Abstract
It is well established that bone forming cells (osteoblasts) secrete proteins with autocrine, paracrine, and endocrine function. However, the identity and functional role for the majority of these secreted and differentially expressed proteins during the osteoblast (OB) differentiation process, is not fully established. To address these questions, we quantified the temporal dynamics of the human stromal (mesenchymal, skeletal) stem cell (hMSC) secretome during ex vivo OB differentiation using stable isotope labeling by amino acids in cell culture (SILAC). In addition, we employed pulsed SILAC labeling to distinguish genuine secreted proteins from intracellular contaminants. We identified 466 potentially secreted proteins that were quantified at 5 time-points during 14-days ex vivo OB differentiation including 41 proteins known to be involved in OB functions. Among these, 315 proteins exhibited more than 2-fold up or down-regulation. The pulsed SILAC method revealed a strong correlation between the fraction of isotope labeling and the subset of proteins known to be secreted and involved in OB differentiation. We verified SILAC data using qRT-PCR analysis of 9 identified potential novel regulators of OB differentiation. Furthermore, we studied the biological effects of one of these proteins, the hormone stanniocalcin 2 (STC2) and demonstrated its autocrine effects in enhancing osteoblastic differentiation of hMSC. In conclusion, combining complete and pulsed SILAC labeling facilitated the identification of novel factors produced by hMSC with potential role in OB differentiation. Our study demonstrates that the secretome of osteoblastic cells is more complex than previously reported and supports the emerging evidence that osteoblastic cells secrete proteins with endocrine functions and regulate cellular processes beyond bone formation.
Bone is a complex tissue that is composed of cells and extracellular matrix. Bone cells can be classified into two main categories; bone forming osteoblastic cells and bone resorbing osteoclastic cells. The osteoblastic cells differentiate from stem cells in the bone marrow stroma called marrow stromal stem cells (also known as skeletal or mesenchymal stem cells, MSC). The main function of the osteoblastic cells is to form bone through secretion of a large number of extracellular collagenous and noncollagenous proteins as well as mediators of bone mineralization. In addition, there is increasing evidence that the osteoblastic cell lineage may function as an “endocrine organ” that regulates a number of functions in addition to bone formation (1). Osteoblastic cells secrete a large number of growth factors and cytokines that control osteoclastic bone resorption (2) and support hematopoiesis (3). In addition, osteoblastic cells interact with the immune system through secretion of immune modulatory factors (4) and participate in overall energy metabolism through secretion of circulating factors e.g. osteocalcin (5).
Several studies have examined secreted proteins produced by osteoblastic cells and employed either ELISA (6, 7) or standard biochemical methods (8, 9). However, these studies are limited in the number of proteins identified because of the nonglobal nature of these approaches and the quantification of only known factors. Recently, quantitative proteomics studies of MSC obtained from a variety of sources have been reported using a 2D-gel approach where only the differentially expressed proteins are analyzed (10, 11). In addition, mass spectrometry-based secretome analyses of human MSC derived from a number of tissues have been reported (8, 12–15). Among these, two studies examined the secretome of MSC during OB differentiation (12, 14) and one of these specifically the secretome of bone marrow derived MSC (14). Part of the proteins secreted by MSC is contained within the exosomes that have recently been reported to participate in a number of biological functions e.g. promote tumor growth in vivo (16) and exert cardioprotective properties (17). A proteomic analysis of the microvesicle proteome isolated from the extracellular matrix and from culture medium of mineralizing murine OB has been described (18). Although these studies have revealed qualitative lists of proteins, it remains to be established for the majority of these proteins that they are genuine secreted factors, are differentially expressed during OB differentiation, and have a functional role in OB biology.
To address these questions, we employed stable isotope labeling by amino acids in cell culture (SILAC)1 to quantify the temporal dynamics of the secretome at five time-points (days 0, 1, 4, 7, and 14) during ex vivo osteoblastic differentiation of hMSC covering the transition from undifferentiated to fully differentiated mineralized matrix producing OB. The SILAC method is one of the most accurate MS-based approaches to globally quantify proteins in cell cultures and is based on culturing cell populations in media containing different isotopic variants of amino acids e.g. lysine and arginine. This is followed by mixing the cell populations allowing for the relative quantification of differentially expressed proteins from various cell states based on the relative intensities of the corresponding peptide m/z signals distinguishable by MS analysis (19–21).
Quantitative proteomic studies of hMSC during lineage-specific differentiation have been hampered by the inability of generating large number of cells from normal hMSC because of donor variation and in vitro replicative senescence-associated growth arrest during long-term culture (22). To overcome these limitations, we employed the hMSC-TERT cell line that over-expresses the catalytic subunit of the human telomerase reverse transcriptase gene (hTERT) (23). hMSC-hTERT cells exhibit standard surface markers characteristics of MSC, maintain their capacity of multi-lineage differentiation both in ex vivo cultures and in vivo (24).
A major concern when analyzing secreted proteins with MS-based methods is serum contamination. One of the benefits of the experimental strategy based on SILAC (Fig. 2A) is that serum contaminants can be discriminated from hMSC-derived proteins by the absence of stable isotope incorporation (25). Serum contamination can reduce the dynamic range of the MS-based detection due to the presence of the highly abundant serum proteins and thereby the number of identifications (26). To minimize the amount of serum contamination, hMSC-hTERT cells were adapted to grow and differentiate in low FBS (2%). Importantly, reducing FBS concentrations seems to be a more appropriate option to minimize cell stress responses otherwise induced by complete serum deprivation (27).
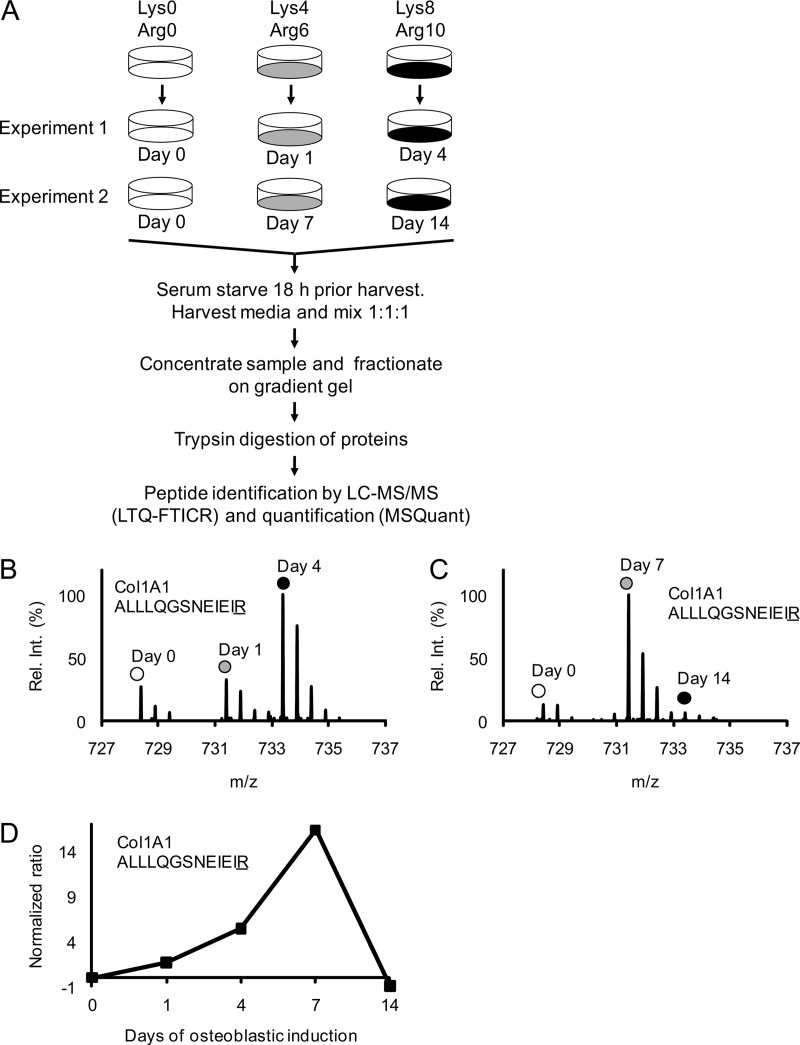
Fig. 2.
Schematic outline of the complete SILAC experiment to quantify the relative abundance changes of proteins secreted from MSC during osteoblastic differentiation. Mass spectrometry-based proteomics were used to quantify changes in the relative abundance of proteins secreted from MSC during osteoblast differentiation. A, Three populations of MSC were prelabeled with different isotope variants of lysine and arginine as indicated. The fully labeled cell populations were then stimulated with osteoblastic inducers for 1 and 4 days in experiment 1 and for 7 and 14 days in experiment 2. The unstimulated “day 0” cells were used as a common reference. Eighteen hours before collecting the conditioned medium with secreted proteins the cells were carefully washed and changed to serum free medium containing osteoblastic inducers. The medium collected from the three cell populations in each experiment was mixed in a ratio of 1:1:1 based on the total protein concentration. Each sample was then concentrated and fractionated on a 1D gradient gel and subjected to in-gel trypsin digestion. The resulting peptide mixtures were analyzed by reverse-phase liquid chromatography coupled on-line to an LTQ-FTICR instrument and quantified using the MSQuant software. B, The mass spectrum of a collagen 1 A1 (ALLLQGSNEIEIR) peptide from experiment 1 illustrates the relative abundance changes during OB differentiation where the same peptide originating from the three different cell populations can be distinguished by the isotope mass differences as indicated. C, The corresponding mass spectrum of a collagen 1 A1 (ALLLQGSNEIEIR) peptide from experiment 2. D, The signal intensity ratios of the collagen 1 A1 peptide from experiment 1 and 2 (Fig. 2B, 2C) were integrated into one temporal protein abundance profile over five time-points. All ratios were normalized to 0 (using day 0 as baseline) meaning that a normalized ratio of ±1 equals a twofold change in abundance.
Despite successful identification of differentially expressed proteins known to be secreted during osteoblastic differentiation, another major challenge is to distinguish whether the newly identified candidate proteins are actively secreted or represents intracellular contaminants released into the medium from damaged or apoptotic cells. Commonly used approaches to discriminate between secreted and nonsecreted proteins are based on bioinformatics prediction tools, e.g. SecretP (28) or Phobius (29), that can be used to annotate proteins as secreted based on sequence features.
In the present study, we explored the additional use of both complete and pulsed SILAC to classify genuine secreted proteins. The pulsed SILAC method has been used to measure protein turnover based on the ratio between the newly synthesized pool of labeled protein and the old pool of unlabeled protein (30). We and others have previously revealed compartment specific protein turnover using pulsed SILAC labeling (31, 32). Thus, the method provides the option to measure differentially aged proteins during translocation and sorting. Importantly, during our experimental strategy to analyze secreted proteins, the old pool of proteins secreted into the conditioned medium is removed. Proteins secreted into the fresh medium are therefore likely to represent newly synthesized proteins with a high fraction of labeling unless they are secreted from intracellular storage compartments or activated by processing of precursor molecules in a latent state. We tested this strategy by the quantitative analysis of proteins secreted at days 0, 1, 4, 7, and 14 during ex vivo osteoblastic differentiation of hMSC covering the transition from undifferentiated to fully differentiated mineralized matrix producing OB. We observed that a high fraction of isotope incorporation support classification of the identified proteins as true secreted proteins. Furthermore, among the identified proteins, we demonstrated that stanniocalcin 2 (STC2) stimulates osteoblast differentiation of hMSC in an autocrine fashion.
EXPERIMENTAL PROCEDURES
Cell Culture, OB Differentiation, and Protein Labeling
As a model for normal human bone marrow-derived multipotent stromal (skeletal or mesenchymal) stem cells (hMSC), we employed hMSC-TERT which is a polyclonal hMSC line created by stable transduction of the catalytic subunit of the human telomerase reverse transcriptase gene (hTERT) (23, 24). The cells were grown in Minimal Essential Media (MEM) without Phenol red and l-glutamine, supplemented with 10% (v/v) fetal bovine serum (FBS), 2 mm Glutamax, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen-Invitrogen, Carlsbad, CA), referred to as standard medium. The cells were divided in three independent populations and shifted to the SILAC labeling medium containing Dulbecco's Modified Eagle Medium (DMEM) without arginine, lysine, and methionine, supplemented with 2% (v/v) dialyzed FBS, 2 mm Glutamax, 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen-Invitrogen, Carlsbad, CA) and 30 mg/L l-methionine. The difference in the medium composition of the three cell populations was the additional presence of 73 mg/L l-lysine HCl and 28 mg/L l-arginine HCl, (Sigma) (for the Lys0/Arg0 cell population), or the corresponding amounts of l-lysine (D4) and l-arginine (13C614N4), (for the Lys4/Arg6 cell population), or l-lysine (13C615N2) and l-arginine (13C615N4), (for the Lys8/Arg10 cell population). All isotope labeled amino acids were purchased from Cambridge Isotope Laboratories (Andover, MA). The labeling medium was prepared and the cells labeled for 5 passages as described previously (19, 33). At 80% confluence the media was shifted to labeled osteoblastic induction medium. The Lys0/Arg0 cell population was employed as unstimulated control. The osteoblastic induction medium contained 10 mm β-glycerophosphate, 50 μg/ml 2-phosphate ascorbate, 10 nm dexamethasone, and 10 nm 1,25-dihydroxyvitamin D3 (34) and new medium was added every third day.
Quantitative Real-time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
qRT-PCR was performed as described (35). The primers used for the OB markers were described previously (35). Primers for the secreted factors are listed in Table I. In short, RNA was isolated using the Trizol reagent (Invitrogen, Carlsbad, CA). The purified RNA was DNase treated using Deoxyribonuclease 1 (Sigma). cDNA was synthesized using RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas). The PCR products were visualized in real-time using SYBR Green I Supermix (Bio-Rad) and an iCycle instrument (Bio-Rad). The quantitative data presented is an average of duplicate or triplicate per independent experiments.
