Abstract
Advances in proteomic analysis of human samples are driving critical aspects of biomarker discovery and the identification of molecular pathways involved in disease etiology. Toward that end, in this report we are the first to use a standardized shotgun proteomic analysis method for in-depth tissue protein profiling of the two major subtypes of nonsmall cell lung cancer and normal lung tissues. We identified 3621 proteins from the analysis of pooled human samples of squamous cell carcinoma, adenocarcinoma, and control specimens. In addition to proteins previously shown to be implicated in lung cancer, we have identified new pathways and multiple new differentially expressed proteins of potential interest as therapeutic targets or diagnostic biomarkers, including some that were not identified by transcriptome profiling. Up-regulation of these proteins was confirmed by multiple reaction monitoring mass spectrometry. A subset of these proteins was found to be detectable and differentially present in the peripheral blood of cases and matched controls. Label-free shotgun proteomic analysis allows definition of lung tumor proteomes, identification of biomarker candidates, and potential targets for therapy.
Lung cancer is one of the deadliest cancers, with ∼200,000 newly diagnosed individuals and 160,000 deaths every year in the United States (1). Despite the most advanced treatments that modern medicine has to offer, the five-year survival rate remains less than 15%. Although a small subset of tumors have been found to be driven by single mutated oncogenes for which active, but still noncurative, therapies are available, the vast majority of patients have complex multifactorial disease with few effective therapeutic options. New early detection strategies and molecular therapeutic targets are urgently needed to improve patient survival.
Genomic analysis has enabled us to measure the sequence, copy number, and expression changes of thousands of genes simultaneously, which can be used to discover transcripts specifically altered or expressed in tumor tissues (2–4). Although genomic studies have given important new insights into the mechanisms of carcinogenesis, therapeutic targets, and most practical biomarkers are their protein products, and the correlation between transcript sequence or level and protein function remains complex and poorly understood. Protein expression, in part, depends on transcript levels, but it is clear that significant translational and post-translational regulation of protein levels and function occurs, adding another level of complexity in the regulation of activity, especially in tumor cells (5, 6). It would be ideal to have a comprehensive understanding of the novel changes in protein expression levels and the modifications of proteins in cancer cells, but the technology to directly study proteomes has lagged behind that to assess genomes and transcriptomes. We and others have used matrix-assisted laser desorption and ionization-time of flight mass spectrometry protein profiling to better understand protein expression pattern alterations and discover biomarkers, but the number of proteins detected is far from satisfactory. Matrix-assisted laser desorption and ionization-time of flight mass spectrometry-based proteomic profiling (7, 8) yields only a couple of hundred anonymous signals predominantly derived from low-molecular weight and high abundance proteins, and identification of the proteins that generate these signals is problematic. Proteome analyses based on an alternative method using two-dimensional gel electrophoresis (9) are difficult to reproduce and typically yield only several hundred proteins that can be adequately compared between phenotypes.
Shotgun proteomic analysis based on multidimensional liquid chromatography-tandem mass spectrometry (LC-MS/MS)1 provides high-throughput peptide sequence identification of complex peptide mixtures (10). This approach has been successfully used for proteomic analysis not only of tissues (11–14), but also of pleural fluid and plasma from lung cancer patients (15, 16). The major advantage of this technique is sensitivity, with thousands of proteins directly identified in typical analyses (13). Detection of low abundance proteins is possible and quantitative information can be obtained from the spectral counts obtained for each peptide sequence (17–19). Recent studies by the National Cancer Institute Clinical Proteomic Technology Assessment for Cancer (CPTAC) network, in which we participate, have demonstrated high reproducibility and sensitivity of shotgun proteomics platforms (20, 21). A related proteomic technology platform, liquid chromatography-multiple reaction monitoring mass spectrometry (LC-MRM-MS) provides targeted quantitative analyses of proteins through sensitive measurements of their component peptides (22).
In this report, we have demonstrated that we can not only efficiently mine the proteome of lung tumors and noninvolved lung tissue specimens with great accuracy and sensitivity (3621 proteins identified, with rigorous definitions), but that we uncovered a potentially important new molecular pathway in lung cancer progression (PAK2). Shotgun proteomic analysis of primary lung tumors was combined with MRM analysis that represents a powerful new approach to identify and quantify proteins and molecular pathways that may be altered in the pathogenesis of lung cancer. Finally we show that we can translate differentially expressed proteins to an ELISA platform and interrogate the plasma of individuals at risk (including patients with lung nodules and COPD) or with lung cancer with reproducible prediction accuracy.
EXPERIMENTAL PROCEDURES
Study Subjects
We selected cases of stage I lung carcinomas in our lung tissue biorepository that were surgically resected with curative intent at the Vanderbilt Medical Center and the Nashville VA Medical Center between January 2001 and February 2007. All patients provided informed consent for participation and this project was approved by the Institutional Review Board at both institutions. All specimens were collected immediately after surgery, snap frozen, and stored in liquid nitrogen until the time of analysis to minimize the effects of storing and handling the tissue. Tissue specimens used in this analysis included cancer tissue from pathological Stage I lung cancer patients with no previous cancer history. In addition, normal lung tissue specimens were obtained from patients undergoing lung resection for suspicion of lung cancer but not carrying a diagnosis of lung or other cancer. We evaluated normal lung from cases resected because of clinical suspicion of lung cancer rather than adjacent normal-appearing lung from cancer resections in order to avoid referring to premalignant “field-cancerization” alterations, known to be present in lung cancer patients, as our “normal” control. These patients had similar demographic characteristics as shown in supplemental Table S1.
Shotgun Analysis Sample Preparation
To increase the efficiency of our analysis, pooled protein lysates from sets of 19 to 20 samples per phenotype were analyzed. Four protein lysate pools were generated: two from noninvolved lung tissue (normal control, n = 20 and n = 19, respectively), one from stage I adenocarcinomas (ADC, n = 20), and one from stage I squamous cell carcinomas (SCC, n = 20). Patient characteristics are described in supplemental Table S1. Hematoxylin and eosin (H&E) stained sections were reviewed by a pathologist (ALG) to identify areas containing tumor cells and to determine tumor percentage. Specimens were selected with at least 80% tumor cells. By aligning the H&E section with the tumor block, macrodissection was able to be performed on the tumor tissue with a razor blade.
Tissue proteins were extracted and digested by the method of Wang et al. (23). Five to 20 macrodissected sections of tissue were suspended in 200 μl of 50% 2,2,2-trifluoroethanol (Acros Organics, Belgium), 50% 50 mm ammonium bicarbonate (Fisher Scientific) (v/v). The tissue lysates were homogenized using sonication with three, 20 s cycles at 30 s intervals, followed by incubation at 60 °C for 1 h with shaking. After the 1-h incubation, the sonication cycle was repeated. After the second sonication cycle, the protein concentration was measured using the BCA protein assay (Pierce Biotechnology, Rockford, IL) of each individual tissue lysate using bovine serum albumin as a protein standard. A total of 1 mg of pooled protein lysate was created by adding equal amounts of protein from each individual sample from each of the four pools in a total volume of 50 μl each. These pooled lysates (1 mg) were digested by diluting with 100 μl of 40 mm tris (2-carboxyethyl) phosphine hydrochloride (Pierce Biotechnology) with 100 mm dithiothreitol (Acros Organics), and incubating at 60 C° for 30 min with shaking. After cooling down the tubes, 100 μl of 200 mm iodoacetamide was added, and incubated 20 min at room temperature in the dark. Samples were diluted with 600 μl of 50 mm ammonium bicarbonate. In order to generate peptides suitable for MS-MS analysis, these pooled lysates were digested by adding trypsin (20 μg, trypsin/protein ratio of 1:50 (w/w), Promega, Madison, WI) and digestion was carried out at 37 °C overnight.
After lyophilizing the resulting peptide mixture, samples were reconstituted by distilled water and applied to Sep-Pak C18 cartridges (Waters, Milford, , MA). After washing the column with 1 ml of distilled water, digested peptides were eluted from the column with 1 ml of 80% acetonitrile. Eluted peptides were evaporated to dryness in a SpeedVac (Thermo-Fisher) and reconstituted with 2.5 ml of 6 m urea for isoelectric focusing (IEF) of peptides.
IEF Fractionation of Peptide Digests and LC-MS/MS Analyses
Four independent IEF peptide separations were performed on aliquots of each pool digest equivalent to 200 μg protein. Immobilized pH gradient (IPG) strips, 24 cm, pI 3.5–4.5 (IPGphor, GE Health Care, NJ), were rehydrated overnight, then loaded and focused using an Ettan IPGphor 3 IEF system (GE Health Care) for 25 h as described previously (13). Immediately after focusing, IPG strips were cut into 20 pieces and peptides were extracted, the extracts were dried down, desalted, dried down again and then reconstituted in 0.1 ml of 0.1% formic acid for LC-MS/MS analysis (13).
LC-MS/MS analyses were performed on an LTQ-Orbitrap hybrid mass spectrometer (Thermo Fisher) equipped with an Eksigent 1D Plus NanoLC pump and autosampler (Dublin, CA). Peptides were separated on a packed capillary tip (Polymicro Technologies, 100 μm X 11 cm) with Jupiter C18 resin (5 μm, 300 Å, Phenomenex) using an in-line solid-phase extraction column (100 μm × 6 cm) packed with the same C18 resin using a frit generated with liquid silicate Kasil 1 (24) similar to that previously described (25). Mobile phase A consisted of 0.1% formic acid and Mobile phase B consisted of 0.1% formic acid in acetonitrile. A 95 min gradient was performed with a 15 min washing period (100% A at a flow rate of 1.5 μl min−1 for the first 10 min followed by a gradient to 98% A at 15 min) to allow for solid-phase extraction and removal of any residual salts. Following the washing period, the flow rate was reduced to 0.6 μl min−1 and the gradient was increased to 25% B by 50 min, followed by an increase to 90% B by 65 min and held for 9 min before returning to the initial conditions. Centroided MS/MS scans were acquired on the LTQ-Orbitrap using an isolation width of 2 m/z, an activation time of 30 ms, an activation q of 0.250 and 30% normalized collision energy using 1 microscan with a max ion time of 100 ms for each MS/MS scan and 1 microscan with a max ion time of 500 for each full MS scan. The mass spectrometer was tuned prior to analysis using the synthetic peptide TpepK (AVAGKAGAR), so that some parameters may have varied slightly from experiment to experiment, but typically the tune parameters were as follows: spray voltage of 2 KV, a capillary temperature of 150 °C, a capillary voltage of 50 V and tube lens of 120 V. The AGC target value was set at 500,000 for the full MS and 10,000 for the MS/MS spectra. A full scan was obtained for eluting peptides in the range of 400–2000 atomic mass unit (amu) was collected on the Orbitrap portion of the instrument at a resolution of 60,000, followed by five data-dependent MS/MS scans on the LTQ portion of the instrument with a minimum threshold of 1000 set to trigger the MS/MS spectra. MS/MS spectra were recorded using dynamic exclusion of previously analyzed precursors for 60 s with a repeat of 1 and a repeat duration of 1.
