Abstract
The molecular networks that control the progression of chronic kidney diseases (CKD) are poorly defined. We have recently shown that the susceptibility to development of renal lesions after nephron reduction is controlled by a locus on mouse chromosome 6 and requires epidermal growth factor receptor (EGFR) activation. Here, we identified microphthalmia-associated transcription factor A (MITF-A), a bHLH-Zip transcription factor, as a modifier of CKD progression. Sequence analysis revealed a strain-specific mutation in the 5′ UTR that decreases MITF-A protein synthesis in lesion-prone friend virus B NIH (FVB/N) mice. More importantly, we dissected the molecular pathway by which MITF-A modulates CKD progression. MITF-A interacts with histone deacetylases to repress the transcription of TGF-α, a ligand of EGFR, and antagonizes transactivation by its related partner, transcription factor E3 (TFE3). Consistent with the key role of this network in CKD, Tgfa gene inactivation protected FVB/N mice from renal deterioration after nephron reduction. These data are relevant to human CKD, as we found that the TFE3/MITF-A ratio was increased in patients with damaged kidneys. Our study uncovers a novel transcriptional network and unveils novel potential prognostic and therapeutic targets for preventing human CKD progression.
Keywords: EGFR, genetic susceptibility, MITF-A, renal lesions, TGF-alpha
INTRODUCTION
Chronic kidney disease (CKD), one of the major public health challenges of the 21st century, is characterized by a progressive decline in renal function to end stage renal failure (ESRF) that can occur irrespective of the cause of the renal damage (diabetes, hypertension, immune diseases, etc.), once a critical number of nephrons are lost. CKD is a worldwide concern: over 7 million people in the European Community are affected by CKD and 300,000 are undergoing renal replacement therapy, either by dialysis or transplantation. Similar rates are found in developing countries. This number is expected to increase steadily by 6–8% each year, hence imposing a major socioeconomic burden on national health care systems (Couser et al, 2011). Despite extended efforts of the health care community, the quality of life and survival of CKD patients remains poor. Faced with this persistently poor ESRD outcome, current clinical research efforts are focused on preventive strategies to slow down the rate of CKD progression. Understanding the pathophysiology of CKD progression is the first step in the development of new therapeutic strategies.
Despite the increasing prevalence of CKD, the mechanisms underlying the inescapable progression of CKD remain poorly defined. Although clinical observations have underscored the importance of environmental factors in the biological processes leading to renal deterioration (Taal and Brenner, 2006), multiple studies have highlighted the critical role played by genetic factors (Schelling et al, 1999). Indeed, epidemiological studies have clearly showed that there is an inter-individual risk of CKD progression and familial aggregation (Satko et al, 2007). More recently, linkage and genome-wide association studies have mapped several susceptibility loci to various chromosomal regions (Boger and Heid, 2011; Keller et al, 2012). However, the genes and the genetic networks that account for the increased susceptibility to CKD progression remain mostly unidentified.
Several experimental animal models have been developed to elucidate the molecular basis of CKD progression. Among them, the remnant kidney model is particularly relevant, since nephron reduction characterizes the progression of most human nephropathies (Remuzzi et al, 2006). Interestingly, the development and the progression of renal lesion after nephron reduction is highly strain-dependent (Esposito et al, 1999; Kren and Hostetter, 1999; Ma and Fogo, 2003). Hence, this model is an ideal tool for the identification of genes, which modify the evolution of CKD. We previously performed systematic screening in six different strains of mice and demonstrated that only one, the friend virus B NIH (FVB/N), develops severe lesions, while the other strains undergo compensatory growth alone at least up to 6 months after 75% nephron reduction (Laouari et al, 2011; Pillebout et al, 2001). We also showed that the susceptibility among inbred strains segregates with a locus that maps to chromosome 6 (Laouari et al, 2011). Additional markers allowed us to reduce the Ckdp1 locus to a 17-centimorgan interval. Notably, the locus includes Tgfa (transforming growth factor alpha, TGF-α), a gene that encodes for a ligand of epidermal growth factor receptor (EGFR). Consistent with the crucial role of this pathway in the renal deterioration process (Zeng et al, 2009), we observed that the expression of TGF-α markedly increased after nephron reduction in the lesion-prone FVB/N strain. However, further molecular analysis ruled out the hypothesis that a cis-acting mutation in the Tgfa gene accounted for the increased susceptibility to develop renal lesions in FVB/N mice (Laouari et al, 2011). Taking all these data together, we hypothesized that another gene within the locus may predispose FVB/N mice to renal deterioration by modulating the expression of Tgfa in trans.
Here, we used in silico, in vitro and in vivo approaches and identified a hypomorphic allele that may confer increased susceptibility to renal lesion development in FVB/N mice. This gene encodes for microphthalmia-associated transcription factor A (MITF-A), a bHLH-Zip transcription factor. Furthermore, we dissected a novel transcriptional network critically involved in kidney disease progression and provide evidence that TGF-α is a crucial transcriptional target of MITF-A during lesion development.
RESULTS
Mitfa, a candidate modifier gene
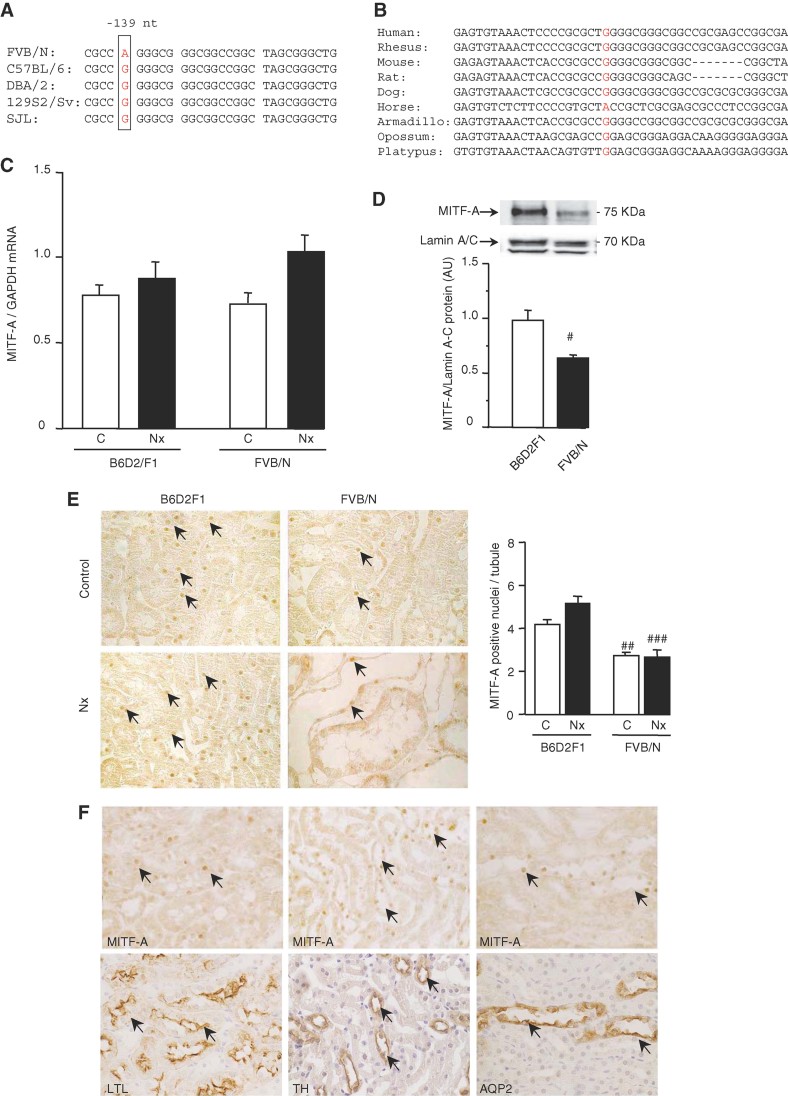
In order to prioritize the analysis of possible candidates, i.e. genes that control the expression of TGF-α, we used an in silico approach (http://www.geneontology.org) and identified the genes within the Ckdp1 confidence interval that encode for transcription factors (Table S1 of Supporting Information). Twenty-three transcription factors were identified. To decrease the number of these candidates, we tried to reduce the interval by performing a haplotype analysis (http://mouse.cs.ucla.edu/perlegen/). The results showed that the Ckdp1 confidence interval is heavily fragmented (117 fragments) in its ancestral origin. Twenty-five Mb have been found identical between the sensitive and at least one of the two resistant strains over a total of 37 Mb. Only 11 Mb were ancestrally different between the FVB/N and the resistant strains. Notably, 8 of the 23 candidate genes encoding for transcription factors were in haplotype regions that were different between the sensitive and resistant strains. Analysis of putative DNA regulatory sequences of Tgfa revealed that among these eight candidates only MITF and BHLHB2 might potentially bind the Tgfa promoter. Both these transcription factors are expressed in kidney (http://symatlas.gnf.org). Intriguingly, it has been shown that Mitf-deficient mice (Steingrimsson et al, 1994) develop microphtalmia, a trait that is also observed in transgenic mice that overexpress TGF-α in the lens (Reneker et al, 1995). This peculiar coincidence suggested a possible functional link between these two genes. Hence, we first sequenced all the promoters and complementary DNA (cDNA) encoding for Mitf, since this gene is expressed as a series of isoforms differing in their first exons and promoters. We found a single nucleotide polymorphism (G/A) located 139 nt upstream of the translation initiation site of the MITF-A isoform, in a sequence particularly well conserved throughout evolution. We observed that the FVB/N strain carries the ‘A’ allele, whereas none of the resistant strains have this variant (Fig 1A). More importantly, we observed that the G/A genotype was the marker with the highest concordance between the observed and the expected phenotype in the G2 progeny. By consequence, this variant corresponded to the lowest p-value of the Ckdp1 locus (Table S2 of Supporting Information). In addition, amongst the nine mammals for which we could compare the sequence, all carried a ‘G’, except the horse that carried an ‘A’ (Fig 1B).
