Background: APOBEC3A can hyperedit nuclear DNA and generates double-stranded DNA breaks.
Results: TRIB3 is an interactor for APOBEC3A and APOBEC3C. TRIB3 is a negative regulator of APOBEC3A.
Conclusion: Through its control of APOBEC3A, TRIB3 is another guardian of genome integrity.
Significance: TRIB3 is part of protein network that involves cell cycle control, cell survival, DNA repair, and genome stability.
Keywords: Cancer, Enzyme Mutation, Mutagenesis, Mutant, Myc, APOBEC3, TRIB3, Cancer Genomics, Cytidine Deaminase, Editing
Abstract
The human polydeoxynucleotide cytidine deaminases APOBEC3A, APOBEC3C, and APOBEC3H are capable of mutating viral DNA in the nucleus, whereas APOBEC3A alone efficiently edits nuclear DNA. Deamination is rapidly followed by excision of uracil residues and can lead to double-stranded breaks. It is not known to which protein networks these DNA mutators belong. Using a yeast two-hybrid screen, we identified the human homolog of Drosophila Tribbles 3, TRIB3, as an interactor for APOBEC3A and APOBEC3C. The interaction was confirmed by co-affinity purification. Co-transfection of APOBEC3A with a TRIB3 expression vector reduced nuclear DNA editing whereas siRNA knockdown of TRIB3 increased the levels of nuclear DNA editing, indicating that TRIB3 functioned as a repressor of A3A. It also repressed A3A-associated γH2AX positive double-stranded breaks. The interaction results in degradation of A3A in a proteasome-independent manner. TRIB3 has been linked to cancer and via its own interactors and links the A3A DNA mutators to the Rb-BRCA1-ATM network. TRIB3 emerges as an important guardian of genome integrity.
Introduction
The human genome encodes eight cytidine deaminases (A1, A3A–C, A3F–H, AID) capable of mutating DNA of which AID, responsible for immunoglobulin class switch recombination and somatic hypermutation, is perhaps the most widely known example (1, 2). Several of the APOBEC3 (A3) cytidine deaminases can target retroviral cDNA intermediates, notably human immunodeficiency virus (HIV), the result being DNA peppered with cytidine to uridine mutations (3–7). Indeed, APOBEC3F and APOBEC3G (A3F and A3G) proved such a barrier to HIV that it evolved an antagonist encoded by the viral gene, vif (8–10). Recently, it was shown that A3D impacted HIV-1 replication (11, 12). Hepatitis B virus (HBV),4 a nonclassical retrovirus, is also susceptible to restriction by A3 enzymes, particularly in late stage disease, with A3G and probably A3C being the major players (13–17). Unlike HIV, HBV does not encode an A3 antagonist.
DNA viral genomes too can be edited, notably human papillomavirus types 1a and 16 (18), as well as herpes simplex virus type 1 (HSV-1), Epstein-Barr virus (19), and transfusion transmitted virus (20). For human papillomaviruses the restricting molecules are A3A, A3C, and A3H whereas for HSV-1 only A3C edits efficiently. Not surprisingly, the single-stranded (ss) DNA genomes of parvoviruses are vulnerable to A3 editing (21, 22). As such, the A3 enzymes constitute a set of viral restriction factors. This notion is supported by the finding that several A3 genes, notably A3A and A3G, are up-regulated by type I and II interferons (23–26). As all these DNA viruses replicate in the nucleus, the intriguing question as to how A3 enzymes in the nucleus distinguish between viral and host nuclear DNA (nuDNA), if at all, comes to the fore.
Recent work has shown that A3 restriction of viral genomes is only part of the picture (26–28). Human mitochondrial DNA (mtDNA) can be hyperedited in vivo and in tissue culture experiments involving A3 enzymes (28). Editing occurred at relatively low levels in the cytoplasm. At least five A3 enzymes were involved, although as A3C is invariably the most abundant A3 enzyme expressed it is likely that much of the editing could be attributed to it (28). By contrast, A3A is singular in that it can hyperedit nuDNA outside of any microbial context (28). Observed levels of both mtDNA and nuDNA editing are rate-limited by uracil DNA-glycosylase (UNG) for, in the absence of UNG, far more hyperedited mt and nuDNA could be detected (28). Apart from mutating DNA, A3A appears to generate double-stranded DNA breaks (DSBs) thus reflecting the two attributes of AID, i.e. class switch recombination and somatic hypermutation (27, 29). These activities are carefully controlled, and AID interacts with a number of proteins to achieve specificity (30, 31). However, AID can edit nuDNA outside of the rearranged IgV locus and has been implicated in the development of cancers (32). Ectopic expression of AICDA has been noted in a number of cancers and tissues unrelated to germinal center B cells (33, 34). Indeed, AICDA transgenic mice develop cancers, the type depending on the cell tropism of promoter used (35, 36). Similar findings were made for transgenic mice bearing the eponymous APOBEC1 gene (37).
Using the AID paradigm, it is probable that the ssDNA A3 mutators interact with numerous proteins. This is indeed the case for A3G, which is particularly present in cytoplasmic P bodies (38). Interactors for the other APOBEC3 deaminases have not yet been reported. Given our interest in the restriction of DNA viruses replicating in the nucleus and the finding that A3A can edit nuDNA, we set out to identify interactors for the A3A, A3C, and A3H deaminases, all of which have cytoplasmic and nuclear localizations. We found that A3A and A3C interacted with the human homolog to the Drosophila protein, Tribbles 3 (TRIB3) (39). Although it shares homology to serine/threonine kinases such as PIM1, it is actually a pseudokinase (40). TRIB3 is an inhibitor of AKT, a prosurvival kinase (41), is implicated in diabetes (41), and is part of G1/S and G2/M checkpoint control (42, 43). siRNA knockdown of TRIB3 resulted in increased A3A editing of nuDNA whereas overexpression reduced editing, indicating that it was a negative regulator of A3A. TRIB3 emerges as another guardian of genome integrity.
EXPERIMENTAL PROCEDURES
Reagents
siTRIB1 (sc-77704), siTRIB2 (sc-94644), siTRIB3 (sc-44426), and control siRNAs (sc-44237) were from Santa Cruz Biotechnology. Epoxomycin and bafilomycin were from Sigma. TRIB3 shRNA and control shRNA lentiviral particles were from Santa Cruz Biotechnology. A3A antibodies (SAB4500753) were from Sigma. Type I IFN-αA (6.8·108 units/mg) were from PBL Biomedical Laboratories. The A3A constructs have been described previously (28). The TRIB1 and TRIB2 constructs were recloned into pCI-neo-3×FLAG vector. The precise amino acid sequences of the 3×FLAG-tagged TRIB constructs as well as the A3A-NLS construct are given in supplemental Fig. S1. The HEK-293T-UGI cell line has been described (26).
Cells
Approximately 450,000 HeLa cells, HEK-293T, and HEK-293T-UGI were seeded in 6-well plates and transfected 24–48 h later by 2 μg of total plasmid using JetPrime (Polyplus TransfectionTM). 100,000 U937 cells were stimulated by IFN IαA (1000 units/ml) and were transduced by TRIB3 shRNA or control shRNA lentiviral particles (Santa Cruz Biotechnology). After 48 h, total DNA was extracted. For imaging, 25,000 HeLa cells were seeded in Lapteck and transfected 36 h later by 100 ng of total plasmid using FuGENE HD Transfection Reagent (Roche Applied Science).
