Abstract
In Dictyostelium, the cytoskeletal proteins Actin binding protein 1 (Abp1) and the class I myosin MyoK directly interact and couple actin dynamics to membrane deformation during phagocytosis. Together with the kinase PakB, they build a regulatory switch that controls the efficiency of uptake of large particles. As a basis for further functional dissection, exhaustive phagosome proteomics was performed and established that about 1300 proteins participate in phagosome biogenesis. Then, quantitative and comparative proteomic analysis of phagosome maturation was performed to investigate the impact of the absence of MyoK or Abp1. Immunoblots and two-dimensional differential gel electrophoresis of phagosomes isolated from myoK-null and abp1-null cells were used to determine the relative abundance of proteins during the course of maturation. Immunoblot profiling showed that absence of Abp1 alters the maturation profile of its direct binding partners such as actin and the Arp2/3 complex, suggesting that Abp1 directly regulates actin dynamics at the phagosome. Comparative two-dimensional differential gel electrophoresis analysis resulted in the quantification of mutant-to-wild type abundance ratios at all stages of maturation for over one hundred identified proteins. Coordinated temporal changes in these ratio profiles determined the classification of identified proteins into functional groups. Ratio profiling revealed that the early delivery of ER proteins to the phagosome was affected by the absence of MyoK and was coupled to a reciprocal imbalance in the delivery of the vacuolar proton pump and Rab11 GTPases. As direct functional consequences, a delayed acidification and a reduced intraphagosomal proteolysis were demonstrated in vivo in myoK-null cells. In conclusion, the absence of MyoK alters the balance of the contributions of the ER and an endo-lysosomal compartment, and slows down phagosome acidification as well as the speed and efficiency of particle degradation inside the phagosome.
Professional phagocytes, ranging from phagotrophic protozoa to specialized cells of the innate immune system such as macrophages, neutrophils, or dendritic cells, ingest and digest large particles (> 250 nm). Indeed, the core machineries acting in phagocytosis have been conserved as its basic purpose evolved from predation and feeding to antigen presentation (1). Particle recognition by the phagocytes initiate the internalization process and triggers remodeling of the plasma membrane and its underlying cytoskeleton, to project a circular cup-shaped lamella around the particle. The phagocytic cup finally encloses the particle into a de novo membrane-bound vacuole, the phagosome. Endomembrane compartments, such as early and late endosomes (2–4) and the endoplasmic reticulum (5), are recruited to provide membrane to form the phagosome. Modifications of the environment within the closed phagosome generates a microbicidal milieu and leads to particle degradation. Maturation of the phagosome is, basically, a linear process driven by a flux of incoming and outgoing vesicular trafficking, and is characterized by a progressive decrease in pH and successive fusions with early endosomes, late endosomes, and lysosomes (6, 7). The trafficking events are controlled by small GTPases and the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) machinery (6, 8), and phagosome acidification is driven by the activity of the vacuolar H+-ATPase proton pump (V-ATPase)1 (4).
In Dictyostelium, a synchronized pulse/chase protocol for the isolation of latex bead-containing phagosomes was developed that covers the entire maturation process (8, 9). Temporal quantitative profiling by immunoblot and two-dimensional electrophoresis (2-DE) was used to characterize the protein content of the phagosome and follow the time-dependent variations driving maturation (8, 10). Five successive phases in the maturation program were clearly identified i.e. uptake, metabolism/ion exchange, late endosomal, digestive, and exocytic stages. The uptake phase was the more complex. Abundant proteins characteristic of this phase were involved in signaling, actin cytoskeleton, membrane dynamics and the emergence of a degradative phase. Although a clear sequence of maturation events could be established, the discontinuity of individual profiles revealed that a protein, required at different steps of maturation, can be retrieved in-between (10). A similar plasticity was observed in macrophages by temporal profiling of isolated phagosomes after stable isotope labeling (11).
Having precisely characterized the maturation process in wild type Dictyostelium, we used proteomic profiling to investigate the impact on phagocytosis of an ablation of either Actin binding protein 1 (Abp1) or the class I myosin, MyoK. Together with the kinase PakB, MyoK and Abp1 build a regulatory switch that regulates the efficiency of uptake of large particles. These two proteins interact and couple actin dynamics to membrane deformation but exert opposite regulatory roles on phagocytic uptake (12, 13). Compared with wild type cells, myoK-null mutants display 20% decrease whereas abp1-null display 35% increase in the rate and extent of uptake of large particles. Despite their direct interaction, MyoK and Abp1 localize independently at the phagocytic cup and on early phagosomes (12). Like other class I myosins, MyoK is essential for the maintenance of resting cortical tension and provides a force to push on negatively curved membranes (12, 13). Although there is no evidence yet that MyoK might share these functions with other class I myosins, MyoB in Dictyostelium (14), and Myosin IB in human (15), for example, are involved in membrane trafficking in the pinocytic pathway, potentially contributing to membrane deformation both at the endosomal level and at the plasma membrane (16). The function of Abp1 itself is not well characterized although its interaction network is well described. It contributes to vesicle uptake, interacting notably with 1) the membrane binding and force generating class I myosins motors such as MyoK in Dictyostelium and Myo5p in yeast, 2) the Arp2/3 actin nucleation complex in yeast, and 3) dynamin, a protein involved in membrane constriction, in Dictyostelium and mammals (12, 17). Although the role of Abp1 in vesicle formation has been mainly studied during uptake at the plasma membrane, it might perform similar tasks on endomembranes. Indeed, Abp1 is the main phagosomal F-actin binding protein (18).
The initial goal of the study was to understand how the removal of one member of a regulatory complex of the actin cytoskeleton remodeling can decrease or increase uptake, and what the downstream consequences are on phagosome maturation. On one hand, the uptake rate and extent are strictly regulated and can be significantly decreased or increased. The Abp1-MyoK-PakB regulatory loop is not the only illustration of this concept. Mutation of the interacting partners CH-Lim or LimF induce respectively an increase or a decrease in phagocytosis and the balance is regulated by the small GTPase Rab21 (19). Increased uptake as a result of a gene knockout might appear counter-intuitive, however, knockout mutants for dynamin A, the actin cross-linking protein, Abp34, and both profilins I and II show increased phagocytosis (20, 21). On the other hand, actin polymerization and Abp1 play an active role in phagosome maturation. Indeed, the formation of a transient actin coat prevents phagosome-lysosome fusion and delays phagocytosis (22) and the presence of Abp1 regulates phagosome binding to actin in vitro (18). Nevertheless, the impact of a regulatory switch of actin polymerization on the downstream maturation process has not been investigated.
As a targeted approach, the maturation profiles of Abp1, MyoK and their direct binding partners were first determined by quantitative immunoblots to delineate the impact of myoK- or abp1-knockouts. Quantitative profiling of maturation by 2D-DIGE was then used to measure mutant to wild type protein abundance ratios of more than one hundred identified phagosomal proteins over the entire maturation process in both myoK-null and abp1-null mutants. Coordinated changes were observed in the temporal profiles of functionally related proteins predicting a delay in phagosome acidification resulting from compensatory trafficking defect in the myoK-null mutant and an alteration of the phagosomal protease composition in abp1-null cells. Alterations in phagosome acidification and proteolysis were observed in vivo as these predictions were verified at a functional level.
EXPERIMENTAL PROCEDURES
Cell Culture
Dictyostelium cells of the parent wild type strain AX-2 were grown at 22 °C in HL-5c medium (Formedium) supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen). myoK null cells (13) and abp1 null mutants were maintained with additional 5 μg/ml blasticidin (12).
Electron Microscopy
Phagosomes directly collected from the gradient were diluted and normalized for concentration in HESES (0.25 m sucrose, 20 mm HEPES-KOH, pH 7.2). Phagosomes in equivalent number for each fraction were mixed 1:1 (v/v) in freshly prepared fixative solution (0.4 m sodium cacodylate, pH 7.2, 4% glutaraldehyde, 0.6% OsO4). Samples were fixed for 1h on a rotating wheel. Phagosomes were pelleted 15 min, 14′000 rpm on a mini-centrifuge and washed in PBS until the supernatant was clear. Phagosome pellets were then contrasted and embedded (23, 24).
Flow Cytometry-based Uptake Assay
Uptake was measured as described (10) with the following modifications. Fluorescent beads of 0.5, 1.0 and 4.5 μm in diameter (Fluorescent YG carboxylated beads, Polyscience) were added to the cell at 800:1, 200:1 and 10:1 ratios, respectively. Flow cytometry was performed with a FACScalibur (Beckton Dickinson). Data were analyzed with the FlowJo software (TreeStar).
Phagosome Isolation
Latex bead-containing phagosomes were isolated via flotation on sucrose step gradients and processed as described (9, 10).
Relative Abundance Profiling by Immunoblotting
Quantitative immunoblots were performed as described (10) with following modifications. The chemiluminescent signal was detected in a UVP EpiChem II Darkroom equipped with a digital camera. Signals were quantified with Labworks 4.0 software. Bands were defined as boxes inside lanes and total box volume was quantified after “filtered profile” background subtraction. Signal intensities were then normalized, setting the maximum signal intensity to 100%, resulting in a relative abundance profile during phagosome maturation time. To measure mutant-to-wild type ratios, phagosomes were isolated in parallel in the mutant and wild type cells at early time-points (5′/0, 15′/0). Immunoblot signal intensities were measured and averaged (n = 3). The signal at 5′/0 in wild type phagosomes was set as 100% and signal intensity values in the mutant were normalized accordingly. Mutant-to-wild type ratios were consistent over the 5′/0 and 15′/0 time-points. Thus, the whole mutant profile was normalized accordingly.
Antibodies
Antibodies raised against the following Dictyostelium antigens were obtained from: (1) actin (mAb, 224-236-1), calreticulin (mAb, 251-67-1) (gift of Dr. G. Gerisch, MPI for Biochemistry, Martinsried); (2) profilin II (mAb, 174-380-3) (gift of Dr. M. Schleicher, Adolf-Butenandt-Institute, Munich); (3) Arp3 (pAb; gift of Dr. R.H. Insall, CR-UK Beatson Institute for Cancer Research, Glasgow), (4) dynamin A (gift of Dr. D. J. Manstein, Hannover Medical School). The anti-MyoK and anti-Abp1 antibodies were described in (12). For immunoblots, goat-anti-rabbit IgGs or goat-anti-mouse IgGs conjugated to HRP (BioRad) were used at dilutions between 1:2,000 and 1:10,000.
