Background: The AAA ATPases of the PAN/Rpt1–6 group regulate access of substrates to the 20S proteasome.
Results: Two groups of AAA proteins, CDC48 and AMA, function as novel proteasomal ATPases in archaea.
Conclusion: This network of regulatory ATPases increases the capacity of proteasomal protein degradation in archaea.
Significance: Diversification at the level of the regulatory ATPase provides a contrast to the fully differentiated 26S proteasome of eukaryotes.
Keywords: Archaea, ATP-dependent Protease, ATPases, Proteasome, Protein Degradation
Abstract
The proteasome is the central machinery for targeted protein degradation in archaea, Actinobacteria, and eukaryotes. In its basic form, it consists of a regulatory ATPase complex and a proteolytic core particle. The interaction between the two is governed by an HbYX motif (where Hb is a hydrophobic residue, Y is tyrosine, and X is any amino acid) at the C terminus of the ATPase subunits, which stimulates gate opening of the proteasomal α-subunits. In archaea, the proteasome-interacting motif is not only found in canonical proteasome-activating nucleotidases of the PAN/ARC/Rpt group, which are absent in major archaeal lineages, but also in proteins of the CDC48/p97/VAT and AMA groups, suggesting a regulatory network of proteasomal ATPases. Indeed, Thermoplasma acidophilum, which lacks PAN, encodes one CDC48 protein that interacts with the 20S proteasome and activates the degradation of model substrates. In contrast, Methanosarcina mazei contains seven AAA proteins, five of which, both PAN proteins, two out of three CDC48 proteins, and the AMA protein, function as proteasomal gatekeepers. The prevalent presence of multiple, distinct proteasomal ATPases in archaea thus results in a network of regulatory ATPases that may widen the substrate spectrum of proteasomal protein degradation.
Introduction
Dynamic regulation of the proteome through protein degradation is a central metabolic strategy for organisms in a changing environment. In archaea, Actinobacteria, and eukaryotes, the proteasome plays a crucial role in protein degradation as part of stress responses and for regulatory purposes (1). Removal of misfolded proteins reduces the burden of potentially toxic proteins (2), and targeted destruction by the proteasome, especially in eukaryotes, is involved in processes such as cell cycle progression (3) and the cellular immune response (4). The proteasome is composed of a regulatory particle, which binds to and unfolds substrate proteins, and a catalytic particle, which fragments the substrates into peptides of 8–11 amino acids in length (5). An additional layer of regulation is provided by the covalent attachment of degradation tags to substrates, SAMP4 in archaea (6), Pup in Actinobacteria (7), and ubiquitin in eukaryotes (8), to direct them to the proteasome.
The catalytic 20S proteasome core particle (CP) forms a cylindrical assembly consisting of four stacked heptameric rings with an α7β7β7α7 stoichiometry (9). This arrangement sequesters the active sites to a central compartment formed by the β-subunits, limiting access to only those proteins that present a degradation signal. The catalytic nucleophile, a threonine residue (10), is the N-terminal residue of β-subunits after autoprocessing of a leader peptide (11). The α-subunits guide substrates to the proteolytic chamber through a central channel, which is occluded in the resting state by their flexible N-terminal ends (12). The regulatory AAA ATPases Rpt1–6 (in eukaryotes), PAN (in archaea), and ARC/Mpa (in Actinobacteria) recognize, unfold, and translocate substrates into the CP (13–15). Members of this orthologous group share a common architecture, which consists of an N-terminal domain, involved in substrate binding and interaction with the degradation tag or with subunits of the regulatory particle (16–18), and an AAA unfoldase module. At their C terminus, the ATPases possess an HbYX motif (where Hb is a hydrophobic residue, Y is tyrosine, and X is any amino acid) (19) that can penetrate into a binding pocket of the α-subunits, thereby stabilizing the open gate conformation of their N-terminal ends (20). The functional importance of the HbYX motif is reflected by the ability of 7-residue C-terminal peptides, isolated or fused to the 11S/PA26 non-ATPase activator, to mimic the biochemical effects of full-length PAN (19–22).
Although preparations of the 26S proteasome from eukaryotes are routinely obtained via fractionation of whole cell lysate, in archaea, the preparation of proteasome ATPase complexes has been notoriously difficult. So far, there is no description of a fractionation approach, and the heterologous complex consisting of PAN from Methanocaldococcus jannaschii and the core particle from Thermoplasma acidophilum, an organism that lacks PAN (23), is the most widely studied archaeal model system and the only one that could be imaged to date (24).
Although the core particle degrades peptides and intrinsically disordered proteins in the absence of regulatory particles (5, 25), the Rpt subunits are essential for Saccharomyces cerevisiae (26), and Mpa is required by Mycobacterium tuberculosis in an infectious context (27), illustrating that the proteasomal ATPases perform a crucial function, in particular for the unfolding and degradation of (mis)-folded polypeptides under stress conditions. Nonetheless, we find that PAN ATPases are absent in a number of archaea (28), which raises the question of how substrate proteins are made available to the proteasome in these organisms.
Detecting a proteasome-interacting motif in the AAA ATPase CDC48 of T. acidophilum prompted us to perform a systematic analysis of archaeal AAA proteins, which uncovered a network of ATPases with a common HbYX motif including CDC48 and AMA proteins. For two model organisms, we provide evidence that these ATPases indeed physically interact with their cognate core particle and show that they stimulate proteasome activity in proteolytic assays, establishing CDC48 and AMA proteins as regulators of the proteasome in archaea.
EXPERIMENTAL PROCEDURES
Bioinformatics
Homologs of archaeal AAA proteins were identified with HHsenser (29) searching the non-redundant database of archaeal proteins (National Center for Biotechnology Information (NCBI), nr_arc) with the AAA+ module of AMA from Methanosarcina mazei (gi 21226406, Mm_0304119–372). Assignment to orthologous groups of full-length sequences was based on cluster analyses using CLANS (30). p values for clustering were selected interactively to achieve formation of orthologous groups. Groups of AAA proteins were distinguished from other members of the AAA+ superfamily using different p value cutoffs and relying on our classification of AAA+ proteins (31). Members of orthologous groups were verified by testing for concordant domain composition with HHpred (32) and MUSCLE (33). The presence or absence of genes was mapped onto the archaeal species tree with iToL (34). C-terminal peptides comprising the last seven residues of AAA proteins were extracted from full-length sequences. Assignment of the HbYX motif (19) was based on the presence of a small or hydrophobic residue in third last and a Phe or Tyr residue in penultimate position.
