Abstract
The orange color of tiger lily (Lolium lancifolium ‘Splendens’) flowers is due, primarily, to the accumulation of two κ-xanthophylls, capsanthin and capsorubin. An enzyme, known as capsanthin-capsorubin synthase (CCS), catalyzes the conversion of antheraxanthin and violaxanthin into capsanthin and capsorubin, respectively. We cloned the gene for capsanthin-capsorubin synthase (Llccs) from flower tepals of L. lancifolium by the rapid amplification of cDNA ends (RACE) with a heterologous non-degenerate primer that was based on the sequence of a gene for lycopene β-cyclase (lcyB). The full-length cDNA of Llccs was 1,785 bp long and contained an open reading frame of 1,425 bp that encoded a polypeptide of 474 amino acids with a predicted N-terminal plastid-targeting sequence. Analysis by reverse transcription–PCR (RT–PCR) revealed that expression of Llccs was spatially and temporally regulated, with expression in flower buds and flowers of L. lancifolium but not in vegetative tissues. Stable overexpression of the Llccs gene in callus tissue of Iris germanica, which accumulates several xanthophylls including violaxanthin, the precursor of capsorubin, resulted in transgenic callus whose color had changed from its normal yellow to red-orange. This novel red-orange coloration was due to the accumulation of two non-native κ-xanthophylls, capsanthin and capsorubin, as confirmed by HPLC and ultraperformance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) analysis with authentic standards. Cloning of the Llccs gene should advance our understanding of the molecular and genetic mechanisms of the biosynthesis of κ-carotenoids in general and in the genus Lilium in particular, and will facilitate transgenic alterations of the colors of flowers and fruits of many plant species.
Keywords: Antheraxanthin, Capsanthin, Capsorubin, Carotenoids, Lycopene β-cyclase, Violaxanthin
The nucleotide sequences reported in this paper have been submitted to GenBank under accession numbers: lycopene β-cyclase (LllcyB) GU471230; capsanthin-capsorubin synthase (Llccs) mRNA JF304153; capsanthin-capsorubin synthase (Llccs) genomic clone GU443955.
Introduction
Flower color, which is due predominantly to the accumulation of three types of secondary metabolite (flavonoids, carotenoids and betalains), is a very important floricultural trait. Carotenoids impart yellow, orange and red pigmentation to flowers and fruits of many plant species and play important roles in plant reproduction via attraction of pollinators and promotion of seed dispersal (Cunningham and Gantt 1998, Tanaka et al. 2008, Ohmiya 2011).
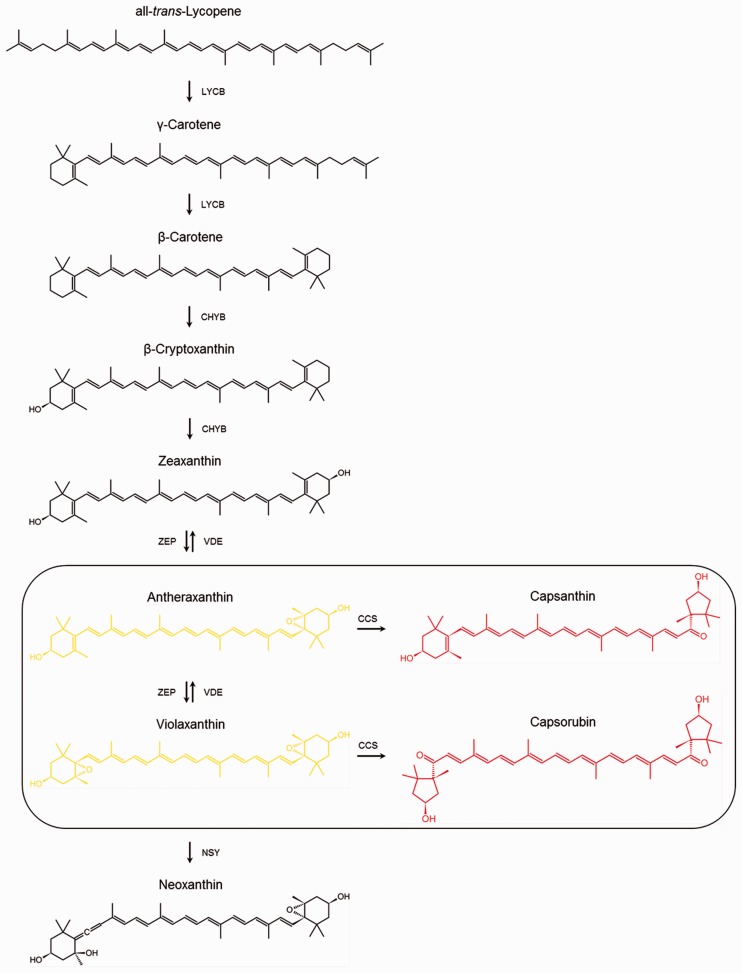
Carotenoids are lipophilic C40 isoprenoids with polyene chains that are synthesized by plants, by photosynthetic and some non-photosynthetic bacteria and by some fungi (Hirschberg et al. 1997, Cunningham 2002, Rodríguez-Concepción 2010). In higher plants, carotenoids are typically synthesized in plastids via the MEP (2-C-methyl-d-erythriol 4-phosphate) pathway which is catalyzed by enzymes encoded by nuclear genes (Cunningham 2002). The head-to-head condensation of two geranylgeranyl diphosphate (GGPP) molecules yields phytoene (C40) and is the first committed step in the biosynthesis of carotenoids. All-trans-lycopene is then formed from 15-cis-phytoene via four desaturation reactions and enzymatic or light-mediated photo-isomerization. Cyclization of the linear all-trans-lycopene occurs at a branching point in the pathway. Cyclohexane rings are formed at both ends of the lycopene molecule to produce (i) α-carotene with one ε- and one β-ionone end group and (ii) β-carotene with two β-ionone end groups. Carotenoids with two ε-ionone groups are rare. Up to this point in the pathway, all the carotenoids mentioned above are exclusively hydrocarbons in nature. Subsequent steps involve the enzymatic addition of oxygen moieties to the carotenes to yield hydroxy, epoxy and oxy derivatives, such as lutein on the β,ε-branch; zeaxanthin, antheraxanthin, violaxanthin and neoxanthin on the β,β-branch (Fig. 1); and lactucaxanthin on the ε,ε-branch of the pathway. The oxygen-containing carotenoids are known collectively as xanthophylls (Bartley and Scolnik 1995, Ladygin 2000).
Fig. 1.
Schematic of the β,β-branch of the carotenoid biosynthesis pathway in plants. Biosynthesis of capsanthin and capsorubin from antheraxanthin and violaxanthin, catalyzed by capsanthin-capsorubin synthase, is circled. LCYB, lycopene β-cyclase; CHYB, β-carotene hydroxylase; ZEP, zeaxanthin epoxidase; VDE, violaxanthin de-epoxidase; CCS, capsanthin-capsorubin synthase; NSY, neoxanthin synthase.
Both β,ε- and β,β-xanthophylls accumulate to very high levels in the flower petals of many plant species and they are responsible for pale yellow and deep yellow-to-orange coloration, which depends on relative levels and the precise composition of these compounds in each flower (Zhu et al. 2010, Ohmiya 2011). Carotenoids are the main determinants of flower color in several species in the genus Lilium. Yellow lily flowers accumulate large amounts of yellow β,ε- and β,β-xanthophylls, such as antheraxanthin, (9Z)-violaxanthin, cis-lutein, violaxanthin and lutein, as well as some β-carotene, in descending order of abundance (Yamagishi et al. 2010). The orange and red-orange flowers of several species in the genus Lilium have very unusual carotenoid profiles that are due to modified carotenoid biosynthetic pathways in these species. The orange and red-orange color of tepals (indistinguishable petals and sepals) and anthers of these species, including L. lancifolium (formerly L. tigrinum), which is known in the vernacular as tiger lily, is due to the accumulation of two red pigments, capsanthin (3,3′-dihydroxy-β,κ-carotene-6′-one) and capsorubin (3,3′-dihydroxy-κ,κ-carotene-6,6′-dione) (Valadon and Mummery 1976, Märki-Fischer and Eugster 1985, Yamagishi et al. 2010). Capsanthin and capsorubin are κ-xanthophylls that contain, respectively, one and two unusual cyclopentane κ-rings. They are the main pigments of red peppers (Capsicum annuum, also referred to hereinafter as pepper) and of the orange and red-orange flowers of several species of Lilium. Capsanthin and/or capsorubin have been identified in the following species in the genus Lilium: L. tigrinum (Karrer and Oswald 1935); L. amabile (Seybold 1953); L. Davidii and L. leichtinii, as well as their hybrid L. Maxwill (Valadon and Mummery 1976); L. pumilum (Partali et al. 1987); and the Asiatic hybrid lily ‘Saija’ (Yamagishi et al. 2010). In the red-orange tepals of L. tigrinum ‘Red Night’ flowers, the main pigments are capsanthin and its Z-isomers [(9′Z)-, (9Z)-, (13′Z)- and (13Z)-capsanthin], which together constitute >53% of the total carotenoids (Märki-Fischer and Eugster 1985), while the main pigment in the red-orange tepals of L. pumilum is capsorubin, which accounts for as much as 80% of the total carotenoids (Partali et al. 1987).
Capsanthin and capsorubin have been identified in only a few species other than C. annuum and Lilium spp. Capsanthin was isolated from anthers of several Aesculus spp., from fruits of seven Berberis spp. and from fruits of Asparagus officinalis (Rüttimann 1982, Deli et al. 2000). Capsorubin was isolated from fruits of A. officinalis, from the integument of Encephalartos altensteinii, from the fruits of Encephalartos villosus, from the petals of Cajophora lateritia and from the anthers of several Aesculus spp. (Rüttimann 1982, Deli et al. 2000).