Peptide Identification from MS/MS Data, Protein Assembly, and Filtering
Captured peak lists from the mass spectral .RAW files were transcoded to mzML version 1.1 format by the ProteoWizard MSConvert tool (26). The software was configured to transcode only tandem mass spectra; MS scans were excluded. MS data was searched using the MyriMatch version 1.6.57 search algorithm (27) against the International Protein Index (IPI) human database version 3.64 supplemented with potential contaminant sequences for a total of 84,079 sequences in forward and reverse orientation. The search results were filtered and assembled using IDPicker version 2.0 (28). Any number of miscleavages was allowed and peptides were allowed to have one nontryptic end. A static modification for carbamidomethylation was defined for cysteines, whereas dynamic modifications reflecting oxidation of methionines and formation of N-terminal pyroglutamines were allowed. Precursor mass tolerance was set at 10 ppm and product ion mass tolerance was set at 0.5 m/z. Peptide identification stringency was set at a maximum of 2.5% reversed peptide identifications (5% overall peptide false discovery rate (FDR)) and a minimum of two unique peptides to identify a given protein within the full data set. IPI database entries that mapped to the identical set of peptide identifications were grouped into “protein groups,” which consist almost exclusively of isoforms or identical proteins resulting from redundancy in the database (29). Because the majority of false identifications occur with low frequency and such low-count identifications are unlikely to yield statistically significant results, an additional filter was applied that removed all protein groups that were identified by 7 or fewer MS spectra in the comparison between normal control and ADC, SCC, by three or fewer in the comparison between ADC and SCC. The full and unfiltered IDPicker output data set is provided as supplementary data (supplemental Table S2, see data sharing below) and includes a complete list of protein IDs and their sequence relationships, the number of distinct peptides and peptide coverage observed per protein, the number of spectra observed per protein, and full peptide sequences.
Comparison of Proteome Inventories Based on MS/MS Spectral Count Data
Previous results have shown that a frequency-based analysis approach using the number of observed spectra (spectral counting) provides a measure of protein concentration in complex protein mixtures, especially for more abundant proteins (17, 30). We developed a statistical method to model and compare different shotgun data sets for proteins that were likely to be present at different levels in the samples analyzed (31). To account for the specific properties of spectral count data, this method uses a quasi-likelihood model (32), which has no restriction on distribution assumptions. This approach also accounts for the type of overdispersion and/or underdispersion usually observed in shotgun data. To protect against multiple comparison issues while simultaneously testing thousands of proteins, we applied the FDR method (33). Normalization between different runs was achieved by adding the number of confident identifications into the model as the offset. This serves as the size variable, which determines the number of opportunities for proteins to occur. The model generates quasi-p values for each of the protein entries in the data set and estimates an average spectral count (λ) across the replicate analyses. Three separate comparisons were performed: a) one using the combined spectral counts from the two control groups compared with either the pooled ADC and SCC data sets, and b) one using the pooled ADC data set compared with the pooled SCC data set. Proteins were considered statistically different if their spectral counts log ratio was more than 3 (in SCC versus normal comparison) or 2 (in ADC versus normal comparison, ADC versus SCC comparison) and if their quasi p value was less than 0.01. The log ratio is the logarithm transformation of the ratio between the peptide counts of a protein in one pool over the same protein in the other pool.
MRM Analysis
LC-MRM-MS Analyses
Extracts from unfractionated tryptic peptide digests from lung tissues were prepared as described above and 2 μl aliquots were analyzed on a TSQ Quantum Ultra mass spectrometer (Thermo-Fisher) equipped with a Thermo Surveyor solvent delivery system, autosampler, and a microelectrospray source and equipped with a 6 cm (150 μm inner diameter) fused silica capillary precolumn packed with C18 resin (5 μm, 300Å) (Phenomenex Inc., Torrance, CA). Peptides were loaded on the precolumn and desalted for 6 min with 1% acetonitrile/0.1% formic acid at 1.8 μl min−1 and then resolved by reversed-phase chromatography on an 11 cm fused silica capillary column (100 μm inner diameter) packed with same C18 media at a flow rate of 1 μl min−1. The mobile phase consisted of 0.1% formic acid in either HPLC grade water (A) or acetonitrile (B). Peptides were eluted with a linear gradient from 98% A at 6 min to 75% A at 45 min, then programmed to 50% A at 55 min to 10% A from 65–76 min and returned to 99% A from 76–85 min. For MRM analysis, 3–4 optimized transitions for each peptide from the corresponding proteins were monitored. Instrument parameters included Q2 gas 1.5 mTorr, scan width 0.002 m/z, scan time 5 ms, Q1 and Q3 FWHM resolution were 0.2 and 0.7 respectively. Collision energy was continuously adjusted according to the relationship CE = 3.314 + 0.034 × precursor m/z. The integrated chromatographic peak areas for the transitions of each targeted peptide were summed. When data for multiple peptides were collected for a candidate protein, the reported values are for the peptide yielding the highest mean signal.
Peptide Selection and MRM Transitions
For each protein candidate, up to four proteotypic peptides from the shotgun data set having the highest spectral counts for a given charge state were selected for MRM analysis. The MS/MS spectrum with the greatest overall signal intensity for each selected peptide was extracted and the 3–5 most intense y ions were selected. Observed y ions were required to be within 1.0 m/z of the predicted values and ions within a window below 20 m/z of the precursor were excluded. Precursor m/z, selected fragment m/z and computed collision energies were saved in a text file, which was imported into the Xcalibur method MRM setting tables. Peptide sequences used for analysis are provided in a separate table (supplemental Table 3).
Webgestalt Cellular Component Analysis and GO Enrichment Analysis
To categorize the cellular compartments from which the identified proteins in our data set were derived we used Webgestalt (34) cellular component analysis. To do this, we first had to transform the IPI accession to Ensembl gene identifiers before applying the Webgestalt program. The second analysis we performed was to characterize those proteins differentially expressed between the SCC and ADC pools. GOTM (35) was used for this GO enrichment analysis. For the analysis of proteins differentially expressed between ADC and SCC, we selected 54 proteins using a cutoff log ratio of more than two and a quasi p value of less than 0.01. We used all identified proteins in our data set as reference data set. Enrichment analysis was performed with the statistical cutoff of 0.01(adjusted p value, by Benjamini & Hochberg (33)).
PAK2 Expression in Nonsmall Cell Lung Cancers (NSCLCs)
A tissue microarray (TMA) of NSCLC was prepared from paraffin-embedded formalin-fixed tumor tissues from the patients whose frozen tissues were used for proteomic analyses. Paraffin-embedded formalin-fixed tissue blocks representing 20 normal lung, 19 adenocarcinoma, and 19 squamous cell carcinoma were used to construct the TMA following protocols described earlier (36). For all tissue blocks, H&E-stained sections were reviewed and the areas to be punched for array production were carefully marked. One millimeter diameter cores were punched in duplicate from the selected area of each specimen and inserted into a recipient paraffin block for a total of 116 cores from 58 patients. Five micron sections were cut from the TMA block and mounted onto charged slides. Immunohistochemical staining was performed by the avidin-biotin complex method using the Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA) as described previously (36). Slides were deparaffinized in xylene and hydrated with successive 100 and 95% ethanol washes (v/v). Antigen retrieval was performed in citrate buffer (DAKO, Carpinteria, CA) with heating by microwave for 10 min followed by 20 min at room temperature. Slides were washed with distilled water twice and placed in 3% H2O2 blocking solution for 10 min to inhibit endogenous peroxidase activity. After washing with distilled water, blocking serum was applied for 1 h. Slides were incubated with PAK2 antibody (diluted 1:100, Epitomics, Burlingame, CA) overnight at 4 °C. Universal secondary antibody from the kit and the ABC Elite reagent was applied for 30 min each, followed by washing of the slides with TBST (1%Tween-20). Reactions were developed using diaminobenzidine (DAKO) and counterstaining with hematoxylin.
A pathologist (ALG) scored each of the tissues represented on the TMA and each of the cores was classified as either positive or negative. A sample was defined as partially positive if one core was positive and the other core was negative. The immunostained TMA slide was also analyzed by an Ariol SL-50 automated slide scanner (Applied Imaging, San Jose, CA). Because the PAK2 antibody demonstrated primarily cytoplasmic staining, we trained the Ariol scanner to accurately distinguish positive areas (DAB staining) and whole core (counter staining). The positive area divided by whole core (percent positive area) was recorded. We manually excluded inappropriate areas such as connective tissue and folded tissue. The data was analyzed using the restricted residual maximum likelihood (REML) based mixed effect model.
SDS-PAGE and Immunoblot Analysis
Tumor and normal tissues were lysed with RIPA buffer (Sigma-Aldrich) containing Complete Protease Inhibitor (Roche Diagnostics, Indianapolis, IN). The tissue lysate was sonicated three times for 20 s at 30-s intervals. The protein concentration was measured, and 25 μg of protein from each sample was separated on SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA). The membranes were then blocked with 5% milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) and were incubated with the primary antibody anterior gradient homolog (AGR2), PTGES3 (Abcam, Cambridge, MA), STRAP (Becton-Dickenson, Franklin Lakes, NJ), AKR1B10 (Abcam, Cambridge, MA), and beta-actin (Sigma-Aldrich, St. Louis, MO) overnight. After washing the membrane with TBST, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody and was developed using the chemiluminescent detection kit (Pierce, Rockville, IL).
Down-regulation of PAK2 by shRNA
The MISSION shRNA expression constructs (TRC0000002118) and control (nontarget) were purchased from Sigma-Aldrich (St Louis, MO) as glycerol stocks. The shRNA vector and packaging vectors (pMD2.G and pCMV dR7.74ps PAX2) were cotransfected into 293FT cells by Fugene 6 according to the manufacturer's protocol (Roche Diagnostics, IN). The culture medium was changed 7 h after transfection. Two days post-transfection, virus-containing culture medium was collected, centrifuged to remove cells and filtered with a 45-μm pore size membrane. Target cells were prepared in a six-well plate at a confluency of 70–90%. Transduction was performed by replacing the culture medium with lentivirus-containing media for 6 h. After transduction, the culture medium was replaced, and the cells incubated for 4 days before the experiments were conducted. Four days after transduction, the cells were harvested for the colony formation assay and the growth rate assay. For the colony formation assay, 103 cells were seeded in each well of a six-well plate and were incubated for 7 days. Cells were fixed with 4% paraformaldehyde in PBS (v/v) and were stained with crystal violet. For the growth rate assay, 1000 cells were seeded in each well of a 96-well plate, and cell viability (the number of living cells) was measured by WST-1 reagent (Clontech, Mountain View, CA) in triplicate each day for 4 days according to the manufacturer's protocol. The PAK2 knockdown level was confirmed by immunoblot analysis of the cells after 4 days of transduction.
Transmembrane Migration Assay With a PAK Inhibitor (IPA-3)
Cells were serum-starved for 18–24 h then seeded into a chamber containing an 8.0 μm porous membrane (24-well plate format) at a density of 2 × 104 H1299 cells and 4 × 104 A549 cells. Wells contained serum free medium with a DMSO control or IPA-3 at 1 or 10 μm. After a 45 min preincubation in inhibitor, each chamber was moved into a well containing either 0.1% FBS, 10% FBS+DMSO, 10% FBS+ 1 μm IPA-3, or 10% FBS+ 10 μm IPA-3. After 6 h (H1299) or 12 h (A549) of incubation, cells were fixed and stained with the Diff quick Stain kit (SIEMENS, Munich, Germany). Cells not passing through the membrane were removed by wiping with a cotton swab. Cells in eight randomly selected fields were counted under a microscope using the 20× objective lens.
Matrigel Invasion Assay With a PAK Inhibitor (IPA-3)
Cells were prepared and seeded onto Matrigel invasion chambers (Becton Dickinson, Franklin Lakes, NJ) in the same manner as for the migration assay with the exception that 25 × 103 cells were seeded for both H1299 and A549. Cells on the chamber membrane were fixed and stained after 24 h (H1299) or 48 h (A549) of incubation. Cell counts were obtained as described for the transmembrane migration assay. For statistical analysis, mixed-effect model was used to assess the effect of 4 treatment groups (0.1% FBS, 10% FBS+DMSO, 10% FBS+ 1 μm IPA-3, or 10% FBS+ 10 μm IPA-3) on cell count where square root transformation was applied to the cell count data to meet the normal assumption of mixed-effect model.