Plasmids
cDNAs encoding for APOBEC3A, APOBEC3C, APOBEC3G, APOBEC3H, TRIB1, TRIB2, or TRIB3 were amplified by standard PCR (Taq Platinum; Invitrogen) and cloned by in vitro recombination in pDONR207 using Gateway technology (Invitrogen). PCR primers displayed 20–30 specific nucleotides matching ORF extremities so that their Tm is close to 60 °C. The 5′ ends of Gateway forward primers were fused to attB1.1 recombination sequence 5′-GGGGACAACTTTGTACAAAAAAGTTGGCATG-3′, whereas reverse primers were fused to attB2.1 recombination sequence 5′-GGGGACAACTTTGTACAAGAAAGTTGGTTA-3′. Recombination of PCR products into pDONR207 was performed following the manufacturer's recommendation (BP cloning reaction; Invitrogen). All constructs were transformed and amplified in Escherichia coli DH5α strain.
Yeast Two-hybrid Screen
Yeast culture media were prepared as described previously (44). A3A and A3C coding sequences were transferred by in vitro recombination (LR cloning reaction, Gateway technology) from pDONR207 into yeast two-hybrid vector pDEST32 (Invitrogen) to be expressed in fusion downstream of Gal4 binding domain (Gal4-BD). Bait constructs were transformed into AH109 yeast strain (Clontech) using a standard lithium acetate procedure. Spontaneous transactivation of the HIS3 reporter gene was not observed in yeast cells expressing Gal4-BD-APOBEC3C. Consequently, the screen was performed on a synthetic medium lacking histidine (−His medium) and not supplemented with 3-amino-1,2,4-triazole. A mating strategy was used for screening a human spleen cDNA library cloned in the Gal4-AD pPC86 vector (Invitrogen) and previously established into the Y187 yeast strain (Clontech). After 6 days of culture on −His selective medium, [His+] colonies were selected and purified over 3 weeks by culture on selective medium to eliminate false positives. AD-cDNAs were amplified by PCR from zymolase-treated yeast colonies using primers that hybridize within the pPC86 regions flanking cDNA inserts. PCR products were sequenced, and cellular interactors were identified by multiparallel BLAST analysis.
Co-affinity Purification
To perform co-affinity purification experiments, cloned ORFs were transferred from pDONR207 to pDEST27 expression vector (Invitrogen) to achieve GST fusion, and to pCI-neo-3×FLAG vector (45) for 3×FLAG fusion. Tagged proteins were expressed in human HEK-293T cells maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) containing 10% fetal bovine serum, penicillin, and streptomycin at 37 °C and 5% CO2. Cell transfections were achieved with JetPrime reagent following the manufacturer's recommendations. Briefly, 5·105 HEK-293T cells were dispensed in each well of a 6-well plate and transfected 24 h later with 300 ng of the GST constructs and 100 ng of the 3×FLAG constructs. Two days after transfection, HEK-293T cells were washed in phosphate-buffered saline (PBS) and then resuspended in lysis buffer (0.5% Nonidet P-40, 20 mm Tris-HCl, pH 7.4, 120 mm NaCl, and 1 mm EDTA) supplemented with Complete Protease Inhibitor Mixture (Roche Applied Science). Cell lysates were incubated on ice for 20 min and then clarified by centrifugation at 14,000 × g for 30 min. For pulldown analysis, 400 μg of protein extracts were incubated for 1 h at 4 °C with 25 μl of glutathione-Sepharose beads (Amersham Biosciences) to purify GST-tagged proteins. Beads were then washed three times in ice-cold lysis buffer, and proteins were recovered by boiling in denaturing loading buffer (Invitrogen). Purified complexes and protein extracts were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 4–12% NuPAGE Bis-Tris gels with MOPS running buffer (Invitrogen), and transferred to a nitrocellulose membrane. 3×FLAG and GST-tagged proteins were detected using standard immunoblotting techniques. Membranes were blotted with a mouse monoclonal HRP conjugated anti-3×FLAG antibody (M2; Sigma) or a rabbit polyclonal anti-GST antibody (Sigma). Secondary anti-rabbit HRP-conjugated antibody was from Amersham Biosciences.
To detect the physiological interaction between A3A and TRIB3, protein extracts were precleared for 1 h with protein G-Sepharose Fast Flow (Sigma). The supernatant without the sticky protein was incubated for 1 h with TRIB3 antibody (Santa Cruz Biotechnology) or with an antibody with the same isotype of TRIB3 (IgG2b) or without antibody. The complex was incubated for 1 h in presence of protein G-Sepharose Fast Flow. Western blotting was performed in presence of a rabbit anti-A3A antibody at 1/1000 dilution (Sigma) or a mouse anti-TRIB3 antibody at 1/1000 dilution (Santa Cruz Biotechnology) followed by a secondary antibody anti-rabbit or anti-mouse at 1/10,000 (GE Healthcare).
PCR and 3D-PCR
For amplification of human CMYC, the first-round reaction parameters were 95 °C for 5 min followed by 42 cycles (95 °C for 1 min, 57 °C for 45 s, and 72 °C for 1 min) and finally 10 min at 72 °C. Second-round three-dimensional PCR were performed using the equivalent of 0.5 μg of the first-round reaction as input using Eppendorf gradient Mastercycler S. The reaction parameters were 91–95 °C for 5 min followed by 42 cycles (91–95 °C for 1 min, 57 °C for 1 min, and 72 °C for 2 min), and finally 10 min at 72 °C. All DNAs were extracted using the Epicenter kit. All amplifications were performed using first-round standard PCR followed by nested three-dimensional PCR. PCR was performed with 2.5 units of Taq (Bioline) DNA polymerase per reaction. PCR products were cloned using the TOPO vector, and sequencing was outsourced to GATC. The human CMYC primers have been described (28).
Confocal Microscopy
After PBS washing, cell were fixed in 50/50 methanol/acetone for 20 min at room temperature. The anti-V5 antibody (Invitrogen) and anti-3×FLAG antibody (Sigma) were incubated at 1/200 for 1 h at room temperature (Sigma) followed by incubation with fluorescein-conjugated secondary antibody for 1 h at room temperature. Slides were mounted with Vectashield-DAPI. Confocal Imaging was performed using a Zeiss AxioImagerZ2 LSM700. HCX PL APOS63 × 1.4 OIL optics was used.
Western Blotting
Total protein was recovered 24 h after transfection. Western blot analysis was carried out according to standard procedures by using a mouse monoclonal specific for the V5 epitope (Invitrogen) applied for 1 h. After incubation with an anti-mouse IgG horseradish peroxidase-coupled secondary antibody (Amersham Biosciences), the membrane was subjected to detection by enhanced chemiluminescence (Pierce).
PBMC, CD4, and Neutrophil Purification
Ficoll-purified peripheral blood mononuclear cells (PBMCs) were stimulated with phytohemagglutinin (Sigma), IL2 (Sigma) and IFNα (PBL, Biomedical Laboratories). CD4 isolation has been performed with antibody-coated magnetic beads (Miltenyl Biotec). Human neutrophils were isolated from peripheral blood of healthy donors using the human neutrophils enrichment kit (negative selection EasySep Kit), according to the manufacturer's instructions (StemCell Technologies).
Transcriptomes
For the cirrhosis samples the methods have been described (17). For the cell lines and PBMCs, all extractions were performed with RNA EXTRACT RNeasy® Plus Mini Kit (Qiagen). The APOBEC3 primers and probes have been described (25). The TRIB3 oligonucleotides were designed using Roche Applied Science probe finder version 2.43: TRIB3, 5′-GTCTTCGCTGACCGTGAGA; TRIB3, 3′-CAGTCAGCAGCGAGGAGTC; probe 67, TGCTGGAG.
FACS Analysis of Double-stranded Breaks
24 h after transfection, cells were washed with PBS, fixed in 2–4% ice-cold paraformaldehyde (Electron Microscopy Sciences) for 10 min, and permeabilized in 90% ice-cold methanol (Sigma) for 30 min. After washing with PBS, cells were incubated with 1:200 diluted mouse anti-V5 antibody (Invitrogen) for 1 h followed by incubation with 1:500 diluted Alexa Fluor 633 F(ab′)2 fragment of goat anti-mouse IgG (H+L) (Invitrogen) for 45 min. DNA double-stranded breaks were analyzed by staining for 1 h with 1:50 diluted Alexa Fluor 488-conjugated rabbit monoclonal anti-γH2AX (20E3) antibody (Cell Signaling). All incubation steps were performed on ice. 104 cells were analyzed on FACSCalibur using CellQuest Pro (BD Biosciences, version 5.2) and FlowJo software (Tree Star, Inc., version 8.7.1).
