Abstract
HIV infection is initiated by binding of the viral glycoprotein gp120, to the cellular receptor CD4. Upon CD4 binding, gp120 undergoes conformational change, permitting binding to the chemokine receptor. Crystal structures of gp120 ternary complex reveal the CD4 bound conformation of gp120. We report here the application of Gaussian Network Model (GNM) to the crystal structures of gp120 bound to CD4 or CD4 mimic and 17b, to study the collective motions of the gp120 core and determine the communication propensities of the residue network. The GNM fluctuation profiles identify residues in the inner domain and outer domain that may facilitate conformational change or stability, respectively. Communication propensities delineate a residue network that is topologically suited for signal propagation from the Phe43 cavity throughout the gp120 outer domain. . These results provide a new context for interpreting gp120 core envelope structure-function relationships.
Keywords: HIV, gp120, Cd4 binding, Chemokine Receptor, Gaussian Network Model, Molecular Dynamics, Communication Propensities, Commute Times, Slow Mode, Signal Propagation
INTRODUCTION
The Human Immunodeficiency Virus (HIV) causes the development of acquired immunodeficiency syndrome (AIDS) by depletion of CD4+ lymphocytes of an infected individual 1,2. The infection is mediated by a series of attachment events initiated by the HIV viral coat glycoprotein, gp160 which is cleaved into its two components, gp120 and gp41 3. The HIV glycoproteins, gp120 and gp41 are assembled as a trimer 4,5. The infection process of HIV in human T-cell lymphocytes occurs via binding of gp120, to the host T-cell CD4 receptor 6,7 followed by gp120 restructuring 8,9. This conformational change exposes the binding site of the chemokine receptor, allowing binding of gp120 to either CCR5 or CXCR4. Chemokine binding is the second obligatory event to viral entry 10-13 and is followed by insertion of the gp41 fusion-peptide in the host cell membrane allowing fusion and viral entry 14-17.
Several X-ray structures of the gp120:CD4:17b-antibody complex elucidate the conformation of gp120 after binding CD4 and 17b 18,19, Figure 1. These X-ray structures of the CD4 bound gp120 form, reveal three key domains- the inner domain, outer domain and a bridging sheet- that fold to form a large binding cavity. Two key CD4 residues, Phe43 and Arg59 bind in the gp120 cavity 18 and to Asp 368 on an adjacent alpha-helix, respectively. However, when Phe43 and Arg59 are mutated to Ala, CD4 no longer binds gp120 20. The core gp120 protein from various strains has also been crystallized with several antibodies 21 revealing the structure of the previously undescribed third variable (V3) loop. In addition, several mini-protein CD4 mimetics have also been solved, bound to gp120 22 showing a biphenyl group bound deep in the CD4 cavity. A recent structure of gp120 containing the gp41 interacting region 23 when compared to the CD4-binding site antibody, F105-gp120 complex 24 with a disassociated bridging sheet, reveals the plasticity of the inner and bridging sheet domains. Furthermore, modification of CD4 Phe 43 with various derivatized bromo-acetamide conjugates indicates that the gp120 cavity has the flexibility to accommodate ligands much larger than the CD4 Phe43 25. While a crystal structure of the unliganded HIV gp120 does not exist, that of the unbound SIV gp120 has been solved 26. The structure of SIV gp120, which has 35% sequence identity with HIV gp120, indicates an invariant outer domain, with conformational changes occurring in both the bridging sheet and inner domain 26.
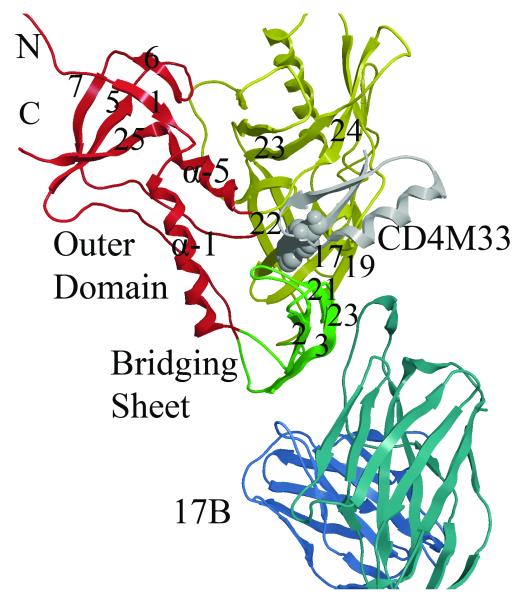
Figure 1. Structural details of gp120 core.
Ribbon diagram of the crystal structure (2I5Y) from the YU2 strain showing gp120 inner, outer and bridging sheet colored red, yellow and green, respectively. The mini-protein CD4 mimetic (CD4M33) containing a biphenyl group (purple) binds in the cavity formed at the junction of the three domains. The D1 domain of the 17b Fab is shown in blue. (Rendered with MOE70 )
Thermodynamic studies indicate that CD4 binding to gp120 results in a highly favorable binding enthalpy (ΔH = −63 kcal/mol) balanced with a highly unfavorable molecular ordering (−ΔTS = 52 kcal/mol) 27, 28. The large conformational change may be accounted for by the restructuring of up to 100 amino-acid residues or the burying of 10, 000 Å2 of surface area. Furthermore, titration calorimetry from two mutant gp120 envelope proteins (S375W and I432P) suggests two distinct conformational states, a CD4-bound state represented by the crystal structure of the CD4-gp120-17b complex and a S375W mutant and a non-CD4 bound state represented by an I423P mutation 29, 30. A third cavity stabilizing mutation T257S has been characterized and the double mutant T257S/S375W has increased CD4 affinity 28. The conformational change is thought to play a role in formation of the cytokine receptor binding interface, which spans portions of both gp120 and CD4. A series of compounds (denoted NBD) discovered by Zhao et al in database screening 31 have been shown to induce the CD4 stimulated conformational change in a manner similar to CD4 binding and enhance viral infection on CD4 deficient target cells 32. These compounds compete with CD4 binding to gp120 and enhance binding of CD4:gp120 to the chemokine receptor CCR5 32. Mutational analysis of gp120 cavity residues has shown that several mutations increased NBD compound affinity and enhanced viral infectivity on CD4 deficient cells 33. Furthermore, second generation compounds 33, 34 with improved affinity have shown to cause a rapid inactivation of the virus, establishing it as a potential entry inhibitor and antiviral therapeutic agent.
The dynamics of CD4 35 suggested that the gp120-binding loop has a higher mobility, aiding the CD4 molecule in binding to target molecules. The dynamics of gp120 proteins also have been examined computationally 36, 37, 38, 39, 40, 41. Molecular Dynamics (MD) simulations of wild-type gp120 and S375W mutant 40 indicated significant differences in protein flexibilities. The S375W mutant form preferred the bound-like conformation while the wild-type diverged from this conformation, with partial unfolding of some of the β- strands Non-equilibrium steered MD, in which the bridging sheet β-2/3 or β-20/21 were pulled away from α-helix-1 of the inner-domain 39 indicated that the β-2/3 had more flexibility than the β-20/21, which favored interaction with the inner-domain α-helix, keeping a conformation close to the CD4-bound-form . Liu et al 38 examined the dynamic domains of homology models of HIV-1 gp120 core, in the presence and absence of CD4 and the effects of mutation on these motions. They generated an ensemble of conformations for different gp120 models and performed an essential dynamics analyses to identify the principal modes of motion. Interestingly, their results also indicated that the S375W mutation favors the CD4 bound conformation, while the I423P conformation prefers the unliganded conformation. MD studies by Hsu et al. 36, 37 indicate that there are concerted loop motions in the vestibule of the CD4 cavity, stabilization of the bridging sheet and a coalescing of the bridging sheet and V3 loop to form the co-receptor binding site. Binding entropies extracted from these MD trajectories 37 suggest that the large entropy loss associated with CD4 binding is derived from hydrophobic interactions from CD4 Phe43 insertion into the cavity, the formation of a hydrogen bonding network and the restructuring of the bridging sheet. Temperature accelerated MD was used for large scale sampling of gp120 motions by Abrams and Van den-Eijnden 42 and predicted a counter rotation between the inner and outer domains as well as disruption of the bridging sheet occurs in the unbound form of gp120. Tan and Rader 41 have used a program that implements graph theory (Floppy Inclusion and Rigid Substructure Topography (FIRST)) 43 to analyze the flexibility and rigidity of all known gp120 structures. A flexibility index describing the extent and distribution of flexible and rigid regions in the inner, outer and bridging sheet domains of the twenty-two gp120 structures with various ligands indicated the inner domain and bridging sheet domains to be more flexible while the outer domain to be more rigid. Gp120 proteins bound with CD4 exhibited less flexibility in the inner domain than gp120 bound to mini-protein mimetics. Increasing inner domain flexibility accompanied by increasing outer domain rigidity was also observed for the b12 antibody bound gp120 complex. Comparison of these various structures (CD4 bound, b12 bound and apo gp120) led to identification of a universal rigid region on α-2 helix (residues 335-352) which is proposed to be a potential initial recognition site associated with co-receptor binding. A consensus rigid cluster on a β-sheet located on the co-receptor binding surface was also identified 41.
