Abstract
Peptide sequencing by computational assignment of tandem mass spectra to a database of putative protein sequences provides an independent approach to confirming or refuting protein predictions based on large-scale DNA and RNA sequencing efforts. This use of mass spectrometrically-derived sequence data for testing and refining predicted gene models has been termed proteogenomics. We report herein the application of proteogenomic methodology to a database of 10.9 million tandem mass spectra collected over a period of two years from proteolytically generated peptides isolated from the model legume Medicago truncatula. These spectra were searched against a database of predicted M. truncatula protein sequences generated from public databases, in silico gene model predictions, and a whole-genome six-frame translation. This search identified 78,647 distinct peptide sequences, and a comparison with the publicly available proteome from the recently published M. truncatula genome supported translation of 9,843 existing gene models and identified 1,568 novel peptides suggesting corrections or additions to the current annotations. Each supporting and novel peptide was independently validated using mRNA-derived deep sequencing coverage and an overall correlation of 93% between the two data types was observed. We have additionally highlighted examples of several aspects of structural annotation for which tandem MS provides unique evidence not easily obtainable through typical DNA or RNA sequencing. Proteogenomic analysis is a valuable and unique source of information for the structural annotation of genomes and should be included in such efforts to ensure that the genome models used by biologists mirror as accurately as possible what is present in the cell.
Many analyses in systems biology rely on an annotated genomic sequence as a starting point, and the quality of the genome sequence and annotation directly affects the reliability of the resulting conclusions. Improving the accuracy of the structural and functional annotation should therefore be a major focus in the study of any model organism, and many sources of data are available which can be used to assist in this effort. Common sources of experimental evidence used to improve in silico gene model predictions include the sequences of full-length cDNA clones and expressed sequence tag (EST)1 libraries, alignment of homologous sequences from related organisms, and, more recently, the deep sequencing of mRNA-derived cDNA libraries using next-generation platforms (RNA-Seq). The use of information from these sources can significantly improve the results of automated gene-calling efforts, but all operate at the transcript level and are unable to differentiate between coding and non-coding sequences. The field of proteogenomics has recently emerged in response to this perceived gap. Broadly defined, proteogenomics is the use of proteomics data and methodology to assist in the annotation of genome sequences. This typically involves the “sequencing” of an organism's proteome using tandem mass spectrometry (MS/MS) with a greatly expanded search database consisting of published protein sequences, possible splice variants, and a six-frame translation of the entire genome. The identified peptide sequences are then mapped back to the genome, and these peptide/genome mappings are used to confirm, refute, or add to existing gene annotations. They can also be included directly in the annotation pipeline alongside other sources of evidence. Proteogenomics, along with other recent developments such as ribosome profiling (1, 2), can thus provide an additional layer of information to assist in delineating transcript coding regions and reading frames.
Recently the draft sequence of the Medicago truncatula genome was released (3). M. truncatula, a relative of the important agricultural crop alfalfa, serves as a model organism for the legume family and is the focus of much research to understand the mechanisms of symbiosis between the plant and soil microbes that result in fixation of atmospheric nitrogen. Although publication of the draft sequence is an important step forward for Medicago researchers, efforts to improve the genomic assembly and structural and functional annotations are ongoing. To assess the quality of the published annotations and establish an independent source for improving them, we have evaluated the use of existing MS/MS data to confirm or correct current gene models and discover possible novel, unannotated genes in the M. truncatula genome. Similar work performed in other sequenced organisms (4–9) has shown the potential for this type of analysis, and proteogenomic data for the model organism Arabidopsis is being incorporated directly into the structural annotation process (10). MS/MS data can confirm expression of current gene models, help to correct errors in splice sites and reading frames, suggest missing exons and alternative splicing, and provide evidence for novel genes missing from the current annotations. We used a database of 10.9 million MS/MS spectra generated from ongoing proteomic and phosphoproteomic studies to test the utility of this approach in the model legume. Although the vast majority of identified peptides supported existing gene models, there is evidence for the need for further work to improve the Medicago annotations. Conclusions based on mapped peptide evidence were independently validated using a database of 341 million RNA-Seq reads taken from ongoing transcriptomics experiments. The results show the validity of the use of MS/MS data to improve the quality of existing structural annotations, particularly in cases in which peptide data provides evidence not derivable from other sources. In practice, all available sources of information (MS/MS, RNA-Seq, EST databases, etc) should be used simultaneously to guide the construction of accurate gene models both by automated gene calling and, where feasible, by manual curation.
EXPERIMENTAL PROCEDURES
Sample Preparation and MS/MS
The data used in this study were generated from tissue of M. truncatula 'Jemalong A17′ wild type as well as C31 and TRV25 mutants in 11 different experiments using multiple growth conditions, treatments and protein isolation procedures. Aeroponic and hydroponic plants were grown as described previously (11, 12). Additionally, seedlings were sown on 23 × 23 cm2 plates containing modified Fahraeus medium overlaid with moist sterile germination paper and grown at room temperature in the dark for 5 days. All plants were treated for one hour by replacement of the medium with modified Fahraeus medium with and without 10−8 m Nod factors obtained from Sinorhizobium meliloti strain Rm1021 pRmE43 (pTE3:nodD1) as described in (12). Seedlings were harvested after one hour of treatment and either flash frozen in liquid nitrogen or processed using two-phase isolation of membrane fractions as outlined in (13).
Proteins were isolated for MS/MS analysis from whole-cell lysates of flash-frozen root tissue or membrane-enriched fractions with the addition of a variety of phosphatase inhibitors as described previously (37). Protein samples were reduced with DTT at a final concentration of 5 mm and alkylated with 15 mm iodoacetamide before final capping with 5 mm DTT. Proteins were digested with trypsin, derivitized with isobaric labels (TMT 6-plex, iTRAQ 4-plex, or iTRAQ 8-plex according to experiment) (14, 15) and fractionated by strong cation exchange (SCX). For phosphorylation experiments, samples were enriched for the presence of a phosphate group by IMAC chromatography. All samples were analyzed on a LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific). For mRNA sequencing, seedlings from ‘Jemalong A17’ wildtype grown using the plate system were used and root tissue was excised and flash frozen. RNA was isolated using a Qiagen RNeasy Plant Mini kit. Sequencing libraries were prepared using the Illumina TruSeq RNA Sample Preparation Kit (mRNA protocol rev. A) and sequenced on an Illumina HiSeq 2000 system.
Database Generation, Searching, and False Discovery Rate Estimation
The database of protein sequences used for spectral searching was generated from several sources. Protein sequences from the published version of the M. truncatula genome annotations (Mt3.5v4) were downloaded from the JCVI Medicago FTP server (ftp://ftp.jcvi.org/pub/data/m_truncatula/Mt3.5/Mt3.5v4/). The Mt3.5 genome sequence from the same site was used to generate a six-frame translation of the entire genome, discarding open readings frames of fewer than 30 putative amino acids (a.a.) between stop codons to minimize the database size. Lastly, the gene-finding program AUGUSTUS (16) was used to generate a de novo gene model prediction for the Mt3.5 genome based on Arabidopsis training parameters. Parameters for AUGUSTUS were set to be liberal in intron/exon prediction and to report all possible predicted splice variants for each gene model to maximize the search space for discovering novel peptides, as described in Castellana et al. (4). The three sets of predicted protein sequences were reduced to a non-redundant database using in-house software to remove duplicate protein entries. Redundant proteins were determined by comparing amino acid sequences (ignoring I/L ambiguity) for an exact match against all other proteins. A decoy database of reversed sequences was added for the purpose of false discovery rate (FDR) estimation as described previously (17).