Capsanthin and capsorubin are synthesized from two ubiquitous 5,6-epoxy-xanthophylls, namely antheraxanthin and violaxanthin, respectively (Fig. 1). The synthesis of capsanthin and capsorubin is catalyzed by the same enzyme, capsanthin-capsorubin synthase (CCS) (Bouvier et al. 1994).
A search of the NCBI GenBank yielded several sequences from five plant species that were identified as genes for CCS. These genes had been cloned from Citrus sinensis (GenBank accession No. AF169241.1), C. annuum (multiple GenBank accession Nos., e.g. X76165.1), Ricinus communis (GenBank accession No. XP_002523837.1), Daucus carota (GenBank accession Nos. ABB52072.1 and AF208530.1) and Medicago truncatula (GenBank accession No. AES67093.1). It is likely that all of the above-mentioned genes, with the exception of the ccs gene from pepper, were identified tentatively as ccs on the bases of homology searches alone. Furthermore, all of the cited species with the exception of C. annuum have never been proven to produce and/or accumulate capsanthin and capsorubin. Thus, to date, only the ccs gene from red bell pepper (C. annuum) has been cloned and characterized (Bouvier et al. 1994, Kumagai et al. 1998, Mialoundama et al. 2010).
Lilies are very important ornamental plants and are used worldwide as cut flowers and as potted and garden-grown plants. The genus Lilium comprises about 85 species and is classified into seven sections (Comber 1949, de Jong 1974). Carotenoids play a very important role in the pigmentation of lily flowers, and genes for carotenoid biosynthesis encoding phytoene synthase (LhPSY), phytoene desaturase (LhPDS), ζ-carotene desaturase (LhZDS), carotenoid isomerase (LhCRTISO), lycopene β-cyclase (LhLCYB), β-carotene hydroxylase (LhHYB) and lycopene ε-cyclase (LhLCYE) have recently been identified and their patterns of expression studied in flowers of several cultivars of Asiatic hybrid lilies (Yamagishi et al. 2010). However, the gene that encodes the CCS responsible for the orange and red-orange colors of Lilium flowers had not yet been characterized.
Our preliminary attempts to clone the ccs gene from L. lancifolium using heterologous non-degenerate primers based on the sequence of the ccs gene from C. annuum failed. This was most probably due to the differences in codon usage between monocots, such as L. lancifolium, and dicots, such as C. annuum (Campbell and Gowri 1990, Kawabe and Miyashita 2003).
Using a different cloning strategy, we successfully cloned and functionally characterized the gene for CCS from L. lancifolium (Llccs). In addition, we also successfully cloned the promoter region of this gene.
Results and Discussion
Molecular cloning and sequence analysis of the Llccs gene
Our strategy for cloning the Llccs gene from L. lancifolium was based on the fact that lycopene cyclase (LCY), neoxanthin synthase (NSY) and CCS have strong sequence homology and similar putative catalytic mechanisms (Bouvier et al. 2000, Mialoundama et al. 2010). We decided, first, to try to obtain a partial cDNA sequence of the gene for lycopene β-cyclase (LllcyB) that was located between two adjacent regions that are conserved in both the lcyB gene and the ccs gene from pepper and then to use the resultant sequence information to synthesize a number of primers for 3′ rapid amplification of cDNA ends (RACE). We postulated that this approach (3′RACE) would allow us to clone the 3′-proximal end of LllcyB. However, because of the presumed strong similarity of the LllcyB to Llccs and the matching codon usage, we hoped that some of the primers might preferentially amplify Llccs instead of LllcyB or might, at least, generate mixed PCRs with amplicons of both genes. Therefore, we decided to clone both LllcyB and Llccs candidate genes and then to determine which one was the true Llccs gene by sequence analysis and functional characterization in a suitable test system.
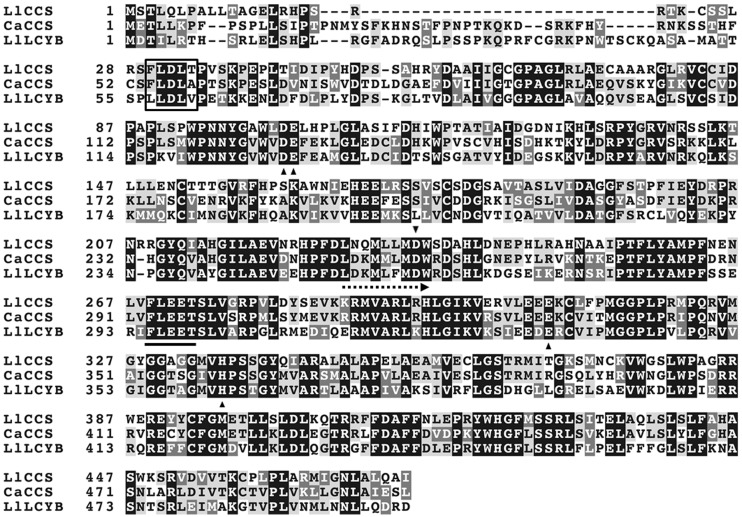
A pair of heterologous, non-degenerate primers, forward (Cs-F) and reverse (Cs-R), amplified a DNA fragment (∼400 bp) which encoded a deduced amino acid sequence with strong similarity to that of LCYB. We designed a number of forward primers that were based on this newly cloned sequence and used them in combination with the 3′-outer primer from the RACE kit to try and clone the 3′-proximal ends of both LllcyB and Llccs by 3′RACE. Several of the products of PCR that we obtained were identical to one another; however, one of the products of PCR was very similar to the others but was, nonetheless, different. Using this approach, we were able to identify two different genes with strong similarity to both lcyB and pepper ccs. Two multisequence alignments showing the position and homology of Sc-F and Sc-R primers relative to LllcyB, Llccs and C. annuum ccs (Caccs) are presented in Supplementary Fig. S1a, b.
We cloned one of these two genes using the forward primer lcyB-3Rc in combination with the 3′-outer primer from the RACE kit. We generated full-length LllcyB cDNA of 1,986 bp by aligning and splicing DNA fragments obtained by 3′ and 5′RACE. It contained an open reading frame (ORF) of 1,500 bp, a 5′ untranslated region (UTR) of 375 bp and a 3′ UTR with a poly(A) tail downstream of the stop codon of 108 bp. Based on homology analyses, we tentatively identified it as a gene for LCYB (GenBank accession No. GU471230).
We succeeded in cloning the second gene, identified later as Llccs (GenBank accession No. JF304153), using the forward primer ccs-3Rc in combination with the 3′-outer primer from the RACE kit. The position and homology of the ccs-3Rc primer relative to LllcyB, Llccs and Caccs is presented in Fig. 2 and Supplementary Fig. S1b. We generated a full-length cDNA sequence of 1,758 bp by aligning and splicing DNA fragments obtained from 3′ and 5′RACE. The full-length cDNA included an ORF of 1,425 bp, a 5′ UTR of 94 bp and a 3′ UTR of 239 bp with a poly(A) tail downstream of the termination codon. The predicted protein was 474 amino acid residues long with a calculated molecular mass of 52.97 kDa and a theoretical pI of 8.4. Amplification of the Llccs gene by PCR, with genomic DNA as a template, yielded a DNA fragment of approximately the same size as the Llccs cDNA. Sequencing of this fragment revealed that Llccs did not include any introns.
Fig. 2.
Alignment of the deduced amino acid sequences of capsanthin-capsorubin synthase (LlCCS) from L. lancifolium (GenBank accession No. JF304153), capsanthin-capsorubin synthase (CaCCS) from C. annuum (GenBank accession No. Q42435.1) and lycopene β-cyclase (LlLCYB) from L. lancifolium (GenBank accession No. GU471230). White letters on a black background indicate globally conserved residues; black letters on a light-gray background indicate identical residues, and white letters on a dark-gray background indicate similar residues. Dashes represent gaps introduced to improve the alignment. The putative cleavage site of the N-terminal plastid transit peptide is boxed. The position of the heterologous non-degenerate primer (ccs-3Rc) used in 3′RACE of the Llccs is indicated by a dashed arrow. The conserved FLEET motif essential for β- and κ-cyclase activity is underlined. Black triangles designate conserved amino acids with important catalytic activity as identified by Mialoundama et al. (2010).
In order to analyze the newly obtained sequence, we first compared the deduced LlCCS protein with other proteins using BLASTp and, as we had anticipated, LlCCS was very similar, in terms of deduced amino acid sequence, to LCYB, NSY and pepper CCS. The strongest similarities were to LCYB from Citrus × paradisi (GenBank accession No. ACR09635.1; 63% identity, 77% positives; query coverage 96%), NSY from Solanum lycopersicum (GenBank accession No. CAB93342.1; (61% identity, 75% positives; query coverage 97%) and CCS from C. annuum (GenBank accession No. Q42435.1; 60% identity, 75% positives; query coverage 95%).
Next, we conducted amino acid sequence alignment of the deduced LlCCS protein, which enabled us to identify an N-terminal dinucleotide-binding motif with the characteristic configuration that is referred to as the Rossmann fold and is characteristic of flavoproteins (Rossmann et al. 1974, Mialoundama et al. 2010). Mialoundama et al. (2010) showed that pepper CCS is a typical flavoprotein with one non-covalently bound FAD that is essential for enzymatic activity in the presence of NADPH as the FAD reductant. The reaction proceeds with no net change in redox potential (Mialoundama et al. 2010). We also identified, a so-called 269-FLEET-273 motif, with two tandem glutamate residues, that is essential for both the β- and the κ-cyclase activities of pepper CCS (Fig. 2; Bouvier et al. 1997, Mialoundama et al. 2010). In addition to the FLEET motif, Mialoundama et al. (2010) identified five other amino acids with important catalytic roles in pepper CCS (Asp127, Asp259, Glu128, Glu332 and His360), all of which are conserved in LlCCS (Asp102, Asp235, Glu103, Glu308 and His336) (Fig. 2).