Table I. RT-PCR primer sequences.
| Gene ID | Gene name | Forward primer (5′–3′) | Reverse primer (5′–3′) | Length (bp) |
|---|---|---|---|---|
| CFH | Complement factor H | CCTGCTCCGAGATGTACCTTGAAA | TGACACGGATGCATCTGGGAGTAG | 391 |
| CTSD | Cathepsin D | CGCGCCAGCACAGAAACAGAGGAG | CCAGGGCGCCCAGGACAGTG | 109 |
| FSTL1 | Follistatin-like 1 | TGCCCCCACCCTGCCACATACTA | GCTTCTTCCCCTGGCTTCGGTCTA | 115 |
| KIAA1199 | Protein KIAA1199 | TCTTTGGGCCACTGCTTCTTCACG | GTCTTGCCTGGGCTTGGGGATGTA | 177 |
| PAI-1 | plasminogen activator inhibitor 1 | CATCATGTGGCCCAACTCTCCTG | CTGGGCACGCATCTGACATTTCTT | 161 |
| PTX3 | pentraxin-related protein | CTTGCGATTCTGTTTTGTGCTCTC | ACGGCGTGGGGTCCTCAGT | 130 |
| STC2 | Stanniocalcin 2 | AAGGCGAGCAAAAAGGAAGAGTGG | CGCGCCGGGTCAAAGGTG | 189 |
| SVEP1 | Selectin-like osteoblast-derived protein | CTGGCCTTTTGTTGCTGGGGTCTG | GGGCGGCGCGGGGATACTC | 138 |
| LGMN | Legumain | CAGACGCGTGCCATGCCTACCAGA | GACTTTGCCGGATCCTATGCCCTTCAC | 230 |
| OC | Osteocalcin | CATGAGAGCCCTCACA | AGAGCGACACCCTAGAC | 310 |
| COL1 | Collagen type I | AGGGCTCCAACGAGATCGAGATCCG | TACAGGAAGCAGACAGGGCCAACGTCG | 223 |
| ALP | Alkaline phosphatase | ACGTGGCTAAGAATGTCATC | CTGGTAGGCGATGTCCTTA | 476 |
| ACTB | Beta-actin | TGTGCCCATCTACGAGGGGTATGC | GGTACATGGTGGTGCCGCCAGACA | 430 |
| B2M | β2-microglobulin | CCTTGAGGCTATCCAGCGT | CCTGCTCAGATACATCAAACATG | 510 |
Collection of Conditioned Medium from SILAC Labeled Cells
Conditioned medium was collected at day 1, 4, 7, and 14 after osteoblastic induction from SILAC labeled cells. Control unstimulated cells were referred to as day 0. At these time points, the cells were washed with DMEM and incubated for 18 h with serum free OB induction medium with either Lys0/Arg0, Lys4/Arg6, or Lys8/Arg10 amino acids (Fig. 2A). The supernatants from six culture dishes (10 cm2) were collected and pooled. Samples from experiment 1 that included day 0, 1, and 4, and from experiment 2 that included day 0, 7, and 14, respectively, were mixed in a ratio of 1:1:1 based on the protein concentration measured by the Bradford method (36) (Bio-Rad, Hercules, CA) to correct for variation in cell numbers.
The experimental design for the pulse labeling SILAC experiments is shown in Fig. 3A. Unlabeled cells were incubated in OB induction medium as mentioned above. At day 0, 1, 4, 7, and 14, the cells were incubated the last 18 h prior to media collection, with serum free medium containing Lys4/Arg6 in the presence or absence (day 0) of osteoblastic induction factors. The samples from the five cell populations were processed and analyzed separately.
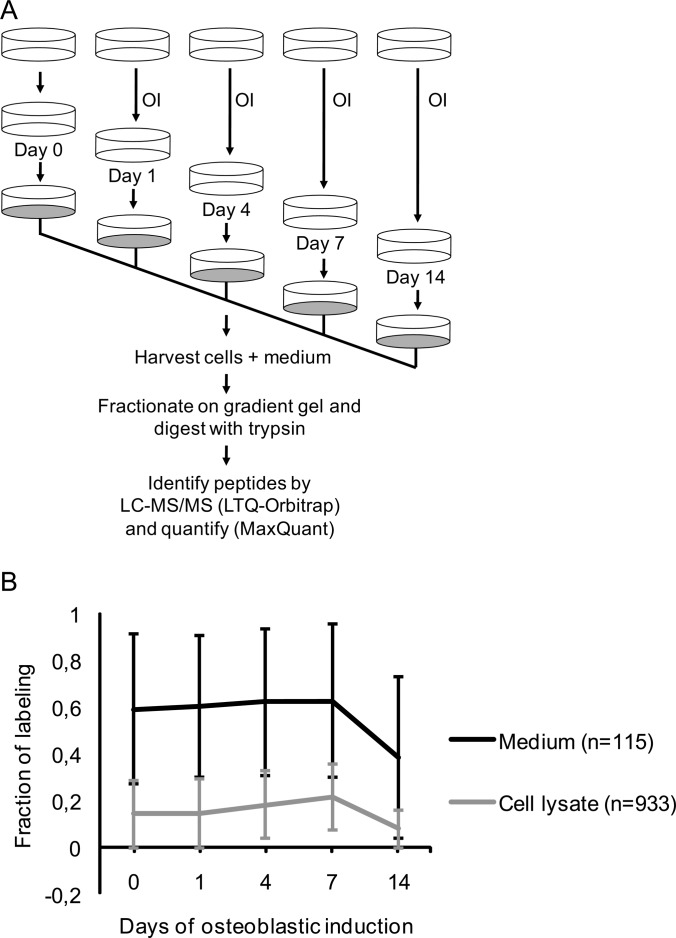
Fig. 3.
Schematic outline of the pulsed SILAC experiment to quantify the fraction of labeling of proteins secreted from MSC during osteoblastic differentiation. To distinguish true secreted proteins from a potential background of intracellular proteins we measured the degree of heavy labeled amino acid incorporation following 18 h incubation of cells in SILAC medium and compared the results for secreted proteins collected from the medium with the results obtained for intracellular proteins from lysed cells. A, The osteoblastic induction (OI) was conducted in unlabeled medium containing 2% FBS for the indicated time-points. Eighteen hours prior to collection of the conditioned medium with secreted proteins and the harvest of cells, the nonlabeling medium was replaced with serum free SILAC medium containing D4 lysine and 13C614N4 arginine as well as osteoblastic inducers. Thus, the pulsed labeling time matches the time the cells secrete protein into the medium. B, Comparison of the average fraction of labeling during osteoblastic induction for proteins from the medium (n = 115) and cell lysate (n = 933), respectively. The standard deviation for each time point is indicated as error bars. The proteins in the medium have a significantly higher incorporation of labeled amino acids than proteins in the cell lysate at all time-points (p < 0.05).
Preparation of Peptide Mixtures for Mass Spectrometric Analyses
The mixed supernatants from experiments 1 (days 0, 1, 4) and experiment 2 (days 0, 7, 14) and the five samples from the pulse labeling experiments were centrifuged for 5 min. at 1200 rpm at 4 °C and filtered through a 0.2 μm filter. The samples were concentrated in CentriCon filters (Millipore) with a molecular mass cutoff of 3000 Da at 4 °C. Forty five μg of each sample was loaded onto a 4–20% TGS gradient gel and separated in one dimension by SDS-PAGE using MES SDS running buffer (NuPAGE, Invitrogen). The gel was fixed and stained with Colloidal Blue (NuPAGE, Invitrogen) according to the manufacturer's description.
The gel lanes from the complete SILAC labeled samples were cut in 16 slices. The gel lanes from the pulsed labeled samples were cut into 10 slices and all slices subjected to reduction in 1 mm DDT (Sigma) for 45 min at 56 °C, S-carbamidomethylation in 5.5 mm iodoacetamide (Sigma) for 30 min in the dark, and in-gel digestion with 10 ng/μl trypsin (sequencing grade, Promega) at 37 °C overnight (37). The resulting peptide mixtures were acidified with 3% (v/v final) trifluoroacetic acid (TFA) followed by desalting and concentration in micro-columns (StageTips) consisting of a disc of C18 reverse-phase material (Empore Disc, 3 m) as described (38). The peptides were washed twice with solvent A (0.5% (v/v) acetic acid) and subsequently eluted directly into a 96-wells plate in solvent B (0.5% (v/v) acetic acid in 80% (v/v) acetonitrile). The organic phase was eliminated by vacuum drying at 45 °C and the remaining liquid was supplemented with 1% TFA to a final volume of 8 μl.
Mass Spectrometric Analyses of Proteins Present in the Conditioned Medium
The mass spectrometric analyses of the complete labeled samples were performed on a 7-tesla LTQ-FT instrument (Thermo Fisher) coupled to an Agilent 1100 nano-flow liquid chromatography (LC) system (Agilent Technologies) using a nanoelectrospray ion source (Proxeon Biosystems). The LC separation of the peptides was carried out on a reverse-phase column packed with ReproSil-Pur 120 C18-AQ 3 μm resin (Dr. Maisch GmbH). The peptides were eluted in a 140 min linear gradient from solvent A to solvent B with a constant flow of 250 nl/min. The LTQ-FT instrument was operated in data-dependent acquisition mode enabling automatic switching between MS and MS/MS mode. The FT-MS full scan MS spectra (m/z 400–1600) were acquired by FTICR with a resolution of 50000 at m/z 400. For accurate mass measurements the three most intense ions in each full scan were isolated for a “selected ion monitoring (SIM) scan” with a resolution of 50000 and a 24 Da mass range. The MS/MS spectra of the three most intense ions were obtained by collision induced dissociation in the linear ion trap with a normalized collision energy of 28%. A second biological replicate of the complete SILAC labeling experiment was performed using an LTQ-Orbitrap Velos (Thermo Fisher) coupled to an EASY nLC system (Thermo Scientific). The MS/MS spectra of the 10 most intense ions were obtained by higher-energy collision dissociation.
The mass spectrometric analyses of the pulse labeling experiments were performed on a LTQ-Orbitrap (Thermo Fisher) instrument coupled to an Agilent 1200 nano-flow LC system (Agilent Technologies) using a nanoelectrospray ion source (Proxeon Biosystems). The full scan MS spectra (m/z 350–1600) were acquired in profile mode in the Orbitrap analyzer with a resolution of 60000 at m/z 400 using real-time internal lock-mass recalibration (39). The MS/MS spectra were acquired in centroid mode in the linear ion-trap. The MS/MS spectra of the 5 most intense ions were obtained by collision induced dissociation in the linear ion-trap with a normalized collision energy of 35%.
Data Processing and Validation
The raw data files from the LTQ-FT (the complete SILAC labeled samples) were converted to the Mascot generic mgf format using an in-house developed tool DTASuperCharge v. 1.17 (http://msquant.sourceforge.net/) using default parameters. The mgf files from experiment 1 (day 0, 1, 4) and experiment 2 (day 0, 7, 14), respectively were combined and the two mgf peak list files were subjected to Mascot MS/MS ion search using the Mascot Search Engine v. 2.2 (Matrix Science, London, UK) (40). The search criteria were as follows: database: MSIPIslim_human (Date: 20071031, 68992 sequences) (41); enzyme: MSIPI_DPTrypsin; allow up to two missed cleavages; fixed modification: Carbamidomethyl (C); variable modifications: Acetyl (Protein N-term), Ammonia-loss (N-term C), Gln → pyro-Glu (N-term Q), Glu → pyro-Glu (N-term E), Oxidation (M), and Oxidaiton (P); quantification: SILAC K+4 K+8 R+6 R+10 [MD]; peptide tol.: ± 10 ppm; MS/MS tol.: 0.8 Da (after recalibration using the in house developed script (MSRecal v. 1.04, http://msquant.sourceforge.net/)).
The day x/day 0 ratios of the arginine and lysine-containing peptides were quantified employing the MSQuant v. 1.4.3a31 program (www.msquant.sourceforge.net) (42). All ratios were normalized to zero using the control day 0 sample as the baseline (normalized ratio = “day x/day 0 ratio” – 1, for ratios ≥ 1 and 1 – (1/“day x/day 0 ratio”), for ratios < 1). Thus a ratio of ± 1 equals a twofold regulation as compared with the unstimulated day 0 state. Furthermore, MSQuant was used to set filters and manually validate both the protein identification and quantification. The criteria used for identification were: peptide mass accuracy: ± 7 ppm, peptide length: at least 7 amino acids, at least two peptides with a Mascot score ≥ 25 or single peptides with a Mascot score ≥ 25 and a MS3 score ≥ 50 (43). Each peptide quantification was manually validated with at least two spectra where all isotopic peaks and SILAC triplets must fit the predicted mass in MSQuant (for single peptide identifications at least three spectra were validated). If these criteria were met for a protein in only one of the dataset (i.e. either experiment 1 day 0, 1, 4 or experiment 2 day 0, 7, 14), an identification of the protein in the other dataset was allowed based on the same criteria but with at least two peptides with a Mascot score ≥ 20 or single peptides with a Macsot score ≥ 20 and an MS3 score ≥ 40. No single peptide identifications were allowed considering the sum of identified peptides in experiment 1 and 2. In addition, the selected threshold Mascot score was tested for false positive by searching a non-sense database: MSIPI-humanwdecoy (41) ensuring a < 0.5% false positive rate on peptide level (44). Thus, all identifications from the complete SILAC labeling experiment were based on two independent experiments 1 and 2. The peptide evidence from the complete SILAC experiment is shown in the output format from MSQuant (42) in supplemental Table S5B. A twofold change in relative abundance was chosen as the initial cut-off for regulated proteins based on two times the average of all protein standard deviations for the time point with the largest average standard deviation. In addition, the MaxQuant/Perseus software was used to calculate significance B, a p value dependent on the protein intensities to determine whether a protein was significantly regulated (45). A plot of the average protein intensities versus the protein ratio is shown in supplemental Fig. S1 with data points color coded according to significance B. The significance A and B values for each protein are shown in supplemental Table S5B.
The raw data files from the LTQ-Orbitrap Velos (the second biological replicate of the complete SILAC labeled samples) were processed using the MaxQuant software v. 1.0.12.23 (45, 46) and the Mascot search engine v. 2.3.02. The Mascot search criteria were as already described (adapted to MaxQuant) except the peptide mass tolerance was set to ± 7 ppm, the MS/MS tolerance was 0.5 Da, the database used was IPIDecoy v. 3.84 (date: 20110601, 180856 sequences) including known nonhuman contaminants), the variable modification Oxidation (P) was omitted and the enzyme was Trypsin/P + DP. The identification criteria were: peptides > 6 amino acid residues; a maximum false positive rate of 1% on peptide level. All protein identifications were required to have at least two (unique + razor) peptide quantified with a mascot score of at least 25. A comparison of the reproducibility of protein identification and quantification between the two replicates of complete SILAC labeling is shown in supplemental Fig. S2.
