Abstract
The stromal ascorbate peroxidase (sAPX) functions as central element of the chloroplast antioxidant defense system. Its expression is under retrograde control of chloroplast signals including redox- and reactive oxygen species-linked cues. The sAPX promoter of Arabidopsis thaliana was dissected in transient reporter assays using mesophyll protoplasts. The study revealed regulatory elements up to –1868 upstream of the start codon. By yeast-one-hybrid screening, the transcription factor ANAC089 was identified to bind to the promoter fragment 2 (–1262 to –1646 bp upstream of translational initiation). Upon mutation of the cis-acting element CACG, binding of ANAC089 was abolished. Expression of a fused fluorescent protein version and comparison with known endomembrane markers localized ANAC089 to the trans-Golgi network and the ER. The transcription factor was released upon treatment with reducing agents and targeted to the nucleus. Transactivation assays using wild type and mutated versions of the promoter showed a partial suppression of reporter expression. The data indicate that ANAC089 functions in a negative retrograde loop, lowering sAPX expression if the cell encounters a highly reducing condition. This conclusion was supported by reciprocal transcript accumulation of ANAC089 and sAPX during acclimation to low, normal, and high light conditions.
Keywords: ascorbate peroxidase, gene expression, redox regulation, retrograde signaling, transcription factor
INTRODUCTION
Photosynthetic metabolism involves light driven electron transfer reactions, production of metabolic intermediates with high reducing potential and the generation of reactive oxygen species (ROS). The photosynthesizing chloroplast is equipped with an intricate redox sensory system, a multilayered antioxidant defense and diverse repair mechanisms to minimize ROS-dependent damage. On the one hand, appropriate sensing allows for using redox and ROS information to adjust photosynthetic metabolism and tune gene expression in the plastids but also in the nuclear genome by retrograde signal transfer from the chloroplast to the nucleus (Baier and Dietz, 2005). Pogson et al. (2008) distinguished retrograde signaling in developmental and operational control. Operational control coordinates nuclear gene expression with the actual needs of photosynthetic metabolism. Analyses of transcriptional dynamics in response to environmental or pharmacological perturbations and elaborated mutant screenings have allowed researchers to pinpoint to signaling cues triggering retrograde signaling in operational control (Leister, 2012). Accumulation of singlet oxygen, changes in redox state of intersystem electron transport chain, redox state of metabolites or proteins as for example thioredoxin downstream of photosystem I, intermediates of chlorophyll synthesis and hormone precursors such as for abscisic acid are linked to specific changes in transcript abundance of nuclear encoded chloroplast proteins (Pfannschmidt, 2010). While ROS generation in the illuminated chloroplast is mostly governed by primary light reactions and counteracted by mechanisms that quench excess excitation energy (Li et al., 2009), ROS are decomposed by high capacity antioxidant systems (Asada, 1999; Dietz et al., 2006).
Superoxide is dismutated by CuZn- and Fe-SOD, and hydrogen peroxide decomposed by thylakoid-bound and stromal ascorbate peroxidase (tAPX, sAPX; Nakano and Asada, 1981) or thiol-dependent peroxidases (Dietz, 2011). It has been estimated that thiol-dependent peroxidase activity reaches about 40% and ascorbate-dependent activity about 60% of total hydrogen peroxide decomposition activity in chloroplasts (Dietz et al., 2006). Both systems are prone to inhibition and thus are subjected to turnover, sAPX and tAPX in particular if ascorbate concentrations are low (Miyake and Asada, 1996). Their expression in the nucleus must be under retrograde control to cope with demand for antioxidant capacity (Oelze et al., 2012).
sAPX and tAPX are encoded by separate nuclear genes in Arabidopsis thaliana. Complete deletion of sAPX and tAPX is compensated in A. thaliana under regular growth conditions (Kangasjärvi et al., 2008). Symptoms of oxidative stress in these plants develop in stressful environment and in particular during early seedling development. sAPX and tAPX transcript levels are regulated in dependence on developmental state of the leaves (Pena-Ahumada et al., 2006) and environmental cues, such as light intensity (Oelze et al., 2012). After transfer of low or normal light acclimated A. thaliana to 100- or 10-fold higher light intensity, respectively, sAPX-mRNA levels start to increase after 30 min and reach a maximum at 3–6 h after transfer to high light (Oelze et al., 2012). In contrast to transcript regulation, sAPX and tAPX protein levels respond much less.
Signaling pathways often innervate transcription factors that modulate target gene expression in response to environmental stimuli (Golldack et al., 2011). However despite the importance of chloroplast ascorbate peroxidases in antioxidant defense, transcription factors involved in their regulation have not been described. Based on transcript regulation it can be assumed, that redox and ROS signals might be involved in regulating tapx and sapx gene expression. Transcriptome analyses have differentiated O2.–/H2O2-dependent regulation from singlet oxygen-dependent regulation (op den Camp et al., 2003). sAPX was not among the significantly regulated transcripts responding to methylviologen or to illumination of protochlorophyllide accumulating flu mutants (op den Camp et al., 2003). Compensatory retrograde regulation is apparent from knock down lines of A. thaliana deficient in the alternative H2O2-detoxifying 2-Cys peroxiredoxin (Baier et al., 2000) where sAPX and tAPX transcripts were up-regulated, and in double knock out of tAPX and sAPX (Kangasjärvi et al., 2008) where 2-Cys peroxiredoxin protein levels were increased. Apparently there is a delicate feedback from the chloroplast antioxidant defense system to nuclear gene expression.
The aim of the study was to approach a better and mechanistic understanding of sapx gene expression regulation. The promoter was analyzed for regulatory regions. The transcription factor ANAC089 identified in a yeast-one-hybrid (Y1H) screening was subjected to a more detailed inspection for binding site and regulatory properties. The specific promoter region sapx2-1 proved to be responsive to oxidative versus reductive cues by ANAC089. The obtained data indicate a role of ANAC089 as repressor of sapx gene activity under highly reducing conditions where the need for sAPX expression or sAPX turnover might be low.
MATERIALS AND METHODS
PLANT GROWTH AND LIGHT TREATMENT
Arabidopsis thaliana was grown in a growth chamber at a photoperiod of 10 h with 80 µmol quanta m–2 s–1 and 21°C, and a dark phase of 14 h at 18°C, both at 50% relative humidity. The pots contained Spezialsubstrat (Stender AG, Schermbeck, Germany), Osmocote Start as fertilizer (Scotts Australia PTY Ltd, Bella Vista, Australia), and one tablet of Lizetan (Bayer, Leverkusen, Germany) per L soil. Three week old plants were either transferred to 8 µmol quanta m–2 s–1 (low light, L-light) or kept at 80 µmol quanta m–2 s–1 (normal light, N-light) for another 10 days. Then the 4.5 week old plants were exposed to high light (H-light, 800 µmol quanta m–2 s–1) 1 h after onset of light (9 am). Control plants were kept in L- or N-light. At 3 p.m. complete rosettes of 4–12 plants from the four treatments were harvested, frozen in liquid nitrogen, and stored at –80°C until further processing.
TRANSCRIPT ANALYSIS
RNA isolation, cDNA synthesis, and semi-quantitative RT-PCR analysis were performed according to Wormuth et al. (2006) using primers as described in Table 1. Equal loading of cDNA was adjusted with ACTIN2 amplificate. Annealing temperatures and amplification cycle numbers were optimized for each target transcript (Oelze et al., 2012).
Table 1.
Oligonucleotide primers used for (a) cloning of the sAPX promotor fragments into the p35SYFP vector using BamHI and NcoI restriction sites and (b) transcript analysis.
| Sequence | |
|---|---|
| Promoter fragment | |
| sAPX-1-for | 5′-AAAAAGGATCCTTTCGGACCTGGAGAG-3′ |
| sAPX-2-for | 5′-AAAAAGGATCCGCACGTCTAGTGAAAGATCC-3′ |
| sAPX-2-mut-for | 5′-AAAAAGGATCCGAAAATCTAGTGAAAGATCC-3′ |
| sAPX-3-for | 5′-AAAAAGGATCCTGTCAACCAAGTCGCCTTG-3′ |
| sAPX-5-for | 5′-AAAAAGGATCCCCCGTCACCATTACCATC-3′ |
| sAPX-6-for | 5′-AAAAAGGATCCCTCTATGGACTTTATTGG-3′ |
| sAPX-rev | 5′-AAAAACCCATGGTTCTGAGGGGTATAATAGTAAT-3′ |
| Transcript analysis | |
| ANAC089-for | 5′-ATGGACACGAAGGCGGTT-3′ |
| ANAC089-DB-rev | 5′-CAATCAGACGGGCTCCCTG-3′ |
| sAPX-for | 5′-ATGCTGCTAACGCTGGTCTT-3′ |
| sAPX-rev | 5′-CCTAACGTGTGAGCACCAGA-3′ |
| Actin-2-for | 5′-TTGGTAGGCCAAGACATCAT-3′ |
| (At3g18780) Actin-2-rev |
5′-GGAGCCTCGGTAAGAAGAAC-3′ |
GENERATION OF YEAST-ONE-HYBRID LIBRARY AND SCREENING
cDNA synthesis, construction of Y1H library and screening were performed using the Clontech Matchmaker system as described in Klein and Dietz (2010). To achieve a wide coverage of conditionally expressed transcripts, RNA was isolated from a set of differentially stress-treated A. thaliana seedlings. The treatments were as follows: (1) Control: 1 h at 120 µmol quanta m–2 s–1; (2) combined oxidative and high light stress: 1 h at 5 mM H2O2 and 1140 µmol quanta m–2 s–1; (3) drought stress and high light: 1 h drought and 1140 µmol quanta m–2 s–1; (4) heat stress: 1 h at 40°C and 84 µmol quanta m–2 s–1; (5) cold and darkness: 1 h at 4°C in darkness; (6) cold in light: 1 h at 4°C and 32 µmol quanta m–2 s–1; (7) UV-illumination: 3 × UV for 10 s each with regeneration for 30 min at 120 µmol quanta m–2 s–1; (8) dark-light transition: 1 h darkness followed by 30 min 120µmol quanta m–2 s–1; (9) high light: 1 h at about 1100 µmol quanta m–2 s–1; and (10) low salt: 1 h at 5 mM NaCl and 1100 µmol quanta m–2 s–1. The bait DNA sequence was cloned into the pHis2 vector using the SmaI and SacI endonucleases, while the pGADT7-Rec2 was used as the prey vector. Both vectors were cotransformed into yeast strain Y187. Interaction between fusion protein and DNA-sequence was scored on SD medium supplemented with the appropriate amino acids, i.e., SD/-His/-Leu/-Trp. To suppress leaky His3 activity, 3-amino-1,2,4-triazol (3-AT) was added, for the promoter fragments sapx2-1 and sapx2-1mut, the 3-AT concentration was adjusted to 15 mM.