Database searching was performed using the Coon OMSSA Proteomic Analysis Software Suite (COMPASS) (18) and using the Open Mass Spectrometry Search Algorithm (OMSSA) version 2.1.8 (19). Proteins were digested in silico by OMSSA using tryptic cleavage specificity. Peptide precursors were searched using a multi-isotopic search (±50 ppm, max 4 isotopes) and product ion mass tolerance was set to ±0.015 Da. Carbamidomethylation of cysteines, isobaric labeling (TMT or iTRAQ) on the N terminus, and isobaric labeling on lysines were included as fixed modifications. Oxidation of methionines and isobaric labeling on tyrosines were included as variable modifications. For all phosphorylation experiments, variable modifications of phosphorylation on threonines, serines, and tyrosines were applied. Results were filtered to a 1% peptide FDR based on decoy database matches using the high resolution tool FDROptimizer from the COMPASS suite. Peptides were further grouped into protein groups and filtered to a 1% protein FDR based on the product of included peptide p values. The highest (worst) p value was used when peptides were observed in multiple spectra. Only peptides belonging to filtered protein groups were used in further analysis.
Peptide Mapping and Analysis
The full list of spectral matches was collapsed to a set of unique peptide sequences. Each peptide was mapped back to its genomic location(s) using a combination of database/peptide and database/genome coordinate tables, taking into account split peptides spanning splice junctions and Leu/Ile ambiguity. This generated a set of expressed peptide tags (EPTs) as originally defined by Savidor et al. (6). Each EPT was subsequently classified by a number of non-exclusive criteria using a combination of the bedtools software package (20), in-house scripts, and manual inspection using the Integrative Genomics Viewer (IGV) (21). Intergenic clusters were generated by single-linkage clustering using bedtools with a distance cutoff of 1500 bp. AUGUSTUS was used to search for alternative gene models for intragenic nEPT loci and for novel gene models at intergenic nEPT clusters, allowing for multiple alternative transcripts per model with a sampling of 100 and using EPT-based hints to guide prediction.
RNA-Seq Validation
A total of 340,622,166 100bp single-end reads from Illumina HiSeq2000 sequencing of M. truncatula ‘Jemalong A17’ poly-A enriched mRNA were mapped to the Mt3.5 genome using Tophat v1.3.2 (22). Parameters for minimum intron size, maximum intron size, and microexon searching were set to 10, 20000, and TRUE, respectively. All other parameters used default settings. A total of 269,109,482 reads were successful mapped to the genome. These alignments were used in both automated validation using bedtools and in-house software as well as visual validation using IGV.
RESULTS
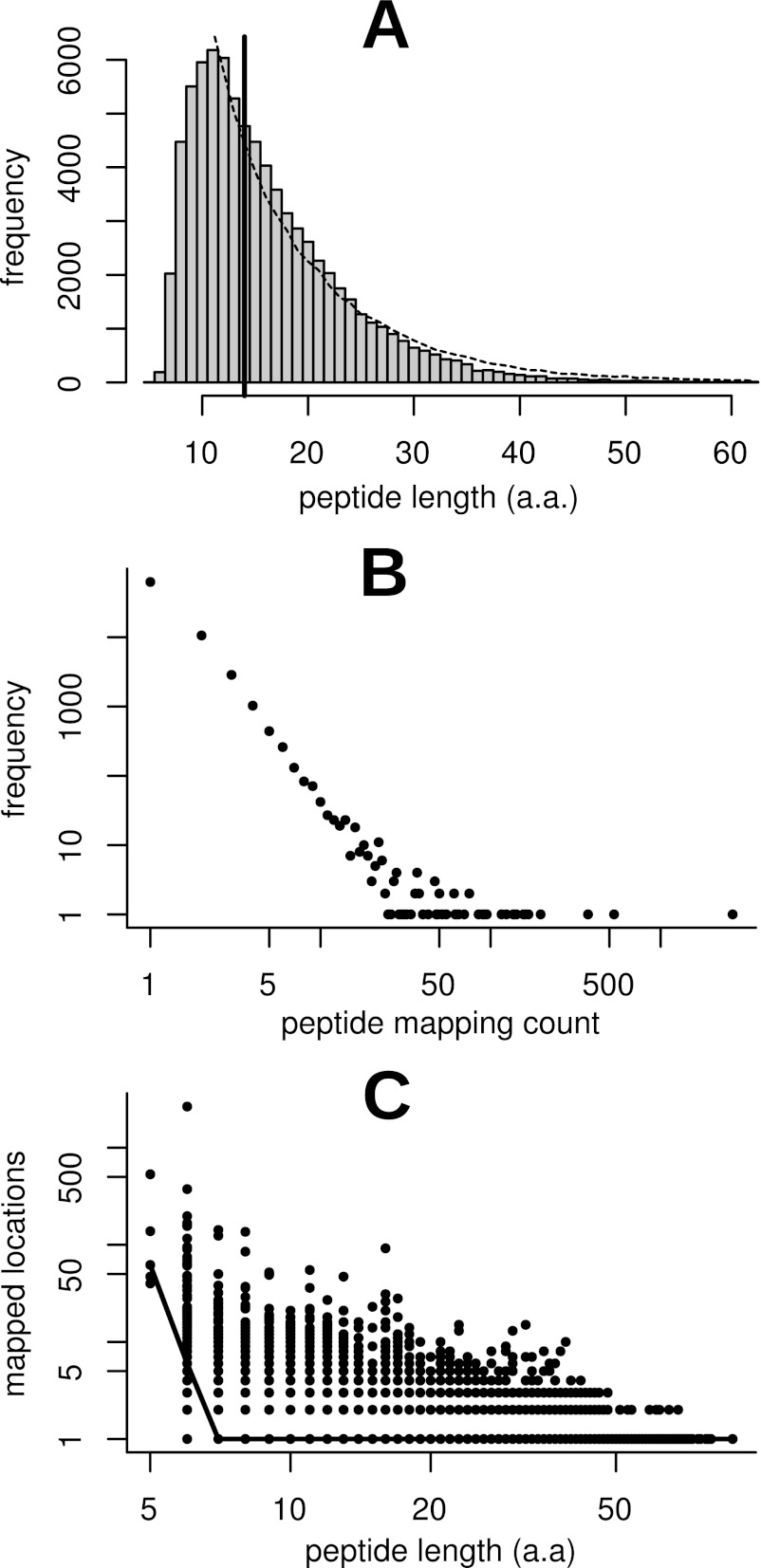
Spectral searching against our M. truncatula protein databases resulted in 1.7 million spectral matches at a 1% protein FDR (supplemental Table S1), representing 78,647 unique peptide sequences. Of these, 78,558 unique peptide sequences were mapped to 112,720 locations on the genome with no mismatches allowed. We refer to these unique peptide sequences as expressed peptide tags or EPTs, as suggested by Savidor et al. (6). The parallel to the terminology of ESTs emphasizes that, for the purposes of proteogenomics, EPTs occupy genome coordinate space. Of these EPTs, 62,802 mapped to unique locations, 10,608 mapped to two locations and 5,148 mapped to three or more locations. The median length of identified EPT sequences was 14 a.a, with a minimum length of 5 a.a. and a maximum of 89 a.a. (Fig. 1A). Although the overwhelming majority of peptides mapped to only one location, a comparison of length against mapping count for each EPT showed distinct non-specificity for short peptides (Fig. 1B, C). The median mapping count for 5 a.a. EPTs is 62 locations, with no 5 a.a. EPT mapping to fewer than 40 locations. The median mapping count for 6 a.a. EPTs is 6, whereas at a length of 7 a.a the median falls to one. Based on this analysis, and to minimize the number of spurious mappings resulting from the probability of any given short peptide occurring randomly in a six-frame translation of the genome, we discarded from consideration any EPTs shorter than 7 a.a. This reduced the number of unique EPTs by 0.2% to 78,362 while reducing the number of mapped locations by 6.0% to 105,973 (Table I). A similar threshold has been used by others in proteogenomic studies (23). EPTs above this threshold were further classified and characterized as described in the following sections and are summarized in Table I.