Cloning
Ta20Sα (Ta1288, gi 16081896), Ta20Sβ (Ta0612, gi 16081708), TaCDC48ΔC (TaCDC481–733 lacking the last 12 residues), and TaCDC48-L745W (containing W466F, W541Y, and L745W mutations) genes were synthesized by GenScript. MmPAN-A (Mm1006, gi 20905437), MmPAN-B (Mm0789, gi 20905207), Mm20Sα (Mm2620, gi 21228722), Mm20Sβ (Mm0694, gi 21226796), and TaCDC48 (Ta0840, gi 16081896) were obtained as gifts from W. Baumeister and P. Zwickl. MmCDC48-A (Mm0248, gi 20904601), MmCDC48-B (Mm0447, gi 20904821), MmCDC48-C (Mm1256, gi 20905716), MmAMA (Mm0304, gi 20904664), and Mm0854 (gi 20905268) ORFs were amplified from genomic DNA of M. mazei strain OCM88 (ATCC number: BAA159) by PCR. GFPssrA fragment was PCR-amplified from pEGFP-N1 plasmid (Clontech) using a reverse primer containing the ssrA tag (AANDENYALAA) sequence. Proteasomal α-subunit DNA fragments were cloned into pET30b expression vector (Novagen); ATPases and GFPssrA were cloned as N-terminally hexahistidine-tagged proteins into pET28b; and proteasomal β-subunits were cloned as C-terminally hexahistidine-tagged proteins into pET22b.
Protein Production and Purification
Plasmids were transformed into Escherichia coli C41(DE3) RIL expression strain. Plasmids encoding the proteasomal α- and β-subunits were co-transformed to assemble the CP proteasome directly inside E. coli cells. Expression was achieved by growing single colonies in LB medium, supplemented with the appropriate antibiotics at 37 °C until an optical density of ∼0.6 was reached followed by induction with 1 mm isopropyl-1-thio-β-d-galactopyranoside and continued culturing overnight at 20 °C. Cell pellets were resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 500 mm NaCl, 10 mm MgCl2, 5% glycerol, 10 mm imidazole, 5 mm β-mercaptoethanol, 0.4 mg/ml lysozyme, 40 μg/ml DNase, and except for the proteasome, EDTA-free protease inhibitor mixture (Roche Applied Science)) and lysed by French press treatment. After centrifugation, lysates were applied consecutively to nickel-nitrilotriacetic acid metal affinity (50 mm Tris-HCl, pH 8.0, 500 mm NaCl, 10 mm MgCl2, 5% glycerol, 10 mm-1 m imidazole gradient, 5 mm β-mercaptoethanol), mono-Q anion-exchange (50 mm Tris-HCl, pH 7.5, 50 mm-1 M NaCl gradient, 10 mm MgCl2, 5% glycerol, 1 mm dithiothreitol (DTT)), and Superose 6 gel filtration (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 1 mm DTT) chromatography columns (GE Healthcare). MmAMA accumulated in inclusion bodies and was purified under denaturing conditions using 4 m urea containing buffers. In parallel, the quality of protein preparations was controlled by electron microscopy, activity assays, and mass spectrometry.
Peptidase and Protease Assays
Proteolytic assays made use of three different fluorescent substrates, namely: quenched LFP (24) (Mca-AKVYPYPME-Dpa(Dnp)-amide; GenScript; excitation, 322 nm; emission, 398 nm) in the assays using 7-residue peptides and quenched BODIPY-TR X casein (35) (Invitrogen; excitation, 590 nm; emission, 645 nm) and GFPssrA (excitation, 488 nm; emission, 507 nm) in the assays using full-length ATPases. Peptides were synthesized by GenScript. LFP assays were performed with 0.6 nm CP, 250 μm 7-residue peptide, and 10 μm LFP in 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 1 mm DTT, for the duration of 120 min at 59 °C for T. acidophilum proteins and 37 °C for M. mazei ones. Full-length protein assays were done with 100 nm CP, 400 nm ATPase, 1 μm substrate, 20 mm ATP, and 10 mm MgCl2. All measurements are the average of three independent experiments and were carried out using a Synergy H4 plate reader (BioTek). For the tryptophan fluorescence shift assay, proteins were excited at 280 nm, and emission spectra of the proteins with or without ATPγS (Sigma-Aldrich) were collected from 330 nm to 380 nm.
Surface Plasmon Resonance
Surface Plasmon Resonance (SPR) experiments were performed at 25 °C with a Biacore 2000 instrument (Biacore) and a data collection rate of 1 Hz. MmCP was immobilized via amine coupling on a CM5 sensor chip (GE Healthcare). The surface of the upstream flow cell was subjected to the same coupling reaction in the absence of protein and was used as a reference. MmPAN-A, MmCDC48-B, and Mm0854 were supplemented with a 100-fold molar excess of ATPγS and then serially diluted into running buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, and 0.005% (v/v) surfactant P20). Analyte samples with five different concentrations (1000–62.5 nm) were injected over the control and experimental surfaces at a flow rate of 10 μl/min. A blank sample, containing only the running buffer supplemented with ATPγS in the respective concentration, was injected under the same conditions to allow for double referencing. Data analysis was carried out with the BIAevaluation software (version 4.1.1).
Pulldowns
100 μg of bait proteins (Ta20S, TaCDC48, and TaCDC48ΔC) were bound to cobalt-nitrilotriacetic acid immobilized beads (GE Healthcare), mixed with T. acidophilum cellular lysate, and incubated for 1 h at 59 °C. The samples were then applied to Mobicol spin columns (MoBiTec) and washed three times with washing buffer (50 mm Tris-HCl, pH 8.0, 250 mm NaCl, 10 mm MgCl2, 5% glycerol, 40 mm imidazole, 5 mm β-mercaptoethanol). Bound proteins were eluted with elution buffer (50 mm Tris-HCl, pH 8.0, 250 mm NaCl, 10 mm MgCl2, 5% glycerol, 500 mm imidazole, 5 mm β-mercaptoethanol), and captured binding partners were identified with mass spectrometry.