Two-dimensional Gel Electrophoresis (2D-DIGE)
Cell were synchronized for growth in suspension over a 3 days period, harvested by centrifugation (5 min, 1200 rpm, GH3.8A) and washed twice in Sørensen buffer. Cell pellets were resuspended in 10 μl of 14x complete protease inhibitors (Roche) water solution, flash frozen in liquid nitrogen and stored at −80 °C. Cells were then lysed in DIGE sample buffer and samples were labeled directly after determination of the protein concentration [≈ 5 μg/μl, 2D-Quant kit (Amersham Biosciences)]. Phagosome pellets were resuspended in a minimal volume of DIGE sample buffer (7 m urea (Amersham Biosciences), 2 m thiourea (Amersham Biosciences), 4% 3-((3-Cholamidopropyl)dimethylammonio)-1-propanesulfonate (CHAPS), 15 mm 1,2-diheptanoyl-sn-glycero-3-phosphatdiyl choline (DHPC) (Avanti Lipids), 30 mm Tris (Sigma Aldrich), pH 8.5). Protein samples (50 μg) were adjusted to the same concentration (≥ 2.5 μg/μl) and labeled in parallel in the DIGE sample buffer with the respective minimal fluorescent CyDye-NHS (Cy5, Cy3, Cy2), according to manufacturer's instructions (Amersham Biosciences). Labeled samples were mixed and co-migrated with additional 200–300 μg of corresponding unlabeled sample. Total protein sample was adjusted to 450 μl in two-dimensional sample buffer (7 m urea, 2 m thiourea, 65 mm DTE 1,4-Dithioerythritol (DTE), 4% CHAPS, 15 mm DHPC, 2% Resolyte ampholytes, Tris 30 mm, pH 8.5). Samples were separated as described (10, 25) with the following modifications. First dimension was performed in NL 3–10, 24 cm strips (Amersham Biosciences) at 15 °C with a current of 75 μA/strip and up to 45 kVh. Second dimension was run on polyacrylamide gel of 12.5%T, 2.6%C (National Diagnostics) at 15 °C, 15 mA/gel, O/N in an Ettan Dalt Twelve chamber (Amersham Biosciences). Gels were poured in a six-gel chamber between low-fluorescence plates according to manufacturer's instructions (Amersham Biosciences). Gels were scanned in an Ettan DIGE imager (Amersham Biosciences). Scan times were kept identical in a comparative gel run and adjusted for each dye to result in the best signal to noise ratio and the most extensive range of signal intensity.
Gel Analysis and Mutant/Wild Type Ratio Profiling
Gels were analyzed with Melanie Image Master 2D Platinum version 6 for DIGE. The initial number of detected spots was set to 2′500 and then spots were filtered according to spot area, percentage volume and saliency depending on gel signal-to noise ratio and manually curated. For phagosome samples, gels corresponding to the same time point were matched pair-wise. Gels were then matched within the pulse/chase series, setting the 5′/0 time point as a reference. To match spot absent from the 5′/0 time point, matched spots were withdrawn from analysis and remaining spots were then matched to the 15′/105′ time point as a reference. Merging of matched pairs and time-series were performed in Excel using the SuperCombine add-in (gift from Marianne Tardif and Jérôme Garin, Institute of Life Sciences Research and Technologies, Grenoble) and resulted in a list containing all matched spots and their respective mutant/WT ratios during maturation i.e. all the spot ratio profiles. In case of conflict, matches associated with a known mutant/WT ratio value and/or the most complete time-series were favored on the final matching table. Differential spots fulfilled three criteria: 1) their differential ratio was higher than ±1.6, 2) standard deviation of the mean should not overlap, 3) spot detection should not introduce any potential bias. For analysis of cell lysates, a ratio of 1.3 (30% difference) was experimentally determined as being a reasonable cut off to consider bona fide differential spots. As the spot ratio variability was slightly higher in phagosome samples compared with cell lysates, the cut off was set twice as high (1.6, 60% difference). Ratio profiles of proteins identified in multiple neighboring spots (i.e. spot groups or spot trains) are very similar (see supplementary Data). Thus, only an averaged profile of all isoforms is shown here (i.e. VatB, VatA, CrtA, PDI1, PDI2, …).
Protein Identification by Mass Spectrometry and Data Analysis
Protein identification was performed essentially as described (10, 26). In brief, proteins were in-gel digested with trypsin and extracted from Coomassie stained one-dimensional or two-dimensional gel pieces in a liquid handling robot (DigestPro MS, Intavis AG) according to standard procedures.
For peptide fingerprinting (PMF) by MALDI-TOF mass spectrometry (Ultraflex, Bruker), samples were analyzed as described (10). Protein sequencing was performed either on an ESI-Q-TOF (QSTAR Pulsar, PE Sciex) (10) or an Orbitrap (LTQ Orbitrap, Thermo Scientific) as described (26). From the MS and MS/MS spectra, peak lists were generated without peak removal after centroiding and deisotoping (analyst QS 1.1; Applied Biosystems; combined with Mascot script version 1.6b13; Matrix Science). The peak list was then applied to a database search against Dictybase, the Dictyostelium protein database (27) (dicty_primary_decoy downloaded on 22.02.2010, 54294 entries), using the Mascot software, version 2.2.4 (Matrix Science). The Scaffold software (Proteome Software Inc.), version 3.0.8, was used for editing of MS/MS identifications. The algorithm was set to use trypsin as enzyme, allowing at maximum for one missed cleavage site. Iodoacetamide derivative of cysteine was specified in Mascot as a fixed modification. Deamidation of asparagine and glutamine and oxidation of methionine were specified in Mascot as variable modifications. For PMF identifications, mass tolerance was set to 100 ppm using already described parameters and threshold for peak-picking (10). Protein hits were considered identified if the Mascot score exceeded the significance level (p ≤ 0.05). For MS/MS protein identifications from two-dimensional spots, mass tolerance was set to 0.1 Da for precursor and fragment ions. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides with a peptide identification probability greater than 20%. The threshold was set to maximize sensitivity and minimize the overlap between correct and incorrect identity score distribution in the Scaffold software. The relative abundance of each protein in the spot was evaluated. The total number of spectra (NSn) of each protein was normalized to the total number of spectra of the most abundant protein in the spot (NS1). Proteins with a relative abundance over 20% of the most abundant protein in the sample (NSn/NS1 > 0.2) were excluded from further analysis because their contribution to the differential fluorescent signal in-gel is minor. For MS/MS protein identifications from one-dimensional bands, mass tolerance was set at 10 ppm for precursor ions and 0.5 Da for fragment ions. Protein identifications were accepted if their protein identification probability was over 99%, their peptide identification probability was greater than 95.0% and they were identified by at least two unique peptides. The protein false discovery rate was 0.1% and the peptide false discovery rate was 5.3% as calculated with a probabilistic method. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were listed without further filtering, except redundant entries for actin (31 entries), which were grouped into one common entry (act15, DDB_G0272520). All protein identifications and the corresponding assigned peptide sequences are listed in supplemental Tables 3 and 4, respectively.
The list of phagosomal protein (supplemental List S1) is compiled from published data (1, 10), 2D-DIGE spot identifications (2D MSMS and 2D PMF, this study) and all proteins identified in the major bands of a phagosomal sample separated on a 10% one-dimensional SDS-PAGE (one-dimensional MSMS) (9). Phagosomal proteins were associated to GO terms and matched in KEGG pathways (http://www.genome.jp/kegg/) (28, 29) using the Orange freeware (http://orange.biolab.si/(30)). A stringent p value was applied to select only significantly represented GO terms (p < 10−5) and KEGG pathways (p < 10−4). Only functionally non-redundant GO terms are represented. Transmembrane domains and signal peptides in phagosomal protein sequences were predicted using different prediction programs including TMHMM (31) and SignalP (32).
Microarray Analysis
Cells were synchronized for growth as for proteomics. 15–20 × 107 cells were lysed by eight passages in a 5 ml Dounce homogenizer with a small void clearance (S, 10–30 μm) pestle and processed with the Qiagen RNA isolation Midi kit following manufacturer's instructions. Three biological replicates of total cell RNA were isolated for each strain. Each mutant strain was compared with its respective wild type in two technical duplicates, yielding six direct wild type-to-mutant comparisons for each strain analyzed. Mutants were compared in two genetic backgrounds (DH1–10 and AX-2) and data were cross-correlated. Samples and data were processed as described (33). Analysis was performed on a total of 9247 probes, representing 8579 D. discoideum specific genes.
Measure of pH and Proteolytic Digestion in Phagosomes
The kinetics of intraphagosomal acidification and proteolytic activity were monitored using a fluorescence plate reader (Synergy Mx, Biotek) over a period of 120 min at intervals of 1 measurement/1.5 min. The indicator beads for the pH measurement were prepared as described (57). Briefly, 3 μm carboxylated silica beads (Kisker Biotech, Steinfurt, Germany) were coupled with BSA and labeled with the pH-sensitive fluorochrome carboxyfluorescein succinimidyl ester (S.E.) and the pH-insensitive internal reference fluorochrome Alexa Fluor 594-S.E. (Molecular Probes). The measurement of proteolytic activity is based on the principle of dye dequenching induced by proteolysis of the carrier protein. The indicator beads were prepared as described (58), In brief, the proteolytic reporter Self-Quenched BODIPY ® Dye Conjugates of Bovine Serum Albumin (DQTM Green BSA, Molecular Probes) and the reference dye Alexa Fluor 594-S.E. were coupled to 3 μm carboxylate-modified silica particles. Dictyostelium cells were plated as a monolayer in clear bottom black wall 96-well dishes (Costar) and allowed to adhere in LoFlo medium (Formedium). The fluorescent indicator beads were added to the cells at a ratio of 1:2 and the plate was centrifuged for 30 s. Non-ingested beads were removed immediately by washing twice with Soerensen-buffer. For pH monitoring, the fluorescent emission of carboxyfluorescein at 520 nm when excited with 490 nm is pH-sensitive, whereas emission at 520 nm, when excited at 450 nm, is largely unaffected by changes in pH. Conversion from the excitation ratio to pH was achieved through polynomial regression of a standard curve generated by calculation of the excitation ratio of the fluorochrome in environments of known pH. For proteolysis monitoring, the emission fluorescence was measured in these buffers at 490 nm and 450 nm excitation. The ratio 490/450 nm reflects the bulk proteolytic activity within the bead-containing phagosomes.