Plasma Collection and Preparation
Our initial signature evaluation set consisted of 45 plasma samples and was obtained from individuals with and without lung cancer from our Lung SPORE repository. Thirty samples were from patients with histology proven lung cancers (stages IA-IIIB) of either squamous cell carcinoma or adenocarcinoma subtypes. Another 15 control plasma samples were obtained from individuals matched for age, gender, and smoking history. Control individuals were proven without evidence of lung cancer at one year follow up. Plasma was prepared following a standard operating procedure (supplemental Table S11), aliquoted, and stored at −80 °C until analysis.
Our independent set consisted of 169 independent blood samples, cases carefully matched to controls for relevant clinical characteristics including COPD, and in addition, the majority of our controls also had small pulmonary nodules thought to be cancer, but ultimately proven not to be cancer. This is one of the most rigorous comparisons possible. The clinical characteristics of this set are given in supplemental Tables S12 and S13.
ELISA Analysis of Plasma Samples
The plasma protein concentration measurements were tested in two phases. First we verified the differential protein expression expected from the shotgun and MRM analysis for 12 candidate proteins in 45 samples consisting of 15 SCC, 15 ADC of predominantly advanced stages and matched controls (supplemental Table S11). Second, we validated a subset of nine candidate biomarkers in a case control study of 75 samples made of 41 cases of SCC and 34 controls (supplemental Table S12) and a subset of six candidates in a second case controls study of 94 samples made of 45 cases and 49 matched controls (supplemental Table S13).
Candidate biomarker protein levels were measured in plasma from controls and patients with NSCLC using commercially available sandwich ELISA kits. The optimal plasma dilutions for each protein that fall within the linear range of the assays detection were determined for each analyte empirically. Samples were diluted in ELISA kit diluents buffer following the manufacturer recommendations. ELISA for Calprotectin (a heterodimer of S100A8 and S100A9) that was used as a surrogate to measure levels of S100A8 and S100A9 was purchased from Hycult biotec (Canton, MA) at plasma dilution of 1:40. Levels of LGAL7, LGAL3A, CSTB, MSLN, and advanced glycosylation end product-specific receptor were measured in plasma using ELISA assay kits purchased from R&D systems (Minneapolis, MN) using plasma dilutions of 1:3, 1:2, 1:10, 1:50, and 1:4 (v/v) respectively. ELISA for PPBP measurements was performed using ELISA kit specific for proteolytic fragment of this protein, (neutrophil activating protein-2) NAPII from R&D systems (Minneapolis, MN, USA) at 1:1000 plasma dilution. ELISA for Krt19 (proteolytic fragments CYFRA 21-1) was purchased from DRG International, Inc (Mountainside, NJ) and used at 1:4 dilutions. Plasma measurements for MMP2, matrix metallopeptidase 10, NAMPT, and IGBP2 were performed at Aushon BioSystems, Inc using chemiluminescence detection system (Billerica, MA). The characteristics of the ELISA assays used the two test sets is summarized in supplemental Table S14.
RESULTS
NSCLC Shotgun Proteomic Analysis Detection, Coverage, and Reproducibility
Proteomic analysis was performed on pools of samples of two lung tumor types, adenocarcinoma (ADC), and squamous cell carcinoma (SCC), with each pool composed of 20 individual tissue specimens. We used two pools of normal lung tissue samples with 20 and 19 tissue specimens respectively as normal controls. Lung cancer specimens were obtained from patients with pathological stage I disease. The control pool consisted of normal lung tissues from patients found not to have lung cancer during resection for suspected lung masses.
Spectral counting was used as our primary quantitative metric to examine protein expression profiles for each tissue type. Shotgun proteomics using LC-MS/MS is essentially a sampling technique, in which probability of detection is a function of protein abundance and quantitation is assessed by counting the numbers of spectra that map to identified proteins. This method has been evaluated in several laboratories and displays robust performance and broad dynamic range (17–19). To reduce the complexity of the mixture for each LC-MS/MS analysis and to improve detection of low abundance proteins we used peptide IEF separation as a prefractionation method (13), followed by LC-MS/MS analysis of each IEF fraction on an LTQ-Orbitrap-hybrid instrument (IEF-LC-MS/MS). Following a database search to identify peptide sequences that match the acquired MS/MS spectra, IDpicker (29) was used to filter the peptide identifications to a uniform FDR and to generate a minimum set of proteins to account for the identified peptide sequences. We refer to these assignments as “protein groups.” Although some peptide sets map to multiple protein database entries that comprise a group, the vast majority of the assigned protein groups map to a single database entry.
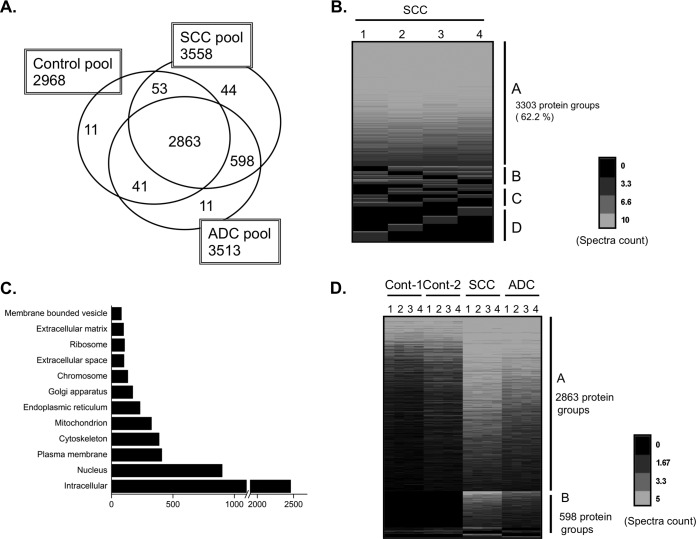
The complete IDpicker data set of proteins identified by at least two distinct peptides consisted of 5923 protein groups, of which eight were classified as “contaminant proteins” (common laboratory contaminant proteins such as trypsin and human epidermal keratins, which may or may not be true contaminants) and 631 as “reverse proteins.” In large data sets, a threshold of two unique peptides per protein identification yields a large number of reverse proteins—in this case leading to an estimated protein-level FDR of 21.3%. We thus required at least eight spectral counts per protein to accept identifications into this data set, which resulted in a calculated 2.3% protein-level FDR. The resulting data set contained 3621 protein groups, including six “contaminant proteins” and 42 “reverse proteins” (Table I). The number of protein groups in the ADC, SCC, and normal pools were 3513, 3558, and 2968, respectively (Table I; Fig. 1A).
Table I. Numbers of discovered proteins.
| Protein groups | |
|---|---|
| Total | 3621 |
| ADC | 3513 |
| SCC | 3558 |
| Normal (2 pools) | 2968 |
Fig. 1.
The shotgun proteomic method identifies large numbers of proteins, reproducibly, from clinical samples. A, Venn diagram of the proteins. The histology and number of protein groups shown for each histology is shown in the box. B, Heatmap view of identified proteins in the SCC pool from four repeated experiments as indicated by the numbers 1–4 shown above each row. Out of the 5310 protein groups identified, 3303 (62.2%) protein groups were observed in every experiment (A), and 487 (9.2%) protein groups were observed in three experiments (B). The number of proteins observed in two experiments (C) and 1 experiment (D) was 597 (11.2%) and 923 (17.3%), respectively. The blue lines in each row represent identified protein groups. The intensity of each line is indicative of the spectra counts for each protein group with the scale shown at the bottom of the figure. C, Gene ontology (GO) cellular component analysis with all identified proteins was performed to determine the distribution of the identified proteins in various subcellular compartments. D, Heatmap view of all identified proteins. Peptides were generated from three different histology pools including two control pools, adenocarcinoma (ADC), and squamous cell carcinoma (SCC) pools. The number of protein groups (2863) across all pools (A), is shown at the upper right side of the panel. Protein groups present only in the SCC and ADC tumor pools (598) (B), is shown at the lower right. Control: Noninvasive lung tissue, SCC: Squamous cell carcinoma, ADC: Adenocarcinoma.
To increase the peptide coverage and to allow assessment of the spectral count distributions within and across groups, we carried out four independent experiments on the same peptide mixture. A heatmap view of the spectra counts from four independent experiments in the SCC pool (Fig. 1B) shows that a high percentage, 62.2%, of all discovered protein groups was observed in all of the experiments, whereas ∼17.3% of the proteins were observed only once.
We subjected all of the identified proteins to gene ontology (GO) cellular component analysis to provide information about the cellular compartments from which the identified proteins came. The analysis showed that whereas most of the proteins were cytoplasmic and nuclear (including chromosomal), proteins associated with the membrane, extracellular matrix, and with organelles such as the ER, Golgi, and mitochondria (Fig. 1C) were also successfully identified. This result shows that shotgun proteomics provides a comprehensive analysis that can identify proteins from every cellular compartment.
Differential Protein Levels in ADC and SCC versus Normal Controls
Our primary goal was to determine the differences in protein expression profiles between tumor tissue and tissue with normal histology. Of the 3621 protein groups, 2863 protein groups (79.1%) were observed across all pools, whereas 598 protein groups (16.5%) were shared among the tumor pools (Fig. 1D). A Venn diagram depicts the number of overlapping and unique protein groups identified in the various tissues (Fig. 1A). The number of protein groups observed in each pool but below the detection threshold in the others was 11 in the combined control pools, 11 in the ADC pool and 44 in the SCC pool (supplemental Tables S4A–4C).
Spectra assigned to some known tumor markers were detected in the tumor pools, but not in normal tissues in our analysis. Carcinoembryonic antigen (CEA) is one of the first tumor markers to be described and is elevated in a number of NSCLCs (37). This protein was detected in our ADC and SCC pools with spectral counts of 14 and 10, respectively (supplemental Table S5). Squamous cell carcinoma antigen (SCC) is also a well-known tumor marker frequently observed in lung and laryngeal cancer patients' sera (38, 39). We observed this molecule with a spectral count of 13 in the SCC pool and three in the ADC pool. Other known tumor markers, CYFRA 21-1(KRT19) and neuron-specific enolase, were also identified in our data set, but were not significantly different from normal tissue (data not shown).
Twenty-five significantly up- or down-regulated proteins are listed along with their spectral counts (Tables IIA, 2B, IIIA, and 3B). No spectra were detected in normal tissue for any of the listed up-regulated proteins in either cancer histology so a log ratio could not be calculated. Of the top 25 up-regulated proteins listed for each histology group, most are shared between the ADC and SCC pools. However, six proteins are unique to ADC (calcitonin-related polypeptide alpha, Chromogranin B, IPI00911047, proprotein convertase subtilisin/kexin type 1, and nerve growth factor inducible) whereas two proteins are found only in the SCC pool (visinin-like 1 and matrix metallopeptidase 10). For the down-regulated proteins, many have peptides that can be detected at some level in tumor tissues. Some of the identified proteins were (as might be expected) hemoglobins from vascular elements. However the presence of embryonic (epsilon) and fetal (gamma) hemoglobin proteins is interesting and not previously reported to be expressed in lung or lung cancer. Only advanced glycosylation end product-specific receptor had no detectable spectra in either cancer histology. A complete list of up-/down-regulated proteins is provided in supplemental Table S6A–6D with spectral counts and, when detected in the normal samples, a log ratio.