Figure 1. The FVB/N variant selectively decreases MITF-A protein expression.
- Analysis of the mouse MITF-A 5′ UTR sequences revealed a specific G/A variant at −139 bp to the ATG in the sensitive FVB/N strain.
- Multi-alignment analysis of the 5′ MITF-A non-coding region from nine mammalians: the G allele is highly conserved among the species.
- MITF-A mRNA expression in kidneys of control (C) and 75% nephrectomized (Nx) B6D2F1 and FVB/N mice, evaluated by real-time quantitative RT-PCR, 2 months after surgery.
- MITF-A protein expression in kidney nuclear extracts of control FVB/N and B6D2F1 mice.
- Immunostaining (left panel) and quantification (right panel) of MITF-A in kidneys of control (C) and 75% nephrectomized (Nx) B6D2F1 and FVB/N mice, 2 months after surgery. X600.
- Colocalization experiments; serial sections stained for MITF-A (upper panels) and lotus tetragonolobus lectin (LTL; left-low panel), a marker of proximal tubules, Tamm–Horsfall (TH, middle-low panel), a marker of ascending limbs of loops of Henle and aquaporin 2 (AQP2, right-low panel), a marker of collecting ducts. X600. Data are means ± SEM; n = 4–6. Statistical analysis: Fig 1D: Student's t-test (n = 6): FVB/N versus B6D2F1: #p < 0.05. Fig 1C and 1E: ANOVA, followed by Tukey–Kramer test (n = 6): FVB/N versus B6D2F1: ##p < 0.01, ###p < 0.001.
Few allele-specific sequence variants were identified in Bhlhb2 gene among the three parental FVB/N, C57 black 6 (C57BL/6) and dilute brown non-agouti/2 (DBA/2) and the referent 129/Sv strains (http://www.informatics.jax.org), but all were silent polymorphisms.
The A variant impairs MITF-A protein translation in FVB/N mice
The high conservation of the G allele suggested that this non-coding sequence might have a functional role in controlling MITF-A expression. Hence, we analyzed MITF-A expression in our experimental model of nephron reduction. Surprisingly, we observed that whereas MITF-A messenger RNA (mRNA) expression was identical in kidneys of FVB/N and cross between C57BL/6 (B6) female and DBA/2 (D2) male (B6D2F1) mice (Fig 1C), MITF-A protein was markedly reduced in FVB/N mice (Fig 1D). Immunohistochemistry confirmed that the proportion of MITF-A-positive nuclei was significantly lower in kidneys of control sham-operated FVB/N mice as compared to B6D2F1 animals, and this difference did not change after nephron reduction (Fig 1E). Colocalization experiments demonstrated that MITF-A is expressed along all the tubular segments of the nephron (Fig 1F), but not in glomeruli (unpublished observation).
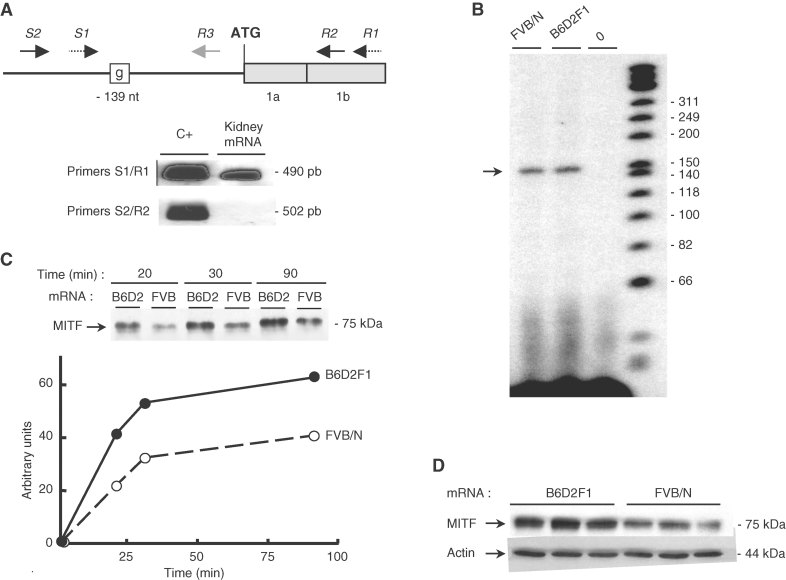
In view of the different patterns of mRNA and protein expression, we wondered if the G/A variant was located in the promoter or in the 5′ UTR sequence. Using two primers designed in the region to generate cDNA from total kidney mRNA, we first determined the extent of the 5′ UTR of Mitfa (Fig 2A). A primer extension assay revealed a major transcription start site located approximately at 160 bp upstream from the ATG codon of the mRNA (Fig 2B), demonstrating that the −139A/G variant lies in the Mitfa 5′ UTR. Since the 5′ UTR sequence is presumed to have an influence on the efficiency of protein translation (Pickering and Willis, 2005), we next investigated whether the G/A variant affected the rate of MITF-A protein synthesis. Using an in vitro-translation approach, we showed that MITF-A protein levels were reduced by about 50% when FVB/N MITF-A mRNA was used as the template (Fig 2C). Similarly, MITF-A protein expression was diminished (around 50%) when renal cortical thick ascending limb cell (CTAL) cells were transfected with a plasmid encoding the FVB/N MITF-A variant (Fig 2D), whereas MITF-A mRNA levels were identical regardless of the expression vector (unpublished observation). Collectively, these results showed that the G/A 5′ UTR variant decreases MITF-A mRNA translational efficiency in the FVB/N strain.
Figure 2. The FVB/N mutation lies in the 5′ UTR of the Mitfa gene and decreases MITF-A translation.
- Upper panel: Organization of the region encompassing 250 bp upstream of the ATG and the first two exons (1a, 1b) of Mitfa mouse gene. Primers used for either RT-PCR (S1/R1, S2/R2) or primer extension (R3) are indicated by arrows. The FVB/N-139 variant is indicated by a box. Lower panel: RT-PCR of mouse kidney mRNA using the primer sets differentially located as respect to the ATG. A vector encoding MITF-A served as positive control (C+).
- Mapping of Mitfa transcription initiation site by primer extension using kidney RNA from FVB/N or B6D2F1 mice. A sample lacking RNA was used as negative control (lane 0).
- In vitro MITF-A translation using in vitro-transcribed mRNA from B6D2F1 and FVB/N cDNA and nuclease-treated rabbit reticulocyte lysates. Protein expression was analyzed 20, 30 and 90 min after incubation.
- Western blot analysis of MITF expression in CTAL cells transfected with the B6D2F1 or the FVB/N MITF-A expression vector. Data are means ± SEM; n = 4–6.
MITF-A binds the Tgfa promoter and inhibits its expression
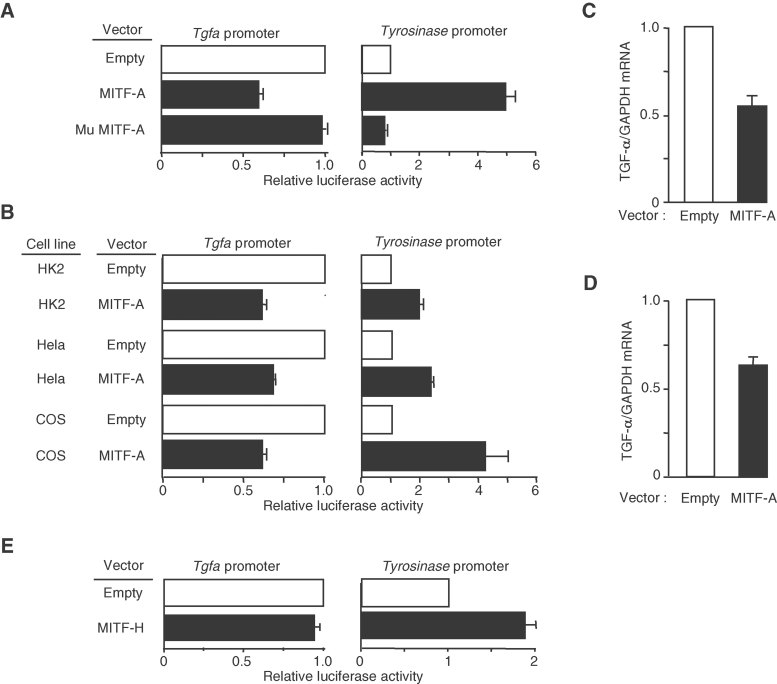
We next investigated if MITF-A modulates mouse Tgfa promoter activity, by cotransfecting renal CTAL cells with a TGF-α-luciferase reporter and a vector expressing MITF-A. The results showed that MITF-A significantly repressed Tgfa promoter activity, whereas it stimulated the tyrosinase-promoter, a well-characterized MITF-A target (Fig 3A). Deletion of the bHLH-Zip domain of MITF-A completely abolished the effect of MITF on both tyrosinase and Tgfa promoter activities (Fig 3A). MITF-A also inhibited Tgfa promoter activity in human Hela and human kidney-2 (HK2) cells as well as in monkey cercopithecusaethiops origin defective SV40 (COS) cells, indicating that the inhibitory effect of MITF-A is independent of species or cell line (Fig 3B). Notably, MITF-A inhibited the transcription of the endogenous Tgfa gene in both MITF-A transfected CTAL cells (Fig 3C) and human renal cells in primary culture (Fig 3D). Moreover, we observed that the inhibitory effect of MITF-A was specific to this isoform. In fact, the expression of MITF-H did not impair TGF-α-luciferase activity in CTAL cells (Fig 3E), indicating distinct functions for each isoform.