RESULTS
Relative Contribution of Protein Classes to the Composition of Pure and Intact Isolated Phagosomes
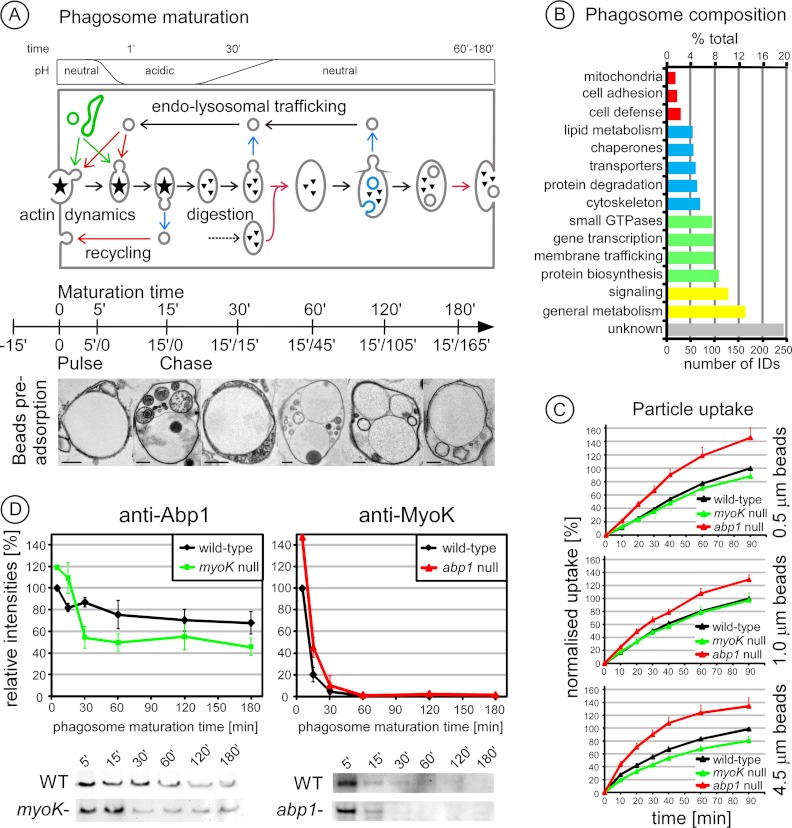
Molecular characterization of the phagosome maturation process in wild type Dictyostelium cells is based on a powerful phagosome purification strategy associated with a synchronized pulse/chase particle feeding protocol (Fig. 1A, scheme) (8, 10). Pre-adsorption of latex beads on the cells in the cold, followed by rapid re-warming, generates a strong and synchronous wave of uptake (9). Phagosomes isolated at 5 min contained single particles surrounded by an intact membrane as shown by electron microscopy and FM4–64 lipid dye staining (supplemental Fig. S1). Phagosome maturation can be followed not only molecularly but also morphologically in isolated phagosomes (Fig. 1A, images). Internal phagosomal vesicles were observed with the successive appearance of electron-lucent vesicles, multi-vesicular bodies and electron-dense vesicles, reminiscent of early endosomes, late endosomes and lysosomes, respectively. Then, morphological complexity diminished so that only electron-lucent internal vesicles were observed at pre-exocytosis time points. Large multi-bead phagosomes, indicative of intense phagosome-phagosome fusion, were also visible, but only at late time-points.
Fig. 1.
Analysis of phagosome maturation in wild type cells and the regulatory roles of Abp1 and MyoK. A, The major steps of phagosome maturation in Dictyostelium are depicted along a time axis that indicate the six time points used to isolate phagosomes following a pulse/chase protocol. In brief, beads are first adsorbed on cells 15′ in the cold, followed by an uptake pulse of 15′ and a chase of up to 165′ for a total of 3 h. Ultrastructural changes were visualized by electron microscopy at the indicated pulse/chase times. The successive formation of electron-lucent internal phagosomal vesicles (5′/0), multi-vesicular bodies (15′/0), and electron-dense vesicles (15′/15′) was observed. At (15′/15′) and (15′/45′), phagosomes contain a complex mix of dense vesicles and multivesicular bodies. Complexity of internal vesicles then decreased. Only electron-lucent vesicles were observed in the last stage of maturation (15′/165′). Large multi-bead phagosomes were observed from (15′/45′) onward. Each panel shows a representative picture of a mixed population. Scale bar, 0.5 μm. B, Functional classification of an exhaustive wild-type phagosome proteome of 1291 proteins identified by a combination of 1D and 2D PAGE separation and MS sequencing (supplemental List S1). The number of identified proteins belonging to each class and its percentage to the total are indicated. The same protein classification terms were used as in our initial study (10). C, Phagocytic uptake of 0.5, 1.0, and 4.5 μm fluorescent latex beads by myoK null (green) and abp1 null (red) cells relative to wild type. Data are expressed as a percentage of beads ingested by wild-type cells at 90 min. D, MyoK and Abp1 have differential temporal profiles during phagosome maturation as shown by immunoblots of phagosomes purified at the indicated times. Immunoblot signals were quantified and normalized to 100% according to the maximum value in wild type. Mutant profiles are normalized according to mutant-to-wild type ratios at early time-points (5′/0, 15′/0) as described in details in the Material and Methods section.
An exhaustive list of proteins identified in phagosomes isolated from wild type cells was built by compiling all our data acquired in this study and others (1, 10). It comprises 1291 proteins identified from two-dimensional and one-dimensional gels (Fig. 1B, supplemental List S1). Its content in predicted transmembrane proteins (18%) and secreted proteins (11% with a signal peptide) as well as in proteins of unknown function (20%) are similar to what is found in the whole Dictyostelium proteome (22% of transmembrane proteins, 12% of proteins with signal peptides, >30% of proteins of unknown function). Therefore, this list offers the most representative picture to date of the diverse biological processes participating in phagosome biogenesis and maturation in Dictyostelium. Protein classification illustrates the central but uncharacterized role of metabolic enzymes in nutrient assimilation (>10%), the importance of signaling cascades (≈ 10%) downstream of a small set of potential receptors (≈ 2%), and the complexity of membrane trafficking, targeting and remodeling factors such as Rab GTPases, SNAREs and cytoskeletal proteins, respectively (≈ 20% in total). Novel categories emerged from the present analysis: 1) lipid metabolism (4.1%), indicative of the intense remodeling of phagosomal membrane lipid composition and the degradation of the prey by lipases, 2) cell defense, including proteins involved in immunity-related functions and particle degradation, possibly representing proteins involved in specific bacteria-sensing and killing machineries (34, 35).
A majority of the identified proteins have a known GO term annotation (1124/1291, 87.1%) or can be matched in the KEGG pathways (1288/1291, 99.8%). Significant GO terms encompass the basic functions of the phagosome i.e. vesicle trafficking, reorganization of the cytoskeleton, energy-dependent proton pumping, mainly because of phagosome acidification by the V-ATPase complex, and killing of the engulfed bacteria (Table I). KEGG pathways stress that phagocytosis is an endocytic process and plays a central role in nutrition and cell metabolism. Unexpected is the significant enrichment of ER-associated protein processing, protein export machineries and the proteasome in the phagosome (Table I). Indeed, the ER-associated degradation (ERAD) machinery (sec61, p97, Ufd1, Dsk2) and ER-Golgi intermediate compartments (ERGIC; ERGIC53, COPII) represent a majority of the proteins contributing to the “protein processing in the ER” pathway (supplemental Fig. S2). In addition, ubiquitin mediated proteolysis, a pathway closely associated with the proteasome, is also highly represented on the phagosome (p = 0.58).
Table I. Number and percentage of identified phagosomal proteins matching Gene Ontology (GO) terms and KEGG pathways.
| GO term: | Phagosome | Whole cell | Enrichment |
|---|---|---|---|
| GO:0015986: ATP synthesis coupled proton transport | 14 (1.25%) | 16 (0.21%) | 5.82 |
| GO:0009617: response to bacterium | 40 (3.56%) | 53 (0.71%) | 5.02 |
| GO:0009144: purine nucleoside triphosphate metabolic process | 29 (2.58%) | 47 (0.63%) | 4.11 |
| GO:0030036: actin cytoskeleton organization | 42 (3.74%) | 107 (1.43%) | 2.61 |
| GO:0006886: intracellular protein transport | 41 (3.65%) | 109 (1.46%) | 2.5 |
| GO:0016192: vesicle-mediated transport | 56 (4.98%) | 157 (2.10%) | 2.37 |
| KEGG pathways: | Phagosome | Whole cell | Enrichment | p value |
|---|---|---|---|---|
| Ribosome | 42 (3.26%) | 86 (0.65%) | 5.05 | <105 |
| Protein processing in endoplasmic reticulum | 22 (1.71%) | 66 (0.50%) | 3.44 | <105 |
| Protein export | 11 (0.85%) | 17 (0.13%) | 6.69 | <105 |
| Phagosome | 18 (1.40%) | 40 (0.30%) | 4.65 | <105 |
| Oxidative phosphorylation | 30 (2.33%) | 66 (0.50%) | 4.70 | <105 |
| Metabolic pathways | 114 (8.85%) | 499 (3.75%) | 2.36 | <105 |
| Endocytosis | 19 (1.48%) | 39 (0.29%) | 5.03 | <105 |
| Citrate cycle (TCA cycle) | 15 (1.16%) | 30 (0.23%) | 5.17 | <105 |
| Biosynthesis of secondary metabolites | 40 (3.11%) | 174 (1.31%) | 2.38 | <105 |
| Alanine, aspartate and glutamate metabolism | 11 (0.85%) | 20 (0.15%) | 5.68 | 1 × 105 |
| Proteasome | 14 (1.09%) | 38 (0.29%) | 3.81 | 3 × 105 |
| Glycolysis/Gluconeogenesis | 12 (0.93%) | 28 (0.21%) | 4.43 | 3 × 105 |
The comparison of our phagosome proteome with the recently published Dictyostelium macropinosome proteome (36) allowed us to identify pathways and proteins unique to the phagosome. Both proteomes share 62% of identified proteins (supplemental List S1) (36). General metabolism (81%), lipid metabolism (75%), protein degradation (75%) and biosynthesis (85%) and membrane trafficking (68%) are mostly shared between the two organelles whereas receptors (47%) and signaling (47%) are less shared than the average. Indeed, both macropinosome and phagosome are used to assimilate endocytosed nutrients but phagosome formation is receptor-mediated, confirming our classification. Interestingly, the KEGG pathways “ER-associated protein processing, protein export machineries” and “proteasome” are also significantly enriched on the macropinosome (p value < 10−5, data not shown).
MyoK and Abp1 have Distinct Roles in Phagosome Maturation
The characterization of the phagosome proteome in wild type Dictyostelium was a prerequisite to analyze phagocytosis mutants at the molecular level. The phagocytic phenotypes of myoK- and abp1-null mutants have been characterized previously using rather big particles (yeasts or 4.5 μm beads) compared with the size of Dictyostelium (10 μm diameter in suspension) (12, 13), but usually, smaller beads have to be used to isolate phagosomes in sufficient quantities for extensive immunoblot and 2D-DIGE analyses. Thus, the ability of both mutants to ingest beads of different sizes was tested (Fig. 1C). In abp1-null cells, uptake of fluorescent latex beads was increased, independently of bead size. In myoK-null cells, uptake was generally decreased for all bead sizes, but only significantly for the 4.5 μm beads. Therefore, it confirms that Abp1 is a negative regulator of phagocytosis whereas MyoK is a positive regulator essential for efficient phagocytosis of large particles (12). Although the absence of MyoK is not limiting for the uptake of smaller beads, our experimental set up might still allow us to observe the signature of a defect in mutant cells. Indeed, the slow uptake of single large particles is likely mimicked by the simultaneous ingestion of a large number of medium-sized beads as is the case during the initial pulse of our phagosome isolation procedure (9).