Table II. Top 25 proteins detected in cancer but not in normal.
| A, ADC versus normal comparison | |||
|---|---|---|---|
| IPI | Spectra count | Symbol | Full name |
| IPI00000914 | 66 | CALCA | calcitonin-related polypeptide alpha |
| IPI00011062 | 51 | CPS1 | carbamoyl-phosphate synthetase 1, mitochondrial |
| IPI00006601 | 45 | CHGB | chromogranin B (secretogranin 1) |
| IPI00911047 | 37 | cdna FLJ58131, highly similar to secretogranin-1 | |
| IPI00011692 | 28 | IVL | involucrin |
| IPI00007427 | 24 | AGR2 | anterior gradient homolog 2 (Xenopus laevis) |
| IPI00179953 | 22 | NASP | nuclear autoantigenic sperm protein (histone-binding) |
| IPI00009790 | 22 | PFKP | phosphofructokinase, platelet |
| IPI00018769 | 21 | THBS2 | thrombospondin 2 |
| IPI00646689 | 21 | TXNDC17 | thioredoxin domain containing 17 |
| IPI00301961 | 20 | PCSK1 | proprotein convertase subtilisin/kexin type 1 |
| IPI00216088 | 20 | CRABP2 | cellular retinoic acid binding protein 2 |
| IPI00009315 | 19 | ACBD3 | acyl-Coenzyme A binding domain containing 3 |
| IPI00028931 | 19 | DSG2 | desmoglein 2 |
| IPI00002255 | 19 | LRBA | LPS-responsive vesicle trafficking, beach and anchor containing |
| IPI00294536 | 18 | STRAP | serine/threonine kinase receptor associated protein |
| IPI00289501 | 17 | VGF | VGF nerve growth factor inducible |
| IPI00294891 | 17 | NOP2 | NOP2 nucleolar protein homolog (yeast) |
| IPI00299547 | 17 | LCN2 | lipocalin 2 |
| IPI00658109 | 16 | CKMT1B | creatine kinase, mitochondrial 1B |
| IPI00105407 | 16 | AKR1B10 | aldo-keto reductase family 1, member B10 |
| IPI00021700 | 16 | PCNA | proliferating cell nuclear antigen |
| IPI00027078 | 16 | CPD | carboxypeptidase D |
| IPI00030243 | 16 | PSME3 | proteasome (prosome, macropain) activator subunit 3 |
| IPI00218852 | 16 | VIL1 | villin 1 |
| B, SCC versus normal comparison | |||
|---|---|---|---|
| IPI | Spectra count | Symbol | Full name |
| IPI00303300 | 64 | FKBP10 | FK506 binding protein 10, 65 kDa |
| IPI00783625 | 54 | SERPINB5 | serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| IPI00000494 | 53 | RPL5 | ribosomal protein L5 |
| IPI00071509 | 49 | PKP1 | plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) |
| IPI00796480 | 49 | 16 kda protein | |
| IPI00398057 | 47 | RPL10 | ribosomal protein L10 |
| IPI00105407 | 43 | AKR1B10 | aldo-keto reductase family 1, member B10 (aldose reductase) |
| IPI00029733 | 43 | AKR1C1 | aldo-keto reductase family 1, member C1 |
| IPI00021700 | 41 | PCNA | proliferating cell nuclear antigen |
| IPI00013485 | 39 | RPS2 | ribosomal protein S2 |
| IPI00291483 | 39 | AKR1C3 | aldo-keto reductase family 1, member C3 |
| IPI00018769 | 38 | THBS2 | thrombospondin 2 |
| IPI00009315 | 37 | ACBD3 | acyl-Coenzyme A binding domain containing 3 |
| IPI00216313 | 36 | VSNL1 | visinin-like 1 |
| IPI00012007 | 36 | AHCY | adenosylhomocysteinase |
| IPI00013405 | 34 | MMP10 | matrix metallopeptidase 10 (stromelysin 2) |
| IPI00419979 | 33 | PAK2 | p21 protein (Cdc42/Rac)-activated kinase 2 |
| IPI00011692 | 32 | IVL | involucrin |
| IPI00644127 | 31 | IARS | isoleucyl-tRNA synthetase |
| IPI00012268 | 31 | PSMD2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| IPI00044587 | 31 | GBP5 | guanylate binding protein 5 |
| IPI00031517 | 30 | MCM6 | minichromosome maintenance complex component 6 |
| IPI00022078 | 30 | NDRG1 | N-myc downstream regulated 1 |
| IPI00006379 | 29 | NOP58 | NOP58 ribonucleoprotein homolog (yeast) |
| IPI00019869 | 29 | S100A2 | S100 calcium binding protein A2 |
Table III. Top 25 proteins higher in normal than tumor.
| A, ADC versus normal comparison | ||||||
|---|---|---|---|---|---|---|
| IPI | Spectra count |
LogRatio | p value | Symbol | Full name | |
| ADC | Normal | |||||
| IPI00014810 | 0 | 47 | AGER | advanced glycosylation end product-specific receptor | ||
| IPI00020017 | 0 | 21 | C10orf116 | chromosome 10 open reading frame 116 | ||
| IPI00019904 | 0 | 15 | ADD2 | adducin 2 (beta) | ||
| IPI00024853 | 1 | 58 | −5.79 | 0.00070 | PRX | periaxin |
| IPI00299404 | 1 | 40 | −5.25 | 0.00096 | LAMB3 | laminin, beta 3 |
| IPI00299301 | 1 | 33 | −4.97 | 0.00375 | SYNM | synemin, intermediate filament protein |
| IPI00220741 | 7 | 226 | −4.94 | 0.00007 | SPTA1 | spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| IPI00216697 | 5 | 130 | −4.63 | 0.00016 | ANK1 | ankyrin 1, erythrocytic |
| IPI00217471 | 23 | 459 | −4.25 | 0.00002 | HBE1 | hemoglobin, epsilon 1 |
| IPI00220706 | 31 | 502 | −3.95 | 0.00001 | HBG1 | hemoglobin, gamma A |
| IPI00215983 | 12 | 191 | −3.92 | 0.00021 | CA1 | carbonic anhydrase I |
| IPI00744907 | 9 | 138 | −3.87 | 0.00008 | TNXB | tenascin XB |
| IPI00015525 | 5 | 66 | −3.65 | 0.00008 | MMRN2 | multimerin 2 |
| IPI00410714 | 162 | 2058 | −3.60 | 0.00000 | HBA1 | hemoglobin, alpha 1 |
| IPI00009236 | 3 | 35 | −3.47 | 0.00034 | CAV1 | caveolin 1, caveolae protein, 22kDa |
| IPI00473011 | 155 | 1805 | −3.47 | 0.00001 | HBB | hemoglobin, beta |
| IPI00856012 | 7 | 81 | −3.46 | 0.00026 | COL6A6 | collagen, type VI, alpha 6 |
| IPI00013912 | 2 | 22 | −3.39 | 0.00707 | C1orf198 | chromosome 1 open reading frame 198 |
| IPI00221328 | 2 | 20 | −3.25 | 0.00423 | CLIC2 | chloride intracellular channel 2 |
| IPI00005809 | 13 | 123 | −3.17 | 0.00020 | SDPR | serum deprivation response |
| IPI00100980 | 5 | 47 | −3.16 | 0.00049 | EHD2 | EH-domain containing 2 |
| IPI00021854 | 8 | 68 | −3.02 | 0.00169 | APOA2 | apolipoprotein A-II |
| IPI00219772 | 2 | 17 | −3.02 | 0.00781 | NDUFB7 | NADH dehydrogenase 1 beta subcomplex, 7, 18kDa |
| IPI00056334 | 2 | 15 | −2.84 | 0.00707 | PRKCDBP | protein kinase C, delta binding protein |
| IPI00377045 | 15 | 112 | −2.83 | 0.00006 | LAMA3 | laminin, alpha 3 |
| B, SCC versus normal comparison | ||||||
|---|---|---|---|---|---|---|
| IPI | Spectra count |
LogRatio | p value | Symbol | Full name | |
| SCC | Normal | |||||
| IPI00329108 | 0 | 63 | SCEL | sciellin | ||
| IPI00024853 | 0 | 58 | PRX | periaxin | ||
| IPI00478672 | 0 | 53 | SCEL | sciellin | ||
| IPI00014810 | 0 | 47 | AGER | advanced glycosylation end product-specific receptor | ||
| IPI00013912 | 0 | 22 | C1orf198 | chromosome 1 open reading frame 198 | ||
| IPI00410506 | 0 | 19 | FLJ45950 | FLJ45950 protein | ||
| IPI00003373 | 0 | 18 | OCLN | occludin | ||
| IPI00021302 | 0 | 17 | SUSD2 | sushi domain containing 2 | ||
| IPI00645815 | 0 | 17 | SLC9A3R2 | solute carrier family 9, member 3 regulator 2 | ||
| IPI00029928 | 0 | 16 | ELN | elastin | ||
| IPI00856012 | 1 | 81 | −6.70 | 0.00012 | COL6A6 | collagen, type VI, alpha 6 |
| IPI00216697 | 3 | 130 | −5.80 | 0.00009 | ANK1 | ankyrin 1, erythrocytic |
| IPI00220741 | 6 | 226 | −5.60 | 0.00001 | SPTA1 | spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| IPI00299301 | 1 | 33 | −5.41 | 0.00154 | SYNM | synemin, intermediate filament protein |
| IPI00217471 | 21 | 459 | −4.81 | 0.00000 | HBE1 | hemoglobin, epsilon 1 |
| IPI00221328 | 1 | 20 | −4.68 | 0.00206 | CLIC2 | chloride intracellular channel 2 |
| IPI00902726 | 1 | 20 | −4.68 | 0.00510 | cdna FLJ39125 highly similar to homo sapiens collagen, type xxi, alpha 1 (col21a1), mrna. | |
| IPI00220706 | 30 | 502 | −4.43 | 0.00000 | HBG1 | hemoglobin, gamma A |
| IPI00553043 | 3 | 48 | −4.36 | 0.00006 | LIMCH1 | LIM and calponin homology domains 1 |
| IPI00744907 | 9 | 138 | −4.30 | 0.00010 | TNXB | tenascin XB |
| IPI00294443 | 2 | 29 | −4.22 | 0.00438 | CLIC5 | chloride intracellular channel 5 |
| IPI00382499 | 1 | 13 | −4.06 | 0.00530 | ig heavy chain v-iii region jon | |
| IPI00473011 | 154 | 1805 | −3.91 | 0.00000 | HBB | hemoglobin, beta |
| IPI00216704 | 22 | 249 | −3.86 | 0.00003 | SPTB | spectrin, beta, erythrocytic |
| IPI00237806 | 22 | 248 | −3.86 | 0.00003 | SPTB | spectrin, beta, erythrocytic |
Verification of Shotgun Data by LC-MRM-MS and Western Blot
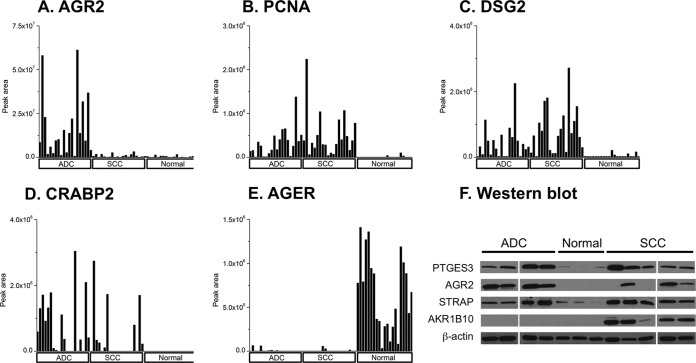
LC-MRM-MS is a targeted, quantitative proteomics method that enables multiplexed quantitation of proteins through measurements of representative peptides in a medium-throughput manner (22). LC-MRM-MS provides a means to more precisely measure levels of biomarker candidate proteins without the use of antibodies. We chose 14 proteins of interest for analysis by LC-MRM-MS. Each protein was quantified by measurements of at least two peptides and each peptide was quantified as the sum of four MRM transitions. When we analyzed the original individual samples making up the pools of 20 ADC, 20 SCC, and 22 normal controls used for the MS-MS analyses, we successfully confirmed the presence and relative expression levels of AGR2 (Fig. 2A), proliferating cell nuclear antigen (PCNA) (Fig. 2B), desmoglein 2 (Fig. 2C), cellular retinoic acid binding protein 2 (Fig. 2D), and advanced glycosylation end product-specific receptor (Fig. 2E). These four up-regulated and one down-regulated proteins showed significant differences based on spectral count data from the shotgun analyses. These differences could be verified by LC-MRM-MS in individual sample analyses based upon the comparison of normalized peak areas from the target peptides. The results of an additional nine up-regulated proteins are shown in supplemental Figs. S1A–S1I. With most of the candidate proteins, there are clear differences in peak areas of the peptides monitored between the tumor and normal samples. However, cathepsin B (CTSB) (supplemental Fig. S1F) and eukaryotic translation initiation factor 5A (supplemental Fig. S1G), while appearing to have higher protein levels in most tumor samples, also have significant levels of protein in the selected normal samples. We also validated protein expression level of several up-regulated proteins by Western blot (Fig. 2F). We examined four proteins that were detected as being highly expressed in tumor pools, which included AGR2 (spectral count ADC 24, SCC 24, normal 0), STRAP (spectral count ADC 18, SCC 20, normal 0), PTGES3 (spectral count ADC 15, SCC 14, normal 0) and AKR1B10 (spectral count ADC 16, SCC 43, normal 0). Immunoblot analysis showed a clear difference in expression level between tumor samples and normal controls, indicating our proteomics analysis can routinely be confirmed by antibody based immunoassays. In some cases; however, there is discordance, requiring further investigation and may be related to the differing specificities of MS proteomic detection and relatively less specific antibody-based methods.
