Abstract
Full-length de novo sequencing from tandem mass (MS/MS) spectra of unknown proteins such as antibodies or proteins from organisms with unsequenced genomes remains a challenging open problem. Conventional algorithms designed to individually sequence each MS/MS spectrum are limited by incomplete peptide fragmentation or low signal to noise ratios and tend to result in short de novo sequences at low sequencing accuracy. Our shotgun protein sequencing (SPS) approach was developed to ameliorate these limitations by first finding groups of unidentified spectra from the same peptides (contigs) and then deriving a consensus de novo sequence for each assembled set of spectra (contig sequences). But whereas SPS enables much more accurate reconstruction of de novo sequences longer than can be recovered from individual MS/MS spectra, it still requires error-tolerant matching to homologous proteins to group smaller contig sequences into full-length protein sequences, thus limiting its effectiveness on sequences from poorly annotated proteins. Using low and high resolution CID and high resolution HCD MS/MS spectra, we address this limitation with a Meta-SPS algorithm designed to overlap and further assemble SPS contigs into Meta-SPS de novo contig sequences extending as long as 100 amino acids at over 97% accuracy without requiring any knowledge of homologous protein sequences. We demonstrate Meta-SPS using distinct MS/MS data sets obtained with separate enzymatic digestions and discuss how the remaining de novo sequencing limitations relate to MS/MS acquisition settings.
Database search tools, such as Sequest (3), Mascot (4), and InsPecT (5), are the most frequently used methods for reliable protein identification in tandem mass (MS/MS) spectrometry based proteomics. These operate by separately matching each MS/MS spectrum to peptide sequences from reference protein databases where all proteins of interest are presumably contained. But this assumption often does not hold true as many important proteins, such as monoclonal antibodies, are not contained in any database because mechanisms of antibody variation (including genetic recombination and somatic hyper-mutation (6)) constantly create new proteins with novel unique sequences. These mechanisms of variation are the foundation of adaptive immune systems and have enabled highly successful antibody-based therapeutic strategies (7, 8). Nevertheless, such variation also means that antibody MS/MS spectra are typically impossible to identify via standard database search techniques whenever the corresponding sequences are not known in advance. An inherent drawback of database search strategies is that they are only as good as the database(s) being searched and incomplete databases often result in proteins being misidentified or left unidentified (9).
Despite the importance of novel protein identification, few high-throughput methods have been developed for de novo sequencing of unknown proteins. Low-throughput Edman degradation is a well-known de novo sequencing approach that can accurately call amino acid sequences in N/C-terminal regions of unknown proteins but has drawbacks that make it unsuitable for sequencing proteins longer than 50 amino acids or proteins with post-translational modifications (10, 11). Many have recognized the potential of tandem mass spectrometry for protein sequencing. For example, in 1987 Johnson and Biemann (12) manually sequenced a complete protein from rabbit bone marrow. Meanwhile, automated de novo sequencing methods that rely on interpretations of individual MS/MS spectra are limited in that they typically cannot reconstruct long (8+ AA) sequences without mis-predicting 1 in 5 AA on average for low accuracy collision-induced dissociation (CID) spectra (13, 14). Recent advances in de novo peptide sequencing have improved sequencing accuracy to over 95% for high resolution higher energy collisional dissociation (HCD)1 spectra (15), but at limited sequence coverage (Chi H et al. report only 55% sequence coverage of peptides identified by database search). In fact, all current per-spectrum de novo sequencing strategies face a significant tradeoff between sequencing accuracy and coverage as spectra exhibiting complete peptide fragmentation rarely cover entire target proteins, yet are required to accurately reconstruct full-length peptide sequences. An alternative approach to separately sequencing individual spectra is to simultaneously interpret multiple MS/MS spectra from overlapping peptides. This Shotgun Protein Sequencing (SPS) paradigm differs from traditional algorithms by deriving consensus sequences from contigs - sets of multiple MS/MS spectra from distinct peptides with overlapping sequences (1, 16). Because SPS aggregates multiple spectra from overlapping peptides, protein sequences extending beyond the length of enzymatically digested peptides can be extracted from spectra with incomplete peptide fragmentation. Furthermore, SPS has been found to generate sequences that frequently cover 90–95+% of the target protein sequence(s) whereas mis-predicting only 1 out of every 20 amino acids on high resolution MS/MS spectra (2). But a remaining limitation of SPS is that it still generates fragmented sequences that do not singularly cover large regions of the target protein sequences, much less complete proteins: SPS sequences have an average length of 10–15 amino acids (depending on input data) and the longest recovered SPS de novo sequence is less than 45 amino acids long (1).
The considerable limitations of de novo sequencing strategies have typically been addressed by attempting to circumvent them using error-tolerant matching to known protein sequences. One such strategy (17) is to generate short de novo sequence tags and then match them exactly to protein databases without requiring matching the N/C-term flanking masses (to allow for unexpected polymorphisms or post-translational modifications). Short sequence tags are usually derived from parts of the spectrum with high signal-to-noise ratios and typically have higher sequencing accuracy than full-length de novo sequences (18). This approach was later extended in MS-Shotgun (19) and continues to be a popular technique for speeding up database search tools (5, 20–22). Homology matching of full length de novo sequences was first explored in CIDentify (23) and later in MS-BLAST (24) by searching de novo sequences using FASTA and WU-BLAST2 (respectively) to find homologous matches to sequences of related proteins; FASTS (25) also approached the problem using a modified version of FASTA. However, common de novo sequencing errors tend to produce sequences that are heavily penalized in pure sequence homology searches. For example, missing peaks in MS/MS spectra may easily cause GA subsequences to be reconstructed as Q or AG (same-mass sequences), thus making subsequent BLAST searches unlikely to succeed. This issue was partially considered in CIDentify and more thoroughly addressed in SPIDER (26) by explicitly modeling de novo sequencing errors together with BLOSUM scores in MS/MS-based sequence homology searches. In addition, OpenSea (27) further explored database matching of de novo sequences for analysis of unexpected post-translational modifications (PTMs). Finally, Shen et al. (28) used short unique de novo sequence tags, called UStags, to discover protein-localized PTMs.
Recent approaches to homology matching of de novo sequences have built on genome assembly and sequencing techniques to achieve database-assisted full-length sequencing of unknown proteins. Comparative Shotgun Protein Sequencing (cSPS) complemented SPS assembly techniques with usage of error tolerant matching of de novo sequences to find overlapping SPS de novo sequences that are then further assembled into full-length protein sequences (2). cSPS was designed to support the sequencing of highly divergent proteins that have regions close enough in homology to transfer matches from a reference. cSPS was shown to enable de novo sequencing of monoclonal antibodies at 95+% sequencing accuracy, while simultaneously tolerating and identifying unexpected PTMs (29). In difference from cSPS, Champs (30) de novo sequences individual spectra to obtain putative peptide sequences, which are then mapped to homologous proteins to correct sequencing errors and reconstruct protein sequences with 100% accuracy and 99% coverage. However, Champs is designed to only map peptides that differ from the reference sequence by one or two amino acids and does not handle PTMs. As such, its sequencing accuracy is not directly comparable to that of cSPS as Champs was not designed to sequence highly divergent proteins (such as monoclonal antibodies) with multiple PTMs, insertions, deletions, and/or recombinations. GenoMS (31) extended the approaches in cSPS/Champs by explicitly modeling protein splice variants as paths in splice graphs where nodes represent translated exon regions (32). MS/MS spectra are first searched for exact sequence matches against all possible protein isoforms. The remaining unidentified MS/MS spectra are then aligned to the matched peptides and de novo sequenced to extend the matched sequences into novel regions. Reported sequences are 97–99% accurate and cover 96–99% of target proteins depending on sequence similarity between the novel and reference sequences (31). However, GenoMS de novo sequences are usually extended less than 3 amino acids beyond matched peptides because sequencing accuracy degrades as sequences are extended, thus preventing the consistent extension of long (10+ AA) sequences. Altogether, the use of homology matching approaches for full-length de novo protein sequencing continues to be limited by 1) requiring the previous knowledge of closely related protein sequences and 2) the inherent difficulties in statistically significant homology-tolerant matching of error-prone short de novo sequences.