RESULTS
Tribbles Homolog 3 Is an Interactor for A3A and A3C
Complete A3A, A3C, and A3H cDNA coding sequences were cloned in a yeast vector to be used as bait in the two-hybrid system. A3 ORFs were expressed as fusion proteins downstream of the Gal4-BD. We failed to obtain yeast colonies with Gal4-BD-A3A, suggesting that A3A is toxic in yeast. This interpretation is supported by viable expression of the A3AC101S catalytic mutant in the same system. By contrast, Gal4-BD-A3C and Gal4-BD-A3H constructs did not interfere with yeast growth and were used as bait to screen a human cDNA library made from human spleen (Invitrogen) for partners. A3H failed to capture any prey proteins. By contrast, numerous yeast colonies were obtained on selective medium with the Gal4-BD-A3C construct, and interacting prey proteins were identified by PCR, sequencing, and multiparallel BLAST analysis. From among 2·107 colonies screened from a library made from human spleen (Invitrogen), we recovered 4 hits for the human TRIB3 first identified for Drosophila. All hits were full-length cDNAs showing two known synonymous polymorphisms at codons 111 and 320. In more than 250 screens performed using different bait proteins of either viral or human origin, this is the first time we have identified TRIB3.
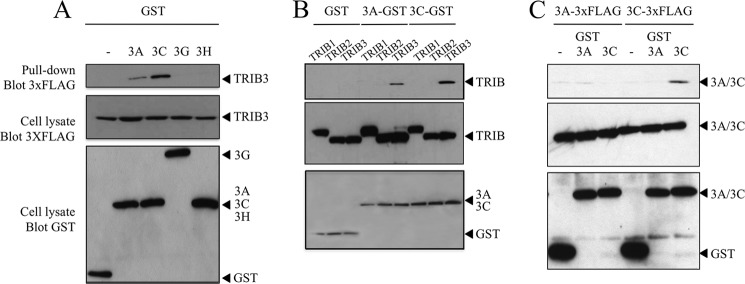
To confirm the interaction, we performed co-affinity purification experiments (GST pulldown). HEK-293T cells were co-transfected by a 5′ 3×FLAG-TRIB3 construct along with GST-A3A, GST-A3C, GST-A3H, and GST-A3G as negative control. As can be seen from Fig. 1A, A3C interacted strongly with TRIB3 whereas for A3A the interaction was weaker. Human A3H and the A3G negative control failed to pull down TRIB3. To confirm the specificity of the A3A and A3C co-affinity purification, we explored the possible interaction with either of the other two human Tribbles homologs, TRIB1 and TRIB2 which show ∼46 and 48% amino acid identity with TRIB3 (supplemental Fig. S1A). No interaction was found with either protein confirming the specificity of the A3A/TRIB3 and A3C/TRIB3 interactions (Fig. 1B). Although TRIB1 is nuclear, no co-localization with A3A was observed whereas for TRIB2 a nonspecific co-localization was noted and follows on from the fact that both TRIB2 and A3A are partitioned between the nucleus and cytoplasm (supplemental Fig. S2A). As some APOBEC3 proteins homo- and heterodimerize, notably A3B, A3F, and A3G (2, 9), we tested the possibility that A3A and A3C could form dimers. In the same co-affinity purification format, A3C formed homodimers but not heterodimers with A3A, whereas A3A failed to do either (Fig. 1C). It is possible that the apparent stronger interaction between TRIB3 and A3C might reflect greater avidity, given A3C dimer formation. To see whether A3C affect the A3A/TRIB3 interaction, HEK-293T cells were transfected with A3A-GST (300 ng) and 3×FLAG-TRIB3 (100 ng) and with an increasing amount of 3×FLAG-A3C (50, 200, 800 ng). After 24 h, cells were lysed with a radioimmune precipitation assay buffer, and total proteins were extracted. We proceeded to a co-purification affinity and observed that an increased concentration of A3C did not affect the A3A/TRIB3 interaction (supplemental Fig. S3A).
FIGURE 1.
APOBEC3A and 3C interact with human TRIB3 protein. A, GST pulldowns were followed by Western blotting with antibodies to 3×FLAG. Below are the TRIB3 and APOBEC3 controls. B, interaction was specific for TRIB3 compared with the paralogous human proteins TRIB1 and TRIB2 (see supplemental Fig. S1A for sequence comparisons). C, APOBEC3C but not APOBEC3A can form multimers.
Tribbles 3 Inhibits A3A Editing of Nuclear DNA
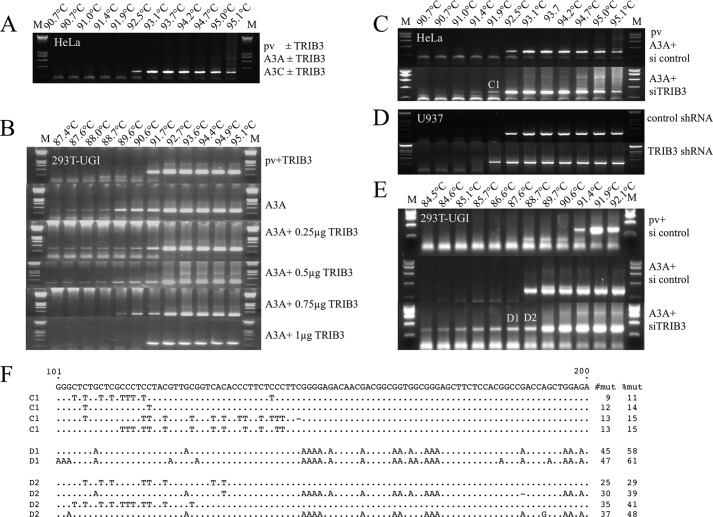
HeLa cells were co-transfected with TRIB3 and V5-tagged A3 expression plasmids with recovery of cytidine deamination of CMYC DNA as readout (28). Normally transfection with A3A alone followed by 3D-PCR (46) fails to recover hyperedited CMYC from HeLa cell DNA (28). Co-transfection with TRIB3 did not change the situation (Fig. 2A). Detection of CMYC editing represents a dynamic between A3A editing and DNA catabolism by a UNG initiated pathway, the latter being rate-limiting. HEK-293T-UGI cells, which constitutively express the UNG inhibitor, UGI (47), allow detection of edited CMYC DNA following A3A transfection (28). Accordingly, HEK-293T-UGI cells were co-transfected by A3 expression plasmids with and without TRIB3. Now, TRIB3 expression proved to inhibit A3A editing of CMYC in a dose-dependent manner (Fig. 2B). As previously noted, transfection with A3C, A3H, and A3G had no impact on CMYC editing, a finding that was not altered either by co-transfection with TRIB3 or with TRIB1 or TRIB2 (supplemental Fig. S2, B and C). Given that two-hybrid screens pick up a subset of protein interactors, the remaining APOBEC3 enzymes were tested, notably A3B and A3F with and without all three TRIB constructs (supplemental Figs. S2, B and C). Again, no evidence of CMYC editing was identified by the nested PCR/3D-PCR protocol.
FIGURE 2.