Fig. 2.
Verification of shotgun proteomic candidate biomarkers by liquid chromatography-multiple reaction monitoring-mass spectrometry (LC-MRM-MS) analysis and Western blot analysis in lung cancer. Twenty ADC, 20 SCC, and 21 normal controls were analyzed by MRM for A, AGR2 (IPI00007427); B, PCNA (IPI00021700); C, desmoglein 2 (IPI00028931); D, cellular retinoic acid binding protein 2 (IPI00216088); and E, AGER (IPI00014810). The bar graph shows the normalized expression level for each sample. Histology is shown at the bottom of the figure. F, Immunoblot analysis with AGR2, STRAP, AKR1B10, PTGES3 antibody. Four ADC, five SCC, and three normal samples were immunoblotted. Beta-actin was used for internal control.
To further assess the concordance between shotgun and MRM analyses of differentially expressed proteins, we analyzed three additional sets of normal lung (n = 21), ADC, (n = 20), and SCC (n = 20) tissues by LC-MRM-MS (supplemental Table S7). The majority of these samples (70%) were stages I-II, whereas the remainder (30%) were stages III–IV. These analyses measured 95 proteins that had been observed in the shotgun data sets for normal, ADC and SCC tissues. A comparison of these MRM measurements with the spectral count data for shotgun analyses of the pooled samples are presented in supplemental Table S8. In the ADC versus normal comparison, out of 50 proteins that were differentially expressed, 44 proteins were verified by MRM (88%). In the SCC versus normal comparison there were also 50 proteins that were differentially expressed and 42 of them were validated by MRM (84%). Thus, most of the protein expression differences measured by shotgun analyses of the normal, ADC and SCC pools were confirmed in an independent set of specimens.
Gene Ontology Enrichment Analysis of ADC versus SCC Reveals Discriminate Proteins Between the Two Histologies
Although they are usually readily distinguishable morphologically, biological differences between ADC and SCC at the molecular level have not been fully defined. To elucidate molecular differences at the protein level, the protein profile for ADC was compared with that derived for SCC. The top 20 proteins significantly differentially expressed between histologies are shown in supplemental Table S9A and S9B. In addition to identifying enriched proteins in each of the histologies, we used GOTM (35), to conduct GO enrichment analysis. GO enrichment analysis identifies functions and pathways that are differently activated between two different histology subtypes (supplemental Fig. S2). For GO functional analysis, we used 54 differentially expressed proteins. All proteins in our data set were used for a reference to identify the differences in two histologies of lung cancer. This analysis identified the biological processes categories of ectoderm development and epidermis development (supplemental Fig. S2A; supplemental Table S10A), which included several keratin family members, KRT5, KRT6A, 6B, and KRT13 enriched in SCC. The ADC histology was enriched in cellular retinoic acid-binding protein, and sciellin (SCEL). In the cellular component analysis (supplemental Fig. S2B; supplemental Table S10B), the extracellular region category was enriched. This category includes SCC enriched matrix metallopeptidase 10, SERPINB2, SERPINB5, as well as ADC enriched proteins such as mucins (MUC1, MUC5B). There are two known ADC markers in this category, surfactant protein B (SFTPB), which was reported as having ADC-specific expression by immunohistochemistry (40), and polymeric immunoglobulin receptor (PIGR). These results show that proteins associated with tissue development and structural molecular activities are differentially expressed in ADC and SCC.
Identification of p21-activated Kinase, PAK2
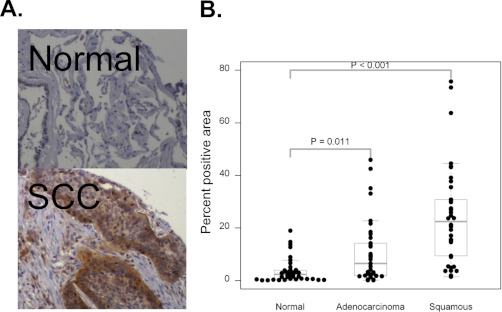
In addition to biomarker identification, our shotgun approach provides insight into potential molecular pathways that are aberrantly regulated in NSCLC. We were especially interested in proteins that are potential therapeutic targets. One group of proteins, the Group I, p21-activated kinases (PAKs), were of interest as PAK2 was identified in each histology, but with a much higher spectral counts in the SCC pool (Table 2B). PAK1 was also present, but was much less abundant than PAK2. The PAK proteins have been reported to be up-regulated in multiple cancer types (41–43) but have not yet been implicated in NSCLC. To confirm increased expression levels of PAK2 in NSCLC in our shotgun proteomics analysis, we stained a tissue microarray from the same patients whose frozen tissue blocks were used for proteomics analysis. After pathologic (ALG) review, it was determined that out of 19 SCC samples, 12 cases were strongly stained, whereas three normal cases were stained out of 14 samples (Table IV). A representative image of normal and squamous cell carcinoma tissue stained with a PAK2-specific antibody shows high protein levels of PAK2 in the tumor (Fig. 3A). In the ADC samples we did not see high expression as frequently as in SCC with only three positives out of 19 samples. We observed a varying degree of cytoplasmic staining of PAK2 in tumor cells, especially in SCC histology where weak staining was observed in some normal type two alveolar cells, inflammatory cells, and vascular endothelial cells. We also quantified positive staining area by image analysis. The distribution of percent PAK2 positive area for each histology is shown in Fig. 3B. This method of analysis supported the previous result of high PAK2 in SCC and also provided evidence of moderately higher PAK2 in ADC when compared with normal tissue.
Table IV. PAK2 Tissue microarray analysis.
| Negative | Partially positive | Positive | Total | |
|---|---|---|---|---|
| ADC | 14 | 2 | 3 | 19 |
| Normal | 5 | 6 | 3 | 14 |
| SCC | 2 | 5 | 12 | 19 |
Fig. 3.
PAK2 overexpression in NSCLC. A, Immunohistochemistry of normal (upper panel) and lung tumor (lower panel) tissues. B, Distribution of percent positive area by quantitative image analysis in tissue microarray.
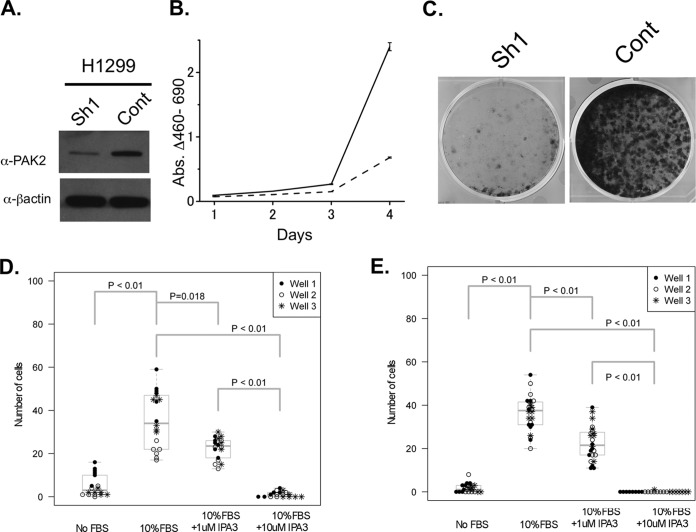
To investigate whether PAK2 is important for cell survival in lung cancer cell lines we knocked down PAK2 with several shRNA expressing lentiviruses in H1299 and A549 cells. Loss of PAK2 protein (Fig. 4A; supplemental Fig. S3A) resulted in a significant reduction in cell growth on plastic and a dramatic reduction in the number of colonies formed (Figs. 4B and 4C; supplemental Figs. S3B and S3C). A small molecule inhibitor of the Group I PAKs, IPA-3, has recently been identified through a screening effort to identify allosteric inhibitors of these proteins (44). This in vitro screen took advantage of the auto-inhibitory domain present in the Group I PAKs and the conformational change that takes place upon CDC42 binding and subsequent activation of the kinase activity. Allosteric inhibitors, unlike ATP mimetics, provide a higher level of specificity because they rely on protein structure rather than the ATP-binding domain that is so similar among the ATP-binding proteins. However, because of the structural similarity of the group I PAK proteins, IPA-3 will inhibit PAKs 1, 2, and 3 (44). To assess the ability of IPA-3 to interfere with migration and invasion of NSCLC cell lines we performed migration and invasion assays (Figs. 4D and 4E; supplemental Figs. S3D and S3E). In both analyses, IPA-3 inhibited the migration and invasion abilities in a dose-dependent manner. These observations support the idea that p21-activated kinase may have an important role in lung cancer tumorigenesis and as potential therapeutic targets.
Fig. 4.
PAK2 contributes to cell proliferation, migration and invasion. A, Immunoblot of H1299 cells 4 days after infection with shRNA control virus (Cont) or with shRNA directed against PAK2 (Sh1). B, H1299 cell line growth curve after shRNA infection. Cell viability was measured using the WST-1 reagent for 4 days after seeding the cells in the culture plate. Solid line - control virus; Dashed line - PAK2 shRNA containing virus C, Colony formation assay using H1299 cells seeded 4 days after infection control virus (Cont) or PAK2 shRNA containing virus. (Sh1) D, Transmembrane migration assay results with IPA-3 at the indicated concentrations. E, Matrigel invasion assay results with IPA-3 at the indicated concentrations.
