hybrid nonribosomal peptide-polyketide antitumor antibiotic leinamycin in Streptomyces atroolivaceus S-140. J. Nat. Prod. 69, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Perlova O., Gerth K., Kaiser O., Hans A., Müller R. (2006) Identification and analysis of the chivosazol biosynthetic gene cluster from the myxobacterial model strain Sorangium cellulosum So ce56. J. Biotechnol. 121, 174–191 [DOI] [PubMed] [Google Scholar]
- 80. Zhao C., Coughlin J. M., Ju J., Zhu D., Wendt-Pienkowski E., Zhou X., Wang Z., Shen B., Deng Z. (2010) Oxazolomycin biosynthesis in Streptomyces albus JA3453 featuring an “acyltransferase-less” type I polyketide synthase that incorporates two distinct extender units. J. Biol. Chem. 285, 20097–20108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bumpus S. B., Magarvey N. A., Kelleher N. L., Walsh C. T., Calderone C. T. (2008) Polyunsaturated fatty-acid-like trans-enoyl reductases utilized in polyketide biosynthesis. J. Am. Chem. Soc. 130, 11614–11616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Anderson J. L., Chapman S. K. (2006) Molecular mechanisms of enzyme-catalyzed halogenation. Mol. BioSyst. 2, 350–357 [DOI] [PubMed] [Google Scholar]
- 83. Podzelinska K., Latimer R., Bhattacharya A., Vining L. C., Zechel D. L., Jia Z. (2010) Chloramphenicol biosynthesis. The structure of CmlS, a flavin-dependent halogenase showing a covalent flavin-aspartate bond. J. Mol. Biol. 397, 316–331 [DOI] [PubMed] [Google Scholar]
- 84. Coulthurst S. J., Kurz C. L., Salmond G. P. (2004) luxS mutants of Serratia defective in autoinducer-2-dependent “quorum sensing” show strain-dependent impacts on virulence and production of carbapenem and prodigiosin. Microbiology 150, 1901–1910 [DOI] [PubMed] [Google Scholar]