To delineate the time frame of action of MyoK and its binding partner Abp1 during maturation, their presence was monitored by quantitative immunoblotting of phagosomes isolated from wild type cells following our pulse/chase feeding protocol. To further assess their functional inter-relationships, their relative abundance was measured in phagosomes from myoK-null and abp1-null mutants (Fig. 1D). The maturation profiles of MyoK and Abp1 in phagosomes isolated from wild type cells were similar but distinct. Both proteins were more abundant on early than on later phagosomes but Abp1 remained associated longer with phagosomes. MyoK was significantly more abundant on early phagosomes from abp1-null cells although its overall maturation profile was not drastically altered. Abp1 showed both an increased abundance on early phagosomes from myoK-null cells and a decreased relative abundance at later time points. In conclusion, Abp1 and MyoK are not necessary for their mutual recruitment on the phagosome, but the absence of one increases the relative abundance of the other at early time points, confirming their distinct but mutually dependent roles in phagosome maturation.
Absence of Abp1 Alters the Maturation Profile of its Direct Binding Partners
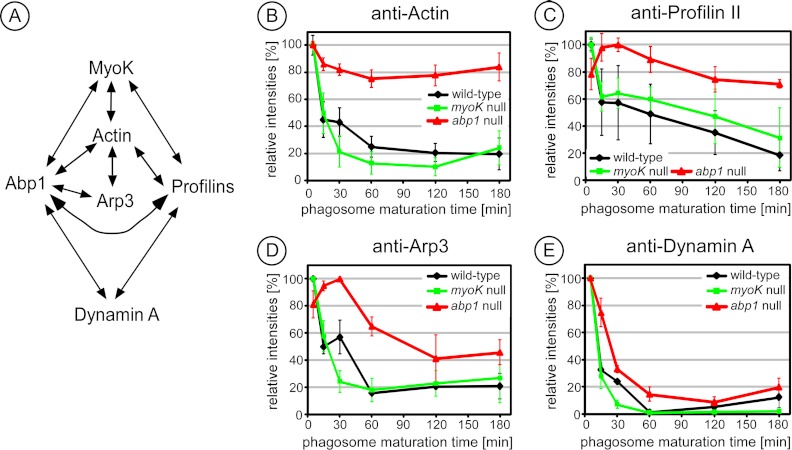
To delineate the impact of myoK- or abp1-null mutations on maturation, the relative abundance profiles in wild type and mutants of the direct binding partners of MyoK and Abp1 (schematically presented in Fig. 2A) were compared by quantitative immunoblotting. Absence of Abp1 but not MyoK markedly altered the maturation profile of its direct binding partners. Wild type phagosomes were highly enriched in actin at early stages only. In contrast, the relative abundance of actin did not vary significantly during maturation in abp1-null cells (Fig. 2B). However, it is not clear if this signal corresponds to actin monomers or polymers, because the signal was under the detection threshold for staining with the fluorescent probes DNase I and phalloidin, binding respectively G- and F-actin, or indirect immunofluorescence against the G-actin binding protein, profilin II. Furthermore, abp1 mutation markedly delayed the maximum peak of relative abundance of profilin II (Fig. 2C) or Arp3 (Fig. 2D). Dynamin A also remained associated longer with abp1-null phagosomes (Fig. 2E). Therefore, the absence of Abp1 directly impacts on the abundance of actin and proteins involved in actin dynamics on the phagosome whereas this is not the case in absence of MyoK.
Fig. 2.
Absence of Abp1 alters the maturation profile of its direct binding partners. A, The MyoK-Abp1 interaction network as characterized in (12) B–E, Maturation profiles of the indicated direct Abp1 binding-partners are affected in the abp1 null (red) cells but not in myoK null (green) cells compared with wild type (black). Phagosomes were purified from the respective genetic background at the indicated times. Immunoblotting signals were quantified and normalized to 100% according to the peak value in wild type. Curves are the average of at least triplicate experiments. Error bars represent ± S.E.
2D-DIGE Reveals An Early Maturation Defect in myoK Null Cells and an Overall Maturation Defect in abp1-null Cells
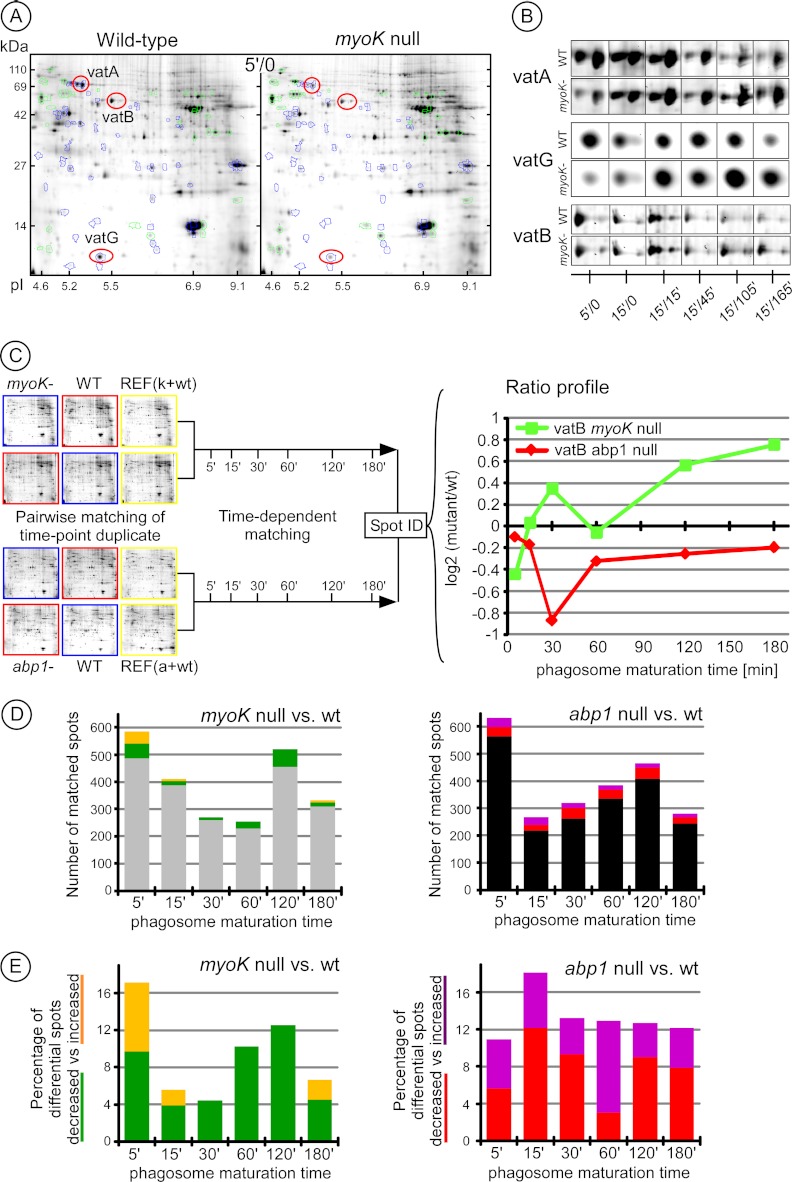
Defects in the maturation profiles of individual proteins were readily apparent using targeted immunoblot profiling. To expand the analysis, in depth characterization of the phagocytic phenotypes of myoK- and abp1-null mutants was carried out with an unbiased 2D-DIGE cross-comparison. Differences in the relative abundance of proteins were visible between phagosomes of wild type and mutant cells. These differences were not only seen early in maturation but were also consistent throughout maturation as, for example, for the individual subunits (VatA, VatG and VatB) of the V-ATPase complex (Figs. 3A, 3B). Phagosomes from each maturation stage were isolated in quadruplicate from wild type cells and in duplicate from each mutant cell line. Thus, each of the six time-points in the wild type was compared in duplicate to its corresponding time point in the mutant, with dye inversion, corresponding to 24 gels in total (Fig. 3C and supplemental Fig. S3). Gels were first matched between biological replicates of identical time-points to obtain a reliable quantitative mutant-to-wild type (wt) spot ratio. Such ratios were determined for about 200 to 600 spots pairs per time point (Fig. 3D). The analysis of myoK-null phagosomes revealed a higher proportion (17%) of differential spot pairs early (5′/0) and fewer differences later during maturation (4–12%, average ∼7%) (Figs. 3B–3D). In contrast, the number of differential spots in abp1-null phagosomes was higher and rather constant (11–18%, average ∼13%) (Fig. 3E). Therefore, 2D-DIGE analyses identify an early maturation defect in myoK-null cells but an overall perturbation of maturation in abp1-null cells.
Fig. 3.
Characterization of the phagocytosis defect of Dictyostelium myoK null and abp1 null mutants by quantitative 2D-DIGE analysis of isolated phagosomes. A, Differences were visible in phagosomes isolated at early times (5′/0) from wild-type compared with myoK null cells. Differential spots with increased or decreased intensity in the mutant are highlighted in green (increased) or blue (decreased). The spots containing the VatA, VatB, and VatG subunits of the V-ATPase are circled in red. B, A representative gallery of these spots highlights variations in spot intensity during maturation in wild type compared with myoK null. C, Workflow of the approach used to quantify mutant-to-wild type spot intensity ratios during the maturation process. D, The relatedness of the phagosome maturation program in wild type and mutants is represented by the number of differential versus nondifferential phagosomal spots at each time point in myoK null (left) and abp1 null mutants (right). Differential spots are in color (detailed view in Fig. 3E) whereas non-differential spots are in gray (myoK null) and black (abp1 null). E, The variation of spots differential during maturation in mutants is expressed as a percentage of total matched spots. The left panel shows spots decreased (green) or increased (orange) in myoK null phagosomes. The right panel shows spots decreased (red) or increased (violet) in abp1 null phagosomes.
In order to identify general cellular defects caused by gene knockout and evaluate their impact on phagosome maturation, total cell lysates of both mutants were compared with wild type by 2D-DIGE. Two spots were differential in the myoK-null mutant, representing less than 0.5% of all spots, and contained two proteins of unknown function (supplemental Table S1). Eight spots were differential in the lysate of abp1-null cells, representing 1% of all spots, and spanned various functions at all levels of cell metabolism (supplemental Table S2). The impact of the mutations on the whole cell is very limited (≤ 1% differential spots) in comparison to the impact on phagosome maturation (4–18% differential spots). Nevertheless, the mutation of abp1 affects more profoundly cell metabolism than does the mutation of myoK, confirming the phagosome analysis.