Translation of the Tissue-based Approach Into Candidate Plasma Biomarkers
To test how the large protein inventories obtained from shotgun proteomics analysis of lung tumors may identify tumor-derived proteins that are elevated in the plasma of early stage lung cancer patients and thus of potential use as candidate diagnostic biomarkers, we tested the concentration of shotgun proteomic candidate biomarkers in the plasma of individuals with and without lung cancer. To prioritize our candidates, we used a combination of statistical and biological criteria including: the expression level in cancer tissues, presence in plasma and other biofluids, and statistical significance using commercial and publicly available bioinformatics tools (including Ingenuity Pathway Analysis (IPA), Webgestalt). The resulting list of candidates was further refined by cross-examination against other publicly available databases such as the Plasma Proteome Project of Human Proteome Organization (PPP-HUPO). This process yielded 164 candidate biomarkers that are differentially expressed in early stage squamous or adenocarcinomas of the lung as compared with matched controls.
The plasma protein concentration measurements were tested in 2 phases (supplemental Fig. S4). First, in a proof of concept experiment, we verified the differential protein expression expected from the shotgun and MRM analysis for 12 candidate proteins for which commercial ELISA kits were available. We tested those in 45 samples consisting of 15 SCC, 15 ADC with predominantly advanced stages and matched controls (supplemental Table S11). The data presented in supplemental Fig. S5 show that all 12 of the selected proteins were detected in the plasma from control, ADC or SCC samples. The concentrations of MMP2 were significantly higher in plasma from SCC patients compared with controls, and KRT19 was nearly significant (p = 0.06), consistent with previously published reports (45, 46). Here we also report for the first time elevated plasma concentrations of LGALS7 in patients with SCC of the lung.
Second, we developed and carefully validated custom-made ELISAs for 30 proteins using in-house raised and purified polyclonal antibodies for detection in plasma. These were first tested in a pilot study for protein concentration in 45 plasma samples of patients with advanced disease and matched controls (data not shown). From these 30, the nine candidate biomarkers that displayed a consistent relationship to cancer phenotypes in this proof of concept phase were then tested by ELISA in a larger data set for confirmation of these findings. The performance characteristics of these nine ELISAs are presented in supplemental Table S14. These nine candidate biomarkers were tested in a case control study of 75 samples comprising 41 cases of SCC and 34 controls (supplemental Table S12). A subset of six candidates in a second case controls study of 94 samples made of 45 cases of ADC and 49 matched controls (supplemental Table S13). The results of this independent and carefully case-control matched set of 135 patients (the sum of all cases and controls from these two data sets 169 (74 + 94) corresponds to a total of 135 individuals because controls overlap between the groups) are shown in supplemental Table S15. A multivariable logistic regression model was built to assess these biomarkers' ability to differentiate cancers from controls. The prediction performance was measured primarily by AUC metrics. The results for prediction of the diagnosis of squamous cell carcinoma (n = 41 cases and 34 matched controls) showed an AUC of 0.72 [95% CI: 0.62, 0.82]. The AUC for the prediction of the diagnosis of the adenocarcinomas (45 cases, 49 matched controls) was 0.59 [95% CI: 0.5, 0.67]. Note that both the reported point estimators and their corresponding 95% confidence interval were bias-corrected by the bootstrap method. We also investigated the support vector machine algorithm to address performance accuracy and obtained very similar results (supplemental Table S15). It is important to note that not only were the cases and controls were very closely matched for clinical characteristics including age, gender, pack year smoking history, and COPD, but the majority of the “controls” were patients who presented for evaluation of pulmonary nodules suspicious for cancer, but were proven not to have cancer after a one year follow up.
DISCUSSION
Lung cancer is by far the largest single cause of cancer death in the western world, responsible for the deaths of more people than the next four most frequent cancers combined. Significant progress has been made in the treatment of this disease with the identification of subsets of lung cancer containing mutations in the epidermal growth factor receptor and fusions of the anaplastic lymphoma kinase gene, but together these account for only about 15% of cases, and in even these cases therapy is palliative and not curative. Technology is becoming available for the practical sequencing of the entire genome or transcriptome, which will undoubtedly identify other potentially targetable lesions, though preliminary data suggest that they will be in even smaller subsets, and many will not have clear therapeutic interventions. It is very clear, however, that genes can also be activated or repressed by mechanisms other than coding region mutations, including methylation and histone acetylation, and significant progress has been made using expression array technology to address the genome-wide alterations in RNA expression that results from these mechanisms.
However, in virtually every case, it is neither DNA or RNA that is the ultimate biologically functional moiety, but protein. Protein expression and activity levels can be affected by another layer of postgenomic regulation, including post-translational modification and alteration in protein degradation. Thus the most complete portrait of the dysregulated functional networks in a cancer cell will very likely be achieved only at the protein level, and a complete knowledge of the state and structure of the proteins in a cancer cell will undoubtedly be more informative than that of the genome. This would have use not only for better understanding the biology of cancer, but also defining new targets for therapy and even practical biomarkers for the early identification of disease while it is still in a readily curable stage. However, previous proteomic technologies have limited analysis to a very small and superficial subset of the cancer proteome.
Although other studies have yielded inventories of selected subsets of proteins, such as phosphopeptides (47), the shotgun proteomic analysis presented here is the most comprehensive analysis of the global cancer proteome in the literature to date. We have identified and assessed the levels of more than 3500 proteins in the two major subtypes of nonsmall cell lung cancer and uninvolved lung tissues, significantly greater than any previous study (48–54). The success of this multidimensional proteomic analysis from trypsin-digested biological specimens has been partly owing to the careful selection of prefractionation technologies. An optimized pre-fractionation method is a key step if shotgun proteomic analysis is to have good sensitivity for the detection of low-abundance peptides and to provide reproducible results. Our previous investigation (13) indicated that the IEF prefractionation method may have an advantage in terms of reproducibility over strong cation exchange-based multidimensional LC-MS/MS (MudPIT) (55) and that four replicate analyses of each pooled sample achieves ∼90% of the identifications that would be made by exhaustive analysis (nine replicates). The other critical element of the analysis is our approach to database searching of the obtained MS/MS spectra, the filtering of results and inference of protein identifications. We used reversed sequence database searching to enforce a peptide identification FDR of 0.05 and required two distinct peptide sequences per protein identification. We also used parsimonious assembly to minimize redundant protein identifications (29) and also required eight or more spectral counts across the pools for protein identifications in the final data set. These latter steps significantly reduced the number of protein identifications, but produced a much more reliable data set, as demonstrated by a 2.3% protein FDR. Thus, although the final data set represents a conservative inventory of protein expression in the tissues, the inventories are sufficiently deep to reflect the biological diversity of both ADC and SCC.
With the approach presented here, we have generated a large data set of identified proteins whose levels were significantly different between lung tumor groups and normal lung tissues. Previous proteomic analyses (e.g. those on two-dimensional gel platforms) provided relatively superficial coverage of the proteome and detect only the most abundant proteins (9). One indication that we have probed deeply enough into the proteome to potentially identify novel tumor biomarkers was that spectra were detected for the known lung tumor biomarkers such as carcinoembryonic antigen in ADC pool, and SCC in the squamous cell carcinoma pool (supplemental Table S5). To further substantiate our shotgun results, a targeted proteomic analysis employing LC-MRM-MS, Western blot, and immunohistochemistry were used to examine individual patient tumor samples for expression of select proteins identified in the shotgun analysis. The high percentage of proteins verified as up or down regulated (11 of 13 up and 1 of 1 down) with separate methods of analysis increases our confidence that differences detected with the initial shotgun analysis provide useful information regarding protein levels in NSCLC. MRM analyses of 95 proteins in three additional sets of normal, ADC and SCC tissues confirmed many additional protein expression differences measured in our shotgun analyses of the pools and provides further evidence of the reliability of our approach.
In this first study of its kind in lung cancer, we decided to pool samples from each of these two subtypes. We fully understand that there is significant heterogeneity within these morphologically defined subtypes (e.g. 10% frequency of EGFR mutations in adenocarcinomas), but our intent was to attempt to uncover common alterations in these two major categories that would be detectable by a pooled approach. This is particularly important when seeking to define candidate diagnostic biomarkers applicable to patients with suspected tumors. In addition, a pooled approach allows “signal averaging” to emphasize the dominant signals and reduce noise. The efficacy of our approach is demonstrated by the MRM data showing significant heterogeneity in single samples for our marker proteins, but clear confirmation of the histologic class distinctions. Single sample analyses are underway, but these will take a long time to achieve statistically significant cohort sizes, especially within these heterogeneous subgroups.
The decision not to use adjacent “normal” lung as our normal but rather patients who underwent resection for suspected lung cancer but were found to have benign lesions was also deliberate. Lung cancers have clear molecular abnormalities that have been observed in adjacent normal-appearing tissue (“field effect”) that are clearly found in cancer patients and not matched normal (56). With our intent to discover differences between cancer and noncancer normal, and for biomarker discovery, we felt that clinically matched normal controls were the optimal choice, especially for a pooled strategy.
Among the proteins differentially expressed between normal and tumor, several have already been shown to be up-regulated in lung cancer. These would include serine/threonine kinase receptor associated protein (STRAP), aldo-keto reductase family 1, member B10 (AKR1B10), and proliferating cell nuclear antigen (PCNA), which has been routinely used as a proliferation marker in many cancer types (57, 58). AKR1B10 is involved in retinoid metabolism and may be responsible for the alterations in retinoid signaling known to be important in lung carcinogenesis (59). Maspin (SERPINB5) was higher in our SCC pool and is a well-described marker of the SCC histology (60). This is supported by data from the public immunohistochemistry database, the Human Protein Atlas (61), which shows high levels of maspin in SCCs of the lung. Proteins up-regulated in our study not previously associated with lung cancer are creatine kinase, mitochondrial 1B (CKMT1B), which was found to be up-regulated in breast cancer by expression arrays and associated with poor prognosis (62), and FAM3C (63), a secreted interleukin-like protein that was identified in proteomic screens for secreted proteins from pancreatic cancer cells (62, 64). More recently FAM3C was discovered as a translationally controlled protein implicated in EMT and breast cancer progression (63). Also discovered through a proteomics approach in CRC was proteasome (prosome, macropain) activator subunit 3, a proteasome-associated protein that was recently identified as a novel serum tumor marker in this cancer type (65). The expression of calcitonin-related polypeptide alpha, a peptide associated with increased angiogenesis and endothelial cell proliferation in placental development, but not previously associated with cancer, was higher in ADC versus normal tissue (66). Notably, the mini-chromosome maintenance (MCM) proteins, MCM2, MCM3, MCM4, MCM5, and MCM6, were also present in our list of up-regulated proteins. These proteins are reported to exist in a heterohexameric complex with MCM7 (67). The fact that five of the six proteins in this complex were detected in our analysis is further support that this methodology is robust. The MCM proteins, and MCM2 in particular, have been investigated as proliferation markers in many types of cancer because they appear to be more reliable than Ki67 at detecting dysplastic cells and correlating with survival in lung cancer (68, 69).
Our analysis also identified a potential drug target not previously reported to be important in lung cancer. The p21-activated kinases (PAKs) were identified in this study as being dysregulated in NSCLC and we demonstrated that knockdown or drug inhibition of the PAKs has significant functional consequences in vitro. The PAKs have been studied in multiple cancer types, but have not yet been examined for their role in lung cancer. The group I PAKs are known to bind to and phosphorylate multiple substrates that have been shown to be involved in proliferation, survival, and cytoskeletal remodeling (70). Ample evidence exists for the involvement of the Group I PAKs in breast, colon, prostate, as well as many other cancers (70). With regard to lung cancer, indirect evidence exists for the involvement of the PAKs in tumor development and progression. In a mouse model of KRAS-induced lung tumors, Rac1, an upstream activator of the PAKs, was necessary for tumor formation and progression (71). Our group's finding through a proteomic analysis that PAKs are over expressed in NSCLC, and that knockdown or drug inhibition affects the tumorigenic properties of lung cancer cell lines, would be the first to directly implicate PAKs as having a role in lung tumor biology, and suggest their potential as therapeutic targets.