Figure 3. MITF-A inhibits TGF-α expression in different cell lines.
- Luciferase assay of CTAL cells transfected with the empty, MITF-A or the mutated MITF-A (Mu MITF-A) vector and either TGF-α (left panel) or tyrosinase (right panel) luciferase reporter vector.
- Luciferase assay of HK2, Hela and COS cells transfected with the empty or MITF-A expression vector and either TGF-α (left panel) or tyrosinase (right panel) luciferase reporter vector.
- TGF-α mRNA expression in CTAL cells transfected with the empty or MITF-A vector.
- MITF-A inhibits TGF-α expression also in primary human renal tubular cells transfected with either the empty or MITF-A expression vector.
- Luciferase assay of CTAL cells transfected with the empty or MITF-H expression vector and either TGF-α (left panel) or tyrosinase (right panel) luciferase reporter vector indicates that the inhibitory effect of MITF-A is isoform-specific. Data are means ± SEM; n = 4–6.
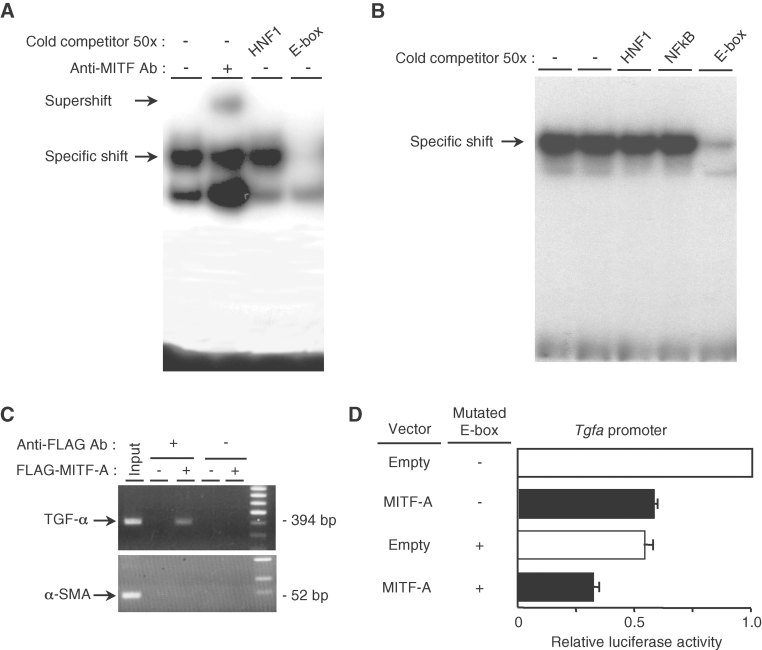
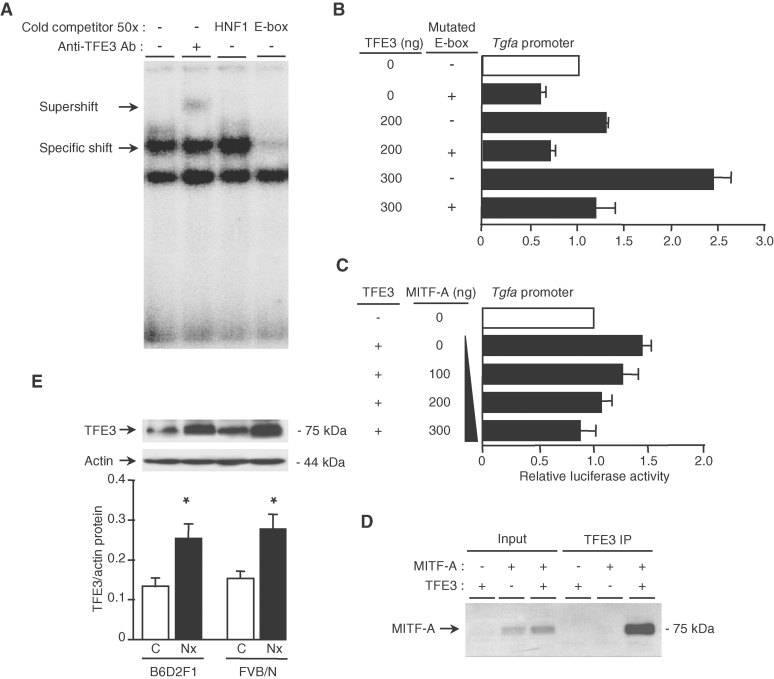
Sequencing of the Tgfa promoter revealed several potential E-box motifs, among which one in particular (nt −2360 −2330) was a perfect match to the classical consensus binding site for MITF (CACGTG). Electrophoretic mobility shift assay with nuclear extracts from either MITF-A transfected cells (Fig 4A) or mouse kidneys (Fig 4B) demonstrated that MITF-A specifically bound this region of the Tgfa promoter. More importantly, chromatin immunoprecipitation experiments confirmed the binding of MITF-A to the endogenous Tgfa promoter (Fig 4C). To prove the functional significance of this E-box, we tested the ability of MITF-A to transactivate a mutated TGF-α-luciferase reporter containing four substitutions within the E-box sequence (gAatTc). Surprisingly, in CTAL cells transfected with only the mutated reporter vector, the Tgfa promoter activity was markedly reduced and the decrease was comparable to that induced by MITF-A (Fig 4D). This suggested that the deleted element had a positive function on the transactivation of this promoter in basal conditions. A modest further decrease was observed when the mutated reporter was cotransfected with MITF-A. Together, these data suggest that MITF-A might act by antagonizing the stimulatory effect of another transcription factor that binds to the same E-box.
Figure 4. MITF-A binds the Tgfa promoter.
- Electrophoretic mobility shift assay (EMSA) using a specific mouse 32P-labelled TGF-α E probe and nuclear protein extracts from MITF-A transfected CTAL cells. Complexes were surpershifted with an anti-MITF antibody.
- EMSA using a specific mouse 32P-labelled TGF-α E probe and nuclear extracts from mouse kidneys.
- Chromatin immunoprecipitation assay in cells transfected with the empty or the FLAG-MITF-A expression vector.
- Luciferase assay of CTAL cells transfected with the wild type or the E box-mutated TGF-α reporter vector together with the empty or MITF-A expression vector. Data are means ± SEM; n = 4–6.
TFE3 activates TGF-α expression and increases after nephron reduction
Microphthalmia-associated transcription factor regulates gene expression by binding DNA as either a homodimer or a heterodimer with other related family members, including transcription factor E3 (TFE3), transcription factor EB (TFEB) and transcription factor EC (TFEC) (Steingrimsson et al, 2004). In particular, TFE3 binds to the same CACGTG consensus sequence and transactivates several MITF target genes. Our results demonstrated that TFE3 specifically bound the E-box of the Tgfa promoter (Fig 5A) and stimulated the Tgfa promoter activity in a dose-dependent manner (Fig 5B). This effect was suppressed when the E-box was mutated (Fig 5B). More importantly, the stimulatory effect of TFE3 was abolished by increasing amounts of MITF-A (Fig 5C), suggesting a functional interaction of these factors on the Tgfa promoter. In agreement with this idea, we observed that TFE3 co-immunoprecipitated with MITF-A (Fig 5D).
Figure 5. TFE3 activates TGF-α, interacts with MITF-A and increases after nephron reduction.
- EMSA using a specific mouse 32P-labelled TGF-α E probe and nuclear extracts from TFE3 transfected CTAL cells. Complexes were surpershifted with an anti-TFE3 antibody.
- Luciferase assay of CTAL cells transfected with the wild-type or the E box-mutated TGF-α reporter vector together with the empty or increasing amount of TFE3 expression vector.
- Increasing amounts of MITF-A prevent TFE3-induced TGF-α luciferase activity in CTAL cells transfected with TGF-α reporter, MITF-A, and either the empty or TFE3 vector.
- MITF-A after immunoprecipitation with anti-FLAG antibodies (FLAG-TFE3 IP) in cells transfected with FLAG-TFE3, MITF-A or both MITF-A and FLAG-TFE3.
- TFE3 expression in control (C) and 75% nephrectomized (Nx) B6D2F1 and FVB/N mice, 2 months after surgery. Data are means ± SEM; n = 4–6. Statistical analysis: ANOVA, followed by Tukey–Kramer test (n = 6): Nx versus control: *p < 0.05.