The Meta-SPS approach proposed here seeks to de novo sequence complete proteins, or long protein regions, without any use of a database. Meta-SPS builds upon SPS by treating SPS de novo sequences (contig sequences) as input spectra and further assembling them into longer de novo sequences (meta-contig sequences). We show that Meta-SPS extends de novo sequences to lengths over 100 AA while boosting sequencing accuracy to only 1 mistake per 40 amino acid predictions, thus enabling database-free de novo sequencing of completely novel proteins while also allowing error-tolerant matching approaches to support higher-divergence homologies (by searching longer, more accurate de novo sequences). Meta-SPS algorithms are demonstrated on CID and HCD MS/MS spectra and its limitations are discussed in relation to the underlying limitations of bottom-up tandem mass spectrometry.
EXPERIMENTAL PROCEDURES
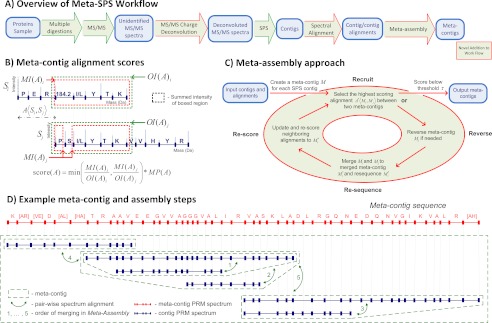
The Meta-SPS workflow is illustrated in Fig. 1A. In brief, because Meta-SPS relies upon the interpretation of MS/MS spectra from overlapping peptides, sample proteins were digested with multiple enzymes. Following MS/MS acquisition, MS/MS Charge Deconvolution was performed to convert all MS/MS fragment peaks to charge one (see supplemental Materials - MS/MS Charge Deconvolution) and Shotgun Protein Sequencing (SPS) (1) was used to assemble unidentified MS/MS spectra into contigs—sets of aligned spectra from peptides with overlapping sequences. SPS contigs were then aligned to each other using Spectral Alignment and further assembled into meta-contig sequences in the Meta-Assembly step. Two data sets were used to develop and benchmark Meta-SPS: a mixture of 6 known proteins (6-prot) and a previously described data set from a purified monoclonal antibody raised against the B- and T- lymphocyte attenuator molecule (aBTLA) (2). Briefly, the aBTLA data set consisted of 44,985 MS/MS spectra from the heavy chain and 39,135 MS/MS spectra from the light chain acquired on a Thermo LTQ XL instrument either in the Linear trap (low MS/MS mass accuracy) or in the Orbitrap (high MS/MS mass accuracy). Heavy-chain samples were prepared using five different protease digestions (trypsin, chymotrypsin, pepsin, Glu-C, and AspN) and light-chain samples were prepared with four different protease digestions (trypsin, chymotrypsin, pepsin, and AspN).
Fig. 1.
Meta-SPS Procedures. A, Green arrows denote procedures previously described in (1) and red arrows denote procedures described here. The SPS step involves spectral clustering by MSCluster (34), PepNovo+ PRM scoring (35), and assembly of mass spectra into contigs (1). B, An alignment between two PRM spectra is represented as the shift of the second spectrum wrt the first that yields the highest possible score. The displayed scoring function takes the minimum matched/overlapping intensity ratio and multiplies by the number of matching peaks (denoted by MP(A) for alignment A〈Si,Sj〉 between contig PRM spectra Si and Sj). Matched and overlapping intensities for each spectrum are displayed as red and blue boxed regions, respectively. Sequences are not known in advance; shown only for illustration purposes. C, Here aligned SPS contigs are assembled into meta-contigs by iteratively merging the highest scoring alignment until remaining alignments have a low score. By merging the highest scoring alignment at every iteration, it is guaranteed that all inconsistent alignments that were removed have a lower score. D, Green arrows denote merged alignments and numbers correspond to the order in which they alignments are merged. Initially, every contig was in its own meta-contig. The 6 meta-contigs were then merged by five alignments, yielding a single meta-contig PRM spectrum and its meta-contig sequence.
6-prot Data Acquisition
For the 6-prot sample, first an equimolar mixture of six proteins was prepared. After reduction and alkylation of cysteines, aliquots were digested by different means to produce sets of overlapping peptides. Bovine (6.5 kDa, catalog # A-4529) purified from lung, recombinant murine leptin (16 kDa, catalog # l-3772) expressed in E. coli, horse heart myoglobin (17 kDa, catalog # M-1882) purified from heart, and horseradish peroxidase (39 kDa, catalog # P-6782) purified from horseradish roots were purchased from Sigma-Aldrich. E. coli GroEL (57 kDa, catalog # G8976) purified from an E. coli strain overexpressing GroEL was purchased from United States Biological (Swampscott, MA). Human prostate-specific antigen also known as kallikrein-related peptidase (29 kDa, catalog # P0725) purified from seminal fluid was purchased from Scripps Laboratories (San Diego, CA). The 252 μg total protein mixture was prepared in 100 mm NH4HCO3 then reduced with 5 mm dithiothreitol, and the cysteines were alkylated with 20 mm iodoacetamide. The proteins that had not already precipitated were further precipitated with 60% ice-cold ethanol. After centrifugation, the supernatant was removed and discarded. The pellet was washed several times with 95% cold ethanol and then resuspended in 0.04% Rapigest (Waters Corp. Milford, MA) an acid-labile SDS-like detergent. Seven 32 μg aliquots were created. Three aliquots were diluted to 0.085% Rapigest at pH 8.0 in 100 mm NH4HCO3 and digested for 6 h. with trypsin 1:150, Lys-C 1:300, or Glu-C 1:150. Three aliquots were diluted to 0.01% Rapigest at pH 8.0 in 100 mm NH4HCO3 and digested for 6 h. with Asp-N 1:300, Chymotrypsin 1:150, or Arg-C 1:150. Digestions were stopped, and the detergent was cleaved by acidifying with 1% trifluoro acetic acid (TFA), pH 2. The 7th aliquot was acidified and precipitated with 60% ice-cold ethanol, washed with 95% cold ethanol, dried and digested with cyanogen bromide (CNBr) using 70% TFA for 36 h before drying in a SpeedVac and resuspending in 0.1% TFA. Digests were stored at −80′C prior to LC-MS/MS.
Digests were analyzed with an automated nano LC-MS/MS system, consisting of an Agilent 1100 nano-LC system (Agilent Technologies, Wilmington, DE) coupled to an LTQ-Orbitrap Fourier transform mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with a nanoflow ionization source (James A. Hill Instrument Services, Arlington, MA). Peptides were eluted from a 10 cm column (Picofrit 75 μm ID, New Objectives) packed in-house with ReproSil-Pur C18-AQ 3 μm reversed phase resin (Dr. Maisch, Ammerbuch Germany) using a 95 min acetonitrile/0.1% formic acid gradient at a flow rate of 200 nl/min to yield ∼20 s peak widths. Solvent A was 0.1% formic acid and solvent B was 90% acetonitrile/0.1% formic acid. The elution portion of the LC gradient was 3–7% solvent B in 1 min, 67–37% in 60 min, 37–90% in 6 min, and held at 90% solvent B for 5 min. Data-dependent LC-MS/MS spectra were acquired in ∼3 s cycles; each cycle was of the following form: one full Orbitrap MS scan at 60,000 resolution followed by five MS/MS scans in the orbitrap at 7,500 or 15,000 resolution on the most abundant precursor ions using an isolation width of 2.0 or 2.5 m/z. Dynamic exclusion was enabled with a mass width of ± 25 ppm, a repeat count of 1 and an exclusion duration of 8 s. Charge state screening was enabled along with monoisotopic precursor selection and nonpeptide monoisotopic recognition to prevent triggering of MS/MS on precursor ions with unassigned charge or a charge state of 1. For CID fragmentation the normalized collision energy was set to 30 with an activation Q of 0.25 and activation time of 30 ms. For HCD fragmentation the normalized collision energy was set to 60 (first generation, “software HCD” with 1 segment of black restrictor capillary tubing removed to elevate the ion gauge operating pressure to 1.6 e-5 Torr).
Spectrum Preprocessing and Notation
A total of 11010 high resolution CID and 14040 high resolution HCD 6-prot spectra were obtained after quality filtering by SpectrumMill. All 6-prot spectra were then deconvoluted using MS/MS Charge Deconvolution (see supplemental Materials) and searched with MS-GFDB (33) against the six target proteins and known contaminants with a spectrum-level false discovery rate of 1%; resulting peptide IDs covered 87% of the target proteins. See supplemental Materials for parameters used for SpectrumMill, MS/MS Charge Deconvolution, and MS-GFDB.