Fig. 1.
Statistical analysis of peptide length and mapping occurrence distributions. Histogram (A) depicts frequency of unique peptide length in the dataset. The median peptide length was 14 a.a., as indicated by the solid vertical bar. The minimum length was 5 a.a. and the maximum was 89 a.a. The dashed line represents the median-normalized frequency polygon of an in silico tryptic digest of annotated proteins and correlates well with the observed distribution. The histogram is clipped on the right at the 999th permille for easier viewing. Plot (B) depicts the number of mapped locations per peptide versus frequency. The minimum and median number of mapped locations was one and the maximum was 2,649. Both axes are plotted on a log scale to facilitate viewing. Plot (C) depicts peptide length versus the number of genomic locations the peptide mapped to. Each point represents a unique sequence in the database, and the solid line connects median mapping counts for each peptide length. It is clear that very short peptides map to a large number of locations in the genome and hence are unreliable as indicators of expression at a locus. Based on this analysis we settled on a minimum length cutoff of 7 a.a., the point at which the median number of mapped locations drops to one.
Table I. Summary of the mapping of a database of 10.9 m MS/MS spectra against the M. truncatula genome sequence.
All spectral matches were considered and mapped to locations on the Mt3.5v4 genome sequence if possible. After filtering out peptides < 7 aa in length, the remaining EPTs were classified into initial categories as shown. Unique EPT counts represent unique sequences, while location counts represent unique loci on the genome to which EPTs map. Intragenic EPTs are considered to be those which have any overlap with current gene models, including coding sequences, introns, and untranslated regions.
| Initial category | Unique EPTs | Locations | Unspliced locations | Spliced locations |
|---|---|---|---|---|
| Mapped to Mt3.5 genome | 78,362 | 105,973 | 90,393 | 15,580 |
| Support Mt3.5v4 gene models | 76,505 | 95,633 | 80,541 | 15,092 |
| Explained by Mt3.5v4 TE models | 289 | 304 | 276 | 28 |
| Locations ignored due to gene model match | NA | 8,305 | 7,954 | 351 |
| Novel peptides | 1,568 | 1,731 | 1,622 | 109 |
| Category within novel peptides | ||||
| Intergenic | 1,060 | 1,134 | 1118 | 16 |
| Intragenic | 552 | 597 | 504 | 93 |
Supporting Peptides
The Mt3.5v4 release of the M. truncatula gene models contains 64,152 predicted protein-coding models, although many of the models located in short-read sequencing scaffolds are expected to be partial fragments of the same genes because of the relatively small size of the assembled contigs (Illumina N50 = 2364; N80 = 1095). A total of 76,505 EPTs were mapped to 95,633 locations that were in agreement with the published gene models of Mt3.5v4. These supporting EPTs (sEPTs) provide evidence for translation of the existing gene models. Of the 64,152 current Mt3.5v4 gene models, 160 were covered by sEPTs over > 80% of their coding sequence length, 2205 had > 50% sEPT coverage, and 15,541 contained at least one sEPT as evidence of translation (Table II). A more stringent evaluation considering only gene models containing two or more sEPTs with at least one uniquely mapped at that genomic locus provides evidence for the translation of 9,843 gene models (supplemental Table S2).
Table II. Summary of supporting evidence for existing gene models.
For each annotated gene model, the fraction of transcript length covered by at least one EPT at each base position was calculated using bedtools. R (38) was used to transform the resulting coverage counts into a cumulative frequency table. Fractions listed represent the lower edge of bins, exclusive. The total number of predicted gene models is likely to be artificially high due to splitting of genes over multiple small short read contigs.
| Fraction of transcript covered | Cumulative abs. frequency | Cumulative percent |
|---|---|---|
| 0.9 | 18 | 0.03 |
| 0.8 | 160 | 0.25 |
| 0.7 | 533 | 0.83 |
| 0.6 | 1261 | 1.97 |
| 0.5 | 2205 | 3.44 |
| 0.4 | 3475 | 5.42 |
| 0.3 | 5031 | 7.84 |
| 0.2 | 7164 | 11.17 |
| 0.1 | 10436 | 16.27 |
| >0.0 | 15541 | 24.23 |
| All | 64152 | 100.00 |
N-Terminal Modification
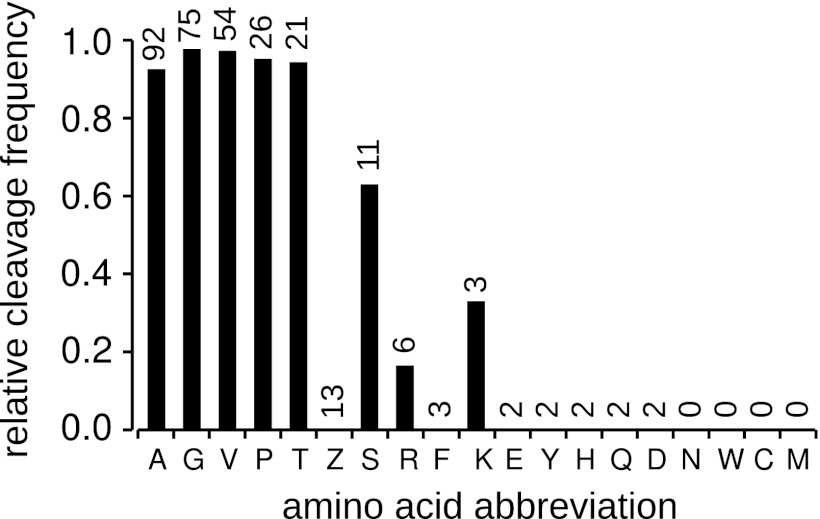
To take advantage of the unique nature of MS/MS data as applied to structural annotation, we analyzed the sEPT data set to look for evidence of N-terminal methionine excision (NME). A total of 352 sEPTs were identified as being N-terminal (starting at position 1 or 2 of the protein sequence). Thirty-eight of these were filtered out as being contained within longer peptides in the full set of EPTs, suggesting possible degradation. Of the remaining sEPTs, 47 mapped to position +1 of the protein sequence and 267 mapped to position +2 (evidence of N-terminal methionine excision). This suggests a cleavage rate of 85%, in agreement with research in other organisms finding that the majority of both cytoplasmic and organellar proteins undergo NME in plants as well as other eukaryotes and prokaryotes (24–27). The frequency of occurrence of each of the 19 possible amino acids (Leu and Ile are indistinguishable in MS/MS) was determined at the +1 position in cleaved and +2 position in uncleaved N-terminal sEPTs (Fig. 2). The activity of the MAP peptidases involved in NME is thought to be specific to small second position amino acids, typically one of [GAPCSTV] (24, 25, 28). Our results show a high MAP specificity for [GAPTV] in the +2 position, all of which result in > 93% cleavage frequency. Serine showed a slightly lower frequency of cleavage, and cysteine was never observed at the +1 position in any of the identified N-terminal sEPTs. It should be noted that another common post-translation modification, N-alpha-acetylation (NAA), has also recently been shown to occur in a large portion of the proteome of Arabidopsis (27). We did not specifically search for acetylation modifications, so it is possible that the ratio of cleaved to non-cleaved termini could be affected by the lack of acetylated identifications if acetylation occurs more or less frequently in cleaved versus non-cleaved proteins.