AMPLIFICATION OF sapx PROMOTER FRAGMENTS
sapx (At4g08390) promoter fragments were amplified by polymerase chain reaction using the primers listed in Table 1. Amplification products were separated by agarose gel electrophoresis, eluted and used for cloning. For transient reporter assays they were fused to EYFP as described in Shaikhali et al. (2008). As reference construct, CFP was fused downstream of the p35S-promoter (Shaikhali et al., 2008). Correctness of constructs was confirmed by DNA sequencing (MWG Biotech, Eberswalde or CeBiTec, Bielefeld, Germany).
RECOMBINANT PRODUCTION OF ANAC089 PROTEIN AND ELECTROPHORETIC MOBILITY SHIFT ASSAY
The anac089 coding sequence was amplified using forward (ANAC089-for: 5′-AAAAAAGGATCCATGGACACGAAGGCGGTTG-3′) and reverse primers (ANAC089-rev: 5′-AAAAACTCGAGTTCTAGATAAAACAACATTG-3′) and directionally cloned into pET28a vector using BamHI and XhoI restriction sites. The vector was transformed into BL21 (DE3) pLysS Escherichia coli cells. Following inoculation with preculture the main culture was grown to OD = 0.6, induced with 400 µM isopropyl-β-D-thiogalactopyranoside and further incubated for 4 h. Cells were sedimented, frozen, and thawn, resuspended in lysis buffer (50 mM Na-phosphate buffer, pH 8, 300 mM NaCl, 10 mM imidazole, 1 mg/ml lysozyme), sonified, incubated and cell debris sedimented by centrifugation. The His6-tagged ANAC089 protein was purified by Ni-nitrilotriacetic acid chromatography (Qiagen, Hilden, Germany). Following washing of loosely bound proteins, ANAC089 protein was eluted with elution buffer (50 mM Na-phosphate buffer, pH 8, 300 mM NaCl, 250 mM imidazole). Recombinant protein was dialyzed against 40 mM K-phosphate buffer, pH 7, using dialysis tubing with 10 kDa cutoff. Protein was quantified with BioRad reagent (BioRad Laboratories, München, Germany) with bovine serum albumin as standard. An electrophoretic mobility shift assay (EMSA) was performed with the DIG Gel Shift Kit (2nd generation, Roche, Mannheim, Germany): target DNA was the sAPX promoter fragment 2-1 at an amount of 8 ng per lane. As indicated unlabeled competitor DNA was added at 2 µg concentration and ANAC089 protein at 100 ng. An additional label free EMSA was performed as well. Here recombinant ANAC089 protein equivalent to 360 µM concentration was incubated with 25 ng promotor fragment sapx-2 (or sapx-2mut) in 20 µl EMSA buffer (100 mM HEPES, pH 7.5, 500 mM KCl, 25% glycerol, 5 mM dithiothreitol) at 22°C for 30 min. The assay mix was subsequently loaded on 4% agarose gels and the DNA separated by electrophoresis at 80 V. DNA was visualized by ethidium bromide staining and documented.
CONSTRUCTS FOR FLUORESCENCE MICROSCOPY
ANAC089 was fused to fluorescent proteins for various cell imaging experiments. In each case the enhanced variants of CFP and YFP (ECFP, EYFP) were used for the construction of the vectors. In the following, the short versions CFP and YFP are used in the text. In the transactivation analysis of Figure 7, mCherry was fused to ANAC089 (Seidel et al., 2010). The constructs were p35S:CFP:ANAC089, p35S:YFP:ANAC089, p35S:ANAC089:CFP, p35S:ANAC089:YFP, p35S:YFP:ANAC089:CFP, and p35S:mCherry:ANAC089. The cloning strategy included the insertion of the anac089 gene upstream of the fluorophore gene into the p35S:CFP-NOST and the p35S:YFP-NOST vector using the ANAC089-for (5′-AAAAAAGGATCCAATGGACACGAAGGCGGTTG-3′) and ANAC089-rev (5′-AAAAAACCGGTTCTAGATAAAACAACATT-GC-3′) primers for gene amplification. The anac089 gene was then fused to the vector utilizing the BamHI and AgeI endonucleases. The p35S:CFP:ANAC089 and p35S:YFP:ANAC089 constructs were created using the ANAC089-for (5′-AAAAAGCGGCCGCATGGACACGAAGGCGGTT-3′) and ANAC089-rev (5′-AAAAAGAATTCTTATTCTAGATAAAACAACA-3′) primers for the gene amplification which was inserted into the vector using the NotI and EcoRI endonucleases. The generation of the p35S:YFP:ANAC089:CFP construct was performed in two steps. First the anac089:cfp hybrid gene was amplified using the p35S:ANAC089:CFP vector as DNA template. The ANAC089-for (5′-AAAAAAGCGGCCGCATGGACACGAAGGCGGTT-3′) and CFP-rev primer (5′-AAAAAGAATTCTTACTTGTACAGCTCGTC-3′) allowed to insert the amplified hybrid gene into the NotI and EcoRI restriction sites of the p35S:YFP-C vector, resulting in the p35S:YFP:ANAC089:CFP construct. The various psAPX-YFP promoter fragments were generated using the primer combinations from Table 1. The deletion fragments were finally cloned into the pEYFP vector. Gos12 was cloned into 35S-CFP-Nost using the oligonucleotides Gos12-BamHI-for (5′-AAAAGGAT-CCAATGACAGAATCGAGTCTGGAT-3′) and Gos12-AgeI-rev (5′-AAAAACCGGTGATTTTGAGAGCCAGTAGATGAT-3′). Sar1 (Sar1-BamHI-for 5′-AAAAGGATCCAATGTTCCTGGTGGATT-GG-3′; Sar1-AgeI-rev 5′-TTTTACCGGTCCGTCGATATATTGA-GA-3′), and Clathrin light chain (ClathrinLC-BamHI-for 5′-AAAAGGATCCAATGTCGTCAACCTTGAGC-3′; ClathrinLC-AgeI-rev 5′-TTTT-ACCGGTCCCAACTTCTCTGTAAC-3′) were cloned into 35S-CFP-NosT and 35S-YFP-NosT using BamHI and AgeI restriction sites and plasma membrane ATPase AHA1 (AHA1-BamHI-For 5′-AAAAGCGGCCGCATGTCAGGTCTCGAAGAT-3′; AHA1-EcoRI-rev 5′-TTTTTGAATTCTACACAGTGTAGTGA-TG-3′) was cloned into 35S-CFP-C and 35S-YFP-C, respectively, using NotI and EcoRI restriction sites. Emissions of fused fluorescent protein in cotransfection or transactivation experiments were quantified in the appropriate detector channel of the confocal laser scanning microscope as described in Shaikhali et al. (2008).
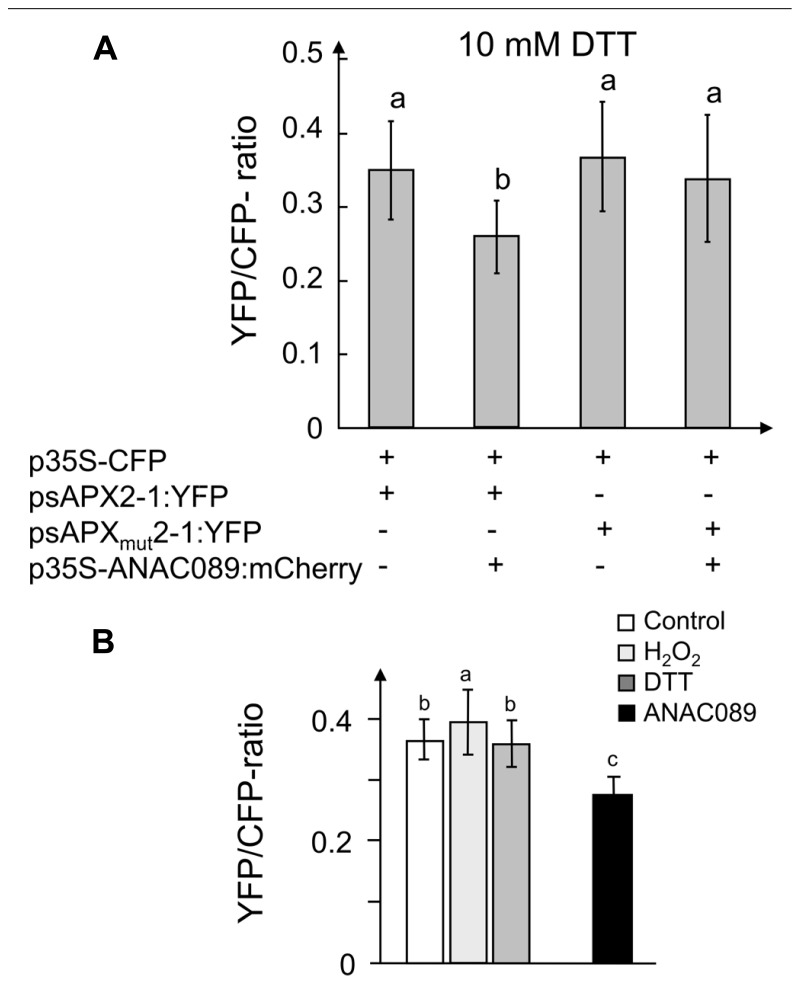
FIGURE 7.