High resolution aBTLA MS/MS spectra were also deconvoluted using our approach and repeated spectra were detected and converted to consensus spectra using MS-Cluster (34) separately for low and high resolution spectra. This resulted in 8328 high resolution and 13,863 low resolution clustered CID spectra from the aBTLA light chain, as well as 13,261 high resolution and 14,424 low resolution clustered CID spectra form the aBTLA heavy chain. Spectra were then searched using MS-GFDB at 1% spectrum-level FDR and the resulting peptide identifications covered 99% of the aBTLA protein sequence. We note that peptide identifications were only used for benchmarking the accuracy and coverage of de novo sequences. The following notation is used below: a peptide MS/MS spectrum S is defined as a collection of peaks where each peak p ∈ S corresponds to an ion with mass m[p], charge z[p], intensity i[p], and where p = m[p]/z[p]. The parent mass P[S] is the cumulative mass of all residues in the peptide sequence plus the mass of H2O and the precursor charge Z[S] is the charge of the peptide ion.
Shotgun Protein Sequencing
SPS uses MS-Cluster (34) to cluster deconvoluted spectra from the same peptide and uses PepNovo+ (35) to convert clustered MS/MS spectra into PRM (prefix residue mass) spectra where peak intensities are replaced with log-likelihood scores. Ideal PRM spectra have peaks only at prefix residue masses (PRMs, cumulative amino acid masses of N-term prefixes of the peptide sequence) and peak scores combining evidence supporting the presence of b/y-ions, such as peak intensity, neutral losses (e.g. loss of H2O) and b/y-ion complementarities, and contrasting it with the estimated level of noise (13, 36). But in actuality, PRM scoring procedures cannot perfectly differentiate between prefix residue masses and suffix residue masses (SRMs, cumulative amino acid masses of C-term suffixes of the peptide sequence plus the mass of H2O) when complementary b and y ion series are present in a spectrum. PRM and SRM peaks typically receive high scores relative to other peaks whereas PRM peaks usually explain a higher percentage of a spectrum's total score.
SPS then aligns PRM spectra to each other in an all-to-all comparison. For each pair of overlapped spectra, PRM and SRM peaks are separated by two complementary alignments, which can be visualized as complementary paths in an alignment matrix (supplemental Fig. S1). PRM spectrum alignments are retained if their scores are above a certain threshold: SPS fits a Gaussian distribution to spectra alignment scores and chooses score thresholds corresponding to a given p value (0.045); an alignment between two spectra is retained if it passes the significance threshold for both aligned spectra. Because MS/MS spectra from different acquisition modes have different ion statistics, PRM spectra from different acquisition modes were run separately through SPS. Because the alignments are symmetric because of the b/y-ion and PRM/SRM complementarities, SPS cannot tell which peaks are PRMs and which peaks are SRMs, only differentiate between the two. Therefore, contig sequences can assemble either aligned PRM peaks or aligned SRM peaks with the majority (∼70%) of sequences assembling PRM peaks as they typically receive higher scores than SRM peaks (1). Contig sequences assembling SRM peaks must be reversed to match the target protein sequence in the correct orientation.
Finally, SPS assembles aligned PRM spectra into contigs, which are sets of aligned spectra from overlapping peptides (1). Each contig has a corresponding de novo contig sequence, which is the sequence of amino acids and mass gaps (masses that do not match the mass of a single amino acid) that best explains the overlapping peaks in the assembled spectra (supplemental Fig. S1). Each contig sequence returned by SPS is represented as a contig PRM spectrum, which is a spectrum S with PM[S] equal to the cumulative mass of all residues and gaps in the contig sequence. Each prefix of the contig sequence corresponds to a contig PRM peak and the score of each contig PRM is the summed score of its assembled spectrum PRMs.
Spectral Alignment
Overlaps between contig PRM spectra were computed using a modified version of the spectral alignment technique introduced in SPS (16). An alignment between two PRM spectra Si and Sj is a set of matched PRM pairs imposed by the shift A〈Si,Sj〉 (defined below) such that for each matched PRM pair (pi,pj), pi ∈ Si, pj ∈ Sj, and pi = pj + A〈Si,Sj〉. Because some contig sequences may be reversed wrt each other, the highest scoring alignment of Si and Sj may be between Si and the reversed orientation of Sj. Reversing the orientation of a PRM spectrum S involves simply converting all of S's masses to SRMs by subtracting each PRM mass from the parent mass. Thus, SR represents the reversed orientation of spectrum S with PRMs {p′ = PM[S] − p, ∀ p ∈ S}. The definitions in the table below are illustrated in Fig. 1B.
| A〈Si,Sj〉 | Mass shift (in Da) of Sj wrt Si that yields the maximum score(A) |
| MP(A) | Number of matched peak pairs between Si and Sj |
| MI(A)i | Summed intensity of all peaks in Si that match peaks in Sj |
| MI(A)j | Summed intensity of all peaks in Sj that match peaks in Si |
| OI(A)i | Summed intensity of all peaks in the m/z range of Si that overlaps with the aligned m/z range of Sj |
| OI(A)j | Summed intensity of all PRMs in the m/z range of Sj that overlaps with the aligned m/z range of Si |
| score(A) | |
| R[A] | true if score(A〈Si,SjR〉) > score(A〈Si,Sj〉), false otherwise |
For each unique pair of contig PRM spectra (Si,Sj), all possible shifts of Sj wrt Si and SjR wrt Si that yielded at least 6 matching peaks were considered and the shifts A〈Si,Sj〉 and A〈Si,SjR〉 were set. Of these two shifts, the shift with the highest score was reported for the pair. If score(A〈Si,SjR〉) > score(A〈Si,Sj〉), the reverse state of A, R[A], was set to true in order to indicate that Sj should be reversed wrt Si (R[A] ← false otherwise). Given an input minimum score τ, alignments were then discarded if score(A) < τ. The parameter τ is also enforced in Meta-Assembly and was separately trained for low mass accuracy contig PRM spectra (0.5 Da peak tolerance) and high mass accuracy contig PRM spectra (0.05 Da peak tolerance).
Meta-Assembly
Similar to the SPS assembly of aligned PRM spectra into contigs, Meta-Assembly groups aligned contig PRM spectra into meta-contigs. Similar to the relationship between a contig and a contig PRM spectrum, every meta-contig also has a meta-contig PRM spectrum. Each meta-contig initially contains one contig PRM spectrum. As illustrated in Fig. 1C, Meta-assembly then iterates over the following steps: (1) Recruit → (2) Reverse → (3) Re-sequence → (4) Re-score. Step 1 finds the highest scoring aligned pair of meta-contigs A*〈Mi,Mj〉 and stops if the score is below threshold τ; Step 2 reverses Mj if required by the alignment; Step 3 merges Mi and Mj into Mi* and determines the updated meta-contig PRM spectrum; Step 4 transfers and re-scores alignments from Mi and Mj to Mi* and returns to Step 1.
The problem addressed by Meta-Assembly is in the context of an overlap graph (16), where each vertex is a meta-contig Mi initialized to SPS contig Si and meta-contig vertices are connected by scored alignment edges labeled with shifts A〈Mi,Mj〉, scores score(A〈Mi,Mj〉), and reverse states R[A〈Mi,Mj〉] all initialized using alignments between the corresponding contigs, as described above. In a perfect graph, all connected meta-contigs can be aggregated by merging every alignment edge. However, even though contig PRM spectral alignments are much more reliable than alignments between PRM spectra derived directly from MS/MS spectra, there are still incorrect edges in the graph. There are two types of incorrect edges: inconsistent edges disagree on the shift of meta-contigs wrt each other and incoherent edges disagree on the orientation of meta-contigs wrt each other. For example, there may be three alignment edges A1〈Mi,Mj〉, A2〈Mj,Mk〉, and A3〈Mi,Mk〉 between three meta-contigs Mi, Mj, and Mk such that A1 + A2 ≠ A3. Here the path from Mi to Mk following A1 and A2 imposes a transitive shift (a shift imposed by two or more pair-wise alignments) between Mi and Mk that is not consistent with A3. It may also be the case that R[A3] = true while R[A1] = R[A2] = false. Here the edges are incoherent because A1 and A2 indicate that Mi, Mj, and Mk are in the same orientation whereas A1 and A3 indicate that Mk is reversed wrt Mi and Mj. The meta-contig assembly problem is that of finding and merging the maximal scoring subset of consistent and coherent alignment edges such that every contig PRM spectrum can be aligned to its meta-contig PRM spectrum with score at least τ. It has been shown that finding the maximal scoring subset of consistent and coherent edges is a hard problem (1). Thus, we propose an iterative algorithm to approach the optimal solution. See Supplemental Meta-Assembly for a detailed description of Meta-Assembly steps.