Fig. 2.
Relative frequency of N-terminal methionine cleavage for each of the 19 possible amino acids in the +2 position. Frequencies are calculated as the number of observed cleavage events for the amino acid divided by the sum of the observed cleaved and uncleaved termini with that amino acid in the +2 position [cleaved/(cleaved + uncleaved)]. Numbers above the bars are the total number of observed termini with this amino acid in the +2 position for uncleaved or +1 position for cleaved proteins. The amino acid code “Z” represents either Leu or Ile, which are indistinguishable in the technology used. In general the trends agree with research published on the phenomenon in other organisms, with cleavage occurring in the presence of small amino acids in the +2 position.
Intragenic Novel Peptides
The remainder of the analysis dealt with so-called “novel” EPTs (nEPTs) - peptides that could not be explained by any existing gene models. Of the 1,568 novel peptides identified, 552 were intragenic (i.e., overlapped existing gene models, including introns and UTRs) at 597 locations in the genome. These were classified according to a number of criteria to roughly quantify the types of evidence they provide. A total of 79 spliced nEPTs were identified which suggested novel splice junctions at 75 genomic locations. A visual inspection of these locations indicated that 64 were supported by RNA-Seq alignments as being the only or predominant splice form, suggesting corrections to the gene models. These were classified as corrections to donor sites (n = 14), acceptor sites (n = 15), both donor and acceptor sites (n = 4), extraneous exons/introns (n = 19), missing exons/introns (n = 11), and incorrectly split genes (n = 1). Seven sites were covered by EPTs with both annotated and novel splice forms, giving strong evidence for alternative splicing at these locations. Five of these nEPTs were the minor form based on spectral match counts and two were major forms. One additional nEPT was suggested to be a minor splice form by RNA-Seq alignments alone, and three novel splice sites were unsupported by RNA-Seq data. Overall, of the 75 novel splice junctions, 71 (95%) were supported by 10 or more (median = 500) spliced mRNA reads with identical donor and acceptor sites, indicating a high degree of correlation between EPT and RNA-Seq evidence for splice site correction.
The 504 unspliced intragenic nEPT locations were classified as either completely within exons (33%), completely within introns (20%), or overlapping exon boundaries (47%). These locations were further examined by visual inspection alongside RNA-Seq alignments to assign putative explanatory categories (Table III). A total of 201 locations suggested splice corrections. Of these, 86 suggested removal of extra introns at the 3′ end, one of the more consistent patterns observed in the nEPT data. Frameshifts resulting from small insertions or deletions (indels) in the genomic sequence that are clearly visible in the RNA-Seq alignments accounted for another 56 intragenic nEPT locations. These 1–2 bp indels generally resulted in either a truncation of the gene model or the insertion of an erroneous intron to correct for the frameshift (Fig. 3). Although nEPT data itself cannot confirm the presence of these errors, it can provide strong supporting evidence for possible mistakes identified using more direct evidence such as DNA or RNA sequencing. An additional 99 nEPT locations overlapping the ends of gene models located near the ends of short genomic contigs are assumed to be because of the incomplete nature of the gene models and would likely be resolved with an improved genome assembly. The remainder of the unspliced intragenic nEPTs were classified into a number of smaller categories, including alternate ATG usage, strand/frame disagreements, gene fusion corrections, and possible noncanonical start codon usage as described below.
Table III. Classification of unspliced intragenic nEPTs.
Each nEPT location was classified by visual inspection of mapped EPT on the genome alongside the published gene models, six reading frame translations, and RNA-Seq read alignments using IGV. EPTs were grouped into common categories based on the most likely type of addition or correction suggested.
| Category | Count |
|---|---|
| Other splice fix | 115 |
| Missing gene end | 99 |
| Splice fix at 3′ exon | 86 |
| Insertion/deletion | 56 |
| Different strand/frame | 53 |
| Alternative splicing | 18 |
| Retained intron | 15 |
| Gene fusion correction | 11 |
| Alternate start codon | 11 |
| Noncanonical start codon | 7 |
| Other/unexplained | 33 |
| Total: | 504 |
Fig. 3.
Insertion at locus AC225449_6.1 of M. truncatula. Shown are length of depicted region (in base pairs), contig ID, location of mapped EPTs, the Mt3.5v4 gene model and 3-frame translation, and depth of RNA-Seq coverage (scale in reads per bp shown at left). An apparent 1-bp insertion in the genomic sequence causes the gene caller to insert an erroneous intron to correct for the frameshift. EPT_046967 (8 PSMs) overlapping the intron justified a closer look at the genomic locus, and RNA-Seq data of ∼1700-fold coverage clearly showed an insertion in the genomic sequence at the location of the intron (black arrow). Such indels were observed numerous times in both BAC-based pseudomolecule constructs as well as short read contigs.
Gene Model Refinement Based on Intragenic nEPTs
AUGUSTUS was used to search for refined gene models for each intragenic nEPT locus. A region of 10,000 bp on either side of each intragenic nEPT was defined, and overlapping regions were merged using bedtools. Each region was searched using AUGUSTUS for all predicted transcript variants. Each intragenic nEPT was re-mapped to the resulting protein sequences and classified as either explained by or not explained by these predicted models, and a minimal set of predicted gene models explaining all possible nEPTs was generated. Of the 552 intragenic nEPT locations initially identified, 390 were explained by a minimal set of 293 refined gene models.
Noncanonical Translation Initiation
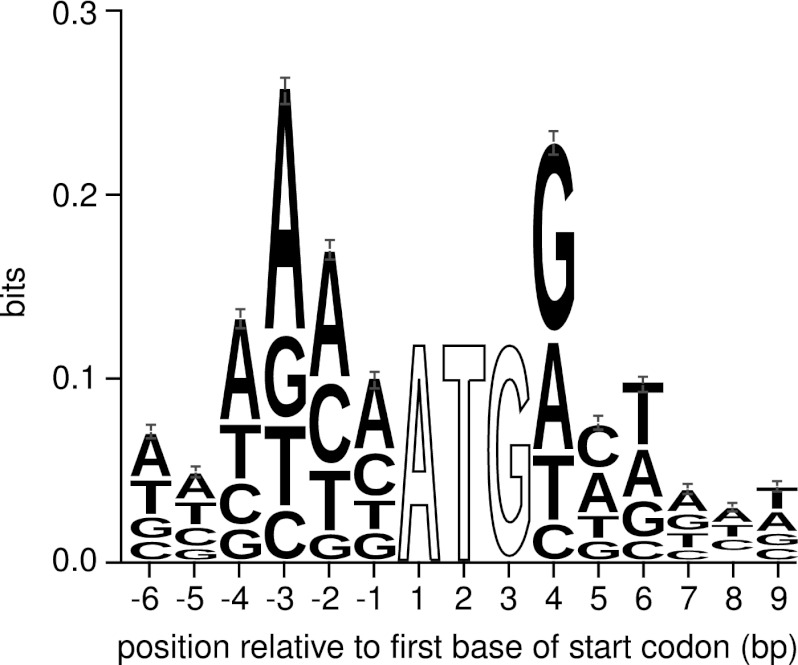
The recurring identification of nEPTs with no upstream in-frame canonical (ATG) start codons led us to investigate the possibility of noncanonical start codon usage in M. truncatula. Non-canonical start codon usage, or the use of a codon other than ATG for translation initiation, is not uncommon in prokaryotes, occurring in ∼17% of Escherichia coli genes (29). Evidence for non-canonical usage has also been found in eukaryotic organisms for a small number of genes (30–35), and in silico homology-based analysis can be used to identify further potential candidates (36). The emerging technique of ribosome profiling has provided further evidence that the phenomenon may be more common in eukaryotes than previously thought (1, 2). Non-canonical start codons in other eukaryotic organisms tend to have a single base difference from the canonical ATG codon, as well as having optimal or near-optimal surrounding sequence context. For example, in plants there is an increased frequency of A or G at the −3 position and G at the +4 position (34). Analysis of the surrounding sequence context of annotated translation start sites in M. truncatula shows a similar trend (Fig. 4). This information allows for a qualitative analysis of the likelihood of a given non-canonical codon being used for translation initiation when EPT evidence suggests such an event.
