Abstract
Advances in genome technology have facilitated a new understanding of the historical and genetic processes crucial to rapid phenotypic evolution under domestication1,2. To understand the process of dog diversification better, we conducted an extensive genome-wide survey of more than 48,000 single nucleotide polymorphisms in dogs and their wild progenitor, the grey wolf. Here we show that dog breeds share a higher proportion of multi-locus haplotypes unique to grey wolves from the Middle East, indicating that they are a dominant source of genetic diversity for dogs rather than wolves from east Asia, as suggested by mitochondrial DNA sequence data3. Furthermore, we find a surprising correspondence between genetic and phenotypic/functional breed groupings but there are exceptions that suggest phenotypic diversification depended in part on the repeated crossing of individuals with novel phenotypes. Our results show that Middle Eastern wolves were a critical source of genome diversity, although interbreeding with local wolf populations clearly occurred elsewhere in the early history of specific lineages. More recently, the evolution of modern dog breeds seems to have been an iterative process that drew on a limited genetic toolkit to create remarkable phenotypic diversity.
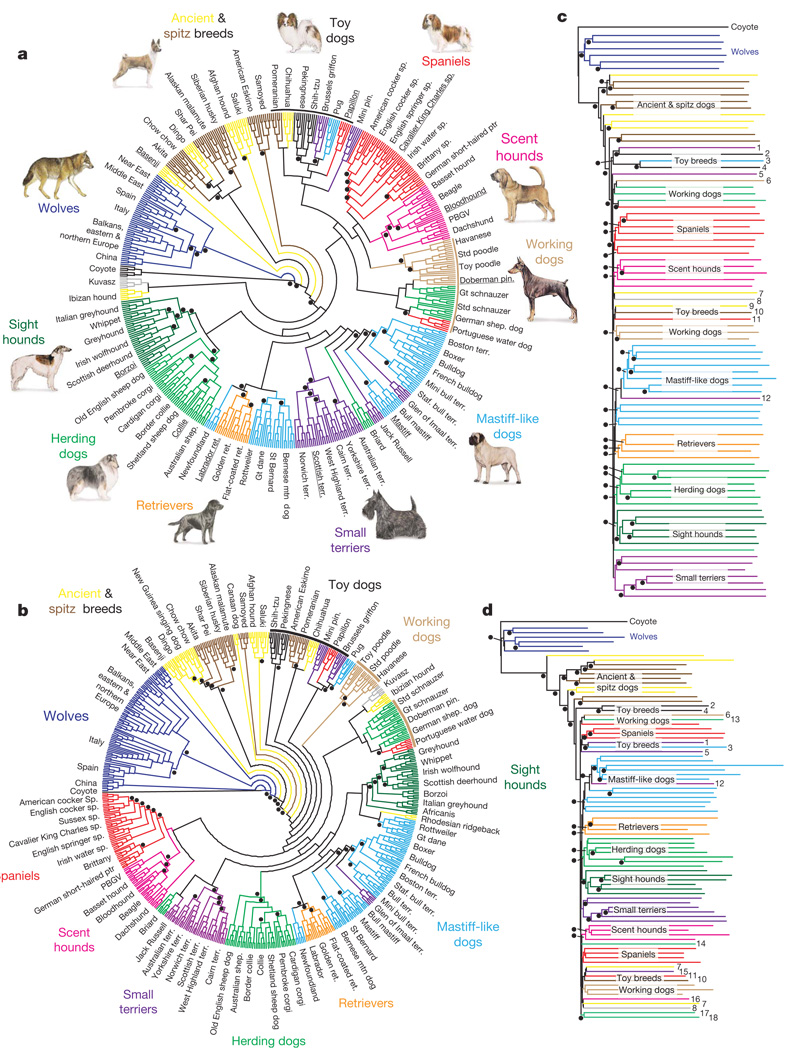
The dog is a striking example of variation under domestication, yet the evolutionary processes underlying the genesis of this diversity are poorly understood. To understand the geographic and evolutionary context for phenotypic diversification better, we analysed more than 48,000 single nucleotide polymorphisms (SNPs) typed in a panel of 912 dogs from 85 breeds as well as an extensive sample of 225 grey wolves (the ancestor of the domestic dog4,5) from 11 globally distributed populations (Supplementary Table 1). We constructed neighbour-joining trees using individuals and populations as units of analysis based on both individual SNP and haplotype similarity (Fig. 1 and Supplementary Note A). All trees identify dogs as a distinct cluster. Moreover, using as few as 20 diagnostic SNPs, all dog and wolf samples can be correctly assigned to species of origin with high confidence (see Supplementary Table 2, Supplementary Figs 1–4 and Supplementary Note B). Applying the Bayesian clustering method implemented in STRUCTURE6, we found strong