TRIB3 protects whereas TRIB3 siRNA knockdown increases A3A deamination of nuDNA. A, no apparent deamination of CMYC DNA by A3A or A3C ± TRIB3 in HeLa cells was due to rate-limiting UNG degradation. B, in HEK-293T-UGI cells TRIB3 protects nuDNA from editing by A3A in a dose-response manner. DNA concentrations were 2 μg throughout and were compensated for by plasmid vector. C, co-transfection of A3A with 1 μg of TRIB3 siRNA knockdown only results in recovery of hyperedited CMYC DNA in HeLa cells. D, transduction of IFN IαA stimulated U937 cells with TRIB3 shRNA or control shRNA lentiviral particles. E, co-transfection of A3A with 1 μg of TRIB3 siRNA knockdown only results in recovery of enhanced hyperediting of CMYC DNA in 293T-UGI cells. F, a collection of hyperedited CMYC sequences was recovered from samples C1, D1, and D2 (C and E). Only differences are shown for 100 bases of a total of 241 bp. To the right are the number mutations per sequence and the percentages of Cs or Gs (Cs on opposite strand) edited.
If TRIB3 was restricting A3A activity then TRIB3 siRNA knockdown should enhance A3A editing of CMYC. This is, indeed the case for HeLa cells (Fig. 2, C and F), or for IFN IαA-stimulated U937 cells transduced with TRIB3 shRNA or control shRNA lentiviral particles (Fig. 2D). Hyperedited CMYC was obtained at restrictive temperature of 91.4 °C and 91.9 °C in the presence of TRIB3 shRNA (Fig. 2D). For the HEK-293T-UGI cell line co-transfection of A3A+siTRIB3 allowed recovery of hyperedited CMYC DNA down to 84.5 °C, the lowest CMYC 3D-PCR denaturation temperature from any experimental conditions identified to date (Fig. 2, E and F). Efficiency of TRIB3 siRNA was performed and presented in supplemental Fig. S3H. A collection of hyperedited CMYC sequences recovered from samples C1, D1, and D2 is presented in Fig. 2F. This result is worth emphasizing for it is the first time we have recovered edited CMYC DNA without recourse to either UNG knockdown, or UNG−/− cell lines (28). TRIB3 siRNA knockdown failed to reveal any A3C or A3H-associated nuDNA editing (supplemental Fig. S2, B and C) confirming our previous negative findings even with UGI (28). Indeed, co-transfection of HEK-293T-UGI cells by A3C or A3H along with siRNAs for TRIB3, TRIB2 or TRIB1 failed to yield hyperedited CMYC DNA, emphasizing the singularity of the A3A interaction with TRIB3 (supplemental Fig. S2C).
Cell Localization of A3A, A3C, and TRIB3
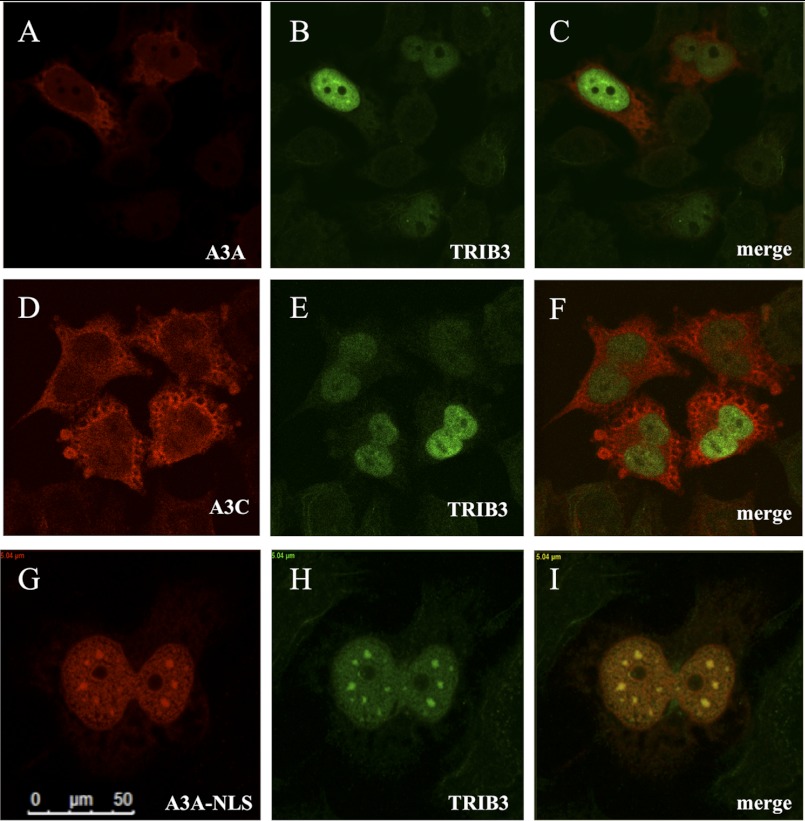
The localization of overexpressed A3A-V5 and A3C-V5 in HeLa cells is mainly cytoplasmic with a nuclear component, whereas 3×FLAG-TRIB3 is exclusively nuclear (supplemental Fig. S4B) in keeping with published reports (21, 27, 48). Co-transfection of A3A+TRIB3 showed neither substantial relocalization of A3A to the nucleus nor strong co-localization within the nucleus, indicating that TRIB3 is not transporting A3A to the nucleus (Fig. 3, A–C). The same was true for A3C (Fig. 3, D–F). To force the system, we tagged A3A with the SV40 TAg nuclear localization signal (NLS, residues PPKKKRKV). Now, most A3A-NLS-V5 was located in the nucleus and co-localized with 3×FLAG-TRIB3 in a punctuate distribution (Fig. 3, G–I). As expected, A3A-NLS hyperedited CMYC in the HEK-293T-UGI cell line to a similar degree compared with A3A, whereas co-transfection with TRIB3 reduced the degree of editing (Fig. 4B and supplemental Fig. S4A).
FIGURE 3.
Cell localization of the A3A and TRIB3. A–C, confocal microscopy yielding scant evidence of cytoplasmic and slightly nuclear A3A-V5 co-localizing with a nuclear 3×FLAG-TRIB3 in HeLa cells 36 h after transfection. D–F, A3C, TRIB3, and merge. G–I, addition of the SV40 NLS (residues PPKKKRKV) to the carboxyl terminus of A3A co-localization with TRIB3 is readily observed.
FIGURE 4.
The A3A interaction involves the amino terminus of TRIB3. A, immunofluorescence of TRIB3, its cytoplasmic ΔNLS derivative, and the murine homolog, mTRIB3. B, CMYC editing by A3A, A3A-NLS, and modulation by an amino-terminal 36-residue deletion of hTRIB3. C, GST pulldown analyses for the A3A+TRIB3, +TRIB3ΔNLS, and +mTRIB3 constructs. WB, Western blot.
The TRIB3 Amino Terminus Modulates the Interaction with A3A
TRIB3 encodes numerous basic residues at its amino terminus that resemble a bipartite nuclear localization signal (supplemental Fig. S1B). If correct, we reasoned that a 3×FLAG-TRIB3ΔNLS construct should no longer be capable of inhibiting A3A deamination of nuDNA. Deletion of the first 36 residues yielded a protein that was strictly cytoplasmic (Fig. 4A). Co-transfection of HEK-293T-UGI cells with A3A showed that the 3×FLAG-TRIB3ΔNLS abrogated A3A editing on nuDNA (Fig. 4B and supplemental Fig. S4A), an experiment performed independently four times with the same result suggesting that A3A was functionally restricted by 3×FLAG-TRIB3ΔNLS. In agreement with this, GST pulldown experiments revealed a greatly reduced interaction between the two partners (Fig. 4C). Murine TRIB3 shows ∼74% overall homology to its human TRIB3 counterpart (supplemental Fig. S1B) and is also exclusively nuclear (Fig. 4A). Interestingly, co-transfection of HEK-293T-UGI cells with mTRIB3 and A3A abrogated A3A editing of CMYC DNA like its human counterpart (Fig. 4B and supplemental Fig. S4A). Finally, by co-affinity purification, we have also demonstrated the interaction between A3A and mTRIB3 (Fig. 4C).