Fig. 4.
Sequence logo of the context surrounding the annotated start codons of Mt3.5v4 gene models. Only gene models from the pseudomolecule assemblies were used to generate the logo, because an abundance of partial gene models in the short read contigs may add artificial noise to any possible patterns. We observed a strong preference for adenine in the −3 position and guanine in the +4 position, in agreement with other plant species. Positions 1–3 were 100% conserved - they are not shown to scale. Logo was generated using WebLogo 3.2 (39) from a TRANSFAC motif file generated in-house.
After removing from consideration loci with clear RNA-Seq evidence for missing 5′ exons, as well as loci within 5000 bp of the 5′ end of a contig (which may also have missing 5′ exons) and loci without any supporting RNA-Seq evidence, we evaluated 7 loci for the potential use of non-canonical start codons. To be considered as a possible start site, a codon must have both optimal nucleic acid residues at positions −3 and +4 and have no more than a one base difference from the canonical ATG codon. Of the 7 loci considered, 5 contained upstream in-frame codons satisfying these criteria. Highlighted in Figs. 5 and 6 are two examples with particularly strong supporting evidence based on homology to other published work. As the N termini of proteins often direct cellular localization, it is important that gene models contain accurate coding sequence boundaries, and proteogenomics is one of only a handful of tools able to provide evidence for possible mistakes in start codon annotation. It should be noted, however, that because of the low expected frequency of noncanonical usage events compared with the inherent 1% error rate in peptide assignments, evaluation of such EPT evidence is of more use in manual curation than in automated gene calling pipelines and would require additional supporting evidence such as provided for the two examples given.
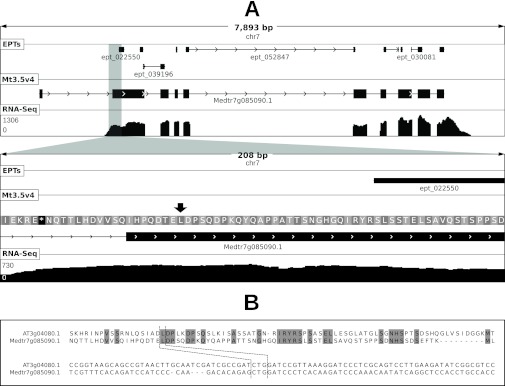
Fig. 5.
Noncanonical start codon usage at locus Medtr7g085090 of M. truncatula. Panel (A) shows a graphical depiction of the locus at two levels of resolution. Shown in each sub-panel are length of depicted region (in base pairs), chromosome ID, location of mapped EPTs, the Mt3.5v4 gene model and frame translation (in secondary panel), and depth of RNA-Seq coverage (scale in reads per bp shown at left). RNA-Seq evidence indicates that the first exon in the gene model is erroneous. EPT_022550 (11 PSMs) maps to a genomic location upstream of the first in-frame start codon. The proposed true noncanonical start codon is shown by a black arrow. Panel (B) shows a multiple alignment of the protein and nucleotide sequences of the region surrounding the proposed true start codon in M. truncatula and that of the homolog in Arabidopsis, which has been shown to use the noncanonical codon highlighted by dashed lines to initiate translation (34).
Fig. 6.
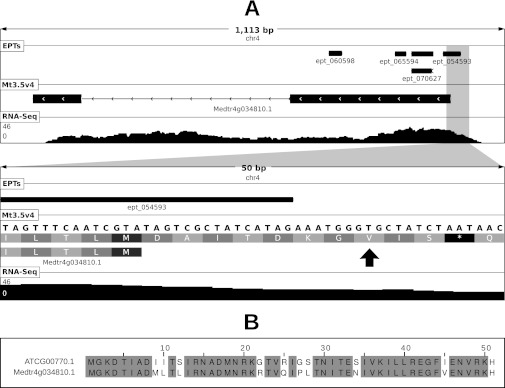
Noncanonical start codon usage at locus Medtr4g034810 of M. truncatula. Panel (A) shows a graphical depiction of the locus at two levels of resolution. Shown in each sub-panel are length of depicted region (in base pairs), chromosome ID, location of mapped EPTs, the Mt3.5v4 gene model and frame translation (in secondary panel), and depth of RNA-Seq coverage (scale in reads per bp shown at left). EPT_054593 (9 PSMs) maps to a genomic location upstream of the annotated start codon. Note the optimal sequence context at positions −3 and +4 relative to the proposed start codon. Panel (B) shows a multiple alignment of the protein sequences between the putative ribosomal protein S8 encoded by Medtr4g034810.1 (using the proposed noncanonical start codon) and its closest homolog in Arabidopsis which uses a canonical ATG start codon.
Intergenic Novel Peptides
The remaining 1,060 novel EPTs mapped to 1,134 intergenic locations on the Mt3.5 genome. For our purposes, “intergenic” is defined as having no overlap with existing gene models, including annotated UTRs. These nEPTs generally fall into two categories - evidence for novel gene models and evidence for the extension of existing gene models. Initial classification was done by single-linkage clustering, both to other novel EPTs and to existing gene models. Choosing an appropriate distance for clustering is complicated in eukaryotic organisms in which exons from a single gene can be separated by large distances. An increase in distance results in an increased likelihood of EPTs from adjacent genes being clustered together, whereas a decrease in the distance cutoff results in an increased likelihood of single-gene EPTs being clustered separately. In practice, the impact of the first type of error is minimized during the gene modeling stage. EPTs incorrectly clustered together are filtered out when predicted models are tested for inclusion of all clustered peptides. On the other hand, the second type of error may result in missed novel genes, as a gene containing two novel EPTs that are incorrectly clustered separately would not pass the minimum EPT count filtering. We therefore chose a distance cutoff of 1500 bp, which represents the 96th percentile of the intron length distribution and the 36th percentile of the intergenic distance distribution in Mt3.5v4, to minimize the second error type. Using this distance, nEPTs clustering with existing models were considered as likely evidence of gene model extensions, and the remaining clusters were considered as likely evidence of novel unannotated genes.
The intergenic nEPT locations clustered into 209 novel clusters of two or more peptides and 118 clusters with existing genes, with 227 nEPTs remaining as singletons. Clusters containing existing gene models were considered to be evidence for extension of the current models. Novel clusters were further filtered to remove those not containing at least one uniquely mapped peptide, leaving 201 clusters considered as possible evidence for novel genes. Each cluster, along with 10,000 bp of genomic sequence on either side, was analyzed with the AUGUSTUS gene finding software to look for predicted gene models. For 190 of the novel clusters, gene models were predicted which contained all of the clustered peptides. Gene models were predicted for an additional two clusters that contained at least one unique and two total peptides but did not contain all clustered peptides, possibly because of incorrect clustering. The translation products of these novel genes were searched against the RefSeq protein database using NCBI BLAST (37) with an E-value cutoff of 1e−20, and a list of the top hit and relevant scores for each of the 133 clusters with RefSeq hits can be found in supplemental Table S3.