Effect of overexpressed ANAC089 on reporter gene expression and confirmation of involved cis-element CACG. (A) (1st bar) Co-transformation of fragment psapx2-1:YFP with 35S-CFP. (2nd bar) Co-transformation of psapx2-1:YFP with 35S-ANAC089-mCherry and 35S-CFP. (3rd bar) Co-transformation of psapx2-1mut:YFP with 35S-CFP. (4th bar) Co-transformation of psapx2-1mut with 35S-ANAC089-mCherry and 35S-CFP. Prior to the measurement protoplasts were incubated with 10 mM DTT for 2 h. The result represent means ± SD from two experiments with a total n > 30 protoplasts per construct. (B) A similar effect was seen if the full length promoter (–1 to –1868) was used. The first three columns are as in Figure1 (expression in control, H2O2, or DTT-treated protoplasts), the set of three columns on the left hand side represent reporter strength under control conditions, in the presence of H2O2 and DTT, respectively, the fourth column shows the result with co-expressed p35S:ANAC089. The data are means ± SD from 68, 56, 56, 72, 74, and 83 protoplasts (from left to right column). Different letters indicate significant difference at p < 0.05 (Student’s t-test).
REDOX REGULATION OF THE sAPX PROMOTER
Protoplasts were prepared from At7 cell suspension culture of A. thaliana (Seidel et al., 2010). Five days after passage to new medium, cells were sedimented, washed in 50 ml 240 mM CaCl2, digested and transfected with sapx:YFP promoter constructs as described in Seidel et al. (2004). After incubation at 26°C in darkness for 16 h the different batches were adjusted to the final H2O2 concentration of 5 mM and DTT of 10 mM, respectively, or left as controls and incubated for further 2 h in darkness at 26°C for 16 h. The relative YFP and CFP emission intensities were measured using the confocal laser scanning microscope (Leica SP2 Heidelberg, Germany). Finally, out of those three different samples the YFP/CFP was calculated.
CELL IMAGING OF SUBCELLULAR ANAC089 LOCALIZATION AND PROCESSING
Protoplasts prepared from the At7 cell suspension culture were transfected either with the construct combination 35S:ANAC089:CFP and 35S:ANAC089:YFP or 35S:CFP:ANAC089 and 35S:YFP:ANAC089. Homodimerization of ANAC089 in vivo was analyzed by Förster resonance energy transfer (FRET; Seidel et al., 2005). Fluorescence of single protoplasts was imaged with a confocal laser scanning microscope (Leica SP2, Heidelberg, Germany). Calculation of FRET efficiency was done as described in Seidel et al. (2005).
BIOINFORMATIC ANALYSIS OF PROMOTER SEQUENCES AND AMINO ACID SEQUENCES
sapx promoter sequences were searched for the presence of putative cis-elements with the program Matinspector1. The promoters of 27416 A. thaliana genes with a length of 3000 bp were downloaded from TAIR2 and screened for the ATGCACGTC motif allowing for 1 bp mismatch with the program CLC Main Workbench3 . Search for transcripts co-expressed with sAPX was performed using the online tool offered at http://www.arabidopsis.leeds.ac.uk/act/coexpanalyser.php. Cleavage site prediction of ANAC089 was performed with ProP1.04 and resulted in two hits, a peculiar Arg/Lys-specific site at amino acid position 163 and a second site at position 297 just close to the membrane spanning α-helix based on prediction from mammalian protease processing sites.
RESULTS
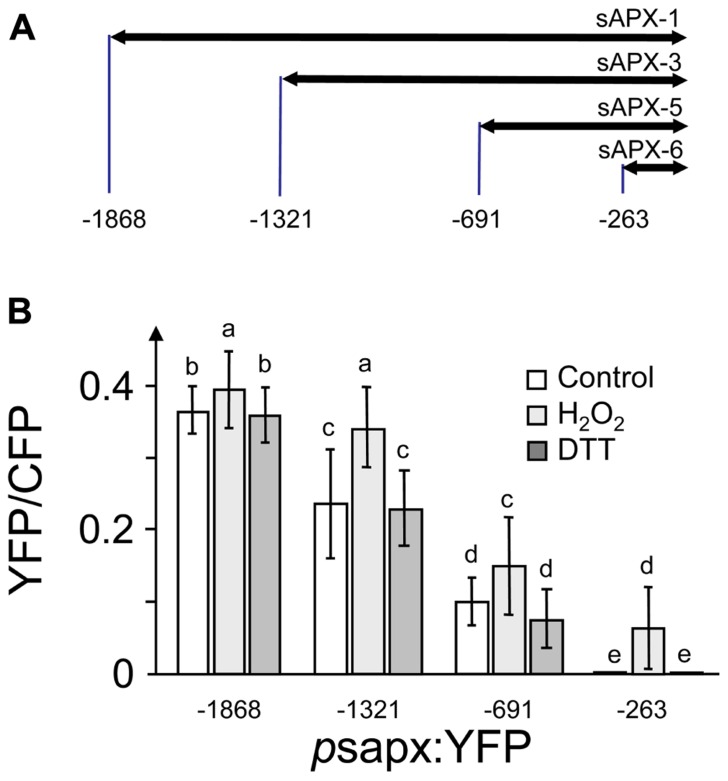
The promoter region of sapx was cloned as assumed full length or truncated form and fused to enhanced yellow fluorescent protein (YFP) as reporter gene. The selected sapx1 DNA sequence started from –1868 bp upstream of the translational initiation site down to –1, sapx3 from –1321 to –1, sapx5 from –691 to –1 and sapx6 from –263 to –1. At7-protoplasts were co-transfected with these EYFP reporter constructs simultaneously with a p35S:CFP construct as reference to correct for variable expression levels. The normalized ratios of YFP/CFP were calculated and plotted against the DNA fragment length (Figure 1). Transcriptional enhancers were present in all segments since reporter activity decreased with each truncation. H2O2 stimulated expression driven by each fragment significantly.
FIGURE 1.
FIGURE 1. Deletion fragments of the sapx promoter constructed for promoter activation assay in A. thaliana protoplasts. (A) Schematics of the sapx promoter fragments. (B) Expression of the YFP reporter gene fused downstream to the sapx promoter deletion fragments under control, oxidizing (5 mM H2O2) and reducing conditions (10 mM DTT) normalized to CFP accumulation under control of the 35S promoter. (1) Promoter fragment psapx1; (2) promoter fragment psapx3; (3) promoter fragment psapx5; and (4) promoter fragment psapx6. Intensities of CFP and YFP were quantified in the respective emission channels of the CLSM (Shaikhali et al., 2008). The results are means ± SD of 60–80 protoplasts. Different letters mark groups of significant difference with p < 0.05 (Student’s t-test).
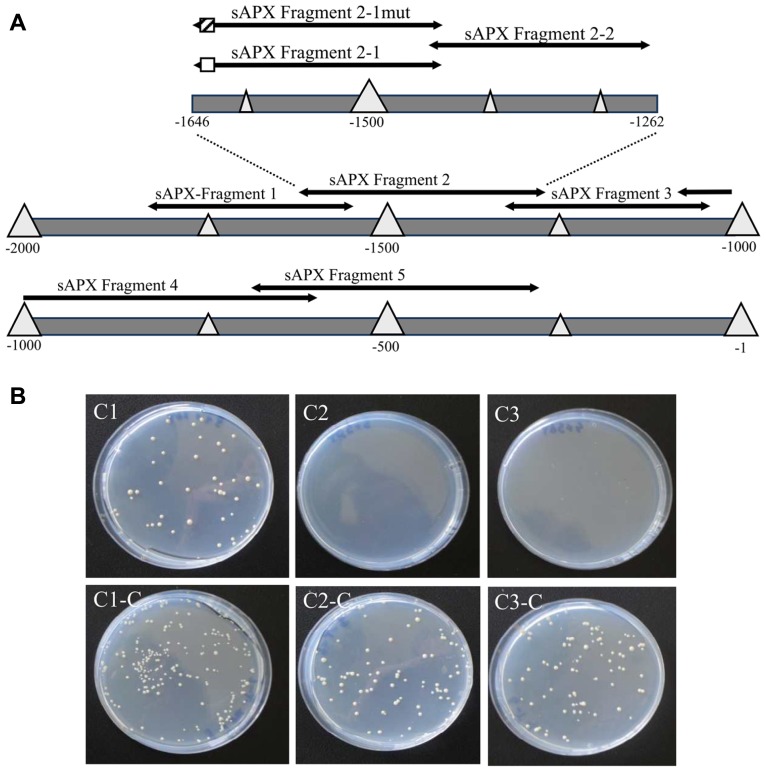
Since the data of Figure 1 did not reveal a clear and restricted redox regulation site, the sapx promoter was fragmented into five overlapping segments (Figure 2) and used for a Y1H screening. 3-AT concentrations to suppress for leaky His3-expression without transactivator were adjusted for each construct and were within recommended concentration range for sapx1 and sapx2 with 15–20 mM, and already quite high for sapx3 and sapx4 (80 and 110 mM, respectively). The sapx5 fragment remained leaky even at >120 mM 3-AT. This may explain why reliable and specific binding of promising candidates could not be observed for sapx3 to 5. Among the many positive clones only the gene encoding ANAC089 (At5g22290) which was identified as activator of sapx2 fragment driven reporter expression in yeast appeared promising for further analysis. Retransformation of yeast cells with isolated bait and prey vectors reproduced the interaction between ANAC089 and sapx2. Bioinformatic analysis identified two potential binding sites for NAC transcription factors in sapx2 (sequence position –1646 till –1431, original sequence: GCACGTCTAGTGAAAGATCC, mutated sequence: GAAAATCTAGTGAAAGATCC; second possible binding site was the motive CATCCC at –1627 till –1622). Subcloning confined the transactivation to the sapx2-1 fragment of 216 bp length (Figure 2B, C1) and mutation of the upstream located CACG motif to AAAA (position –1645 to –1642) abolished transactivation of HIS3 reporter in the sapx2-1mut:HIS3 reporter construct after cotransfection with ANAC089 (Figure 2, C3).