To monitor the variation of the mutant-to-wt ratio during maturation, spots pairs were then matched across the whole time-course resulting in time-resolved ratio profiles (Fig. 3C). 70% of all spot pairs (≥ 1000) matched between biological replicates were matched to at least another pair across the time-course. The 150 spot pairs were matched to at least five time-points resulting in a complete ratio profile. Between 200 (myoK-null) and 300 additional (abp1-null) pairs were matched across at least three time-points. Gel references were chosen in the early time-points (5′/0) to match myoK-null to abp1-null gel series so that mutant-to-wt ratio profiles could be compared across mutants. Abundant spots and most differential spots were picked from the gel and identified by mass spectrometry-based protein sequencing. The 104 proteins were identified, representing an average of 220 spot pairs with known ratio profiles over the two mutant-to-wt series.
myoK-null Phagosomes Display an Early Imbalance in the Abundance of ER Components and the V-ATPase Complex
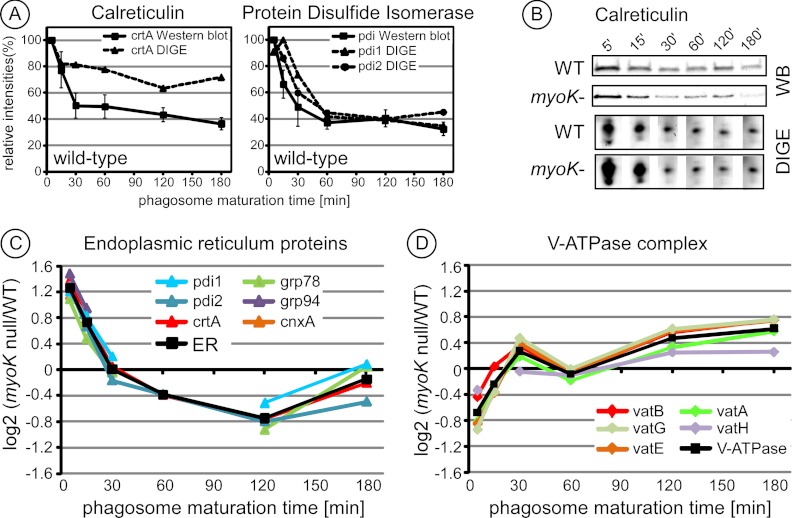
2D-DIGE intensity profiles were first confirmed by immunoblot profiling. Using both detection techniques, the wild type profiles of the ER proteins, calreticulin and protein disulfide isomerase, showed a maximum enrichment in early phagosomes and then a progressive decrease during maturation (Figs. 4A, 4B). These profiles implied that ER-derived membranes fuse with the phagosome just after cup closure and ER-associated proteins are then removed later during maturation.
Fig. 4.
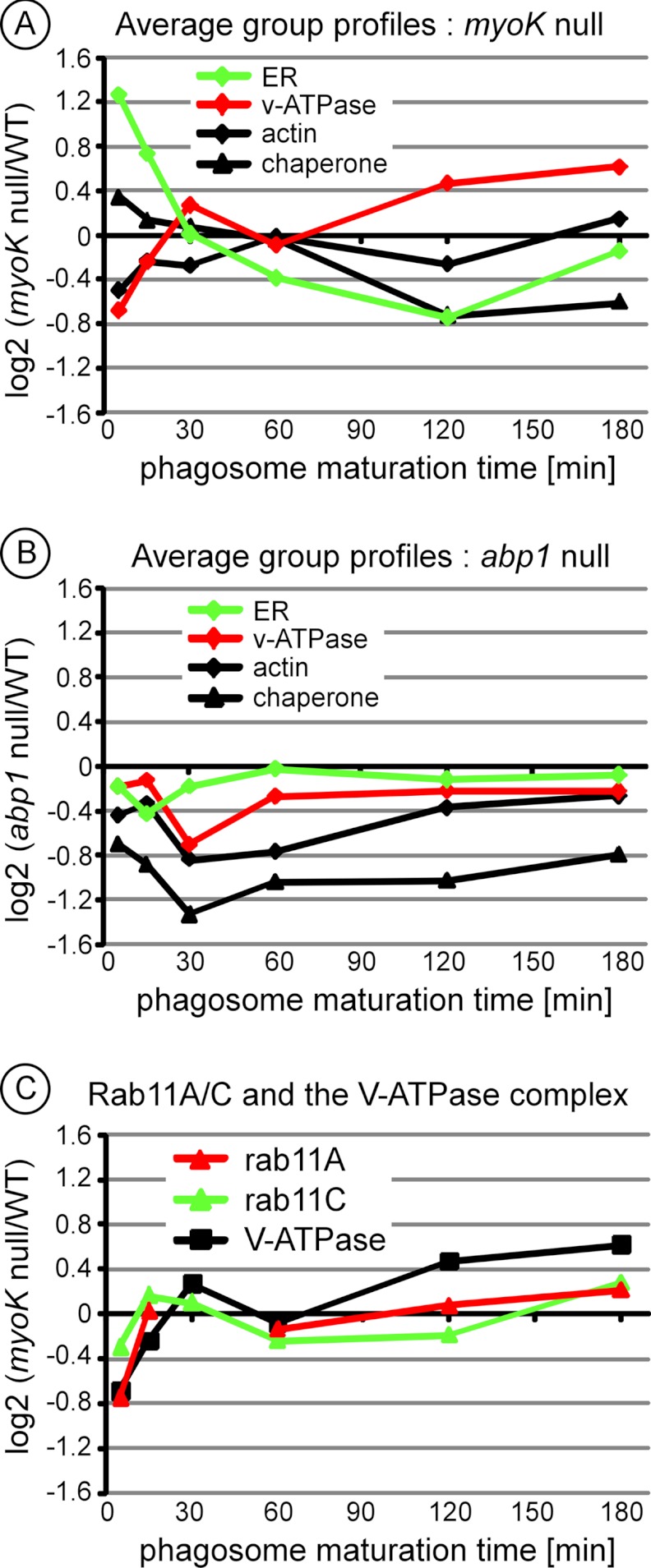
The ratio profiles of ER components and V-ATPase subunits are inversely correlated when comparing phagosomes from myoK null and wild type cells. A, Curves from both quantitative immunoblotting and 2D-DIGE intensity profiling of calreticulin (crtA) and protein disulfide isomerase (pdi) isoforms in phagosomes from wild-type cells were highly similar, with a maximum early in maturation (5′/0). For immunoblot and 2D-DIGE, the peak intensity value of the profile was set to 100% and the rest of the curve was normalized accordingly. In immunoblots, curves are based on triplicates and error bars represent ± S.E. 2D-DIGE profiles represent duplicates. B, Immunoblot profiles and 2D-DIGE spot images of the maturation sequence of calreticulin in phagosomes from wild-type and myoK null cells. C, D, myoK null/wild-type ratio profiles in log2 scale of normalized spot intensities of identified ER proteins (C) and of subunits of the v-ATPase complex (D). The profiles of all ER proteins and V-ATPase subunits identified in myoK-null phagosomes are shown here. The profiles were so similar that an averaged profile of the corresponding functional groups was computed and drawn in black. Only gene names (www.dictybase.org) are indicated in the legend. ER proteins: crtA, calreticulin A; cnxA, calnexin A; grp78 and grp94, glucose regulated protein of 78 kDa and 94 kDa; pdi1 and 2, protein disulfide isomerase isoform 1 and 2; soluble v-ATPase complex subunits vatA, B, E, G, H.
When comparing phagosomes from wild type and myoK-null cells, prominent differences were seen in the abundance profiles of both ER proteins and subunits of the V-ATPase complex (Figs. 3B and 4B). Measured ratios of the quantified spot intensities confirmed these observations. Mutant-to-wt ratio profiles of all identified ER proteins (Fig. 4C) were sufficiently homogenous to unequivocally group together. Similar grouping was also obvious for the ratio profiles of the soluble subunits of the V-ATPase complex (Fig. 4D). Ratio profiles of individual proteins fitted so well that a representative average ratio profile could be drawn for both functional groups (black line). A significant increase (1.6-fold, log2 > 0.68) in the abundance of ER proteins and a significant decrease (1.6-fold, log2 < 0.68) in the abundance of the V-ATPase subunits were observed in early phagosomes (5′/0 min.) from myoK-null cells. Strengthening this early deviation from the wild type maturation profile, the ratio profile of both groups showed reciprocal variations during maturation. This revealed that early imbalance of ER versus V-ATPase proteins was then progressively compensated during maturation. Other proteins were also functionally grouped according to their ratio profile, like the phagosomal chaperones and the different actin isoforms (Figs. 5A, 5B, details in supplemental Fig. S4). Unlike the ER and V-ATPase functional groups, the actin and chaperones groups were not significantly differential in early phagosomes and their profile was not linked either to the ER or V-ATPase group in the myoK-null background (Fig. 5A). Neither ER nor V-ATPase proteins were differential in abp1-null phagosomes. Therefore, significant differences in early abundance of ER and V-ATPase proteins on the phagosome and their reciprocal ratio profile variation were specific to the myoK-null to wild type comparison.
Fig. 5.
The early trafficking imbalance of ER components versus V-ATPase subunits is specific to myoK-null phagosomes and might be regulated by Rab11 small GTPases. A, B, myoK null/wild type (A) or abp1 null/wild type (B) averaged ratio profiles in log2 scale for ER proteins, V-ATPase, actin and hsp40/hsp70 chaperones show that the profiles of ER versus V-ATPase are only inversely correlated when comparing myoK null to wild type phagosomes. Proteins included in the ER and V-ATPase groups are listed in Figs. 4C and 4D, respectively. The chaperone group includes all hsp40/hsp70 chaperones identified by 2D DIGE (hsp40-like chaperones, ddj1 & dnaJC7; hsc70-like chaperone, hspE). The actin group includes all the spots where actin isoforms were identified, excluding degradation products. C, Averaged ratio profiles for Rab11A and Rab11C small GTPases isoforms anticipate changes in the averaged profile of the V-ATPase complex, in both time and amplitude on myoK null phagosomes. The profiles of all Rab11A and Rab11C isoforms have been averaged (details in supplemental Fig. S5).
Strikingly, changes in the myoK-null to wild type ratio profile of the small GTPases Rab11A and Rab11C anticipate changes in the profile of the V-ATPase complex, in both time and amplitude changes (Fig. 5). Furthermore, the regulation or the kinetics of these small GTPases on myoK-null phagosomes might depend on their post-translational modifications (supplemental Fig. S5).
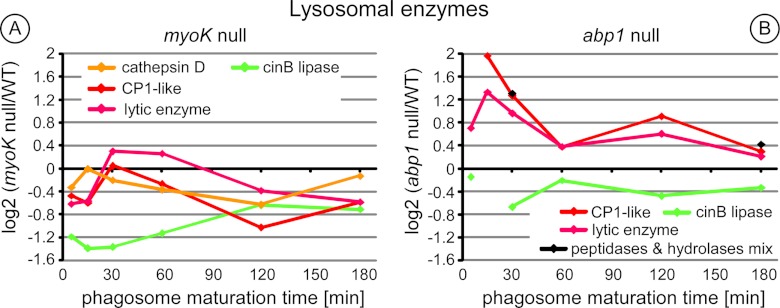
The Relative Abundance of Lysosomal Enzymes is Altered in Early abp1-null Phagosomes
Relative abundance of lysosomal enzymes was specifically increased in early (15′/0) abp1-null phagosomes (2.5–3.9-fold, Fig. 6). This increase was transient as minimal difference was observed at later time-points. Chaperones of the hsp70 family (HspE) and hsp40 family (ddj1, DnaJC7) were also clearly differential (1.7–3.1-fold) from the 15′/15′ time point across the whole phagosomes maturation process in abp1-null cells (Fig. 5B, details in supplemental Fig. S4).