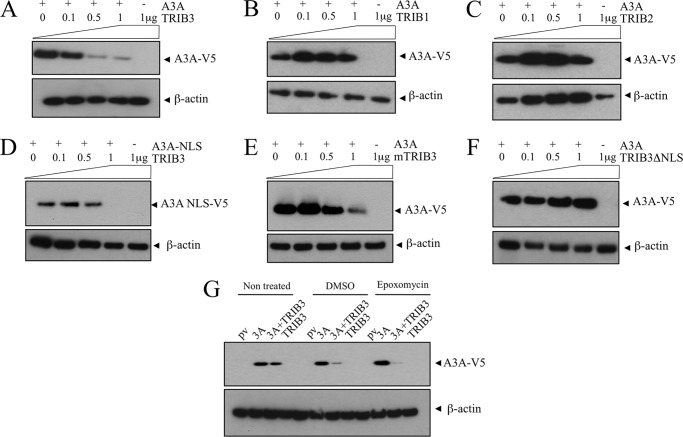
Tribbles 3 Reduces Steady-state Levels of A3A
Clearly TRIB3 and A3A interact but apparently do not co-localize extensively. The amino-terminal pseudo-NADPH binding domain of TRIB3 is proline-rich (19/143, 13% versus carboxyl-terminal 15/215, 7%) which is reminiscent of some highly labile proteins. Indeed, mouse TRIB3 has an extraordinarily short half-life compared with other mouse proteins (49). To see whether TRIB3 expression had any impact on A3A protein levels, HEK-293T cells were co-transfected by V5-tagged A3A and increasing amounts of 3×FLAG-TRIB3. A dose-dependent reduction of A3A steady-state levels was noted whereby at approximately equimolar plasmid concentrations A3A was reduced compared with A3A alone (Fig. 5A). This destabilization was specific to TRIB3 compared with TRIB1 and TRIB2 taken as controls (Fig. 5, B and C). The same experiment was performed with A3C with increased amounts of 3×FLAG-TRIB3 (0, 0.1, 0.5, 1 μg). Similarly to A3A, a dose-dependent reduction of A3C steady-state levels was noted and was specific to TRIB3 compared with TRIB1 and TRIB2 (supplemental Fig. S3, B–D).
FIGURE 5.
TRIB3 interaction with A3A results in lower steady-state levels. A, 293T cells were transfected by A3A, and increasing concentrations of TRIB3 and steady-state levels of A3A were determined by Western blotting at 24 h. The β-actin loading controls are shown below. B and C, TRIB1 and TRIB2 do not degrade A3A. D, the A3A-NLS construct is similarly degraded. E, mTRIB3 is capable of degrading hA3A. F, by contrast, deletion of the amino-terminal 36 residues of TRIB3 which include a NLS abrogates A3A degradation. G, epoxomycin treatment of 293T cells fails to abrogate A3A degradation by TRIB3.
It is worth noting that this situation (1 μg of each plasmid) corresponds to that in Fig. 3, A–C, where there was little co-localization of A3A and TRIB3. The same phenomenon occurs for the A3A-NLS construct (Fig. 5D), and to a lesser degree for mouse TRIB3 (Fig. 5E). However, with deletion of the TRIB3 NLS motif, A3A degradation ceased (Fig. 5F). Treatment of HEK-293T cells with 10 μm epoxomycin, a proteasome inhibitor, resulted in more V5-tagged A3A as previously noted (Fig. 5G) (50). By contrast, A3A levels decreased when co-expressed with TRIB3, indicating that the A3A-TRIB3 complex is not degraded by the proteasome (Fig. 5G), the same phenomenon was observed with A3C-TRIB3 (supplemental Fig. S3E). Moreover treatment with 100 nm bafilomycin, a specific inhibitor of vacuolar-type H+-ATPase, failed to abrogate A3A or A3C degradation by TRIB3 (supplemental Fig. S3F).
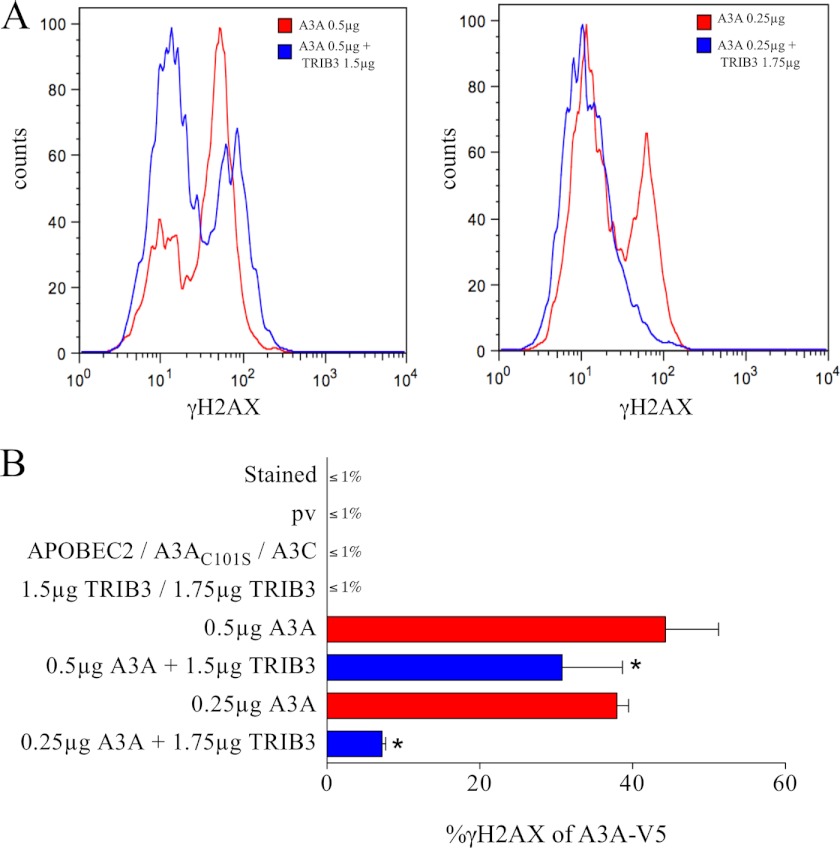
TRIB3 Protects the Cell against A3A-induced Double-stranded DNA Breaks
Cytidine deamination of DNA can also result in DSBs and follows on from UNG excision of uracil bases and subsequent APE1 cleavage of the DNA backbone at abasic sites (29, 51). This is the case for AID and more recently A3A (27, 52, 53). The γ-phosphorylated form of H2AX is an excellent marker for DSBs (54). As can be seen from Fig. 6, transfection of HeLa cells by A3A resulted in numerous DSBs as evidenced by γH2AX as a function of A3A-V5-positive cells. By contrast, co-transfection of A3A with TRIB3 resulted in lower numbers of γH2AX-positive cells and then with lower mean γH2AX intensity. Overexpression of the A3AC101S catalytic mutant, TRIB3 alone, APOBEC2, devoid of any cytidine deaminase activity, or the vector control failed to induce DSBs (Fig. 6B). The greater the ratio of TRIB3 to A3A, the smaller the fraction of DSBs. That A3C did not generate DSBs is consistent with its failure to deaminate nuDNA efficiently (28 and Fig. 6B).
FIGURE 6.
A3A-induced DSBs can be countered by TRIB3. A, superposition of FACS analysis of γH2AX-positive HeLa cells gated on the V5-tagged A3A with and without TRIB3 at two different A3A:TRIB3 ratios. B, means and standard deviations γH2AX-positive cell frequencies for quadruplicate transfections, DNA concentrations were always 2 μg and compensated for by plasmid vector. * indicates a statistically significant difference between two observed percentages (p < 0.05).