The raw data files from the LTQ-Orbitrap (the pulsed SILAC labeled samples) were processed using the MaxQuant software v. 1.0.12.16 (45, 46). The Mascot search criteria were as already described for MaxQuant except the MS/MS tolerance was 0.6 Da, the database used was IPIHumanDecoy (date: 20080506, 152616 sequences including known non-human contaminants) and the enzyme was Trypsin/P + DP. The identification criteria were: peptides > 6 amino acid residues; a maximum false positive rate of 1% on peptide level. All protein identifications were required to have at least one (unique + razor) peptide quantified in each of the five time-points for clustering based on the pulsed labeling analysis. However, all protein identifications used for clustering were required to have at least two unique peptides in total. All heavy/light ratios were transformed into a fraction of labeling by the following equation: fraction of labeling = 1/((1/ratio)+1).
To evaluate the accuracy and reproducibility of quantification, the pulsed SILAC experiment was repeated in a second biological replicate and analyzed as described except the use of in-solution digestion instead of in-gel digestion. Because of the high correlation of the quantified values (supplemental Fig. S3) the pulsed SILAC data from the experiments using in-gel and in-solution digestion the two data sets were combined to improve the number of proteins quantified at all time-points. Thus, all identifications from the pulsed SILAC labeling used for temporal profiling were based on at least five independent experiments. The peptide evidence from the pulsed SILAC experiments is shown in the output format from MaxQuant (46) in supplemental Table 5A.
All of the reported protein accession numbers were based on the leading protein according to the output of the MSQuant and MaxQuant software. The statistics for assessing the accuracy and significance of the reported quantified values were based on the output from MSQuant and MaxQuant as described (42, 45, 46).
Bioinformatics Analysis
ProteinCenter® (Thermo Fisher Scientific) was used to identify individual members of a protein family, to remove redundant identifications, and to compare subsets of proteins. The database for annotation, visualization and integrated discovery (DAVID) was used for further functional clustering and annotation (47). Clustering of the identified proteins from the pulsed SILAC labeling was performed based on the temporal profiles using the MultiExperiment Viewer (MeV 4 v. 4.1.02) software by K-means clustering applying Euclidean Distance as distance metric (48, 49). Four clusters were the optimal number of clusters as determined by the Fig. of Merit (FOM) function in the MeV software that is a measure of fit of the expression patterns for the clusters produced by the K-means algorithm (50). Manual tests of various algorithms, distance metrics, and number of clusters in the MeV software, confirmed that these settings resulted in the most well defined clusters with least outliers and approximately the same number of proteins in each cluster. The following stringent criteria was used for the definition of predicted secreted proteins: The proteins should contain a signal peptide for the classical secretion pathway but no transmembrane domains as predicted by the Phobius tool http://phobius.cbr.su.se/(29). Proteins without a signal peptide or transmembrane domains but predicted to be secreted by the non-classical pathway by the SecretomeP tool (51) with a NN score > 0.5, were accepted as predicted secreted proteins as well. The proteins were validated as secreted based on annotations from the literature retrieved by a combination of database search using UniProt http://www.uniprot.org/ (when possible entries from the Swiss-Prot database) and manually literature mining using iHOP http://www.ihop-net.org/UniPub/iHOP/ (52). The same tools were used to search the literature for evidence of a direct involvement in OB differentiation. The GProX software, http://sourceforge.net/projects/gprox/files/ (53) was used to compare the biological replicates and for the preparation of several figures.
Effects of Human Stanniocalcin 2 on Osteoblast Differentiation of hMSC
hMSC-TERT were seeded in a 24-well plate (Corning), and maintained in control medium consisting of DMEM-high glucose with 10% FBS (Hyclone) and 100 μg/ml penicillin-streptomycin (Invitrogen). At 70% confluence the cells were incubated with osteoblastic induction medium as described above. Recombinant human stanniocalcin-2 (hSTC2) (Prospec, Israel) was added in concentrations ranging from 0 to 200 ng/ml. The medium was replaced every 3 days. Identification of osteoblast markers and gene expression were performed on days 3, 7, 10, and 13.
Alkaline Phosphatase Cytochemical and Alizarin Red S Histochemical Staining
Alkaline phosphatase (ALP) cytochemical staining was performed on day 7 where the cells were rinsed with PBS and fixed in acetone/citrate (1.5:1, v:v) buffer (pH 4.2) for 5 min at room temperature. The cells were incubated with buffer containing 0.2 mg/ml naphthol AS-TR phosphate for 1 h at 37 °C. For Alizarin red S histochemistry of mineralized extracellular matrix, the cells were stained on day 13 after fixation in 70% ethanol for 1 h at −20 °C and staining with 40 mm Alizarin red S pH 4.2 (Sigma). For quantitative analysis, Alizarin red dye was eluted by 10% (w/v) cetylpyridinium chloride for 1 h, and its OD570 nm measured in a spectrophotometer.
Alkaline Phosphatase Enzymatic Activity
The enzymatic activity of ALP was quantified together with cell viability at day 7 in a 96-well plate. The media were aspirated and the cell layers were incubated in 100 μl medium and 20 μl of the CellTiter-Blue solution (Promega). The plates were incubated for 2 h and cell viability was determined by measuring the fluorescent intensity at wavelength of 579Ex/584Em. For ALP activity the cell layers were rinsed and incubated in 100 μl substrate containing 50 mm NaHCO3, 1 mm MgCl2, and 1 mg/ml of p-nitrophenyl phosphate (Sigma) at 37 °C for 20 min. Absorbance was measured at 405 nm in an ELISA plate reader. ALP Enzymatic activity was normalized to cell number.
Western Blotting Analysis
For analysis of cell lysates the hMSC-TERT cells were washed in PBS and lysed in RIPA buffer (Invitrogen) supplemented with protease inhibitors (Roche). After 1h incubation at 4 °C under rotation, samples were centrifuged for 15 min at 14,000 rpm, 4 °C. Samples from conditioned medium were added protease inhibitors and concentrated in CentriCon filters (Millipore) with a molecular mass cutoff of 3000 Da at 4 °C. Protein concentration was determined with a BCA kit (Bio-Rad), and equal amounts of proteins were loaded on a 10% polyacrylamide gel (Invitrogen). Blotted nitrocellulose membranes were incubated overnight with polyclonal rabbit anti human stanniocalcin 2 (Abcam, Cambridge, UK). The blots were developed after 1 h incubation with secondary anti-goat horseradish peroxidase-conjugated antibody (Santa Cruz Biotechnology, CA) using an ECL Western blotting kit (GE Healthcare) and Kodak films.
Statistical Analysis
Comparison between groups was performed using 2-tailed unpaired Student's t test. p < 0.05 was considered significant.
RESULTS
hMSC Cells Maintain Their Ex Vivo OB Differentiation Capacity in SILAC Medium
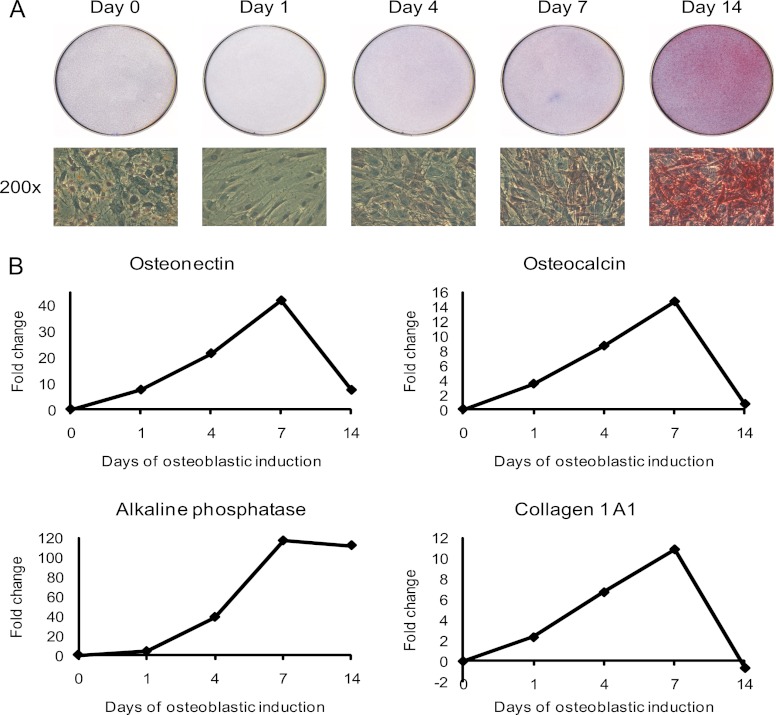
To identify and quantify changes of proteins secreted by hMSC during OB differentiation, we employed hMSC-TERT as a model system (23). hMSC-TERT fulfill the standard criteria of the hMSC phenotype: they differentiate into osteoblasts in ex vivo cultures and form heterotopic bone in vivo (24). To confirm the maintenance of OB differentiation capacity in SILAC medium, hMSC-TERT were stimulated with osteoblast induction mixture in SILAC medium. As seen in Fig. 1A, staining for alkaline phosphatase (ALP) as a marker for the OB differentiation phenotype, revealed a progressive increase in the number of ALP+ cells during the course of 14-days ex vivo differentiation. Moreover, RT-PCR gene expression analysis of four osteoblastic markers (ALP, collagen type I, osteonectin, and osteocalcin) during the course of OB differentiation confirmed successful ex vivo OB differentiation in SILAC medium (Fig. 1B). These results also corroborate our previous findings where hMSC-TERT cells were employed to identify growth factor-induced phosphotyrosine signaling in hMSC (54).
Fig. 1.
Induction of ex vivo osteoblastic differentiation of hMSC. Osteoblastic differentiation of the human marrow stromal (mesenchymal) stem cell line (hMSC-TERT) in SILAC medium was evaluated by the level of alkaline phosphatase activity and the expression of OB differentiation marker proteins. A, Cytochemical staining for alkaline phosphatase (upper part). The lower part shows the bright field images of the corresponding cells at 200× magnification. The red staining indicates alkaline phosphatase activity and the induction of OB differentiation in SILAC medium. B, Quantification of osteoblastic gene expression of osteonectin, osteocalcin, alkaline phosphatase, and collagen type I during the time course of OB differentiation using qRT-PCR. The data is expressed as fold changes using β-2-microglobulin as internal standard. All results are average of at least two independent experiments.
Quantification of Secreted Proteins During Ex Vivo OB Differentiation
To study the temporal changes of the relative abundance of secreted proteins during OB differentiation, we performed two three-plex SILAC experiments, covering the time-points of day 0, 1, 4, 7, and 14 (Fig. 2A). The method of quantification is illustrated by mass spectra of the collagen type 1 A1 peptide ALLLQGSNEIEIR acquired from experiment 1 (Fig. 2B) and experiment 2 (Fig. 2C). The marked peaks represent the three independent cell populations in each experiment that were distinguishable by their peptide mass differences. As seen in Figs. 2B and 2C, there was an increase in the signal intensities of collagen type 1 A1 on day 4 and day 7 relative to day 0. The ratios of day x/day 0 from the two experiments were integrated into a temporal profile and formed the basis for protein quantification (Fig. 2D).
A total of 466 proteins were quantified at 5 time-points during the course of ex vivo OB differentiation of hMSC and included 41 proteins (9%) associated with the OB differentiation phenotype (supplemental Table S1). Literature mining revealed that 27% of the proteins have previously been reported as secreted and were termed “validated secreted” (supplemental Table S2). In addition, “Phobius” was used to predict the presence of signal peptides and the absence of trans-membrane domains. “SecretomeP” predicted the nonclassical secreted proteins. We termed these proteins “predicted secreted” and found that 60% of the identified proteins were included in this category (supplemental Table S3). The identification and quantification of secreted proteins were supported by the analysis of an independent biological replicate (supplemental Fig. S2). Proteins that were neither validated secreted nor predicted secreted were considered potential contaminants that were released into the conditioned media because of cell damage (supplemental Table S4).
Identification of Secreted Proteins Using Temporal Pulsed SILAC Labeling
To evaluate whether the identified proteins were truly secreted, we employed a pulsed SILAC method. Newly synthesized proteins were metabolically labeled during the last 18 h prior to medium collection (Fig. 3A). The fraction of labeling was determined for both the cellular pool of proteins and for those secreted into the conditioned medium. We observed that proteins present in the conditioned medium on average had a significantly higher fraction of labeling compared with proteins from the cell lysates throughout the course of OB differentiation (p < 0.05, Fig. 3B). An independent biological replicate demonstrated reproducible values for the fraction of labeling of the proteins in the conditioned medium at all time-points (R2 values between 0.91 and 0.97, supplemental Fig. S3).
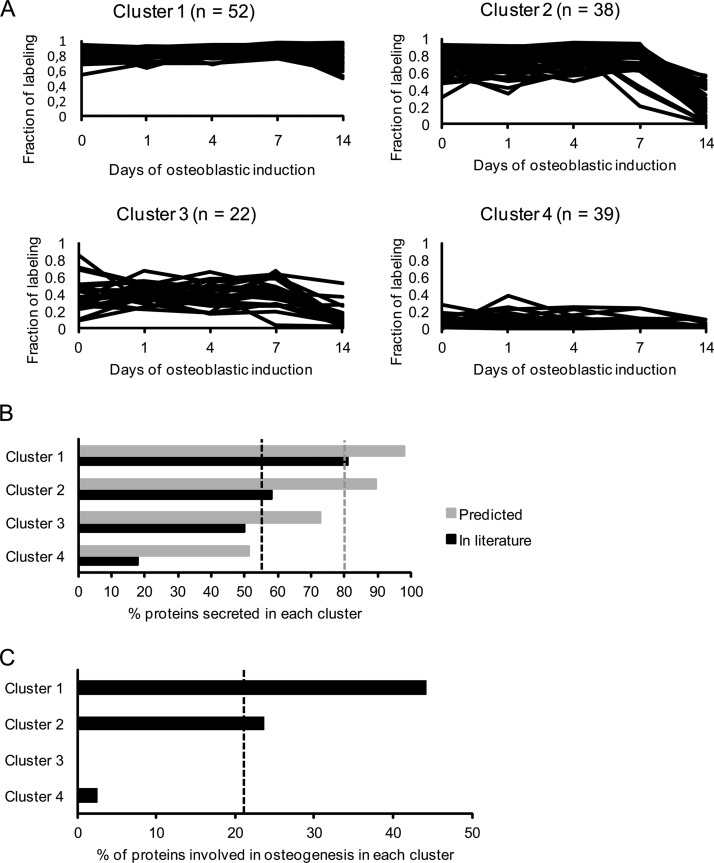
This suggests that the fraction of labeling can be used as a parameter to distinguish true secreted proteins from intracellular contaminants released into the medium from e.g. lysed cells. To further evaluate this hypothesis, we clustered the identified proteins into four groups based on their temporal fraction of labeling profiles and examined the relative enrichment of proteins predicted secreted, validated secreted, or known to be involved in OB differentiation for each of these clusters (Fig. 4A). We included 151 proteins that were detected at all time-points in the pulsed SILAC and in at least three time-points in the complete SILAC experiments. Proteins in cluster 1 had a high fraction of labeling at all time-points, proteins in cluster 2 had a high fraction of labeling at day 0, 1, 4, and 7 followed by a decrease at day 14, proteins in cluster 3 had an overall intermediate fraction of labeling at all time-points, whereas proteins in cluster 4 had a low fraction of labeling at all time-points (Fig. 4A).