evidence for admixture with wolves only in a minority of breeds (Supplementary Fig. 5). Neighbour-joining trees reveal that most of these breeds (basenji, Afghan hound, Samoyed, saluki, Canaan dog, New Guinea singing dog, dingo, chow chow, Chinese Shar Pei, Akita, Alaskan malamute, Siberian husky and American Eskimo dog) are highly divergent from other dog breeds (Fig. 1 and Supplementary Figs 6–11). These highly divergent breeds have been identified previously and termed ‘ancient’ breeds (as opposed to ‘modern’)4 because, consistent with their high levels of divergence, historical information suggests that most have ancient origins (>500 years ago)7–9. The limitation of evidence for admixture to only a few breeds is striking given that backcrossing between dogs and wolves is known to occur10 and dogs and wolves coexist widely. Given that modern breeds are the products of controlled breeding practices of the Victorian era (circa 1830–1900)4,7–9, the lack of detectable admixture with wolves is consistent with the strict breeding regimes recently implemented by humans.
Figure 1. Neighbour-joining trees of domestic dogs and grey wolves.
Branch colour indicates the phenotypic/functional designation used by dog breeders8,9. A dot indicates ≥95% bootstrap support from 1,000 replicates. a, Haplotype-sharing cladogram for 10-SNP windows (n = 6 for each breed and wolf population). b, Allele-sharing cladogram of individuals based on individual SNP loci. c, Haplotype-sharing phylogram based on 10-SNP windows of breeds and wolf populations. d, Allele-sharing phylogram of individual SNPs for breeds and wolf populations. For c and d, we note breeds where genetic assignments conflict with phenotypic/functional designations as follows: 1, Brussels griffon; 2, Pekingese; 3, pug; 4, Shih-tzu; 5, miniature pinscher; 6, Doberman pinscher; 7, Kuvasz; 8, Ibizian hound; 9, chihuahua; 10, Pomeranian; 11, papillon; 12, Glen of Imaal; 13, German shepherd; 14, Briard; 15, Jack Russell; 16, dachshund; 17, great schnauzer; and 18, standard schnauzer. Gt, great; mtn, mountain; PBGV, petit basset griffon vendeen; pin., pinscher; ptr, pointer; ret., retriever; shep., shepherd; sp., spaniel; Staf., Staffordshire; std, standard; terr., terrier. Canine images not drawn to scale. Wolf image adapted from ref. 31; dog images from the American Kennel Club (http://www.akc.org).