We also used the TargetP 1.1 Server (http://www.cbs.dtu.dk/services/TargetP/) to look for the presence of a transit peptide (TP) sequence in LlCCS, in an effort to predict the subcellular location of the enzyme, and we found an N-terminal pre-sequence that targeted the enzyme to plastids. The sequence, FLDLT, of the putative site of cleavage of the plastid TP is shown in Fig. 2 (Bouvier et al. 2000). The amino acid sequence of LlCCS was very similar to that of pepper CCS with the exception of the N-terminal plastid TP region (Fig. 2).
Additionally, sequence analysis revealed that LlCCS belongs to the lycopene cyclase family of proteins, which also includes LCYB, lycopene ε-cyclase (LCYE) and NSY. An alignment of the deduced amino acid sequences of LlCCS and LlLCYB with C. annuum CCS is presented in Fig. 2. All three enzymes (LCY, NSY and CCS) have strong sequence homology and similar putative catalytic mechanisms (Bouvier et al. 2000, Mialoundama et al. 2010). These similarities suggest that the three genes might be phylogenetically related (Kubasik and Sandmann 2000, Ronen et al. 2000).
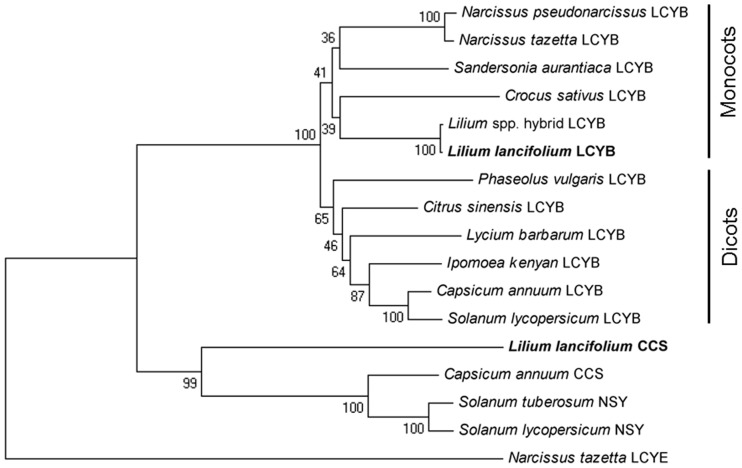
Indeed, phylogenetic analysis of LlCCS compared with CCS, NSY, LCYB and LCYE from other plant species (Fig. 3) showed that NSY and CCS share a common evolutionary ancestor which is most closely related to LCYB, while LCYE forms an outgroup. The LCYB group is further divided into two distinct subgroups consisting of monocotyledonous and dicotyledonous species, respectively (Fig. 3). The percentage consensus support derived from bootstrap testing with 1,000 replications indicates that these groupings are due to strong correlations in ancestry. This supports the hypothesis that ccs and nsy evolved from LCY genes via gene duplication and neofunctionalization (Kubasik and Sandmann 2000, Ronen et al. 2000).
Fig. 3.
Phylogenetic tree based on amino acid sequences of LCYB from six monocotyledonous species [Crocus sativus (ADA82241.1), Narcissus pseudonarcissus (ACT78995.1), Narcissus tazetta (AFH53820.1), Sandersonia aurantiaca (AAL92175.1), Asiatic hybrid lily (Lilium spp.) (BAH10590.1), Lilium lancifolium (JF304153)] and six dicotyledonous species [Capsicum annuum (ADH04271.1), Solanum lycopersicum (NP001234226.1), Ipomoea kenyan (BAI47577.1), Citrus sinensis (AAM21152.1), Phaseolus vulgaris (ADX60450.1), Lycium barbarum (AAW88382.1)], NSY from Solanum tuberosum (Q9M424.1) and S. lycopersicum (CAB93342.1), CCS from C. annuum (Q42435.1) and L. lancifolium (JF304153), and LCYE from N. tazetta (AFG19394.1). LCYE was chosen because it is a closely related lycopene cyclase protein that catalyzes the formation of ε-rings in the β,ε-branch of the carotenoid biosynthesis pathway. The GenBank accession number for each gene is given in parentheses. Values displayed at the nodes indicate the percentage consensus support as calculated using a bootstrapping test with 1,000 replications.
Furthermore, it has been shown previously that CCS is also able to catalyze the synthesis of β-carotene from lycopene with approximately 10–20% efficiency, while no ε-cyclase activity has been found (Hugueney et al. 1995). In addition, BLASTp analysis of LlCCS found that one of the most similar LCYE genes (Narcissus tazetta; GenBank accession No. AFG19394.1) had only 35% identity and 53% positives (89% query coverage), while the most similar LCYB gene (Citrus × paradise; GenBank accession No. ACR09635.1) had 63% identity and 77% positives (query coverage 96%). These findings strongly suggest that CCS is more closely related to LCYB than to LCYE, as is shown in our phylogenetic analysis (Fig. 3).
Computational analyses, including multisequence alignment analyses, of these genes failed to reveal any obvious long motifs or conserved regions that would distinguish LCY from CCS and NSY. It is likely that the distinguishing functional regions between these genes are limited to the change of a single (or a few) amino acid. However, there exists only a very limited data resource (two CCS, including the newly cloned LlCCS, and two NSY genes) with which to carry out computational multisequence alignment analysis and, because neither crystal structure nor homology modeling is possible at this time for LCYB, NSY and CCS (Bouvier et al. 2000), there is currently no way to reliably predict functional region(s) and conserved motif(s) that distinguish these genes. Still, the availability of the Llccs sequence will assist us in designing strategies for cloning additional genes that catalyze κ-xanthophyll formation from other species, which in turn will aid in determining functional regions that differentiate the three genes. Furthermore, the availability of more cloned sequences of LCY, NSY and CCS will facilitate further research of carotenoid cyclase mechanisms, by allowing us to exploit the similarities and differences among these three genes, using strategies such as domain swapping or directed mutagenesis.
Finally, Mialoundama et al. (2010) reported that CCS also catalyzed the synthesis of two minor 3,5,6-trihydroxy xanthophylls, 5,6-diepikarpoxanthin and 5,6-diepilatoxanthin. In addition, it has been hypothesized that CCS also catalyzes the synthesis of several carotenoids with a 6-oxo-κ end group. Namely, it is thought that CCS catalyzes the conversion of β-cryptoxanthin 5′,6′-epoxide to 3′-deoxycapsanthin and 3,4-dehydroxy-3′-deoxycapsanthin in pepper fruits, as well as the conversion of β-carotene 5,6-epoxide to the sapotexanthin in mamey (Pouteria sapota) fruits (Maoka et al. 2004, Maoka 2009, Murillo et al. 2011). Similarly, the synthesis of two recently isolated minor carotenoids, 3′-deoxycapsorubin and 3,3′-dideoxycapsorubin, which also contain a 6-oxo-κ end group, is likely to be catalyzed by CCS as well (Murillo et al. 2012). The synthesis of the above-mentioned carotenoids was not the focus of our current study. However, Llccs-transgenic iris callus should be an excellent tool with which to study the potential role of LlCCS in the synthesis of these rare carotenoids. As mentioned above, it has also been shown that CCS possesses β-cyclase activity and is able to catalyze the synthesis of β-carotene from lycopene with approximately 10–20% efficiency (Hugueney et al. 1995). Therefore, CCS appears to be an enzyme that is capable of utilizing multiple substrates to catalyze the synthesis of the above-mentioned carotenoids in addition to capsanthin and capsorubin.
Expression of the Llccs and LllcyB genes in L. lancifolium
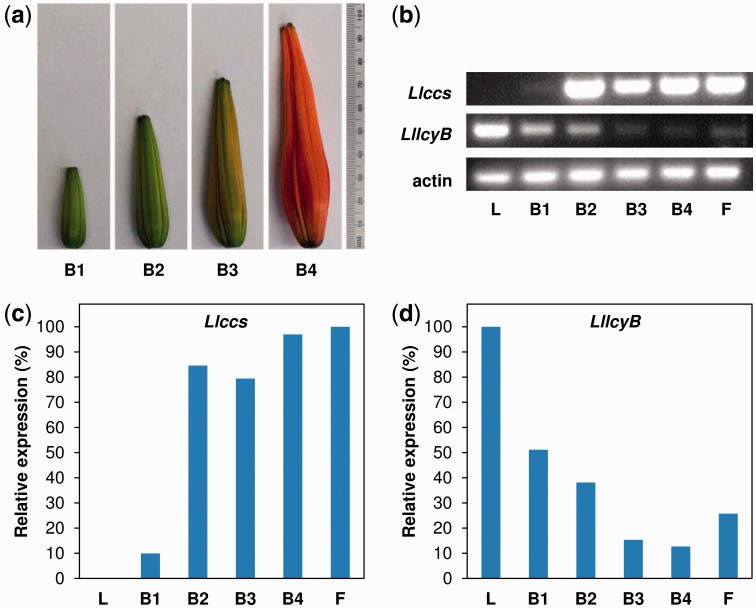
To examine the expression of the LllcyB and Llccs candidate genes in various tissues of lily, we performed reverse transcription–PCR (RT–PCR) using total RNA from the tepals of flower buds (at four developmental stages), the tepals of fully opened flowers, and leaves (Fig. 4a). We found that Llccs was expressed specifically in flower tepals and was not expressed in vegetative tissues, namely leaves (Fig. 4b, c). During flower development, the pattern of expression of Llccs (Fig. 4b, c) resembled that of the ccs gene during the development of pepper fruit, which is characterized by strong induction at early developmental stages and a constant high level of the transcript throughout the development of fruits (Bouvier et al. 1994). Thus, the expression of Llccs was closely synchronized with the development of flower buds and color break, while no Llccs transcripts were detected in leaves, indicating that expression of this gene was flower specific (Fig. 4b, c).