Fig. 6.
Ratio profiles of the lysosomal enzymes in abp1-null phagosomes highlight a trafficking defect in early maturation. Ratio profiles of the lysosomal enzymes (A, B) from phagosomes of myoK-null cells (A) or abp1-null cells (B). Lysosomal enzymes: cathepsin D (ctsD, DDB_G0279411), cysteine protease 1-like (CP1-like, DDB_G0291191), lytic enzyme (DDB_G0293566), a/b hydrolase (DDB_G0287609), mix of putative physaropepsin, acyloxyacyl hydrolase, peptidase V4–7 (DDB_G0290333, DDB_G0272785, DDB_G0281823). Cathepsin D is not present in the gels of one of the time-series of abp1-null phagosomes. Therefore, it does not fulfill our criteria of significance and is not represented in the left panel. CinB is an esterase/lipase with its closest mammalian homolog (UniProt ID: NCEH1_MOUSE) being a cholesterol esterase present in the ER. It is represented here as a counterexample.
Functional Impact of the Trafficking Defects Detected by 2D-DIGE on Phagosomal pH and Proteolytic Activity
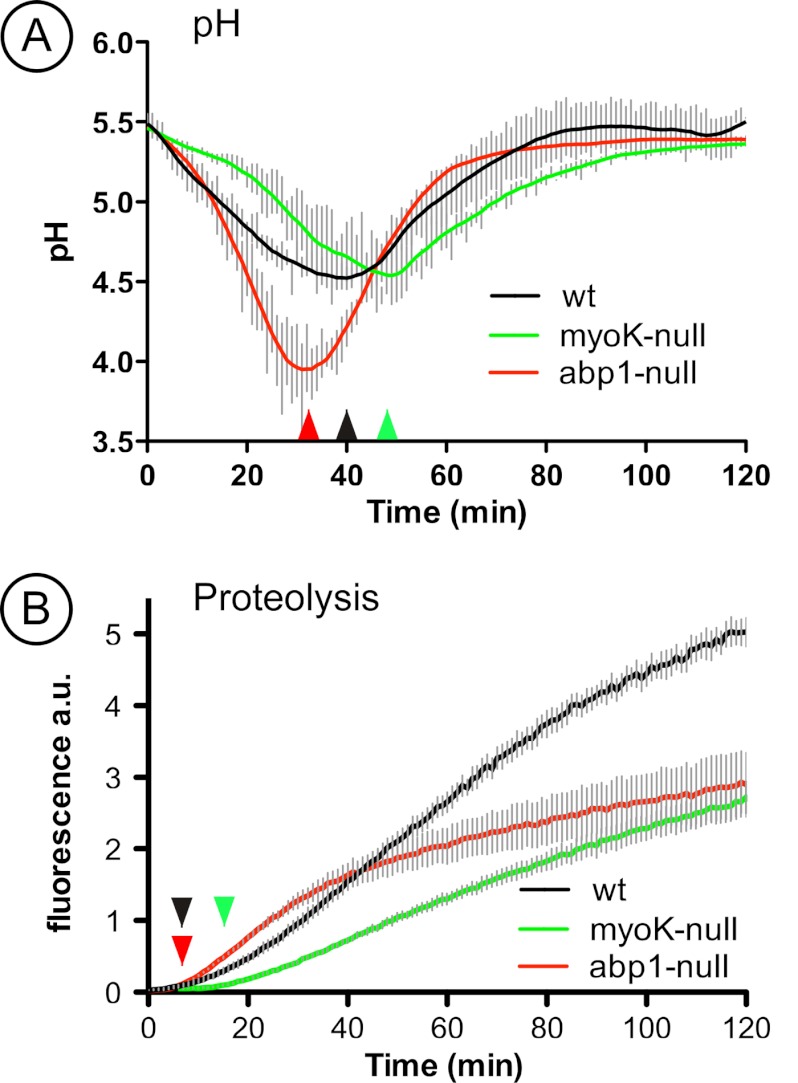
In order to monitor in vivo the functional impact of the severe trafficking defects revealed by ratio profiling, we adapted to Dictyostelium the use of intra-phagosomal biosensors developed by Russell and colleagues (57, 58). Such approaches based on dual wavelength fluorescence intensity ratio measurements allow to quantitatively measuring environmental parameters such as pH and proteolysis. Compared with the acidification profile obtained in wild type cells, in which the pH minimum of ∼4.5 was reached after about 40 min and the re-neutralization plateau reached after about 70 min, the acidification of phagosomes in myoK-null cells was significantly delayed. (Fig. 7A, 7D). It also reached a pH of ∼4.5, but after about 50 min and the neutralization plateau not before about 100 min. As a likely consequence, the onset of proteolytic activity was slightly delayed but it also progressed at a severely reduced but almost linear rate (Figs. 7B, 7D). Note that the proteolytic activity in wild type phagosomes was also subjected to a decrease of rate after about 70 min, corresponding to the time taken to reach the neutralization plateau (Fig. 7C). Strikingly, in abp1-null cells, phagosomes reached a more acidic value of 4.0 earlier, at about 30 min, the re-neutralization phase started earlier but led to a plateau at a time (70 min) similar as in wild type cells (Figs. 7A, 7C). The proteolytic activity monitored in abp1-null cells was strongly affected (Figs. 7B, 7E). The onset of activity occurred slightly earlier than in the two other cell lines, and initially progressed at a higher rate, but after about 30 min, this rate decreased significantly, corresponding to the start of the neutralization phase.
Fig. 7.
myoK-null cells are defective in phagosomal acidification and proteolytic digestion whereas abp1-null cells display a sudden decrease in phagosomal proteolytic digestion during maturation. Measure of pH variation (A) and proteolytic digestion (B) during maturation in phagosomes in vivo in AX-2 (black), myoK-null cells (green), and abp1-null cells (red). Curves are averaged from at least three experiments in duplicates and error bars represent ± S.E.
Overall, these functional results are in agreement with the 2D-DIGE ratio profiling data documenting the transient initial deficit in V-ATPase delivery in myoK-null cells and the transient initial overabundance of lysosomal enzymes in abp1-null cells.
DISCUSSION
Before engaging on an exhaustive comparative and quantitative analysis of phagosome maturation in Dictyostelium myoK-null and abp1-null mutants, we characterized the purity, the morphology and the composition of phagosomes isolated from wild type cells (Fig. 1). Observation of intraphagosomal vesicles and formation of large multi-bead phagosomes during maturation highlight that phagosome plasticity is achieved not only through classical vesicle fission from and fusion to the phagosome, but also via the formation of intralumenal vesicles and phagosome-phagosome fusions. Phagosomes not only shares 62% of their protein composition with macropinosomes (36) but also morphological features with the pinocytic endosomal pathway as changes of intraphagosomal vesicles during maturation are similar to those observed in fluid-phase endosomes (37). Intralumenal vesicles have already been observed in macrophage phagosomes containing latex beads or red blood cells (38). These vesicles might result from the action of the ESCRT pathways (39) or from fusion with autophagosomes containing cytoplasmic material (40).
The analysis of phagosome composition revealed several proteins involved in lipid metabolism, indicating that the phagosome lipid composition might be dynamically modified to participate in membrane remodeling and vesicle trafficking (Fig. 1B). Comparison with the macropinosome proteome (36) emphasized the fact that the pathways for ER-associated protein processing, protein export machineries and the proteasome are significantly enriched in both proteomes (Table I). Given the differences in the purification procedures of both organelles and their significant enrichment, it strongly suggests that ER components and the proteasome are not mere contaminants in the biogenesis of both phagosome and macropinosome.
Detailed analysis of the phagocytic phenotypes of myoK- and abp1-null cells confirmed that Abp1 is a negative regulator of phagocytosis whereas MyoK is a positive regulator determinant for phagocytosis efficiency of large particles only (Fig. 1C). The analysis of maturation profiles shows that Abp1 and MyoK are not necessary for their mutual recruitment on the phagosome and, in addition, that the absence of one increases the relative abundance of the other (Fig. 1D). These observations confirm that MyoK and Abp1 have opposite regulatory roles in phagocytosis (12).
2D-DIGE analysis of phagosomes showed that myoK-null cells display an early maturation defect whereas an overall perturbation of maturation is observed in abp1-null cells. The amplitude of the defects correlates well with their respective abundance during phagosome maturation in wild type cells (Figs. 3D, 3E). These defects also correlated with differences in the cellular transcriptome (data not shown) and proteome (supplemental Tables S1, S2). The impact of the mutations is on average ten times bigger on phagosome maturation than on general cell metabolism, as judged by the number of differential spots in phagosomes compared with whole cell lysates.
We had previously shown that quantitative analysis of maturation profiles, by Western blotting and two-dimensional gels, allowed hierarchical clustering of functional groups based on profile similarities (10). Here, we show that comparative 2D-DIGE analysis of phagosome maturation in mutant cells resulted in the quantitative monitoring of the mutant to wild type ratio profiles of more than one hundred identified proteins. Strikingly, ratio profiles of functionally related proteins showed coordinated changes during maturation. These changes were specific for each of the two mutants examined, thus validating the functional classification of phagosomal proteins.
Our study shows that Abp1 has pleiotropic effects on phagosome maturation. Its absence transiently increases phagosomal proteolytic activity paralleling an early decrease in pH (Fig. 7E) and the transient early accumulation of lysosomal enzymes (Fig. 6B). This increased abundance of lysosomal enzymes in the phagosome is either due to increased delivery or to impaired retrieval. The causes for this early defect in trafficking efficiency could originate from an accumulation of actin and other direct binding partner of Abp1 on the phagosome as shown by immunoblot profiling (Fig. 2). Other data support this hypothesis. Abp1 is the major F-actin binding player on phagosomes isolated from Dictyostelium in vitro and its absence might disorganize the dynamics of the actin network on the phagosome (18). Indeed, an increase in phagocytosis rates in knockdown of human Rab27a correlates with a quicker turnover of F-actin coating and degradation at the phagocytic cup (41). Furthermore, persistence of transient actin flashes on the phagosome prevents fusion with lysosomes (22). Unexpectedly, the early trafficking defect of lysosomal enzymes in abp1-null phagosomes is followed by a decrease in proteolytic activity. Therefore, not only a decrease in pH but also a precisely coordinated trafficking of lysosomal enzymes regulates phagosomal proteolysis.