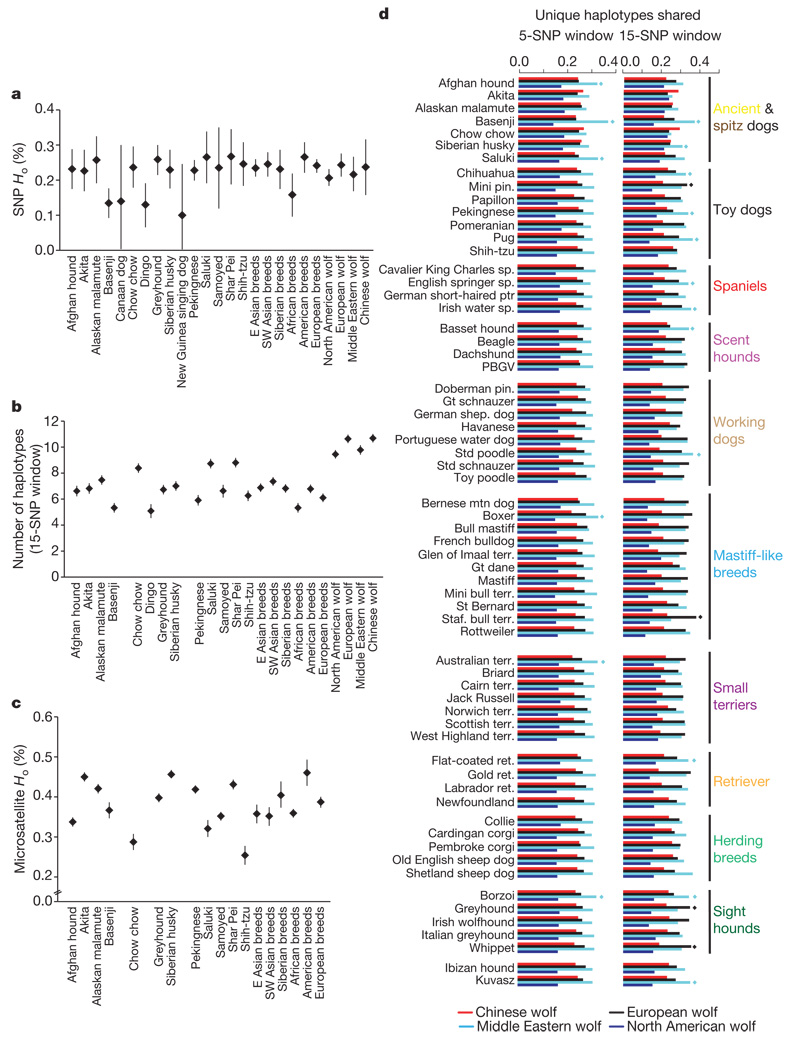
To identify the primary source of genetic diversity for domestic dogs, we used three approaches. First, we assessed whether a single wolf population clustered with dogs in neighbour-joining trees based on allele sharing of SNPs and sharing of 5- and 10-SNP haplotypes for individuals and breed/population groupings (Fig. 1, Supplementary Figs 6–11 and Supplementary Note A). Only in individual SNP and 5-SNP haplotype trees were specific populations of Middle and Near Eastern grey wolves found to be most similar to domestic dogs (Fig. 1b and Supplementary Fig. 10). In all other trees, wolves form a single genetic group and are not informative with regard to the wolf population that is most similar to dogs. We further tested the approach of a previous mitochondrial DNA (mtDNA) sequence study that suggested dogs have an origin in east Asia because diversity was highest in east Asian dog breeds3,11. We find that genetic diversity of dogs does not vary with geography in a consistent pattern. Specifically, breeds of east Asian origin do not have the highest level of nuclear variability, even when the SNP discovery scheme differed or haplotype measures of diversity were used to minimize ascertainment bias12 (Fig. 2a, b and Supplementary Figs 12a and 13). Furthermore, we confirmed an absence of geographic patterns in nuclear variation through a reanalysis of previously published microsatellite data4,7 (Fig. 2c and Supplementary Fig. 12b). For example, the two ancient breeds with highest SNP haplotype diversity, saluki and Chinese Shar Pei, originated in widely different areas (the Middle East and China, respectively8,9; Fig. 2b). However, ancient and island breeds are exceptions in consistently having lower diversity (basenji, Canaan dog, dingo, New Guinea singing dog; Fig. 2a–c). Thus, in contrast to previous mtDNA sequence results, current levels of autosomal diversity do not support an east Asian origin (or any other location). Indeed, if demographic history has varied substantially in dog breeds across geographic regions after domestication, current levels of genetic diversity may not directly reflect the oldest, ancestral population as it does in other species such as humans12–14. In addition, we note that recently, the use of genetic diversity to infer centres of domestication has been questioned by studies of semi-feral village dogs from Africa and Puerto Rico that found levels of mtDNA diversity as high or higher than those in east Asia11,15. High diversity in African dog populations reflects the added contribution of ancient indigenous dogs to the gene pool, which elsewhere is often dominated by modern breeds15.