Since these data suggested that MITF-A modulates TGF-α expression by antagonizing TFE3 transactivation, we next evaluated the expression of TFE3 in our experimental model of CKD. Western blot analysis revealed that TFE3 protein levels markedly increased in remnant kidneys of both FVB/N and B6D2F1 mice, 2 months after nephron reduction (Fig 5E). Hence, it seems that the ratio between MITF-A/TFE3 rather than expression level of each partner is the critical parameter in regulating Tgfa gene expression.
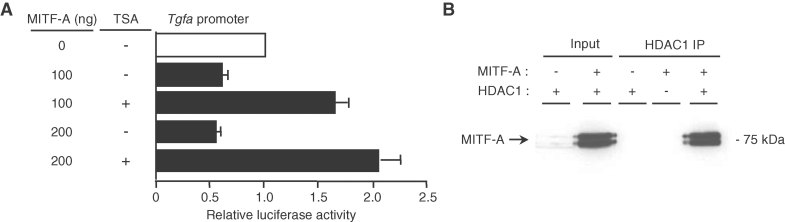
HDAC inhibition prevents MITF-A-mediated repression of Tgfa promoter
It is known that transcriptional factors can recruit histone deacetylases (HDACs) to repress transcription (Yang and Seto, 2007). To explore the role of these corepressors in the MITF-A-induced Tgfa transcriptional inhibition, we studied the effect of Trichostatin A (TSA), an HDAC inhibitor. Our results showed that TSA treatment of MITF-A cotransfected cells led to a marked increase of TGF-α promoter-luciferase activity (Fig 6A), suggesting a specific recruitment of HDAC by MITF-A on the Tgfa promoter. Interestingly, co-immunoprecipitation experiments confirmed that MITF-A physically interacts with HDAC1 in renal CTAL cells (Fig 6B).
Figure 6. HDAC inhibition prevents MITF-A-induced TGF-α repression.
- CTAL cells transfected with TGF-α luciferase reporter and increasing amount of MITF-A expression vector were treated or not by Trichostatin A (TSA).
- MITF-A immunoblot after immunoprecipitation with anti-FLAG antibodies (FLAG-HDAC1 IP) in cells transfected with FLAG-HDAC1, MITF-A or both MITF-A and FLAG-HDAC1. Data are means ± SEM; n = 6.
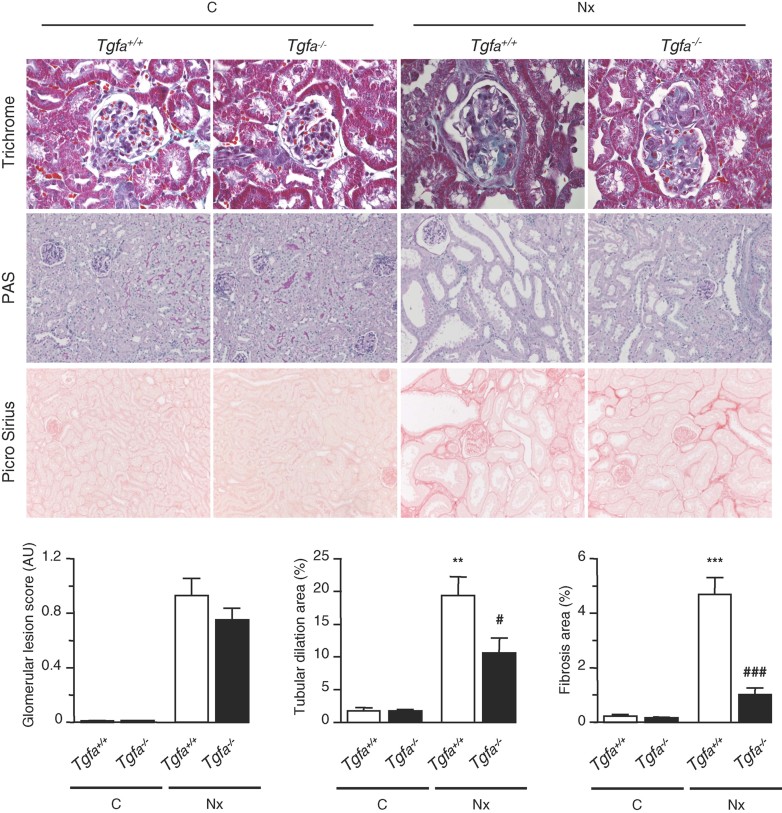
Tgfa gene inactivation prevents lesion development in FVB/N mice
To investigate the importance of this novel molecular network in vivo, and in particular if TGF-α might be the effector, we performed 75% nephron reduction in Tgfa−/− mice (Luetteke et al, 1993). To this end, we first introduced the Tgfa mutated allele in the lesion-prone (FVB/N) background. The Tgfa−/− FVB/N mice reproduced normally and had no apparent phenotype under physiological conditions, with the exception of waved fur and whiskers. This phenotype has been already reported in the mixed 129svXC57BL/6J genetic background (Luetteke et al, 1993). As expected, 2 months after nephron reduction, wild-type FVB/N mice developed severe renal lesions, mainly including glomerulosclerosis, tubular atrophy and cystic dilation and mild interstitial fibrosis (Fig 7). However, the frequency and severity of renal lesions were dramatically reduced in Tgfa−/− mice, despite that these mice have the MitfaFVB hypomorphic allele. Altogether, these results demonstrate that TGF-α is a crucial target of MITF-A in the genetic predisposition to renal deterioration.
Figure 7. Tgfa deficiency prevents tubulo-interstitial lesions in FVB/N mice.
Morphology and lesion scores of kidneys from control (C) and 75% nephrectomized (Nx) Tgfa+/+ and Tgfa−/− mice, 2 months after surgery. Magnification: X600 and X200 for glomerular and tubular/interstitial lesions, respectively. Data are means ± SEM; n = 6–10. Statistical analysis: ANOVA, followed by Tukey–Kramer test (n = 6 for C and n = 10 for Nx)): Nx versus control: **p < 0.01, ***p < 0.001; Tgfa−/− versus Tgfa+/+ #p < 0.05, ###p < 0.001.
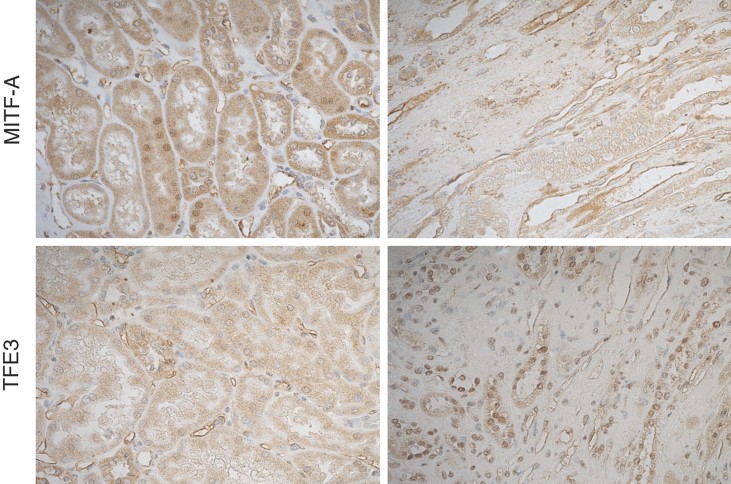
MITF-A and TFE3 expression in human CKD
Finally, to determine whether our experimental observations were relevant to human CKD progression, we analyzed MITF-A and TFE3 expression in a cohort of kidney transplant recipients. We chose this group because renal transplant patients are a human model of nephron number reduction and in our centre, they systematically undergo a surveillance renal transplant biopsy 1 year post-transplantation. We observed that MITF-A was scarcely detectable in most of the sections. However, when expressed, MITF-A was predominantly found in tubular nuclei of kidneys with normal morphology (Fig 8). Remarkably, we observed that the expression of TFE3, barely detectable in transplant biopsies with normal morphology, markedly increased in tubular nuclei of kidneys displaying severe tubulo-interstitial lesions (Fig 8). In addition, immunohistochemical analysis showed that TFE3 and MITF-A were expressed in the same nephron segments, i.e. tubules at the cortico-medullary junction.
Figure 8. MITF-A and TEF3 expression in kidney transplant recipients.
MITF-A (upper panels) and TFE3 (lower panels) staining in kidneys of two groups of patients, one with normal morphology (left panel) and the other one with severe tubulo-interstitial lesions (right panel). Of note, whereas MITF-A was detectable in nuclei of kidneys with normal morphology, TEF3 could be detected mainly in nuclei of kidneys with severe renal lesions. Pictures are representative samples of 12 patients for each group.
DISCUSSION
The genetic networks that control the progression of CKD are largely unknown. By applying genetic and molecular approaches to an experimental model of renal injury, we have identified Mitfa, a gene encoding for a bHLH transcription factor, as a candidate modifier of CKD progression. Sequencing revealed a strain-specific hypomorphic variant in the 5′ UTR of Mitfa that decreased the efficiency of MITF-A translation. Interestingly, MITF-A acts in a complex genetic network in which it inhibits transactivation of Tgfa by TFE3, by recruiting a HDAC. Consistent with the idea that TGF-α is a critical target of MITF-A, we showed that Tgfa gene inactivation prevented lesion development in mice after nephron reduction. These data are relevant to human CKD, as we found that TFE3/MITF-A ratio increased after nephron reduction as shown in renal transplant recipients. Collectively, these data identify a novel transcriptional network and suggest a crucial role for MITF-A/TFE3 balance in modulating the progression of CKD.