Fig. 4.
Cluster analysis of secreted proteins during osteoblastic differentiation based on their fraction of labeling profile followed by enrichment analysis of proteins with a known role in OB differentiation. To evaluate the relevance of proteins with a high fraction of labeling during OB differentiation we clustered the proteins into four groups and compared the relative enrichment of protein predicted or validated to be secreted and proteins with a known role in OB differentiation for each of these clusters. A, Secreted proteins were clustered based on their fraction of labeling during osteoblastic differentiation considering only those quantified at all time-points in the pulsed SILAC labeling experiments and identified in the complete SILAC labeling experiment. The proteins in cluster 1 have a very high fraction of labeling in all time-points during the osteoblastic differentiation. Proteins in cluster 2 have a high fraction of labeling from day 0 to day 7 but are dramatically reduced at day 14. Proteins in cluster 3 have an overall intermediate fraction of labeling at all time-points. Proteins in cluster 4 have a very low fraction of labeling in all time-points during differentiation. B, The percentage of proteins in each of the four clusters that are predicted secreted (based on bioinformatics predication software) (gray) and validated secreted (through manual literature search) (black). The gray and black dashed lines represent the percentage of all the proteins before clustering annotated as predicted secreted (80%) and validated secreted (54%), respectively. Thus, the clustering based on fraction of labeling leads to enrichment for secreted proteins in cluster 1. C, The percentage of proteins known to be associated with OB differentiation in the four clusters. The dashed line indicates the percentage of all proteins before clustering directly involved in osteoblastic differentiation (22%). Thus, there is enrichment from 22% to 44% in cluster 1, while there are none or virtually none in cluster 3 and 4.
As seen in Fig. 4B, predicted and validated secreted proteins were highly enriched in cluster 1 (98 and 81%, respectively) whereas cluster 4 contained only 51% predicted secreted and 18% validated secreted proteins. Moreover, a total of 33 proteins (22%) of the 151 proteins were known to be involved in OB differentiation. This subset of proteins was highly enriched in cluster 1 as compared with cluster 3 and 4. These data support the notion that a high fraction of labeling is indicative of a secreted protein and furthermore suggest that proteins of relevance to OB biology have a high fraction of labeling as well.
Functional Annotation of Proteins in the Four Clusters Suggest That Potential Novel Secreted Regulators of the OB Microenvironment are Most Likely Found in Cluster 1
To gain insight into functions of the proteins detected in each of the four clusters, we manually annotated the proteins into several functional categories. In the following, we initially focused on the proteins detected at all time-points that exhibited at least twofold up- or down-regulation in at least one time point of the complete labeling experiment. A twofold change in the relative abundance was chosen as the cut-off for regulated proteins based on two times the average of all protein standard deviations for the time point with the largest average standard deviation. A particular protein can be present in several functional categories. The resulting functional groups associated with each of the four clusters in Fig. 4A are presented in Table II. Significantly regulated proteins (p < 0.05) are indicated in the table.
Table II. Identifications with fraction of labeling data in all time-points from the pulsed SILAC experiments and a > twofold regulated normalized ratio from the complete SILAC experiments.
Identifications with fraction of labeling data in all time-points from the pulsed SILAC experiments and a > 2-fold regulated normalized ratio from the complete SILAC experiments. The proteins are grouped first based on the four clusters (C) they belong to based on the fraction of labeling profile (Fig 4A). Furthermore, within each cluster the proteins are grouped based on manual functional annotation allowing that each protein can be listed more than once within each cluster. Each protein is listed with name, IPI identifier, and the quantified normalized ratio at days 0, 1, 4, 7, and 14. In addition, the R column indicates (+) if a protein is significantly regulated (p < 0.05) based on a calculated significance B value (supplemental Table S5B).
| Cluster 1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| C | Name |
IPI number | Normalized ratio |
R | ||||
| Growth factor/cytokine | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | |||
| 1 | Connective tissue growth factor | IPI00020977.4 | 0 | 4.59 | 23.36 | 7.91 | 3.92 | + |
| 1 | Transforming growth factor-β-induced | IPI00018219.1 | 0 | 1.4 | 1.89 | –0.5 | –0.14 | + |
| 1 | Insulin-like growth factor binding protein 2 | IPI00297284.1 | 0 | 2.17 | 33.47 | 21.21 | 12.09 | + |
| 1 | Follistatin-like 1 | IPI00029723.1 | 0 | 0.93 | 3.27 | 2.27 | 0.34 | + |
| 1 | Insulin-like growth factor binding protein 7 | IPI00016915.1 | 0 | –0.18 | 1.33 | 1.01 | 0.84 | |
| 1 | Stanniocalcin 2 | IPI00008780.3 | 0 | 1.27 | 2.52 | 0.75 | –0.01 | + |
| 1 | Colony stimulating factor 1 | IPI00556665.1 | 0 | 0.06 | 0.19 | 0.1 | 1.54 | |
| 1 | Insulin-like growth factor binding protein 4 | IPI00305380.3 | 0 | 1.04 | 1.49 | 1.46 | 2.6 | + |
| 1 | Insulin-like growth factor binding protein 6 | IPI00029235.1 | 0 | 0.2 | –0.52 | –1.24 | –5.1 | |
| 1 | Granulin | IPI00296713.4 | 0 | 0.5 | 1.28 | –1.33 | –0.31 | |
| 1 | Bone morphogenetic protein 1 | IPI00009054.1 | 0 | 0.08 | 3.44 | 4.68 | 0.32 | + |
| 1 | Osteonectin | IPI00014572.1 | 0 | 1.07 | 4.2 | 5.38 | 0.11 | + |
| 1 | Insulin-like growth factor binding protein 3 | IPI00444386.2 | 0 | 0.23 | 1.61 | |||
| C | Extracellular matrix | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 1 | Lumican | IPI00020986.2 | 0 | –1.22 | –4.38 | –0.42 | –0.33 | + |
| 1 | Matrix metalloproteinase 2 | IPI00027780.1 | 0 | –1.49 | –3.55 | –3.02 | –0.14 | + |
| 1 | Collagen, type VI, alpha 1 | IPI00291136.4 | 0 | –0.7 | –0.23 | –0.06 | –1.84 | + |
| 1 | Osteonectin | IPI00014572.1 | 0 | 1.07 | 4.2 | 5.38 | 0.11 | + |
| 1 | Extracellular matrix protein 1 | IPI00003351.2 | 0 | –0.44 | –0.5 | –1.12 | –3.05 | + |
| 1 | Fibrillin 1 | IPI00328113.2 | 0 | 0.33 | 1.56 | 1.73 | 0.41 | |
| 1 | Periostin | IPI00410241.2 | 0 | –0.54 | 1.01 | 2.59 | 4.95 | + |
| 1 | Tissue inhibitor of metalloproteinase 1 | IPI00032292.1 | 0 | –0.18 | 0.18 | 0.42 | 2.32 | |
| 1 | Cathepsin L1 | IPI00012887.1 | 0 | 0.14 | 0.46 | –1.75 | –1.25 | + |
| 1 | Fibrillin-like protein | IPI00220813.1 | 0 | 0.27 | 1.76 | 2.13 | 0.45 | |
| 1 | Nidogen-1 | IPI00026944.2 | 0 | 0.29 | 1.66 | 2.34 | 1.2 | |
| 1 | Connective tissue growth factor | IPI00020977.4 | 0 | 4.59 | 23.36 | 7.91 | 3.92 | + |
| 1 | Transforming growth factor-β-induced | IPI00018219.1 | 0 | 1.4 | 1.89 | –0.5 | –0.14 | + |
| C | Protease | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 1 | Cathepsin D | IPI00011229.1 | 0 | 1.19 | 2.92 | 0.69 | 0.75 | + |
| 1 | Legumain | IPI00293303.1 | 0 | 0.89 | 2.92 | 2 | –1.41 | + |
| 1 | Cathepsin B | IPI00295741.4 | 0 | 0.54 | 0.39 | –2.57 | 8.08 | + |
| 1 | Complement component 1, s subcomponent | IPI00017696.1 | 0 | –0.8 | –0.42 | –1.02 | –1.28 | + |
| 1 | Complement C1r subcomponent | IPI00296165.5 | 0 | –1.18 | –0.25 | –0.52 | –1.9 | + |
| 1 | Cathepsin Z | IPI00002745.1 | 0 | 0.49 | 0.94 | 0.73 | –1.77 | |
| 1 | Prosaposin isoform a preproprotein | IPI00012503.1 | 0 | 0.12 | 0.6 | –1.89 | –0.59 | + |
| 1 | Cathepsin L1 | IPI00012887.1 | 0 | 0.14 | 0.46 | –1.75 | –1.25 | + |
| 1 | Matrix metalloproteinase 2 | IPI00027780.1 | 0 | –1.49 | –3.55 | –3.02 | –0.14 | + |
| 1 | Bone morphogenetic protein 1 | IPI00009054.1 | 0 | 0.08 | 3.44 | 4.68 | 0.32 | + |
| 1 | Tetranectin | IPI00009028.1 | 0 | –0.39 | 2.99 | + |
| C | Angiogenesis | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 1 | Pigment epithelium-derived factor | IPI00006114.4 | 0 | –1.98 | –0.94 | –1.69 | –6.94 | + |
| 1 | Plasminogen activator inhibitor-1 | IPI00007118.1 | 0 | 3.22 | 5.04 | 3.74 | 0.97 | + |
| 1 | Connective tissue growth factor | IPI00020977.4 | 0 | 4.59 | 23.36 | 7.91 | 3.92 | + |
| C | Immune response | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 1 | Pentraxin-related protein PTX3 | IPI00029568.1 | 0 | 3.33 | 3.56 | 2.9 | 0.15 | + |
| 1 | Complement component 1 inhibitor | IPI00291866.5 | 0 | –1.26 | 0.85 | 0.42 | –0.27 | + |
| 1 | Complement factor H | IPI00029739.4 | 0 | –0.05 | 4.29 | 10.11 | 1.46 | + |
| C | Immune response | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 1 | Complement component 1, s subcomponent | IPI00017696.1 | 0 | –0.8 | –0.42 | –1.02 | –1.28 | + |
| 1 | Complement C1r subcomponent | IPI00296165.5 | 0 | –1.18 | –0.25 | –0.52 | –1.9 | + |
| C | Other | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 1 | Protein KIAA1199 | IPI00376689.1 | 0 | 0.64 | 3.8 | 0.62 | 0.59 | + |
| 1 | Selectin-like osteoblast-derived protein | IPI00847609.1 | 0 | –0.03 | 2.81 | 2.12 | –1.3 | + |
| 1 | Plasminogen activator inhibitor-1 | IPI00007118.1 | 0 | 3.22 | 5.04 | 3.74 | 0.97 | + |
| 1 | Plasminogen activator inhibitor-1–2 | IPI00009890.1 | 0 | 0.34 | 0.5 | 1.07 | 3.68 | + |
| 1 | Epididymis-specific alpha-mannosidase | IPI00328488.6 | 0 | 0.44 | 1.44 | 0.28 | –1.66 | |
| 1 | Calsyntenin 1 | IPI00413959.2 | 0 | 0.33 | 1.16 | 0.63 | 1.58 | |
| 1 | Dickkopf 1 | IPI00016353.1 | 0 | –0.06 | 1.13 |
| Cluster 2 | ||||||||
|---|---|---|---|---|---|---|---|---|
| C | Name |
IPI number | Normalized Ratio |
R | ||||
| Extracellular matrix | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | |||
| 2 | Isoform c of fibulin-1 | IPI00296537.3 | 0 | –1.18 | 0.3 | 0.44 | 0.54 | + |
| 2 | Dystroglycan | IPI00028911.1 | 0 | 0.07 | 1.03 | 0.01 | –0.3 | |
| 2 | Biglycan | IPI00010790.1 | 0 | 0.23 | 1.77 | 3.76 | 1.41 | |
| 2 | Collagen type I A1 | IPI00297646.4 | 0 | 1.65 | 5.38 | 16.32 | –0.96 | + |
| 2 | Collagen type I A2 | IPI00304962.3 | 0 | 1.95 | 5.66 | 11.26 | –0.79 | + |
| 2 | Decorin | IPI00012119.1 | 0 | –0.43 | –0.18 | –0.24 | –1.34 | |
| 2 | Collagen type VI A3 | IPI00022200.2 | 0 | –0.69 | –1.11 | –2.73 | –2.6 | + |
| 2 | Procollagen C-endopeptidase enhancer 1 | IPI00299738.1 | 0 | –1.03 | –0.78 | –0.65 | –3.61 | + |
| 2 | Collagen type VI A2 | IPI00304840.4 | 0 | –0.83 | –0.77 | –1.71 | –1.96 | + |
| 2 | Cathepsin A | IPI00640525.2 | 0 | 0.21 | –0.06 | –1.95 | –2.98 | + |
| 2 | Laminin subunit alpha-4 | IPI00329482.4 | 0 | –1.22 | –0.01 | –0.01 | –1.3 | + |
| 2 | Laminin subunit gamma-1 | IPI00298281.3 | 0 | –0.46 | 0.66 | 1.26 | –0.09 | |
| 2 | Lysyl oxidase | IPI00002802.1 | 0 | 2.63 | 9.24 | + | ||
| C | Protease inhibitor | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 2 | Tissue factor pathway inhibitor | IPI00021834.1 | 0 | –0.42 | 1.37 | 0.59 | 0.01 | |
| 2 | Thrombospondin 1 | IPI00296099.6 | 0 | 1.63 | 3.65 | 5.27 | –1.99 | + |
| 2 | Complement component 3 | IPI00783987.2 | 0 | –0.8 | 1.15 | 1.07 | –15.13 | + |
| 2 | Collagen type VI A3 | IPI00022200.2 | 0 | –0.69 | –1.11 | –2.73 | –2.6 | + |
| C | Protease | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 2 | N-acetylgalactosamine-6-sulfatase | IPI00029605.1 | 0 | 0.65 | 1.13 | 0.78 | –0.04 | |
| 2 | N-acylsphingosine amidohydrolase 1 | IPI00013698.1 | 0 | 0.77 | 1.8 | 0.19 | –0.25 | |
| 2 | HtrA serine peptidase 1 | IPI00643586.3 | 0 | –0.66 | 1.1 | 1.1 | 0.8 | |