In addition to identification of dysregulated pathways and proteins to better understand NSCLC biology or find new therapeutic targets, our analysis also identified novel candidates as blood biomarkers for the early detection of lung cancer. It is possible that a subset of the proteins found to be over-expressed in lung tumors might be detectable by highly sensitive and specific assays in the peripheral blood and be useful in the clinical setting. In this study, we show results from an early assessment of the plasma levels of candidate biomarkers derived from this shotgun proteomics analysis. Although direct MRM analysis of plasma could have been employed, detection of low abundant target analytes without enrichment techniques would not have afforded the same level of sensitivity as that of the ELISA platform. Therefore, ELISA assays were initially performed to test candidates for which commercial assays for the protein candidates of interest were available. We then developed and carefully characterized in-house ELISAs and tested our most promising candidates in 135 clinical specimens including early stage lung cancers and matched controls with very encouraging performance characteristics. The large inventories of differentially expressed proteins identified by this platform yield many more candidates for diagnostic plasma biomarkers that we are in the process of evaluating.
Proteomics analysis may help identify molecular differences not easily addressed by microarray or genomic analysis, as it is clear that many very important cellular processes are primarily regulated by protein stability or post-translational modification. Further development of this technology will undoubtedly increase the analytic depth and can be extended to catalog not only protein levels but also specific somatic mutant sequences and post-translational modifications adding further dimensions to our understanding of the cancer cell inaccessible to genomic and transcriptomic technologies. Our results demonstrate that a tissue-based in depth proteomic approach allows the identification and validation of candidate dysregulated pathways and diagnostic biomarkers. Future work will be required to validate potential candidate targets and to determine whether the identified plasma biomarkers may add value to the performance of the screening chest CT scans and change clinical management.
Supplementary Material
Acknowledgments
We thank, Sarah Stuart and Nancy Winters for their help with sample preparation and MS analysis. We also would like to thank Harriet Davis, Candace Murphy and Snjezana Zaja-Milatovic for their efforts to maintain our tissue bank and clinical information database, Heidi Chen and Yan Guo for statistical analysis help, David Tabb and Bing Zhang for helpful discussions.
Footnotes
* This work was supported by the Clinical Proteomic Technology Assessment for Cancer program (U24CA126479), SPORE in Lung Cancer (P50CA090949) and SPECS in lung cancer (U01CA114771) from the National Cancer Institute.
 This article contains supplemental Tables S1 to S15 and Figs. S1 to S5.
This article contains supplemental Tables S1 to S15 and Figs. S1 to S5.
Data sharing: The data associated with this manuscript may be downloaded from the Ayers Institute website at http://www.vicc.org/jimayersinstitute/data/. The data include all raw MS datafiles and descriptive information for each.
1 The abbreviations used are:
- LC-MS/MS
- liquid chromatography-tandem MS
- NSCLC
- nonsmall cell lung cancer
- LC-MRM-MS
- liquid chromatography-multiple reaction monitoring-mass spectrometry
- ADC
- adenocarcinoma
- SCC
- squamous cell carcinoma
- FDR
- false discovery rate
- MCM
- minichromosome maintenance
- IEF
- isoeletric focusing
- AGR2
- anterior gradient homolog 2
- CTSB
- cathepsin B (supplemental Fig. 1F).
REFERENCES
- 1. Ries L. A. G., Melbert D., Krapcho M., Stinchcomb D. G., Howlader N., Horner M. J., Mariotto A., Miller B. A., Feuer E. J., Altekruse S. F., Lewis D. R., Clegg L., Eisner M. P., Reichman M., Edwards B. K. (2008) SEER Cancer Statistics Review, 1975–2005 [Google Scholar]
- 2. Bach P. B., Silvestri G. A., Hanger M., Jett J. R. (2007) Screening for lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition). Chest 132, 69S–77S [DOI] [PubMed] [Google Scholar]
- 3. Beer D. G., Kardia S. L., Huang C. C., Giordano T. J., Levin A. M., Misek D. E., Lin L., Chen G., Gharib T. G., Thomas D. G., Lizyness M. L., Kuick R., Hayasaka S., Taylor J. M., Iannettoni M. D., Orringer M. B., Hanash S. (2002) Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 8, 816–824 [DOI] [PubMed] [Google Scholar]
- 4. Bhattacharjee A., Richards W. G., Staunton J., Li C., Monti S., Vasa P., Ladd C., Beheshti J., Bueno R., Gillette M., Loda M., Weber G., Mark E. J., Lander E. S., Wong W., Johnson B. E., Golub T. R., Sugarbaker D. J., Meyerson M. (2001) Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. U.S.A. 98, 13790–13795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kumar M. S., Erkeland S. J., Pester R. E., Chen C. Y., Ebert M. S., Sharp P. A., Jacks T. (2008) Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. U.S.A. 105, 3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu S. L., Chen H. Y., Chang G. C., Chen C. Y., Chen H. W., Singh S., Cheng C. L., Yu C. J., Lee Y. C., Chen H. S., Su T. J., Chiang C. C., Li H. N., Hong Q. S., Su H. Y., Chen C. C., Chen W. J., Liu C. C., Chan W. K., Chen W. J., Li K. C., Chen J. J., Yang P. C. (2008) MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 13, 48–57 [DOI] [PubMed] [Google Scholar]
- 7. Yanagisawa K., Shyr Y., Xu B. J., Massion P. P., Larsen P. H., White B. C., Roberts J. R., Edgerton M., Gonzalez A., Nadaf S., Moore J. H., Caprioli R. M., Carbone D. P. (2003) Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet 362, 433–439 [DOI] [PubMed] [Google Scholar]
- 8. Yanagisawa K., Tomida S., Shimada Y., Yatabe Y., Mitsudomi T., Takahashi T. (2007) A 25-signal proteomic signature and outcome for patients with resected non-small-cell lung cancer. J. Natl. Cancer Inst. 99, 858–867 [DOI] [PubMed] [Google Scholar]
- 9. Chen G., Gharib T. G., Wang H., Huang C. C., Kuick R., Thomas D. G., Shedden K. A., Misek D. E., Taylor J. M., Giordano T. J., Kardia S. L., Iannettoni M. D., Yee J., Hogg P. J., Orringer M. B., Hanash S. M., Beer D. G. (2003) Protein profiles associated with survival in lung adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 100, 13537–13542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 11. Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M. S., Gramolini A. O., Morris Q., Hallett M. T., Rossant J., Hughes T. R., Frey B., Emili A. (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125, 173–186 [DOI] [PubMed] [Google Scholar]
- 12. Fiske W. H., Tanksley J., Nam K. T., Goldenring J. R., Slebos R. J., Liebler D. C., Abtahi A. M., La Fleur B., Ayers G. D., Lind C. D., Washington M. K., Coffey R. J. (2009) Efficacy of cetuximab in the treatment of Menetrier's disease. Sci. Transl. Med. 1, 8ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Slebos R. J., Brock J. W., Winters N. F., Stuart S. R., Martinez M. A., Li M., Chambers M. C., Zimmerman L. J., Ham A. J., Tabb D. L., Liebler D. C. (2008) Evaluation of Strong Cation Exchange versus Isoelectric Focusing of Peptides for Multidimensional Liquid Chromatography-Tandem Mass Spectrometry. J. Proteome Res. 7, 5286–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sprung R. W., Jr., Brock J. W., Tanksley J. P., Li M., Washington M. K., Slebos R. J., Liebler D. C. (2009) Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis. Mol. Cell Proteomics 8, 1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujii K., Nakano T., Kanazawa M., Akimoto S., Hirano T., Kato H., Nishimura T. (2005) Clinical-scale high-throughput human plasma proteome analysis: lung adenocarcinoma. Proteomics 5, 1150–1159 [DOI] [PubMed] [Google Scholar]
- 16. Tyan Y. C., Wu H. Y., Lai W. W., Su W. C., Liao P. C. (2005) Proteomic profiling of human pleural effusion using two-dimensional nano liquid chromatography tandem mass spectrometry. J. Proteome Res. 4, 1274–1286 [DOI] [PubMed] [Google Scholar]
- 17. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Analytical Chemistry 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 18. Gao J., Opiteck G. J., Friedrichs M. S., Dongre A. R., Hefta S. A. (2003) Changes in the protein expression of yeast as a function of carbon source. J. Proteome Res. 2, 643–649 [DOI] [PubMed] [Google Scholar]
- 19. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 20. Paulovich A. G., Billheimer D., Ham A. J., Vega-Montoto L., Rudnick P. A., Tabb D. L., Wang P., Blackman R. K., Bunk D. M., Cardasis H. L., Clauser K. R., Kinsinger C. R., Schilling B., Tegeler T. J., Variyath A. M., Wang M., Whiteaker J. R., Zimmerman L. J., Fenyo D., Carr S. A., Fisher S. J., Gibson B. W., Mesri M., Neubert T. A., Regnier F. E., Rodriguez H., Spiegelman C., Stein S. E., Tempst P., Liebler D. C. (2010) Interlaboratory study characterizing a yeast performance standard for benchmarking LC-MS platform performance. Mol. Cell. Proteomics 9, 242–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tabb D. L., Vega-Montoto L., Rudnick P. A., Variyath A. M., Ham A. J., Bunk D. M., Kilpatrick L. E., Billheimer D. D., Blackman R. K., Cardasis H. L., Carr S. A., Clauser K. R., Jaffe J. D., Kowalski K. A., Neubert T. A., Regnier F. E., Schilling B., Tegeler T. J., Wang M., Wang P., Whiteaker J. R., Zimmerman L. J., Fisher S. J., Gibson B. W., Kinsinger C. R., Mesri M., Rodriguez H., Stein S. E., Tempst P., Paulovich A. G., Liebler D. C., Spiegelman C. (2010) Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 9, 761–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang H., Qian W. J., Chin M. H., Petyuk V. A., Barry R. C., Liu T., Gritsenko M. A., Mottaz H. M., Moore R. J., Camp Ii D. G., Khan A. H., Smith D. J., Smith R. D. (2006) Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J. Proteome Res. 5, 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cortes H. J., P. C. D., Richter B. E., Stevens T. S. (1987) Porous ceramic bed supports for fused silica packed capillary columns used in liquid chromatography. J. High Resolution Chromatog. 10, 446–448 [Google Scholar]
- 25. Licklider L. J., Thoreen C. C., Peng J., Gygi S. P. (2002) Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Analytical Chemistry 74, 3076–3083 [DOI] [PubMed] [Google Scholar]
- 26. Kessner D., Chambers M., Burke R., Agus D., Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tabb D. L., Fernando C. G., Chambers M. C. (2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma Z. Q., Dasari S., Chambers M. C., Litton M. D., Sobecki S. M., Zimmerman L. J., Halvey P. J., Schilling B., Drake P. M., Gibson B. W., Tabb D. L. (2009) IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 8, 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang B., Chambers M. C., Tabb D. L. (2007) Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zybailov B., Coleman M. K., Florens L., Washburn M. P. (2005) Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Analytical Chemistry 77, 6218–6224 [DOI] [PubMed] [Google Scholar]