Figure 2. Genome-wide analysis of SNP variation in domestic dogs and grey wolves.
a, Average observed heterozygosity (Ho). b, Average number of haplotypes per breed or group for phased SNP loci (15-SNP windows). c, Average observed heterozygosity of microsatellite data4,7. d, Fraction of unique haplotypes shared between 64 dog breeds and wolf populations for 5-(left) and 15- (right) SNP windows. Diamonds indicate significant sharing (P<0.05) using permutation test 2 (Supplementary Note A). Six (a–c) or nine (d) individuals represent each breed and wolf population. Error bars indicate s.e.m. E, east; SW, southwest.
Consequently, as a third approach to determine the primary centre of dog domestication, we considered haplotype sharing of modern and ancient dog breeds with specific wolf populations (see Supplementary Note A). Haplotype diversity patterns have been shown to be less sensitive to ascertainment biases12, and the sharing of SNPs that are otherwise private to specific wolf populations provides a unique signal to support ancestry or admixture. We analysed haplotype sharing between 64 well-sampled (n ≥ 9) dog breeds and wolf populations from Europe, the Middle East and China for 500-kilobase (kb) haplotype windows containing 5 and 15 SNPs drawn at random (Fig. 2d and Supplementary Table 3, Supplementary Note A). The Middle East and China have been implicated as centres for dog origination based on the archaeological record or mtDNA diversity3,11,16–18. We also assessed haplotype sharing between dog breeds and North American wolves as a negative control because dogs did not originate there19. Across all breeds, and for both window sizes, levels of sharing between dogs and North American wolves are substantially lower than the analogous comparison with Old World wolves, as expected (Fig. 2d). For 5-SNP haplotype windows, we found that haplotype sharing was uniformly higher between modern dog breeds and Middle Eastern wolves than between other wolf populations (Fig. 2d, left). For 15-SNP windows (Fig. 2d, right), the majority of breeds show the most sharing with Middle Eastern wolves, including some dog breeds of diverse geographic origins (for example, basenji, chihuahua, basset hound and borzoi). Notably, significant sharing with European wolves is found in miniature pinschers, Staffordshire bull terriers, greyhounds and whippets. The increased haplotype sharing between some European breeds and European wolves in the 15-SNP analysis may not be revealed in the 5-SNP windows because the European and Middle Eastern wolf haplotypes are less readily distinguished when based on fewer SNPs. Finally, only two east Asian breeds (Akita and chow chow) had higher sharing with Chinese wolves, although the results were not significant. In an analysis with fewer chromosomes per breed (n ≥ 6), four east Asian breeds—the Akita, Chinese Shar Pei, chow chow and dingo—showed most sharing with Chinese wolves (the latter two breeds showing significantly more sharing than expected), corroborating STRUCTURE clustering results (Supplementary Figs 5 and 14).
Notably, in both 5-SNP and 15-SNP window analyses, the basenji, a breed of Middle Eastern origin, had a greater proportion of shared haplotypes with Middle Eastern wolves than any other domestic dog (Fig. 2d and Supplementary Table 3). This result suggests that basenjis had a larger effective population size early in domestication or that they have more recently backcrossed with wolves. Overall, these data implicate the Middle East as a primary source of genetic variation in the dog, with potential secondary sources of variation from Europe and east Asia. In contrast to the mtDNA results, east Asian wolves are a predominant source of haplotype diversity for only a few east Asian dog breeds that have a long history in that region.
