See related article in EMBO Molecular Medicine http://dx.doi.org/10.1002/emmm.201101105
There is growing evidence that inactivation of the p53 tumour suppressor pathway is required for full-blown tumourigenesis (Levine, 1997). While half of human cancers harbour mutations or deletions of the p53 tumour suppressor locus, the remaining cancers employ alternative mechanisms to subvert the activity of wild-type p53. This clinically relevant pathway has therefore logically become the target for the development of innovative avenues in cancer therapy. For instance, restoration of p53 function has been extensively pursued as a therapeutic modality in cancers in which p53 function is compromised although the TP53 locus remains intact (Brown et al, 2009). Such approach is, however, only applicable to tumours expressing wild-type p53. Because alteration of p53 is so frequent in cancer, identification of synthetic lethal partners of p53 should lead to conceptually simple and attractive approaches to selective targeting of cancer cells (Kaelin, 2005). Inactivation of such a target would in theory be detrimental to virtually all cancer cells irrespective of the mechanisms that led to p53 inactivation. In the present issue, Haley and colleagues (Haley et al, 2012) provide evidence for a synthetic lethal interaction between the p53 and Igf2 pathways.
IGF2 is a growth-promoting hormone during gestation and commonly overexpressed in human cancer through re-expression of the imprinted maternal allele by loss of imprinting (LOI). IGF-2 exerts its effects by binding to the IGF-1 receptor and activating both the PI3K-AKT-mTOR and RAS-RAF-MEK-ERK downstream signalling pathways.
Although previous in vitro studies highlighted several mechanistic pathway interactions between IGF2 and p53 signalling genetic evidence for such interactions was lacking. To search for such genetic links in vivo, Haley and colleagues engineered mice with varying allelic doses of Igf2 and Tp53.
Although a fraction of Tp53-deficient females exhibit exencephaly and die during embryonic development (Armstrong et al, 1995), the vast majority of Tp53-null mice are viable and fertile (Donehower et al, 1992). Unexpectedly, Harley and colleagues found that Igf2 KO females (harbouring a targeted paternal allele and a maternal allele silenced due to imprinting) were not viable, likely due to lung malformations (Fig 1). Interestingly, loss of one p53 allele partly rescued this lethality indicating that an increase in p53 activity may be at least partly responsible for this phenotype (Fig 1). The authors searched for differentially expressed genes that may cause this phenotype. As the phenotype is gender-specific (see below) and p53-dependent, they reasoned that the disease-causing genes should lie on the X-chromosome and possess p53-responsive elements. They identified such a gene signature and among others Fn1 as a possible candidate. However, further studies will be required to establish the specific contribution of the selected genes to the phenotype. Regardless of the underlying molecular mechanism, this finding highlights a first important interaction between the Igf2 and p53 pathways. The data support the view that complete loss of Igf2 in females leads to increase p53 activity in vivo.
Figure 1. Igf2 and Tp53 genetic interactions in mice.
Whereas Igf2-deficient males are viable, Igf2-deficiency is lethal in females. Loss of one Trp53 allele is sufficient to partly rescue this lethality. Loss of both Igf2 and p53 expression is not compatible with life.
»…this finding highlights a first important interaction between the Igf2 and p53 pathways.«
In contrast to the females, Igf2 KO males are viable (Fig 1), although reduced in size compared to controls. Interestingly, the authors find that the combined loss of Igf2 and Tp53 is incompatible with life both in males and females (Fig 1). Igf2/Tp53-double mutants die during embryonic development, at birth or during postnatal life. Surprisingly, histopathological examination did not reveal any obvious developmental abnormalities that could explain the lethality. Nevertheless, although the underlying mechanism remains obscure, this observation indicates that loss of Tp53 leads to lethal developmental defects, which are suppressed by Igf2.
The authors also investigated possible cross talk between these two signalling pathways during tumour development. Tp53 heterozygosity leads to spontaneous tumour development in mice (Jacks et al, 1994). Interestingly, biallelic expression of Igf2, as a result of LOI, accelerated tumour formation in Tp53 heterozygous animals. More importantly, while loss of heterozygosity (LOH) of Tp53 is often seen in tumours of Tp53+/− mice, biallelic expression of Igf2 reduced the selection pressure to inactivate the remaining p53 allele. These data indicate that increased Igf2 signalling may favour tumour development, at least partly, by dampening down the activity of the p53 tumour suppressor pathway. Finally, Haley and colleagues also assessed the impact of Igf2 inactivation on tumour development. Importantly, they find that conditional homozygous deletion of Igf2 (using a newly generated conditional cKO allele) significantly delays the onset of p53-null tumour phenotype. This observation indicates that the development of p53-deficient tumours is at least partly dependent on Igf2 signalling.