In step 1, we recruit the highest scoring edge A*〈Mi,Mj〉 between any two meta-contigs Mi and Mj. If score(A*) < τ, then all remaining edges have a score below the threshold and the merging process ends. Otherwise, Mi and Mj are merged in steps 2–4.
In step 2, Mj is reversed if R[A*] = true. As described in Spectral Alignment, some alignments between contig PRM spectra are in different orientations. Thus, if aligned contig PRM spectra are to be assembled into coherent meta-contigs, some of them will need to be reversed. In step 2, meta-contig Mj is reversed to MjR if R[A*] = true to assure spectra inside Mi and inside Mj are in the same orientation before the meta-contigs are merged. The reversed meta-contig MjR is obtained from Mj by reversing all of its assembled contig PRM spectra and their relative alignments. Given an alignment shift A〈Sa,Sb〉, its reversed alignment shift AR〈SaR,SbR〉 is equal to PM[Sa] − A − PM[Sb]. The final step in reversing Mj is to update the reverse state of alignment edges connected to it. For all alignment edges Ak〈Mj,Mk〉 connecting Mj to other meta-contigs, Ak is also reversed and R[Ak] ← not R[Ak] to indicate whether Mk also needs to be reversed if it is to be merged to Mi and Mj in a subsequent iteration (only Mj is reversed in this iteration).
In step 3, Mi* is created as the union of Mi and Mj and the meta-contig PRM spectrum of Mi* is determined. A* is used as the shift to connect contig PRM spectra in Mi to contig PRM spectra in Mj. So after Mi* ← (Mi ∪ Mj), every contig PRM spectrum Sx ∈ Mi is connected to every contig PRM spectrum Sy ∈ Mj by the transitive shift A〈Sx,Sy〉 = A〈Sx,Si〉 + A* + A〈Sj,Sy〉 where Si and Sj were the first contig PRM spectra in Mi and Mj, respectively. Because only one shift is used to connect contig PRM spectra in Mi and Mj, all assembled alignments between spectra in Mi* are guaranteed to be consistent because Mi and Mj are internally consistent. The contig PRM spectra and their spectral alignments in Mi* are then used as input to SPS to determine its meta-contig PRM spectrum as in (1).
In step 4, alignment edges connected to Mi and Mj are re-scored and moved to Mi*. For every Mk connected to Mi through some alignment edge A1〈Mi,Mk〉, A1*〈Mi*,Mk〉 ← A1 is used to connect Mi* to Mk. If a Mk is connected to Mj through some alignment edge A2〈Mj,Mk〉, A2*〈Mi*,Mk〉 ← A* + A2 is used to connect Mi* to Mk. If a Mk is connected to both Mi and Mj through A1 and A2, A1* is used if score(A1*) > score(A2*) and A2* is used otherwise. After all edges are transferred from Mi and Mj to Mi*, Mi and Mj are removed from the graph. Then the scores of all edges connected to Mi* are updated for recruitment in step 1 of the next iteration.
Fig. 1D illustrates how this approach aggregates contigs connected by high scoring alignments before considering contigs with less reliable alignments. An important benefit of this property is that meta-contig sequences are reliably extended and updated (by merging high scoring alignment edges first) before they are used to re-score less reliable alignments. An alternative approach to further capitalize on this property by discovering new alignments between updated meta-contigs could be to add a step between Re-score and Recruit that re-aligns Mi* to every other meta-contig in the overlap graph. This was attempted, but it significantly increased the running time of the implementation without yielding longer meta-contig sequences.
After iterative merging of meta-contigs, only meta-contigs that assemble at least 2 contig PRM spectra or more are reported. Also, contigs and meta-contigs were required to yield an amino acid sub-sequence of at least five consecutive residues.
RESULTS
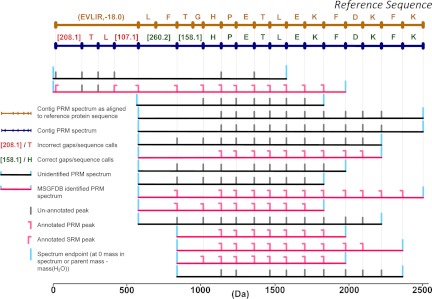
The performance of Meta-SPS and SPS was assessed in reference to target protein sequences and compared with determine the effectiveness of these additions to the SPS workflow. Two separate procedures were used to evaluate the performance of SPS and Meta-SPS, which was mainly measured in terms of de novo sequencing length, coverage, and accuracy. First, PRM spectra identified by MS-GFDB at 1% spectrum-level FDR were used to annotate contig PRM spectra (described in Fig. 2) and determine de novo sequencing accuracy. If a contig assembled at least one identified PRM spectrum, the contig itself was labeled identified. Peptides IDs were then mapped to their corresponding protein IDs and used to annotate peaks in identified PRM spectra as PRMs or SRMs. Mass differences between consecutive peaks in contig PRM spectra (i.e. sequence calls or gaps) were labeled using peaks from the annotated PRM spectra they assembled (Fig. 2). A contig sequence call was labeled annotated if its flanking peaks each assemble a mass from the same identified PRM spectrum. An annotated sequence call was correct if its flanking peaks assemble spectrum masses in the same ion series in the same identified spectrum (i.e. both are identified PRMs or both are identified SRMs) on the same protein. Annotated sequence calls not labeled correct are labeled incorrect. Because meta-contigs assemble contigs, every peak in a meta-contig PRM spectrum also assembles a set of PRM masses. Therefore, meta-contigs are annotated in the same manner as contigs.
Fig. 2.
Annotation of contigs and meta-contigs with MS-GFDB spectrum identifications. The annotation of a SPS contig is shown here but the same procedure applies for meta-contigs. Above the contig PRM spectrum are all sequence calls that align to the reference. Below the contig PRM spectrum are all spectra from overlapping peptides that were assembled to yield the contig PRM spectrum. Only assembled peaks are shown in each assembled PRM spectrum. For a sequence call to be labeled correct, it must be flanked by at least one pair of annotated PRM or SRM peaks in the same ion series that map to the same protein. If a sequence call that is not labeled correct is flanked by at least one pair of peaks from an identified spectrum then it is labeled incorrect. If a sequence call is not flanked by at least one pair of peaks from an identified spectrum then it is labeled un-annotated.
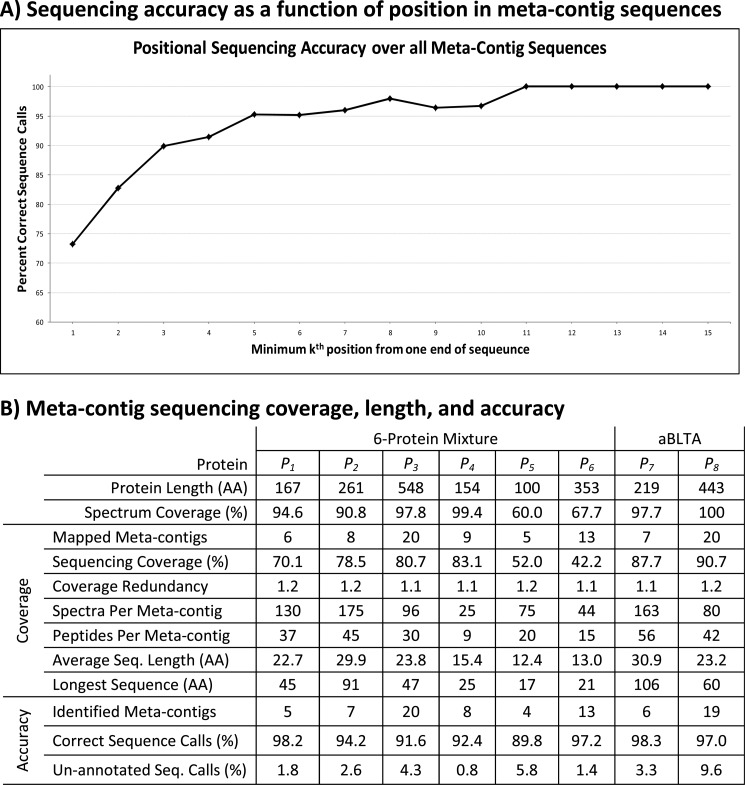
The graph displayed in Fig. 3A demonstrates that sequencing errors are localized toward the ends of sequences and are not distributed randomly. This occurs because often more PRM spectra overlap toward the middle of contig and meta-contig sequences, which gives a stronger consensus sequence. Given that sequence calls at the first or last residue of every meta-contig sequence were 20% less accurate than sequence calls two or more positions in from both ends, we truncated every meta-contig and contig sequence by one sequence call from each end. This post-processing step had the effect of increasing sequencing accuracy by roughly 2% over all contig and meta-contig sequences at a limited loss in sequencing coverage. Meta-contigs were then 94% accurate (1 error per 18 AA) over all 6-prot proteins and 97% (1 error per 35 AA) accurate over the aBTLA antibody (Fig. 3B) whereas SPS contigs were 88% accurate (1 error per 8 AA) over all 6-prot proteins and 96% accurate (1 error per 25 AA) over the aBTLA antibody (supplemental Table S3).