Another coarse-grained approach to protein flexibility is the Gaussian Network Model (GNM) 44-46 which permits an efficient exploration of collective motions of proteins. The GNM approach is based on local packing density and bonded and non-bonded contact topology in a given structure. The decomposition of the vibrational modes elucidates the slowest (global motion) and the fastest (local motion) modes. The slowest mode provides information on the large-scale cooperative movements of large domains, as well as the regions with minimal motion during these collective motions. The regions with restricted motion are considered to play an important role in modulating and monitoring the catalytic activity of enzymes and serve as focal points for modulating collective domain motions or hinge bending 44, 47, 48. GNM has been shown to yield results which are in remarkable agreement with results obtained from models with standard semi-empirical potentials, as far as low-frequency normal modes are concerned 49-51. The collective motions also determine the communication propensities inherent to the protein structure. The communication propensities of residues in a given network can be expressed in terms of the Kirchoff matrix of inter-residue contacts, which is also the underlying theory of GNM 51. In this network model, the catalytic residues are distinguished by their fast and precise communication capabilities 51.
In this study we present the dynamics and communication propensities of the gp120 core residues calculated with GNM, to provide a computational framework for elucidating the residues that are critical in stabilizing the unbound form of gp120 and or that are key communicating gp120 conformational change. We report here the results for three CD4 bound-gp120 crystal structures: the gp120, CD4, 17b complex from HIV strain HXBC2 (PDB ID: 1G9M) 19; the gp120, CD4, 17b complex from HIV strain YU2 (PDB ID: 1G9N) 19 and the gp120, 43 residue scorpion-toxin CD4 mimic, and 17b complex from HIV strainYU2 (PDB ID: 2I5Y) 22. By analysis of gp120 fluctuation dynamics and communication propensities a stable outer domain is revealed with communication network emanating from the Phe43 cavity and poised for signal propagation throughout the gp120 outer domain.
RESULTS AND DISCUSSION
GNM Fluctuation Profiles
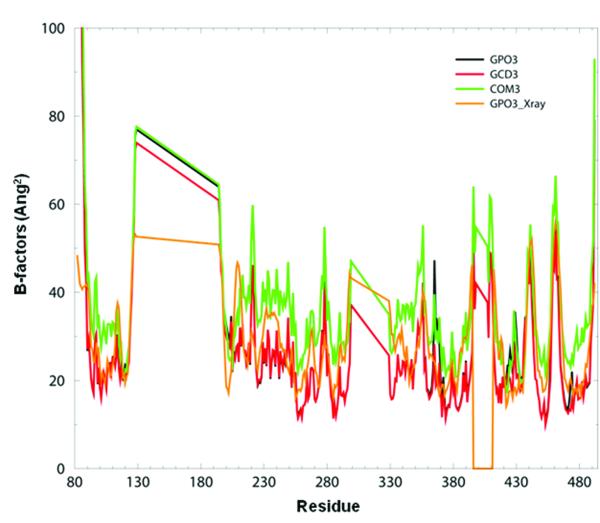
We have adopted a simplified nomenclature to represent the various complexes and components used in this study (Table 1). Briefly, for the HXBC2 strain, the three component complex of gp120, CD4 and 17b is referred to as COM1, the two component complex of gp120 and CD4 is referred to as GCD1, and gp120 alone is referred to as GPO1. Similarly, complexes from the YU2 strain are denoted COM2, GCD2, GPO2 and the third system with the scorpion-toxin CD4 mimic bound to Gp120 and 17b are labeled COM3, GCD3, and GPO3. Residue numbering for the core gp120 crystal structures maintains the numbering from the full-length gp120 sequence (see methods section). We first sought to establish agreement between experimental Debye-Waller temperature factors (or B-factors) and the theoretical B-factors, computed from GNM, for the three systems of study. B-factors provide an experimental measurement of the intrinsic protein dynamics 52 which is defined by the local packing density and is often used to establish a correlation between theory and experiment 53, 54. As shown in Figure 2, there is a reasonable agreement between the experimental and theoretical B-factors for gp120 in COM3, GCD3 and GPO3. The broad flat peaks correspond to the deletions of the V1/V2 (residues 129-194) and V3 (residues 300-329) loops in the gp120 core X-ray structure. This overall agreement between X-ray and GNM B-factors is seen in the other gp120 complexes as well (see supplementary data, Figures S1A, and S1B) and is consistent with earlier observations on other protein systems 45-48, 55.
Table 1.
Nomenclature for the 3 pdb complexes and their ligand components used in GNM and MD studies.
| Strain | PDB | Gp120 | Gp120 + CD4/Min i |
Gp120 + CD4 + 17b |
|
|---|---|---|---|---|---|
| 1 | HXBC2 | 1G9M | GP01 | GCD1 | COM1 |
| 2 | YU2 | 1G9N | GP02 | GCD2 | COM2 |
| 3 | YU2 | 2I5Y | GP03 | GCD3 | COM3 |
Figure 2. Experimental and theoretical B-factors.
The B-factors calculated in GNM for GPO3 (black curve) GCD3 (red curve), COM3 (green curve) and the experimental B-factors obtained from X-ray data for GPO3 (orange curve).
The second question we posed was: does the gp120 core exhibit different fluctuation profile for the two strains, HXBC2 and YU2? The YU2 and HXBC2 sequences of the core region of gp120s have 86% sequence identity 19. Most of the sequence differences observed are in the outer domain and are listed in Table S1 in the supplemental materials. A total of 33 outer domain residues in the region from V275 to E464 show conservative amino acid sequence substitutions.
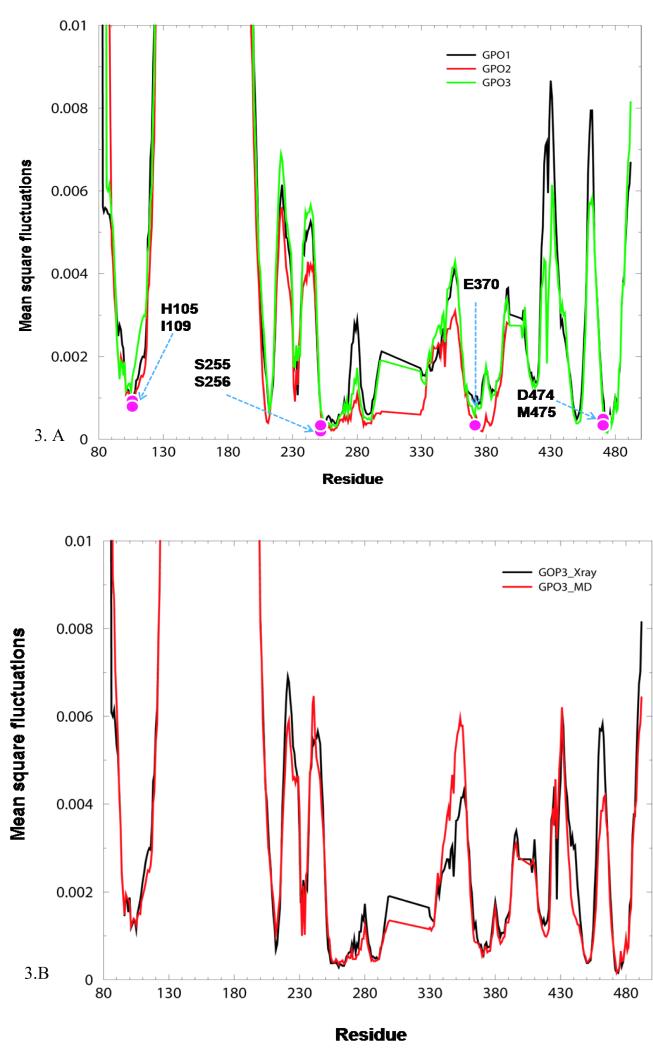
The slow mode profiles in GNM provide information on the intrinsic dynamics of the protein, with the maxima considered as recognition sites and the minima as hinge sites 48. We focus our analysis on the slowest GNM modes which will provide information on the cooperative motions of gp120 domains, to elucidate the components of gp120 flexibility. Figure 3 A, depicts the slow mode profile (or mode shape) for GPO1, GPO2 and GPO3, as a function of the residues. The ordinate represents the normalized mean square (ms) fluctuation of residues, driven by the slowest two modes (2 <, k <3) of motion. Although the maxima in GPO3 are less pronounced than in GPO1 or GPO2, the overall profiles are similar, with both minima and maxima occurring in the same vicinity, for all the three structures. Table S2 presents the residues corresponding to minima and maxima in GPO3. In spite of the modest differences in the amino acid sequence between HXBC2 and YU2 gp120 proteins, the cores exhibit similar dynamics (Figure 3A). This result is consistent with the similar binding affinities and energetic profiles determined by isothermal titration calorimetry and surface plasmon resonance respectively 27,28,31,56.
Figure 3.