Fig. 4.
Analysis of the expression of the Llccs and LllcyB genes in L. lancifolium. (a) Four stages in the development of buds, indicated as B1–B4. (b) Results of analysis by RT–PCR showing relative levels of transcripts of the Llccs and LllcyB genes with the lily gene for actin as a control for normalization, in leaves (L), buds at four developmental stages (B1–B4) and fully opened flowers (F). (c) Relative levels of expression of the Llccs and LllcyB genes normalized with respect to expression of the gene for actin and expressed as percentages relative to the maximal level among individual developmental stages.
In contrast to Llccs, the LllcyB gene was expressed in both leaves and flowers (Fig. 4b, d). The highest level of LllcyB transcript was detected in leaves. The level of the LllcyB transcript was inversely correlated with the coloration of tepals, and the levels of this transcript fell as flower buds developed (Fig. 4b, d).
Since the Llccs candidate gene exhibited stronger similarity to pepper ccs than did LllcyB and since the expression of Llccs coincided spatially and temporally with the synthesis and accumulation of capsanthin and capsorubin in flower buds and opened flowers, we chose this gene for subsequent analysis and functional characterization. Furthermore, because of the flower-specific expression of the Llccs candidate gene, we decided to clone the upstream regulatory region, namely the promoter of this gene, postulating that it might serve as a useful flower-specific promoter for generalized transformation of monocot plants.
Expression of an Llccs transgene in callus tissue of Iris germanica
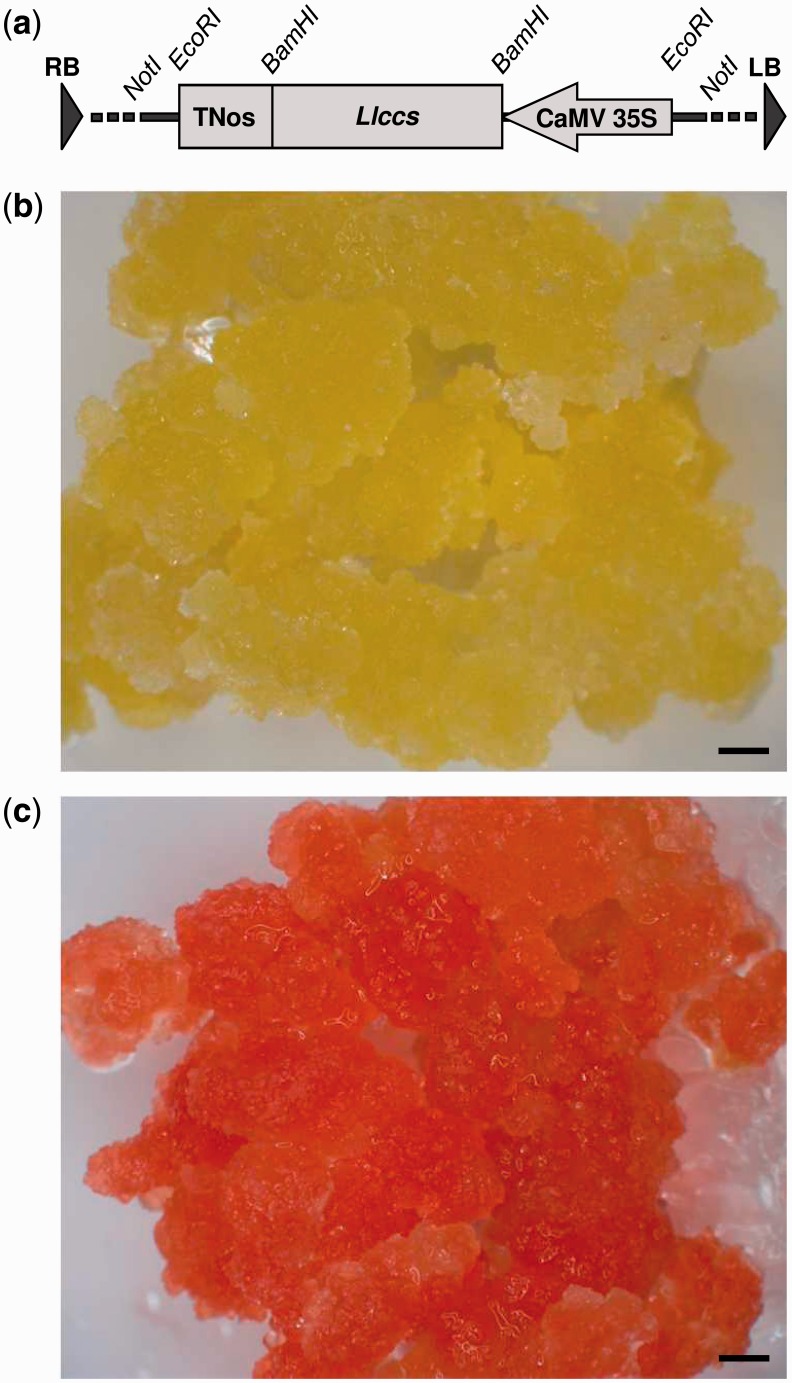
To demonstrate the function of the newly cloned Llccs candidate gene, we decided to use it to stably transform non-embryogenic iris (Iris germanica) callus tissue, whose normal yellow color is due to the accumulation of several xanthophylls, such as violaxanthin, the precursor of capsorubin. We performed Agrobacterium-mediated transformation with the binary vector pWBVec10a/P35S::Llccs::TNos that harbored Llccs under the control of the Cauliflower mosaic virus (CaMV) 35 S constitutive promoter and the nopaline synthase (Nos) terminator (Fig. 5a). We generated a total of seven independent Llccs-transgenic calli and confirmed, by β-glucuronidase (GUS) staining and PCR analysis of the transgene, that they were stably transformed (data not shown). The Llccs-transgenic calli changed color from their normal yellow to varying degrees of red-orange in different independent transgenic lines (Fig. 5b, c). The appearance of the new pigment(s) in the transgenic callus tissue demonstrated that Llccs was a functional gene and was responsible for the development of this novel red-orange color. The results also indicated that the TP had been recognized and processed correctly for targeting LlCCS into chromoplasts. The two independent transgenic calli that showed the most profound change in color were selected for further analysis.
Fig. 5.
(a) A partial map of the T-DNA region of the pWBVec10a/CaMV35S::Llccs::TNos binary transformation vector showing the expression cassette that included the gene for capsanthin-capsorubin synthase (Llccs) under the control of the CaMV 35 S promoter and Nos terminator. (b) Wild-type callus tissue of Iris germanica ‘Hot Property’. (c) A representative picture of multiple independent Llccs-transgenic calli of Iris germanica ‘Hot Property’ that had accumulated capsanthin and capsorubin. RB, right border; LB, left border. Bars represent 1 mm.
Analysis by HPLC of carotenoids in Llccs-transgenic calli of Iris germanica
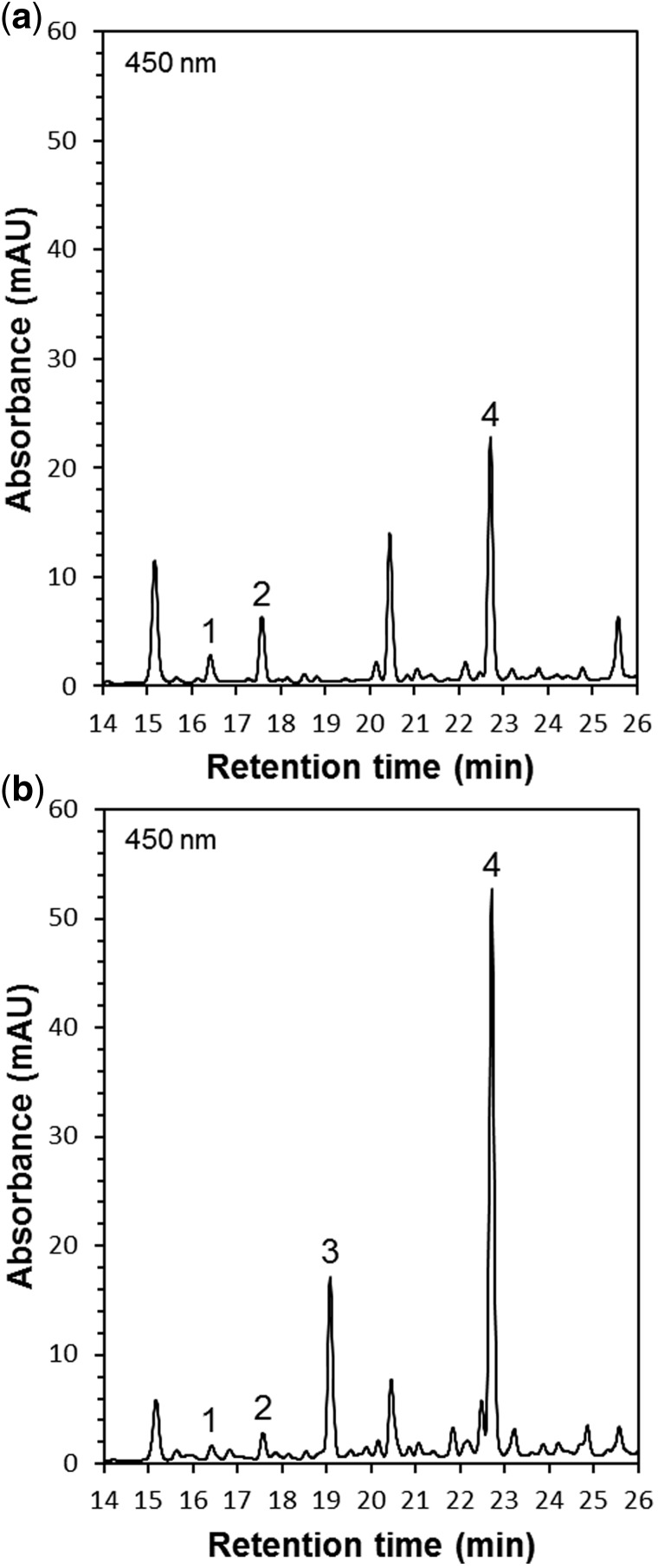
We performed HPLC analysis of carotenoid pigments in non-saponified and saponified extracts of wild-type and two independent Llccs-transgenic iris calli, designated Llccs-1 and Llccs-2 (Fig. 6a, b; Supplementary Table S1). In the analysis of Llccs-transgenic callus, a new carotenoid peak appeared at 19 min on the chromatogram after HPLC (Fig. 6b). The retention time and the spectroscopic properties of this peak in comparison with an authentic capsanthin standard indicated that this carotenoid was capsanthin (Fig. 6b). The new peak was not detected in the analysis of wild-type callus (Fig. 6a). The two independent Llccs-transgenic calli, Llccs-1 and Llccs-2, accumulated capsanthin in its free, non-esterified form at a level of ∼1.8 and ∼2 µg g–1 DW, respectively (Supplementary Table S1). However, the amount of total capsanthin (non-esterified and esterified) in both calli was much higher, ∼10 and ∼13 µg g−1 DW, respectively (Supplementary Table S1). After saponification of carotenoid extracts, the amount of free, non-esterified capsanthin in two independent transgenic lines increased approximately 5- to 7-fold, indicating that ∼80% to ∼85% of capsanthin in the chromoplasts of Llccs-transgenic iris callus was in an esterified form. In contrast to the capsanthin in pepper fruits and in Llccs-transgenic iris callus, Kumagai et al. (1998) found that the capsanthin in leaves of Nicotiana benthamiana plants transfected with an RNA viral vector that harbored the pepper ccs gene was not esterified. These authors speculated that the CCS had been targeted to chloroplasts rather than to chromoplasts. A gradual decrease in levels of free pigments, accompanied by increases in the esterification of xanthophylls with fatty acids, occurs during the ripening of pepper fruit and is directly linked to the transition of chloroplasts into chromoplasts (Camara and Monéger 1978, Márkus et al. 1999). Iris callus grown in darkness contains no chloroplasts, and LlCCS was, apparently, targeted to the chromoplasts. Therefore, the high level (>80%) of esterification of capsanthin in Llccs-transgenic iris callus was probably due to the fact that LlCCS had been targeted to the chromoplasts, as occurs at the later stages of development of pepper fruits and lily flowers. The analysis by HPLC of carotenoids extracted from red-orange Llccs-transgenic iris callus revealed the presence of capsanthin and its esters; however, lacking an authentic standard during our analysis, we were unable to identify capsorubin positively by HPLC.