| 2 | Gamma-glutamyl hydrolase | IPI00023728.1 | 0 | 1.16 | 1.37 | 2.69 | 0.1 | + |
| 2 | Lysophospholipase 3 | IPI00301459.2 | 0 | 0.44 | 0.23 | –1.16 | –2.18 | |
| 2 | Mannosidase alpha 2B member 1 | IPI00644131.1 | 0 | 0.46 | 1.74 | 0.39 | –2.18 | |
| 2 | Cathepsin A | IPI00640525.2 | 0 | 0.21 | –0.06 | –1.95 | –2.98 | + |
| 2 | Glucosamine (N-acetyl)-6-sulfatase | IPI00012102.1 | 0 | 0.5 | 0.45 | –1.63 | –1.72 | + |
| 2 | Fucosidase alpha-l-1 | IPI00745745.2 | 0 | –0.06 | 0.47 | –1.21 | –0.46 | |
| 2 | IGF-dependent IGFBP-4 protease | IPI00001869.2 | 0 | –0.65 | 1.99 |
| C | Other | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 2 | ABI gene family 3 (NESH) binding protein | IPI00440822.1 | 0 | –0.47 | –1.51 | –1.89 | –2.6 | + |
| 2 | Milk fat globule-EGF factor 8 protein | IPI00002236.3 | 0 | 0.51 | 3 | 0.53 | –33.48 | + |
| 2 | Matrix-remodelling associated 8, Limitin | IPI00153049.3 | 0 | –0.52 | 0.77 | 0.13 | 1.4 | |
| 2 | Stress 70 protein chaperone | IPI00299299.3 | 0 | 0.58 | 1.2 | 1.03 | –0.43 | |
| 2 | AXL receptor tyrosine kinase isoform 1 | IPI00296992.7 | 0 | 0.36 | 2.88 | + |
| Cluster 3 | ||||||||
|---|---|---|---|---|---|---|---|---|
| C | Name |
IPI number | Normalized Ratio |
R | ||||
| Extracellular matrix | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | |||
| 3 | Collagen type V A1 | IPI00477611.1 | 0 | 0.75 | 3.3 | 2.27 | –2.42 | + |
| 3 | Isoform d of fibulin-1 | IPI00296534.1 | 0 | –2.52 | –0.28 | –0.35 | –0.27 | + |
| Cluster 3 | ||||||||
|---|---|---|---|---|---|---|---|---|
| C | Name |
IPI number | Normalized Ratio |
R | ||||
| Extracellular matrix | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | |||
| 3 | Perlecan | IPI00024284.4 | 0 | 0.18 | 2.5 | 2.79 | –1.38 | + |
| 3 | Collagen type XII A1 | IPI00827558.2 | 0 | 0.18 | –0.05 | –0.69 | –11.2 | |
| 3 | Collagen type VIII A1 | IPI00219000.3 | 0 | 0.3 | 7.25 | 16.78 | 33.98 | + |
| 3 | Collagen triple helix repeat containing 1 | IPI00060423.4 | 0 | –2.73 | –0.55 | + | ||
| C | Calcium binding | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 3 | Nucleobindin 1 | IPI00295542.5 | 0 | 0.13 | 0.4 | 0.1 | –1.82 | |
| 3 | Ependymin related protein 1 | IPI00657648.1 | 0 | 0.27 | 1.06 | –2.39 | –3.98 | + |
| 3 | Low-density lipoprotein receptor-related 1 | IPI00020557.1 | 0 | –1.14 | 0.07 | –0.04 | 0.46 | + |
| 3 | Gelsolin | IPI00026314.1 | 0 | 0.02 | 2.41 | 2.12 | 0.69 | + |
| 3 | Isoform d of fibulin-1 | IPI00296534.1 | 0 | –2.52 | –0.28 | –0.35 | –0.27 | + |
| C | Other | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 3 | Neogenin | IPI00023814.2 | 0 | –0.22 | 2.25 | 1.72 | 0.64 | + |
| 3 | Plasma glutamate carboxypeptidase | IPI00007664.5 | 0 | 0 | 0.39 | 0.3 | –2.94 | |
| 3 | Steroid-sensitive protein 1 | IPI00260630.6 | 0 | 0.01 | 1.4 | 0.64 | –2.02 | |
| 3 | Arylsulfatase A | IPI00329685.10 | 0 | 0.15 | 1.16 | 0.13 | –2.32 | + |
| 3 | Dipeptidyl-peptidase 2 | IPI00296141.3 | 0 | –0.23 | 0.35 | –1.03 | –1.92 |
| Cluster 4 | ||||||||
|---|---|---|---|---|---|---|---|---|
| C | Name |
IPI number | Normalized Ratio |
R | ||||
| Actin binding/cytoskeleton | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | |||
| 4 | Actin, gamma 1 | IPI00021440.1 | 0 | –0.19 | 1.58 | 0.59 | 0.89 | |
| 4 | Transgelin | IPI00216138.6 | 0 | –0.04 | 4.5 | 2.34 | 2.37 | + |
| 4 | Filamin A, alpha | IPI00302592.2 | 0 | –0.11 | 1.63 | 0.59 | 0.44 | |
| 4 | Ezrin | IPI00843975.1 | 0 | 0.29 | 0.44 | 0.01 | 1.44 | |
| 4 | Actinin alpha 1 | IPI00013508.5 | 0 | 0.17 | 1.32 | 0.55 | 1.34 | |
| 4 | Isoform A of Lamin-A/C | IPI00021405.3 | 0 | 0.21 | 0.47 | –0.22 | –12.51 | |
| C | Metabolism | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 4 | Family with sequence similarity 3 C | IPI00021923.1 | 0 | –0.06 | 1.02 | –0.57 | –3.44 | + |
| 4 | Lysyl hydroxylase 1 | IPI00027192.5 | 0 | –0.8 | –0.38 | –2.82 | –1.93 | + |
| 4 | Lysyl hydroxylase 3 | IPI00030255.1 | 0 | 0.22 | –0.17 | –1.89 | 0.84 | + |
| 4 | Peptidylprolyl isomerase A | IPI00419585.9 | 0 | –0.63 | 0.11 | –1.04 | 1.48 | |
| 4 | Phosphoglycerate kinase 1 | IPI00169383.3 | 0 | –0.66 | –0.17 | –0.7 | 1.19 | |
| 4 | Enolase 1 | IPI00465248.5 | 0 | –0.46 | 0.16 | –0.81 | 2.36 | |
| 4 | Peptidylprolyl isomerase C | IPI00024129.1 | 0 | –0.14 | –0.15 | 0.23 | –3.93 | + |
| 4 | Leukocyte-derived arginine aminopeptidase | IPI00465261.2 | 0 | 0.08 | 0.19 | –1.03 | –0.99 |
| C | Receptor/adhesion | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 4 | Thy-1 cell surface antigen | IPI00555577.1 | 0 | –1.16 | 0.32 | –0.68 | –0.41 | + |
| 4 | Activated leukocyte cell adhesion molecule | IPI00783803.2 | 0 | –0.39 | 1.85 | 1.23 | 0.68 | |
| 4 | Macrophage mannose receptor 2 | IPI00005707.6 | 0 | –0.57 | 1.07 | 0.47 | 0.28 | |
| 4 | CD44 antigen | IPI00827658.1 | 0 | –0.25 | –0.33 | –2.32 | –1.87 | + |
| 4 | CD109 antigen | IPI00795801.1 | 0 | –0.24 | 0.27 | –1.11 | –2.51 | + |
| 4 | Protein C receptor, endothelial | IPI00009276.2 | 0 | –0.42 | 1.94 | 0.26 | 0.58 |
| C | Angiogenesis | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 4 | Membrane alanine aminopeptidase | IPI00221224.5 | 0 | –0.62 | 1.12 | 0.02 | 1.47 | |
| 4 | Thy-1 cell surface antigen | IPI00555577.1 | 0 | –1.16 | 0.32 | –0.68 | –0.41 | + |
| 4 | Chondroitin sulfate proteoglycan 4 | IPI00019157.2 | 0 | –2.8 | –2.03 | + |
| C | Other | IPI number | Day 0 | Day 1 | Day 4 | Day 7 | Day 14 | R |
|---|---|---|---|---|---|---|---|---|
| 4 | Discoidin, CUB and LCCL domain containing 2 | IPI00419836.1 | 0 | –1.58 | –2.94 | –4.35 | –1.04 | + |
| 4 | 14-3-3 protein zeta | IPI00021263.3 | 0 | 0.11 | 0.36 | 0.07 | 2.73 | |
| 4 | Heat shock 70kDa protein 8 | IPI00003865.1 | 0 | 0.17 | 0.44 | –0.34 | 1.73 | |
| 4 | Eukaryotic initiation factor 5A variant A | IPI00376005.2 | 0 | 0.08 | 0.33 | 0 | 1.99 |
Cytokine and growth factors were mainly present in cluster 1 (13 in cluster 1 and 2 in cluster 2) and several members of two growth factor families were detected: insulin-like growth factors (IGF) as well as their binding proteins (IGFBP) and the transforming growth factor β (TGF-β) family. Among this group, several proteins have been reported to exert regulatory functions on OB differentiation, including IGFBP2 (55), IGFBP3 (56), IGFBP4 (57), IGFBP6 (58), IGF-dependent IGFBP-4 protease (59), TGFBI (60), BMP1 (61), Connective tissue growth factor (62), colony stimulating factor 1 (63), osteonectin (64) and HtrA serine peptidase 1 (65). Interestingly, a peptide hormone, stanniocalcin 2 (66) was identified in cluster 1, supporting the notion that OB act as an endocrine organ (1).
A large number of ECM-related proteins were detected especially in cluster 1 and 2, including collagen type I, collagen type IV, collagen type V, collagen type VI, collagen type VIII, and collagen XII. Several noncollagenous proteins were also detected e.g. fibulin 4, fibrillin-1, ECM-1, lumican, and decorin. Other ECM proteins involved in ECM assembly and cell matrix interaction were detected e.g. periostin, nidogen-1, perlecan, and laminin. Several of the ECM-related proteins are known to play a direct role in bone formation i.e. lumican (67), matrix metalloproteinase 2 (68), collagen type VI A1 (69), osteonectin (64), extracellular matrix protein 1 (70), fibrillin 1 (71), periostin (72), tissue inhibitors of metalloproteinase 1 (73), connective tissue growth factor (62), biglycan (74), collagen type I (75), decorin (76, 77), and lysyl oxidase (78). Several proteases identified in cluster 1 and 2 are known for their role in extracellular matrix remodeling and regulation of the activity of ECM-bound proteins e.g. matrix metalloproteinase 2, BMP1, IGF-dependent IGFBP-4 protease, and HtrA serine peptidase 1. Acidic proteases involved in lysosomal degradation of proteins were identified such as the cathepsin group of proteases, cathepsin A, B, D, L1, and Z. The high representation of cathepsins in cluster 1 indicates a role of lysosomal protein degradation during osteoblastic differentiation of MSC. A marked increase in total lysosome organelles and endocytosis in osteoblastic cells as well as implication of lysosomes in procollagen processing has been reported (79).
In addition, a number of proteins found in cluster 1 possess immune response-related functions and included several proteins from the complement system e.g. complement component 1 inhibitor, complement factor H, complement component 1 s subcomponent, and complement C1r subcomponent. Complement component 3 was identified in cluster 2. Previous studies have suggested a role of complement components during OB differentiation (80), which corroborates our findings.
In cluster 1 and 4, we identified proteins related to angiogenesis, e.g. pigment epithelium-derived factor, plasminogen activator inhibitor-1, connective tissue growth factor. It is well established that angiogenesis is essential for bone growth, repair, and remodeling (81, 82). Other proteins in cluster 4: membrane alanine aminopeptidase, thy-1 cell surface antigen, and chondroitin sulfate proteoglycan 4, are all transmembrane proteins and not secreted but may be relevant to hMSC biology. Overall, the proteins in cluster 4 are mainly intracellular proteins with annotated functions such as actin binding, cytoskeletal proteins, metabolism, and membrane bound proteins (receptor/cell adhesion).
Taken together, these analyses suggest that cluster 1 and 2 contain the proteins that are most likely to be secreted and to contain secreted factors with a functional role in osteoblastic differentiation.
We further analyzed our data in relation to exosomes and secretory vesicle-related proteins to study the fraction of labeling of these proteins. We first examined our data for the three evolutionary conserved exosomes' markers (CD81, CD 63, and CD9). Both CD81 and CD63 were identified at all time-points of the “complete SILAC experiments” indicating the presence of exosomes in the conditioned medium of hMSC-TERT. Furthermore, we tested our data for the presence of fingerprints of osteoblast microvesicle proteins based on a previous report (18). Among the 45 reported microvesicle proteins also quantified in at least one time point in the pulsed SILAC experiments, 82% had a very low fraction of labeling corresponding to the characteristic feature of cluster 4. This implies that the proteins in secreted microvesicles from hMSC display a delay in terms of synthesis and secretion.
Identification of Secreted Proteins with a Potential Novel Role in Osteoblastic Differentiation Based on the Differential Expression Profile
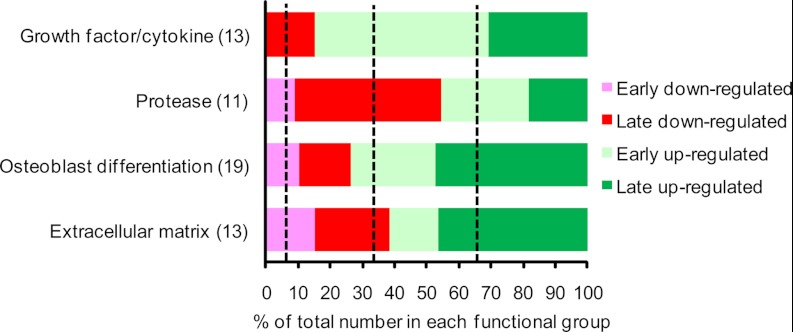
To identify novel proteins with a potential role in osteoblastic differentiation, we focused on the proteins in cluster 1 regulated more than twofold in at least one time point. We classified four major functional groups of proteins based on their temporal expression profile as either “early secreted” (peak secretion observed between day 1–4) or “late secreted” (peak secretion observed between day 7 and 14) (Fig. 5). We observed an enrichment of growth factor/cytokine in the “early up-regulated” group, proteases in the “late down-regulated” group, and OB differentiation-related as well as extracellular matrix proteins in the “late up-regulated” group. The distribution of the proteins with a known direct function in OB differentiation is dominated by early up-regulated and late up-regulated proteins. This is also the case for the growth factor/cytokine and extracellular matrix functional group which are the most relevant with respect to OB differentiation (2). Thus, these observations suggest that the search for novel regulators of OB differentiation should be focused among the early up-regulated and late up-regulated.