Microphthalmia-associated transcription factor is considered to play an important role in pigmented cell development and function (Opdecamp et al, 1997), though it was suggested that it might also play a role in other pathophysiological events, including cardiac growth and hypertrophy (Tshori et al, 2006), B cell homeostasis (Lin et al, 2004) and melanocyte cell proliferation and invasiveness (Carreira et al, 2006). Our results extend these observations by providing the first evidence that a Mitfa hypomorphic mutation might predispose to CKD progression. The hypomorphic mutation is located in the 5′ UTR and partially impaired protein synthesis. It should be pointed out that this sequence is highly conserved among species, indicating an evolutionary pressure. Interestingly, the interval containing the locus is orthologous to the rat chromosome 4 D4Rat95 marker region. This marker has shown significant linkage to QTL that affects the propensity to develop urinary albumin excretion in MWF rats (Schulz et al, 2003), a model of progressive CKD. More importantly, our Ckdp1 locus is syntenic to the human chromosome 3p14-12 and 3q21-22 regions, which have been linked to increased proteinuria in patients with type 1 diabetic nephropathy (Osterholm et al, 2007; Rogus et al, 1998). It is also worth noting that several studies have suggested evidence of linkage to chromosome 3 in type 2 diabetic nephropathy (Karnib et al, 2007). Although future studies are required to determine whether MITF variants may contribute to the complex genetic networks that modulate CKD progression in humans, our results and the concordance of the locus among the three species strongly support a role of MITFA among the potential candidates.
The FVB/N genetic background has been reported to be highly permissive to the onset of renal lesions in other experimental models of CKD, i.e. HIV-1 associated nephropathy (HIVAN) (Gharavi et al, 2004; Papeta et al, 2009; Prakash et al, 2011), diabetic nephropathy (Chua et al, 2010) or congenital cortico-resistant nephrotic syndrome (Ratelade et al, 2008). Mapping studies have identified significant linkage to several loci (Chua et al, 2010; Gharavi et al, 2004; Papeta et al, 2009; Ratelade et al, 2008), including one on chromosome 6 for HIVAN (Prakash et al, 2011). However, all these susceptibility loci map to distinct chromosomal regions, suggesting that different molecular pathways might modulate different forms of CKD. On the other hand, since each locus has been linked to a specific renal phenotype, i.e. the onset of proteinuria, the severity of glomerulosclerosis, the impairment of renal function or the global kidney damage score, we cannot rule out the possibility that different genetic networks trigger these different events. Altogether, these findings highlight the complex genetics of renal disease progression.
Microphthalmia-associated transcription factor A (MITF-A) belongs to a family of transcription factors containing nine distinct isoforms (Hershey and Fisher, 2005) among which five are expressed in kidney (data not shown). These isoforms, that arise from different promoters and differential splicing of nine distinct first exons to a common invariant set of eight downstream exons, differ in their amino termini, but share identical functional domains (Steingrimsson et al, 2004). The fact that MITF-H does not transactivate TGF-α suggests, however, distinct and essential functions for each isoform. In support of this, microarray analyses have revealed that MITF isoforms regulate distinct transcriptomes (Shahlaee et al, 2007). Moreover, Mitf mutant mice display distinct phenotypes consistent with the mutated isoform (Steingrimsson et al, 2004). Notably, cotransfection experiments have shown that the transactivation potential of different MITF isoforms may depend on either their amino-terminal domain (Saito et al, 2003) or the nature of their target genes (Takemoto et al, 2002; Tshori et al, 2007). The observation that target genes are selectively impaired by dominant negative Mitf alleles, and not with the Mitf null allele (Steingrimsson et al, 2004), indicates that the unique MITF amino termini may mediate specific protein-protein interactions and/or alter DNA binding specificities. Hence, as MITF-H has been shown to play a selective role in heart hypertrophy (Tshori et al, 2006), MITF-A seems to be critical in renal pathophysiology.
Microphthalmia-associated transcription factor may control the expression of target genes by interacting with other related family members, namely TFE3, TFEB and TFEC (Hershey and Fisher, 2004). Here, we show that MITF-A represses the transcription of TGF-α by counteracting the transactivation mediated by TFE3. The marked increase of TFE3 after nephron reduction in both mice and humans suggests that the ratio between MITF-A and TFE3 rather than the absolute level of MITF-A protein is the key element in modulating TGF-α expression during CKD progression. In further support to this connection, it deserves special note that genomic translocations leading to TFE3 overexpression have been implicated in renal cell carcinoma (Argani and Ladanyi, 2005), a tumour characterized by intense TGF-α expression. More importantly, a recent paper showed that a SUMOylation-defective germline mutation of MITF predisposes to both melanoma and renal carcinoma (Bertolotto et al, 2011).
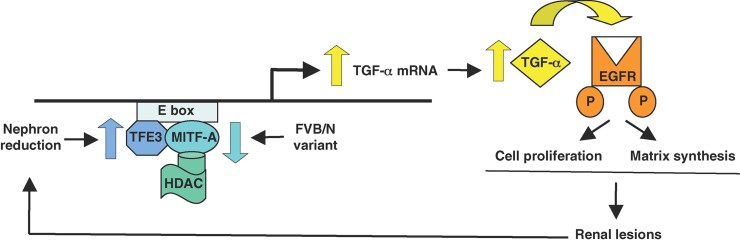
Microphthalmia-associated transcription factor/TFE interactions result mainly in activation of transcription; only a few studies have reported inhibition of target genes (Lin et al, 2004; Pham et al, 2011; Schwahn et al, 2005; Takemoto et al, 2002). It is now largely accepted that transcription factors act as a part of a large complex, recruiting distinct chromatin remodelling and histone tail modifying factors, i.e. histone acetyltransferases (HATs) and HDACs, to, respectively, activate or repress target gene transcription. Our observation that MITF-A interacts with HDAC1 and that TSA switched MITF-A from a repressor to an activator of the Tgfa promoter provides the first evidence that MITF-A recruits HDAC to suppress gene transcription in mammals. Consistently, a recent study showed that MITF-A represses Sox10 gene expression in zebrafish likely by interacting with HDAC1 (Greenhill et al, 2011). Thus, we propose a new transcriptional network in which MITF-A represses TFE3-induced Tgfa transcription by recruiting HDAC activity to the Tgfa promoter (Fig 9).
Figure 9. Model for mechanism of MITF-A action.
MITF-A interacts with HDAC to inhibit transactivation of its related partner TFE3 on the Tgfa promoter. In most mouse strains, high MITF-A levels are sufficient to antagonize the increased expression of TFE3 after nephron reduction. In contrast, in FVB/N nephrectomized mice, the decreased synthesis of MITF-A cannot oppose TFE3 overexpression leading to MITF-A/TFE3 imbalance and TGF-α overexpression. Subsequent EGFR activation leads to lesion development by promoting cell proliferation and matrix accumulation. A vicious circle is then initiated leading to progressive reduction of functional nephrons.
Our results together with recent works elucidate some important molecular mechanisms that modulate CKD progression. The observation that Tgfa gene inactivation in FVB/N mice harbouring the Mitfa hypomorphic allele protected them from lesion development points to TGF-α as the key transcriptional target of MITF-A during lesion development. Supporting this observation, we have recently demonstrated that the pharmacological inhibition of EGFR prevents renal deterioration in the lesion-prone FVB/N strain (Laouari et al, 2011). Previous studies highlighted the key role of EGFR activation in other models of CKD (Zeng et al, 2009). However, little is known about the molecular pathways that trigger EGFR activation during kidney diseases. EGFR is activated by a family of 11 growth factors (Singh and Harris, 2005). Among these, it has been shown that TGF-α is a critical player between angiotensin II (AngII) signalling and EGFR transactivation during AngII-induced nephropathy (Lautrette et al, 2005). Notably, TGF-α also seems to act in the genetic predisposition to CKD progression. In fact, whilst inactivation of Tgfa gene prevented lesion development in the lesion-prone FVB/N mice, inactivation of Jund, a partner of the AP1 transcription complex, promotes the renal deterioration process in resistant strains by stimulating the expression of TGF-α (Pillebout et al, 2003). Hence, it is tempting to speculate that genes encoding molecules involved in regulation of Tgfa gene expression might be candidate modifiers of CKD progression. It is worthy of note that in renal transplant patients, TFE3 expression was found increased in kidneys that displayed severe renal lesions 1 year after transplantation.
In conclusion, our study identifies a new transcriptional network critically involved in the susceptibility to develop renal lesions after nephron reduction. To our knowledge, this is the first demonstration that an hypomorphic variant of a transcription factor might influence CKD progression, by modulating, likely through chromatin modification, the expression of target genes, i.e. Tgfa (Fig 9). Consistent with a modifier role, the ratio among the components of the transcriptional network rather than the dosage of each molecule is critical for the predisposition to lesion development. The fact that the pharmacological inhibition of EGFR may prevent CKD progression in the lesion-prone FVB/N mice (Laouari et al, 2011) points to MITF-A, TFE3 and TGF-α as potential prognostic and therapeutic targets for treating the growing number of individuals with progressive CKD. We suspect that our findings will be critical in other pathological conditions that are also characterized by aberrant growth, such as cancers which demonstrate both intense TGF-α expression and MITF-A/TFE3 deregulation.