A. Mean square fluctuations from GNM slow modes. Mean square fluctuations calculated from the average of the two slowest, most cooperative GNM slow modes (2,<k,<3), considering only the GP120 core, for GPO1 (black curve), GP02 (red curve) and GPO3 (green curve). Pink balls indicate residues that display GNM slow mode minima and are less the 4.5 Å from W427
B. Comparison of GNM Slow mode profiles for X-ray and MD snapshot. Mean square fluctuation of the X-ray structure (black curve) and the MD structure averaged over 5ns for GPO3 (red curve).
We considered two complexes from the YU2 strain with different primary ligands, CD4 and a 43 residue mini-protein derived from scorpion-toxin (GPO2 and GPO3, respectively). The gp120 crystal structures from these complexes differ by a C-α RMSD of 1.55 Å. The normalized mean square fluctuation plots of the two gp120 cores show the same fluctuation profile, despite the difference in primary ligand in the crystal structure complex. The similarity of GNM slow mode profiles regardless of HIV strain and primary ligand indicates a set of conserved collective motions common to all monomeric core gp120 envelop proteins. The residues, at which the GNM slow mode profiles exhibit minima in the three gp120s, are tabulated in the Table 2. We choose to focus the description of the conserved collective motions using the coordinate set for GPO3, since it represents a higher resolution structure from the YU2 strain and the biphenyl group binds more deeply in the gp120 cavity. GPO3 minima are depicted in Figure 4 and listed in Table 2. As illustrated in the Figure 4, most of the minima occur at the interface between the inner and outer domain. Many of the residues that display minima in the GNM mean square fluctuations are also within 4.5 Å of W427 in the Phe43 cavity, Figure 3. These residues span a portion of the Phe43 cavity, the interface between the inner domain (α- helix-5 and loop A) and outer domain (ß-strands 9, 10, 11, 23, 24 and loop outer domain α-helix-3. Majority of the minima observed are in the outer domain (Figure 4) with fewer minima in the inner domain (Figure 4). Notably, none of the residues from the bridging sheet domain display minima in GNM slow modes. The residues at minima occupy critical loci in the global mode and could be viewed as trigger points for controlling gp120 conformational change. These sites may function either in a hinge binding capacity and/ or define a core folding nucleus found in the pre-bound state of unliganded HIV gp120. We note that the core folding nucleus described here, does not imply a folding mechanism, but rather represents a set of residues that may be loosely structured in various pre-bound states, suggesting that other folding events may crystallize around this nucleus. The crystal structure of the unliganded SIV gp120 26 indicates that much of the outer domain shares a common structure with the HIV CD4 bound conformation. Thus, it is reasonable to conclude that minima occurring in the outer domain represent a core folding nucleus found in the pre-bound form of gp120. Furthermore, this conclusion is supported by the observation that most of the residues which occur at a minimum in GNM slow mode, are also conserved across primate and human HIVs (Q258, E370, I371, P470, G471, G472, G473, D474, M475 and D477 (Table 2). Of the eight gp120 residues that are within 4.5 Å of the mini-protein biphenyl in the 2I5Y complex (T257, E370, I371, S375, N425, M475, W427 and G473) six display minima, T257, E370, I371, S375, M475, and G473 (all from the outer domain). Interestingly, a residue that does not display a GNM slow mode minimum is W427. This residue from the bridging sheet is highly conserved across all primate HIV sequences, lines the ligand-binding pocket (Phe43 cavity) and forms hydrophobic interactions with CD4-Phe43 in the gp120-CD4 complexes and the biphenyl in CD4M47 receptor 18, 19, 21. Residues R252 to E275, in the low-fluctuation region, span the inner domain and the outer domain, Figure 4B. Within this span resides the tripeptide, L259-L260-L261 adjacent to the Q258 which is conserved in HIV. A second region of low fluctuation occurs from I449 to L453 which includes a portion of the tripeptide L452-L453- L454. A third low fluctuation region, from residues P470 to M475, contains the tripeptide G471-G472- G473. These three regions are noteworthy because they contain the conserved residues Q258, P470, G471, G472, G473 and M47. This core region of GNM, including regions of low fluctuation and slow mode minima, extends to β-strands on both sides encompassing residues I285-S291, S375 to N377 and F383. As viewed in Figure 4C, the GNM minima form a continuous surface of a stable secondary structure. Interestingly, the GNM minima are only located on one face of the Phe43 cavity, suggesting that this face provides a core structural region that is similarly structured in the CD4 unbound form of gp120.
Table 2.
Residues in GPO3 (gp120 from 2I5Y) that have GNM calculated minima. Residues depicted in bold, italic and underlined fonts are conserved among all primates strains, all human strains and moderately conserved in human strains, respectively.
| Residue | Domain | 2nd Structure |
Residue | Domain | 2nd Structure |
|---|---|---|---|---|---|
| T232 | Inner | Loop A | P470 | Outer | ß-24 |
| N234 | Inner | Loop A | G471 | Outer | ß-24 |
| R252 to V255 |
Inner | Loop B | G472 | Outer | |
| S256 to T257 |
Outer | Loop B | G473 | Outer | |
| Q258 | Outer | Loop B | D474 | Outer | |
| L259 to R273 |
Outer | Loop B, ß-9, Loop 10 |
N475 | Inner | α-5 |
| I285 to S291 |
Outer | ß-11 | D477 | α-5 | |
| E370 | Outer | α-3 | N478 | Inner | α-5 |
| I371 | Outer | α-3 | S481 to E482 |
Inner | α-5 |
| S375 to N377 |
Outer | ß-16 | |||
| F383 | Outer | ß-17 | |||
| I499 to L453 |
Outer | ß-22, ß- 23 |
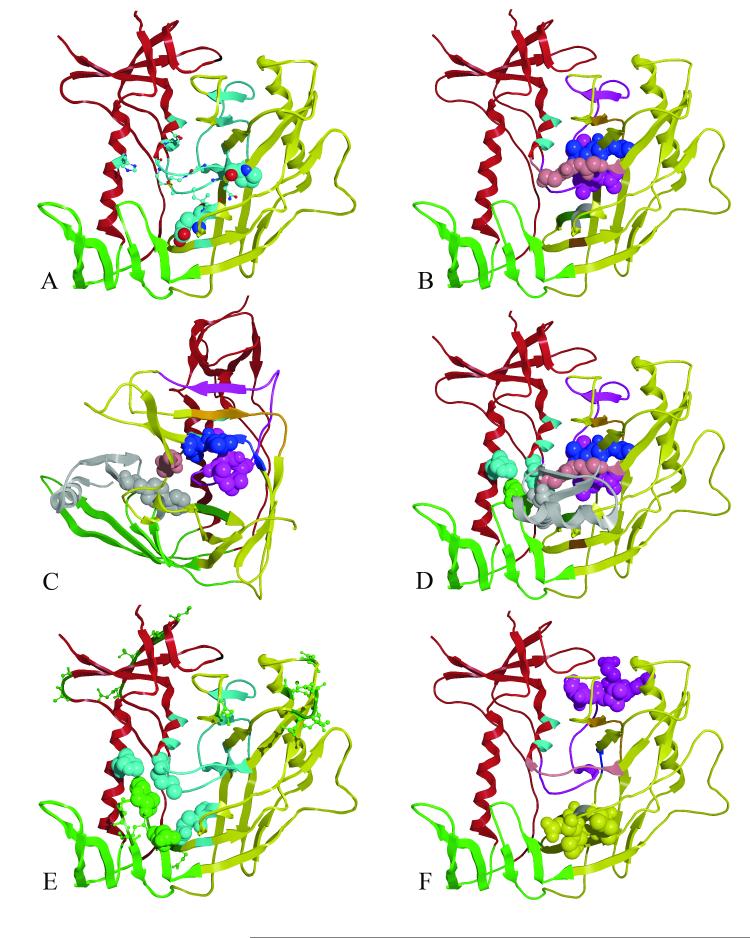
Figure 4. GNM slow mode minima and maxima, mapped to the gp120 core structure.