Fig. 5.
Searching for novel regulators of osteoblastic differentiation among those identified with a high fraction of labeling in the pulsed SILAC experiment and found to be regulated in the complete SILAC experiment. Functional annotation of secreted proteins from cluster 1 that exhibit more than twofold up- or down-regulation during early (day 1 or 4) or late (day 7 or 14) ex vivo OB differentiation of hMSC-TERT cells. The dashed lines between each of the four groups (early down-regulated, late down-regulated, early up-regulated, and late up-regulated) indicate the percentage expected if the distribution of the biological clusters is random. There is an enrichment for early up-regulated proteins among the growth factors, late down-regulated proteins among the proteases, and late up-regulated among the proteins related to both OB differentiation and extracellular matrix.
Verification of the SILAC Profile of Novel Candidate Proteins by Quantitative Real-time PCR
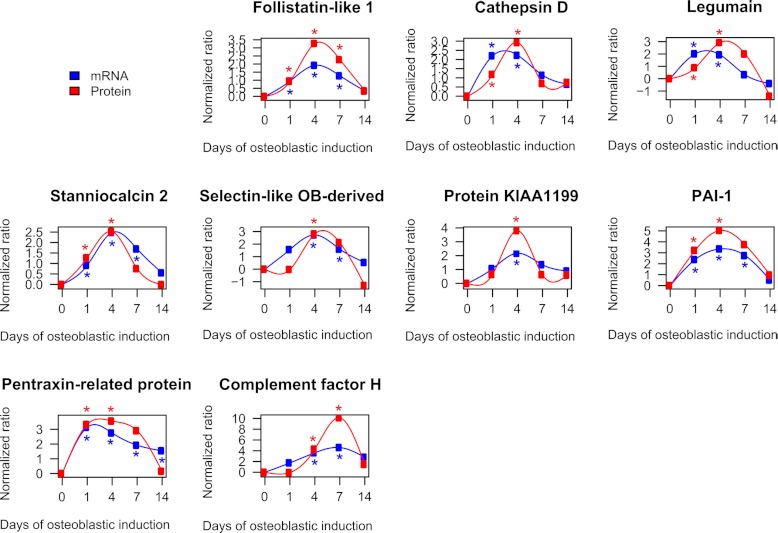
To verify the temporal profiles quantified by the complete SILAC experiments, a direct comparison of the protein profiles and the corresponding mRNA expression profiles of the identified early up-regulated and late up-regulated candidates are shown in Fig. 6. Only the proteins significantly regulated (p < 0.05) based on the calculated significance B value are shown in Fig. 6. Among the early significantly up-regulated proteins we observed the following proteins: follistatin-like 1, cathepsin D, legumain, stanniocalcin 2, selectin-like osteoblast-derived protein, protein KIAA1199, plasminogen activator inhibitor 1, and pentaxin-related protein PTX3. Follistatin-like 1 exhibits structural similarity to follistatin a known inhibitor of BMP2 signaling and possibly OB differentiation (83). Follistatin-like 1 has previously been identified in the secretome of hMSC (8, 12). Cathepsin D is a lysosomal protease. Because cathepsin B (84) and K (85) are directly involved in OB differentiation, it is plausible that cathepsin D plays a similar role. Legumain is a lysosomal protease shown to inhibit osteoclast formation and bone resorption (86). Stanniocalcin 2 is a hormone and shares structural similarity to stanniocalcin 1 which stimulates OB differentiation (87). Stanniocalcin 2 has been identified previously in the secretome from hMSC (8) and its expression is induced by vitamin K2 in osteoblastic cells in a protein kinase A-dependent manner (88). The selectin-like osteoblast-derived protein is virtually only expressed in placenta (89), skeletal tissue in vivo and in osteoblasts in vitro (90). It is regulated by estrogen and has functions related to cell adhesion (91). The function of protein KIAA1199 is currently unclear. Plasminogen activator inhibitor-1 (PAI-1) is an inhibitor of tissue plasminogen activator and urokinase is negatively regulating the degradation of blood clots. Pentaxin-related protein PTX3 (PTX3) is involved in immune response and is rapidly induced by IL-1 and tumor necrosis factor (92). Complement factor H was the only significantly up-regulated protein candidate associated with the late phase of osteoblastic differentiation. Complement factor H has been shown to bind to bone sialoprotein and osteopontin, two ECM proteins important for OB differentiation, thereby preventing complement-mediated cell lysis (93).
Fig. 6.
Direct comparison of the temporal expression profiles of significantly regulated candidate proteins as quantified by SILAC and qRT-PCR. The figure compares the temporal profiles of candidate proteins and their corresponding gene expression quantified by qRT-PCR. The early up-regulated candidate proteins are follistatin-like 1, cathepsin D, legumain, stanniocalcin 2, selectin-like osteoblast-derived protein, protein KIAA1199, plasminogen activator activator inhibitor-1 (PAI-1), and pentraxin-related protein PTX3. Complement factor H exhibits significantly late up-regulation during the course of ex vivo differentiation of hMSC-TERT cells. The data was expressed as a fold change normalized to the unstimulated day 0. All quantified mRNA values represent the average of at least two independent experiments. A significantly regulated protein ratio was indicated with red * (p < 0.05). The blue * indicate a significant regulation on mRNA level (p < 0.05).
Validation of Stanniocalcin 2 (STC2) as an Autocrine Regulator for Osteoblast Differentiation for hMSC
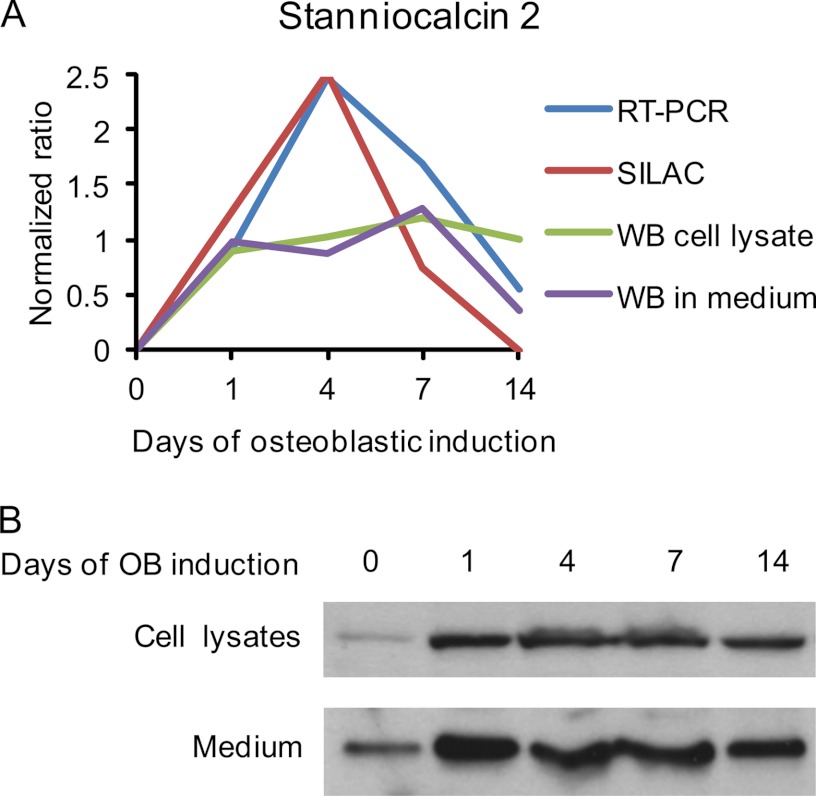
Among the significantly regulated candidates in Cluster 1 (Fig. 6), we chose STC2 for detailed studies. The temporal expression of STC2 during osteoblastic differentiation of hMSCs revealed by qRT-PCR, SILAC, Western blotting from cell lysate, and Western blotting from medium, was compared directly by log2 normalizing all data to day 0 (Fig. 7A). The Western blot analysis of cell lysate as well as its secretion in conditional medium (Fig 7B) coincided with the protein levels quantified by SILAC.
Fig. 7.
Production of stanniocalcin 2 by hMSC. A, Direct comparison of the STC2 expression and secretion during osteoblast differentiation of hMSC as measured by qRT-PCR, SILAC (from conditioned medium), Western blot from cell lysates and Western blot from conditioned medium. All data were log2 normalized into a normalized ratio with respect to the values of unstimulated day 0. B, Western blot analysis of STC2 protein levels in cell lysates (upper lane, 50 μg/well) and conditioned medium (lower lane, 25 μg/well).
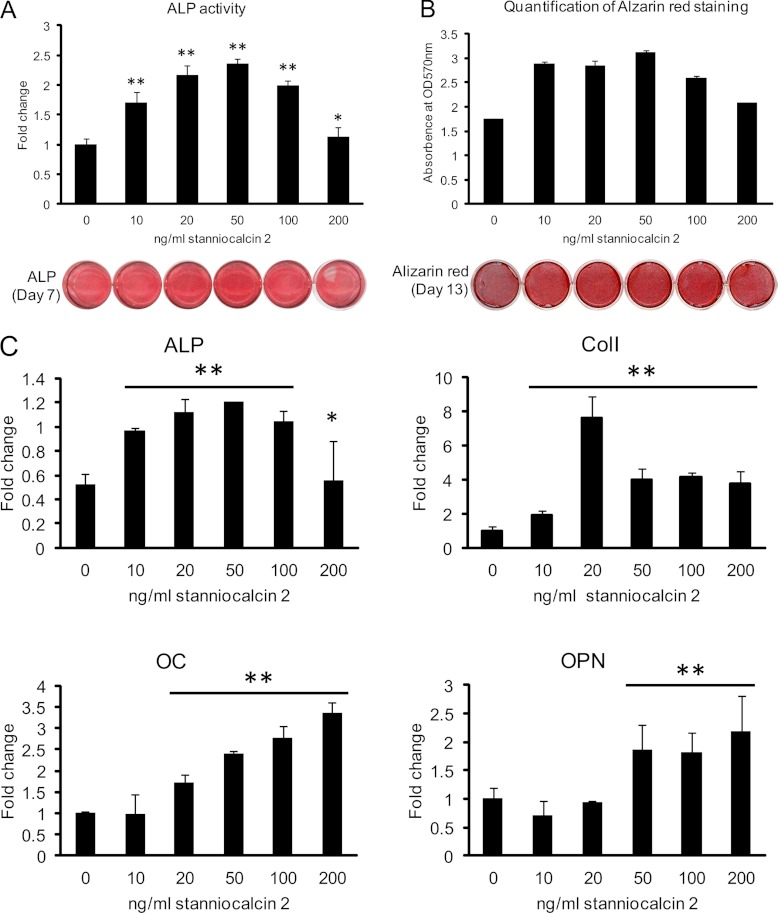
To further study the biological function of STC2, we examined its effects on osteoblastic differentiation in hMSC. Human recombinant STC2 significantly increased ALP activity, staining intensity (Fig 8A), and the formation of mineralized matrix evidenced by Alizarin red S staining (Fig. 8B) in a dose-dependent fashion with maximal effects achieved at 20–50 ng/ml. Similarly, STC2 significantly increased osteoblast-specific gene expression: ALP, collagen type I (Col I), osteocalcin (OC) and osteopontin (OPN) (Fig. 8C).
Fig. 8.
Human recombinant stanniocalcin 2 stimulates osteoblastic differentiation of hMSC. Cells were cultured in 24-well plate and induced to osteoblastic differentiation in absence or presence of different concentrations of recombinant human STC2 protein. A, Alkaline phosphatase (ALP) activity and staining were performed after 7 days induction. B, Mineralized matrix formation was demonstrated and quantified by Alizarin red S staining after 13 days treatment. C, Expression of specific osteoblastic marker genes as quantified by qRT-PCR. ALP: alkaline phosphatase; Col I: collagen type I; OC: osteocalcin; OPN: osteopontin. Data were represented as mean ± S.D. from four independent experiments. * indicate p < 0.05, ** indicate p < 0.001.
To confirm these results we carried out siRNA-based knock down and over-expression of STC2 in hMSC-TERT cells and examined the biological effects on osteoblastic differentiation. As demonstrated in supplemental Fig. S4, deficiency of STC2 decreased expression of osteoblastic markers as well as mineralized matrix formation. The opposite effects were observed upon STC2 over-expression (supplemental Fig. S5).
DISCUSSION
In the present study we employed quantitative proteome analysis using SILAC labeling to provide, for the first time, a global quantitative analysis of proteins secreted during ex vivo OB differentiation of bone marrow-derived hMSC. We identified several known and potentially novel proteins associated with the OB differentiation process. These proteins are involved in several biological processes and thus support the hypothesis that the secretome of hMSC exerts a multitude of functions including local skeletal tissue regeneration, immune modulation, and endocrine functions. Secreted proteins are usually identified through either literature searches (termed validated secreted) or by theoretical prediction using bioinformatics tools such as Phobius or SecretomeP (termed predicted secreted). None of these methods are expected to give the correct subset of bona fide secreted proteins. We combined both methods and employed strict prediction criteria that included the presence of a signal peptide for secretion through the classical or nonclassical pathway and the absence of transmembrane domains. Comparing our results with results obtained in other secretome studies of ex vivo cultured stem cells that employed less stringent criteria, revealed better identification of secreted proteins in our study (12, 13, 94, 95).