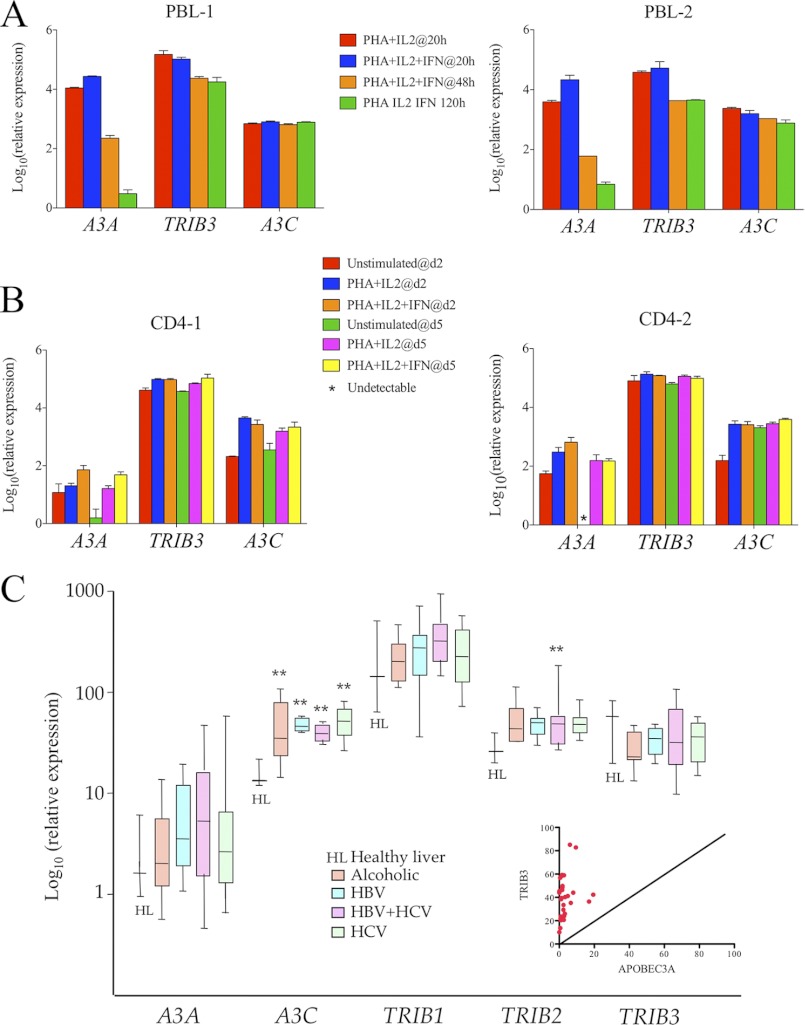
Relative A3A and TRIB3 Expression Levels
To appreciate the relative expression levels of A3A and TRIB3, a TaqMan transcriptional study was made of A3 and TRIB3 genes using the RPL13A gene as reference (25, 26). In four established cell lines A3A levels were essentially zero excluding any meaningful comparison. Those for A3C were always >TRIB3 (supplemental Fig. S2D). In two donors, activated human PBMCs IFNα treatment up-regulated A3A (Fig. 7A) as previously noted (24–26). However, TRIB3 levels always exceeded those of A3A. The striking loss of A3A levels over time could reflect the loss of polymorphonuclear and myeloid cells which invariably express high levels of A3A rather than specific down-regulation of the gene. By contrast, in purified CD4+ T cells from two donors, gene levels were far more stable, and once again TRIB3 levels always exceeded those of A3A (Fig. 7B). To prove that A3A/TRIB3 interaction is physiological, we immunoprecipitated TRIB3 from human neutrophils isolated from peripheral blood of healthy donors, and A3A was detected with anti-A3A antibody. As control, anti-TRIB3 was also detected from the same complex (supplemental Fig. S3G). Hence, A3A/TRIB3 detection in neutrophil granulocytes suggests that this endogenous interaction is physiologically relevant (supplemental Fig. S3G).
FIGURE 7.
Transcriptome analysis of A3A, A3C, and TRIB3 in hematopoietic cells and peritumoral cirrhotic tissues. A, TaqMan analysis of PBMC from two healthy donors at 20, 48, and 120 h after stimulation. B, TaqMan analysis of positively selected CD4+ T cells from two healthy donors at days 2 and 5. C, ABI chip analysis of A3A, A3C, TRIB1–3 in peritumoral tissues from patients with alcoholic, HBV-, HBV+hepatitic C virus (HCV)-, and HCV-associated cirrhosis. The inset gives paired A3A and TRIB3 levels from the same samples. **, statistically significant difference compared with healthy liver (p < 0.01). Expression levels are all normalized to RPL13A levels.
To compare the A3A, A3C, and TRIB1–3 expression in inflamed tissue, a TaqMan PCR chip was performed on 41 cirrhotic samples and was compared with 4 healthy livers (17). The RPL13A gene was as used as reference. Fig. 7C provides the TRIB1, TRIB2, and TRIB3 transcriptomes along with those for A3A and A3C. Although A3C was strongly up-regulated, there were few overall changes in any on the three Tribbles gene levels (Fig. 7D). When the A3A and TRIB3 levels were analyzed for individual samples, TRIB3 levels were always greater than those of A3A (Fig. 7D inset).
DISCUSSION
TRIB3 was identified as an interactor for both A3A and A3C. Given that A3A alone of all A3 enzymes can edit nuclear DNA (28), we concentrated on the A3A/TRIB3 interaction and its consequences. TRIB3, which is essentially nuclear, effectively prevents A3A from editing nuDNA, essentially mopping up A3A being translocated to the nucleus, so protecting the genome. Nonetheless, some editing occurs when A3A is overexpressed as revealed by using the HEK-293-UGI cell line or co-transfection with siTRIB3. This suggests that A3A editing of nuDNA depends to some extent on the relative levels of A3A and TRIB3. In established cell lines, A3A levels are very low indeed whereas TRIB3 levels are comparable with those in some in vivo settings. A3A gene expression can be up-regulated by phorbol esters, poly(I:C), and interferon (24–26). In turn, TRIB3 is induced by endoplasmic reticulum stress (55) whereas Helicobacter pylori lipopolysaccharide TLR-2 mediated signal transduction can down-regulate TRIB3 (56). By contrast, overall TRIB3 levels were not altered in four types of human cirrhosis (Fig. 7C). This complex situation between relative levels of A3A and TRIB3 expression is probably further impacted by A3C levels for A3C strongly interacts with TRIB3 (Fig. 1C). Although the A3C gene is insensitive to type I interferons, its levels are 3–5-fold increased in liver cirrhosis (17). By titrating out TRIB3 elevated A3C levels may allow A3A to mutate nuDNA.
From a mutational perspective, however, the situation is rather simple: a balance in favor of TRIB3 would maintain genome stability, the status quo, whereas the converse would impact the genome. Indeed, fluctuations in the balance could result in waves of A3A editing. Although A3A nuDNA hyperediting may result in cell death, we have argued that low levels, hypoediting, may result in DNA repair with only a handful of mutations remaining, selection acting on the resulting mutant population. Many forms of diabetes are considered to be an inflammatory disease, whereas there is a higher incidence of cancer on a background of diabetes (57, 58). It would be interesting to assess A3A, A3C, TRIB3 gene expression in a number of chronic inflammatory diseases in cross-sectional and longitudinal senses. By contrast, if tumor tissue was the reference tissue, it may be that negative selection against a DNA mutator phenotype at this advanced stage may be advantageous. With this in mind, it is interesting to note that A3A levels in established cell lines are almost inexistent, which fits with the observation that high level, constitutively expressing A3A cell lines have not been obtained.