MATERIALS AND METHODS
Animals
Mice used for these studies were FVB/N and C57BL/6xDBA2/F1 (B6D2F1) (Charles River). The G2 cohort was generated as previously described (Laouari et al, 2011). Tgfa−/− mice (Luetteke et al, 1993) were bred onto the FVB/N genetic background for at least 10 generations. To separate the 129/Sv Tgfa allele from the FVB/N Mitf allele, additional microsatellite markers or single-nucleotide polymorphisms (SNP) susceptible to differentiate FVB/N from 129/Sv alleles were studied as described (Laouari et al, 2011). Animals were fed ad libitum and housed at constant ambient temperature in a 12 h light cycle. Animal procedures were approved by the Departmental Director of ‘Services Vétérinaires de la Préfecture de Police de Paris’ and by the ethical committee of Paris Descartes University.
All experiments were performed on 9-week-old mice. Mice were subjected to either 75% nephrectomy (Nx; n = 6–10) or sham-operation (control; n = 4–6), as previously described (Terzi et al, 2000a). After surgery, mice were fed a defined diet containing 30% casein and 0.5% sodium. At the time of sacrifice (2 months after surgery), the kidney was removed for morphological, protein and mRNA studies.
Cell lines
Cortical thick ascending limb cells derived from microdissected loops of Henle of the Tg(SV40E) Bri7 mouse were grown as described previously (Igarashi et al, 1996), with minor modifications (20% O2 in atmosphere). COS and Hela cells (ATCC) were grown in Dulbecco's modified Eagle medium (DMEM) with 10% foetal calf serum, whereas HK2 cells (ATCC) were grown in Roswell Park Memorial Institute (RPMI). Human renal epithelial cells in primary culture were cultured as indicated (Anglicheau et al, 2006). Cells were transfected with 400 ng/well of promoter–reporter construct and 100–300 ng/well of expression vectors using lipofectamine (Invitrogen), together with a CMV-βgal vector (100 ng/well) to evaluate transfection efficiency. Twenty-fours hours after transfection, cells were lysed and firefly luciferase and βgal activities were assayed. For HDAC inhibition, cells were treated with 160 nM TSA (Sigma–Aldrich) for 24 h. All transfection experiments were performed in duplicate and repeated at least three times.
Clinical samples
The study was conducted on renal biopsies from 24 renal transplant recipients being followed-up at the Renal Transplant Department of Necker Hospital. Surveillance biopsies are routinely performed 1 year after transplantation. The patients were divided into two groups: (i) patients with normal renal morphology (n = 12, mean age = 46 ± 12, male/female = 7/5); (ii) patients with severe tubulo-interstitial lesions (n = 12, mean age = 44 ± 14, male/female = 3/9).
This protocol was approved by the Institutional Review Board of Necker Hospital; informed written consent was obtained from each patient.
Vectors
Microphthalmia-associated transcription factor A and MITF-H expression vectors and the tyrosinase-luciferase reporter were provided by Shigeki Shibahara (Tohoku University, Sendai, Japan). FLAG-TFE3 and FLAG-HDAC1 expression vectors and CMV-βgal reporter vector were a gift from Clifford Takemoto (Dana-Farber Cancer Institute, Cambridge, MA, US), Edward Seto (H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, US) and Marc Lombes (Inserm U693, Faculté de Médecine Paris sud, Paris, France), respectively. The MITF-A vector was mutated by excision of exons 4–9 using the ClaI restriction sites (Mu MITF-A). To generate the TGF-α-luciferase reporter vector, the mouse Tgfa promoter (GeneBank accession number U64873) was amplified from genomic murine DNA by polymerase chain reaction (PCR) using the primers detailed in Table S3 of Supporting Information. A single 3330 bp fragment was obtained that was first ligated into pCRII (Invitrogen), then cloned into pGL3 (Promega) upstream of the luciferase reporter. The E-box element at −2326 nt to the ATG was mutated, from CACGTG into gAatTc, by site-directed mutagenesis using the Stratagene QuickChange system.
To isolate murine MITF-A cDNA with the 5′-UTR, two MITF-A fragments (GeneBank accession numbers NT039353 and AC021060) spanning nucleotides −60 to +1609 and nucleotides −216 to +254 were amplified by PCR. To generate the first fragment (1669 bp), cDNA from B6D2F1 or FVB/N kidneys was amplified using the primers detailed in Table S3 of Supporting Information. The second fragment (470 bp) was amplified using genomic DNA from B6D2F1 or FVB/N kidney (Table S3 of Supporting Information for primers). The two fragments were successively inserted in-frame into pCRII and the reconstituted MITF-A cDNA with the 5′ UTR was subcloned into pcDNA3 vector (Invitrogen) at the XbaI and HindIII restriction sites. To generate the FLAG-MITF-A expression vector, a FLAG sequence was inserted at the SmaI and HindIII restriction sites.
Gene sequencing and linkage analysis
All the promoters and cDNA encoding for Mitf isoforms were sequenced in the three parental FVB/N, DBA/2 and C57BL/6 strain. The G2 cohort (Laouari et al, 2011) was genotyped for the G/A polymorphism (rs31000275 marker) using a Biotage pyrosequencer PSQ96MA. Linkage was estimated by the Fischer's exact test for the fit for the expected renal phenotype (lesions vs. no-lesions), using Prism Software.
MITF-A 5′ UTR analysis
To define the 5′ UTR sequence of murine MITF-A, two forward and two reverse primers (Table S3 of Supporting Information) were used to generate cDNA from total kidney mRNA. Transcription initiation site of MITF-A was defined using Primer Extension System-AMV Reverse Transcriptase from Promega. A primer complementary to the 5′-UTR of the MITF-A cDNA (R3, Table S3 of Supporting Information) and mouse kidney RNA were used for reverse transcription.
In vitro transcription/translation
Mouse FVB/N or B6D2F1 MITF-A expression vectors were transcribed using T7 RNA polymerase. Purified RNA (4 µg) was translated using nuclease-treated rabbit reticulocyte lysates for 20, 30 and 90 min according to the supplier's recommendations (Promega). The translated products were analyzed by western blot.
Morphological analysis
Mouse kidneys were fixed in 4% paraformaldehyde, paraffin embedded, and sections were stained with periodic acid Schiff (PAS), Masson's trichrome, haematoxylin and eosin (H&E) and picro-sirius red. The degree of renal lesions was evaluated as previously described (Viau et al, 2010).
Human transplant biopsies were fixed in alcohol–formalin–acetic acid solution (AFA) and embedded in paraffin. Sections (4 µm) were stained with PAS, Masson's trichrome and H&E. The degree of renal lesions was evaluated according to the Banff's 07 classification (Racusen et al, 1999).
The paper explained
PROBLEM:
Chronic kidney disease is characterized by the progressive decline of renal function to end stage renal disease (ERSD) that can occur, irrespective of the cause of the renal damage (diabetes, hypertension, ischemia or immune diseases), once a critical number of nephrons has been lost. Despite the efforts extended by the health care community, the survival and quality of life of ESRD patients remain poor. Hence, understanding the pathophysiology of CKD progression is a key challenge for public health. On the other hand, since epidemiological studies have shown that the evolution of CKD varies considerably among patients, there is an urgent need to identify the genetic factors that predispose individual to faster progression.
RESULTS:
In the present study, we identified a strain-specific variant in the 5′ UTR of MITF-A, a bHLHZip transcription factor, that decreases the efficiency of MITF-A protein translation and accelerates the development of renal lesions in FVB/N mice. Cell culture studies demonstrated that MITF-A acts in a complex genetic network in which it inhibits transactivation of TGF-α, a ligand of EGFR, by TFE3, by recruiting a HDAC. Consistent with the key role of this network in lesion progression, inactivation of Tgfa gene protected FVB/N mice from renal deterioration after nephron reduction. These date are relevant to human CKD, as we found that TFE3/MITF-A ratio was increased in patients with damaged kidneys. Collectively, our data identify a novel transcriptional network critically involved in the development of renal lesions and reveal a critical role for MITF-A/TFE3 balance in CKD susceptibility.
IMPACT:
The observation that both the pharmacological inhibition of EGFR and, more specifically, the molecular inhibition of TGF-α prevent CKD progression in the lesion-prone FVB/N mice points to these molecules as promising therapeutic targets for treating the growing number of individuals with progressive CKD. The identification of a gene variant that predisposes mice to renal deterioration after nephron reduction provides a candidate modifier for the elucidation of the complex genetic network that modulate human CKD progression and the design of prognostic biomarkers.
Immunohistochemical analysis
For mouse samples, kidneys were fixed in 4% paraformaldehyde, paraffin embedded, and 4-µm sections were treated overnight at 4°C with a rabbit anti-MITF-A antibody 1/200, then incubated with a secondary biotinylated anti-rabbit antibody (vector) 1/500, followed by streptavidine-peroxidase (Dako) 1/500, for 30 min at room temperature. Peroxidase activity was revealed by 3-3′-diamino-benzidine-tetrahydrochloride (DAB, Dako). The number of MITF-A labelled tubular nuclei was determined in all the kidney section (X600) and corrected for the number of tubular sections.
For mouse colocalization experiments, 4-µm serial sections were incubated with either MITF-A or the specific nephron segment antibody. The following antibodies were used: (i) a goat anti-Tamm–Horsfall antibody (Biogenesis) 1/200, followed by a biotinylated goat antibody (Dako) 1/500 and streptavidin/horseradish peroxidase (Dako) 1/500, (ii) a rabbit anti-aquaporin 2 antibody (Sigma–Aldrich) 1/400, followed by a peroxidase-conjugated donkey anti-rabbit antibody (Amersham) 1/300. Lotus tetragonolobus lectin (LTL) was detected using a biotinylated LTL (vector) 1/50, followed by streptavidin/horseradish peroxidase 1/500. Peroxidase activity was revealed by DAB.