- 31. Li M., Gray W., Zhang H., Chung C. H., Billheimer D., Yarbrough W. G., Liebler D. C., Shyr Y., Slebos R. J. (2010) Comparative shotgun proteomics using spectral count data and quasi-likelihood modeling. J. Proteome Res. 9, 4295–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Breslow N. (1990) Test of hypotheses in overdispersed Poisson regression and other quasi-likelihood models. J. Am. Statistical Assoc. 85, 565–571 [Google Scholar]
- 33. Benjamini Y., Hochberg Y. (1995) Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. Roy. Statistical Soc. 57, 289–300 [Google Scholar]
- 34. Zhang B., Kirov S., Snoddy J. (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang B., Schmoyer D., Kirov S., Snoddy J. (2004) GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Massion P. P., Taflan P. M., Jamshedur Rahman S. M., Yildiz P., Shyr Y., Edgerton M. E., Westfall M. D., Roberts J. R., Pietenpol J. A., Carbone D. P., Gonzalez A. L. (2003) Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 63, 7113–7121 [PubMed] [Google Scholar]
- 37. Vincent R. G., Chu T. M., Lane W. W. (1979) The value of carcinoembryonic antigen in patients with carcinoma of the lung. Cancer 44, 685–691 [DOI] [PubMed] [Google Scholar]
- 38. Nikliński J., Furman M., Laudański J., Kozlowski M. (1992) Evaluation of squamous cell carcinoma antigen (SCC-Ag) in the diagnosis and follow-up of patients with non-small cell lung carcinoma. Neoplasma 39, 279–282 [PubMed] [Google Scholar]
- 39. Lachowicz M. A., Hassmann-Poznańska E., Kozlowski M. D., Rzewnicki I. (1999) Squamous cell carcinoma antigen in patients with cancer of the larynx. Clin. Otolaryngol. Allied Sci. 24, 270–273 [DOI] [PubMed] [Google Scholar]
- 40. Borczuk A. C., Gorenstein L., Walter K. L., Assaad A. A., Wang L., Powell C. A. (2003) Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am. J. Pathol. 163, 1949–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Balasenthil S., Sahin A. A., Barnes C. J., Wang R. A., Pestell R. G., Vadlamudi R. K., Kumar R. (2004) p21-activated kinase-1 signaling mediates cyclin D1 expression in mammary epithelial and cancer cells. J. Biol. Chem. 279, 1422–1428 [DOI] [PubMed] [Google Scholar]
- 42. Carter J. H., Douglass L. E., Deddens J. A., Colligan B. M., Bhatt T. R., Pemberton J. O., Konicek S., Hom J., Marshall M., Graff J. R. (2004) Pak-1 expression increases with progression of colorectal carcinomas to metastasis. Clin. Cancer Res. 10, 3448–3456 [DOI] [PubMed] [Google Scholar]
- 43. Holm C., Rayala S., Jirström K., Stål O., Kumar R., Landberg G. (2006) Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J. Natl. Cancer Inst. 98, 671–680 [DOI] [PubMed] [Google Scholar]
- 44. Deacon S. W., Beeser A., Fukui J. A., Rennefahrt U. E., Myers C., Chernoff J., Peterson J. R. (2008) An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem. Biol. 15, 322–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pujol J. L., Grenier J., Daurès J. P., Daver A., Pujol H., Michel F. B. (1993) Serum fragment of cytokeratin subunit 19 measured by CYFRA 21–1 immunoradiometric assay as a marker of lung cancer. Cancer Res. 53, 61–66 [PubMed] [Google Scholar]
- 46. Sasaki H., Kiriyama M., Fukai I., Yamakawa Y., Fujii Y. (2002) Elevated serum pro-mMP2 levels in patients with advanced lung cancer are not suitable as a prognostic marker. Surg. Today 32, 93–95 [DOI] [PubMed] [Google Scholar]
- 47. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 48. Bergman A. C., Benjamin T., Alaiya A., Waltham M., Sakaguchi K., Franzén B., Linder S., Bergman T., Auer G., Appella E., Wirth P. J., Jornvall H. (2000) Identification of gel-separated tumor marker proteins by mass spectrometry. Electrophoresis 21, 679–686 [DOI] [PubMed] [Google Scholar]
- 49. Chen R., Tan Y., Wang M., Wang F., Yao Z., Dong L., Ye M., Wang H., Zou H. (2011) Development of glycoprotein capture-based label-free method for the high-throughput screening of differential glycoproteins in hepatocellular carcinoma. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.006445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li C., Zhan X., Li M., Wu X., Li F., Li J., Xiao Z., Chen Z., Feng X., Chen P., Xie J., Liang S. (2003) Proteomic comparison of two-dimensional gel electrophoresis profiles from human lung squamous carcinoma and normal bronchial epithelial tissues. Gen. Proteomics Bioinformat. 1, 58–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Poschmann G., Sitek B., Sipos B., Ulrich A., Wiese S., Stephan C., Warscheid B., Kloppel G., Vander Borght A., Ramaekers F. C., Meyer H. E., Stuhler K. (2009) Identification of proteomic differences between squamous cell carcinoma of the lung and bronchial epithelium. Mol. Cell. Proteomics 8, 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Soltermann A., Ossola R., Kilgus-Hawelski S., von Eckardstein A., Suter T., Aebersold R., Moch H. (2008) N-glycoprotein profiling of lung adenocarcinoma pleural effusions by shotgun proteomics. Cancer 114, 124–133 [DOI] [PubMed] [Google Scholar]
- 53. Zeng X., Hood B. L., Sun M., Conrads T. P., Day R. S., Weissfeld J. L., Siegfried J. M., Bigbee W. L. (2010) Lung cancer serum biomarker discovery using glycoprotein capture and liquid chromatography mass spectrometry. J. Proteome Res. 9, 6440–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zeng X., Hood B. L., Zhao T., Conrads T. P., Sun M., Gopalakrishnan V., Grover H., Day R. S., Weissfeld J. L., Wilson D. O., Siegfried J. M., Bigbee W. L. (2011) Lung cancer serum biomarker discovery using label-free liquid chromatography-tandem mass spectrometry. J. Thorac. Oncol. 6, 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu H., Lin D., Yates J. R., 3rd (2002) Multidimensional separations for protein/peptide analysis in the post-genomic era. BioTechniques 32, 898, 900, 902 passim [DOI] [PubMed] [Google Scholar]
- 56. Spira A., Beane J. E., Shah V., Steiling K., Liu G., Schembri F., Gilman S., Dumas Y. M., Calner P., Sebastiani P., Sridhar S., Beamis J., Lamb C., Anderson T., Gerry N., Keane J., Lenburg M. E., Brody J. S. (2007) Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat. Med. 13, 361–366 [DOI] [PubMed] [Google Scholar]
- 57. Halder S. K., Anumanthan G., Maddula R., Mann J., Chytil A., Gonzalez A. L., Washington M. K., Moses H. L., Beauchamp R. D., Datta P. K. (2006) Oncogenic function of a novel WD-domain protein, STRAP, in human carcinogenesis. Cancer Res. 66, 6156–6166 [DOI] [PubMed] [Google Scholar]
- 58. Fukumoto S., Yamauchi N., Moriguchi H., Hippo Y., Watanabe A., Shibahara J., Taniguchi H., Ishikawa S., Ito H., Yamamoto S., Iwanari H., Hironaka M., Ishikawa Y., Niki T., Sohara Y., Kodama T., Nishimura M., Fukayama M., Dosaka-Akita H., Aburatani H. (2005) Overexpression of the aldo-keto reductase family protein AKR1B10 is highly correlated with smokers' non-small cell lung carcinomas. Clin. Cancer Res. 11, 1776–1785 [DOI] [PubMed] [Google Scholar]
- 59. Brabender J., Metzger R., Salonga D., Danenberg K. D., Danenberg P. V., Hölscher A. H., Schneider P. M. (2005) Comprehensive expression analysis of retinoic acid receptors and retinoid X receptors in non-small cell lung cancer: implications for tumor development and prognosis. Carcinogenesis 26, 525–530 [DOI] [PubMed] [Google Scholar]
- 60. Katakura H., Takenaka K., Nakagawa M., Sonobe M., Adachi M., Ito S., Wada H., Tanaka F. (2006) Maspin gene expression is a significant prognostic factor in resected non-small cell lung cancer (NSCLC). Maspin in NSCLC. Lung Cancer 51, 323–328 [DOI] [PubMed] [Google Scholar]
- 61. Berglund L., Björling E., Oksvold P., Fagerberg L., Asplund A., Szigyarto C. A., Persson A., Ottosson J., Wernerus H., Nilsson P., Lundberg E., Sivertsson A., Navani S., Wester K., Kampf C., Hober S., Pontén F., Uhlén M. (2008) A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol. Cell. Proteomics 7, 2019–2027 [DOI] [PubMed] [Google Scholar]
- 62. Cimino D., Fuso L., Sfiligoi C., Biglia N., Ponzone R., Maggiorotto F., Russo G., Cicatiello L., Weisz A., Taverna D., Sismondi P., De Bortoli M. (2008) Identification of new genes associated with breast cancer progression by gene expression analysis of predefined sets of neoplastic tissues. Int. J. Cancer 123, 1327–1338 [DOI] [PubMed] [Google Scholar]
- 63. Waerner T., Alacakaptan M., Tamir I., Oberauer R., Gal A., Brabletz T., Schreiber M., Jechlinger M., Beug H. (2006) ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 10, 227–239 [DOI] [PubMed] [Google Scholar]
- 64. Grønborg M., Kristiansen T. Z., Iwahori A., Chang R., Reddy R., Sato N., Molina H., Jensen O. N., Hruban R. H., Goggins M. G., Maitra A., Pandey A. (2006) Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol. Cell. Proteomics 5, 157–171 [DOI] [PubMed] [Google Scholar]
- 65. Roessler M., Rollinger W., Mantovani-Endl L., Hagmann M. L., Palme S., Berndt P., Engel A. M., Pfeffer M., Karl J., Bodenmüller H., Rüschoff J., Henkel T., Rohr G., Rossol S., Rosch W., Langen H., Zolg W., Tacke M. (2006) Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol. Cell. Proteomics 5, 2092–2101 [DOI] [PubMed] [Google Scholar]
- 66. Dong Y. L., Reddy D. M., Green K. E., Chauhan M. S., Wang H. Q., Nagamani M., Hankins G. D., Yallampalli C. (2007) Calcitonin gene-related peptide (CALCA) is a proangiogenic growth factor in the human placental development. Biol. Reprod. 76, 892–899 [DOI] [PubMed] [Google Scholar]
- 67. Forsburg S. L. (2004) Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 68, 109–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tan D. F., Huberman J. A., Hyland A., Loewen G. M., Brooks J. S., Beck A. F., Todorov I. T., Bepler G. (2001) MCM2–a promising marker for premalignant lesions of the lung: a cohort study. BMC Cancer 1, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ramnath N., Hernandez F. J., Tan D. F., Huberman J. A., Natarajan N., Beck A. F., Hyland A., Todorov I. T., Brooks J. S., Bepler G. (2001) MCM2 is an independent predictor of survival in patients with non-small-cell lung cancer. J. Clin. Oncol. 19, 4259–4266 [DOI] [PubMed] [Google Scholar]
- 70. Dummler B., Ohshiro K., Kumar R., Field J. (2009) Pak protein kinases and their role in cancer. Cancer Metastasis Rev. 28, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kissil J. L., Walmsley M. J., Hanlon L., Haigis K. M., Bender Kim C. F., Sweet-Cordero A., Eckman M. S., Tuveson D. A., Capobianco A. J., Tybulewicz V. L., Jacks T. (2007) Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer Res. 67, 8089–8094 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.