FIGURE 2.
Schematics of the promoter of sapx used for yeast-one-hybrid screening and binding to the specific cis-element. (A) Fragment psapx1 (–1563 to –1868 bp); psapx2 (–1262 to –1646 bp); psapx3 (–954 to –1321 bp); psapx4 (–609 to –1018 bp); psapx5 (–246 to –691 bp); psapx6 (–1 to –263 bp). Division of psapx2 for localization of the ANAC-binding cis-element: pHis2:sapx2-1/mut (–1432 bis –1646), pHis2:sapx2-2 (–1432 bis –1451). The potential target element for binding of ANAC089 is marked as hatched and the mutagenized region in white box. (B) Experimental identification of the binding site of ANAC089 within promoter segment pHis2:sapx2 using the Y1H system on SD/-Trp/-His/-Leu/+3 AT selective medium (C1, C2, C3) and for control on SD/-Trp/-Leu selective medium (C1-C, C2-C, C3-C). (C1) p His2:sapx2-1; (C2) pHis2:sapx2-1mut; and (C3) p His2:sapx2-2. All cells grew under non-stringent selection showing their viability, while under stringent conditions only the cells grew that were harboring the wild type sapx2-1 fragment and expressing ANAC089.
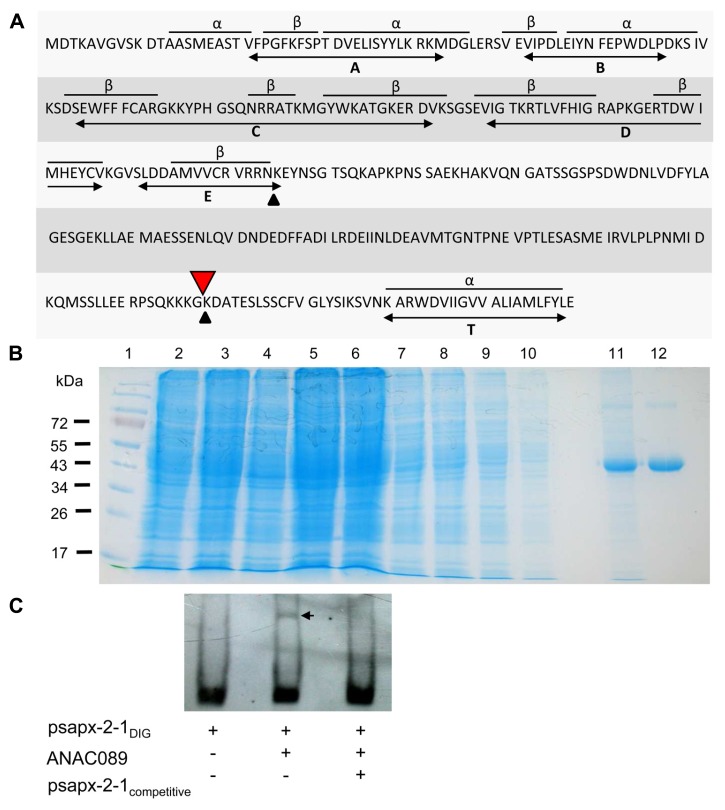
Analysis of the in silico translated cDNA allowed the identification of the N-terminal NAC domain and a putative C-terminal membrane anchor (Figure 3A). ANAC089 protein was recombinantly expressed in E. coli as His6-tagged protein and purified by Ni-affinity chromatography (Figure 3B). Binding of His6-ANAC089 to sapx2-1 promoter fragment in vitro was studied using the EMSA (Figure 3C). A shift was seen upon addition ANAC089 protein and the shift was abolished upon addition of 250-fold excess competitor DNA. Mutation of the binding site abolished the ANAC089-dependent gel shift (not shown).
FIGURE 3.
Structure of ANAC089, generation of recombinant His 6-ANAC089 and EMSA. (A) Primary structure of the ANAC089 transcription factor. The N-terminal ANAC domain consists of three α-helices and eight β-sheets in conserved arrangement (A–E). The bioinformatically predicted transmembrane α-helix (T) of the ANAC089 protein is localized at amino acid position 319–339. Putative protein cleavage sites are indicated with arrows. His6-ANAC089 was expressed in E. coli and purified by Ni-NTA affinity chromatography. (B) Heterologous expression and purification of His6-tagged ANAC089. The recombinant ANAC089 protein was expressed in BL21 (DE3) pLysS E. coli cells which were induced with 400 µM isopropyl-β-D-thiogalactopyranoside for 4 h. By SDS PAGE the time-dependent expression of the recombinant ANAC089 protein, as well as the purification process was analyzed. (1) Prestained protein ladder (Fermentas); (2) 0 h after IPTG induction; (3) 2 h after IPTG induction; (4) 4 h after IPTG induction; (5) cell extract; (6) cell pellet; (7–10) column wash fractions; (11) first fraction of ANAC089 elution; and (12) second fraction of ANAC089 elution. (C) Electrophoretic mobility shift assay: ANAC089 promoter fragment interaction in vitro using the DIG Gel Shift Kit (2nd generation, Roche). Verification of the specificity of ANAC089 binding to the cis-element within the sAPX promoter fragment 2-1. Promoter fragment psapx2-1 was loaded at 8 ng DNA. The amount of unlabeled competitor was 2 µg DNA. ANAC089 protein was added at 100 ng. The arrow head marks the shifted band.
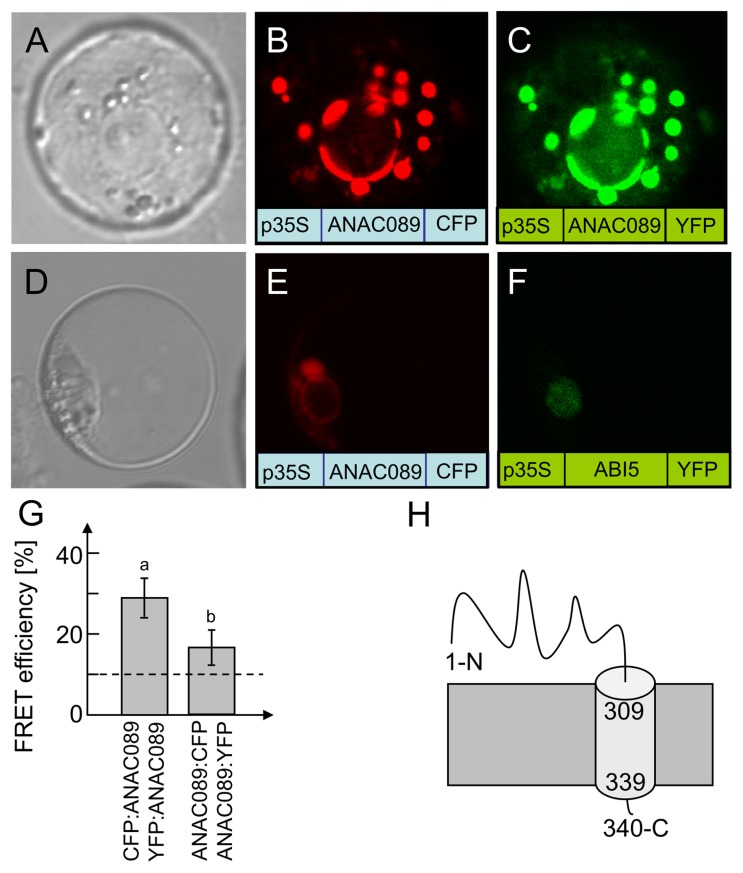
At7 protoplasts were transfected with constructs of p35S:ANAC089:CFP and p35S:ANAC089:YFP (Figure 4). Both constructs were co-expressed and the reporter fluorescence was observed in the plasma membrane and membrane vesicles (Figures 4B,C). Analysis for protein–protein interaction by FRET between the CFP- and YFP-fused ANAC089 revealed homooligomer formation (Figure 4G). FRET efficiency was significantly above the threshold which was defined as 10% from transfection results with free CFP and YFP (Seidel et al., 2005). FRET was higher if CFP and YFP were fused to the N-terminus of ANAC089. In this case, the fluorophore must have rested in the cytosol. Apparently, also the membrane-associated transcription factor is able to oligomerize. FRET between CFP:ANAC089 and YFP:ANAC089 was at about 28%, whereas the fusion the CFP and YFP fluorophores to the C-terminus of the ANAC089 resulted in 14% FRET efficiency, a value also above the threshold. Lower FRET might be explained by steric constraints, higher distance, or less stable interaction at the C-terminus of ANAC089 and cannot be distinguished by this in vivo method.
FIGURE 4.