For human samples, 4-µm sections were first retrieved at 95°C for 40 min in 10 mM EDTA buffer, pH 8, then incubated overnight at 4°C with a rabbit polyclonal anti-TFE3 antibody (Abcam) 1/50 or a rabbit polyclonal anti-MITF-A antibody 1/50. For TEF3 immunostaining, the sections were then treated with a secondary biotinylated anti-rabbit antibody (vector) 1/200, followed by streptavidine-peroxidase (Dako) 1/5000, for 60 min at room temperature. For MITF-A immunostaining, the sections were then incubated with a secondary HRP anti-rabbit antibody (GE Healthcare) 1/200, for 1 h at room temperature. Peroxidase activity was revealed by DAB.
To produce antibodies directed specifically against the MITF-A isoform, the first exon of MITF-A spanning nucleotides −6 to +104 relative to ATG was amplified by PCR and inserted into the pGEX4-T1 vector (Amersham). The GST fusion protein was expressed in E. coli and purified on glutathione resin. Polyclonal antibodies were generated by immunizing rabbits (Covalab) and the anti-MITF-A IgG antibodies were affinity-purified using the fusion protein.
Western blot analysis
Immunoblotting on nuclear protein extracts from FVB/N and B6D2F1 kidneys were performed using a rabbit anti-MITF-A antibody 1/300. Immunoblotting of MITF-A translated products and MITF-A transfected cells were performed with 1 µg/ml mouse monoclonal C5 anti-MITF antibody (Chemicon). TFE3 immunoblotting of mouse kidneys was performed using 1 µg/ml anti-TFE3 monoclonal antibody (BD Pharmingen). Proteins from MITF-A and either FLAG-TFE3 or FLAG-HDAC1 cotransfected cells were immunoprecipitated with an anti-FLAG antibody (FLAG-agarose M2, Sigma). Eluted proteins were immunoblotted with a rabbit anti-MITF-A antibody. Mouse monoclonal anti-β-actin (Sigma–Aldrich) and rabbit monoclonal anti-laminin A/C (Epitomics) antibodies were used as controls.
Real-time RT-PCR
Microphthalmia-associated transcription factor A and TGF-α mRNA from kidneys and cell lysates were quantified by real-time polymerase chain reaction (after reverse transcription) (RT-PCR) using specific primers (Table S3 of Supporting Information).
DNA-binding assays
Electrophoretic mobility shift assays were performed on nuclear extracts from MITF-A or TFE3 transfected cells or from mouse kidneys with a 32P-labelled oligonucleotide TGF-α E-box probe (Table S3 of Supporting Information) as previously described with minor modifications (Terzi et al, 2000b). To assess the specificity of the binding the labelled probe was used in competition with 50-fold excess of cold probes containing either E box or two non-relevant boxes, hepatocyte nuclear factor 1 (HNF1) or nuclear factor-kappa B (NFκB). For supershift experiments, mouse monoclonal antibodies against MITF or TFE3 were added to the reaction mixture 1 h before the addition of the probe.
Chromatin immunoprecipitation assay
ChiP assays were performed using a Chromatin Immunoprecipitation assay kit (Upstate Biotechnology), with minor modifications. In brief, CTAL cells were transfected with either the FLAG-MITF-A or the empty vector, then treated with 1% formaldehyde. The reaction was stopped by 0.125 M glycine and cell lysates were sonicated, cleared by centrifugation and dialyzed to eliminate sodium dodecyl sulfate (SDS). The dialyzed samples were incubated with anti-FLAG-agarose M2 antibody (Sigma) or the protein A/protein G-agarose mixture at 4°C overnight. Immunoprecipitated complexes were eluted, heated at 65°C for 4 h in 200 mM NaCl, then treated with proteinase K. The DNA recovered was subjected to PCR amplification, using primers surrounding the E box on Tgfa promoter (Table S3 of Supporting Information). Smooth muscle alpha-actin 2 served as the control (Table S3 of Supporting Information for primers).
Data analysis and statistics
Data are expressed as means ± SEM. Differences between the experimental groups were evaluated using ANOVA, followed when significant (p < 0.05), by the Tukey–Kramer test. When only two groups were compared, the Student's t-test was used.
Acknowledgments
We thank Sophie Berissi and Clément Nguyen for technical assistance. We are grateful to Shigeki Shibahara, Clifford Takemoto, Edward Seto and Marc Lombes for MITF-A/MITF-H/Tyrosinase-luciferase, TFE3, HDAC1 and βgal expression vectors, respectively. We thank Mordi Muorah for English editing. This work was supported by INSERM, Université Paris Descartes, Assistance Publique - Hôpitaux de Paris, AURA Paris, Fondation de la Recherche Médicale, Agence Nationale de la Recherche, Fondation Bettencourt-Schueller (Prix Coup d'Elan), Roche Laboratories.
Supporting Information is available at EMBO Molecular Medicine online.
The authors declare that they have no conflict of interest.
Author contributions
DL performed most of the in vitro experiments (cell culture, vector designs, gene expression analysis and molecular studies), developed MITF-A antibody, and analyzed the data; MB performed most of the in vivo experiments, the genetic studies and the bioinformatics analysis; AP performed the immunohistochemical studies; FB performed the morphological studies in experimental models; LHN performed the morphological studies in humans; DCL analyzed the data; CL followed the patients in the clinical department; GF analyzed the data; MP supervised part of the molecular studies and wrote the paper; FT supervised the project, provided funding and wrote the paper.
Supplementaary material
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
References
- Anglicheau D, Pallet N, Rabant M, Marquet P, Cassinat B, Meria P, Beaune P, Legendre C, Thervet E. Role of P-glycoprotein in cyclosporine cytotoxicity in the cyclosporine-sirolimus interaction. Kidney Int. 2006;70:1019–1025. doi: 10.1038/sj.ki.5001649. [DOI] [PubMed] [Google Scholar]
- Argani P, Ladanyi M. Translocation carcinomas of the kidney. Clin Lab Med. 2005;25:363–378. doi: 10.1016/j.cll.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, d'Hayer B, Mohamdi H, Remenieras A, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- Boger CA, Heid IM. Chronic kidney disease: novel insights from genome-wide association studies. Kidney Blood Press Res. 2011;34:225–234. doi: 10.1159/000326901. [DOI] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua S, Jr, Li Y, Liu SM, Liu R, Chan KT, Martino J, Zheng Z, Susztak K, D'Agati VD, Gharavi AG. A susceptibility gene for kidney disease in an obese mouse model of type II diabetes maps to chromosome 8. Kidney Int. 2010;78:453–462. doi: 10.1038/ki.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couser WG, Remuzzi G, Mendis S, Tonelli M. The contribution of chronic kidney disease to the global burden of major noncommunicable diseases. Kidney Int. 2011;80:1258–1270. doi: 10.1038/ki.2011.368. [DOI] [PubMed] [Google Scholar]
- Esposito C, He CJ, Striker GE, Zalups RK, Striker LJ. Nature and severity of the glomerular response to nephron reduction is strain-dependent in mice. Am J Pathol. 1999;154:891–897. doi: 10.1016/S0002-9440(10)65336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharavi AG, Ahmad T, Wong RD, Hooshyar R, Vaughn J, Oller S, Frankel RZ, Bruggeman LA, D'Agati VD, Klotman PE, et al. Mapping a locus for susceptibility to HIV-1-associated nephropathy to mouse chromosome 3. Proc Natl Acad Sci USA. 2004;101:2488–2493. doi: 10.1073/pnas.0308649100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhill ER, Rocco A, Vibert L, Nikaido M, Kelsh RN. An iterative genetic and dynamical modelling approach identifies novel features of the gene regulatory network underlying melanocyte development. PLoS Genet. 2011;7:e1002265. doi: 10.1371/journal.pgen.1002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey CL, Fisher DE. Mitf and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function. Bone. 2004;34:689–696. doi: 10.1016/j.bone.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Hershey CL, Fisher DE. Genomic analysis of the microphthalmia locus and identification of the MITF-J/Mitf-J isoform. Gene. 2005;347:73–82. doi: 10.1016/j.gene.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Igarashi P, Whyte DA, Li K, Nagami GT. Cloning and kidney cell-specific activity of the promoter of the murine renal Na-K-C1 cotransporter gene. J Biol Chem. 1996;271:9666–9674. doi: 10.1074/jbc.271.16.9666. [DOI] [PubMed] [Google Scholar]
- Karnib HH, Chua S, Gharavi AG. Genes for diabetic nephropathy: sweet prospects on the horizon. Kidney Int. 2007;71:94–96. doi: 10.1038/sj.ki.5002022. [DOI] [PubMed] [Google Scholar]
- Keller BJ, Martini S, Sedor JR, Kretzler M. A systems view of genetics in chronic kidney disease. Kidney Int. 2012;81:14–21. doi: 10.1038/ki.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kren S, Hostetter TH. The course of the remnant kidney model in mice. Kidney Int. 1999;56:333–337. doi: 10.1046/j.1523-1755.1999.00527.x. [DOI] [PubMed] [Google Scholar]