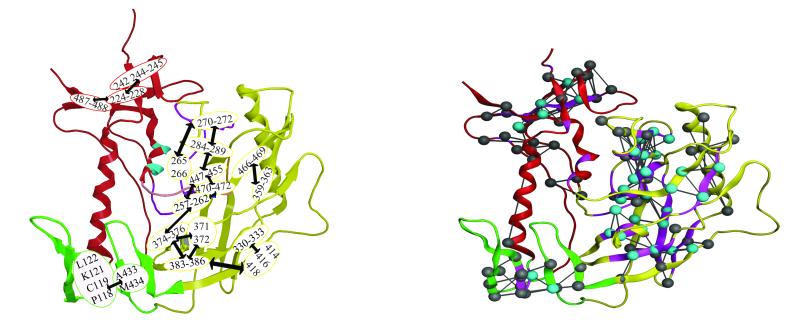
(A)The core gp120 inner (red), outer (yellow) and bridging sheets (green) domains are depicted as a ribbon diagram for GPO3 (pdb code, 2I5Y). Residues that have GNM minima are colored light blue. Residues with GNM minima which are conserved in all HIV gp120s are shown in spacing filling model (E370 and P470) and residues conserved in human HIV gp120s are shown as ball-and-stick (Q258, I371, G472, G473, M475, D477). ( B) Residues 252-275(purple) span the inner and outer domain and form the largest continuous stretch of GNM slow mode minima with the tripeptide L259-L260-L261 shown as space filling model. Adjacent GNM slow mode minima, L452, L453, L454: G471, G472, G473: I285, V286, Q287, L288, N289, E280, S291, S37 F376, N377 and P370, I371 are colored blue, pink, orange, dark green and gray respectively. (C) As in B but rotated 90 degrees with surface structural elements removed to depict proximity of the core GNM slow mode folding nucleus proximal to the 2I5Y mini-protein, biphenyl (gray) bound in the Phe 43 cavity. (D) GNM slow mode minima in the inner domain H105 (cyan) and M475 (cyan) are shown straddling W427 (green space filling model). (E) GNM slow mode minima and maxima are show in cyan and green ball and stick, respectively. Residues W427 and N425 (space filling green) are conserved residues locating on the bridging sheet. These GNM maxima likely for the recognition features that for ligand binding and interact with GNM minima (residues H105, M475, and E370, light blue space filling) which provide binding site stability. F) Residues E268, E269, I272, R273 and S274 (purple space filling model) have correlated GNM fluctuations with residues D368, P369, V372, T373 and S375 (yellow space filling model) that are more that 10.0 Å apart. (Rendered with MOE70)
Several GNM slow mode minima are also exhibited by residues in the inner domain, H105, E211, P212, I213, K232, N234, R252-V255, M475, D477, N478, N481 and E482. Of these, H105 and M475 are conserved residues and are located in the inner domain on α-helix 1 and 5, respectively (Figure 4D). Both these residues interact with the highly conserved W427 in the Phe43 cavity. In the X-ray structure, W427 is closely interacting with a conserved residue, I109 which is one-turn away from H105 on α-helix 1. These residues (H105 and M4745) seem to stabilize the Phe43 cavity by maintaining tight interactions with W427. In addition, M475 also interacts with G473 of the G471-G471-G473 tripeptide, in the outer domain. Thus, it is tempting to speculate that H105 and M475 serve as hinge residues that help drive the conformational change that forms the Phe43 cavity. Notably, many of the residues initially characterized by mutagenesis as essential for gp120-CD4 binding 3,57,58 are observed to either be at a minimum or in its close proximity in GNM slow mode, in this study (S256-T257, L259, A266-E267, E269, E370, Y384, P470, D477, E482).
While the residues at minima are known to be critical in catalytic or binding sites, those at the maxima have been observed to be good recognition sites 55. The entire loop between bridging sheet strands β-20 and β-21(residues N425 to K432) exhibits a maximum in the GNM slow mode profile, underlying the significance of this region as a recognition site for CD4 binding. This includes the highly conserved W427 which forms key hydrophobic interactions with the biphenyl of mini-protein (GCD3) (Figure 4C) and CD4 Phe43 (GCD1 and GCD2). The position of W427 on the bridging sheet in the Phe43 cavity suggests it might indeed be playing a critical role in recognition of the CD4 receptor, and has in fact been known to be critical in receptor-protein interactions 57. The less conserved N425 (a maximum) on β-sheet 20 of the bridging sheet interacts with the highly conserved E370 (a minimum) (Figure 4E), a GNM minima adjacent to the Phe43 cavity in the outer-domain. These two residues together provide rigidity (minimum) as well as flexibility (maximum), at the interface of the bridging sheet and outer domain. This juxta-positioning of a rigid region with a flexible region is ideal for ligand binding- the flexible region enhances ligand-recognition, while the rigid-region enhances ligand-binding stability. The residues N425 and W427 are in direct contact with the CD4-Phe 43 ligand, in the complexed structure (GCD2), as well as with the mini-protein biphenyl (GCD3), thus further supporting their importance as recognition sites.
A preliminary energy minimization, followed by a short molecular dynamics simulation in a solvated environment, is expected to relax the X-ray structure, driving it closer to its true minimum. We thus performed MD simulations of solvated GPO3 structure, and analyzed the dynamics. (See supplementary material for simulation protocols). Five nanosecond (ns) simulation trajectories for GPO3 were generated, using GROMACS 59, 60 in a fully solvated environment, at 310K and under constant NPT conditions. The root mean square fluctuations (rmsf) of the GP120 from MD simulations are in excellent agreement with the mean square fluctuations observed with GNM (Figure S2). The former is the mean square fluctuation from the 5 ns average of the simulation trajectory, while the latter is the mean square fluctuation (B-factor), calculated with GNM, for the mean structure, averaged over 5ns simulation trajectory. Interestingly, these two quantities also agree well with the experimentally determined B-factors of the GPO3 X-ray structure (Figure S2).
As shown in Figure 3B, the GNM mean square fluctuation profile for the MD relaxed GPO3 coordinate set closely mirrors the profile from the GPO3 X-ray structure. Comparing the GNM fluctuation profiles produced from MD snapshots for GPO3 and GCD3 and GCD2, Supplemental S3, also indicates that MD relaxation of the gp120-CD4 (or mini-protein) bound complexes reproduces a fluctuation profile similar to the MD relaxed GPO3 (relaxed in the absence of ligand). The GNM dynamics calculated with a simple, Hookean potential are thus indeed robust and comparable to that calculated from an atomic level force-field potential, used in MD simulations suggesting that relaxation of an X-ray structure by MD simulations does not significantly alter the GNM dynamics.
Communication propensities
The GNM slow mode profiles, have elucidated regions of fluctuation minima and maxima in the gp120 proteins. As noted by Chennubhotla and Bahar 51, equilibrium motions exhibited by the protein also determine the communication mechanism inherent in the protein residue network. Moreover, residues which display a high coordination number in a given protein network, are also effective communicators, providing a topological basis for communication propensities (CP) of residues in a protein. This communication propensity analysis, based on elastic network models, directly relates residue fluctuation to their CP’s 51, 61, i.e., residues whose distances fluctuate with low intensity communicate with a higher efficiency than residues with larger fluctuations. In Figure 5, the commute times for GPO1, GPO2 and GPO3 are plotted. We choose to use a cutoff value of commute times (C (i, j) < 0.21 as criteria for a residue pair (i, j) which are at a distance of at least 10 Å. or more, as having good CPs’. While there is no direct correlation between commute times and physical distances, there are some residue pairs which communicate efficiently, in spite of their long physical distances. As previously noted, the GNM slow modes of GPO1, GPO2, and GPO3 display a similar dynamic profile. The communication profiles of GPO1, GPO2, and GPO3 (Figure 5) also exhibit a similar pattern of residue communication propensities. Inspection of Figure 5 reveals differences in communicating pairs of residues are concentrated in the inner domain and bridging sheet domains (the region spanning residues 80-230 in Figure 5) for GPO1, GPO2 and GPO3. The amino-acid sequence as well as secondary and tertiary structures are conserved between HXBC2 (GPO1) and YU2 (GPO2 and GPO3) strains. Tan and Rader 41 also observed a variation in the domain flexibility among HIV strain and bound CD4 ligands in a study of relative domain flexibility for twenty CD4 and scyllatoxin CD4 mimic -gp120 crystal structures. In a broad sense, the set of conserved communication hubs identified in this study serve as a basis for hypothesis generation and further experimentation. Furthermore, recent structure reports 23,62 indicate that the inner domain contains three structurally distinct layers that define gp120 mobility accounting for the scarcity of GNM slow mode minimia and residues with good communication propensities in this region.
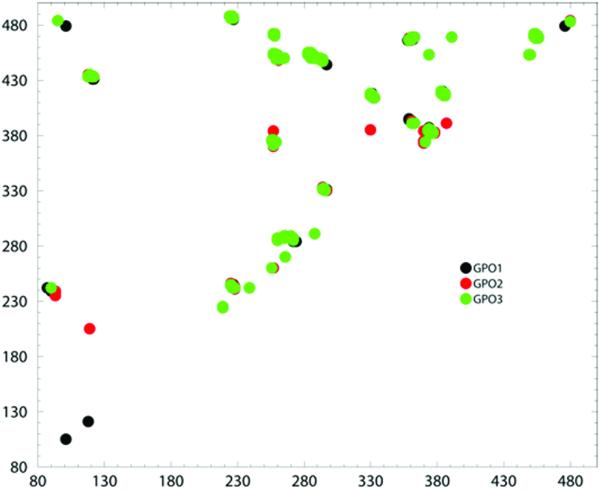
Figure 5. Communication propensities for gp120 core envelop.