Nano LC-MS/MS Analysis
Tryptic digestion of the samples and analyses of the peptides by LC-MS/MS and subsequent data processing were done as described previously with slight modifications (36). Analyses of the peptides were performed on a Proxeon Easy-nLC system (Proxeon Biosystems) coupled to an LTQ-Orbitrap-XL (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Proxeon Biosystems). The injected peptide mixtures were subsequently eluted with a 47-min segmented gradient of 2–80% HPLC solvent B (80% acetonitrile in 0.5% acetic acid) at a flow rate of 200 nl/min. Mass spectra were analyzed using the software suite MaxQuant, version 1.0.14.3 (37). The data were searched against a target-decoy M. mazei database containing 5645 forward protein sequences and 262 frequently observed contaminants. Trypsin was set as protease in which two missed cleavage sites were allowed. Acetylation at the N terminus and oxidation of methionine were set as variable modifications. Carbamidomethylation of cysteine was set as fixed modification. Initial precursor mass tolerance was set to 7 ppm at the precursor ion and 0.5 Da at the fragment ion level. False discovery rates were set to 1% at peptide and protein group level.
RESULTS
Prediction of Novel Proteasomal ATPases in the Archaeal Kingdom
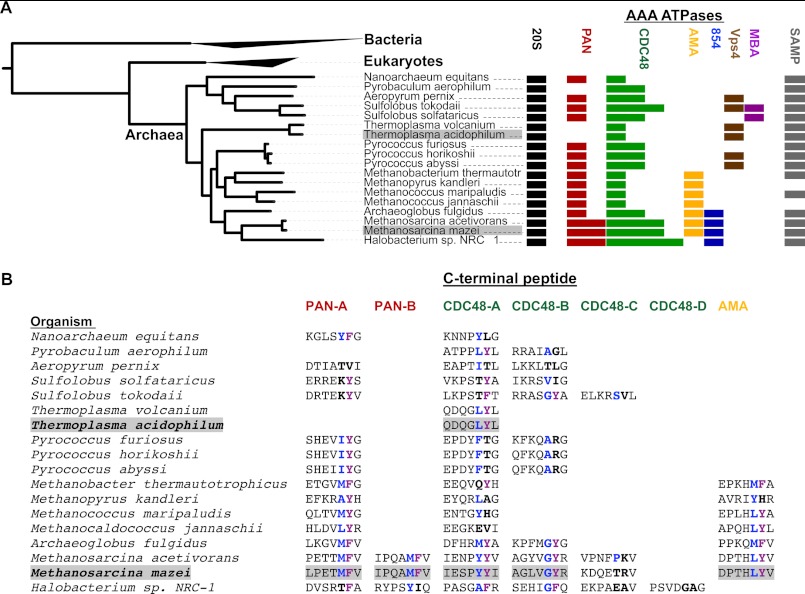
AAA proteins form a distinct group within the AAA+ superfamily, whose unifying activity is the energy-dependent remodeling of proteins and nucleic acids (31, 38). In a cluster analysis of archaeal AAA proteins (supplemental Fig. S1), we obtained separate clusters for Vps4 (39), membrane-bound AAA (MBA) (40), an uncharacterized group exemplified by open reading frame number 854 in the M. mazei genome (Mm0854), AMA (41, 42), the proteasome-activating nucleotidase PAN (14), and CDC48 (43). Comparative analysis across 81 sequenced archaeal genomes showed that CDC48 is the sole subgroup found in all archaea and also has the highest degree of paralogy (Fig. 1A). The proteasome-activating nucleotidase PAN, however, is absent from major archaeal groups, including all Thermoplasmata, Thermoproteales, and the deep-branching Thaumarchaeota and Korarchaeota (supplemental Fig. S2).
FIGURE 1.
Archaeal AAA proteins and the prediction of novel proteasome regulators. A, the distribution of AAA ATPases is mapped onto a tree of selected archaeal species. Analysis of the C termini of archaeal AAA proteins indicates the presence of the HbYX motif in CDC48 and AMA, as well as in PAN proteins. CDC48 is ubiquitous and shows the highest degree of paralogy, whereas PAN is absent in Thermoproteales and Thermoplasmata and in deep-branching Korarchaeota and Thaumarchaeota, suggesting subgroup-specific loss rather than lateral gene transfer. AMA most likely originated in the ancestor of Archaeoglobales and methanogens, and was lost in Halobacteria. For comparison, the occurrence of the 20S proteasome and SAMP proteins is shown. Panel A was generated with iToL (34) (MBA, membrane-bound AAA). B, C-terminal sequences of PAN, CDC48, and AMA proteins. The species and sequences are shown in the same order as in panel A and illustrate the presence of the HbYX motif (hydrophobic/small residue in blue, Tyr/Phe in magenta). Species lacking PAN contain at least one CDC48 ortholog with the HbYX motif. All AMA proteins contain the HbYX motif. The number of putative proteasomal ATPases ranges from one to five, suggesting a regulatory network in many archaea. T. acidophilum and M. mazei, which were selected for experimental study, are highlighted.
The C-terminal HbYX motif required for interaction with the 20S proteasome is not only present in the known proteasomal regulators of the PAN group, but also in all AMA proteins and in many CDC48 proteins. All archaeal organisms sequenced to date have at least one AAA protein with an HbYX motif, supporting the notion that ATPase-mediated proteasomal regulation is an essential function (supplemental Fig. S3). Thus, organisms lacking PAN encode at least one CDC48 protein with a canonical HbYX motif.
The number of putative proteasomal ATPases varies from one in Thermoplasmata to five in some Methanosarcinales (Fig. 1B). We therefore selected AAA proteins and cognate proteasome core particles of T. acidophilum and M. mazei to explore the idea that archaea employ multiple AAA proteins to regulate proteasomal activity. T. acidophilum encodes only one CDC48 protein and no PAN ortholog, and its proteasomal gatekeeper is still elusive despite the fact that the core particle (TaCP) has served as a long-standing model system together with the heterologous PAN protein of M. jannaschii (19–22, 24). In contrast, M. mazei contains seven AAA proteins, five of which have the proteasome-interacting motif, including two CDC48 proteins and a member of the AMA group (Fig. 1).