Fig. 3.
De novo sequencing length, coverage, and accuracy. A, The x axis plots the minimum distance (k) a sequence call or gap is from one end of a meta-contig sequence and the y axis plots the average sequencing accuracy over all annotated calls at each k-distance. Over all annotated calls reported more than 8 positions from their closest end, there were a total of 3 incorrect sequence calls at k = 20, 21, and 22 of a single meta-contig aligned to the aBTLA heavy chain (discussed in the Results section of Supplementary Materials). B, Protein identifiers are: P1 - leptin precursor, P2 - kallikrein-related peptidase, P3 - GroEL, P4 - myoglobin, P5 - aprotinin, P6 - peroxidase, P7 - aBTLA light chain, and P8 - aBTLA heavy chain. Protein Length is the length of each reference protein in amino acid residues. Spectrum Coverage is the percent of each protein covered by peptides identified MS-GFDB with 1% FDR. Coverage is taken over all mapped contigs and Accuracy is taken over all identified meta-contigs. Mapped meta-contigs must be aligned to a reference protein as described in the text whereas identified meta-contigs must assemble at least one identified spectrum whose peptide sequence is a substring of a reference protein. Sequencing Coverage is the percent of amino acids in each protein covered by at least one mapped meta-contig sequence. Coverage Redundancy is the average number of mapped meta-contig sequences covering each amino acid residue that is covered by at least one meta-contig sequence. Spectra Per Meta-contig is the average number of spectra assembled by each mapped meta-contig whereas Peptides Per Meta-contig is the average number of peptides (spectra with distinct parent masses) assembled by each mapped meta-contig. Average Seq. Length is the average number of amino acid residues covered by each mapped meta-contig and Longest Sequence is the maximum number of amino acid residues covered by a mapped meta-contig. Correct Sequence Calls is the percentage of annotated sequence calls that were correct in identified meta-contigs. Un-annotated Seq. Calls is the percentage of sequence calls that were un-annotated in identified meta-contigs.
MS-GFDB IDs could also have been used to evaluate sequencing coverage and length, but because less than 45% of spectra assembled into contigs and meta-contigs were identified in both data sets, such an approach would ignore many contigs that assemble unidentified spectra. Thus, contig and meta-contig PRM spectra were also directly mapped to reference proteins to evaluate de novo sequencing coverage and length. Contig spectra were aligned to protein sequences using an algorithm similar to MS-Alignment (37, 38). The protein sequences were first converted to perfect, unmodified PRM spectra and they were aligned (as in supplemental Fig. S1) to contig and meta-contig PRM spectra requiring at least seven matching peaks. Alignments of contig PRM spectra were allowed with one modification to capture PTMs and meta-contig PRM spectra were allowed with at most two modifications because of their increased length. A contig or meta-contig that was aligned to a reference protein in this manner is termed mapped. Roughly 50% more SPS contigs were mapped than were identified over both data sets, which is expected as many contigs assemble low-quality MS/MS spectra that are often left unidentified at 1% FDR. Only about 10% more meta-contigs were mapped than were identified, which is also expected as ∼5X more spectra were assembled per meta-contig than for SPS contigs. To evaluate the accuracy of the alignment mappings, the mapped residue locations of aligned contig and meta-contig PRM peaks were compared with those of assembled annotated peaks in MS-GFDB identified spectra. Over all aligned contig and meta-contig PRM peaks that assembled at least one mass from an identified spectrum, greater than 95% were aligned to the same residue as at least one their assembled masses. 593 of 666 (89%) 6-prot SPS contigs were mapped to target or contaminant proteins (482 mapped to target proteins) whereas for 6-prot, all 68 meta-contigs were mapped (64 mapped to target proteins). Similarly, 290 of 329 (88%) aBTLA SPS contigs were mapped (192 mapped to the antibody sequence) whereas all 43 aBTLA meta-contigs were mapped (27 mapped to the antibody sequence).
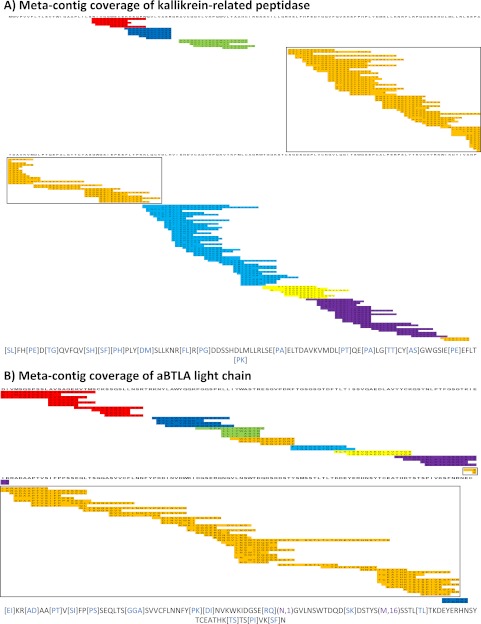
Figs. 3A and 3B illustrate the resulting meta-contig coverage for kallikrein-related peptidase and aBTLA light chain, respectively. Supplemental Figs. S5–S10 materials illustrate meta-contig coverage for remaining 6-prot proteins as well as the aBTLA heavy-chain. The largest meta-contig in Fig. 4A (colored red) corresponds to a 91 AA meta-contig sequence covering more than one third of the protein. The yellow meta-contig in Fig. 4A appears to have sufficient overlap with neighboring blue and purple meta-contigs to combine them, but the ends of the three meta-contig sequences contained too many gaps (seven missing PRMs) and incorrect sequence calls (two incorrect PRMs) to exceed the current acceptance threshold of sharing six or more matching peaks (supplemental Fig. S3). Such gaps and errors stem from incomplete MS/MS peptide fragmentation. In the discussion section we describe foreseeable data acquisition and algorithmic adjustments that could either generate data with higher sequence content and/or enable reducing the acceptance threshold without diminishing sequencing accuracy. In Fig. 4B, the largest meta-contig (colored orange) corresponds to a 106 AA meta-contig sequence covering more than one half of the target protein. See Fig. 3B for meta-contig coverage statistics on all proteins and see supplemental Materials for SPS contig coverage statistics in the same format. De novo sequencing gave 83% of MS-GFDB coverage between both data sets and we observe much higher sequence coverage of the purified aBTLA antibody (89%) compared with 6-prot proteins (42–83%). Because the heavy and light chains of the aBTLA antibody were purified prior to MS/MS analysis, higher aBTLA sequencing coverage is expected as more spectra from distinct peptides were identified by MS-GFDB per target protein in the aBTLA sample compared with the 6-prot sample (Fig. 3A). This is not an algorithmic limitation of our approach, but rather limitations of subcellular protein processing and MS/MS data acquisition. The lack of coverage of certain regions of the kallikrein-related peptidase in Fig. 4A is expected. The commercially obtained protein used in these studies was purified from human seminal fluid. Thus, it can be expected to lack the N-terminal region 1–24 because of prior cleavage of the signal peptide, residues 1–17, and activation by cleavage of the propeptide, residues 17–24. Furthermore, N-linked glycosylation is known to occur at residue 69. The subsequent sugar micro-heterogeneity at that position should render any individual proteolytically-generated peptide containing that residue much less concentrated in the digestion mixture, and if subjected to MS/MS much less likely to yield interpretable fragmentation.
Fig. 4.