Neighbour-joining trees based on SNP data provide an explicit framework for investigating hypotheses of breed history and the genesis of phenotypic diversity. Consistent with previous microsatellite results4,7, topological analyses often define three well-supported groups of highly divergent, ancient breeds: an Asian group (dingo, New Guinea singing dog, chow chow, Akita and Chinese Shar Pei), a Middle Eastern group (Afghan hound and saluki) and a northern group (Alaskan malamute and Siberian husky) as being distinct from modern domestic dogs (Fig. 1a, b and Supplementary Figs 6–11). In addition, we find that the basenji often appears as the most divergent breed in allele- and haplotype-sharing trees (Fig. 1a, b and Supplementary Figs 6–11). This finding and high haplotype sharing, as well as a long recorded history8,9, suggest that this breed is one of the most ancient extant dog breeds.
The radiation of modern dog breeds has been difficult to resolve because most have originated recently and lack deep, detailed histories8,9. Consequently, the evolutionary process underlying the genesis of phenotypic/functional groupings is obscure. Specifically, many breeds have been documented as originating through crosses of genealogically or geographically distant stocks9 and thus, parallel evolution and genetic heterogeneity within phenotypic/functional breed groupings is expected. Nonetheless, we discern distinct genetic clusters within modern dogs that largely correspond to those based on phenotype or function, including spaniels, scent hounds, mastiff-like breeds, small terriers, retrievers, herding dogs and sight hounds (see Fig. 1). Most genetic groups have short internodes and often low bootstrap support, reflecting the rapid formation of modern breeds in the Victorian era8,9. Notably, toy and working dogs have a more varied relationship to genetic groupings, which is consistent with their known histories involving crosses between breeds from divergent genetic lineages (Supplementary Table 4). The heterogeneous composition of toy breeds may specifically indicate their frequent origin as a cross between a larger dog from a distinct breed grouping and a toy or dwarfed breed (Supplementary Table 4). Finally, within each breed, there is a remarkable concordance with known origin as all dogs are correctly assigned to the breed or population from which they were sampled, with one exception (bull terrier and miniature bull terrier; Fig. 1a, b). The contribution of these groupings to genetic variation was assessed by an analysis of molecular variance (AMOVA; Supplementary Table 5) which showed that 65% of the variation is due to variation within dog breeds, and 31% is due to variation within breed groups, similar to that reported for microsatellite data4,7. However, our analysis also showed that 3.8% of the variation is between phenotypic/functional breed groups (P<0.001). Consequently, although most variation is within breeds, phenotypic/functional breed groups represent a relatively small but significant component of variation.
The process of domestication involves strong selection of specific phenotypes; therefore, a signal of this selection should be evident in the genome20. Given the genome-wide coverage of our panel of SNPs, we searched for genomic regions that might contain adaptive substitutions due to positive selection during the initial phase of dog domestication (rather than breed formation, see Supplementary Note C). For each SNP, we calculated the fixation index (FST) and cross-population extended haplotype homozygosity (XP-EHH) values between non-admixed wolves and modern dogs and considered SNPs with extreme values as candidates for recent positive selection20. These statistics measure population differentiation and relative levels of genetic diversity, both of which are robust indicators of positive selection for recently domesticated species21. We found that SNPs within the top 5% of FST values and SNPs within the highest 1% of XP-EHH values are each significantly enriched for SNPs in genic regions (P = 0.04 for FST, P = 0.02 for XP-EHH, one-sided exact conditional test, controlling for the ascertainment panel; Supplementary Fig. 15). This result is consistent with a history of adaptive divergence in genic regions. To identify specific regions that are candidates for recent adaptive evolution, we normalized FST and XP-EHH values within ascertainment categories, and targeted regions that have several SNPs with extreme values for both statistics (Supplementary Table 6, see Supplementary Note C for results). Notably, two of our top three signals are near genes that have been implicated in memory formation and/or behavioural sensitization in mouse or human studies (ryanodine receptor 3 (OMIM accession 180903; ref. 22), adenylate cyclase 8 (OMIM accession 103070; Supplementary Note C)). Furthermore, we observed a single SNP with a high FST value located near the WBSCR17 gene responsible for Williams–Beuren syndrome in humans (OMIM accession 194050; Supplementary Fig. 16), which is characterized by social traits such as exceptional gregariousness. These outlier SNPs provide specific candidate regions for fine-scale mapping of genes that are important in the early domestication of dogs.