CDC48 Is the Proteasomal ATPase of T. acidophilum
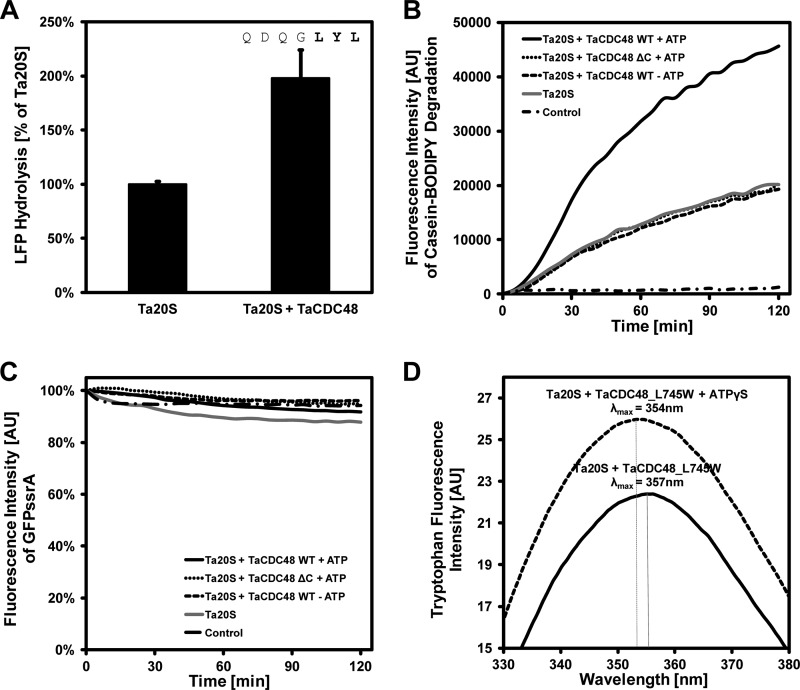
Because the C-terminal peptides of PAN and Rpt ATPases have been shown to mimic efficiently the activation of the proteasome core particle by the full-length proteins (19), we used the C-terminal peptide of TaCDC48, whose last three residues are Leu-Tyr-Leu, to stimulate the hydrolysis of the fluorogenic nonapeptide LFP by TaCP. Although the proteasome can degrade LFP on its own, we observed a 2-fold higher fluorescence signal in the presence of C-terminal peptide as compared with TaCP alone (Fig. 2A), providing initial support for our hypothesis that CDC48 is the proteasomal gatekeeper in T. acidophilum.
FIGURE 2.
CDC48 binds to and activates the proteasome of T. acidophilum. A, the addition of the C-terminal peptide of TaCDC48 results in a 2-fold increase in proteasomal degradation of the fluorogenic nonapeptide LFP, measured as gain in fluorescence intensity. Error bars indicate S.D. B, full-length TaCDC48 enhances the degradation of BODIPY-labeled casein by TaCP in the presence of ATP. A C-terminal deletion mutant of TaCDC48 (TaCDC48ΔC) lacking the last 12 residues of TaCDC48 does not stimulate casein degradation in comparison with TaCP alone. C, eGFP-ssrA is not degraded by TaCP in the presence of TaCDC48 and ATP. AU, absorbance units. D, in the presence of TaCP, the fluorescence emission spectrum of a C-terminal tryptophan variant of TaCDC48 (TaCDC48-L745W) moves from an aqueous to a more hydrophobic environment upon the addition of ATPγS, indicated by a blue shift of the emission maximum and a decreased intensity.
In the next step, we tested the ability of full-length TaCDC48 to activate the T. acidophilum proteasome, using BODIPY-casein as a substrate. TaCP alone is able to degrade casein, and TaCDC48 could stimulate this activity 2.5-fold, as judged by the gain in fluorescence intensity of BODIPY fluorophores (Fig. 2B). This stimulation was entirely dependent on the HbYX motif as a C-terminal deletion mutant of TaCDC48 lacking the last 12 residues (TaCDC48ΔC) was unable to activate the proteasome to any measurable extent (Fig. 2B).
Finally, we attempted to stimulate the degradation of GFPssrA using full-length TaCDC48. GFPssrA is an established model substrate for PAN and proteasome from M. jannaschii, as well as for Mpa and proteasome from the actinobacterium M. tuberculosis (44, 45). However, we did not detect its degradation by the TaCDC48-TaCP system (Fig. 2C). It has previously been reported that GFPssrA is unfolded by TaCDC48 at high magnesium concentrations and can then be degraded by a highly active, gateless proteasome mutant, in which the 12 N-terminal residues of α-subunits have been removed (46). We attribute the need for a gateless proteasome in that experiment to the fact that the TaCDC48 construct contained a C-terminal His tag and could not activate the proteasome via its C-terminal HbYX motif. We propose that this uncoupling and the use of a constitutively active proteasome accounts for the different outcome of the experiments. As we will show in the next section, the ability to degrade GFPssrA appears to be limited to a subset of active ATPase-proteasome tandems.
To probe the physical interaction of TaCDC48 and TaCP, we changed the last residue of the ATPase from leucine to a tryptophan in a tryptophan-free mutant background of TaCDC48, TaCDC48-L745W, in analogy to an experiment conducted by Goldberg and colleagues (19). In fluorescence emission spectra, upon the addition of ATPγS and in the presence of tryptophan-free TaCP, the C-terminal tryptophan of TaCDC48-L745W experiences a blue shift and a decrease in fluorescence intensity (Fig. 2D). This is the expected outcome if binding of the exposed C-terminal residues into the binding pocket of the proteasomal α-subunit moved the peptide tail from an aqueous to a more hydrophobic environment. To collect further evidence for an interaction of TaCDC48 and TaCP, we performed pulldown experiments with either TaCDC48 or TaCP as bait in cellular lysate of T. acidophilum. Both CDC48 and TaCP return each other as one of the most significant binding partners, but the C-terminal deletion mutant TaCDC48ΔC does not retrieve TaCP (Table 1, supplemental Table 1). In summary, we find that TaCDC48 directly interacts with and stimulates TaCP and that its C-terminal residues are crucial for this activity.