Mapped Meta-contigs. Meta-contig PRM spectra were aligned to reference proteins to evaluate de novo sequencing coverage. Every colored row corresponds to a contig PRM spectrum as separately mapped to the target protein sequence (information not used by Meta-SPS). Every set of overlapping contigs of the same color corresponds to a meta-contig; sets of contigs of the same color with no overlap indicate separate meta-contigs. Below each coverage map is the longest meta-contig sequence of the boxed meta-contig for the corresponding protein. Purple gaps correspond to mapped sequence calls with PTMs verified by MS-GFDB; blue gaps correspond to mapped gaps that span 2 or more residues in the reference. Remaining un-colored residues represent sequence calls that map to reference amino acid masses. A, Meta-contig coverage of kallikrein-related peptidase from the 6-prot sample is displayed here; 8 meta-contigs covered 78% of the 261 AA protein with the longest sequence spanning 94 AA. B, Meta-contig coverage of the aBTLA light chain is displayed here; 9 meta-contigs covered 87% of the 219 AA protein with the longest sequence spanning 107 AA.
SPS contig alignments were also used to train the minimum spectrum alignment score τ to impose in Spectral Alignment and Meta-Assembly. τ was trained such that at least 97% of transitive alignments (alignments induced by two or more pair-wise alignments) between mapped contig PRM spectra in the same meta-contig were correct (a correct alignment is one whose observed shift matches the theoretical shift within the mass of a PTM). Over both data sets, 91% of all correct alignments were retained between pairs of mapped contig PRM spectra with at least 6 matching peaks. τ was trained to be 2.8 for 6-prot data, 3.0 for aBTLA, and can be estimated for any data set using a subset of identified spectra. After alignments with scores less than τ were removed (just prior to Meta-Assembly), 99% of all pair-wise alignments between mapped contig PRM spectra with at least 6 matching peaks were reported at 90% accuracy. But if transitive alignments were also considered, only 23% of alignments were correct because of the incorrect alignments reported by Spectral Alignment, those between components of multiple aligned contigs induced many more incorrect transitive alignments. The iterative merging procedure of Meta-Assembly was effective at discarding such incorrect alignments as 97% of transitive alignments ultimately reported were correct (supplemental Fig. S4).
The efficiency of Meta-SPS merging of SPS contigs is indicated by the decrease in coverage redundancy from 3.7 (supplemental Table S3) to 1.1 (Fig. 3A) as contigs covering the same regions were aggregated into meta-contigs. But because meta-contigs must assemble at least two contigs, meta-contigs do not cover regions missed by SPS contigs (i.e. coverage can only decrease from contigs to meta-contigs). Meta-contigs covered roughly 10% less of the 6-prot proteins than SPS contigs and 2% less of the aBTLA antibody. Thus, we generally observed a drop in coverage as a trade-off for Meta-SPS's higher sequencing accuracy. Coverage can be recovered by using “leftover” SPS contigs that were not merged by Meta-SPS, although lower sequencing accuracy is to be expected for certain applications (supplemental Table S2).
Meta-SPS also had the effect of doubling the average length of SPS contig sequences (to 20 AA in 6-prot meta-contigs and 25 AA in aBTLA meta-contigs) and tripling their maximum length (to 91 AA over 6-prot meta-contigs and 106 AA over aBTLA meta-contigs). Furthermore, the longest meta-contigs yielded the highest sequencing accuracy as the 91 AA and 106 AA de novo sequences displayed in Figs. 3A and 3B, respectively, were 100% annotated and correct. Although one peak in the 91 AA sequence incorrectly assembled masses mapping to different residues, this error was not reflected in the final sequence because the majority of the peak's assembled masses mapped to the correct residue.
The running time of Spectral Alignment and Meta-Assembly was found to be minor (< 9 min for the 6-prot data set) in comparison to that of SPS, which requires an all-to-all alignment of PRM spectra (see supplemental Materials for a more detailed description). All SPS contigs, meta-contigs, input MS/MS spectra, identified spectra, and annotated de novo sequences associated with this paper may be downloaded from Tranche/ProteomeCommons.org at the following hash:
s+8iy5TbHHydsOPmTf9yqotRGvkxeJPF8BXJxMxxZOnCRXqbje8wbn+Orpxr51YR3L0S2sZBTYljUdHUF35LjfTqeukAAAAAAv6xWQ==. This link also contains de novo sequencing reports that visualize how MS/MS spectra from each data set were used to generate de novo protein sequences. A subset of these reports detailing all 6-prot meta-contigs can also be found directly at http://proteomics.ucsd.edu/Software/MetaSPS/6-prot_meta-contigs/index.html. in supplemental Fig. S11 materials provides a description of how to interpret these reports in relation to algorithmic steps outlined Fig. 1A.
Although the 6-prot sample contained a mixture of proteins, applications of de novo protein sequencing are often targeted toward specific proteins within a larger mixture. To test how Meta-SPS performance might be impacted by such samples, we combined the 6-prot CID MS/MS spectra with the high resolution CID MS/MS spectra from the aBTLA sample and executed the algorithmic steps outlined in Fig. 1A on the combined set of MS/MS spectra. Here, the proteins of interest were the heavy and light chain of aBTLA antibody and the background mixture was represented by the 6-prot data. Because the high resolution CID spectra from the aBTLA and 6prot samples were acquired on a similar model of instrument (LTQ Orbitrap XL and LTQ Orbitrap, respectively), the low-resolution aBTLA spectra were excluded from this experiment to better simulate high resolution data acquisition of an aBTLA/6-prot mixture sample. Although this does not rigorously simulate the expected loss in MS/MS coverage one might expect from such a mixture (because of incomplete peptide sampling by the instrument), it is still a fair approximation of the algorithmic challenges associated with sequencing a small subset of proteins within the background of higher complexity. In practice, one would simply extend the LC gradient time or collect the data on a faster scanning instrument in order to maintain adequate peptide sampling. Compared with sequencing results on the aBTLA high resolution spectra, Meta-SPS produced the same sequencing accuracy (98.1% compared with 98.6%) and average length (18 AA compared with 17 AA) of the aBTLA antibody from the combined aBTLA/6-prot set of MS/MS spectra at the cost of reduced sequencing coverage (58% compared with 71%) and shorter maximum sequence length (35 AA compared with 45 AA). Compared with SPS, Meta-SPS generated de novo sequences 100% longer on average from the combined set with ∼2x as many correct sequence calls per incorrect sequence call. We note that in a real MS experiment mixing the 6-prot and aBTLA samples, the absence of a faster spectral acquisition rate and/or extended peptide separation time could diminish protein sequence coverage by MS/MS spectra and thus further limit the overall sequencing length and coverage.
DISCUSSION
Shotgun protein sequencing with meta-contig assembly is a modification-tolerant method for de novo protein sequence reconstruction. We demonstrate that extensive and accurate protein sequencing can be achieved without the use of a database, meaning more can be gained from experimental MS/MS data before mapping to a reference database. Compared with any other automated approach, our method provides the longest and most accurate de novo sequences without requiring any sequence homology steps. Furthermore, we demonstrate that de novo sequences which extend beyond 90 amino acids can be assembled with 100% accuracy. In the shorter sequences we report sequencing errors that are not distributed randomly, but located overwhelmingly toward the ends of sequences (Fig. 3 A).
Meta-SPS offers an effective improvement to Shotgun Protein Sequencing by doubling the average length of SPS de novo sequences, tripling their maximum sequence length, reducing sequence coverage redundancy ∼4X, and increasing sequencing accuracy 4–5%. There was only one protein, myoglobin, whose meta-contig sequences were less accurate (by 3%) than its SPS contig sequences (supplementary Table S3). In this case there were no sequencing errors introduced in myoglobin's meta-contig sequences that were not already present in its SPS contig sequences. But rather there was little overlap between incorrect and correct SPS sequence calls. When incorrect SPS contig sequence calls overlap with multiple correct contig sequences at multiple positions in the protein sequence, Meta-SPS can repair the incorrect sequence calls in meta-contigs at those positions if the correct calls are the consensus. But if such overlaps occur with limited frequency, as in the case of myoglobin, the reduced percentage of correct sequence calls (because of SPS contig redundancy) is greater than the reduced percentage of incorrect sequence calls in meta-contig sequences. This has the effect of lowering the observed percentage of correct calls from contig to meta-contig sequences.