Subcellular localization of ANAC089 in A. thaliana protoplasts. (A) Protoplast in bright field. Fluorescence image of a protoplast expressing the C-terminally CFP-tagged ANAC089 (B) or the C-terminally YFP-tagged ANAC089 (C). Comparative localization of ANAC089 und ABI5 with C-terminally fused fluorophores in A. thaliana protoplasts. (D) Bright field control picture. (E) ANAC089-CFP-signal detected in vesicles, endoplasmic reticulum, and plasmamembrane. (F) ABI5-YFP-signal in cell nucleus. (G) Dimerization detection by FRET analysis: protoplasts were co-transfected with either CFP:ANAC089 and YFP:ANAC089:YFP (left column) or ANAC089:CFP and ANAC089:YFP (right). In both cases significant FRET above the threshold of co-expressed free CFP and YFP could be detected (mean ± SD, n > 60 protoplasts, three transfections). (H) Putative topology of ANAC089 in the membrane.
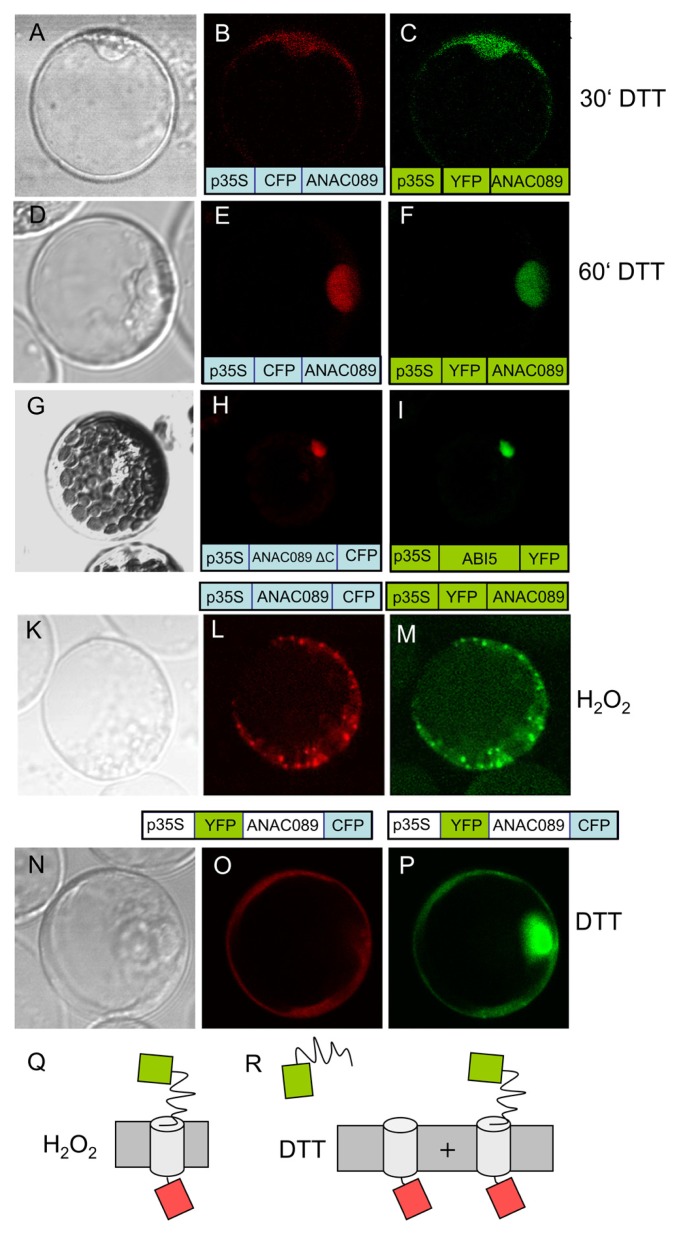
ABI5, a transcription factor with established localization in the nucleus (Lopez-Molina et al., 2003), was used as a control (Figure 4F) and accumulated at a site distinct to ANAC089. Analysis of the coding region with the program TMHMM Server v. 2.0 at http://www.cbs.dtu.dk/services/TMHMM-2.0/ allowed for the identification of a putative transmembrane domain at the C-terminus (Figure 4H, see also Figure 3A). ANAC089 has been reported before to be a member of a subgroup within the family of NAC transcription factors which is tethered to membranes (Kim et al., 2007). Incubation of transfected protoplasts with dithiothreitol (10 mM) showed time-dependent release of ANAC089 from the membrane and translocation to the nucleus when the fluorophore was fused to the N-terminus of the transcription factor (Figures 5A–F). ANAC089 lacking the C-terminal membrane domain localized to the nucleus similar to the nuclear transcription factor ABI5 (Figures 5G–I). ANAC089 remained tethered to the membrane under oxidizing conditions established by addition of 10 mM H2O2 to the protoplast suspension (Figures 5K–M). The next experiment addressed the question whether the carboxyterminus rests at the peripheral membranes after cleavage. The protoplasts were transfected with the triple fusion construct YFP:ANAC089:CFP (Figures 5N–P). Following treatment with DTT, the released YFP:ANAC protein accumulated in the nucleus, while the CFP fused to the carboxyterminus of ANAC stayed at the membrane despite the reductive treatment of the protoplasts.
FIGURE 5.
Redox-dependent dynamics of ANAC089 localization. (A–F) Time-dependent localization of ANAC089 was documented following a treatment with dithiothreitol (10 mM DTT). Images were taken at t = 30 min (A–C) and t = 60 min (D–F). (A, D) Bright field image. (B, E) Fluorescence signal of N-terminally fused CFP (ANAC089-CFP). (C, F) Fluorescence signal of N-terminally fused YFP (ANAC089-YFP). (G–I) Nuclear accumulation of ANAC089 lacking the transmembrane domain. Protoplasts were transformed with the 3′-truncated ANAC089 coding for a variant that lacks the C-terminal membrane domain (total remaining length 298 aa, see Figure 3A, red arrow). In parallel the protoplasts were transformed with ABI5 expression construct. Both fluorophore-fused proteins localized to the nucleus. (K–M) Localization of N-terminally fused fluorophores in response to H2O2. Protoplasts of A. thaliana were treated with 5 mM H2O2 for t = 60 min. (K) Bright field. (L) ANAC089:CFP-signal. (M) ANAC089:YFP-signal. (N–P) Translocation of ANAC089 to the nucleus following reductive activation with 10 mM DTT for 60 min. In this case a double fusion was used, namely YFP:ANAC089:CFP under control of the p35S. (N) Bright field image. (O) CFP signal of the membrane-associated domain. (P) YFP signal of the cleaved N-terminal domain of the ANAC089-YFP in the nucleus. The transfection experiments were repeated three times with 70–90% of the successfully transformed protoplasts showing the same result. (Q,R) Schematics of underlying model explaining release of YFP to the nucleus following cleavage: CFP in each case rests at the membrane due to its fusion to the membrane-residing helix. The N-terminally fused YFP also rests at the membrane under oxidizing conditions (M), or is partially or fully released with time under reducing conditions which cause cleavage of the ANAC089 domain from the membrane anchor (P).
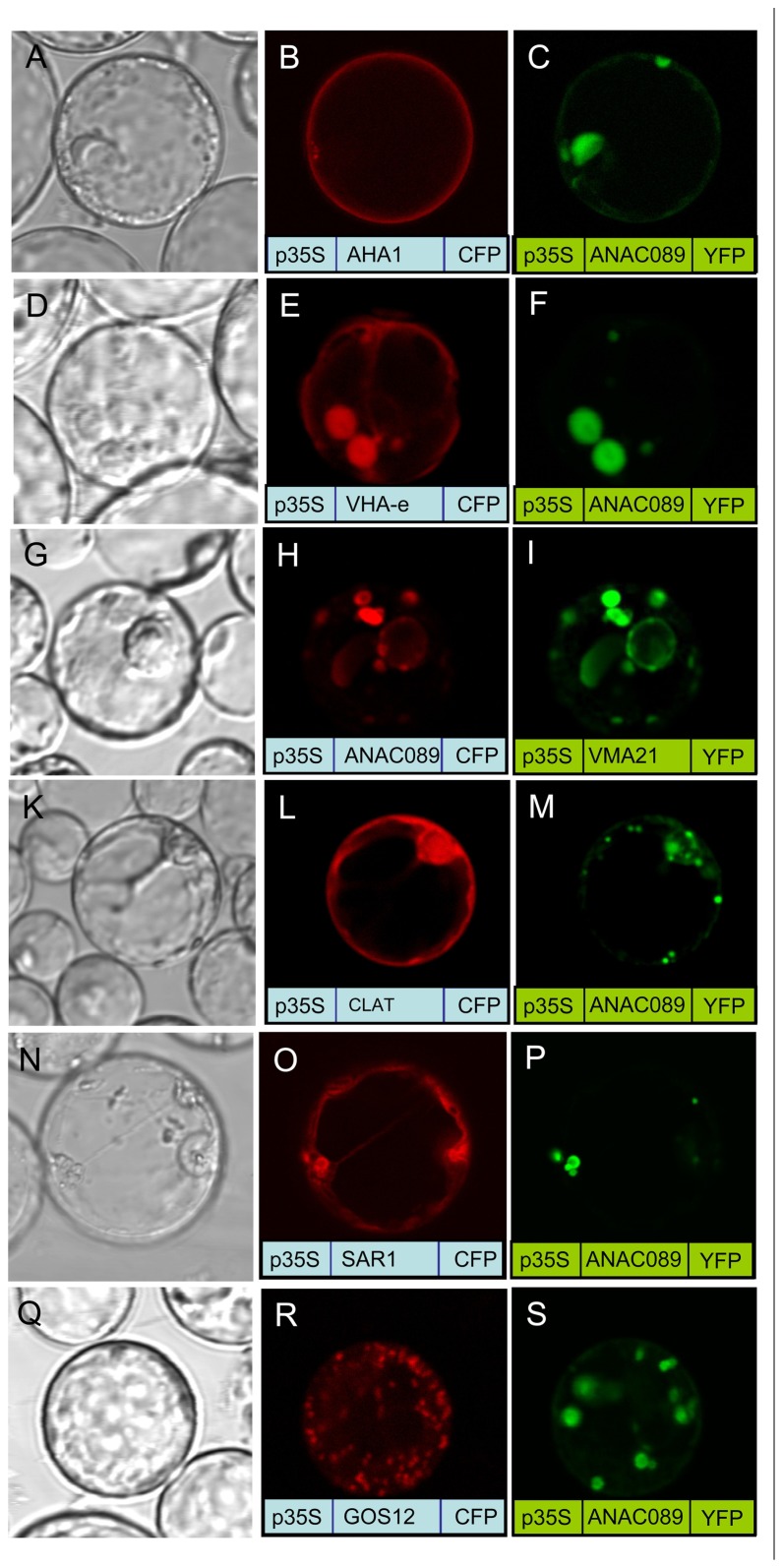
The membrane localization of the membrane-anchored ANAC089 was investigated in more detail in protoplasts co-transfected with a set of marker proteins for compartments of the secretory pathway (Uemura et al., 2004; Hanton et al., 2008; Neubert et al., 2008; Seidel et al., 2008; Hwang and Robinson, 2009; Figure 6). No co-localization was seen with the plasmamembrane H+-ATPase and the Golgi SNARE Gos 12 (Figures 6A–C,Q–S), while VHA-e and Vma21a (Figures 6G–I) showed a high degree of co-localization with ANAC089, respectively (Figures 6D–F). All other marker proteins, the light chain of Clathrin (CLAT; Figures 6K–M) and Sar1 (Figures 6N–P) showed only partial co-localization. The data are consistent with a preferred localization of ANAC089 in the trans-Golgi network and in the ER.