TABLE 1.
TaCDC48 interacts with TaCP
Table 1 summarizes the identification of the α- and β-subunits of TaCP and TaCDC48 in pulldown experiments using three different bait proteins (TaCP, TaCDC48, TaCDC48ΔC) in cellular lysate of T. acidophilum. The complete list of identified proteins is shown in supplemental Table 1.
| Bait proteinsa |
||||
|---|---|---|---|---|
| Control | TaCP | TaCDC48 | TaCDC48ΔC | |
| TaCDC48 | 0 | 13 | 117 | 120 |
| α-Subunit | 10 | 46 | 17 | 8 |
| β-Subunit | 2 | 20 | 8 | 2 |
a Number represents the number of identified peptides.
A Network of Five AAA Proteins Regulates the Proteasome of M. mazei
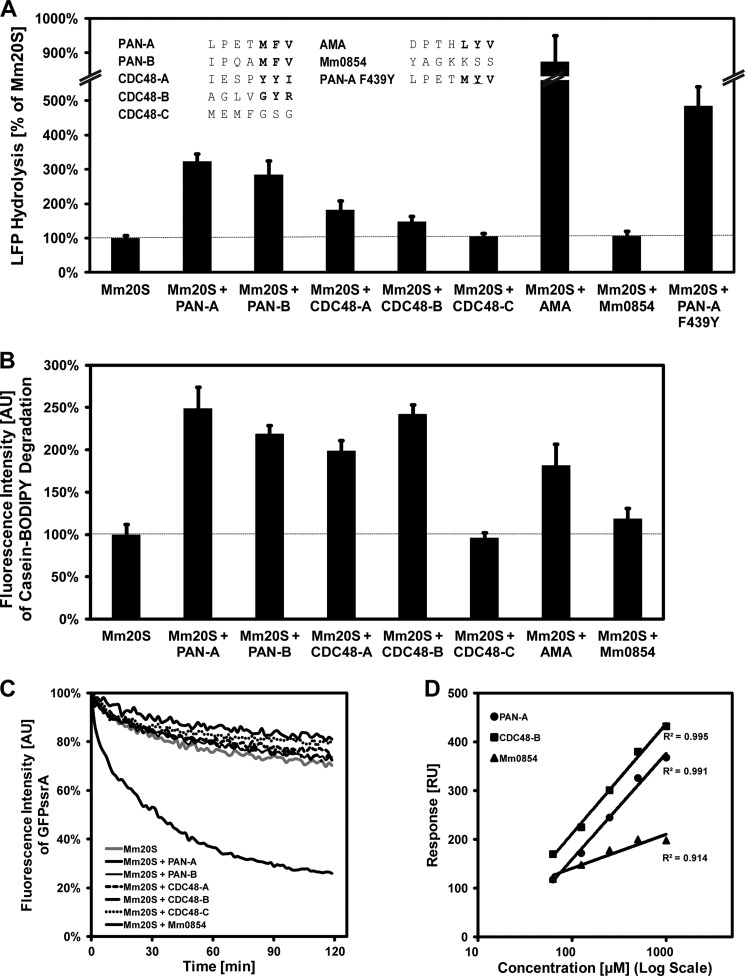
In contrast to T. acidophilum, M. mazei encodes seven AAA proteins, of which we predict five to function as proteasomal ATPases. This is the largest number of ATPases in the putative regulatory network among sequenced archaea. To collect evidence for such a network, we tested whether the 7-residue peptides corresponding to the C termini of the ATPases could stimulate LFP peptidase activity of the M. mazei 20S proteasome (MmCP). In agreement with our prediction, we found that peptides from the five AAA proteins that have the HbYX motif increased the hydrolysis of the fluorogenic LFP. These are the two PAN proteins (35), two out of three CDC48 proteins, and the AMA protein (Fig. 3A). The C-terminal peptides of CDC48-C and of Mm0854, which lack the motif, had no effect. The degree of stimulation ranged from 1.5-fold for CDC48-B to ∼9-fold for AMA. We also found that a PAN-A variant, in which the naturally occurring Phe of the HbYX motif was mutated to Tyr, was a much stronger activator than wild type, in line with results showing that the hydroxyl group of Tyr, which is the most frequent residue in this position, stabilizes the open gate conformation more efficiently (21).
FIGURE 3.
A network of AAA ATPases regulates the proteasome of M. mazei. A, C-terminal peptides of AAA proteins from M. mazei containing the HbYX motif enhance LFP hydrolysis. The PAN-A F439Y mutant is a stronger activator of MmCP than PAN-A with the wild-type HbYX motif. Error bars indicate S.D. B, full-length ATPases with the HbYX motif stimulate degradation of BODIPY-labeled casein by MmCP. Error bars indicate S.D. AU, absorbance units. C, PAN-A is the only AAA protein of M. mazei that mediates degradation of eGFP-ssrA by MmCP. D, SPR sensorgrams show that MmPAN-A and MmCDC48-B physically interact with MmCP in a concentration-dependent manner, whereas Mm0854 shows a much weaker interaction. RU, response units.
Next, we extended our analysis to the seven full-length ATPases, using BODIPY-casein as a substrate in protease assays. The five ATPases with HbYX motifs stimulated proteasome activity from 2-fold for CDC48-A to 2.5-fold for PAN-A (Fig. 3B). Again, CDC48-C and Mm0854, which do not contain the HbYX motif, did not show any effect. Our results show that full-length proteins stimulate the proteasome, albeit not as strongly as isolated peptides. Although peptides are able to bind independently and in maximal number, in full-length proteins, only a subset of C-terminal tails can be expected to bind at a given time due to steric constraints (symmetry mismatch).
Remarkably, of the five ATPases able to activate the proteasome, only PAN-A was able to stimulate the degradation of GFPssrA (Fig. 3C). This suggests that the ATPases may also have differing substrate specificities in a cellular context, possibly mediated in part by their different N-terminal substrate recognition domains. Nevertheless, it is worth noting that differences are apparent even in orthologous ATPases with highly similar N-domains, as seen by the inability of PAN-B to mediate GFPssrA degradation.