Fig. 6.
Chromatograms after HPLC of unsaponified carotenoid extracts from wild-type (a) and Llccs-transgenic (b) callus tissues of Iris germanica ‘Hot Property’. The peak of capsanthin (3) at 19 min in chromatogram (b) was identified from a comparison of its retention time and spectral properties with those of an authentic capsanthin standard. Peaks: (1) neoxanthin; (2) violaxanthin; (3) capsanthin; and (4) lutein.
Analysis by UPLC-MS/MS of carotenoids in Llccs-transgenic calli of Iris germanica
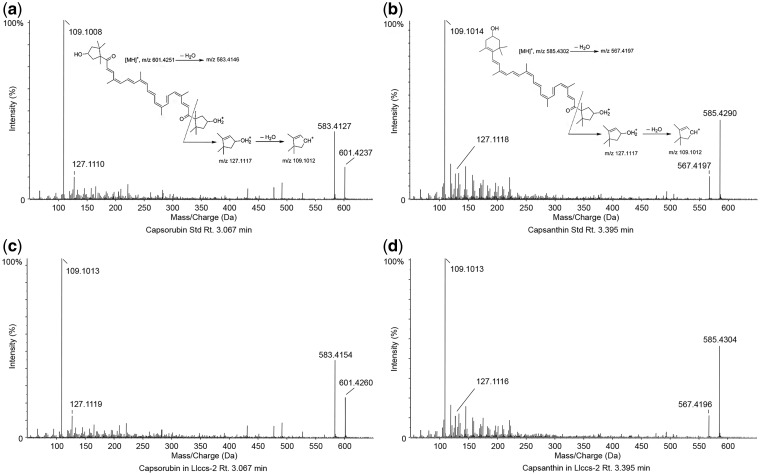
We purchased capsanthin and capsorubin standards from CarotNature and used them for ultraperformance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) analysis of carotenoids extracted from Llccs-transgenic iris callus. Using an ABSciex 5600 Triple ToF, fast-scanning high-resolution mass spectrometer interfaced with a Shimadzu Prominence LC-30AD UPLC system, we confirmed the masses and retention times as well as the predicted fragmentation patterns of the capsanthin and capsorubin standards and compared the results with those obtained with an extract of red-orange Llccs-transgenic iris callus (Fig. 7a–d). The retention time and accurate mass measurement for capsorubin (C40H56O4)H+ in Llccs-transgenic iris callus were 3.067 min and an m/z of 601.4260 + 0.5 p.p.m. (Fig. 7c). The retention time and accurate mass measurement for capsanthin (C40H56O3)H+ in Llccs-transgenic iris callus were 3.395 min and an m/z of 585.4304 –0.6 p.p.m. (Fig. 7d). The results of UPLC-MS/MS analysis for capsorubin and capsanthin from Llccs-transgenic samples were in agreement with the results obtained for the authentic capsorubin and capsanthin standards. The UPLC-MS/MS spectra for authentic standards and the Llccs-transgenic iris callus, as well as the predicted fragmentation patterns for capsorubin and capsanthin, are presented in Fig. 7.
Fig. 7.
Identification of capsorubin and capsanthin in Llccs-transgenic iris callus by UPLC-MS/MS. Upper panels: UPLC-MS/MS spectra representing the fragmentation pattern determined for capsorubin (a) and capsanthin (b) standards. Bottom panels: corresponding UPLC-MS/MS spectra representing the fragmentation pattern determined for capsorubin (c) and capsanthin (d) in the Llccs-transgenic callus of Iris germanica ‘Hot Property’ (Llccs-2).
Thus, the UPLC-MS/MS analysis revealed that both capsanthin and capsorubin accumulated in the red-orange Llccs-transgenic iris callus (Fig. 7). The reported analysis by thin-layer chromatography (TLC) of carotenoid extracts from leaves of N. benthamiana that had been transfected with an RNA viral vector that harbored the pepper ccs gene revealed the presence of capsanthin; however, no capsorubin was detected by TLC, even though its precursor, violaxanthin, was observed (Kumagai et al. 1998). This is the first time, to our knowledge, that the in vivo synthesis of capsorubin, catalyzed by a heterologous ccs gene, has been confirmed in any plant species. Also, this is the only example of a stable transformation with a ccs gene whereby the carotenogenic pathway has been successfully re-routed to yield capsanthin and capsorubin.
Cloning of the Llccs promoter
Cloning of the Llccs gene allowed us also to clone its upstream regulatory region, namely its promoter region (1,877 bp long). After preliminary sequencing, we chose a product of long-distance inverse-PCR (LDI-PCR) generated from the BamHI-digested sample because it was between 6,000 and 7,000 bp long and was most likely to contain the complete Llccs locus. After sequencing, we designed a pair of primers (L-Llccs-F and L-Llccs-R) for amplification of the Llccs locus that encompassed the promoter region of 1,877 bp upstream of the site of initiation of transcription, the 5′ UTR of 94 bp, the Llccs ORF of 1,425 bp and the 3′ UTR of 425 bp with a poly(A) tail and a predicted poly(A) signal downstream of the termination codon (GenBank accession No. GU443955). As discussed above, the analysis of the expression of Llccs showed that the promoter region was responsible for the flower-specific expression of Llccs in L. lancifolium (Fig. 4b, c). The expression of monocot genes, driven by their respective native promoters, in dicot species is not always appropriately regulated (Shimamoto 1994), and the converse is probably also true. Even though comprehensive functional analysis of the Llccs promoter has yet to be completed, the results of expression analysis of Llccs in L. lancifolium indicate that the Llccs promoter could still be a useful tool for directing flower-specific expression of Llccs, or any other gene, in transgenic monocots.
Conclusions
We have successfully cloned the gene for capsanthin-capsorubin synthase (Llccs) from tiger lily (L. lancifolium). The gene is expressed only in floral tissues and not in leaves. We demonstrated that Llccs is a functional gene capable of catalyzing the synthesis of both capsanthin and capsorubin in vivo.
There are several potential applications for the newly cloned Llccs gene. First, most plants do not produce capsanthin and capsorubin, and the natural food sources of capsanthin and capsorubin are basically limited to pepper fruits and their products. Both capsanthin and capsorubin have a strong singlet oxygen-quenching activity and can inhibit the peroxidation of lipids that is induced by free radicals (Matsufuji et al. 1998, Maoka et al. 2001a). They also exhibit strong anti-tumor activity in vitro (Maoka et al. 2001b). Because of the limited distribution of these compounds in nature, they are excellent targets for genetic engineering of other plant species to improve their nutritional value. Secondly, genetic modification of the carotenoid biosynthetic pathway by the introduction of the Llccs gene into ornamental plants will allow the development of novel orange and red colors that have previously been absent from these species. Thirdly, the availability of the Llccs sequence and that of pepper ccs, the only two functionally characterized ccs genes, will help us design strategies for cloning additional ccs genes from other species that synthesize and accumulate the above-mentioned κ-xanthophylls, which in turn will aid in determining the functional regions and sequence motifs that distinguish CCS from LCYB and NSY. Fourthly, the promoter region of the Llccs gene should serve as a useful tool for flower-specific expression of genes in monocot plants. In short, the cloning of the Llccs gene will advance our understanding of molecular and genetic mechanisms of κ-carotenoid biosynthesis and will also facilitate transgenic conversion of yellow flowers and fruits into orange and red ones in different plant species.