FIGURE 6.
Co-localization of fluorophore-fused ANAC089 with marker proteins. Protoplasts were co-transfected with ANAC089 fused to either CFP or YFP as indicated and a marker protein as follows: (A–C) AHA1, the plasmamembrane H+-ATPase, (D–F) VHA-e, a subunit of the V0 domain of V-ATPase that localizes to TGN/EE, (G–I) VMA21a, an ER-resident assembly factor of the V-ATPas, (K–M) Clathrin, the light chain of the clathrin coat, (N–P) Sar1, the small GTPase of COP II vesicle transport anterograde transport between ER and Golgi, and (Q–S) GOS12, a Golgi SNARE.
The regulatory effect of ANAC089 on sapx gene expression was studied in transactivation assays (Figure 7). A. thaliana mesophyll protoplasts were transfected with (i) a reference constructs expressing CFP under the control of the 35S-promoter, (ii) the yfp gene placed under control of the psap2-1 and psap2-1mut promoter sequences as transactivation reporter, and (iii) and the construct ANAC089:mCherry as transcriptional modifier. mCherry was fused to the C-terminus of the ANAC089 to check for expression of the anac089-gene in transfected protoplasts. The ratio of YFP to CFP was constant in protoplasts transfected with all combinations except for the cells containing the wild type psap2-1 promoter and expressing ANAC089. Here a significant suppression of YFP reporter intensity by 25% was observed. This suppression was not detected in protoplasts transfected with the mutated promoter proving the importance of the CACG-element in the suppressive regulation. The same suppressive effect was detected when using the full length sapx promoter in the absence or presence of co-expressed ANAC089 (Figure 7B).
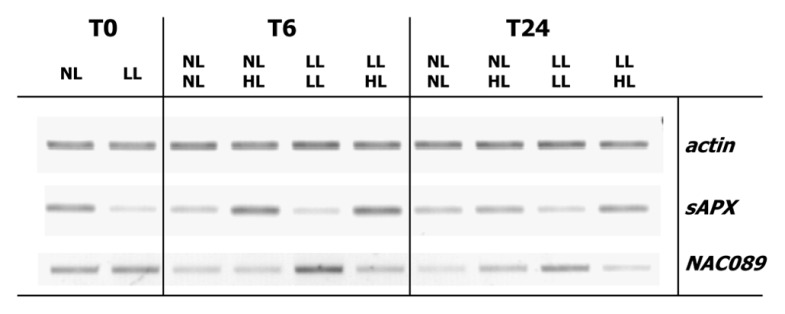
Finally the relationship between anac089 and sapx gene expression was studied in A. thaliana rosettes subjected to a light shift treatment similar to the experiment described in Oelze et al. (2012). Plants fully acclimated to a shady growth condition for 10 days (8 µmol quanta.m–2 s–1) or grown under normal growth conditions (80 µmol quanta.m–2 s–1) were transferred to high light (800 µmol quanta.m–2 s–1) for 6 h. This treatment is known to induce strong regulation of sAPX transcript amounts (Oelze et al., 2012). sAPX and ANAC089 transcript amounts were semiquantified and normalized to ACTIN2 transcript amounts (Figure 8). While sAPX-mRNA levels were higher in normal light grown plants than in low light acclimated ones, they increased upon transfer to high light. The response was stronger 6 h after transfer than after 24 h. ANAC089 mRNA levels behaved in a roughly inverse manner. They were high when sAPX transcript levels were low and vice versa.
FIGURE 8.
Regulation of sAPX and ANAC089 transcript amounts in a light shift experiment. A. thaliana were grown for 3 weeks in normal light, either maintained under this condition for another 10 days (80 µmol quanta m–2 s–1, N-light) or acclimated to very low light (8 µmol quanta.m–2 s–1, L-light) for 10 days. The plants were transferred to high light (800 µmol quanta m–2 s–1, H-light). Rosettes were harvested at t = 0, after 6 or 24 h. RNA was isolated and processed as described by Oelze et al. (2012). Semiquantitative RT-PCR was performed following cDNA synthesis. DNA products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Band intensity indicates relative abundance of transcripts.
In the last step, the presence of the cis-element binding ANAC089 in sAPX promoter was searched in the whole A. thaliana genome. To this end the frequency of the putative ANAC cis-element ATGCACGTC identified by Matinspector with a score of 0.979 was investigated in the whole A. thaliana genome. The promoters of 27416 A. thaliana genes with a length of 3000 bp were downloaded from TAIR5 and screened for the ATGCACGTC motif with the program CLC Main Workbench6. The fully conserved ATGCACGTC motif was found in 196 promoters, either in forward or reverse orientation. In 6903 promoters the motif was present with a single nucleotide exchange. sAPX co-expression was analyzed at http://www.arabidopsis.leeds.ac.uk/act/coexpanalyser.php. and the 1400 most co-expressed genes were compared with the identified ANAC cis-element containing promoters. The obtained list contains candidate genes that might be partially co-regulated with sAPX based on the presence of the ATGCACGTC motif (Table 2). Among the genes containing the ATGCACGTC motif (one mismatch allowed) with a co-expression p-value > 0.5 are many candidates that show a functional link to stress acclimation and redox metabolism like glutathione reductase, flavonoid biosynthesis enzymes or mitochondrial function.
Table 2.
Bioinformatic identification of genes that carry the ATGCACGTC motif in their promoter and co-expressed with sAPX.
| Identifier | p-value | r-value | Function |
|---|---|---|---|
| AT4G33520 | 0.628337 | 8.9e–37 | Metal-transporting P-type |
| ATPase AT3G29200 | 0.611953 | 1.8e–34 | Chorismate mutase, chloroplast (CM1) |
| AT3G55120 | 0.594613 | 3.6e–32 | Chalcone–flavanone isomerase |
| AT3G24170 | 0.592559 | 6.6e–32 | Glutathione reductase, putative |
| AT2G45440 | 0.585632 | 4.9e–31 | Dihydrodipicolinate synthase 2 (DHDPS2) |
| AT1G63290 | 0.569233 | 4.8e–29 | Ribulose-phosphate 3-epimerase, cytosolic, putative |
| AT4G16265 | 0.560068 | 5.5e–28 | DNA-directed RNA polymerase II, putative |
| AT1G18320 | 0.559598 | 6.3e–28 | Mitochondrial import inner membrane translocase subunit Tim17 |
| AT3G17609 | 0.551960 | 4.5e–27 | bZIP transcription factor/HY5-like protein (HYH) |
| AT3G06680 | 0.544932 | 2.7e–26 | 60S ribosomal protein L29 (RPL29B) |
| AT3G49180 | 0.530617 | 8.8e–25 | Transducin family protein/WD-40 repeat family |
| AT5G44160 | 0.528320 | 1.5e–24 | Zinc finger (C2H2 type) family protein |
| AT2G20450 | 0.523633 | 4.5e–24 | 60S ribosomal protein L14 (RPL14A) |
| AT2G26380 | 0.522760 | 5.6e–24 | Disease resistance protein-related/LRR protein |
| AT5G59850 | 0.519257 | 1.3e–23 | 40S ribosomal protein S15A |
| AT3G10530 | 0.519230 | 1.3e–23 | Transducin family protein/WD-40 repeat family |
| AT5G20600 | 0.514334 | 3.8e–23 | Expressed protein |
| AT5G50810 | 0.513390 | 4.7e–23 | Mitochondrial import inner membrane translocase TIM8 |
| AT5G15910 | 0.512829 | 5.4e–23 | NAD(P)-dependent steroid dehydrogenase, related |
| AT2G23350 | 0.512790 | 5.4e–23 | Polyadenylate-binding protein, putative/PABP, putative |
| AT2G17270 | 0.510441 | 9.2e–23 | Mitochondrial substrate carrier family protein |
| AT4G36020 | 0.508859 | 1.3e–22 | Cold-shock DNA-binding family |
| AT1G60850 | 0.507399 | 1.8e–22 | DNA-directed RNA polymerase, putative |
Following a genome-wide search with the ATGCACGTC motif, the hits were compared with the promoters of sAPX-co-regulated transcripts. The list gives the results with a p-value >0.5.