To probe the physical interaction between the ATPases and MmCP, we selected three for analysis by surface plasmon resonance (SPR), based on the degree of stimulation and ease of handling (PAN-A, CDC48-B, and Mm0854). Mm0854 showed only a background level of binding to MmCP, which correlates with the absence of an HbYX motif in this protein and its inability to stimulate MmCP in degradation assays. PAN-A and CDC48-B, in contrast, showed a clear concentration-dependent response, providing evidence that a direct interaction of ATPase and CP is responsible for their ability to activate the proteasome (Fig. 3D).
DISCUSSION
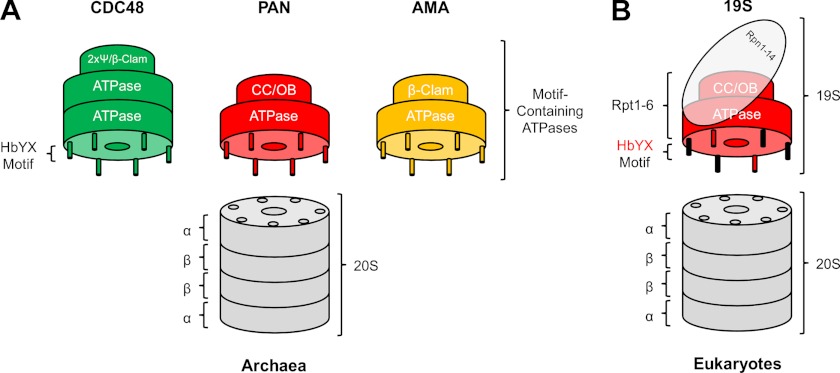
We have defined a regulatory network of proteasomal ATPases across the archaeal kingdom and validated their interactions with the proteasome through in vitro studies with proteins from two archaeal organisms that mark extremes in the number of genomically encoded AAA ATPases. This network features, in addition to known proteasome-activating nucleotidases of the PAN type, two groups of AAA proteins, AMA and CDC48, that hitherto have not been seen directly associated with the 20S proteasome (Fig. 4).
FIGURE 4.
The network of proteasomal AAA ATPases. A, in archaea, PAN, CDC48, and AMA proteins constitute a regulatory network of proteasomal ATPases. Interaction is governed by the C-terminal HbYX motif; different N-domain folds in the ATPases are denoted. 2xpsi, double-psi barrel; CC/OB, coiled-coil and oligosaccaride-binding domain. B, in eukaryotes, the heterooligomeric PAN-like Rpt1–6 subunits function within the 19S particle as the regulatory ATPase of the proteasome. The HbYX motifs of Rpt2 and Rpt5 are important for gate opening.
Archaea-wide Significance of the Network Model
The interaction of AAA ATPases with the proteasome is determined by the presence of HbYX motifs at the very C terminus of ATPase subunits. However, not only AAA ATPases contain this motif. We find the HbYX motif (pattern: [LVIMGAPFYW][FY]x>) at the C terminus of 63 proteins (of 1484 proteins coding genes) in the T. acidophilum genome, one of which is CDC48. For M. mazei, we detect this motif in 133 (of 3368) proteins, five of which are the AAA proteins that are part of the regulatory network we describe. We assume that only very few of these proteins form ring-shaped complexes and offer this interaction motif in a sufficiently high local concentration and extended conformation to stimulate gate opening of the core particle. Furthermore, there are other interaction partners of the proteasome with this motif, for instance, the proteasome assembly chaperones PbaA and PbaB, which are also conserved in archaea (47), and there might be more remaining to be found. Nevertheless, our analysis specifically and comprehensively tracks this motif in ATP-dependent proteasome regulators of the family of AAA unfoldases.
The conservation of residues forming the binding pocket for the HbYX motif (invariant residues proline 17 and glycine 19) in the proteasome α-subunits indicate that all archaeal proteasomes are most likely activated by the same mechanism. Although we observe peculiar or highly divergent C termini that do not allow an unambiguous decision regarding the presence of the motif, all archaea encode at least one AAA protein that contains the HbYX motif in canonical form, emphasizing the importance of proteasomal regulation by AAA proteins. Even if PAN orthologs are absent in a genome, there is at least one member of the CDC48 family with a clear HbYX motif, defining these members as proteasomal ATPases in the context of our degradation and interaction assays. Likewise, as all members of the AMA group contain the HbYX motif, we conclude that gatekeeping of the proteasome is the main cellular function of this group, thereby increasing the repertoire of proteasome regulators.
Functional Implications
Components of the archaeal proteasome machinery generally improve survival rate under stress conditions (48) and are essential for growth (49). Functional diversification of the core particle through gene duplication is known for Haloferax volcanii, in which a second type of α-subunit (and PAN-B) is up-regulated during starvation (50), and for Pyrococcus furiosus, where the β1-subunit is up-regulated under heat stress, resulting in more thermostable PfCP particles (51). Accordingly, for the dynamic proteome of a cell living in changing environments, an adaptable network of regulators of the central protein destruction machinery conceivably confers advantages. The general capabilities of the proteasomal system for targeted degradation would be increased through the participation of different N-terminal substrate recognition domains of the regulatory ATPases. As a consequence, a greater variety of substrates could be recognized with higher affinity and specificity. In addition to a more sophisticated regulation of proteolysis, the presence of multiple proteasomal ATPases may also generate some redundancy. H. volcanii, for example, tolerates a gene knock-out of both of its PAN proteins under normal growth conditions (6), potentially because it still has two CDC48 proteins with an HbYX motif.
Currently, it is unclear how this regulatory network of ATPases is integrated with the SAMP-based tagging system, whose conjugates are increased (SAMP1) or decreased (SAMP2) in a genetic background depleted of the PAN proteins (6). It remains to be studied whether motif-containing CDC48 proteins participate in the SAMPylation system and whether they are able to compensate for a deletion of PAN proteins. Importantly, there is no correlation between the presence of the SAMP proteins and specific proteasomal ATPases (Fig. 1A).