Materials and Methods
Plant material
Lilium lancifolium ‘Splendens’ (formerly L. tigrinum) bulbs were purchased from Brent and Becky’s Bulbs. Bulbs were grown in 4 liter pots that contained a growth medium composed of three parts peat, two parts pumice and one part sandy loam (by vol.) in a greenhouse, with 16 h days at 25 ± 3°C and 8 h nights at 20 ± 3°C. Natural light was supplemented with light from high-pressure sodium lamps (Energy Technics) to provide a photosynthetic photon flux density (PPFD) of 400–500 µmol m–2 s–1 at the surface of the growth medium. Later, for studies of gene expression, L. lancifolium ‘Splendens’ bulbs were purchased from BulbsDirect.com and grown as described above.
Isolation of total RNA from flowers of L. lancifolium and synthesis of cDNA
Total RNA was isolated from tepals of orange flower buds with TRIazol® reagent (Invitrogen) or an RNeasy Plant Mini Kit (QIAGEN Inc.) in accordance with the manufacturer’s instructions. cDNA was synthesized by M-MLV reverse transcriptase (Promega) according to the manufacturer’s recommendations.
Molecular cloning of LllcyB and Llccs
In an attempt to clone a DNA fragment of the lcyB gene located between two adjacent regions that are conserved in both lcyB and pepper ccs, we used various heterologous, non-degenerate primers. The DNA fragment (∼400 bp) obtained with Cs-F (forward) and Cs-R (reverse) primers had strong similarity in terms of deduced amino acid sequence to lcyB from several plant species. This sequence was then used to design a number of forward primers for cloning of the 3′-proximal ends of LllcyB and Llccs by 3′RACE. The list of all primers used in this study and their sequences are presented in Table 1.
Table 1.
The primers used for cloning and expression analysis by RT–PCR of genes for lycopene β-cyclase (LllcyB) and capsanthin-capsorubin synthase (Llccs) from Lilium lancifolium
| Primer | Sequence (5′ → 3′) |
|---|---|
| Cs-F | TGGAGTTTGGGTTGATGAGT |
| Cs-R | TGGGAATCTCTCCAATCCAT |
| ccs-3Rc | TCTGGATAAGATGCTCTTCATGGATTGGAG |
| ccs-In | ATGCCAATACCTCGGTTCCAGA |
| ccs-Out | CTCTTCCAAGACGCATGAGCAA |
| Llccs-F | GGATCCACCGTCCAGCCGCCTCCTTG |
| Llccs-R | GGATCCATGTGAATTGCATTATATAA |
| lcyB-3Rc | AGAAGAGACTTCTTTGGTTGCTC |
| lcyB-In | TCTCCAATCCATGAAGAGCATCTTATCCAG |
| lcyB-Out | GCCGAGCAACCAAAGAAG |
| P-ccs-R1 | CAGCGGACCTCCCATTGGAAACAAACACTT |
| P-ccs-R2 | TTGGAATAGCAGCGTTGTGAGCACGGTGTT |
| P-ccs-F1 | ATTGGCATGGGTTTATGTCGTCGAGGCTGT |
| P-ccs-F2 | GATGTTGTGACCAAGTGCCCTTTGCCTTTG |
| L-Llccs-F | GCAGCAAACCATGAACCTTTG |
| L-Llccs-R | ATTAAGATATGTTTCGTGAT |
| E-Llccs-F | CTCCTCCGCCCACCGCTAC |
| E-Llccs-R | CGGAGCAGGAGACGGAGGAG |
| E-LllcyB-F | GGGATTCTCGCTGAGGTGGAA |
| E-LllcyB-R | CCCAAAAGGCCATGGTCAGA |
| E-LhAct-F | ATGTATGTTGCAATCCAGGCTGTGC |
| E-LhAct-R | ATACCAGTAGCTTCCATTCCAACCA |
Cloning of LllcyB was achieved with forward primer lcyB-3Rc in combination with the 3′-outer primer from the RACE kit. After cloning and sequencing, a pair of nested primers (lcyB-Out and lcyB-In) was designed on the basis of the newly cloned sequence and used for 5′RACE.
Cloning of Llccs was achieved with forward primer ccs-3Rc in combination with the 3-outer primer from the RACE kit. After cloning and sequencing, a pair of nested primers (ccs-Out and ccs-In) was designed on the basis of the newly cloned sequence and used for 5′RACE.
The 3′RACE and 5′RACE of the corresponding proximal ends of the LllcyB and Llccs genes were performed with a FirstChoice® RLM-RACE Kit (Ambion Inc.) according to the manufacturer’s recommendations.
Cloning of the promoter region of the Llccs gene
The upstream promoter region of the Llccs gene was cloned by LDI-PCR as described by Willis et al. (1997), with some modifications. Total genomic DNA was extracted from young, newly sprouted leaves with a DNeasy Plant Mini Kit (QIAGEN Inc.). Aliquots of genomic DNA (1 µg) were digested separately with different restriction enzymes, which included: HindIII, EcoRI, BamHI, XbaI, SpeI, AvrII, XmaI, PstI, SalI and SphI. Digestion was allowed to proceed overnight in a water bath at 37°C. To remove residual enzymatic activity, all samples were processed with a QIAprep Spin Miniprep Kit (QIAGEN Inc.) as described in the QIAquick PCR Purification Kit Protocol. Samples were eluted twice with 250 µl of TE buffer to ensure maximum recovery of DNA fragments. DNA was then ligated at 15°C overnight in a total volume of 500 µl with 12 U of T4 DNA ligase (Invitrogen). The ligated DNA was purified with a QIAprep Spin Miniprep Kit and eluted in 50 µl of EB buffer. The first amplification by PCR was performed under the following conditions: initial denaturation at 94°C/1 min, followed by 30 cycles of two-step PCR at 98°C/10 s and 68°C/6 min, and a final extension at 68°C/7 min with Hot Start ExTaq-HS Taq polymerase (Takara Shuzo Co., Ltd.). The reaction mixture (25 µl) contained 1 µl of ligated DNA (approximately 20 ng). The product of the first PCR (1 µl) was then re-amplified for 30 cycles with nested primers under identical conditions. To verify the results of LDI-PCR, 5–10 µl of each PCR product were fractionated by agarose gel electrophoresis (0.8% agarose). The DNA fragments of interest were extracted and cloned into the pGEM T-Easy vector (Promega) and sequenced. The sets of forward (P-ccs-F1 and P-ccs-F2) and reverse (P-ccs-R1 and P-ccs-R2) nested primers used in LDI-PCR are shown in Table 1. After sequencing, a pair of primers (L-Llccs-F and L-Llccs-R) was designed to amplify the Llccs locus in the correct (as distinct from the inverse) orientation. The Llccs locus was cloned into the pGEM T-Easy vector and sequenced again to confirm its identity.
Manipulation of DNA and bioinformatic analysis
DNA was extracted from gels with standard or MinElute QIAquick Gel Extraction Kits (QIAGEN Inc.). All PCR products were subcloned into the pGEM T-Easy vector, and DNA sequencing was performed at the Central Service Laboratory at the Center for Genome Research and Biocomputing, Oregon State University, Corvallis, OR. The ORF Finder tool at the NCBI (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi) was used to predict ORFs within newly cloned sequences. Molecular masses and theoretical isoelectric points of predicted proteins were calculated at the ExPASy Bioinformatics Resource Portal using the Compute pI/Mw tool (http://web.expasy.org/compute_pi/). TargetP 1.1 Server (http://www.cbs.dtu.dk/services/TargetP/) was used to search for N-terminal TP sequences. Multiple sequence alignments and homology analyses of nucleotide and deduced amino acid sequences were performed using Probabilistic Consistency-based Multiple Alignment of Amino Acid Sequences (ProbCons) at http://toolkit.tuebingen.mpg.de/probcons and the Basic Local Alignment Tool (BLAST) at http://www.ncbi.nlm.nih.gov. Analysis of gel images after RT–PCR was carried out with ImageJ 1.45 s at http://imagej.nih.gov/ij/. Phylogenetic analysis was carried out by aligning amino acid sequences of various LCYBs, NSYs, CSSs and one LCYE with ClustalW2 version 2.1 (Larkin et al. 2007). The aligned sequences were used to construct a phylogenetic tree using the Neighbor–Joining methodology in MEGA 4 (Tamura et al. 2007). The values displayed at the nodes are percentage consensus support, calculated using a bootstrapping test with 1,000 replications.
Expression analysis
Analysis of the expression of Llccs and LllcyB in tepals of flower buds, in tepals of fully open flowers and in leaves was performed by RT–PCR. Buds at four developmental stages, which were based on the length of buds, were used: stage B1 (3–4 cm), stage B2 (5–6 cm), stage B3 (7–8 cm) and stage B4 (>9 cm; Fig. 4a). For RT–PCR, we used 1 µg of RNA as template for synthesis of first-strand cDNA with an Omniscript® Reverse Transcription kit (QIAGEN Inc.). Primers used for analysis by RT–PCR of the expression of Llccs (E-Llccs-F and E-Llccs-R), LllcyB (E-LllcyB-F and E-LllcyB-R) and the gene for actin (E-LhAct-F and E-LhAct-R) are shown in Table 1. The conditions for PCR were as follows: initial denaturation at 95°C/3 min, followed by 33 cycles of three-step PCR (95°C/30 s, 60°C/30 s, 72°C/1 min) and a final extension at 72°C/7 min with GoTaq® Hot Start Polymerase (0.625U) in Green GoTaq® Flexi Buffer (Promega).