Distribution plot of residue pairs having a communication time less than 0.2 and which are more than 10 Å apart, for GPO1(black spheres), GPO2(red spheres) and GPO3(green spheres)
Of the 234 communicating pairs with Cij values < than 0.21 in the three structures studied (supplemental Table 3) 47% are conserved between GPO1, GPO2 and GPO3 and another 20% are conserved among two of the three structures. In this analysis we focus on the conserved set residue of pairs with low communication propensity observed in GPO1, GPO2 and GPO3 and listed in Table 3. Residues pairs with low communication propensity for GCD1, GCD2, GCD3, COM1, COM2, and COM3 are tabulated in supplemental Table S4 and S5, respectively, and did not indicate a significant difference in intra-domain gp120 communication. Pairs of good communicators between the inner, outer and bridging domains are not observed in any of the three gp120 structures, GPO1, GPO2, and GPO3. Communication hubs in gp120 are only observed between intra-domain residues. In the bridging sheet domain, one communication hub exists between residues P118, C118, K121, L122 on β-strand 2 and adjacent β-strand 22. In the inner domain, residues A224-C228 form good communication with residues V242, T244, V245 and also K487 and V488. This set of residues resides in the β-sheet portion of the inner domain. However, the most extensive network of communicating hubs occurs in the outer domain. Many of these outer domain residue pairs with low commute times and which also display GNM slow mode minima are residues within β-strand secondary elements Table 3. However, only two residues with low commute times (V242 and T244) exhibit GNM slow mode maxima. The dominant communication hub in the outer domain is anchored by residues T257-N262 with low commute times to one hub at residues H374-F376 and a second hub at residues I449-L454, and a third at residues P470 - G472. Other core hubs of communication occur with residues Q287-N289 and I270-I272. The full web of inter-connecting residues pairs for outer domain residues can be delineated from residues in Table 3 and is shown via cartoon diagram in Figure 6.
Table 3.
Residues pairs that have low commute times, (C(i,j)) < 0.20, in the GPO1, GPO2, and GPO3 X-ray structures as determined by GNM fluctuation profiles are sorted by communication hub in the bridging sheet, inner domain and outer domain. Residues that also exhibit GNM slow mode minima or maxima are in bold or underlined font, respectively. Residues that are conserved in HIV sequences are indicated in italics (G258, P270, R469, G471, G472, T458. Residues pairs with efficient communication propensities as determined by MD simulation are colored in blue. The amino-acid residue is given for YU2/GPO3 only.
| Hub | (i) | Hub | (j) 1 | Hub | (j) 2 | Hub | (j) 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Bridging
Sheet |
P118 | C119 | K121 | L122 | A433 | M434 | ||||||||||||
|
Inner
Domain |
A219 | A224 | I225 | |||||||||||||||
| A224 | I225 | L226 | K227 | C228 | V242 | T244 | V245 | K487 | V488 | |||||||||
| C239 | V242 | |||||||||||||||||
|
Outer
Domain |
T257 | Q258 | L259 | L260 | L261 | N262 | H374 | S375 | F376 | I449 | T450 | L453 | L454 | P470 | G471 | G472 | ||
| T257 | I371 | |||||||||||||||||
| L265 | A266 | Q287 | L288 | N289 | ||||||||||||||
| A266 | I270 | |||||||||||||||||
| I270 | V271 | I272 | Q287 | L288 | N289 | |||||||||||||
| I284 | I285 | V286 | Q287 | L288 | T450 | G451 | L453 | L454 | T455 | |||||||||
| V293 | I449 | |||||||||||||||||
| I294 | N295 | N332 | S447 | N448 | I449 | |||||||||||||
| N295 | N332 | |||||||||||||||||
| C296 | H330 | |||||||||||||||||
| H330 | C385 | |||||||||||||||||
| H330 | C331 | N332 | L333 | I414 | L416 | C418 | ||||||||||||
| I359 | I360 | F361 | N362 | E466 | I467 | F468 | R469 | |||||||||||
| F361 | N362 | P363 | F391 | |||||||||||||||
| I371 | T373 | H374 | S375 | |||||||||||||||
| H374 | S375 | F376 | F383 | Y384 | C385 | N386 | ||||||||||||
| F383 | Y384 | C385 | L416 | C418 | I420 | |||||||||||||
| L453 | L454 | T455 | R456 | F468 | R469 | P470 | G471 | G472 |
Figure 6. Consistency between GNM and MD calculated communication propensities.
A) Communication hubs in gp120 core envelope, as determined by GNM Ribbon diagram of the gp120 protein, highlighting the residues identified as communication hubs in gpo1, gpo2 and gpo3. The hubs, determined by network of pairs of residues with communication propensity less than 0.2 are labeled and encircled. The arrows between encircled residues indicate hubs that communicate readily. B) Residues pairs with efficient communication propensities as calculated from the gpo3 17ns MD trajectory, are drawn as gray and cyan balls and connected with a line. Residues pairs demonstrating low communication propensity in both GNM and MD calculations are colored cyan. Residues exhibiting low communication propensity determined by GNM are drawn as purple ribbon.
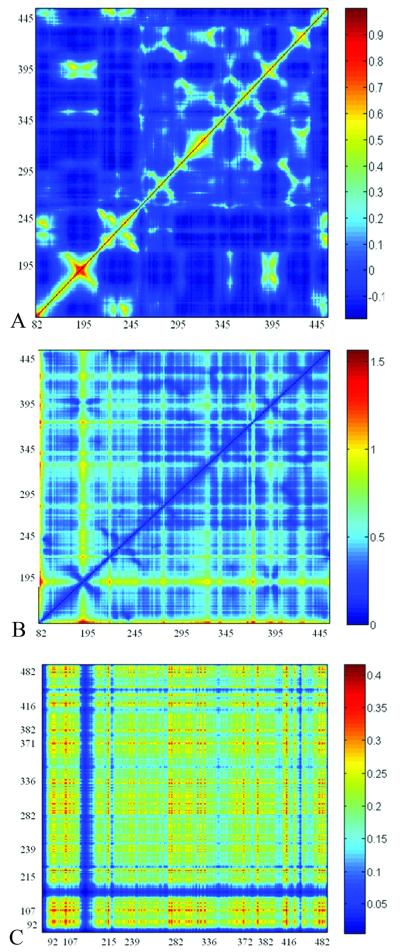
To corroborate this set of communication hubs we examined all atom MD simulation of GPO3. The minimum distances between c-α atoms of communicating hubs was calculated from a 17ns trajectory (Supplemental Figure 4). These minimum distances are maintained over the course of the MD simulation indicating that these residues indeed exhibit low mobility and that signal propagation and equilibrium fluctuations are related 51. A comparison of the cross-correlation matrices for GNM fluctuations, GNM communication propensities and the motions of C-α atoms around the averaged position from MD simulation (Figure 7) shows similarities between regions of low communication propensity with both GNM and MD correlated motions. Several regions with low communication propensities suggested as communication hubs display a positive correlation of C-α motion, suggesting that the local packing density is reflected in the communication propensities, albeit, indirectly. We note that there is not a one to one correspondence between slow mode minimia and communication propensity. While most of the residues which are minima in a given slow mode, may communicate efficiently, there are also residues which have a high mobility, and yet are good communicators, due to the positive correlation between the fluctuations. The commute times reflect the correlation between the fluctuations in the inter-residue distances, whereas the minima in a given slow mode reflect low mobility at a given residue, in that particular slow mode.
Figure 7. Comparison of Cross-Correlations Matrices.
A)GNM residues fluctuations B)GNM communication propensity from MD averaged snapshot C) Cross-correlation of motions of C-α atoms around the averaged position from MD simulation.
To further elaborate the communication network based on MD simulation we calculated residue communication propensity from the trajectory of GPO3 as defined as by Morra et al.61. Residues with low communication propensities are listed in supplemental Table 3. The two methods reveal a set of overlapping residues (Table 3) that communicate efficiently. As the two methods sample protein motion of different magnitudes (0.1nm versus 1nm) agreement of overlap between the two sets reveal the most efficient residue communicators, Table 3. As shown in Figure 8, GNM determined CP define broader regions of communication pairs of residues while the MD communication propensities provide detail on which residues pairs communicate the most efficiently. Residues pairs that residues have positively correlated motions across distance regions of Gp120
Two residues identified by both methods as good communicators which are located within 4.5 Å of the biphenyl group of the mini-protein are T257, and S375. Furthermore, we note a correspondence between residues that have been identified as good communicators and have been shown by mutagenesis 3, 57, 58 to be essential for CD4 binding are residues V120, K121, K227, T257, L259, N262 and Y384, (these residues all participate in key communication hubs). Mutation of the residues to alanine may affect the local packing density which may in turn influence the propensity to communicate efficiently with neighboring residues. Analysis of the cross-correlation matrix of the C-α atomic motion from the average structure produced from the GPO3 MD trajectory (Figure 7C) confirms a positive correlation in motion between several of the PHE43 cavity residues (371 to 383-386 to 330-330 to 414-418) to supporting the notion of signal propagation from the Phe 43 cavity to adjacent regions of the outer domain.
DISCUSSION
We have used Gaussian Network Model, molecular dynamics and the Markovian stochastic model of information diffusion to analyze the equilibrium fluctuations and the communication network of the HIV viral envelop protein, gp120, using the existing X-ray structures available in the CD4 bound form. The gp120 core demonstrates a large conformational change that accompanies molecular recognition of the CD4 and Chemokine receptors. The rationale for focusing computation on the CD4 bound form is based on the observation that the NBD class of small molecule molecules drives the structuring of gp120 to the same extent as CD4 receptor binding 32.