Most novel clusters (92%) were found in short read contigs, suggesting a strong possibility that they are only partial models. To determine probable full-length models, we used two additional filtering criteria. AUGUSTUS was set to allow partial gene models at the ends of genomic sequences, and only 70 of the 192 gene models it built contained both annotated start and stop codons. These were further filtered during the BLAST search by comparing the length of the predicted protein to that of the best RefSeq hit. Predicted proteins which were at least 80% of the length of the best hit and which aligned to the best hit starting within 20 a.a. of the N terminus were classified as being likely to be full-length models. This is a rather simplistic approach and relies on the robustness of the RefSeq database to deduce the expected length of the protein, but it gives an initial estimation of the quality of the predicted novel genes. The 34 cluster models passing this filtering are listed in Table IV, along with the total number of nEPTs in the cluster, the number of uniquely mapping nEPTs, and the description, percent identity, and E-value of the best RefSeq hit for each associated gene model.
Table IV. Novel EPT clusters with full-length gene model predictions.
Genomic regions containing clustered nEPTs and 10,000 bp of flanking sequence were fed into the AUGUSTUS gene prediction software along with EPT-based coding hints. Predicted gene models were then searched against the RefSeq database using NCBI BLAST. Shown are gene models predicted by AUGUSTUS with both annotated start and stop codons and similarity in overall size to the top BLAST hit. Gene models were reported by AUGUSTUS for an additional 99 novel EPT clusters but are likely to be incomplete due to missing genomic sequence.
| Locus | Contained uniquely mapped nEPTs | All contained nEPTs | Description of top RefSeq hit | Percent identity | E-value |
|---|---|---|---|---|---|
| Cluster_149 | 29 | 29 | Nephrocystin-3-like [Glycine max] | 87.1 | 0.00E+000 |
| Cluster_189 | 24 | 24 | Subtilisin-like protease-like [Glycine max] | 82.3 | 0.00E+000 |
| Cluster_162 | 17 | 17 | Reticuline oxidase [Medicago truncatula] | 78.8 | 0.00E+000 |
| Cluster_102 | 11 | 11 | Conserved oligomeric Golgi complex subunit 1-like [Glycine max] | 84.9 | 0.00E+000 |
| Cluster_033 | 10 | 10 | UDP-glycosyltransferase 84B1-like [Glycine max] | 71.5 | 0.00E+000 |
| Cluster_141 | 8 | 8 | S-adenosylmethionine synthase-like isoform 1 [Glycine max]# | 96.4 | 0.00E+000 |
| Cluster_167 | 8 | 8 | Probable glutathione S-transferase-like [Glycine max] | 77.4 | 4.00E−128 |
| Cluster_184 | 8 | 8 | ruBisCO large subunit-binding protein subunit alpha, chloroplastic-like [Glycine max] | 91.1 | 0.00E+000 |
| Cluster_043 | 7 | 7 | Uncharacterized protein LOC100306450 [Glycine max] | 80.2 | 9.00E−053 |
| Cluster_010 | 6 | 6 | Ubiquinone biosynthesis protein COQ9, mitochondrial-like [Glycine max] | 75.6 | 1.00E−161 |
| Cluster_140 | 6 | 6 | Uncharacterized protein LOC100818804 [Glycine max] | 72.6 | 1.00E−101 |
| Cluster_006 | 5 | 5 | NADP-dependent malic enzyme, chloroplastic-like [Glycine max] | 89.3 | 0.00E+000 |
| Cluster_173 | 5 | 5 | Uncharacterized protein LOC100244411 [Vitis vinifera] | 49.7 | 3.00E−076 |
| Cluster_007 | 4 | 5 | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial-like [Glycine max] | 89.8 | 0.00E+000 |
| Cluster_001 | 4 | 4 | Uncharacterized protein LOC100788250 [Glycine max] | 81.2 | 0.00E+000 |
| Cluster_039 | 4 | 4 | Uncharacterized protein LOC100527685 [Glycine max] | 65.8 | 1.00E−021 |
| Cluster_081 | 4 | 4 | Uncharacterized protein LOC100805605 [Glycine max] | 61.8 | 3.00E−129 |
| Cluster_094 | 4 | 4 | Poly(A) polymerase-like [Glycine max] | 80.2 | 0.00E+000 |
| Cluster_096 | 4 | 4 | Chlorophyll a-b binding protein 21, chloroplastic-like [Glycine max] | 91.7 | 6.00E−177 |
| Cluster_169 | 4 | 4 | Predicted protein [Populus trichocarpa] | 69.4 | 2.00E−114 |
| Cluster_160 | 2 | 4 | Probable methyltransferase PMT8-like [Glycine max] | 82.9 | 0.00E+000 |
| Cluster_121 | 3 | 3 | Expansin-A4-like [Glycine max] | 88.1 | 4.00E−171 |
| Cluster_134 | 3 | 3 | Em-like protein GEA1-like [Glycine max] | 79.1 | 3.00E−048 |
| Cluster_029 | 2 | 2 | Uncharacterized protein LOC100306283 isoform 2 [Glycine max] | 73.0 | 1.00E−029 |
| Cluster_067 | 2 | 2 | Hypothetical protein MTR_6g034800 [Medicago truncatula] | 36.2 | 1.00E−033 |
| Cluster_097 | 2 | 2 | LRR receptor-like serine/threonine-protein kinase FLS2-like [Glycine max] | 78.1 | 0.00E+000 |
| Cluster_111 | 2 | 2 | Uncharacterized protein LOC100527746 [Glycine max] | 80.0 | 8.00E−065 |
| Cluster_129 | 2 | 2 | Uncharacterized protein LOC100811471 isoform 1 [Glycine max] | 85.4 | 0.00E+000 |
| Cluster_136 | 2 | 2 | Uncharacterized protein LOC100794459 [Glycine max] | 62.2 | 7.00E−053 |
| Cluster_138 | 2 | 2 | Zinc finger CCCH domain-containing protein 32-like [Glycine max] | 75.5 | 0.00E+000 |
| Cluster_168 | 2 | 2 | Transcription factor RF2b-like [Glycine max] | 73.7 | 0.00E+000 |
| Cluster_186 | 2 | 2 | Octanoyltransferase-like [Glycine max] | 86.2 | 8.00E−138 |
| Cluster_194 | 2 | 2 | Uncharacterized protein LOC100526970 precursor [Glycine max] | 68.3 | 1.00E−101 |
| Cluster_002 | 1 | 2 | Uncharacterized protein LOC100788250 [Glycine max] | 81.2 | 0.00E+000 |
Full Validation Using RNA-Seq Alignments
To evaluate the overall validity of the EPT mapping, the minimum read coverage over the length of the mapped tag for both supporting and novel EPTs was calculated using a set of 269 million 100 bp Illumina reads generated from M. truncatula ‘Jemalong A17’ mRNA and mapped to the Mt3.5 genome. Of all intergenic and intronic regions (based on the Mt3.5v4 annotations), 78.3% were not covered by any mapped reads, indicting little genomic DNA contamination in the sequencing samples. We used a minimum coverage of 5x (the 90th percentile of intergenic/intronic region coverage) over the entire length of the peptide to classify an EPT as confirmed by RNA-Seq. Based on this criteria, 90,843 out of 95,633 sEPT mappings (95%) were confirmed and 1495 out of 1731 nEPTs (86%) were confirmed. Of all EPT locations combined, 93% met the criteria for positive RNA-Seq validation.