We employed pulsed SILAC labeling to quantify the fraction of labeling (protein synthesis rate) because newly synthesized secreted proteins were expected to display a high fraction of labeling. The validity of the pulsed SILAC labeling approach to selectively discriminate secreted from contaminating proteins in the conditioned media is based on several observations. First, comparing the fraction of labeling in conditioned medium and cell lysates revealed a significantly higher fraction of labeling of proteins present in the conditioned medium compared with cell lysates. Second, the pulsed SILAC labeling could be used as a parameter to distinguish the secreted proteins. Among the 151 proteins quantified based on both complete and pulsed SILAC labeling, 80% were classified as predicted secreted and 54% as validated secreted proteins versus only 60 and 27%, respectively, among the proteins identified in the complete SILAC labeling experiments. Third, cluster analysis of the proteins identified based on pulsed SILAC labeling revealed a correlation between the percentage of proteins classified as validated or predicted secreted and the fraction of labeling. Finally, others have reported similar findings that secreted proteins have high label incorporation as compared with intracellular proteins when performing pulsed SILAC labeling e.g. adipose tissue cells (96). The pulsed SILAC method has some limitations. Intracellular and membrane bound proteins with a high fraction of labeling may leak into the conditioned medium. Also, some secreted proteins have a lower fraction of labeling such as ECM-associated proteins or secretory vesicles released into the medium from storages of latent secreted proteins. These factors can explain the presence of validated secreted proteins in cluster 2–4.
The perspectives for the use of pulsed SILAC labeling to study secreted proteins are intriguing and could in principle allow the study of the secretory pathway of the various proteins by compartment-specific analyses of the fraction of labeling as demonstrated in another context (31) or to study the fraction of secreted proteins synthesized on-demand as compared with pre-synthesized dormant molecules.
There was a strong correlation between high fraction of labeling and proteins directly involved in OB differentiation. Interestingly, we observed a labeling collapse at day 14 for the proteins in cluster 2 suggesting that the synthesis of these proteins is regulated and may have an important role in the early phase of osteoblastic differentiation. The temporal profile quantified by complete SILAC labeling confirmed a reduced relative abundance of these proteins at day 14 as compared with the other time-points.
Validation of the gene expression of the protein candidates by qRT-PCR and Western blot analysis revealed a general good concordance between these methods. In addition, we have demonstrated the biological relevance of our findings by studying one of the identified factors: STC2 in a detailed fashion. STC2 and its closely related homolog STC1 are glycoproteins identified as a calcium/phosphate regulating hormones (97). Our study demonstrates that both hMSC and osteoblastic cells produce significant amounts of STC2 and that this protein may play a role in hMSC biology in addition to its presumed role in calcium and phosphate homeostasis. We found that STC2 enhances osteoblastic differentiation of hMSC suggesting a role as an autocrine/paracrine factor in the bone marrow microenvironment. Our study corroborates a recent study (98) demonstrating a role of STC2 in ectopic calcification and that inorganic phosphate stimulates production of STC2. It is thus plausible that STC2 in this context may enhance differentiation of resident MSC to osteoblastic cells. However, detailed understanding of STC2 functions awaits identification of its cognate receptor (97).
In conclusion, the presented data demonstrate a novel global quantitative proteomics-based approach for selective identification of relevant secreted proteins with novel roles in skeletal stem cell biology and OB differentiation. The combination of pulsed and complete SILAC labeling allowed identification of truly secreted proteins and the hormone STC2 as a novel regulator of OB differentiation. Such methods are important for large scale identification of proteins with biological and clinical relevance. The functional annotations of the secreted proteins revealed the complex nature of OB and extend their biological functions to other areas beyond bone formation.
Supplementary Material
Acknowledgments
We thank Tina Nielsen for technical help.
Footnotes
* L.P.K. was supported by a grant from the region of Southern Denmark and L.P.K., J.S.A. and M.K. from the EU FP6 project (LSH-2004-1.2.4-6/Osteocord). The studies were supported by grants from the Novo Nordisk Foundation, the Lundbeck foundation, and Karen Elise Jensen's foundation.
 This article contains supplemental Tables S1 to S5 and Figs. S1 to S5.
This article contains supplemental Tables S1 to S5 and Figs. S1 to S5.
1 The abbreviations used are:
- SILAC
- stable isotope labeling by amino acids in cell culture
- hMSC
- human mesenchymal stem cell
- hTERT
- human telomerase reverse transcriptase
- ECM
- extracellular matrix
- FBS
- fetal bovine serum
- SCID
- severe combined immunodeficiency disease
- IGF
- insulin-like growth factor
- IGFBP
- insulin-like growth factor binding protein
- TGF-β
- transforming growth factor-β
- BMP1
- bone morphogenetic protein 1
- MGP
- matrix gla protein
- CFH
- complement factor H
- DMEM
- Dulbecco's modified Eagle medium
- DAVID
- the database for annotation visualization and integrated discovery
- OB
- osteoblast
- GO
- Gene ontology
- ALP
- alkaline phosphatase
- SVEP1
- selectin-like osteoblast-derived protein
- STC2
- stanniocalcin 2.
REFERENCES
- 1. Fukumoto S., Martin T. J. (2009) Bone as an endocrine organ. Trends Endocrinol. Metab. 20, 230–236 [DOI] [PubMed] [Google Scholar]
- 2. Kobayashi Y., Udagawa N., Takahashi N. (2009) Action of RANKL and OPG for osteoclastogenesis. Crit. Rev. Eukaryot. Gene Exp. 19, 61–72 [DOI] [PubMed] [Google Scholar]
- 3. Porter R. L., Calvi L. M. (2008) Communications between bone cells and hematopoietic stem cells. Arch. Biochem. Biophys. 473, 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lorenzo J., Horowitz M., Choi Y. (2008) Osteoimmunology: interactions of the bone and immune system. Endocr. Rev. 29, 403–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee N. K., Sowa H., Hinoi E., Ferron M., Ahn J. D., Confavreux C., Dacquin R., Mee P. J., McKee M. D., Jung D. Y., Zhang Z., Kim J. K., Mauvais-Jarvis F., Ducy P., Karsenty G. (2007) Endocrine regulation of energy metabolism by the skeleton. Cell 130, 456–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kilroy G. E., Foster S. J., Wu X., Ruiz J., Sherwood S., Heifetz A., Ludlow J. W., Stricker D. M., Potiny S., Green P., Halvorsen Y. D., Cheatham B., Storms R. W., Gimble J. M. (2007) Cytokine profile of human adipose-derived stem cells: expression of angiogenic, hematopoietic, and pro-inflammatory factors. J. Cell. Physiol. 212, 702–709 [DOI] [PubMed] [Google Scholar]
- 7. Haynesworth S. E., Baber M. A., Caplan A. I. (1996) Cytokine expression by human marrow-derived mesenchymal progenitor cells in vitro: effects of dexamethasone and IL-1 alpha. J. Cell. Physiol. 166, 585–592 [DOI] [PubMed] [Google Scholar]
- 8. Sze S. K., de Kleijn D. P., Lai R. C., Khia Way Tan E., Zhao H., Yeo K. S., Low T. Y., Lian Q., Lee C. N., Mitchell W., El Oakley R. M., Lim S. K. (2007) Elucidating the secretion proteome of human embryonic stem cell-derived mesenchymal stem cells. Mol. Cell. Proteomics 6, 1680–1689 [DOI] [PubMed] [Google Scholar]
- 9. Penolazzi L., Lambertini E., Tavanti E., Torreggiani E., Vesce F., Gambari R., Piva R. (2008) Evaluation of chemokine and cytokine profiles in osteoblast progenitors from umbilical cord blood stem cells by BIO-PLEX technology. Cell Biol. Int. 32, 320–325 [DOI] [PubMed] [Google Scholar]
- 10. Celebi B., Elcin A. E., Elcin Y. M. (2010) Proteome analysis of rat bone marrow mesenchymal stem cell differentiation. J. Proteome Res. 9, 5217–5227 [DOI] [PubMed] [Google Scholar]
- 11. Kim S., Min W. K., Chun S., Lee W., Chung H. J., Choi S. J., Yang S. E., Yang Y. S., Yoo J. I. (2010) Protein expression profiles during osteogenic differentiation of mesenchymal stem cells derived from human umbilical cord blood. Tohoku J. Exp. Med. 221, 141–150 [DOI] [PubMed] [Google Scholar]
- 12. Chiellini C., Cochet O., Negroni L., Samson M., Poggi M., Ailhaud G., Alessi M. C., Dani C., Amri E. Z. (2008) Characterization of human mesenchymal stem cell secretome at early steps of adipocyte and osteoblast differentiation. BMC Mol. Biol. 9, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zvonic S., Lefevre M., Kilroy G., Floyd Z. E., DeLany J. P., Kheterpal I., Gravois A., Dow R., White A., Wu X., Gimble J. M. (2007) Secretome of primary cultures of human adipose-derived stem cells: modulation of serpins by adipogenesis. Mol. Cell. Proteomics 6, 18–28 [DOI] [PubMed] [Google Scholar]
- 14. Choi Y. A., Lim J., Kim K. M., Acharya B., Cho J. Y., Bae Y. C., Shin H. I., Kim S. Y., Park E. K. (2010) Secretome Analysis of Human BMSCs and Identification of SMOC1 as an Important ECM Protein in Osteoblast Differentiation. J. Proteome Res. 9, 2949–2956 [DOI] [PubMed] [Google Scholar]
- 15. Lee M. J., Kim J., Kim M. Y., Bae Y. S., Ryu S. H., Lee T. G., Kim J. H. (2010) Proteomic analysis of tumor necrosis factor-alpha-induced secretome of human adipose tissue-derived mesenchymal stem cells. J. Proteome Res. 9, 1754–1762 [DOI] [PubMed] [Google Scholar]
- 16. Zhu W., Huang L., Li Y., Zhang X., Gu J., Yan Y., Xu X., Wang M., Qian H., Xu W. (2012) Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett. 315, 28–37 [DOI] [PubMed] [Google Scholar]
- 17. Lai R. C., Chen T. S., Lim S. K. (2011) Mesenchymal stem cell exosome: a novel stem cell-based therapy for cardiovascular disease. Regen. Med. 6, 481–492 [DOI] [PubMed] [Google Scholar]
- 18. Xiao Z., Camalier C. E., Nagashima K., Chan K. C., Lucas D. A., de la Cruz M. J., Gignac M., Lockett S., Issaq H. J., Veenstra T. D., Conrads T. P., Beck G. R., Jr. (2007) Analysis of the extracellular matrix vesicle proteome in mineralizing osteoblasts. J. Cell. Physiol. 210, 325–335 [DOI] [PubMed] [Google Scholar]
- 19. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 20. Ong S. E., Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 21. Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 22. Stenderup K., Justesen J., Clausen C., Kassem M. (2003) Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone 33, 919–926 [DOI] [PubMed] [Google Scholar]
- 23. Simonsen J. L., Rosada C., Serakinci N., Justesen J., Stenderup K., Rattan S. I., Jensen T. G., Kassem M. (2002) Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat. Biotechnol. 20, 592–596 [DOI] [PubMed] [Google Scholar]
- 24. Abdallah B. M., Haack-Sorensen M., Burns J. S., Elsnab B., Jakob F., Hokland P., Kassem M. (2005) Maintenance of differentiation potential of human bone marrow mesenchymal stem cells immortalized by human telomerase reverse transcriptase gene despite [corrected] extensive proliferation. Biochem. Biophys. Res. Commun. 326, 527–538 [DOI] [PubMed] [Google Scholar]
- 25. Alvarez-Llamas G., Szalowska E., de Vries M. P., Weening D., Landman K., Hoek A., Wolffenbuttel B. H., Roelofsen H., Vonk R. J. (2007) Characterization of the human visceral adipose tissue secretome. Mol. Cell. Proteomics 6, 589–600 [DOI] [PubMed] [Google Scholar]
- 26. Anderson N. L., Anderson N. G. (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867 [DOI] [PubMed] [Google Scholar]
- 27. Kristensen A. R., Schandorff S., Høyer-Hansen M., Nielsen M. O., Jäättelä M., Dengjel J., Andersen J. S. (2008) Ordered organelle degradation during starvation-induced autophagy. Mol. Cell. Proteomics 7, 2419–2428 [DOI] [PubMed] [Google Scholar]
- 28. Yu L., Guo Y., Zhang Z., Li Y., Li M., Li G., Xiong W., Zeng Y. (2010) SecretP: a new method for predicting mammalian secreted proteins. Peptides 31, 574–578 [DOI] [PubMed] [Google Scholar]
- 29. Käll L., Krogh A., Sonnhammer E. L. (2007) Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 35, W429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doherty M. K., Hammond D. E., Clague M. J., Gaskell S. J., Beynon R. J. (2009) Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome Res. 8, 104–112 [DOI] [PubMed] [Google Scholar]
- 31. Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boisvert F. M., Ahmad Y., Gierlinski M., Charriere F., Lamont D., Scott M., Barton G., Lamond A. I. (2011) A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol. Cell. Proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blagoev B., Mann M. (2006) Quantitative proteomics to study mitogen-activated protein kinases. Methods 40, 243–250 [DOI] [PubMed] [Google Scholar]
- 34. Abdallah B. M., Haack-Sørensen M., Fink T., Kassem M. (2006) Inhibition of osteoblast differentiation but not adipocyte differentiation of mesenchymal stem cells by sera obtained from aged females. Bone 39, 181–188 [DOI] [PubMed] [Google Scholar]
- 35. Zou L., Zou X., Chen L., Li H., Mygind T., Kassem M., Bünger C. (2008) Multilineage differentiation of porcine bone marrow stromal cells associated with specific gene expression pattern. J. Orthop. Res. 26, 56–64 [DOI] [PubMed] [Google Scholar]
- 36. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 37. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 38. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 39. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 40. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 41. Schandorff S., Olsen J. V., Bunkenborg J., Blagoev B., Zhang Y., Andersen J. S., Mann M. (2007) A mass spectrometry-friendly database for cSNP identification. Nat. Methods 4, 465–466 [DOI] [PubMed] [Google Scholar]