In the myoK-null mutant, 2D-DIGE temporal ratio profiling clearly shows an early imbalance of ER components versus V-ATPase subunits (Figs. 4C, 4D, Fig. 5A). In addition, ratio profiles of ER proteins and the V-ATPase complex are reciprocal. According to wild type intensity profiles, ER proteins are delivered early to the phagosome to be recycled later suggesting that ER fusion occurs just after phagosome closure (Figs. 4A, 4B). ER proteins and the V-ATPase complex are not delivered from the same organelle, because the largest amount of V-ATPase complexes is localized on the contractile vacuole and V-ATPase complexes are delivered from endosomes and uncharacterized vesicles in Dictyostelium (4). Thus, we interpret this imbalance in protein abundance as an early trafficking imbalance to the phagosome in myoK-null cells.
Our data raise the following essential questions. Which proteins regulate the delivery of the V-ATPase complex to the phagosome? Why are ER proteins delivered early to the phagosome in Dictyostelium and do our data correlate with existing hypotheses? Is an imbalance in the delivery of ER and V-ATPase to the phagosome a conserved phenomenon and how is MyoK implicated in this trafficking event?
Potential candidates regulating the trafficking of the V-ATPase complex are the canonical endosomal GTPases Rab7A and Rab5A. Both proteins have been identified in phagosomal samples (this study) and could participate in the delivery of the V-ATPase from endolysosomes. On the other hand, our 2D-DIGE ratio profiling study suggests that the small GTPases Rab11A, Rab11C and RabC could participate in the delivery of the V-ATPase from an uncharacterized vesicle pool. Changes in the myoK-null to wild type ratio profiles of the Rab11A, Rab11C and RabC small GTPases as well as calmodulin precede, in time and amplitude, the changes in the ratio profile of the V-ATPase complex (supplemental Fig. S5). Like the V-ATPase complex and calmodulin, Rab11A and Rab11C are mainly localized to the contractile vacuole (4, 42–44), and Rab11A is important for the maintenance of the contractile vacuole function and morphology (43). Rab11C and RabC have not been functionally characterized and RabC has no close homolog in any other organism (45). The small GTPase Rab14 was also identified on phagosomes (this study) and might be additionally involved in this process. Indeed, Rab14 localizes both to the contractile vacuole and the endosomal system, where it controls lysosome and phagosomes homotypic fusion (46).
The fusion with ER-derived membranes is an early event of phagosome maturation, and these components are then either recycled or degraded during maturation (Fig. 4, (47, 48)). However, the contribution of ER-derived proteins to phagosome function and the mechanisms driving their delivery are still unclear both in Dictyostelium and animal phagocytes (5, 49). Three working hypotheses might be invoked. First, focal exocytosis of endomembranes including the ER might be crucial to compensate for the loss of cell surface area during particle uptake (7, 50). Our data do not fit with a contribution of ER membranes to enhance efficiency of uptake, because the myoK-null mutant which displays an increase in the delivery of ER components does not exhibit increased uptake of any big-sized particles (Fig. 1C) (13). Second, cortical or phagosomal ER patches could trigger a periphagosomal increase in free calcium concentration, regulating focal exocytosis or local signaling cascades (6, 51, 52). The decreased phagocytic uptake of calnexin/calreticulin double knock out mutants was interpreted as the involvement of calcium signaling in efficient uptake (53). Nevertheless, enhancing contact of the ER with the phagosome does not enhance uptake in myoK-null mutants. Therefore, the extent and duration of the contact between the ER and the phagosome and how long this contact takes place has to be tightly regulated to optimize the efficiency of uptake. Finally, recruitment of ER proteins to the phagosome in macrophages and dendritic cells parallels an increase in cross-presentation of exogenous antigens, a process controlled by Sec22b, a SNARE present on ERGICs (5, 48, 54). This increase in cross-presentation is linked to a slow initial decrease in pH in dendritic cells (55). Although amoebae use phagocytosis to feed and do not express antigen-presenting molecules, molecular machines essential for cross-presentation like ER chaperones, ERGIC and ERAD components are enriched in the phagosome of Dictyostelium (Table I) and have been identified in a recent comparative proteomic analysis of the phagosome in mouse, Drosophila and Dictyostelium, showing that these components have been conserved throughout evolution as parts of the phagosome proteome core (1). It was recently proposed that, during evolution from prokaryotes to eukaryotes, the ER, the ERAD and the proteasome machineries primarily served a function linked to feeding, i.e. import and cleavage of polypeptides resulting from extracellular digestion (56). Thus, ER components could have been first used to assist feeding and then co-opted to maximize efficiency of antigen cross-presentation.
The balance in the delivery of ER components and of the V-ATPase complex to the phagosome can be genetically altered in Dictyostelium by knocking out myoK. This balance can be also altered pharmacologically in macrophages. Specific inhibition of the V-ATPase complex by bafilomycin or inhibition of early membrane trafficking by 3-methyladenine and wortmannin increases interaction of the ER with the phagosome (5). Thus, reducing V-ATPase activity or blocking early membrane delivery might increase ER fusion. In dendritic cells, the balance between ER fusion and lysosome delivery can be altered genetically. Absence of Sec22b delays fusion of lysosomes to latex bead containing phagosomes and reduces phagosomal proteolysis (48). In the myoK-null mutant, the imbalance is likely because of a slowed delivery of V-ATPase versus ER membranes as the abundance of the V-ATPase in the mutant reaches wild type levels at 15′/15′ and even exceeds it beyond this time point (Fig. 4D). Because MyoK has been localized at the phagocytic cup and on early phagosomes [(12) and Fig. 1D], we suggest that it might directly control retrieval of ER components rather than V-ATPase delivery. In analogy, another Dictyostelium class I myosin, MyoB, controls the retrieval of plasma membrane components from the endosomes (14). Alternatively, MyoK could indirectly alter membrane trafficking. MyoK is enriched at sites of cup closure and its absence lowers phagocytic rates (12) (Fig. 1C). Therefore, its absence potentially slows down cup closure, which would increase contact of ER membranes with the phagocytic cup. This might in turn hinder contact and delay fusion with a Rab11-positive endosomal compartment delivering the V-ATPase. In the myoK-null mutant, the reduced delivery of V-ATPase correlates with a reduced phagosomal acidification in vivo (Fig. 7A). As a likely consequence, the intra-phagosomal proteolytic activity is severely decreased (Fig. 7B). Thus, we propose that the efficiency of particle digestion and killing reflects this balance between fusion with the ER and delivery of the V-ATPase complex.
In conclusion, quantitative ratio profiling by immunoblots and 2D-DIGE is a powerful tool to measure mutant to wild type protein abundance ratios over the complete course of the maturation process. Targeted immunoblot profiling pinpointed the central role of Abp1 in organizing actin dynamics on the phagosome. Quantitative and comparative 2D-DIGE profiling was a reliable discovery tool to highlight the proteins that deviate from the wild type maturation profiles induced by genetic ablations. Clustering of such deviant profiles revealed concerted alterations in the abundance of functional protein groups. As a proof of concept, this approach exposed for the first time that the participation of ER components is part of the primordial process of maturation and that a reciprocal imbalance in the delivery of ER and V-ATPase components is induced genetically by knocking out the myosin IK and, consequently, impacts on the kinetic of acidification and proteolysis. Our study brings a genetic and proteomic demonstration that the ER participation in phagocytosis is an evolutionary conserved process regulated by proteins at the interface between cytoskeleton and membrane traffic.
Supplementary Material
Acknowledgments
We thank A. Bosserhof and M. Ellis for help with mass spectrometry, N. Gopaldass and N. Sattler with the pH and proteolysis measurement assays, and M. Hagedorn with electron microscopy. A special thank you goes to P. Gaudet, M.-C. Blatter, and E. de Castro from the SIB Swiss Institute of Bioinformatics for their help with bioinformatic analyses. We are also grateful to the whole team of DictyBase for their support. We thank Matthias Trost, Jonathan Boulais, and Florence Niedergang for careful reading of the manuscript and thoughtful suggestions.
Footnotes
* This work was supported by the Max-Planck Society, the Deutsche Forschungsgemeinschaft, the UK BBSRC, and the Swiss National Science Foundation.
 This article contains supplemental Figs. S1 to S5 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S5 and Tables S1 to S4.
1 The abbreviations used are:
- V-ATPase
- vacuolar H+-ATPase proton pump
- Abp1
- Actin binding protein 1
- MyoK
- myosin IK
- 2D-DIGE
- two-dimensional fluorescent differential gel electrophoresis
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation machinery
- ERGIC
- ER-Golgi intermediate compartments.