Our results show domestic dogs have genetic structure on three fundamental levels resulting from distinct evolutionary processes. First, within dog breeds, nearly all dogs are assigned to a breed of origin. This result is supported by previous microsatellite research4,7 and reflects the limited number of founders, inbreeding and small effective population size characteristic of many breeds23,24. Second, breed groupings are evident at a finer scale than previously described, and mirror breed classification based on form and function. We propose that this result reflects the tendency of dog breeders to develop new breeds by crossing individuals within specific functional and phenotypic groups to enhance abilities such as retrieving and herding, or further develop specific morphological traits4. However, heterogeneity within toy breeds and other breed groupings suggests the importance of discrete phenotypic mutations in the evolution of phenotypic diversity in the dog. Recent genetic studies have established that variation in coat colour25 and texture26, body size2, relative leg length27 and body proportions (A.R.B. et al., manuscript submitted) in different dog breeds are due to variation in shared genes of large phenotypic effect. For example, at least 19 distinct dog breeds with foreshortened limbs all uniquely share the same retrotransposed version of Fgf4 that is strongly implicated as the genetic basis for this phenotype27. Once such discrete mutations are fixed in a breed they can readily be crossed into unrelated lineages and thus enhance the process of phenotypic diversification. This process has perhaps produced more phenotypic diversity in dogs than other domesticated species because they are selected for many functions of value to humans (for example, defence, herding, retrieving, hunting, speed and companionship) as well as for novelty, which culminated in the ‘fancy breeds’ of the Victorian era8,9. Last, we identify divergent lineages of dogs distinct from those breeds that radiated during the nineteenth century and that probably derive from ancient geographically indigenous breeds. This finding mirrors recent genetic discoveries in sheep28 and cattle29 and suggests that some canine lineages may have persisted from antiquity or have more recently admixed with wolves. The latter seems unlikely given that some of these breeds have known ancient histories, exist in areas where wolves are absent, and are phenotypically highly derived8,9. For example, the chow chow originated more than 2,000 years ago8,9. Similarly, the dingo and New Guinea singing dog were probably established over 4,000 years ago and exist in areas without wolves. However, given their close proximity to extensive wolf populations, divergent northern breeds such as the Alaskan malamute, Siberian husky and American Eskimo dog may be better candidates for recent admixture.
Our haplotype sharing analysis evaluates the contribution of specific wolf populations to the genome of dogs, and reveals significant Middle Eastern and, for certain breeds, European ancestry. This result is consistent with the archaeological record that identified the earliest dog remains in the Middle East (12,000 years ago)16, Belgium (31,000 years ago)17, and the Bryansk region in western Russia (15,000 years ago)17, as well as the finding of high mtDNA diversity in ancient Italian dogs18. However, some ancient east Asian breeds show affinity with Chinese wolves, which suggests that they were derived from Chinese wolves or admixed with them after domestication10. The domestic dog seems comparable to other domestic species in containing several sources of variation from wild relatives. This dynamic process enriched the dog genome through interbreeding with wolves early in the domestication process. Similarly, mutations that have occurred since domestication, such as the mutation responsible for black coat colour, have been transferred to grey wolves30. Our genome-wide SNP analysis provides a new evolutionary framework for understanding the rapid phenotypic diversification unique to the domestic dog.
METHODS SUMMARY
SNP genotyping
Genomic DNA was isolated from blood samples of domestic dogs (Canis familiaris, n = 912) and from tissue and blood samples of grey wolves (C. lupus, n = 225) and coyotes (C. latrans, n = 60; see Supplementary Methods and Supplementary Table 1). The samples were genotyped and quality control filters were applied (see A.R.B. et al., manuscript submitted) to obtain high-quality genotypes from 48,036 autosomal SNP loci.