Evolutionary Implications
Comparative analysis of the content of putative proteasomal ATPases in archaeal genomes suggests a complex ancestral state, which is likely to include two CDC48 proteins and one PAN protein. The pattern of conservation suggests that PAN has been lost in certain lineages rather than laterally transferred (23), whereas AMA most likely originated in a common ancestor of methanogens and Archaeoglobales. The number of putative proteasomal ATPases varies in extant archaea between one and five, pointing to clade-specific solutions for the functional task of targeted protein degradation. Nevertheless, the approximate number of three proteasomal ATPases in the ancestor of all archaea (one PAN, two CDC48 proteins) suggests that it would have already relied on a network of AAA proteins to regulate its central proteolytic machinery.
The employment of different AAA proteins as regulators of the proteasome in the majority of archaea contrasts with the situation in eukaryotes, where the heterooligomeric PAN orthologs Rpt1–6 act exclusively as ATPase regulators (the regulators Blm10 and 11S are not ATPases). Although AMA proteins are not found in eukaryotes, CDC48 (p97) is conserved and fulfills functions in a plethora of pathways. However, there is only evidence for a direct interaction of CDC48 with the 19S regulatory particle, rather than the 20S core particle (52), serving as a gearbox that segregates ubiquitylated from unmodified substrate proteins (53). Therefore, the fully differentiated 26S proteasome is, with the notable exception of immunological responses in vertebrates, basically fixed in its composition. Diversity in the ubiquitin-proteasome system of eukaryotes mainly takes place at the level of E3 ligases, which confer specificity in the tagging of substrates. Their radiating number, ranging from ∼80 in yeast to ∼600 in humans (54), enables specific involvement of the proteasome in the regulation of numerous cellular processes. Despite the fact that E3-like ligases for SAMPylation in archaea have not been discovered yet, a network of regulatory ATPases constitutes an additional strategy to expand the capacity of proteasomal protein degradation.
Acknowledgments
We thank Heinz Schwarz for electron microscopy, Peter Zwickl and Wolfgang Baumeister for plasmids, Thilo Stehle for support with SPR, Boris Macek for support with MS, and Volkmar Braun, Jörn Engelmann, and Remco Sprangers for helpful discussions.
This work was supported by institutional funds of the Max Planck Society.

This article contains supplemental Figs. S1–S3 and Table 1.
- SAMP
- small archaeal modifier protein
- CP
- core particle
- PAN
- proteasome-activating nucleotidase
- ATPγS
- adenosine-5′-(γ-thio)-triphosphate
- Mm
- M. mazei
- Ta
- T. acidophilum
- Pup
- prokaryotic ubiquitin-like protein
- Rpt
- regulatory particle triple-A ATPases
- ARC
- AAA ATPase forming ring-shaped complexes
- Mpa
- mycobacterial proteasome ATPase
- VAT
- VCP-like ATPase of Thermoplasma acidophilum
- AMA
- AAA protein of methanogenic archaea and Archaeoglobus
- Mca
- (7-Methoxycoumarin-4-yl)acetyl
- Dpa
- N-3-(2,4-Dinitrophenyl)-L-2,3-diaminopropionyl
- Dnp
- 2,4-dinitrophenyl.
REFERENCES
- 1. Baumeister W., Walz J., Zühl F., Seemüller E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell 92, 367–380 [DOI] [PubMed] [Google Scholar]
- 2. Wickner S., Maurizi M. R., Gottesman S. (1999) Posttranslational quality control: folding, refolding, and degrading proteins. Science 286, 1888–1893 [DOI] [PubMed] [Google Scholar]
- 3. Pagano M., Tam S. W., Theodoras A. M., Beer-Romero P., Del Sal G., Chau V., Yew P. R., Draetta G. F., Rolfe M. (1995) Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269, 682–685 [DOI] [PubMed] [Google Scholar]
- 4. Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771 [DOI] [PubMed] [Google Scholar]
- 5. Kisselev A. F., Akopian T. N., Goldberg A. L. (1998) Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J. Biol. Chem. 273, 1982–1989 [DOI] [PubMed] [Google Scholar]
- 6. Humbard M. A., Miranda H. V., Lim J. M., Krause D. J., Pritz J. R., Zhou G., Chen S., Wells L., Maupin-Furlow J. A. (2010) Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 463, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pearce M. J., Mintseris J., Ferreyra J., Gygi S. P., Darwin K. H. (2008) Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322, 1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 9. Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Ä resolution. Science 268, 533–539 [DOI] [PubMed] [Google Scholar]
- 10. Seemüller E., Lupas A., Stock D., Löwe J., Huber R., Baumeister W. (1995) Proteasome from Thermoplasma acidophilum: a threonine protease. Science 268, 579–582 [DOI] [PubMed] [Google Scholar]
- 11. Seemuller E., Lupas A., Baumeister W. (1996) Autocatalytic processing of the 20S proteasome. Nature 382, 468–471 [DOI] [PubMed] [Google Scholar]
- 12. Rabl J., Smith D. M., Yu Y., Chang S. C., Goldberg A. L., Cheng Y. (2008) Mechanism of gate opening in the 20S proteasome by the proteasomal atpases. Mol. Cell 30, 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rubin D. M., Glickman M. H., Larsen C. N., Dhruvakumar S., Finley D. (1998) Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 17, 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zwickl P., Ng D., Woo K. M., Klenk H. P., Goldberg A. L. (1999) An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J. Biol. Chem. 274, 26008–26014 [DOI] [PubMed] [Google Scholar]
- 15. Wolf S., Nagy I., Lupas A., Pfeifer G., Cejka Z., Müller S. A., Engel A., De Mot R., Baumeister W. (1998) Characterization of ARC, a divergent member of the AAA ATPase family from Rhodococcus erythropolis. J. Mol. Biol. 277, 13–25 [DOI] [PubMed] [Google Scholar]
- 16. Wang T., Darwin K. H., Li H. (2010) Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation. Nat. Struct. Mol. Biol. 17, 1352–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imkamp F., Striebel F., Sutter M., Ozcelik D., Zimmermann N., Sander P., Weber-Ban E. (2010) Dop functions as a depupylase in the prokaryotic ubiquitin-like modification pathway. EMBO Rep. 11, 791–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lander G. C., Estrin E., Matyskiela M. E., Bashore C., Nogales E., Martin A. (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., Goldberg A. L. (2007) Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's α ring opens the gate for substrate entry. Mol. Cell 27, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Religa T. L., Sprangers R., Kay L. E. (2010) Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science 328, 98–102 [DOI] [PubMed] [Google Scholar]