- 42. Mortensen P., Gouw J. W., Olsen J. V., Ong S. E., Rigbolt K. T., Bunkenborg J., Cox J., Foster L. J., Heck A. J., Blagoev B., Andersen J. S., Mann M. (2010) MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J. Proteome Res. 9, 393–403 [DOI] [PubMed] [Google Scholar]
- 43. Olsen J. V., Mann M. (2004) Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc. Natl. Acad. Sci. U.S.A. 101, 13417–13422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 45. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 46. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 47. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 48. Dopazo J., Carazo J. M. (1997) Phylogenetic reconstruction using an unsupervised growing neural network that adopts the topology of a phylogenetic tree. J. Mol. Evolution 44, 226–233 [DOI] [PubMed] [Google Scholar]
- 49. Herrero J., Valencia A., Dopazo J. (2001) A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics 17, 126–136 [DOI] [PubMed] [Google Scholar]
- 50. Yeung K. Y., Haynor D. R., Ruzzo W. L. (2001) Validating clustering for gene expression data. Bioinformatics 17, 309–318 [DOI] [PubMed] [Google Scholar]
- 51. Bendtsen J. D., Jensen L. J., Blom N., Von Heijne G., Brunak S. (2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356 [DOI] [PubMed] [Google Scholar]
- 52. Hoffmann R., Valencia A. (2004) A gene network for navigating the literature. Nat. Genet. 36, 664. [DOI] [PubMed] [Google Scholar]
- 53. Rigbolt K. T., Vanselow J. T., Blagoev B. (2011) GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol. Cell. Proteomics 10, O110.007450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kratchmarova I., Blagoev B., Haack-Sorensen M., Kassem M., Mann M. (2005) Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science 308, 1472–1477 [DOI] [PubMed] [Google Scholar]
- 55. Palermo C., Manduca P., Gazzerro E., Foppiani L., Segat D., Barreca A. (2004) Potentiating role of IGFBP-2 on IGF-II-stimulated alkaline phosphatase activity in differentiating osteoblasts. Am. J. Physiol. Endocrinol. Metab. 286, E648–657 [DOI] [PubMed] [Google Scholar]
- 56. Silha J. V., Mishra S., Rosen C. J., Beamer W. G., Turner R. T., Powell D. R., Murphy L. J. (2003) Perturbations in bone formation and resorption in insulin-like growth factor binding protein-3 transgenic mice. J. Bone Miner. Res. 18, 1834–1841 [DOI] [PubMed] [Google Scholar]
- 57. Kassem M., Okazaki R., De León D., Harris S. A., Robinson J. A., Spelsberg T. C., Conover C. A., Riggs B. L. (1996) Potential mechanism of estrogen-mediated decrease in bone formation: estrogen increases production of inhibitory insulin-like growth factor-binding protein-4. Proc. Assoc. Am. Physicians 108, 155–164 [PubMed] [Google Scholar]
- 58. Kiefer M. C., Schmid C., Waldvogel M., Schläpfer I., Futo E., Masiarz F. R., Green K., Barr P. J., Zapf J. (1992) Characterization of recombinant human insulin-like growth factor binding proteins 4, 5, and 6 produced in yeast. J. Biol. Chem. 267, 12692–12699 [PubMed] [Google Scholar]
- 59. Qin X., Wergedal J. E., Rehage M., Tran K., Newton J., Lam P., Baylink D. J., Mohan S. (2006) Pregnancy-associated plasma protein-A increases osteoblast proliferation in vitro and bone formation in vivo. Endocrinology 147, 5653–5661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim J. E., Kim S. J., Lee B. H., Park R. W., Kim K. S., Kim I. S. (2000) Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, betaig-h3. J. Biol. Chem. 275, 30907–30915 [DOI] [PubMed] [Google Scholar]
- 61. Scott I. C., Blitz I. L., Pappano W. N., Imamura Y., Clark T. G., Steiglitz B. M., Thomas C. L., Maas S. A., Takahara K., Cho K. W., Greenspan D. S. (1999) Mammalian BMP-1/Tolloid-related metalloproteinases, including novel family member mammalian Tolloid-like 2, have differential enzymatic activities and distributions of expression relevant to patterning and skeletogenesis. Develop. Biol. 213, 283–300 [DOI] [PubMed] [Google Scholar]
- 62. Safadi F. F., Xu J., Smock S. L., Kanaan R. A., Selim A. H., Odgren P. R., Marks S. C., Jr., Owen T. A., Popoff S. N. (2003) Expression of connective tissue growth factor in bone: its role in osteoblast proliferation and differentiation in vitro and bone formation in vivo. J. Cell. Physiol. 196, 51–62 [DOI] [PubMed] [Google Scholar]
- 63. Wiktor-Jedrzejczak W., Bartocci A., Ferrante A. W., Jr., Ahmed-Ansari A., Sell K. W., Pollard J. W., Stanley E. R. (1990) Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc. Natl. Acad. Sci. U.S.A. 87, 4828–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Delany A. M., Kalajzic I., Bradshaw A. D., Sage E. H., Canalis E. (2003) Osteonectin-null mutation compromises osteoblast formation, maturation, and survival. Endocrinology 144, 2588–2596 [DOI] [PubMed] [Google Scholar]
- 65. Hadfield K. D., Rock C. F., Inkson C. A., Dallas S. L., Sudre L., Wallis G. A., Boot-Handford R. P., Canfield A. E. (2008) HtrA1 inhibits mineral deposition by osteoblasts: requirement for the protease and PDZ domains. J. Biol. Chem. 283, 5928–5938 [DOI] [PubMed] [Google Scholar]
- 66. Jellinek D. A., Chang A. C., Larsen M. R., Wang X., Robinson P. J., Reddel R. R. (2000) Stanniocalcin 1 and 2 are secreted as phosphoproteins from human fibrosarcoma cells. Biochem. J. 350 Pt 2, 453–461 [PMC free article] [PubMed] [Google Scholar]
- 67. Raouf A., Ganss B., McMahon C., Vary C., Roughley P. J., Seth A. (2002) Lumican is a major proteoglycan component of the bone matrix. Matrix Biol. 21, 361–367 [DOI] [PubMed] [Google Scholar]
- 68. Mosig R. A., Dowling O., DiFeo A., Ramirez M. C., Parker I. C., Abe E., Diouri J., Aqeel A. A., Wylie J. D., Oblander S. A., Madri J., Bianco P., Apte S. S., Zaidi M., Doty S. B., Majeska R. J., Schaffler M. B., Martignetti J. A. (2007) Loss of MMP-2 disrupts skeletal and craniofacial development and results in decreased bone mineralization, joint erosion and defects in osteoblast and osteoclast growth. Human Mol. Genetics 16, 1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tanaka T., Ikari K., Furushima K., Okada A., Tanaka H., Furukawa K., Yoshida K., Ikeda T., Ikegawa S., Hunt S. C., Takeda J., Toh S., Harata S., Nakajima T., Inoue I. (2003) Genomewide linkage and linkage disequilibrium analyses identify COL6A1, on chromosome 21, as the locus for ossification of the posterior longitudinal ligament of the spine. Am. J. Human Gen. 73, 812–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Deckers M. M., Smits P., Karperien M., Ni J., Tylzanowski P., Feng P., Parmelee D., Zhang J., Bouffard E., Gentz R., Löwik C. W., Merregaert J. (2001) Recombinant human extracellular matrix protein 1 inhibits alkaline phosphatase activity and mineralization of mouse embryonic metatarsals in vitro. Bone 28, 14–20 [DOI] [PubMed] [Google Scholar]
- 71. Barisic-Dujmovic T., Boban I., Adams D. J., Clark S. H. (2007) Marfan-like skeletal phenotype in the tight skin (Tsk) mouse. Calcified Tissue Int. 81, 305–315 [DOI] [PubMed] [Google Scholar]
- 72. Coutu D. L., Wu J. H., Monette A., Rivard G. E., Blostein M. D., Galipeau J. (2008) Periostin, a member of a novel family of vitamin K-dependent proteins, is expressed by mesenchymal stromal cells. J. Biol. Chem. 283, 17991–18001 [DOI] [PubMed] [Google Scholar]
- 73. Gomez D. E., Alonso D. F., Yoshiji H., Thorgeirsson U. P. (1997) Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur. J. Cell Biol. 74, 111–122 [PubMed] [Google Scholar]
- 74. Bi Y., Stuelten C. H., Kilts T., Wadhwa S., Iozzo R. V., Robey P. G., Chen X. D., Young M. F. (2005) Extracellular matrix proteoglycans control the fate of bone marrow stromal cells. J. Biol. Chem. 280, 30481–30489 [DOI] [PubMed] [Google Scholar]
- 75. Marini J. C., Forlino A., Cabral W. A., Barnes A. M., San Antonio J. D., Milgrom S., Hyland J. C., Körkkö J., Prockop D. J., De Paepe A., Coucke P., Symoens S., Glorieux F. H., Roughley P. J., Lund A. M., Kuurila-Svahn K., Hartikka H., Cohn D. H., Krakow D., Mottes M., Schwarze U., Chen D., Yang K., Kuslich C., Troendle J., Dalgleish R., Byers P. H. (2007) Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Human Mutation 28, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mochida Y., Duarte W. R., Tanzawa H., Paschalis E. P., Yamauchi M. (2003) Decorin modulates matrix mineralization in vitro. Biochem. Biophys. Res. Commun. 305, 6–9 [DOI] [PubMed] [Google Scholar]
- 77. Mochida Y., Parisuthiman D., Pornprasertsuk-Damrongsri S., Atsawasuwan P., Sricholpech M., Boskey A. L., Yamauchi M. (2009) Decorin modulates collagen matrix assembly and mineralization. Matrix Biol. 28, 44–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kaku M., Mochida Y., Atsawasuwan P., Parisuthiman D., Yamauchi M. (2007) Post-translational modifications of collagen upon BMP-induced osteoblast differentiation. Biochem. Biophys. Res. Commun. 359, 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nabavi N., Urukova Y., Cardelli M., Aubin J. E., Harrison R. E. (2008) Lysosome dispersion in osteoblasts accommodates enhanced collagen production during differentiation. J. Biol. Chem. 283, 19678–19690 [DOI] [PubMed] [Google Scholar]
- 80. Billiard J., Moran R. A., Whitley M. Z., Chatterjee-Kishore M., Gillis K., Brown E. L., Komm B. S., Bodine P. V. (2003) Transcriptional profiling of human osteoblast differentiation. J. Cell. Biochem. 89, 389–400 [DOI] [PubMed] [Google Scholar]
- 81. Carano R. A., Filvaroff E. H. (2003) Angiogenesis and bone repair. Drug Discovery Today 8, 980–989 [DOI] [PubMed] [Google Scholar]
- 82. Gerber H. P., Ferrara N. (2000) Angiogenesis and bone growth. Trends Cardiovasc. Med. 10, 223–228 [DOI] [PubMed] [Google Scholar]
- 83. Abe Y., Abe T., Aida Y., Hara Y., Maeda K. (2004) Follistatin restricts bone morphogenetic protein (BMP)-2 action on the differentiation of osteoblasts in fetal rat mandibular cells. J. Bone Miner. Res. 19, 1302–1307 [DOI] [PubMed] [Google Scholar]
- 84. Aisa M. C., Beccari T., Costanzi E., Maggio D. (2003) Cathepsin B in osteoblasts. Biochim. Biophys. Acta 1621, 149–159 [DOI] [PubMed] [Google Scholar]
- 85. Morko J., Kiviranta R., Mulari M. T., Ivaska K. K., Väänänen H. K., Vuorio E., Laitala-Leinonen T. (2009) Overexpression of cathepsin K accelerates the resorption cycle and osteoblast differentiation in vitro. Bone 44, 717–728 [DOI] [PubMed] [Google Scholar]
- 86. Choi S. J., Reddy S. V., Devlin R. D., Menaa C., Chung H., Boyce B. F., Roodman G. D. (1999) Identification of human asparaginyl endopeptidase (legumain) as an inhibitor of osteoclast formation and bone resorption. J. Biol. Chem. 274, 27747–27753 [DOI] [PubMed] [Google Scholar]
- 87. Yoshiko Y., Maeda N., Aubin J. E. (2003) Stanniocalcin 1 stimulates osteoblast differentiation in rat calvaria cell cultures. Endocrinology 144, 4134–4143 [DOI] [PubMed] [Google Scholar]
- 88. Ichikawa T., Horie-Inoue K., Ikeda K., Blumberg B., Inoue S. (2007) Vitamin K2 induces phosphorylation of protein kinase A and expression of novel target genes in osteoblastic cells. J. Mol. Endocrinol. 39, 239–247 [DOI] [PubMed] [Google Scholar]
- 89. Gilges D., Vinit M. A., Callebaut I., Coulombel L., Cacheux V., Romeo P. H., Vigon I. (2000) Polydom: a secreted protein with pentraxin, complement control protein, epidermal growth factor and von Willebrand factor A domains. Biochem. J. 352 Pt 1, 49–59 [PMC free article] [PubMed] [Google Scholar]
- 90. Shur I., Socher R., Hameiri M., Fried A., Benayahu D. (2006) Molecular and cellular characterization of SEL-OB/SVEP1 in osteogenic cells in vivo and in vitro. J. Cell. Physiol. 206, 420–427 [DOI] [PubMed] [Google Scholar]
- 91. Shur I., Zemer-Tov E., Socher R., Benayahu D. (2007) SVEP1 expression is regulated in estrogen-dependent manner. J. Cell. Physiol. 210, 732–739 [DOI] [PubMed] [Google Scholar]
- 92. Lee G. W., Lee T. H., Vilcek J. (1993) TSG-14, a tumor necrosis factor- and IL-1-inducible protein, is a novel member of the pentaxin family of acute phase proteins. J. Immunol. 150, 1804–1812 [PubMed] [Google Scholar]
- 93. Fedarko N. S., Fohr B., Robey P. G., Young M. F., Fisher L. W. (2000) Factor H binding to bone sialoprotein and osteopontin enables tumor cell evasion of complement-mediated attack. J. Biol. Chem. 275, 16666–16672 [DOI] [PubMed] [Google Scholar]
- 94. Estrada R., Li N., Sarojini H., An J., Lee M. J., Wang E. (2009) Secretome from mesenchymal stem cells induces angiogenesis via Cyr61. J. Cell. Physiol. 219, 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Molina H., Yang Y., Ruch T., Kim J. W., Mortensen P., Otto T., Nalli A., Tang Q. Q., Lane M. D., Chaerkady R., Pandey A. (2009) Temporal profiling of the adipocyte proteome during differentiation using a five-plex SILAC based strategy. J. Proteome Res. 8, 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Roelofsen H., Dijkstra M., Weening D., de Vries M. P., Hoek A., Vonk R. J. (2009) Comparison of isotope-labeled amino acid incorporation rates (CILAIR) provides a quantitative method to study tissue secretomes. Mol. Cell. Proteomics 8, 316–324 [DOI] [PubMed] [Google Scholar]
- 97. Yeung B. H., Law A. Y., Wong C. K. (2012) Evolution and roles of stanniocalcin. Mol. Cell. Endocrinol. 349, 272–280 [DOI] [PubMed] [Google Scholar]
- 98. Takei Y., Yamamoto H., Sato T., Otani A., Kozai M., Masuda M., Taketani Y., Muto-Sato K., Lanske B., Takeda E. (2012) Stanniocalcin 2 is associated with ectopic calcification in alpha-klotho mutant mice and inhibits hyperphosphatemia-induced calcification in aortic vascular smooth muscle cells. Bone 50, 998–1005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.