REFERENCES
- 1. Boulais J., Trost M., Landry C. R., Dieckmann R., Levy E. D., Soldati T., Michnick S. W., Thibault P., Desjardins M. (2010) Molecular characterization of the evolution of phagosomes. Mol. Syst. Biol. 6, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bajno L., Peng X. R., Schreiber A. D., Moore H. P., Trimble W. S., Grinstein S. (2000) Focal exocytosis of VAMP3-containing vesicles at sites of phagosome formation. J. Cell Biol. 149, 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Braun V., Fraisier V., Raposo G., Hurbain I., Sibarita J. B., Chavrier P., Galli T., Niedergang F. (2004) TI-VAMP/VAMP7 is required for optimal phagocytosis of opsonised particles in macrophages. EMBO J. 23, 4166–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clarke M., Köhler J., Arana Q., Liu T., Heuser J., Gerisch G. (2002) Dynamics of the vacuolar H(+)-ATPase in the contractile vacuole complex and the endosomal pathway of Dictyostelium cells. J. Cell Sci. 115, 2893–2905 [DOI] [PubMed] [Google Scholar]
- 5. Gagnon E., Duclos S., Rondeau C., Chevet E., Cameron P. H., Steele-Mortimer O., Paiement J., Bergeron J. J., Desjardins M. (2002) Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110, 119–131 [DOI] [PubMed] [Google Scholar]
- 6. Bozzaro S., Bucci C., Steinert M. (2008) Phagocytosis and host-pathogen interactions in Dictyostelium with a look at macrophages. Int. Rev. Cell Mol. Biol. 271, 253–300 [DOI] [PubMed] [Google Scholar]
- 7. Flannagan R. S., Cosío G., Grinstein S. (2009) Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7, 355–366 [DOI] [PubMed] [Google Scholar]
- 8. Gotthardt D., Warnatz H. J., Henschel O., Brückert F., Schleicher M., Soldati T. (2002) High-resolution dissection of phagosome maturation reveals distinct membrane trafficking phases. Mol. Biol. Cell 13, 3508–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dieckmann R., Gopaldass N., Escalera C., Soldati T. (2008) Monitoring time-dependent maturation changes in purified phagosomes from Dictyostelium discoideum. Methods Mol. Biol. 445, 327–337 [DOI] [PubMed] [Google Scholar]
- 10. Gotthardt D., Blancheteau V., Bosserhoff A., Ruppert T., Delorenzi M., Soldati T. (2006) Proteomics fingerprinting of phagosome maturation and evidence for the role of a Galpha during uptake. Mol. Cell. Proteomics 5, 2228–2243 [DOI] [PubMed] [Google Scholar]
- 11. Rogers L. D., Foster L. J. (2007) The dynamic phagosomal proteome and the contribution of the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 104, 18520–18525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dieckmann R., von Heyden Y., Kistler C., Gopaldass N., Hausherr S., Crawley S. W., Schwarz E. C., Diensthuber R. P., Côté G. P., Tsiavaliaris G., Soldati T. (2010) A myosin IK-Abp1-PakB circuit acts as a switch to regulate phagocytosis efficiency. Mol. Biol. Cell 21, 1505–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwarz E. C., Neuhaus E. M., Kistler C., Henkel A. W., Soldati T. (2000) Dictyostelium myosin IK is involved in the maintenance of cortical tension and affects motility and phagocytosis. J. Cell Sci. 113, 621–633 [DOI] [PubMed] [Google Scholar]
- 14. Neuhaus E. M., Soldati T. (2000) A myosin I is involved in membrane recycling from early endosomes. J. Cell Biol. 150, 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salas-Cortes L., Ye F., Tenza D., Wilhelm C., Theos A., Louvard D., Raposo G., Coudrier E. (2005) Myosin Ib modulates the morphology and the protein transport within multi-vesicular sorting endosomes. J. Cell Sci. 118, 4823–4832 [DOI] [PubMed] [Google Scholar]
- 16. Soldati T., Schliwa M. (2006) Powering membrane traffic in endocytosis and recycling. Nat. Rev. Mol. Cell Biol. 7, 897–908 [DOI] [PubMed] [Google Scholar]
- 17. Robertson A. S., Smythe E., Ayscough K. R. (2009) Functions of actin in endocytosis. Cell Mol. Life Sci. 66, 2049–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gopaldass N., Patel D., Kratzke R., Dieckmann R., Hausherr S., Hagedorn M., Neuhaus E., Hofmann E., Hille K., Kuznetsov S., Soldati T. (2011) Dynamin A, Myosin IB and Abp1 couple phagosome maturation to F-actin binding. Traffic 13, 120–130 [DOI] [PubMed] [Google Scholar]
- 19. Khurana T., Brzostowski J. A., Kimmel A. R. (2005) A Rab21/LIM-only/CH-LIM complex regulates phagocytosis via both activating and inhibitory mechanisms. EMBO J. 24, 2254–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rivero F. (2008) Endocytosis and the actin cytoskeleton in Dictyostelium discoideum. Int. Rev. Cell Mol. Biol. 267, 343–397 [DOI] [PubMed] [Google Scholar]
- 21. Wienke D. C., Knetsch M. L., Neuhaus E. M., Reedy M. C., Manstein D. J. (1999) Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum. Mol. Biol. Cell 10, 225–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liebl D., Griffiths G. (2009) Transient assembly of F-actin by phagosomes delays phagosome fusion with lysosomes in cargo-overloaded macrophages. J. Cell Sci. 122, 2935–2945 [DOI] [PubMed] [Google Scholar]
- 23. Hagedorn M., Rohde K. H., Russell D. G., Soldati T. (2009) Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science 323, 1729–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orci L., Like A. A., Amherdt M., Blondel B., Kanazawa Y., Marliss E. B., Lambert A. E., Wollheim C. B., Renold A. E. (1973) Monolayer cell culture of neonatal rat pancreas: an ultrastructural and biochemical study of functioning endocrine cells. J. Ultrastruct. Res. 43, 270–297 [DOI] [PubMed] [Google Scholar]
- 25. Carrette O., Burkhard P. R., Sanchez J. C., Hochstrasser D. F. (2006) State-of-the-art two-dimensional gel electrophoresis: a key tool of proteomics research. Nat. Protoc. 1, 812–823 [DOI] [PubMed] [Google Scholar]
- 26. Yusenko M. V., Ruppert T., Kovacs G. (2010) Analysis of differentially expressed mitochondrial proteins in chromophobe renal cell carcinomas and renal oncocytomas by 2-D gel electrophoresis. Int. J. Biol. Sci. 6, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fey P., Gaudet P., Curk T., Zupan B., Just E. M., Basu S., Merchant S. N., Bushmanova Y. A., Shaulsky G., Kibbe W. A., Chisholm R. L. (2009) dictyBase–a Dictyostelium bioinformatics resource update. Nucleic Acids Res. 37, D515–D519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 27, 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Curk T., Demsar J., Xu Q., Leban G., Petrovic U., Bratko I., Shaulsky G., Zupan B. (2005) Microarray data mining with visual programming. Bioinformatics 21, 396–398 [DOI] [PubMed] [Google Scholar]
- 31. Sonnhammer E. L., von Heijne G., Krogh A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 [PubMed] [Google Scholar]
- 32. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 33. Bloomfield G., Tanaka Y., Skelton J., Ivens A., Kay R. R. (2008) Widespread duplications in the genomes of laboratory stocks of Dictyostelium discoideum. Genome Biol. 9, R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cosson P., Soldati T. (2008) Eat, kill or die: when amoeba meets bacteria. Curr. Opin. Microbiol. 11, 271–276 [DOI] [PubMed] [Google Scholar]
- 35. Sillo A., Bloomfield G., Balest A., Balbo A., Pergolizzi B., Peracino B., Skelton J., Ivens A., Bozzaro S. (2008) Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium. BMC Genomics 9, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Journet A., Klein G., Brugière S., Vandenbrouck Y., Chapel A., Kieffer S., Bruley C., Masselon C., Aubry L. (2012) Investigating the macropinocytic proteome of Dictyostelium amoebae by high-resolution mass spectrometry. Proteomics 12, 241–245 [DOI] [PubMed] [Google Scholar]
- 37. Neuhaus E. M., Almers W., Soldati T. (2002) Morphology and dynamics of the endocytic pathway in Dictyostelium discoideum. Mol. Biol. Cell. 13, 1390–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee W. L., Kim M. K., Schreiber A. D., Grinstein S. (2005) Role of ubiquitin and proteasomes in phagosome maturation. Mol. Biol. Cell. 16, 2077–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vieira O. V., Harrison R. E., Scott C. C., Stenmark H., Alexander D., Liu J., Gruenberg J., Schreiber A. D., Grinstein S. (2004) Acquisition of Hrs, an essential component of phagosomal maturation, is impaired by mycobacteria. Mol. Cell. Biol. 24, 4593–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shui W., Sheu L., Liu J., Smart B., Petzold C. J., Hsieh T. Y., Pitcher A., Keasling J. D., Bertozzi C. R. (2008) Membrane proteomics of phagosomes suggests a connection to autophagy. Proc. Natl. Acad. Sci. U.S.A. 105, 16952–16957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yokoyama K., Kaji H., He J., Tanaka C., Hazama R., Kamigaki T., Ku Y., Tohyama K., Tohyama Y. (2011) Rab27a negatively regulates phagocytosis by prolongation of the actin-coating stage around phagosomes. J. Biol. Chem. 286, 5375–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Du F., Edwards K., Shen Z., Sun B., De Lozanne A., Briggs S., Firtel R. A. (2008) Regulation of contractile vacuole formation and activity in Dictyostelium. EMBO J. 27, 2064–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harris E., Yoshida K., Cardelli J., Bush J. (2001) Rab11-like GTPase associates with and regulates the structure and function of the contractile vacuole system in dictyostelium. J. Cell Sci. 114, 3035–3045 [DOI] [PubMed] [Google Scholar]
- 44. Zhu Q., Clarke M. (1992) Association of calmodulin and an unconventional myosin with the contractile vacuole complex of Dictyostelium discoideum. J. Cell Biol. 118, 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bush J., Franek K., Daniel J., Spiegelman G. B., Weeks G., Cardelli J. (1993) Cloning and characterization of five novel Dictyostelium discoideum rab-related genes. Gene 136, 55–60 [DOI] [PubMed] [Google Scholar]
- 46. Harris E., Cardelli J. (2002) RabD, a Dictyostelium Rab14-related GTPase, regulates phagocytosis and homotypic phagosome and lysosome fusion. J. Cell Sci. 115, 3703–3713 [DOI] [PubMed] [Google Scholar]
- 47. Campbell-Valois F. X., Trost M., Chemali M., Dill B. D., Laplante A., Duclos S., Sadeghi S., Rondeau C., Morrow I. C., Bell C., Hatsuzawa K., Thibault P., Desjardins M. (2012) Quantitative proteomics reveals that only a subset of the endoplasmic reticulum contributes to the phagosome. Mol. Cell. Proteomics 10.1074/mcp.M111.016378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cebrian I., Visentin G., Blanchard N., Jouve M., Bobard A., Moita C., Enninga J., Moita L. F., Amigorena S., Savina A. (2011) Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell 147, 1355–1368 [DOI] [PubMed] [Google Scholar]
- 49. Touret N., Paroutis P., Grinstein S. (2005) The nature of the phagosomal membrane: endoplasmic reticulum versus plasmalemma. J. Leukoc. Biol. 77, 878–885 [DOI] [PubMed] [Google Scholar]
- 50. Becker T., Volchuk A., Rothman J. E. (2005) Differential use of endoplasmic reticulum membrane for phagocytosis in J774 macrophages. Proc. Natl. Acad. Sci. U.S.A. 102, 4022–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Orci L., Ravazzola M., Le Coadic M., Shen W. W., Demaurex N., Cosson P. (2009) STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 106, 19358–19362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vieira O. V., Botelho R. J., Grinstein S. (2002) Phagosome maturation: aging gracefully. Biochem. J. 366, 689–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Müller-Taubenberger A., Lupas A. N., Li H., Ecke M., Simmeth E., Gerisch G. (2001) Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 20, 6772–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guermonprez P., Saveanu L., Kleijmeer M., Davoust J., Van Endert P., Amigorena S. (2003) ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature 425, 397–402 [DOI] [PubMed] [Google Scholar]
- 55. Amigorena S., Savina A. (2010) Intracellular mechanisms of antigen cross presentation in dendritic cells. Cur. Opin. Immunol. 22, 109–117 [DOI] [PubMed] [Google Scholar]
- 56. Cavalier-Smith T. (2009) Predation and eukaryote cell origins: a coevolutionary perspective. Int. J. Biochem. Cell Biol. 41, 307–322 [DOI] [PubMed] [Google Scholar]
- 57. Yates R. M., Hermetter A., Russel D. G. (2005) The kinetics of phagosome maturation as a function of phagosome/lysosome fusion and acquisition of hydrolytic activity. Traffic 6, 413–420 [DOI] [PubMed] [Google Scholar]
- 58. Yates R. M., Hermetter A., Russell D. G. (2009) Recording phagosome maturation through the real-time spectroflucrometric measurement of hydrolytic activities. Methods. Mol. Biol. 531, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.