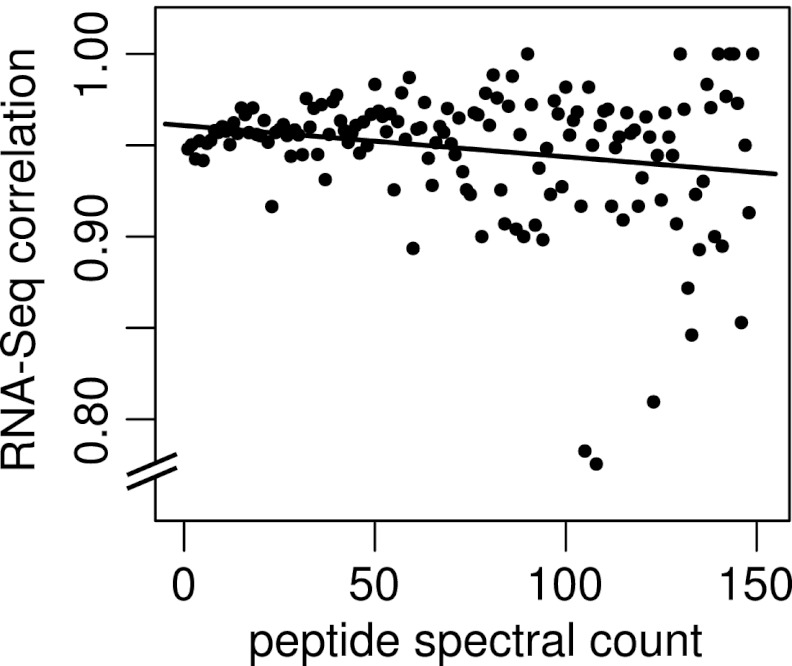
It is sometimes assumed that peptides with higher spectral match counts (peptide spectral matches - PSMs) are more reliable. For our purposes, a PSM is defined as an independent observation of a given spectrum/peptide match and the PSM count is the number of individual spectra matching a given peptide sequence. To test this hypothesis, we repeated the above analysis on sEPTs grouped by PSM and calculated the percent of sEPTs confirmed at each PSM level. Fig. 7 shows a plot of spectral match count against percent RNA-Seq correlation for PSMs in the range of 1–150. The slope of the linear model fitted to this plot is near zero, suggesting little or no correlation between spectral count and the reliability of peptide identification as evidenced by RNA-Seq correlation.
Fig. 7.
Correlation of spectral match counts to RNA-Seq evidence for supporting EPTs. Peptide spectral match counts (PSMs) are plotted against the fraction of sEPTs at that PSM with supporting RNA-Seq evidence. The fitted linear model indicated by the solid line (slope = –0.0002) suggests little correlation between peptide PSM count and validity as estimated by RNA-Seq correlation. Spectral match counts are the number of captured spectra assigned to a given peptide sequence. RNA-Seq correlation values represent the fraction of supporting EPTs with a minimum of 5× RNA-Seq coverage across their entire length. At PSMs above 150 (not shown), the RNA-Seq correlation values become highly discrete because of low representation and cluster at 0 and 1.
Data Availability
All raw spectra and relevant analysis files are available at the Medicago Omics Repository (http://more.biotech.wisc.edu).
DISCUSSION
Proteogenomics is a valuable tool for enhancing existing structural annotations of sequenced organisms. We have endeavored to apply this technique to the model legume Medicago truncatula to gain a further understanding of the state of the published genome and annotations. The purpose of this effort was primarily exploratory in nature. In practice, expressed peptide tag data should be incorporated directly into gene model prediction software alongside other forms of evidence such as EST, RNA-Seq, and homologous protein data. Some existing gene modeling tools, such as AUGUSTUS and Maker, already have this capability. EPT data can also be used for manual curation of individual genes, either by research groups for their gene(s) of interest or in the course of systematic manual curation for a full genome. Both types of analysis are facilitated by the aggregation of MS/MS data by genome working groups who can process it and provide it to end users as EPTs either by download or within genome browsers such as GBrowse and IGV. We are providing our full set of existing EPT data to the International Medicago Genome Annotation Group to assist in their efforts and envision similar collaborations in the future as more MS/MS data is generated.
Although MS/MS and EPT data can serve a unique role in any genome annotation effort, some aspects of structural annotation are just as easily and in some cases more reliably deduced based on other types of evidence. This includes the correct identification of splice sites during mRNA processing, which are a source of common errors in structural annotations based on computer prediction. We have shown the ability of proteogenomics to locate such errors, but also shown a high correlation between EPT evidence and that provided by RNA-Seq alignments. Because of this overlap, and the typically higher level of coverage across any given transcriptome that RNA-Seq provides compared with MS/MS, some researchers in the genomics community have questioned the usefulness of proteogenomic analyses. However, our analysis has shown the reliability of EPT data as an additional tool for genomic work. We have shown that novel peptides are supported by RNA-Seq alignments at only slightly lower levels than supporting peptides, and peptides with low spectral match counts do not appear to be less reliable than those with higher counts based on the same RNA-Seq correlation. We conclude, therefore, that when such data is generated in the course of other experiments it can and should be used as an additional source of information for genome annotation. EPT data can play a role in supporting other evidence and increase the confidence level of a gene model. Its most useful role, however, is in providing evidence that cannot be readily deduced from other common sources of data. This includes confirming translation of questionable coding sequence (such as short ORFs or annotated pseudogenes), correcting reading frames for gene models with several viable alternatives, distinguishing precursor mRNA from retained introns in RNA-Seq and EST sequencing evidence, and providing evidence for rare but potentially important events such as non-canonical start codon usage. Along with other recent developments such as ribosome profiling, it can provide structural information at the coding sequence and even codon level, and can also provide clues to the prevalence of post-translational modifications such as N-terminal methionine excision, provided the spectral search methods used allow for such detection.
Of the 78,362 filtered unique peptides identified in this study, only 1568 (2.0%) were novel. This contrasts with the 12.5% of 144,079 peptides identified as novel in a similar study in Arabidopsis (4). Assuming that we are using similar definitions of identified and novel peptides, the significantly lower proportion of novel peptides identified in M. truncatula is surprising. The Arabidopsis genome sequence and annotations are typically considered to be of high quality, whereas the Medicago draft sequence was only recently published and annotation efforts are still in the early stages. It is possible that the increased number of novel peptides observed in the Arabidopsis study is a consequence of the larger spectral library used. It is also possible that recent genome sequencing efforts in plants have benefited from the substantial work performed in Arabidopsis to improve the quality of annotations. In either case, the MS/MS data analyzed in this study largely support the latest Medicago annotations. However, we have provided examples demonstrating the unique role proteogenomic analysis can play in building the most accurate and descriptive structural annotations possible, and there is a need for continued improvements to existing gene models, annotation of missing genes, and corrections to the genome sequence itself to provide Medicago researchers with the accurate representation of the Medicago genome, transcriptome, and proteome on which their research relies.
Supplementary Material
Footnotes
* This work was supported by a grant from the National Science Foundation (NSF#0701846) to M. R. S., J. J. C., and J. M. A. C. M. R. was funded by an NSF Graduate Research Fellowship and NIH Traineeship (T32GM008505).
 This article contains supplemental Tables S1 and S2.
This article contains supplemental Tables S1 and S2.
1 The abbreviations used are:
- EST
- expressed sequence tags
- EPT
- expressed peptide tag
- FDR
- false discovery rate
- MS/MS
- tandem mass spectrometry
- NME
- N-terminal methionine excision.