We aimed to analyze the equilibrium fluctuations and signal propagation as a means of understanding which amino acid residues play a role in signal transduction in gp120 with the intent of delineating important components of the pre-structured regions of gp120 in the CD4 bound form. GNM slow mode profiles identified key residues with low mean square fluctuations, in the inner and outer domains. The residues at minima in the outer domain are hypothesized to form a core folding nucleus, while those in the inner domain serve as hinge sites driving conformational change. These results derived from GNM calculation agree overall with those reported by Tan et al., 41 suggesting that for 17b antibody binding gp120 cores, the inner domain and bridging sheet are more flexible domains then the outer domain. However, results differ at the residue-level. A rigid core for α-2 helix (residues 335-352) identified by FIRST is not identified by a specific GNM slow mode minimum. Tan and Rader 41 identified this core via comparison to CD4 unliganded gp120s (SIV and b12 bound) whereas our study focused on CD4 and CD4 mimic bound gp120s. They also observed that the flexibility index for the domains is dependent upon either CD4 or CD4 mimic bound to gp120 with increased flexibility of the inner domain and decreased flexibility for the outer domain when scyllatoxin CD4 mimic is bound, as compared to when CD4 is bound, to gp120. Minor differences in slow mode minima and maxima on a residue-level are delineated by the GNM method for GPO2 versus GPO3. A recent structure report of a CD4 unliganded YU2 gp120 24, reveals a less structured bridging sheet where the β2/β3 and β21/β22 strands are dissociated from the interface of the inner and outer domain. The β21/β22 strands contain the longest continuous region of GNM slow mode maxima observed in GPO1, GPO2 and GPO3. Moreover, all the regions of GNM slow mode minimia reported here do not exhibit any conformational changes in the unliganded YU2 gp120.
Analysis of communication propensities based on the elastic network model, yielded hubs of residues which facilitate communication between residues, as reflected by their low commute times. Many of the good communicating pairs revealed with the Gaussian network model are also observed in calculations from MD trajectories. Furthermore, few residues exhibit both GNM maxima and good communication propensities. In contrast, many residues that exhibit slow mode minima also demonstrate good communication propensities and reside on β-strands of the outer-domain. This implies, in the case of gp120, that the more structurally stable portions of the protein are equipped for more efficient communication. We also observe that positively correlated GNM fluctuations across large distances in the outer domain are facilitated by interleaving hubs of good communicators. These results are consistent with the correlated motions observed in four regions of Gp120 using covariance web analysis from Essential Dynamics Calculations 38, 63. This study broadly identified correlated motions in the (the β-bundle at the termini-proximal end of the inner domain;, the gp120 bridging sheet domain, the V3 domain, a region composed of the LC (265-270), LD(278-283), LE (350-357), V5 (459-463), α-2 (334-348)and β-bundle (358-362, 374-379, 463-470) in proximal end of the outer domain. These broadly defined regions overlap with specific communications hubs identified in Figure 6. The study of Liu et al. 38, also describes the major relative motions of the domains in the unbound form of Gp120 namely, twisting of the inner domain and bridging sheet relative the outer domain and a twisting of the outer domain relative to the inner domain, bridging sheet and V3 domain. This relative motion of the inner and outer domains correlates with the observation in this study that most of the regions of GNM slow mode minimum occur in the outer domain along the interface of the inner domain.
We have elucidated the communication hubs in each gp120 domain. The present findings provide us with a topological basis for efficient mediators of communication in the gp120 molecule. The lack of inter-domain communication in gp120 was at first, unexpected. However, communication propensity is delineated by the underlying protein structural network. Given the large re-arrangement of the bridging sheet and inner domain revealed in recent structures 24,23, effective communication among the domains is unlikely. The two communicating hubs in the bridging sheet (residues 118-122 to 433-434) between the adjacent anti-parallel β-strands (β-2/ β-3 and β-20/ β-21) suggests that in the CD4 bound form, a signal may be transmitted by the association of these two previously unstructured regions. Indeed, steered MD studies indicate that the β-2/ β-3 and β-20/ β-21 are conformationally independent 39. The three communication hubs in the inner domain are not exclusively associated with GNM slow mode minima. Nonetheless, two of these residues, V242 and T244, do exhibit GNM slow mode maxima. The three communication hubs, 224-228, 242,244-245 and 487-488 are located on the five stranded β-sandwich located in the N-and C-terminal portion of the inner domain. The molecular arrangement of the gp120 trimer as revealed by electron-tomography indicates that the β-sandwich portion of the inner domain would project towards the center of the trimer axes and the interface of gp41 64. Pancera et al. 23 recently describe the architecture of the inner domain as an invariant β-sandwich anchoring three mobile layers as exhibited various CD4 bound and unbound forms of gp120 complexes. It would be premature to speculate that the three inner domain communication hubs may facilitate communication with other components of the viral spike, but they may have physiological significance.
Characterizing both GNM fluctuation and residue communication profiles may aid the interpretation of mutagenesis data. Residues that are characterized with GNM slow mode minimum reflect the structural topology of the protein based on its underlying packing density. Thus, residues with GNM slow mode minimia would be fairly sensitive to mutations that alter the under lying protein topology. Comparing the consistency of GNM results with mutation data yields credence to the interpretation of the gp120 fluctuation profile revealed in this study. Many gp120 mutations that affect CD4 binding 3,57 are both good communicators and have GNM slow mode minima, such as T257, L259, N262, Y384, suggesting good overlap between computational and biological data. Madani et al 33 have demonstrated that the NBD class of inhibitors has varying sensitivity to Phe43 cavity mutants (V255, T257, S375) depending on the halogen substitution pattern on the NBD-phenyl ring. Both T257 and S375 display a GNM slow mode minimum and good communication propensities as calculated from GNM and MD simulations. Mutation of T257 to either A or S negatively impacted relative viral enhancement compared to wild-type. Given the importance of T257 as a communication hub and its potential interaction with NBD phenyl ring, it is not surprising that even the conservative change to serine would disrupt the communication network embedded in the topological fold in the cavity. Mutation of S375 to G reduced HIV sensitivity to enhancement by NBD compounds while the S375A mutation increased HIV sensitivity to all but the largest phenyl substituted analogues. This suggests that S375 and T257 serve as a fine tuned sensor in the Phe43 cavity and that communication propensity and fluctuation minima are plausible explanations for the roles of these residues in inducing and/or stabilizing the CD4 bound conformation of gp120.
The site of antibody 17b and CCR5 co-receptor binding, overlap at the interface of the bridging sheet and outer-domain including the V3 loop (not included in this study) 21. The co-receptor N-terminal and the second extracellular loop bind to gp120 at the bridging sheet and at the junction of the V3 loop and outer domain. GNM slow mode minima and maxima do not map within the 17b and CCR5 co-receptor binding sites, as defined by the 1QAD crystal structure. However, communication hubs are exhibited at this co-receptor binding site. In the absence of a gp120- CD4-CCR5 crystal structure a direct pathway for signal propagation between the PHE43 cavity and the chemokine receptor site could only be inferred. Yet, residues that may exhibit efficient communication propensities in the general chemokine receptor site binding region have been identified in this study. On the bridging sheet, the communicating network between β-2/ β-3 and β-20/ β-21 (residues 118-122 to 433-434) forms contacts with the antibody interface. Residues K121, R419, K421 andQ422 have also been shown to be important for CCR5 interactions 65. Residues, N295, H330, and N332, located in the outer domain near the base of the V3 loop are also posed for efficient communication. Analysis of the cross-correlation matrix of the C-α atomic motion from the average structure produced from the GPO3 MD trajectory confirms a positive correlation in motion between several of these residues (371 to 383-386 to 330-330 to 414-418) supporting the notion of signal propagation from the Phe 43 cavity
Morra et al. have calculated communication propensity from Hsp90 ligand bound and unbound MD trajectories and inferring signal propagation between distant domains 61. Furthermore, they also report that the correlation matrix from GNM is consistent with that obtained by MD and confirm that a signal propagation pathway extracted from GNM is similar to that inferred from MD. Certainly, further work on the gp120 system would entail MD derived communication propensities using the set of mutant gp120s with various ligands reported by Madani et al. as a basis for identifying a pathway for signal propagation 33. Experimentally, the dynamic and energetic coupling of the communication hubs could also be investigated by in silico alanine mutagenesis analysis 66 where each residue is mutated to alanine successively, and the effect on the communication propensities and energetic coupling can be calculated, providing a measure of the contribution of that particular residue towards stability of the communication network.