Although Meta-SPS fell short of fully reconstructing a protein sequence in either data set, it assembled de novo sequences up to 91 AA long for a protein mixture and 106 AA long for a purified antibody, which are the longest confirmed de novo sequences ever obtained from the automated analysis of unidentified MS/MS spectra. Furthermore, 11 sequences from 6-prot and 6 sequences from aBTLA were extended beyond 40 AA. Sequencing accuracy was 95% for aBTLA and 6-prot samples, whereas the 91 AA and 106 AA sequences were 100% accurate. If we remove the first and last two residues or gaps of every sequence (where there was weaker consensus on average), sequencing accuracy improves to 96% over 6-prot proteins and 98% over the aBTLA antibody. Increased accuracy and reduced coverage redundancy of meta-contigs compared with SPS contigs was achieved at the cost of reduced meta-contig coverage (10% less coverage of 6-prot proteins and 2.5% less coverage of the aBTLA antibody).
Full reconstruction of protein sequence encoded by the genome is subject to limitations of sub cellular protein processing and posttranslational modification. When a protein is purified from its biological source it can be expected to have N and C-terminal signal peptides and pre-pro activation sequences already cleaved off (39). Although these can be predicted from a gene sequence, when a protein isolated from an organism with an un-sequenced genome is sequenced by the process described here, one would not be certain of having obtained the protein termini, unless they were chemically labeled prior to digestion (40). Furthermore, in higher organisms N-linked glycosylation can occur at NX(S/T) motifs, particularly for secreted and extracellular membrane proteins (41). Unless, the protein is de-glycosylated with an enzyme like PNGase-F, prior to proteolytic digestion, the sugar micro-heterogeneity at those sites should render any individual proteolytically-generated peptides containing the Asn residue from the motif much less concentrated in the digestion mixture, and if subjected to MS/MS much less likely to yield interpretable fragmentation.
As with SPS sequencing, Meta-SPS also faces limitations related to proteomics mass spectrometry, such as incomplete enzyme digestion, peptide sampling bias, and ambiguous amino acid masses (17). Cleaved peptides are not equally sampled by MS/MS instrumentation (e.g. hydrophobicity, ionizability, location of basic residues, etc.), leading to biased peptide coverage of target proteins. Furthermore, certain combinations of amino acids have identical masses and may lead to ambiguity in the final sequences (Ile = Leu = 113, GG = Asn = 114, and GA = Gln = 128). Because we require large sets of MS/MS spectra from overlapping peptides covering an entire protein to generate long sequences, we also face limitations when analyzing complex mixtures of proteins and proteins with related sequences. Our method is currently optimized for small mixtures of unrelated proteins or purified proteins, as we observe that coverage and sequence length degrade as fewer quality spectra are acquired per protein in the sample (Fig. 3B). Nonetheless, even in the background of the 6prot MS/MS spectra, Meta-SPS still improved upon SPS sequencing accuracy (from 97% to 98%), average sequence length (from 11 AA to 20 AA), and maximum sequence length (from 25 AA to 35 AA) for the aBTLA antibody. Analyzing more complex mixtures with greater effectiveness may require faster spectral acquisition rates or extended peptide separations to generate enough spectra to cover all proteins in a sample.
To enable assembling longer meta-contigs and achieve higher protein coverage a few adjustments to both data acquisition and algorithmic strategies are currently foreseeable. Compared with the use of CID and/or HCD fragmentation, it has been shown that electron transfer dissociation (ETD) can yield more interpretable MS/MS spectra from more unique peptides and greatly increase the number of interpretable spectra from longer peptides (with precursor charge 3+ or higher) (42, 43). The high resolution CID and HCD spectra described were collected in separate LC-MS/MS runs on a first generation LTQ Orbitrap that is not equipped with ETD. However, the duty cycle on newer LTQ Velos Orbitrap instruments is more than twice as fast. Thus in nearly equivalent chromatographic run time the newer instruments can subject each precursor ion to CID, HCD, and ETD fragmentation in 3 consecutive high resolution MS/MS spectra to provide information that is not only overlapping and complementary, but also all 3 can be directly attributed to the same peptide sequence. To support the combined processing of ETD, CID, and HCD spectra, spectral alignment steps in SPS will have to support alignments between b/c and y/z-type ions all the way from detection of pairs of spectra from overlapping peptides, through assembly of pairwise alignments into multiple alignments, and finally during the consensus interpretation of assembled ABruijn contigs.
Furthermore, high resolution MS/MS spectra allow for more accurate determination of true amino acid mass differences between MS/MS peaks and helps distinguish those from incorrect amino acid predictions in de novo sequencing applications (15). Although most results described here were achieved with high resolution MS/MS spectra acquired with ±15 ppm fragment mass accuracy, a fixed 0.05 Da fragment tolerance was imposed as SPS was not originally designed to support ppm tolerance. The 15 ppm is equivalent to 0.0015 Da at mass 100 and 0.06 Da at mass 4000. Implementing ppm tolerance in the Meta-SPS pipeline will impose much tighter tolerances in the mid-low mass range (0∼2000 Da) and allow alignments of N- and C- terminal fragment peaks in MS/MS spectra from overlapping peptides to be more reliably separated from random alignments of noise peaks. This should also improve the separation of correct and incorrect contig/contig alignments in the Meta-Assembly step. In particular, Meta-SPS currently requires six or more matching peaks to confidently align PRM spectra of overlapping peptides but implementation of ppm tolerance could enable decreasing this threshold without diminishing sequencing accuracy.
The six matching peak requirement further translates into a five consecutive amino acid minimum overlap requirement in pair-wise peptide alignments. However, the proteolytic enzymes currently in common usage have overlapping specificities. For example trypsin cleaves at the C-terminal side of Lys and Arg, whereas Lys-C cleaves only at Lys and Arg-C cleaves only at Arg. Thus in a combined data set peptide triplets often result where two shorter peptides are present that when concatenated are the equivalent of a longer peptide that is also present, but our current algorithmic approach makes only pairwise comparisons. Thus we expect to better capitalize on the enzyme specificity by introducing a step that attempts to concatenate the PRM spectra of 2 smaller peptides prior to comparison to the PRM spectrum of a larger peptide when the sum of the 2 precursor masses matches the larger one after adjusting for precursor charge and the mass difference because of terminal groups added upon peptide bond cleavage. Consequently, we foresee these data acquisition and algorithmic strategy improvements will most likely yield longer, more accurate meta-contig sequences and higher protein coverage.
Supplementary Material
Footnotes
* This work was partially supported by the National Institutes of Health Grant 1-P41-RR024851 from the National Center for Research Resources. This work was also supported in part by a grant to Steven A. Carr from the NCI, National Institutes of Health (1U24 CA126476-02), part of NCI's Clinical Proteomic Technologies Initiative.
 This article contains supplemental Tables S1 to S4 and Figs. S1 to S10.
This article contains supplemental Tables S1 to S4 and Figs. S1 to S10.
1 The abbreviations used are:
- HCD
- higher-energy collisional dissociation
- SPS
- Shotgun Protein Sequencing approach to de novo protein sequencing (1)
- cSPS
- Comparative Shotgun Protein Sequencing approach (2)
- Meta-SPS
- SPS with assembly of contigs into meta-contigs (described here)
- PTM
- post-translational modification
- ETD
- electron transfer dissociation
- PRM
- prefix-residue mass
- SRM
- suffix-residue mass
- FDR
- false discovery rate
- MS/MS
- tandem mass spectrometry
- LC
- liquid chromatography
- aBTLA
- antibody raised against the B- and T- lymphocyte attenuator molecule (described in (2))
- 6-prot
- mixture of six proteins: bovine purified from lung, recombinant murine leptin expressed in E. coli, horse heart myoglobin purified from heart, horseradish purified from horseradish roots, E. coli GroEL purified from an E. coli strain overexpressing GroEL, and human prostate-specific antigen (also known as kallikrein-related peptidase) purified from seminal fluid.