REFERENCES
- 1. Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ingolia N. T., Lareau L. F., Weissman J. S. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Young N. D., Debellé F., Oldroyd G. E. D., Geurts R., Cannon S. B., Udvardi M. K., Benedito V. A., Mayer K. F. X., Gouzy J., Schoof H., Peer Y. V. de, Proost S., Cook D. R., Meyers B. C., Spannagl M., Cheung F., Mita S. D., Krishnakumar V., Gundlach H., Zhou S., Mudge J., Bharti A. K., Murray J. D., Naoumkina M. A., Rosen B., Silverstein K. A. T., Tang H., Rombauts S., Zhao P. X., Zhou P., Barbe V., Bardou P., Bechner M., Bellec A., Berger A., Bergès H., Bidwell S., Bisseling T., Choisne N., Couloux A., Denny R., Deshpande S., Dai X., Doyle J. J., Dudez A.-M., Farmer A. D., Fouteau S., Franken C., Gibelin C., Gish J., Goldstein S., González A. J., Green P. J., Hallab A., Hartog M., Hua A., Humphray S. J., Jeong D.-H., Jing Y., Jöcker A., Kenton S. M., Kim D.-J., Klee K., Lai H., Lang C., Lin S., Macmil S. L., Magdelenat G., Matthews L., McCorrison J., Monaghan E. L., Mun J.-H., Najar F. Z., Nicholson C., Noirot C., O'Bleness M., Paule C. R., Poulain J., Prion F., Qin B., Qu C., Retzel E. F., Riddle C., Sallet E., Samain S., Samson N., Sanders I., Saurat O., Scarpelli C., Schiex T., Segurens B., Severin A. J., Sherrier D. J., Shi R., Sims S., Singer S. R., Sinharoy S., Sterck L., Viollet A., Wang B.-B., Wang K., Wang M., Wang X., Warfsmann J., Weissenbach J., White D. D., White J. D., Wiley G. B., Wincker P., Xing Y., Yang L., Yao Z., Ying F., Zhai J., Zhou L., Zuber A., Dénarié J., Dixon R. A., May G. D., Schwartz D. C., Rogers J., Quétier F., Town C. D., Roe B. A. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Castellana N. E., Payne S. H., Shen Z., Stanke M., Bafna V., Briggs S. P. (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. U.S.A. 105, 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wright J. C., Sugden D., Francis-McIntyre S., Riba-Garcia I., Gaskell S. J., Grigoriev I. V., Baker S. E., Beynon R. J., Hubbard S. J. (2009) Exploiting proteomic data for genome annotation and gene model validation in Aspergillus niger. BMC Genomics 10, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Savidor A., Donahoo R. S., Hurtado-Gonzales O., Verberkmoes N. C., Shah M. B., Lamour K. H., McDonald W. H. (2006) Expressed peptide tags: an additional layer of data for genome annotation. J. Proteome Res. 5, 3048–3058 [DOI] [PubMed] [Google Scholar]
- 7. Tanner S., Shen Z., Ng J., Florea L., Guigó R., Briggs S. P., Bafna V. (2007) Improving gene annotation using peptide mass spectrometry. Genome Res. 17, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baerenfaller K., Hirsch-Hoffmann M., Svozil J., Hull R., Russenberger D., Bischof S., Lu Q., Gruissem W., Baginsky S. (2011) pep2pro: a new tool for comprehensive proteome data analysis to reveal information about organ-specific proteomes in Arabidopsis thaliana. Integr. Biol. 3, 225. [DOI] [PubMed] [Google Scholar]
- 9. Baerenfaller K., Grossmann J., Grobei M. A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320, 938–941 [DOI] [PubMed] [Google Scholar]
- 10. Lamesch P., Berardini T. Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D. L., Garcia-Hernandez M., Karthikeyan A. S., Lee C. H., Nelson W. D., Ploetz L., Singh S., Wensel A., Huala E. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grimsrud P. A., den Os D., Wenger C. D., Swaney D. L., Schwartz D., Sussman M. R., Ané J. M., Coon J. J. (2010) Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 152, 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Catoira R., Galera C., de Billy F., Penmetsa R. V., Journet E. P., Maillet F., Rosenberg C., Cook D., Gough C., Dénarié J. (2000) Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12, 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walter H., Larsson C. (1994) Partitioning procedures and techniques: cells, organelles, and membranes. Methods Enzymol. 228, 42–63 [DOI] [PubMed] [Google Scholar]
- 14. Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 15. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 16. Stanke M., Morgenstern B. (2005) AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33, W465–W467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 18. Wenger C. D., Phanstiel D. H., Lee M. V., Bailey D. J., Coon J. J. (2011) COMPASS: A suite of pre- and post-search proteomics software tools for OMSSA. Proteomics 11, 1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open Mass Spectrometry Search Algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 20. Quinlan A. R., Hall I. M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P. (2011) Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trapnell C., Pachter L., Salzberg S. L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gupta N., Tanner S., Jaitly N., Adkins J. N., Lipton M., Edwards R., Romine M., Osterman A., Bafna V., Smith R. D., Pevzner P. A. (2007) Whole proteome analysis of post-translational modifications: Applications of mass-spectrometry for proteogenomic annotation. Genome Res. 17, 1362–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giglione C., Meinnel T. (2001) Organellar peptide deformylases: universality of the N-terminal methionine cleavage mechanism. Trends Plant Sci. 6, 566–572 [DOI] [PubMed] [Google Scholar]
- 25. Giglione C., Vallon O., Meinnel T. (2003) Control of protein life-span by N-terminal methionine excision. EMBO J. 22, 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ross S., Giglione C., Pierre M., Espagne C., Meinnel T. (2005) Functional and developmental impact of cytosolic protein N-terminal methionine excision in Arabidopsis. Plant Physiol. 137, 623–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bienvenut W. V., Sumpton D., Martinez A., Lilla S., Espagne C., Meinnel T., Giglione C. (2012) Comparative large-scale characterisation of plant vs. mammal proteins reveals similar and idiosyncratic N-alpha acetylation features. Mol. Cell. Proteomics 11, doi:10.1074/mcp.M111.015131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sherman F., Stewart J. W., Tsunasawa S. (1985) Methionine or not methionine at the beginning of a protein. BioEssays 3, 27–31 [DOI] [PubMed] [Google Scholar]
- 29. Blattner F. R., Plunkett G., 3rd, Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Glasner J. D., Rode C. K., Mayhew G. F., Gregor J., Davis N. W., Kirkpatrick H. A., Goeden M. A., Rose D. J., Mau B., Shao Y. (1997) The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 [DOI] [PubMed] [Google Scholar]
- 30. Beerman R. W., Jongens T. A. (2011) A non-canonical start codon in the Drosophila fragile X gene yields two functional isoforms. Neuroscience 181, 48–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gerashchenko M. V., Su D., Gladyshev V. N. (2010) CUG start codon generates thioredoxin/glutathione reductase isoforms in mouse testes. J. Biol. Chem. 285, 4595–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A. C., Vagner S. (2003) Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell 95, 169–178 [DOI] [PubMed] [Google Scholar]
- 33. Riechmann J. L., Ito T., Meyerowitz E. M. (1999) Non-AUG initiation of AGAMOUS mRNA translation in Arabidopsis thaliana. Mol. Cell. Biol. 19, 8505–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simpson G. G., Laurie R. E., Dijkwel P. P., Quesada V., Stockwell P. A., Dean C., Macknight R. C. (2010) Noncanonical translation initiation of the Arabidopsis flowering time and alternative polyadenylation regulator FCA. Plant Cell 22, 3764–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schmitz J., Prüfer D., Rohde W., Tacke E. (1996) Non-canonical translation mechanisms in plants: efficient in vitro and in planta initiation at AUU codons of the tobacco mosaic virus enhancer sequence. Nucleic Acids Res. 24, 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ivanov I. P., Firth A. E., Michel A. M., Atkins J. F., Baranov P. V. (2011) Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. doi:10.1093/nar/gkr007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 38. R Development Core Team (2008) R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 39. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw spectra and relevant analysis files are available at the Medicago Omics Repository (http://more.biotech.wisc.edu).