DISCUSSION
This work identified and describes the transcription factor ANAC089 as a negative regulator of sapx expression (Figure 7). The A. thaliana genome encodes close to 2,000 putative transcription factors (Iida et al., 2005). The plant-specific NAC transcription factor family comprises about 110 genes in A. thaliana. Their usually N-terminally conserved NAC domain of about 60 amino acids was first identified in the three representatives named NAM (no apical meristem), ATAF1/2 and CUC2 (cup shaped cotyledon; Jensen et al., 2010). The NAC domain contains the DNA binding domain, dimerization domain and nuclear localization sequence (Ernst et al., 2004). ANAC089 possesses all these features in its N-terminus.
NAC transcription factors have been shown to be involved in developmental processes, hormonal signaling and responses to abiotic and biotic stresses (Hu et al., 2010). Interestingly, several members of the NAC transcription factor family possess transmembrane anchors. They belong to the membrane-tethered transcription factors (MTTFs). The membrane-associated form is inactive by immobilization. Their proteolytic processing releases the functional TF. The transcript level of NAC-MTTFs often is up-regulated under conditions of abiotic stress or in the presence of hormones (Kim et al., 2006). Here it is shown that ANAC089 fusion protein localizes to vesicle-like structures and peripheral membranes. This is in line with the report by Li et al. (2010) showing similar localization. Upon its reductive release from the membrane ANAC089 localized to the nucleus (Figure 5). Thus our experiments provide evidence for a signaling cue that triggers its release and gives evidence for process dynamics. The almost exclusive nuclear localization after 60 min in this study was identical to images recorded after expressing a C-terminally truncated variant, ANAC089δC containing amino acids 1 to 310 (Li et al., 2010) and could be confirmed in our cell system after expressing a C-terminally truncated version of ANAC089 lacking the transmembrane domain. Our data indicate that a reductive stimulus is involved in release of the functional TF.
A critical element in the proposed signaling pathway is the protease that releases ANAC089 from the membrane. Conditional proteolysis which is hypothetical at present would allow for rapid transcriptional response to a changing environmental or developmental cue. Up to now, the molecular entities shedding the TFs from their membrane in plants are unknown. In the case of another NAC-MTTF named NTM1, the calpain inhibitor ALLN prevented cleavage from the membrane. Since ALLN specificity is not high, the precise nature of the involved protease remains to be established (Kim et al., 2006). The ANAC089 sequence contains two predicted cleavage sites; a peculiar Arg/Lys-specific site at amino acid position 163 (VVCRVRR|NK) with a high probability and a second site with lower score at position 297 (RPSQKKK|GK) just close to the membrane spanning α-helix based on mammalian protease processing sites (Figure 3). About 3% of all A. thaliana genes code for proteases (Garcia-Lorenzo et al., 2006) most of which have not been analyzed in any detail. The chloroplast serine Deg1 protease is activated under reductive conditions. It is suggested to localize to the thylakoid membrane and contains many Cys residue, e.g., 10 Cys in A. thaliana (Str"o, 2008). Deg proteases function as ATP-independent serine endopeptidases and are found in different organelles and the nucleolus (Deg9) but not in the cytosol or at the endomembranes (Schuhmann and Adamska, 2012). Thus they may serve as examples of principle but unlikely are involved in releasing ANAC089 from their endomembrane attachment site. Three Arabidopsis Cys proteases have been shown to be inhibited by binding to protein disulfide isomerase and activated in vitro after addition of DTT. The active Cys proteases are involved in cell type-specific programmed cell death in the endothelium of developing seeds (Ondzighi et al., 2008). Thus in addition to the more frequently described oxidative activation of proteases, the given examples reveal that reductive activation of specific proteases as hypothesized here occurs in planta as well. The cleavage sites in ANAC089 are predicted for serine proteases of the furin type which are also found in plants. In addition to redox-dependent activation of the protease by disulfide reduction or release from binding partners, a third type of redox regulation might be achieved by burying the cleavage domain of the substrate protein ANAC089 either by conformational changes or posttranslational modifications, e.g., by glutathionylation. ANAC089 contains four Cys residues which may be involved in such a mechanism (Figure 3A).
Recently, ANAC089 was linked to sugar sensing in a screen for fructose-insensitive A. thaliana mutants. One of the selected mutants FSQ6 carries an anac089 allele with premature translational stop in the third exon which enabled growth in the presence of 6.5% (w/v) fructose (Li et al., 2011). This C-terminally truncated ANAC089 in FSQ6 lacks the membrane spanning helix, thus is not tethered to membranes and immediately translocates to the nucleus upon its synthesis. The expressed ANAC089 variant in FSQ6 is considered as gain-of function mutation with constitutive ANAC089-dependent target gene expression (Li et al., 2011). Our results demonstrate release of the N-terminus of ANAC089 upon treatment with reductant (Figure 5). In their comprehensive search and cataloguing of NAC transcription factors in the rice and A. thaliana genomes Ooka et al. (2003) sorted ANAC089 in the group of OsNAC08-related NACs together with ANAC060 and ANAC040. Membrane-tethering of at least 13 ANACs was reported by Kim et al. (2007) the transcript levels of which were often up-regulated under cold, salinity, or heat stress. This contrasts expression of ANAC089. Its mRNA level was high at the end of the dark phase and under low light conditions (Figure 8) and down-regulated under high light. High light exposure of normal or low light grown plants imposes excess excitation energy stress and induces a profound transcriptional acclimation response (Khandelwal et al., 2008; Oelze et al., 2012 and unpublished). High light induces oxidative stress and ROS-related signaling (Bechtold et al., 2008). Thus ANAC089 showed a regulation tentatively reciprocal to the ROS level.
First reports have described cues that lead to the release of membrane-tethered NAC transcription factor. Cytokinin-dependent signaling involves the membrane-anchored ANAC transcription factor NTM1 which regulates cell division (Kim et al., 2006). The MTTF-NAC (At3g49530) mediates part of the cold stress response. Cold temperatures lead to release of the functional NAC-TF from the plasmamembrane over a long time period between 0.5 and 15 h (Seo et al., 2010). In neither of these cases a biochemical cue could be proposed that triggers the shedding. Thus the here reported redox-dependent release offers a first mechanistic interpretation of the processing by intracellular redox signals.
ANAC089 has been shown to be expressed in cotyledons, germinating seedlings, in the vasculature of hypocotyls, roots, rosette and cauline leaves, but also in the interveinal region of the rosette leaves (Li et al., 2010). ANAC089 overexpression delayed flowering time possibly by affecting expression of flower regulatory genes such as CONSTANS (CO) and FLOWERING LOCUS T (FT) whose transcript was suppressed and FLOWERING LOCUS C (FLC) whose transcript accumulation was enhanced (Li et al., 2010). Apparently ANAC089 has multiple functions.
This study assigns an additional and novel role to ANAC089, namely in regulating the expression of sAPX, an important player of the chloroplast antioxidant system. The chloroplast antioxidant system in photosynthesizing cells is expressed at a high level under most conditions (Foyer and Noctor, 2005). This realizes a strong antioxidant defense in a variable environment. However leaves may encounter environmental conditions that prevent photosynthesis-related oxidative stress. Such a reduced state of the glutathione system (Noctor and Foyer, 1998) and the thiol-disulfide redox regulatory network (Dietz, 2008) are likely established if leaves are permanently shaded (Oelze et al., 2012). Then down-regulation of the defense system might save resources for other important synthetic activities. Shedding of ANAC089 from the cytosol-facing side of endomembranes by a thiol redox-regulated process may then allow for translocation of the released functional ANAC089-protein to the nucleus and will subsequently lower sapx gene expression. Comparing protein amounts and transcript levels in A. thaliana plants acclimated to low, normal, and high light indicates that low levels of sAPX transcript are sufficient to maintain high sAPX protein amounts under such conditions probably due to reduced turnover. Such a role of ANAC089 is tentatively supported by the antiparallel transcript regulation of ANAC089 and sAPX in rosettes acclimated and maintained in low and normal light or subsequently transferred to high light (Figure 8). According to this scenario, ANAC089 functions in a negative retrograde loop, lowering sAPX expression if the cell encounters highly reducing conditions. In this case the retrograde signal would be a thiol-disulfide redox cue. The putative binding site ATGCACGTC of ANAC089 in the sapx promoter is present in many genes tentatively co-expressed with sAPX. It will be interesting to investigate the effect of ANAC089 on the expression of these candidate targets including sapx by use of transgenic A. thaliana lacking or overexpressing ANAC089.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge support by Prof. Margarete Baier (FU Berlin, Germany) in the very early phase of the project particularly in the Y1H-library construction. This work was conducted within the framework of the FOR804 on retrograde signaling (Di346) funded by the Deutsche Forschungsgemeinschaft.
Abbreviations
- 3-AT
3-amino-1,2,4-triazol
- DTT
dithiothreitol
- CFP
(enhanced) cyan fluorescent protein
- EMSA
electrophoretic mobility shift assay
- FRET
Förster/fluorescence resonance energy transfer
- MTTF
membrane-tethered transcription factors
- ROS
reactive oxygen species
- sAPX
stromal ascorbate peroxidase
- SOD
superoxide dismutase
- TF
transcription factor
- YFP
(enhanced) yellow fluorescent protein.