- Laouari D, Burtin M, Phelep A, Martino C, Pillebout E, Montagutelli X, Friedlander G, Terzi F. TGF-alpha mediates genetic susceptibility to chronic kidney disease. J Am Soc Nephrol. 2011;22:327–335. doi: 10.1681/ASN.2010040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautrette A, Li S, Alili R, Sunnarborg SW, Burtin M, Lee DC, Friedlander G, Terzi F. Angiotensin II and EGF receptor cross-talk in chronic kidney diseases: a new therapeutic approach. Nat Med. 2005;11:867–874. doi: 10.1038/nm1275. [DOI] [PubMed] [Google Scholar]
- Lin L, Gerth AJ, Peng SL. Active inhibition of plasma cell development in resting B cells by microphthalmia-associated transcription factor. J Exp Med. 2004;200:115–122. doi: 10.1084/jem.20040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luetteke NC, Qiu TH, Peiffer RL, Oliver P, Smithies O, Lee DC. TGF alpha deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell. 1993;73:263–278. doi: 10.1016/0092-8674(93)90228-i. [DOI] [PubMed] [Google Scholar]
- Ma LJ, Fogo AB. Model of robust induction of glomerulosclerosis in mice: importance of genetic background. Kidney Int. 2003;64:350–355. doi: 10.1046/j.1523-1755.2003.00058.x. [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Nakayama A, Nguyen MT, Hodgkinson CA, Pavan WJ, Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development. 1997;124:2377–2386. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- Osterholm AM, He B, Pitkaniemi J, Albinsson L, Berg T, Sarti C, Tuomilehto J, Tryggvason K. Genome-wide scan for type 1 diabetic nephropathy in the Finnish population reveals suggestive linkage to a single locus on chromosome 3q. Kidney Int. 2007;71:140–145. doi: 10.1038/sj.ki.5001933. [DOI] [PubMed] [Google Scholar]
- Papeta N, Chan KT, Prakash S, Martino J, Kiryluk K, Ballard D, Bruggeman LA, Frankel R, Zheng Z, Klotman PE, et al. Susceptibility loci for murine HIV-associated nephropathy encode trans-regulators of podocyte gene expression. J Clin Invest. 2009;119:1178–1188. doi: 10.1172/JCI37131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham L, Kaiser B, Romsa A, Schwarz T, Gopalakrishnan R, Jensen ED, Mansky KC. HDAC3 and HDAC7 have opposite effects on osteoclast differentiation. J Biol Chem. 2011;286:12056–12065. doi: 10.1074/jbc.M110.216853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering BM, Willis AE. The implications of structured 5' untranslated regions on translation and disease. Semin Cell Dev Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Pillebout E, Burtin M, Yuan HT, Briand P, Woolf AS, Friedlander G, Terzi F. Proliferation and remodeling of the peritubular microcirculation after nephron reduction: association with the progression of renal lesions. Am J Pathol. 2001;159:547–560. doi: 10.1016/S0002-9440(10)61726-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillebout E, Weitzman JB, Burtin M, Martino C, Federici P, Yaniv M, Friedlander G, Terzi F. JunD protects against chronic kidney disease by regulating paracrine mitogens. J Clin Invest. 2003;112:843–852. doi: 10.1172/JCI17647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Papeta N, Sterken R, Zheng Z, Thomas RL, Wu Z, Sedor JR, D'Agati VD, Bruggeman LA, Gharavi AG. Identification of the nephropathy-susceptibility locus HIVAN4. J Am Soc Nephrol. 2011;22:1497–1504. doi: 10.1681/ASN.2011020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racusen LC, Solez K, Colvin RB, Bonsib SM, Castro MC, Cavallo T, Croker BP, Demetris AJ, Drachenberg CB, Fogo AB, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55:713–723. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- Ratelade J, Lavin TA, Muda AO, Morisset L, Mollet G, Boyer O, Chen DS, Henger A, Kretzler M, Hubner N, et al. Maternal environment interacts with modifier genes to influence progression of nephrotic syndrome. J Am Soc Nephrol. 2008;19:1491–1499. doi: 10.1681/ASN.2007111268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remuzzi G, Benigni A, Remuzzi A. Mechanisms of progression and regression of renal lesions of chronic nephropathies and diabetes. J Clin Invest. 2006;116:288–296. doi: 10.1172/JCI27699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneker LW, Silversides DW, Patel K, Overbeek PA. TGF alpha can act as a chemoattractant to perioptic mesenchymal cells in developing mouse eyes. Development. 1995;121:1669–1680. doi: 10.1242/dev.121.6.1669. [DOI] [PubMed] [Google Scholar]
- Rogus JJ, Moczulski D, Freire MB, Yang Y, Warram JH, Krolewski AS. Diabetic nephropathy is associated with AGT polymorphism T235: results of a family-based study. Hypertension. 1998;31:627–631. doi: 10.1161/01.hyp.31.2.627. [DOI] [PubMed] [Google Scholar]
- Saito H, Takeda K, Yasumoto K, Ohtani H, Watanabe K, Takahashi K, Fukuzaki A, Arai Y, Yamamoto H, Shibahara S. Germ cell-specific expression of microphthalmia-associated transcription factor mRNA in mouse testis. J Biochem (Tokyo) 2003;134:143–150. doi: 10.1093/jb/mvg122. [DOI] [PubMed] [Google Scholar]
- Satko SG, Sedor JR, Iyengar SK, Freedman BI. Familial clustering of chronic kidney disease. Semin Dial. 2007;20:229–236. doi: 10.1111/j.1525-139X.2007.00282.x. [DOI] [PubMed] [Google Scholar]
- Schelling JR, Zarif L, Sehgal A, Iyengar S, Sedor JR. Genetic susceptibility to end-stage renal disease. Curr Opin Nephrol Hypertens. 1999;8:465–472. doi: 10.1097/00041552-199907000-00011. [DOI] [PubMed] [Google Scholar]
- Schulz A, Standke D, Kovacevic L, Mostler M, Kossmehl P, Stoll M, Kreutz R. A major gene locus links early onset albuminuria with renal interstitial fibrosis in the MWF rat with polygenetic albuminuria. J Am Soc Nephrol. 2003;14:3081–3089. doi: 10.1097/01.asn.0000100126.62370.25. [DOI] [PubMed] [Google Scholar]
- Schwahn DJ, Timchenko NA, Shibahara S, Medrano EE. Dynamic regulation of the human dopachrome tautomerase promoter by MITF, ER-alpha and chromatin remodelers during proliferation and senescence of human melanocytes. Pigment Cell Res. 2005;18:203–213. doi: 10.1111/j.1600-0749.2005.00229.x. [DOI] [PubMed] [Google Scholar]
- Shahlaee AH, Brandal S, Lee YN, Jie C, Takemoto CM. Distinct and shared transcriptomes are regulated by microphthalmia-associated transcription factor isoforms in mast cells. J Immunol. 2007;178:378–388. doi: 10.4049/jimmunol.178.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AB, Harris RC. Autocrine, paracrine and juxtacrine signaling by EGFR ligands. Cell Signal. 2005;17:1183–1193. doi: 10.1016/j.cellsig.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D'Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Taal MW, Brenner BM. Predicting initiation and progression of chronic kidney disease: developing renal risk scores. Kidney Int. 2006;70:1694–1705. doi: 10.1038/sj.ki.5001794. [DOI] [PubMed] [Google Scholar]
- Takemoto CM, Yoon YJ, Fisher DE. The identification and functional characterization of a novel mast cell isoform of the microphthalmia-associated transcription factor. J Biol Chem. 2002;277:30244–30252. doi: 10.1074/jbc.M201441200. [DOI] [PubMed] [Google Scholar]
- Terzi F, Burtin M, Hekmati M, Federici P, Grimber G, Briand P, Friedlander G. Targeted expression of a dominant-negative EGF-R in the kidney reduces tubulo-interstitial lesions after renal injury. J Clin Invest. 2000a;106:225–234. doi: 10.1172/JCI8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzi F, Burtin M, Hekmati M, Jouanneau C, Beaufils H, Friedlander G. Sodium restriction decreases AP-1 activation after nephron reduction in the rat: role in the progression of renal lesions. Exp Nephrol. 2000b;8:104–114. doi: 10.1159/000020656. [DOI] [PubMed] [Google Scholar]
- Tshori S, Gilon D, Beeri R, Nechushtan H, Kaluzhny D, Pikarsky E, Razin E. Transcription factor MITF regulates cardiac growth and hypertrophy. J Clin Invest. 2006;116:2673–2681. doi: 10.1172/JCI27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tshori S, Sonnenblick A, Yannay-Cohen N, Kay G, Nechushtan H, Razin E. Microphthalmia transcription factor isoforms in mast cells and the heart. Mol Cell Biol. 2007;27:3911–3919. doi: 10.1128/MCB.01455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viau A, El Karoui K, Laouari D, Burtin M, Nguyen C, Mori K, Pillebout E, Berger T, Mak TW, Knebelmann B, et al. Lipocalin 2 is essential for chronic kidney disease progression in mice and humans. J Clin Invest. 2010;120:4065–4076. doi: 10.1172/JCI42004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- Zeng F, Singh AB, Harris RC. The role of the EGF family of ligands and receptors in renal development, physiology and pathophysiology. Exp Cell Res. 2009;315:602–610. doi: 10.1016/j.yexcr.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.