A wealth of biological and structural data is available for the gp120/CD4/co-receptor complex elucidating structure function relationships. A new context for this information is provided by delineating points of minima and maxima in the fluctuation profile and translating topological features to communication propensities. Residues identified as GNM minima in the outer domain may represent a core folding nucleus found in the pre-bound form of gp120. With respect to the CD4 cavity, residues that are likely to be recognized in the unbound form and which may facilitate signal propagation have been identified (T257, L259, N262, Y384). Furthermore, regions of GNM maxima that contact the biphenyl in the cavity indicate flexible regions that may be amenable to accommodating a larger ligand. We hypothesize that minima in the inner domain may represent hinge regions, driving the structuring of the CD4 bound form of gp120. We conclude from GNM profiles that W427, a GNM maximum, surrounded by two residues, H105 and M475, of GNM minima, form a key structural element that stabilizes the formation of the CD4 cavity. Podesta et al. have demonstrated that the boundary between dynamic regions generalize to regions of catalytic activity in enzymes 67. On the opposite face of the Phe43 cavity from W427, are a number of GNM minima in the outer domain that comprise communication hubs which are topologically suited to transmit signal of cavity structuring to the chemokine receptor site. Our conclusions from this study of CD4 bound forms of gp120 are consistent with recently published gp120 structures revealing plastic regions of the inner domain and bridging sheet. GNM has enabled us to deduce differences in conformational dynamics, which might not be otherwise deciphered from the crystal structure of a CD4 bound form of gp120, and provided insights in elucidating and interpretation of biological data of protein-ligand interactions for the complex process of HIV gp120 envelop recognition, cell attachment and viral entry.
MATERIALS AND METHODS
Protein modeling
Three X-ray crystal structures were prepared for normal mode and molecular dynamic calculations: CD4-bound HIV-1 gp120 core strain HXBC2: PDB code 1G9MCD4-bound HIV-1 gp120 core strain YU2: PDB code 1G9N and the scyllatoxin mini-protein (CD4M47A) bound HIV-1 gp120 core strain YU2: PDB code 2I5Y. The deglycosylated core gp120 construction of envelop protein in the three crystal structures, has a 19- and a 52- amino acid residue deletions from both the N- and C- termini respectively, is devoid of the V3 variable loop and contains a tripeptide substitution (Gly-Ala-Gly) for the 67 residues of the V1/V2 loop. Residue numbering in the crystal structures and in this study conforms to the numbering used in the full-length gp120 sequence. A gap in the numbering exists from residues (127 to 194 and 296 to 330) corresponding to the substitution of the V1/V2 loops with the tripeptide and the deletion the V3 loops. For, 1G9M and 2I5Y, the missing V4 loop was added from the 1G9N crystal structure and minimized. Hydrogen atoms were added and tautomeric states and orientations of Asn, Gln and His residues were determined with Molprobity (http://molprobity.biochem.duke.edu/) 68,69. Hydrogens were added to crystallographic waters using MOE 70. The OPLS-AA force field 71 in MOE 70 was used and all hydrogens were minimized to a root mean square (rms) gradient of 0.01, holding the heavy atoms fixed. A stepwise minimization followed for all atoms, using a quadratic force constant (100) to tether the atoms to their starting geometries; for each subsequent minimization, the force constant was reduced by a half until 0.25. This was followed by a final cycle of unrestrained minimization. Water molecules, Asn-linked acetyl-D-glucosamine, 2-(acetylamino)-2-deoxy-a-d-glucopyranose and small molecules were removed prior to GNM and MD calculations.
Gaussian Network Model
In Gaussian Network Model (GNM), the protein is modeled as a network of residues, with each residue being represented by its α-carbon atom. Bonded and non-bonded pairs of residues located within an interaction cutoff distance Rc (7.0 Å) are assumed to be connected by springs (or harmonic potentials) with a uniform spring constant γ, which is the single parameter (force constant) of the Hookean potential, proposed by Tirion 72. The topology of the structure is fully defined by the Kirchhoff matrix of inter-residue contacts, Γ, also known as the connectivity matrix, which in turn fully defines the equilibrium dynamics of the structure, or the most likely deformations near the folded state. For a network of N residues, the elements of the Kirchoff matrix Γ are defined as:
| (1) |
The cross-correlations between the fluctuations ΔRi and ΔRj of the nodes i and j are given by 55:
| (2) |
where kB is the Boltzmann constant, T is the absolute temperature and γ is a uniform spring constant. The inverse of Γ is expressed in terms of the nonzero eigenvalues λk (1 ≤ k ≤ N-1) and corresponding eigenvectors uk of Γ as 45:
| (3) |
which permits the mean square (ms) fluctuations of a given residue to be expressed as a sum over the contributions of all modes:
| (4) |
where uk and λk are the respective kth eigenvector and eigenvalue of the Kirchoff matrix, Γ. The ith element of uk reflects the mobility of residue i in the kth mode. λk scales with the frequency of mode k, and λK−1 is a statistical weight, which suitably rescales the contribution of mode k. The slowest mode thus has the largest contribution to the observed dynamics and the highest degree of cooperativity 55. The shapes of the slow mode profile reveal the mechanism of the cooperative of global motions. The most constrained residues in these modes play critical mechanical role such as acting as hinge centers 44-48,55. The fastest modes, on the other hand, are localized to single residues that are usually tightly packed in the folded state. Being the most constrained residues, they are the latest to evolve (or reconfigure) and do not contribute significantly towards the global motion. In equation 4, the subscript ii designates the ith diagonal element of the matrix enclosed in parenthesis. The X-ray crystallographic temperature factors (or B-factors) can be compared to the theoretical mean square (ms) fluctuations by the equation:
| (5) |
Communication Propensities
The residue fluctuations in GNM also determine the communication propensities between residues 51. An expression for commute time C (i, j) between residue i and j, in terms of inverse of Khirchoff matrix, Γ−1 is given by:
| (6) |
which reduces to:
| (7) |
where the constants are as in equation 4, and dk is the local interaction density at residue k. The commute time between residues i and j, is thus directly proportional to the fluctuation in the distance between these two residues, since the term in parentheses in the above equation is a constant for all pairs of residues. The mean square fluctuations play a dominant role in determining the communication propensities of a given pair of residues- larger the mean square fluctuation, longer the commute time.
Supplementary Material
Acknowledgements
The authors appreciate the useful suggestions and discussions with Ivet Bahar in analyzing the GNM results and also thank Martha S. Head for suggesting this study. IHS acknowledges NIH for support (R01LM007994-06). This work was supported by a grant from the National Institutes of Health GM56550 (JL).
Abbreviations
- GNM
Gaussian Network Model
- HIV
Human Immunodeficiency Virus
- CCR5 or CXCR4
- V3
third variable
- MD
Molecular Dynamics
- FIRST
Floppy Inclusion and Rigid Substructure Topography; For the HXBC2 strain three component complex of gp120, CD4 and 17b is referred to as COM1, the two component complex of gp120 and CD4 is referred to as GCD1, and gp120 alone is referred to as GPO1. Similarly, complexes from the YU2 strain are denoted COM2, GCD2, GPO2 and the third system with the scorpion-toxin CD4 mimic bound to Gp120 and 17b are labeled COM3, GCD3, and GPO3
- B-factors
temperature factors
- CP
communication propensities
- Fab
fragment, antigen binding
- C
commute times
References
- 1.Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220(4599):868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Gallo RC, Salahuddin SZ, Popovic M, Shearer GM, Kaplan M, Haynes BF, Palker TJ, Redfield R, Oleske J, Safai B, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224(4648):500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 3.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh WC, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237(4820):1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 4.Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2(12):1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 5.Blacklow SC, Lu M, Kim PS. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry. 1995;34(46):14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- 6.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312(5996):763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 7.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381(6584):667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 8.Sattentau QJ, Moore JP. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174(2):407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sattentau QJ, Moore JP, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67(12):7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272(5263):872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 11.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381(6584):661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 12.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85(7):1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 13.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85(7):1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 14.Bosch ML, Earl PL, Fargnoli K, Picciafuoco S, Giombini F, Wong-Staal F, Franchini G. Identification of the fusion peptide of primate immunodeficiency viruses. Science. 1989;244(4905):694–697. doi: 10.1126/science.2541505. [DOI] [PubMed] [Google Scholar]
- 15.Brasseur R, Cornet B, Burny A, Vandenbranden M, Ruysschaert JM. Mode of insertion into a lipid membrane of the N-terminal HIV gp41 peptide segment. AIDS Res Hum Retroviruses. 1988;4(2):83–90. doi: 10.1089/aid.1988.4.83. [DOI] [PubMed] [Google Scholar]