Construction of the binary plasmid vector for transformation
A pair of primers (Llccs-F and Llccs-R) was designed for introduction of a BamHI restriction site at each end of the Llccs gene. The Llccs gene was amplified by PCR, cloned into the pGEM-T Easy vector and sequenced. The amplified DNA fragment encompassed an ‘A’ that was identified as a site of initiation of transcription for the Llccs, the complete 5′ UTR, the Llccs ORF and part of the 3′ UTR (extending 131 bp beyond the termination codon). To flank the Llccs gene with the CaMV 35 S promoter and a Nos terminator, we excised the codA gene from plasmid pUC/codA (Hayashi et al. 1997) with XbaI and SacI and replaced it with a poly-linker (XBSmPS) that contained XbaI, BamHI, SmaI, PstI and SacI restriction sites. This intermediate vector was then digested with HindIII, ends were blunted by the Klenow fragment, and an EcoRI linker was used to introduce an EcoRI restriction site. The resulting plasmid was designated pUC119/E-XBSmPS-E. After sequencing, as noted above, Llccs was excised with BamHI and ligated into plasmid pUC119/E-XBSmPS-E that had previously been digested with BamHI and dephosphorylated with shrimp alkaline phosphatase (UBS) to prevent self-ligation. The resultant plasmid was designated pUC119/P35S::Llccs::TNos. To introduce a NotI restriction site flanking P35S::LTccs::TNos, we excised the expression cassette from pUC119/P35S::Llccs::TNos with EcoRI and transferred it to pGEM-T Easy that had previously been digested with EcoRI and dephosphorylated, as above, to prevent self-ligation. The EcoRI site in pGEM-T Easy vector is flanked by NotI. Finally, the P35S::LTccs::TNos expression cassette was excised with NotI and transferred to pWBVec10a (Wang et al. 1998) that had previously been digested with NotI and dephosphorylated as above. The orientation of the expression cassette inside the binary vector was determined by sequencing. The resulting vector was designated pWBVec10a/P35S::Llccs::TNos. The binary vector pWBVec10a contains a hygromycin resistance gene (PCaMV35S-hpt-T35S) and a GUS gene (PUbi1-uidA-TNOS) expression cassette within the T-DNA borders. Both genes contain an intron to prevent expression in Agrobacterium tumefaciens (Wang et al. 1998).
Agrobacterium-mediated stable transformation of callus tissue of Iris germanica
Agrobacterium tumefaciens LBA4404 (Hoekema et al. 1983) was transformed with the plasmid vector pWBVec10a/P35S::Llccs::TNos using a chemically based procedure, as described by Walkerpeach and Velten (1994). Then non-embryogenic callus tissue of I. germanica ‘Hot Property’ was transformed as described previously (Jeknić et al. 1999) with minor modifications. A. tumefaciens was grown in YEP broth with appropriate antibiotics at 28°C, overnight, with shaking at 250 r.p.m. The bacteria were harvested by centrifugation and resuspended in 2 vols. of MS-Inf medium (Murashige and Skoog basic medium supplemented with 50 g l–1 sucrose, 10 g l–1 glucose, pH 5.2, containing 100 µM acetosyringone). Bacteria were incubated in this medium for 1–2 h with shaking at 100 r.p.m. prior to transformation of callus. Clumps of hygromycin-resistant cells were generated in medium that contained 10 mg l–1 hygromycin and assayed for GUS activity as described by Jefferson (1987). Stable integration of the Llccs transgene was confirmed by PCR.
Analysis of carotenoids by HPLC
Carotenoids were extracted from wild-type and two independent Llccs-transgenic calli of I. germanica (Llccs-1 and Llccs-2) that accumulated high amounts of red-orange pigment(s) as described by Fraser et al. (2000). In each case, we used 100 mg of freeze-dried callus tissue. For HPLC analysis, a 200 µl aliquot of each sample was evaporated to dryness under a stream of nitrogen and the residue was resuspended in 200 µl of 90% acetone with an internal standard (vitamin E). HPLC was performed as described by Van Heukelem and Thomas (2005). Samples were mixed on a vortex mixer and placed in the cooling rack of the HPLC system. Then 357 µl of buffer and 143 µl of extract were injected into the system (LC-10A HPLC system with LCsolution software; Shimadzu) using a pre-treatment program and mixing in the loop prior to injection.
Analysis of carotenoids by UPLC-MS/MS
Total carotenoids were extracted from 2 g of fresh Llccs-transgenic iris callus tissue as follows. The callus tissue was powdered in liquid nitrogen with a mortar and pestle and transferred to a 50 ml tube that contained 20 ml of extraction solvent, which consisted of hexane, acetone and ethanol (50 : 25 : 25, by vol.). The suspension was mixed and agitated for 15 min. Then 5 ml of 50 mM Tris–HCl (pH 8.0) containing 1 M NaCl were added and the suspension was mixed by inversion for an additional 5 min. A clear partition was achieved by centrifugation at 3,500 r.p.m. for 5 min in a clinical centrifuge (NH-SII; IEC). The hexane extract was removed with a Pasteur pipet and evaporated to dryness under a stream of nitrogen. The residue was suspended in 2 ml of methyl-tert-butyl ether (MTBE). Saponification was performed overnight as described by Lee et al. (2001). After three rinses with water, the MTBE layer was filtered through an Isolute® sodium sulfate (Na2SO4) drying cartridge (Biotage, LLC) and evaporated to dryness under a stream of nitrogen. The residue was suspended in 1 ml of ethanol. The sample was then filtered through a PTFE 0.45 µm filter and subjected to UPLC-MS/MS analysis. Capsanthin and capsorubin standards were purchased from CaroteNature. To prepare the reference solutions, we dissolved 1 mg of capsanthin and 1 mg of capsorubin separately in 50 ml of ethanol and filtered each solution through a PTFE 0.45 µm filter.
An ABSciex 5600 TripleToF fast-scanning high-resolution quadrupole time of flight (Q-ToF) system (ABSciex) coupled to a Prominence LC-30AD UPLC system (Shimadzu) was used for accurate mass analysis. A C18, 2.1 mm × 50 mm, 1.9 µm particle size UPLC column (Perkin Elmer) was used for UPLC separation, with LC-MS grade solvents. Solvent A was water (EMD Chemicals), solvent B was acetonitrile (EMD Chemicals), and 0.1% formic acid (Sigma Aldrich) was added to both solvents. The flow rate was 0.7 ml min–1. The following gradient was employed: initial conditions, 20% solvent B ramped to 90% solvent B over the course of 3 min; holding for 2 min at 90% solvent B; and re-equilibration for 1 min at 20% solvent B.
The mass spectrometer was operated in positive ToF-MS/MS mode at the following settings: scan time, 0.25 s per scan; the declustering potential, 80; curtain gas, 25; gas1, 50; gas2, 40; source temperature, 550°C; spray voltage, 5,500 Vs; and an applied collision energy, 35 to induce fragmentation. The m/z scan range was 100–1,000 amu. The instrument was calibrated with calibration solution from ABSciex before each analysis.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Cooley’s Gardens Inc., Silverton, OR [Grant: ARF3711]; the National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health (NIH) [Award Nos. P30ES000210 and S10RR027878]; the Ministry of Education and Science, Republic of Serbia [Grant: TR31019].
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Ming-Bo Wang (CSIRO Plant Industry, Black Mountain, Australia) for providing the pWBVec10a binary vector used in this study. Custom analysis of carotenoids by HPLC was performed by DHI (Høersholm, Denmark). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIEHS or NIH. The authors acknowledge the Biomolecular Mass Spectrometry Core of the Environmental Health Sciences Core Center at Oregon State University.
Glossary
Abbreviations
- CaMV
Cauliflower mosaic virus
- CCS
capsanthin-capsorubin synthase
- GUS
β-glucuronidase
- LCYB
lycopene β-cyclase
- LDI-PCR
long-distance inverse-PCR
- MTBE
methyl-tert-butyl ether
- NSY
neoxanthin synthase
- Nos
nopaline synthase
- ORF
open reading frame
- PTFE
polytetrafluoroethylene
- PPFD
photosynthetic photon flux density
- Q-ToF
quadrupole time of flight
- RACE
rapid amplification of cDNA ends
- RT–PCR
reverse transcription–PCR
- TLC
thin-layer chromatography
- TP
transit peptide
- UPLC-MS/MS
ultraperformance liquid chromatography–tandem mass spectrometry
- UTR
untranslated region.