- 21. Stadtmueller B. M., Ferrell K., Whitby F. G., Heroux A., Robinson H., Myszka D. G., Hill C. P. (2010) Structural models for interactions between the 20S proteasome and its PAN/19S activators. J. Biol. Chem. 285, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu Y., Smith D. M., Kim H. M., Rodriguez V., Goldberg A. L., Cheng Y. (2010) Interactions of PAN's C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 29, 692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruepp A., Rockel B., Gutsche I., Baumeister W., Lupas A. N. (2001) The chaperones of the archaeon Thermoplasma acidophilum. J. Struct. Biol. 135, 126–138 [DOI] [PubMed] [Google Scholar]
- 24. Smith D. M., Kafri G., Cheng Y., Ng D., Walz T., Goldberg A. L. (2005) ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell 20, 687–698 [DOI] [PubMed] [Google Scholar]
- 25. Liu C. W., Corboy M. J., DeMartino G. N., Thomas P. J. (2003) Endoproteolytic activity of the proteasome. Science 299, 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bajorek M., Finley D., Glickman M. H. (2003) Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr. Biol. 13, 1140–1144 [DOI] [PubMed] [Google Scholar]
- 27. Darwin K. H., Ehrt S., Gutierrez-Ramos J. C., Weich N., Nathan C. F. (2003) The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302, 1963–1966 [DOI] [PubMed] [Google Scholar]
- 28. Maupin-Furlow J. (2012) Proteasomes and protein conjugation across domains of life. Nat. Rev. Microbiol. 10, 100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Söding J., Remmert M., Biegert A., Lupas A. N. (2006) HHsenser: exhaustive transitive profile search using HMM-HMM comparison. Nucleic Acids Res. 34, W374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frickey T., Lupas A. (2004) CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704 [DOI] [PubMed] [Google Scholar]
- 31. Ammelburg M., Frickey T., Lupas A. N. (2006) Classification of AAA+ proteins. J. Struct. Biol. 156, 2–11 [DOI] [PubMed] [Google Scholar]
- 32. Söding J., Biegert A., Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Letunic I., Bork P. (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Medalia N., Sharon M., Martinez-Arias R., Mihalache O., Robinson C. V., Medalia O., Zwickl P. (2006) Functional and structural characterization of the Methanosarcina mazei proteasome and PAN complexes. J. Struct. Biol. 156, 84–92 [DOI] [PubMed] [Google Scholar]
- 36. Borchert N., Dieterich C., Krug K., Schütz W., Jung S., Nordheim A., Sommer R. J., Macek B. (2010) Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res. 20, 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 38. Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. (1999) AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43 [PubMed] [Google Scholar]
- 39. Makarova K. S., Yutin N., Bell S. D., Koonin E. V. (2010) Evolution of diverse cell division and vesicle formation systems in Archaea. Nat. Rev. Microbiol. 8, 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Serek-Heuberger J., Hobel C. F., Dunin-Horkawicz S., Rockel B., Martin J., Lupas A. N. (2009) Two unique membrane-bound AAA proteins from Sulfolobus solfataricus. Biochem. Soc. Trans. 37, 118–122 [DOI] [PubMed] [Google Scholar]
- 41. Djuranovic S., Rockel B., Lupas A. N., Martin J. (2006) Characterization of AMA, a new AAA protein from Archaeoglobus and methanogenic archaea. J. Struct. Biol. 156, 130–138 [DOI] [PubMed] [Google Scholar]
- 42. Summer H., Bruderer R., Weber-Ban E. (2006) Characterization of a new AAA+ protein from archaea. J. Struct. Biol. 156, 120–129 [DOI] [PubMed] [Google Scholar]
- 43. Pamnani V., Tamura T., Lupas A., Peters J., Cejka Z., Ashraf W., Baumeister W. (1997) Cloning, sequencing, and expression of VAT, a CDC48/p97 ATPase homologue from the archaeon Thermoplasma acidophilum. FEBS Lett. 404, 263–268 [DOI] [PubMed] [Google Scholar]
- 44. Striebel F., Hunkeler M., Summer H., Weber-Ban E. (2010) The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup's N-terminus. EMBO J. 29, 1262–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang F., Wu Z., Zhang P., Tian G., Finley D., Shi Y. (2009) Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 34, 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gerega A., Rockel B., Peters J., Tamura T., Baumeister W., Zwickl P. (2005) VAT, the thermoplasma homolog of mammalian p97/VCP, is an N domain-regulated protein unfoldase. J. Biol. Chem. 280, 42856–42862 [DOI] [PubMed] [Google Scholar]
- 47. Kusmierczyk A. R., Kunjappu M. J., Kim R. Y., Hochstrasser M. (2011) A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nat. Struct. Mol. Biol. 18, 622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruepp A., Eckerskorn C., Bogyo M., Baumeister W. (1998) Proteasome function is dispensable under normal but not under heat shock conditions in Thermoplasma acidophilum. FEBS Lett. 425, 87–90 [DOI] [PubMed] [Google Scholar]
- 49. Zhou G., Kowalczyk D., Humbard M. A., Rohatgi S., Maupin-Furlow J. A. (2008) Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J. Bacteriol. 190, 8096–8105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reuter C. J., Kaczowka S. J., Maupin-Furlow J. A. (2004) Differential regulation of the PanA and PanB proteasome-activating nucleotidase and 20S proteasomal proteins of the haloarchaeon Haloferax volcanii. J. Bacteriol. 186, 7763–7772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Madding L. S., Michel J. K., Shockley K. R., Conners S. B., Epting K. L., Johnson M. R., Kelly R. M. (2007) Role of the β1 subunit in the function and stability of the 20S proteasome in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 189, 583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Isakov E., Stanhill A. (2011) Stalled proteasomes are directly relieved by P97 recruitment. J. Biol. Chem. 286, 30274–30283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jentsch S., Rumpf S. (2007) Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem. Sci. 32, 6–11 [DOI] [PubMed] [Google Scholar]
- 54. Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. (2008) Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One 3, e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]