Cluster analysis
To visualize genetic relationships suggested by our SNP data we used principal component analysis (PCA) (ndog_breed = 2) and STRUCTURE6 (ndog_breed = 1). For tree reconstruction, we analysed two data sets. First, for individual-based allele-sharing distance analyses, we used 574 individuals (ndogs = 490; nOld_World_wolves = 84). This data set consisted of 75 dog breeds where six individuals were genotyped from each breed and an additional five dog breeds where five or fewer individuals were genotyped. The second data set was created for the population-level and haplotype-sharing distance-based analyses and used a subset of 530 individuals to provide comparable sample sizes from 79 dog breeds (nper_breed = 6) or wolf populations from China (n = 6), Middle East (n = 7), central Asia (n = 6) and Europe (n = 31). Coyotes from the western United States (n = 6) were used for rooting.
SNP haplotype analysis
From phased genotypes, we divided the genome into 500-kb windows to identify haplotypes and estimated haplotype diversity. The level of haplotype sharing was assessed between a dog breed (nindividuals ≥ 9 per breed, nbreeds = 64) and each wolf population (China, Europe, Middle East and North America).
Detecting selection
Population differentiation and extended haplotype homozygosity test statistics were calculated between modern dog breeds and grey wolves. We identified outlier SNP loci based on normalized scores and ranking in the 95th and 99th percentile.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgements
Grants from NSF and NIH (R.K.W.; C.D.B. and J.N.), the Polish Ministry of Science and Higher Education (M.P. and W.J.), European Nature Heritage Fund EURONATUR (W.J.), National Basic Research Program of China (Y.-p.Z.), and Chinese Academy of Sciences (Y.-p.Z.) supported this research. J.N. was supported by the Searle Scholars Program. B.M.vH. was supported by a NIH Training Grant in Genomic Analysis and Interpretation. K.E.L. was supported by a NSF Graduate Research Fellowship. E.A.O., D.S.M., T.C.S., A.E. and H.G.P. are supported by the intramural program of the National Human Genome Research Institute. M.P. was supported by the Foundation for Polish Science. Wolf samples from central and eastern Europe and Turkey were collected as a result of a continuing project on genetic differentiation in Eurasian wolves. We thank the project participants (B. Jedrzejewska, V. E. Sidorovich, M. Shkvyrya, I. Dikiy, E. Tsingarskaya and S. Nowak) for their permission to use 72 samples for this study. We acknowledge R. Hefner and the Zoological collection at Tel Aviv University for Israeli wolf samples. We thank the American Kennel Club (AKC) for the dog images reproduced in Fig. 1. We also gratefully acknowledge the dog owners who generously provided samples, the AKC Canine Health Foundation, and Affymetrix Corporation. We thank B. Van Valkenburgh, K.-P. Koepfli, D. Stahler and D. Smith for reviewing the manuscript.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Samples were contributed by E.G., M.P., W.J., C.G., E.R., D.B., A.W., J.S., M.M., E.A.O. and R.K.W. The experiment was designed and carried out with the help of B.M.vH., J.P.P., H.G.P., P.Q., D.S.M., T.C.S., A.E., A.W., J.S., M.C., P.G.J., Z.Q., W.H., Z.-L.D., Y.-p.Z., C.D.B., E.A.O. and R.K.W. The genotyping program was written by A.R.B., A.A., A.R., K.B., A.B. and C.D.B. and further programming was completed by K.E.L., J.D.D., D.A.E., E.H. and J.N. The analyses were conducted by B.M.vH., J.P.P., K.E.L., E.H., H.G.P., J.D.D., A.R.B., D.A.E., A.A., A.R., J.C.K. and J.N. The manuscript was written by B.M.vH., K.E.L., C.D.B., E.A.O., J.N. and R.K.W.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutter N, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- 4.Parker H, et al. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 5.Vilà C, Maldonado J, Wayne R. Phylogenetic relationships, evolution, and genetic diversity of the domestic dog. J. Hered. 1999;90:71–77. doi: 10.1093/jhered/90.1.71. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker H, et al. Breed relationships facilitate fine-mapping studies: a 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds. Genome Res. 2007;17:1562–1571. doi: 10.1101/gr.6772807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilcox B, Walkowicz C. The Atlas of Dog Breeds of the World. 5th edn. TFH Publications; 1995. [Google Scholar]