References
- Bartley GE, Scolnik PA. Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell. 1995;7:1027–1038. doi: 10.1105/tpc.7.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, d’Harlingue A, Backhous RA, Kumagai MH, Camara B. Identification of neoxanthin synthase as a carotenoid cyclase paralog. Eur. J. Biochem. 2000;267:6346–6352. doi: 10.1046/j.1432-1327.2000.01722.x. [DOI] [PubMed] [Google Scholar]
- Bouvier F, d’Harlingue A, Camara B. Molecular analysis of carotenoid cyclase inhibition. Arch. Biochem. Biophys. 1997;346:53–64. doi: 10.1006/abbi.1997.0278. [DOI] [PubMed] [Google Scholar]
- Bouvier F, Hugueney P, d’Harlingue A, Kuntz M, Camara B. Xanthophyll biosynthesis in chromoplasts: isolation and molecular cloning of an enzyme catalyzing the conversion of 5,6-epoxycarotenoid into ketocarotenoid. Plant J. 1994;6:45–54. doi: 10.1046/j.1365-313x.1994.6010045.x. [DOI] [PubMed] [Google Scholar]
- Camara B, Monéger R. Free and esterified carotenoids in green and red fruits of Capsicum annuum. Phytochemistry. 1978;17:91–93. [Google Scholar]
- Campbell WH, Gowri G. Codon usage in higher plants, green algae, and cyanobacteria. Plant Physiol. 1990;92:1–11. doi: 10.1104/pp.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comber HF. A new classification of the genus Lilium. Lily Yearbook, Royal Hort. Soc. 1949;13:86–105. [Google Scholar]
- Cunningham FX., Jr Regulation of carotenoid synthesis and accumulation in plants. Pure Appl. Chem. 2002;74:1409–1417. [Google Scholar]
- Cunningham FX, Jr., Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- De Jong PC. Some notes on the evolution of lilies. North Amer. Lily Yearb. 1974;27:23–28. [Google Scholar]
- Deli J, Matus Z, Tóth G. Carotenoid composition in the fruits of Asparagus officinalis. J. Agric. Food Chem. 2000;48:2793–2796. doi: 10.1021/jf991243h. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Elisabete M, Pinto S, Holloway DE, Bramley PM. Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. 2000;24:551–558. doi: 10.1046/j.1365-313x.2000.00896.x. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Alia, Mustardy L, Deshnium P, Ida M, Murata N. Transformation of Arabidopsis thaliana with the codA gene for choline oxidase: accumulation of glycinebetaine and enhanced tolerance to salt and cold stress. Plant J. 1997;12:133–142. doi: 10.1046/j.1365-313x.1997.12010133.x. [DOI] [PubMed] [Google Scholar]
- Hirschberg J, Cohen M, Harker M, Lotan T, Mann V, Pecker I. Molecular genetics of the carotenoid biosynthesis pathway in plants and algae. Pure Appl. Chem. 1997;69:2151–2158. [Google Scholar]
- Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Hugueney P, Badillo A, Chen H-C, Klein A, Hirschberg J, Camara B, et al. Metabolism of cyclic carotenoids: a model for the alteration of this biosynthetic pathway in Capsicum annuum chromoplasts. Plant J. 1995;8:417–424. doi: 10.1046/j.1365-313x.1995.08030417.x. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 1987;5:387–405. [Google Scholar]
- Jeknić Z, Lee SP, Davis J, Ernst RC, Chen THH. Genetic transformation of Iris germanica mediated by Agrobacterium tumefaciens. J. Amer. Soc. Hort. Sci. 1999;124:575–580. [Google Scholar]
- Karrer P, Oswald A. Carotinoide aus den staubbeuteln von Lilium tigrinum. Ein neues carotenoid: anthetaxanthin. Helv. Chim. Acta. 1935;13:1303–1305. [Google Scholar]
- Kawabe A, Miyashita N. Patterns of codon usage bias in three dicot and four monocot plant species. Genes Genet. Syst. 2003;78:343–352. doi: 10.1266/ggs.78.343. [DOI] [PubMed] [Google Scholar]
- Kubasik P, Sandmann G. Molecular evaluation of lycopene cyclases involved in the formation of carotenoids with ionone end groups. Biochem. Soc. Trans. 2000;28:806–810. [PubMed] [Google Scholar]
- Kumagai MH, Keller Y, Bouvier F, Clry D, Camara B. Functional integration of non-native carotenoids into chloroplasts by viral-derived expression of capsanthin–capsorubin synthase in Nicotiana benthamiana. Plant J. 1998;14:305–315. doi: 10.1046/j.1365-313x.1998.00128.x. [DOI] [PubMed] [Google Scholar]
- Ladygin VG. Biosynthesis of carotenoids in plastids of plants. Biochemistry (Mosc.) 2000;65:1113–1128. Translated from Biokhimiya 65: 1317–1333. [PubMed] [Google Scholar]
- Larkin. MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee HS, Castle WS, Coates GA. High-performance liquid chromatography for the characterization of carotenoids in the new sweet orange (Earlygold) grown in Florida, USA. J. Chromatogr. A. 2001;913:371–377. doi: 10.1016/s0021-9673(00)01029-3. [DOI] [PubMed] [Google Scholar]
- Maoka T. Recent progress in structural studies of carotenoids in animals and plants. Arch. Biochem. Biophys. 2009;483:191–195. doi: 10.1016/j.abb.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Maoka T, Akimoto N, Fujiwara Y, Hashimoto K. Structure of new carotenoids with the 6-oxo-κ end group from the fruits of paprika, Capsicum annuum. J. Nat. Prod. 2004;67:115–117. doi: 10.1021/np030400a. [DOI] [PubMed] [Google Scholar]
- Maoka T, Goto Y, Isobe K, Fujiwara Y, Hashimoto K, Mochida K. Antioxidative activity of capsorubin and related compounds from paprika (Capsicum annuum) J. Oleo. Sci. 2001a;50:663–665. [Google Scholar]
- Maoka T, Mochida K, Kozuka M, Ito Y, Fujiwara Y, Hashimoto K, et al. Cancer chemopreventive activity of carotenoids in the fruits of red paprika Capsicum annuum L. Cancer Lett. 2001b;172:103–109. doi: 10.1016/s0304-3835(01)00635-8. [DOI] [PubMed] [Google Scholar]
- Märki-Fischer E, Eugster CH. Das carotinoidspektrum der antheren und petalen von Lilium tigrinum cv. ‘Red Night’. Helv. Chim. Acta. 1985;68:1708–1715. [Google Scholar]
- Márkus F, Daood HG, Kapitány J, Biacs PA. Change in the carotenoid and antioxidant content of spice red pepper (paprika) as a function of ripening and some technological factors. J. Agric. Food Chem. 1999;47:100–107. doi: 10.1021/jf980485z. [DOI] [PubMed] [Google Scholar]
- Matsufuji H, Nakamura H, Chino M, Takeda M. Antioxidant activity of capsanthin and the fatty acid esters in paprika (Capsicum annuum) J. Agric. Food Chem. 1998;46:3468–3472. [Google Scholar]
- Mialoundama AS, Heintz D, Jadid N, Nkeng P, Rahier A, Deli J, et al. Characterization of plant carotenoid cyclases as members of the flavoprotein family functioning with no net redox change. Plant Physiol. 2010;153:970–979. doi: 10.1104/pp.110.155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo E, McLean R, Britton G, Agócs A, Nagy V, Deli J. Sapotexanthin, an A-provitamin carotenoid from red mamey (Pouteria sapota) J. Nat. Prod. 2011;74:283–285. doi: 10.1021/np1006982. [DOI] [PubMed] [Google Scholar]
- Murillo E, Mosquera Y, Kurtán T, Gulyás-Feket G, Nagy V, Deli J. Isolation and characterization of novel capsorubin-like carotenoids from the red mamey (Pouteria sapota) Helv. Chim. Acta. 2012;95:983–988. [Google Scholar]
- Ohmiya A. Diversity of carotenoid composition in flower petals. JARQ-Jpn. Agric. Res. Q. 2011;45:163–171. [Google Scholar]
- Partali V, Liaaen-Jensen S, Huneck S, Khaidav T. Carotenoids from the flowers of Lilium pumilum. Die Pharmazie. 1987;42:208. [Google Scholar]
- Rodríguez-Concepción M. Supply of precursors for carotenoid biosynthesis in plants. Arch. Biochem. Biophys. 2010;504:118–122. doi: 10.1016/j.abb.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Ronen G, Carmel-Gorent L, Zamirt D, Hirschberg J. An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc. Natl Acad. Sci. USA. 2000;97:11102–11107. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Moras D, Olsen KW. Chemical and biological evolution of a nucleotide-binding protein. Nature. 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- Rüttimann A. Synthesis and stereochemistry of red pepper carotenoids. In: Britton G, Goodwin TW, editors. Carotenoid Chemistry and Biochemistry. Pergamon: Oxford; 1982. pp. 71–86. [Google Scholar]
- Seybold A. Untersuchungen über den farbwechsel von blumenblättern, früchten und samenschalen. Sber. Heidelb. Akad. Wiss. Math.-Naturwiss. Kl. 1953;4:31–124. [Google Scholar]
- Shimamoto K. Gene expression in transgenic monocots. Curr. Opin. Biotechnol. 1994;5:158–162. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- Valadon LRG, Mummery RS. Carotenoids of lilies and of red pepper: biogenesis of capsanthin and capsorubin. Z. Pflanzenphysiol. 1976;82:407–416. [Google Scholar]
- Van Heukelem L, Thomas CS. The HPLC method. In: Hooker S, Van Heukelem L, Thomas CS, Claustre H, Ras J, Barlow R, et al., editors. The Second SeaWiFS HPLC Analysis Round-Robin Experiment (SeaHARRE-2) 2005. pp. 86–92. NASA Technical Memorandum 2005-212785. [Google Scholar]
- Walkerpeach CR, Velten J. Agrobacterium-mediated gene transfer to plant cells: cointegrate and binary vector systems. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. 2nd edn. The Netherlands: Kluwer Academic, Dordrecht; 1994. pp. B1/1–B1/19. [Google Scholar]
- Wang MB, Li ZY, Matthews PR, Upadhyaya NM, Waterhouse PM. Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. Acta Hortic. 1998;461:401–407. [Google Scholar]
- Willis TG, Jadayel DM, Coignet LJA, Abdul-Rauf M, Treleaven JG, Catovsky D, et al. Rapid molecular cloning of rearrangements of the IGHJ locus using long-distance inverse polymerase chain reaction. Blood. 1997;90:2456–2464. [PubMed] [Google Scholar]
- Yamagishi M, Kishimoto S, Nakayama M. Carotenoid composition and changes in expression of carotenoid biosynthetic genes in tepals of Asiatic hybrid lily. Plant Breed. 2010;129:100–107. [Google Scholar]
- Zhu C, Bai C, Sanahuja G, Yuan D, Farré G, Naqvi S, et al. The regulation of carotenoid pigmentation in flowers. Arch. Biochem. Biophys. 2010;504:132–141. doi: 10.1016/j.abb.2010.07.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.