TRIB3 interacts with a wide variety of proteins, the majority being connected with DNA repair, or coupled to proteins linked to the maintenance of DNA stability and DNA repair (40). For example, TRIB3 is a G1/S and G2/M checkpoint protein (42, 43). Several studies have pointed to a role for TRIB3 in human cancer to the point that it is considered as a tumor susceptibility gene. Importantly, TRIB3 interacts with CtIP, a nuclear cofactor of the transcriptional repressor CtBP (59). CtIP interacts with tumor suppressors such as the Rb family members and BRCA1 through binding sites that are frequently mutated in human cancers (60). In short, TRIB3 is part of the broad CtIP-Rb-BRCA1-ATM protein network that involves cell cycle control, cell survival, DNA repair, and genome stability. Finally the mixed lineage kinase MLK3 has been shown to interact with TRIB3, and attenuation of either compromised mitochondrial integrity and suppressed cellular survival mechanisms via TRB3-dependent inhibition of Akt (61). Three nonsense polymorphisms of TRIB3 have been noted, namely T27stop, Q40stop, and Q84stop, all mapping to the amino-terminal pseudo-NADPH-binding domain (supplemental Fig. S1B). So far there are no reports of TRIB3−/− or TRIB3+/− associated with cancer, but any depletion of TRIB3 levels could increase albeit slightly the frequency of nuDNA hypoediting. In conclusion, through its control of APOBEC3A the present findings identify TRIB3 as another guardian of genome integrity.
Acknowledgments
We thank Drs. Endre Kiss-Toth for human TRIB1 and 2 and murine TRIB3 plasmids, Emmanuel Birlouez for three APOBEC3 plasmids, and Agnès Marchio for formatting Fig. 7C.
This work was supported by grants from Institut Pasteur and the CNRS. The Molecular Retrovirology Unit is équipe labellisé par la Ligue pour la Recherche contre le Cancer.

This article contains supplemental supplemental Figs. S1–S4.
- HBV
- hepatitis B virus
- Bis-Tris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- DSB
- double-stranded break
- Gal4-BD
- Gal4-binding domain
- mtDNA
- mitochondrial DNA
- NLS
- nuclear localization signal
- nuDNA
- nuclear DNA
- PBMC
- peripheral blood mononuclear cell
- TRIB3
- Tribbles 3
- UNG
- uracil DNA-glycosylase.
REFERENCES
- 1. Di Noia J. M., Neuberger M. S. (2007) Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76, 1–22 [DOI] [PubMed] [Google Scholar]
- 2. Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N. (2002) An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79, 285–296 [DOI] [PubMed] [Google Scholar]
- 3. Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. (2003) DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809 [DOI] [PubMed] [Google Scholar]
- 4. Lecossier D., Bouchonnet F., Clavel F., Hance A. J. (2003) Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300, 1112. [DOI] [PubMed] [Google Scholar]
- 5. Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. (2003) Broad antiretroviral defense by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 [DOI] [PubMed] [Google Scholar]
- 6. Mariani R., Chen D., Schröfelbauer B., Navarro F., König R., Bollman B., Münk C., Nymark-McMahon H., Landau N. R. (2003) Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31 [DOI] [PubMed] [Google Scholar]
- 7. Zhang H., Yang B., Pomerantz R. J., Zhang C., Arunachalam S. C., Gao L. (2003) The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424, 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002) Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 [DOI] [PubMed] [Google Scholar]
- 9. Wiegand H. L., Doehle B. P., Bogerd H. P., Cullen B. R. (2004) A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23, 2451–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng Y. H., Irwin D., Kurosu T., Tokunaga K., Sata T., Peterlin B. M. (2004) Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78, 6073–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hultquist J. F., Lengyel J. A., Refsland E. W., LaRue R. S., Lackey L., Brown W. L., Harris R. S. (2011) Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J. Virol. 85, 11220–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Refsland E. W., Hultquist J. F., Harris R. S. (2012) Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 8, e1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Noguchi C., Ishino H., Tsuge M., Fujimoto Y., Imamura M., Takahashi S., Chayama K. (2005) G to A hypermutation of hepatitis B virus. Hepatology 41, 626–633 [DOI] [PubMed] [Google Scholar]
- 14. Rösler C., Köck J., Kann M., Malim M. H., Blum H. E., Baumert T. F., von Weizsäcker F. (2005) APOBEC-mediated interference with hepadnavirus production. Hepatology 42, 301–309 [DOI] [PubMed] [Google Scholar]
- 15. Suspène R., Guétard D., Henry M., Sommer P., Wain-Hobson S., Vartanian J. P. (2005) Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 8321–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Turelli P., Mangeat B., Jost S., Vianin S., Trono D. (2004) Inhibition of hepatitis B virus replication by APOBEC3G. Science 303, 1829. [DOI] [PubMed] [Google Scholar]
- 17. Vartanian J. P., Henry M., Marchio A., Suspène R., Aynaud M. M., Guétard D., Cervantes-Gonzalez M., Battiston C., Mazzaferro V., Pineau P., Dejean A., Wain-Hobson S. (2010) Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog. 6, e1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vartanian J. P., Guétard D., Henry M., Wain-Hobson S. (2008) Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 320, 230–233 [DOI] [PubMed] [Google Scholar]
- 19. Suspène R., Aynaud M. M., Koch S., Pasdeloup D., Labetoulle M., Gaertner B., Vartanian J. P., Meyerhans A., Wain-Hobson S. (2011) Genetic editing of herpes simplex virus 1 and Epstein-Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo. J. Virol. 85, 7594–7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsuge M., Noguchi C., Akiyama R., Matsushita M., Kunihiro K., Tanaka S., Abe H., Mitsui F., Kitamura S., Hatakeyama T., Kimura T., Miki D., Hiraga N., Imamura M., Takahashi S., Hayses C. N., Chayama K. (2010) G to A hypermutation of TT virus. Virus Res. 149, 211–216 [DOI] [PubMed] [Google Scholar]
- 21. Bulliard Y., Narvaiza I., Bertero A., Peddi S., Röhrig U. F., Ortiz M., Zoete V., Castro-Díaz N., Turelli P., Telenti A., Michielin O., Weitzman M. D., Trono D. (2011) Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J. Virol. 85, 1765–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen H., Lilley C. E., Yu Q., Lee D. V., Chou J., Narvaiza I., Landau N. R., Weitzman M. D. (2006) APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16, 480–485 [DOI] [PubMed] [Google Scholar]
- 23. Bonvin M., Achermann F., Greeve I., Stroka D., Keogh A., Inderbitzin D., Candinas D., Sommer P., Wain-Hobson S., Vartanian J. P., Greeve J. (2006) Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43, 1364–1374 [DOI] [PubMed] [Google Scholar]
- 24. Koning F. A., Newman E. N., Kim E. Y., Kunstman K. J., Wolinsky S. M., Malim M. H. (2009) Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83, 9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Refsland E. W., Stenglein M. D., Shindo K., Albin J. S., Brown W. L., Harris R. S. (2010) Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 38, 4274–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stenglein M. D., Burns M. B., Li M., Lengyel J., Harris R. S. (2010) APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 17, 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Landry S., Narvaiza I., Linfesty D. C., Weitzman M. D. (2011) APOBEC3A can activate the DNA damage response and cause cell cycle arrest. EMBO Rep. 12, 444–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suspène R., Aynaud M. M., Guétard D., Henry M., Eckhoff G., Marchio A., Pineau P., Dejean A., Vartanian J. P., Wain-Hobson S. (2011) Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc. Natl. Acad. Sci. U.S.A. 108, 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bross L., Muramatsu M., Kinoshita K., Honjo T., Jacobs H. (2002) DNA double-strand breaks: prior to but not sufficient in targeting hypermutation. J. Exp. Med. 195, 1187–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Conticello S. G., Ganesh K., Xue K., Lu M., Rada C., Neuberger M. S. (2008) Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol. Cell 31, 474–484 [DOI] [PubMed] [Google Scholar]
