Abstract
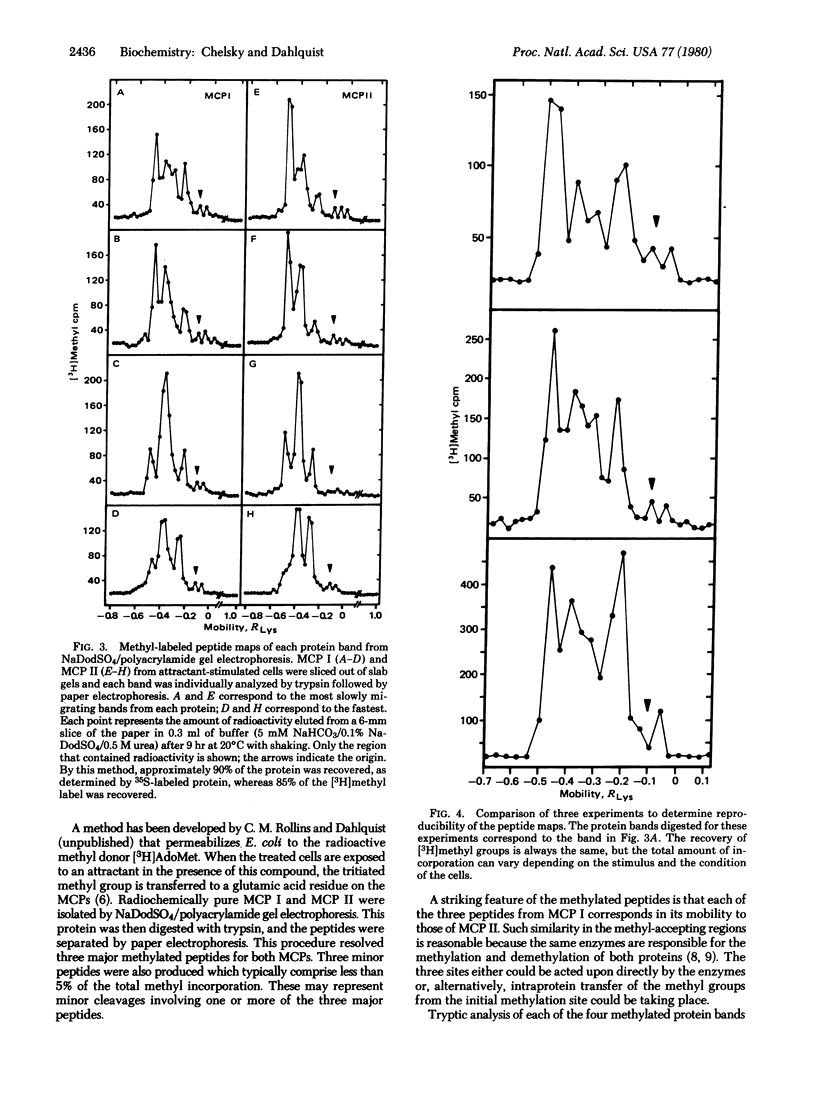
Two-dimensional analysis of tryptic peptides from [35S]methionine-labeled methyl-accepting chemotaxis proteins, MCP I and MCP II, demonstrates a high degree of homology between the two proteins. After the methylation sites were labeled with S-adenosyl-L-methyl-3H]methionine, peptides of three distinct migrations in each protein were found to carry a methyl group. These multiple methylations appear to be responsible in part for the observed multiple banding patterns on sodium dodecyl sulfate/polyacrylamide slab gels.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler J. Chemotaxis in bacteria. Science. 1966 Aug 12;153(3737):708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- DeFranco A. L., Koshland D. E., Jr Multiple methylation in processing of sensory signals during bacterial chemotaxis. Proc Natl Acad Sci U S A. 1980 May;77(5):2429–2433. doi: 10.1073/pnas.77.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Koiwai O., Kozuka M. Studies on bacterial chemotaxis. II. Effect of cheB and cheZ mutations on the methylation of methyl-accepting chemotaxis protein of Escherichia coli. J Biochem. 1979 May;85(5):1213–1223. [PubMed] [Google Scholar]
- Jaskunas S. R., Burgess R. R., Nomura M. Identification of a gene for the alpha-subunit of RNA polymerase at the str-spc region of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1975 Dec;72(12):5036–5040. doi: 10.1073/pnas.72.12.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H., Ball C. B., Adler J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kort E. N., Goy M. F., Larsen S. H., Adler J. Methylation of a membrane protein involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3939–3943. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maley F., Guarino D. U. Differential binding of sodium dodecyl sulfate to amino acids as evidenced by elution from Sephadex. Biochem Biophys Res Commun. 1977 Aug 22;77(4):1425–1430. doi: 10.1016/s0006-291x(77)80138-1. [DOI] [PubMed] [Google Scholar]
- Mattice W. L., Riser J. M., Clark D. S. Conformational properties of the complexes formed by proteins and sodium dodecyl sulfate. Biochemistry. 1976 Sep 21;15(19):4264–4272. doi: 10.1021/bi00664a020. [DOI] [PubMed] [Google Scholar]
- Noel D., Nikaido K., Ames G. F. A single amino acid substitution in a histidine-transport protein drastically alters its mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry. 1979 Sep 18;18(19):4159–4165. doi: 10.1021/bi00586a017. [DOI] [PubMed] [Google Scholar]
- Oishi M., Smith C. L. Inactivation of phage repressor in a permeable cell system: role of recBC DNase in induction. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3569–3573. doi: 10.1073/pnas.75.8.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Chemotaxis in Escherichia coli: methylation of che gene products. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3317–3321. doi: 10.1073/pnas.74.8.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Identification of polypeptides necessary for chemotaxis in Escherichia coli. J Bacteriol. 1977 Jun;130(3):1317–1325. doi: 10.1128/jb.130.3.1317-1325.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer W. R., Koshland D. E., Jr Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci U S A. 1977 Feb;74(2):533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Koshland D. E., Jr A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Werf P., Koshland D. E., Jr Identification of a gamma-glutamyl methyl ester in bacterial membrane protein involved in chemotaxis. J Biol Chem. 1977 Apr 25;252(8):2793–2795. [PubMed] [Google Scholar]