Footnotes
REFERENCES
- Asada K. (1999). The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 601–639 [DOI] [PubMed] [Google Scholar]
- Baier M., Dietz K. J. (2005). Chloroplasts as source and target of cellular redox regulation: a discussion on chloroplast redox signals in the context of plant physiology. J. Exp. Bot. 56 1449–1462 [DOI] [PubMed] [Google Scholar]
- Baier M., Noctor G., Foyer C. H., Dietz K. J. (2000). Antisense suppression of 2-Cys peroxiredoxin in Arabidopsis thaliana specifically enhances the activities and expression of enzymes associated with ascorbate metabolism, but not glutathione metabolism. Plant Physiol. 124 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U., Richard O., Zamboni A., Gapper C., Geisler M., Pogson B. J., et al. (2008). Impact of chloroplastic- and extracellular-sourced ROS on high light-responsive gene expression in Arabidopsis. J. Exp. Bot. 59 121–133 [DOI] [PubMed] [Google Scholar]
- Dietz K. J. (2008). Redox signal integration: from stimulus to networks and genes. Physiol. Plant. 133 459–468 [DOI] [PubMed] [Google Scholar]
- Dietz K. J. (2011). Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox Signal. 15 1129–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K. J., Jacob S., Oelze M. L., Laxa M., Tognetti V., de Miranda S. M. N., et al. (2006). The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 57 1697–1709 [DOI] [PubMed] [Google Scholar]
- Ernst H. A., Olsen A. N., Larsen S, Lo Leggio L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C. H., Noctor G. (2005). Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Physiol. 28 1056–1071 [Google Scholar]
- Garcia-Lorenzo M., Sjödin A., Jansson S., Funk C. (2006). Protease gene families in Populus and Arabidopsis. BMC Plant Biol. 6 30 10.1186/1471-2229-6-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D., Lüking I., Yang O. (2011). Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 30 1383–1391 [DOI] [PubMed] [Google Scholar]
- Hanton S. L., Chatre L., Matheson L. A., Rossi M., Held M. A., Brandizzi F. (2008). Plant Sar1 isoforms with near-identical protein sequences exhibit different localizations and effects on secretion. Plant Mol. Biol. 67 283–294 [DOI] [PubMed] [Google Scholar]
- Hu R., Qi G., Kong Y., Kong D., Gao Q., Zhou G. (2010). Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 10 145 10.1186/1471-2229-10-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Robinson D. G. (2009). Transport vesicle formation in plant cells. Curr. Opin. Plant Biol. 12 660–669 [DOI] [PubMed] [Google Scholar]
- Iida K., Seki M., Sakurai T., Satou M., Akiyama K., Toyoda T., et al. (2005). RARTF: database and tools for complete sets of Arabidopsis transcription factors. DNA Res. 12 247–256 [DOI] [PubMed] [Google Scholar]
- Jensen M. K., Kjaersgaard T., Nielsen M. M., Galberg P., Petersen K., O’Shea C., et al. (2010). The Arabidopsis thaliana NAC transcription factor family: structure–function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426 183–196 [DOI] [PubMed] [Google Scholar]
- Kangasjärvi S., Lepisto A., Hannikainen K., Piippo M., Luomala E. M., Aro E. M., et al. (2008). Diverse roles for chloroplast stromal and thylakoid-bound ascorbate peroxidases in plant stress responses. Biochem. J. 412 275–285 [DOI] [PubMed] [Google Scholar]
- Khandelwal A., Elvitigala T., Ghosh B., Quatrano R. S. (2008). Arabidopsis transcriptome reveals control circuits regulating redox homeostasis and the role of an AP2 transcription factor. Plant Physiol. 148 2050–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Kim S. G., Kim Y. S., Seo P. J., Bae M., Yoon H. K., et al. (2007). Exploring membrane-associated NAC transcription factors in Arabidopsis: implications of membrane biology in genome regulation. Nucleic Acid Res. 35 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. S., Kim S. G., Park J. E., Park H. Y., Lim M. H., Chua N. H., et al. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18 3132–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Dietz K. J. (2010). Identification of DNA-binding proteins and protein–protein interactions by yeast one-hybrid and yeast two-hybrid screen. Methods Mol. Biol. 639 171–192 [DOI] [PubMed] [Google Scholar]
- Leister D. (2012). Retrograde signaling in plants: from simple to complex scenarios. Front. Plant Sci. 3: 135. 10.3389/fpls.2012.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Q., Zhang J. A., Wang X. C., Chen J. (2010). A membrane-tethered transcription factor ANAC089 negatively regulates floral initiation in Arabidopsis thaliana. Sci. China Life Sci. 53 1299–1306 [DOI] [PubMed] [Google Scholar]
- Li P., Wind J. J., Shi X., Zhang H., Hanson J., Smeekens S. C. (2011). Fructose sensitivity is suppressed in Arabidopsis by the transcription factor ANAC089 lacking the membrane-bound domain. Proc. Natl. Acad. Sci. U.S.A. 108 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. R., Wakao S., Fischer B. B., Niyogi K. K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60 239–260 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Kinoshita N., Chua N.-H. (2003). AFP is a novel negative regulator of ABA signalling that promotes ABI5 protein degradation. Genes Dev. 17 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake C., Asada K. (1996). Inactivation mechanism of ascorbate peroxidase at low concentrations of ascorbate; hydrogen peroxide decomposes compound I of ascorbate peroxidase. Plant Cell Physiol. 37 423–430 [Google Scholar]
- Nakano Y., Asada K. (1981). Hydrogen peroxide is scavenged by ascorbate specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 22 867–880 [Google Scholar]
- Neubert C., Graham L., Black-Maier E. W., Liu T. Y., Stierhof Y. D., Seidel T., et al. (2008). Arabidopsis has two functional orthologs of the yeast V-ATPase assembly factor Vma21p. Traffic 9 1618–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Foyer C. H. (1998). Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 249–279 [DOI] [PubMed] [Google Scholar]
- Oelze M. L., Vogel M. O., Alsharafa K., Kahmann U., Viehhauser A., Maurino V. G., et al. (2012). Efficient acclimation of the chloroplast antioxidant defence of Arabidopsis thaliana leaves in response to a 10- or 100-fold light increment and the possible involvement of retrograde signals. J. Exp. Bot. 63 1297–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondzighi C. A., Christopher D. A., Cho E. J., Chang S. C., Staehelin L. A. (2008). Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell 20 2205–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka H., Satoh K., Doi K., Nagata T., Otomo Y., Murakami K., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10 239–247 [DOI] [PubMed] [Google Scholar]
- op den Camp R. G. L., Przybyla D., Ochsenbein C., Laloi C., Kim C., Danon A., et al. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Ahumada A., Kahmann U., Dietz K. J., Baier M. (2006). Regulation of peroxiredoxin expression versus expression of Halliwell-Asada-Cycle enzymes during early seedling development of Arabidopsis thaliana. Photosynth. Res. 89 99–112 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T. (2010). Plastidial retrograde signaling – a true “plastid factor” or just metabolite signatures? Trends Plant Sci. 15 427–435 [DOI] [PubMed] [Google Scholar]
- Pogson B. J., Nick S. W., Förster B., Small I. (2008). Plastid signaling to the nucleus and beyond. Trends Plant Sci. 13 602–609 [DOI] [PubMed] [Google Scholar]
- Schuhmann H., Adamska I. (2012). Deg proteases and their role in protein quality control and processing in different subcellular compartments of the plant cell. Physiol. Plant. 145 224–234 [DOI] [PubMed] [Google Scholar]
- Seidel T., Golldack D., Dietz K. J. (2005). Mapping of C-termini of V-ATPase subunits by in vivo-FRET measurements. FEBS Lett. 579 4374–4382 [DOI] [PubMed] [Google Scholar]
- Seidel T., Kluge C., Hanitzsch M., Ross J., Sauer M., Dietz K. J., et al. (2004). Colocalization and FRET-analysis of subunits c and a of the vacuolar H+-ATPase in living plant cells. J. Biotechnol. 112 165–175 [DOI] [PubMed] [Google Scholar]
- Seidel T., Schnitzer D., Golldack D., Dietz K. J. (2008). Organelle-specific isoenzymes of plant V-ATPase as revealed by in vivo-FRET analysis. BMC Cell Biol. 9 28 10.1186/1471-2121-9-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel T., Seefeldt B., Sauer M., Dietz K. J. (2010) In vivo analysis of the 2-Cys peroxiredoxin oligomeric state by two-step FRET. J. Biotechnol. 149 272–279 [DOI] [PubMed] [Google Scholar]
- Seo P. J., Kim M. J., Song J. S., Kim Y. S., Kim H. J., Park C. M. (2010). Proteolytic processing of an Arabidopsis membrane-bound NAC transcription factor is triggered by cold-induced changes in membrane fluidity. Biochem. J. 427 359–367 [DOI] [PubMed] [Google Scholar]
- Shaikhali J., Heiber I., Seidel T., Ströher E., Hiltscher H., Birkmann S., et al. (2008). The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-Cys peroxiredoxin A and other chloroplast antioxidant enzymes. BMC Plant Biol. 8 48 10.1186/1471-2229-8-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ströher E., Dietz K. J. (2008). The dynamic thiol-disulphide redox proteome of the Arabidopsis thaliana chloroplast as revealed by differential electrophoretic mobility. Physiol. Plant. 133 566–583 [DOI] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R. L., Nakano A., Takeyasu K., Sato M. H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi Network in plant cells. Cell Struct. Funct. 29 49–65 [DOI] [PubMed] [Google Scholar]
- Wormuth D., Baier M., Kandlbinder A., Scheibe R., Hartung W., Dietz K. J. (2006). Regulation of nuclear and chloroplast transcript levels by photosynthetic signals triggered through modified CO2 availability. BMC Plant Biol. 6 15 10.1186/1471-2229-6-15 [DOI] [PMC free article] [PubMed] [Google Scholar]