- 16.Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W, Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J Virol. 1990;64(12):6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins fusogens antigens and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 18.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure Fold Des. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. 8. [DOI] [PubMed] [Google Scholar]
- 20.Moebius U, Clayton LK, Abraham S, Harrison SC, Reinherz EL. The human immunodeficiency virus gp120 binding site on CD4: delineation by quantitative equilibrium and kinetic binding studies of mutants in conjunction with a high-resolution CD4 atomic structure. J Exp Med. 1992;176(2):507–517. doi: 10.1084/jem.176.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang CC, Stricher F, Martin L, Decker JM, Majeed S, Barthe P, Hendrickson WA, Robinson J, Roumestand C, Sodroski J, Wyatt R, Shaw GM, Vita C, Kwong PD. Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth. Structure. 2005;13(5):755–768. doi: 10.1016/j.str.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A. 2010;107(3):1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Kwon YD, Zhou T, Wu X, O’Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang Z-Y, Zhang M-Y, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. Structural Basis of Immune Evasion at the Site of CD4 Attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie H, Ng D, Savinov SN, Dey B, Kwong PD, Wyatt R, Smith AB, 3rd, Hendrickson WA. Structure-activity relationships in the binding of chemically derivatized CD4 to gp120 from human immunodeficiency virus. J Med Chem. 2007;50(20):4898–4908. doi: 10.1021/jm070564e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433(7028):834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 27.Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci U S A. 2000;97(16):9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dey B, Pancera M, Svehla K, Shu Y, Xiang SH, Vainshtein J, Li Y, Sodroski J, Kwong PD, Mascola JR, Wyatt R. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J Virol. 2007;81(11):5579–5593. doi: 10.1128/JVI.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, Chaiken I, Freire E, Smith AB, 3rd, Sodroski JG. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci U S A. 2004;101(14):5036–5041. doi: 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madani N, Perdigoto AL, Srinivasan K, Cox JM, Chruma JJ, LaLonde J, Head M, Smith AB, 3rd, Sodroski JG. Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J Virol. 2004;78(7):3742–3752. doi: 10.1128/JVI.78.7.3742-3752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Q, Ma L, Jiang S, Lu H, Liu S, He Y, Strick N, Neamati N, Debnath AK. Identification of N-phenyl-N’-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology. 2005;339(2):213–225. doi: 10.1016/j.virol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB, 3rd, Sodroski J, Freire E. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry. 2006;45(36):10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madani N, Schon A, Princiotto AM, Lalonde JM, Courter JR, Soeta T, Ng D, Wang L, Brower ET, Xiang SH, Kwon YD, Huang CC, Wyatt R, Kwong PD, Freire E, Smith AB, 3rd, Sodroski J. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure. 2008;16(11):1689–1701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, Gabuzda D, Smith AB, 3rd, Sodroski J. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 2009;5(4):e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ptasek LM, Vijayakumar S, Ravishanker C, Beveridge DL. Molecular Dynamics Studies of the Human CD4 Protein. Biopolymers. 1994;34:1145–1153. doi: 10.1002/bip.360340904. [DOI] [PubMed] [Google Scholar]
- 36.Hsu S-TD, Bonvin AMJJ. Atomic insight into the CD4 binding-induced conformational changes in HIV-1 gp120. Proteins: Struc Funct & Bioinformatics. 2004;55:582–593. doi: 10.1002/prot.20061. [DOI] [PubMed] [Google Scholar]
- 37.Hsu S-TD, Peter D, van Gunsteren WF, Bonvin AMJJ. Entropy Calculation of HIV-1 Env gp120, its Receptor CD4, and their Complex: An Analysis of Configurational Entropy Changes upon Complexation. Biophysical Journal. 2005;88:14–24. doi: 10.1529/biophysj.104.044933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu SQ, Liu SX, Fu YX. Molecular motions of human HIV-1 gp120 envelope glycoproteins. J Mol Model. 2008;14(9):857–870. doi: 10.1007/s00894-008-0327-7. [DOI] [PubMed] [Google Scholar]
- 39.Pan Y, Ma B, Nussinov R. CD4 binding partially locks the bridging sheet in gp120 but leaves the beta2/3 strands flexible. J Mol Biol. 2005;350(3):514–527. doi: 10.1016/j.jmb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Pan Y, Ma B, Keskin O, Nussinov R. Characterization of the conformational state and flexibility of HIV-1 glycoprotein gp120 core domain. J Biol Chem. 2004;279(29):30523–30530. doi: 10.1074/jbc.M404364200. [DOI] [PubMed] [Google Scholar]
- 41.Tan H, Rader AJ. Identification of putative, stable binding regions through flexibility analysis of HIV-1 gp120. Proteins. 2009 doi: 10.1002/prot.22196. [DOI] [PubMed] [Google Scholar]
- 42.Abrams CF, Vanden-Eijnden E. Large-scale conformational sampling of proteins using temperature-accelerated molecular dynamics. Proc Natl Acad Sci U S A. 2010;107(11):4961–4966. doi: 10.1073/pnas.0914540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobs DJ, Rader AJ, Kuhn LA, Thorpe MF. Protein flexibility predictions using graph theory. Proteins. 2001;44:150–165. doi: 10.1002/prot.1081. [DOI] [PubMed] [Google Scholar]
- 44.Bahar I, Atilgan AR, Demirel MC, Erman B. Vibrational dynamics of folded proteins: significance of slow and fast modes in relation to function and stability. Phys Rev Letters. 1998;80:2733–2736. [Google Scholar]
- 45.Haliloglu T, Bahar I, Erman B. Gaussian dynamics of folded proteins. Phys Rev Letters. 1997;79:3090–3093. [Google Scholar]
- 46.Bahar I, Wallqvist DG, Covell DG, Jernigan RL. Correlation between native-state hydrogen exchange and cooperative residue fluctuations from a simple model. Biochemistry. 1998;37:1067–1075. doi: 10.1021/bi9720641. [DOI] [PubMed] [Google Scholar]
- 47.Bahar I, Jernigan RL. Vibrational dynamics of transfer RNAs: a comparison of the free synthetase bound forms. J Mol Biol. 1998;281:871–884. doi: 10.1006/jmbi.1998.1978. [DOI] [PubMed] [Google Scholar]
- 48.Yang LW, Bahar I. Coupling between catalytic site and collective Dynamics: A requirement for mechanochemical activity of Enzymes. Structure. 2005;13:893–904. doi: 10.1016/j.str.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tama F, Wriggers W, Brooks CL., 3rd Exploring global distortions of biological macromolecules and assemblies from low-resolution structural information and elastic network theory. J Mol Biol. 2002;321(2):297–305. doi: 10.1016/s0022-2836(02)00627-7. [DOI] [PubMed] [Google Scholar]
- 50.Chacon P, Tama F, Wriggers W. Mega-Dalton biomolecular motion captured from electron microscopy reconstructions. J Mol Biol. 2003;326(2):485–492. doi: 10.1016/s0022-2836(02)01426-2. [DOI] [PubMed] [Google Scholar]
- 51.Chennubhotla C, Bahar I. Signal propagation in proteins and relation to equilibrium fluctuations. PLoS Comput Biol. 2007;3(9):1716–1726. doi: 10.1371/journal.pcbi.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halle B. Flexibility and packing in proteins. Proc Nat Acad Sci. 2002;99(3):1274–1279. doi: 10.1073/pnas.032522499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinsen K. Structural flexibility in proteins: impact of the crystal environment. Bioinformatics. 2008;24(4):521–528. doi: 10.1093/bioinformatics/btm625. [DOI] [PubMed] [Google Scholar]
- 54.Eyal E, Yang LW, Bahar I. Anisotropic network model: systematic evaluation and a new web interface. Bioinformatics. 2006;22(21):2619–2627. doi: 10.1093/bioinformatics/btl448. [DOI] [PubMed] [Google Scholar]
- 55.Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctations in protein, using a single parameter harmonic potentia. Fold & Des. 1997;2:173–181. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 56.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olshevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J Virol. 1990;64(12):5701–5707. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cordonnier A, Montagnier L, Emerman M. Single amino-acid changes in HIV envelope affect viral tropism and receptor binding. Nature. 1989;340:571–574. doi: 10.1038/340571a0. [DOI] [PubMed] [Google Scholar]
- 59.VanDerSpoel E, Lindahl B, Hess G, Groenhof AM, Berendsen H. GROMACS Fast Flexible and Free. J Comp Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 60.Scott WRP, Hunenberger PH, Tironi IG, Mark AE, Billeter SR, Fennen J, Torda AE, Huber T, Kruger P, van Gunsteren WF. The GROMOS biomolecular simulation program package. J Phys Chem A. 1999;103:3596–3607. [Google Scholar]
- 61.Morra G, Verkhivker C, Colombo G. Modeling singal propogation mechanism and ligand-based conformational dynamics of the Hsp90 molecular chaperone full-length dimer. PLoS Comp Biol. 2009;5:e1000323. doi: 10.1371/journal.pcbi.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiang SH, Finzi A, Pacheco B, Alexander K, Yuan W, Rizzuto C, Huang CC, Kwong PD, Sodroski J. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J Virol. 2010;84(7):3147–3161. doi: 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amadei A, Linssen A, Berendsen H. Essential dynamics of proteins. Proteins. 1993;17:412–415. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 64.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:w109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280(5371):1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 66.Haliloglu T, Ben-Tal N. Cooperative transition between open and closed conformations in potassium channels. PLoS Comp Biol. 2008;41000164(e1000164) doi: 10.1371/journal.pcbi.1000164. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Potestio R, Pontiggia F, Michelett C. Coarse-Grained Description of Protein Internal Dynamics: An Optimal Strategy for Decomposing Proteins in Rigid Subunits. Biophysical Journal. 2009;96:4993–5002. doi: 10.1016/j.bpj.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol. 1999;285(4):1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 69.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 70.MOE Molecular Operating Environment Chemical Computing Group; Montreal Canada: http//www.chemcomp.com/ [Google Scholar]
- 71.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;117:11225–11236. [Google Scholar]
- 72.Tirion MM. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys Rev Letters. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.