- 31. Ta V. T., Nagaoka H., Catalan N., Durandy A., Fischer A., Imai K., Nonoyama S., Tashiro J., Ikegawa M., Ito S., Kinoshita K., Muramatsu M., Honjo T. (2003) AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat. Immunol. 4, 843–848 [DOI] [PubMed] [Google Scholar]
- 32. Kou T., Marusawa H., Kinoshita K., Endo Y., Okazaki I. M., Ueda Y., Kodama Y., Haga H., Ikai I., Chiba T. (2007) Expression of activation-induced cytidine deaminase in human hepatocytes during hepatocarcinogenesis. Int. J. Cancer 120, 469–476 [DOI] [PubMed] [Google Scholar]
- 33. Komori J., Marusawa H., Machimoto T., Endo Y., Kinoshita K., Kou T., Haga H., Ikai I., Uemoto S., Chiba T. (2008) Activation-induced cytidine deaminase links bile duct inflammation to human cholangiocarcinoma. Hepatology 47, 888–896 [DOI] [PubMed] [Google Scholar]
- 34. Matsumoto Y., Marusawa H., Kinoshita K., Endo Y., Kou T., Morisawa T., Azuma T., Okazaki I. M., Honjo T., Chiba T. (2007) Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med. 13, 470–476 [DOI] [PubMed] [Google Scholar]
- 35. Okazaki I. M., Hiai H., Kakazu N., Yamada S., Muramatsu M., Kinoshita K., Honjo T. (2003) Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 197, 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takai A., Toyoshima T., Uemura M., Kitawaki Y., Marusawa H., Hiai H., Yamada S., Okazaki I. M., Honjo T., Chiba T., Kinoshita K. (2009) A novel mouse model of hepatocarcinogenesis triggered by AID-causing deleterious p53 mutations. Oncogene 28, 469–478 [DOI] [PubMed] [Google Scholar]
- 37. Yamanaka S., Balestra M. E., Ferrell L. D., Fan J., Arnold K. S., Taylor S., Taylor J. M., Innerarity T. L. (1995) Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc. Natl. Acad. Sci. U.S.A. 92, 8483–8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gallois-Montbrun S., Kramer B., Swanson C. M., Byers H., Lynham S., Ward M., Malim M. H. (2007) Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81, 2165–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mayumi-Matsuda K., Kojima S., Suzuki H., Sakata T. (1999) Identification of a novel kinase-like gene induced during neuronal cell death. Biochem. Biophys. Res. Commun. 258, 260–264 [DOI] [PubMed] [Google Scholar]
- 40. Hegedus Z., Czibula A., Kiss-Toth E. (2007) Tribbles: a family of kinase-like proteins with potent signaling regulatory function. Cell. Signal. 19, 238–250 [DOI] [PubMed] [Google Scholar]
- 41. Du K., Herzig S., Kulkarni R. N., Montminy M. (2003) TRB3: a Tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 42. Kiss-Toth E., Bagstaff S. M., Sung H. Y., Jozsa V., Dempsey C., Caunt J. C., Oxley K. M., Wyllie D. H., Polgar T., Harte M., O'neill L. A., Qwarnstrom E. E., Dower S. K. (2004) Human Tribbles, a protein family controlling mitogen-activated protein kinase cascades. J. Biol. Chem. 279, 42703–42708 [DOI] [PubMed] [Google Scholar]
- 43. Sakai S., Ohoka N., Onozaki K., Kitagawa M., Nakanishi M., Hayashi H. (2010) Dual mode of regulation of cell division cycle 25 A protein by TRB3. Biol. Pharm. Bull. 33, 1112–1116 [DOI] [PubMed] [Google Scholar]
- 44. Walhout A. J., Vidal M. (2001) High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24, 297–306 [DOI] [PubMed] [Google Scholar]
- 45. Mendoza J. A., Jacob Y., Cassonnet P., Favre M. (2006) Human papillomavirus type 5 E6 oncoprotein represses the transforming growth factor β signaling pathway by binding to SMAD3. J. Virol. 80, 12420–12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suspène R., Henry M., Guillot S., Wain-Hobson S., Vartanian J. P. (2005) Recovery of APOBEC3-edited human immunodeficiency virus G → A hypermutants by differential DNA denaturation PCR. J. Gen. Virol. 86, 125–129 [DOI] [PubMed] [Google Scholar]
- 47. Wang Z., Mosbaugh D. W. (1988) Uracil-DNA glycosylase inhibitor of bacteriophage PBS2: cloning and effects of expression of the inhibitor gene in Escherichia coli. J. Bacteriol. 170, 1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bogerd H. P., Wiegand H. L., Hulme A. E., Garcia-Perez J. L., O'Shea K. S., Moran J. V., Cullen B. R. (2006) Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 103, 8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yacoub Wasef S. Z., Robinson K. A., Berkaw M. N., Buse M. G. (2006) Glucose, dexamethasone, and the unfolded protein response regulate TRB3 mRNA expression in 3T3-L1 adipocytes and L6 myotubes. Am. J. Physiol. Endocrinol. Metab. 291, E1274–1280 [DOI] [PubMed] [Google Scholar]
- 50. Berger G., Durand S., Fargier G., Nguyen X. N., Cordeil S., Bouaziz S., Muriaux D., Darlix J. L., Cimarelli A. (2011) APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 7, e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schrader C. E., Linehan E. K., Mochegova S. N., Woodland R. T., Stavnezer J. (2005) Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J. Exp. Med. 202, 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chiarle R., Zhang Y., Frock R. L., Lewis S. M., Molinie B., Ho Y. J., Myers D. R., Choi V. W., Compagno M., Malkin D. J., Neuberg D., Monti S., Giallourakis C. C., Gostissa M., Alt F. W. (2011) Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hasham M. G., Donghia N. M., Coffey E., Maynard J., Snow K. J., Ames J., Wilpan R. Y., He Y., King B. L., Mills K. D. (2010) Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat. Immunol. 11, 820–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 55. Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. (2005) TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 24, 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith S. M., Moran A. P., Duggan S. P., Ahmed S. E., Mohamed A. S., Windle H. J., O'Neill L. A., Kelleher D. P. (2011) Tribbles 3: a novel regulator of TLR2-mediated signaling in response to Helicobacter pylori lipopolysaccharide. J. Immunol. 186, 2462–2471 [DOI] [PubMed] [Google Scholar]
- 57. Johnson J. A., Carstensen B., Witte D., Bowker S. L., Lipscombe L., Renehan A. G. (2012) Diabetes and cancer. 1. Evaluating the temporal relationship between type 2 diabetes and cancer incidence. Diabetologia 55, 1607–1618 [DOI] [PubMed] [Google Scholar]
- 58. Renehan A. G., Yeh H. C., Johnson J. A., Wild S. H., Gale E. A., Møller H., and Diabetes and Cancer Research Consortium (2012) Diabetes and cancer. 2. Evaluating the impact of diabetes on mortality in patients with cancer. Diabetologia 55, 1619–1632 [DOI] [PubMed] [Google Scholar]
- 59. Xu J., Lv S., Qin Y., Shu F., Xu Y., Chen J., Xu B. E., Sun X., Wu J. (2007) TRB3 interacts with CtIP and is overexpressed in certain cancers. Biochim. Biophys. Acta 1770, 273–278 [DOI] [PubMed] [Google Scholar]
- 60. Chinnadurai G. (2006) CtIP, a candidate tumor susceptibility gene, is a team player with luminaries. Biochim. Biophys. Acta 1765, 67–73 [DOI] [PubMed] [Google Scholar]
- 61. Humphrey R. K., Newcomb C. J., Yu S. M., Hao E., Yu D., Krajewski S., Du K., Jhala U. S. (2010) Mixed lineage kinase-3 stabilizes and functionally cooperates with TRIBBLES-3 to compromise mitochondrial integrity in cytokine-induced death of pancreatic β cells. J. Biol. Chem. 285, 22426–22436 [DOI] [PMC free article] [PubMed] [Google Scholar]