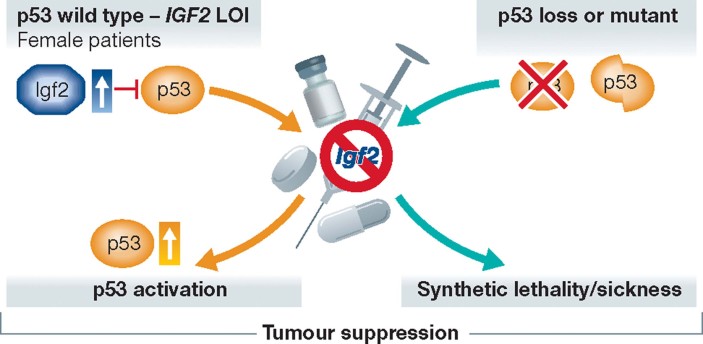
These observations may have important therapeutic implications (Fig 2): IGF2 overexpression might be one important mechanism that leads to p53 inactivation in human tumours that retain wild-type p53. Consequently, IGF2 targeting should be explored as a putative therapeutic avenue for reactivation of p53 tumour suppressor function in cancers with IGF2 LOI. The conclusions drawn from these studies should, however, be considered cautiously. First, increased Igf2 only favours tumour development in females. The reason for this gender-specific phenotype is unknown and severely limits the therapeutic implications of this finding. Second, the model used to mimic IGF2 LOI in mice is imperfect. The authors used mice carrying deletion of the H19 locus, which results in bi-allelic expression of Igf2 but also loss of function of the H19 ncRNA and associated miR-675 and upregulation of miR-483 located in the Igf2 locus (Gabory et al, 2009). It will therefore be important to further investigate whether the effects on tumour development observed in these mice are solely attributable to Igf2 LOI and/or to deregulation of the above-mentioned ncRNAs.
Figure 2. Targeting the Igf2 pathway in human tumours.
The p53 pathway malfunctions in virtually all human cancers. Data reported in Haley et al. indicate that IGF2 LOI promotes tumourigenesis by inactivating p53 in tumours that retain wild-type TP53. Targeting IGF2 signalling in this context may lead to reactivation of p53 tumour suppressor function. Loss of Igf2 significantly affects the progression of p53-deficient tumours in mice. This observation predicts that IGF2 targeting will reduce the fitness of tumours in which p53 function is compromised as a result of TP53 inactivating mutations or deletions.
»…the Igf2 tumour addiction may be caused by a synthetic lethal/sickness between the p53 and Igf2 signalling pathways.«
The observations that Igf2 is required for the survival of Tp53 KO mice and for the development of Tp53-deficient tumours are intriguing from a therapeutic point of view. Although Igf2 dependency of tumours has already been described in other mouse models (Christofori et al, 1994, 1995; Ho et al, 2009), the mechanism underlying this dependency has remained elusive. Although more work is needed to establish this possibility formally and gain more insights into the underlying mechanism, the data reported by Haley et al. indicate that the Igf2 tumour addiction may be caused by a synthetic lethal/sickness between the p53 and Igf2 signalling pathways. Given that the p53 pathway is inactivated in most human cancers, these findings indicate that IGF2 targeting may become a pharmacological mode of tumour-type-specific intervention that could theoretically be applicable to a wide range of cancers. The experiments of Haley et al. are limited to lymphoma and sarcoma, so a confirmation of their findings in other tumour models is advisable, especially given the epithelial origin of most human tumours. It is also imperative to test whether the p53/Igf2 synthetic lethal/sickness interaction occurs irrespective of the various mechanisms selected by tumour cells to alter p53 function (i.e. p53 inactivating mutations or MDM2/MDMX overexpression). In this context it is interesting to note that adrenal carcinoma and osteosarcoma from patients harbouring TP53 mutations (Li-Fraumeni syndrome) had already been shown to be dependent on IGF2 (Avnet et al, 2012).
Together the mouse genetic data reported in Haley et al. provide the rationale for the use of targeted IGF1-receptor antagonists to treat both tumours with IGF2 LOI and/or p53 inactivating mutations in future clinical trials.
Acknowledgments
The authors declare that they have no conflict of interest.
References
- Armstrong JF, Kaufman MH, Harrison DJ, Clarke AR. High-frequency developmental abnormalities in p53-deficient mice. Curr Biol. 1995;5:931–936. doi: 10.1016/s0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- Avnet S, Perut F, Salerno M, Sciacca L, Baldini N. Insulin receptor isoforms are differently expressed during human osteoblastogenesis. Differentiation. 2012;83:242–248. doi: 10.1016/j.diff.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- Christofori G, Naik P, Hanahan D. A second signal supplied by insulin-like growth factor II in oncogene-induced tumorigenesis. Nature. 1994;369(6479):414–418. doi: 10.1038/369414a0. [DOI] [PubMed] [Google Scholar]
- Christofori G, Naik P, Hanahan D. Deregulation of both imprinted and expressed alleles of the insulin-like growth factor 2 gene during beta-cell tumorigenesis. Nat Genet. 1995;10:196–201. doi: 10.1038/ng0695-196. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- Haley VL, Barnes DJ, Sandovici I, Constancia M, Graham CF, Pezzella F, Bühnemann C, Carter EJ, Hassan AB. Igf2 pathway dependency of the Trp53 developmental and tumour phenotypes. EMBO Mol Med. 2012 doi: 10.1002/emmm.201101105. DOI: 10.1002/emmm.201101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Stojanovski A, Whetstone H, Wei QX, Mau E, Wunder JS, Alman B. Gli2 and p53 cooperate to regulate IGFBP-3-mediated chondrocyte apoptosis in the progression from benign to malignant cartilage tumors. Cancer Cell. 2009;16:126–136. doi: 10.1016/j.ccr.2009.05.013. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]