REFERENCES
- 1. Bandeira N., Clauser K. R., Pevzner P. A. (2007) Shotgun protein sequencing: assembly of peptide tandem mass spectra from mixtures of modified proteins. Mol. Cell. Proteomics 6, 1123–1134 [DOI] [PubMed] [Google Scholar]
- 2. Bandeira N., Pham V., Pevzner P., Arnott D., Lill J. R. (2008) Automated de novo protein sequencing of monoclonal antibodies. Nat. Biotechnol. 26, 1336–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yates J. R., 3rd, Eng J. K., McCormack A. L., Schieltz D. (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 4. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 5. Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) InsPecT: Identification of Posttranslationally Modified Peptides from Tandem Mass Spectra. Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 6. Di Noia J. M., Neuberger M. S. (2007) Molecular mechanisms of antibody somatic hypermutation. Ann. Rev. Biochem. 76, 1–22 [DOI] [PubMed] [Google Scholar]
- 7. Maggon K. (2007) Monoclonal antibody “gold rush”. Curr. Med. Chem. 14, 1978–1987 [DOI] [PubMed] [Google Scholar]
- 8. Haurum J. S. (2006) Recombinant polyclonal antibodies: the next generation of antibody therapeutics? Drug Discovery Today 11, 655–660 [DOI] [PubMed] [Google Scholar]
- 9. Duncan M. W., Aebersold R., Caprioli R. M. (2010) The pros and cons of peptide-centric proteomics. Nat. Biotechnol. 28, 659–664 [DOI] [PubMed] [Google Scholar]
- 10. Thoma R. S., Smith J. S., Sandoval W., Leone J. W., Hunziker P., Hampton B., Linse K. D., Denslow N. D. (2009) The ABRF Edman Sequencing Research Group 2008 Study: investigation into homopolymeric amino acid N-terminal sequence tags and their effects on automated Edman degradation. J. Biomol. Tech. 20, 216–225 [PMC free article] [PubMed] [Google Scholar]
- 11. Xiang B., Walters J., Mawuenyega K., Simpson J., Sandoval W., Smith J. S., Hunziker P. (2010) Results of the PSRG 2010 Study: Edman and Mass Spectrometric Terminal Sequencing of a Monoclonal Antibody. J. Biomol. Tech. 21, S18 [Google Scholar]
- 12. Johnson R. S., Biemann K. (1987) The primary structure of thioredoxin from Chromatium vinosum determined by high-performance tandem mass spectrometry. Biochemistry 26, 1209–1214 [DOI] [PubMed] [Google Scholar]
- 13. Frank A., Pevzner P. A. (2005) PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem. 77, 964–973 [DOI] [PubMed] [Google Scholar]
- 14. Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., Lajoie G. (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 2337–2342 [DOI] [PubMed] [Google Scholar]
- 15. Chi H., Sun R. X., Yang B., Song C. Q., Wang L. H., Liu C., Fu Y., Yuan Z. F., Wang H. P., He S. M., Dong M. Q. (2010) pNovo: de novo peptide sequencing and identification using HCD spectra. J. Proteome Res. 9, 2713–2724 [DOI] [PubMed] [Google Scholar]
- 16. Bandeira N., Tang H., Bafna V., Pevzner P. (2004) Shotgun protein sequencing by tandem mass spectra assembly. Anal. Chemi. 76, 7221–7233 [DOI] [PubMed] [Google Scholar]
- 17. Mann M., Wilm M. (1994) Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem 66, 4390–4399 [DOI] [PubMed] [Google Scholar]
- 18. Frank A., Tanner S., Bafna V., Pevzner P. (2005) Peptide Sequence Tags for Fast Database Search in Mass-Spectrometry. J. Proteome Res. 4, 1287–1295 [DOI] [PubMed] [Google Scholar]
- 19. Huang L., Jacob R. J., Pegg S. C., Baldwin M. A., Wang C. C., Burlingame A. L., Babbitt P. C. (2001) Functional assignment of the 20 S proteasome from Trypanosoma brucei using mass spectrometry and new bioinformatics approaches. J. Biol. Chem. 276, 28327–28339 [DOI] [PubMed] [Google Scholar]
- 20. Kim S., Na S., Sim J. W., Park H., Jeong J., Kim H., Seo Y., Seo J., Lee K. J., Paek E. (2006) MODi: a powerful and convenient web server for identifying multiple post-translational peptide modifications from tandem mass spectra. Nucleic Acids Res. 34, W258–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dasari S., Chambers M. C., Slebos R. J., Zimmerman L. J., Ham A. J., Tabb D. L. (2010) TagRecon: high-throughput mutation identification through sequence tagging. J. Proteome Res. 9, 1716–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 23. Taylor J. A., Johnson R. S. (1997) Sequence database searches via de novo peptide sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 11, 1067–1075 [DOI] [PubMed] [Google Scholar]
- 24. Shevchenko A., Sunyaev S., Loboda A., Bork P., Ens W., Standing K. G. (2001) Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 73, 1917–1926 [DOI] [PubMed] [Google Scholar]
- 25. Mackey A. J. (2001) Getting More from Less: Algorithms for Rapid Protein Identification with Multiple Short Peptide Sequences. Mol. Cell. Proteomics 1, 139–147 [DOI] [PubMed] [Google Scholar]
- 26. Han Y., Ma B., Zhang K. (2005) SPIDER: software for protein identification from sequence tags with de novo sequencing error. J. Bioinform. Comput. Biol. 3, 697–716 [DOI] [PubMed] [Google Scholar]
- 27. Searle B. C., Dasari S., Wilmarth P. A, Turner M., Reddy A. P., David L. L., Nagalla S. R. (2005) Identification of protein modifications using MS/MS de novo sequencing and the OpenSea alignment algorithm. J. Proteome Res. 4, 546–554 [DOI] [PubMed] [Google Scholar]
- 28. Shen Y., Tolić N., Hixson K. K., Purvine S. O., Anderson G. A., Smith R. D. (2008) De novo sequencing of unique sequence tags for discovery of post-translational modifications of proteins. Anal. Chem. 80, 7742–7754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bandeira N., Tsur D., Frank A., Pevzner P. A. (2007) Protein identification by spectral networks analysis. Proc. Natl. Acad. Sci. U. S. A. 104, 6140–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu X., Han Y., Yuen D., Ma B. (2009) Automated protein (re)sequencing with MS/MS and a homologous database yields almost full coverage and accuracy. Bioinformatics 25, 2174–2180 [DOI] [PubMed] [Google Scholar]
- 31. Castellana N. E., Pham V., Arnott D., Lill J. R., Bafna V. (2010) Template proteogenomics: sequencing whole proteins using an imperfect database. Mol. Cell. Proteomics 9, 1260–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Castellana N. E., Payne S. H., Shen Z., Stanke M., Bafna V., Briggs S. P. (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. U. S. A. 105, 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim S., Mischerikow N., Bandeira N., Navarro J. D., Wich L., Mohammed S., Heck A. J., Pevzner P. A. (2010) The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol. Cell. Proteomics 9, 2840–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frank A. M., Bandeira N., Shen Z., Tanner S., Briggs S. P., Smith R. D., Pevzner P. A. (2008) Clustering millions of tandem mass spectra. J. Proteome Res. 7, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frank A. M., Savitski M. M., Nielsen M. L., Zubarev R. A., Pevzner P. A. (2007) De novo peptide sequencing and identification with precision mass spectrometry. J. Proteome Res. 6, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dancík V., Addona T. A., Clauser K. R., Vath J. E., Pevzner P. A. (1999) De novo peptide sequencing via tandem mass spectrometry. J. Computational Biol. 6, 327–342 [DOI] [PubMed] [Google Scholar]
- 37. Pevzner P. A., Dancíkm V., Tang C. L. (2000) Mutation-tolerant protein identification by mass spectrometry. J. Computational Biol. 7, 777–787 [DOI] [PubMed] [Google Scholar]
- 38. Tsur D., Tanner S., Zandi E., Bafna V., Pevzner P. A. (2005) Identification of post-translational modifications by blind search of mass spectra. Nat. Biotechnol. 23, 1562–1567 [DOI] [PubMed] [Google Scholar]
- 39. Leversen N. A., DeSouza G. A., Målen H., Prasad S., Jonassen I., Wiker H. G. (2009) Evaluation of signal peptide prediction algorithms for identification of mycobacterial signal peptides using sequence data from proteomic methods. Microbiology 155, 2375–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takahashi T., Muneoka Y., Lohmann J., Lopez, de Haro M. S., Solleder G., Bosch T. C., David C. N., Bode H. R., Koizumi O., Shimizu H., Hatta M., Fujisawa T., Sugiyama T. (1997) Systematic isolation of peptide signal molecules regulating development in hydra: LWamide and PW families. Proc. Natl. Acad. Sci. U. S. A. 94, 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larkin A., Imperiali B. (2011) The expanding horizons of asparagine-linked glycosylation. Biochemistry 50, 4411–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frese C. K., Altelaarm A. F., Hennrich M. L., Nolting D., Zeller M., Griep-Raming J., Heck A. J., Mohammed S. (2011) Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J. Proteome Res. 10, 2377–2388 [DOI] [PubMed] [Google Scholar]
- 43. Hart S. R., Lau K. W., Gaskell S. J., Hubbard S. J. (2011) Distributions of ion series in ETD and CID spectra: making a comparison. Meth. Mol. Biol. 696, 327–337 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.