- 9.American Kennel Club. The Complete Dog Book. 20th edn. Ballantine Books; 2006. [Google Scholar]
- 10.Vilà C, Seddon J, Ellegren H. Genes of domestic mammals augmented by backcrossing with wild ancestors. Trends Genet. 2005;21:214–218. doi: 10.1016/j.tig.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Pang J, et al. mtDNA data indicate a single origin for dogs south of Yangtze river, less then 16,300 years ago, from numerous wolves. Mol. Biol. Evol. 2009;26:2849–2864. doi: 10.1093/molbev/msp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad D, et al. A worldwide survey of haplotypes variation and linkage disequilibrium in the human genome. Nature Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- 13.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 14.Li J, et al. Worldwide human relationships inferred from genome-wide patterns of varition. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 15.Boyko A, et al. Complex population structure in African village dogs and its implication for inferring dog domestication history. Proc. Natl Acad. Sci. USA. 2009;106:13903–13908. doi: 10.1073/pnas.0902129106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dayan T. Early domesticated dogs of the Near East. J. Archaeol. Sci. 1999;21:633–640. [Google Scholar]
- 17.Germonpre M, et al. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes. J. Archaeol. Sci. 2009;36:473–490. [Google Scholar]
- 18.Verginelli F, et al. Mitochondrial DNA from prehistoric canids highlights relationships between dogs and South-East European wolves. Mol. Biol. Evol. 2005;22:2541–2551. doi: 10.1093/molbev/msi248. [DOI] [PubMed] [Google Scholar]
- 19.Leonard J, et al. Ancient DNA evidence for Old World origin of New World dogs. Science. 2002;298:1613–1616. doi: 10.1126/science.1076980. [DOI] [PubMed] [Google Scholar]
- 20.Sabeti P, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Innan H, Kim Y. Pattern of polymorphism after strong artificial selection in a domestication event. Proc. Natl Acad. Sci. USA. 2004;101:10667–10672. doi: 10.1073/pnas.0401720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balschun D, et al. Deletion of the ryanodine receptor type 3 (RyR3) impairs forms of synaptic plasticity and spatial learning. EMBO J. 1999;18:5264–5273. doi: 10.1093/emboj/18.19.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cruz F, Vilà C, Webster M. The legacy of domestication: accumulation of deleterious mutations in the dog genome. Mol. Biol. Evol. 2008;25:2331–2336. doi: 10.1093/molbev/msn177. [DOI] [PubMed] [Google Scholar]
- 24.Gray M, et al. Linkage disequilibrium and demographic history of wild and domestic canids. Genetics. 2009;181:1493–1505. doi: 10.1534/genetics.108.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Candille S, et al. A β-defensin mutation causes black coat color in domestic dogs. Science. 2007;318:1418–1423. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cadieu E, et al. Coat variation in the domestic dog is governed by variants in three genes. Science. 2009;326:150–153. doi: 10.1126/science.1177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker H, et al. An expressed Fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedrosa S, et al. Evidence of three maternal lineages in near eastern sheep supporting multiple domestication events. Proc. R. Soc. Lond. B. 2005;272:2211–2217. doi: 10.1098/rspb.2005.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elsik C, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324:528–532. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson T, et al. Molecular and evolutionary history of melanism in North American gray wolves. Science. 2009;323:1339–1343. doi: 10.1126/science.1165448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macdonald DW, Barrett P. Mammals of Britain and Europe. Collins; 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.