Abstract
Head and neck squamous cell carcinomas (HNSCC) can arise from the oral cavity, oropharynx, larynx or hypopharynx, and is the sixth leading cancer by incidence worldwide. The 5-year survival rate of HNSCC patients remains static at 40–60%. Hence, biomarkers which can improve detection of HNSCC or early recurrences should improve clinical outcome. Mass spectrometry-based proteomics methods have emerged as promising approaches for biomarker discovery. As one approach, mass-spectrometric identification of proteins shed or secreted from cancer cells can contribute to the identification of potential biomarkers for HNSCC and our understanding of tumor behavior. In the current study, mass spectrometry-based proteomic profiling was performed on the conditioned media (i.e. secretome) of head and neck cancer (HNC) cell lines (FaDu, UTSCC8 and UTSCC42a) in addition to gene expression microarrays to identify over-expressed transcripts in the HNSCC cells in comparison to a normal control cell line. This integrated data set was systematically mined using publicly available resources (Human Protein Atlas and published proteomic/transcriptomic data) to prioritize putative candidates for validation. Subsequently, quantitative real-time PCR (qRT-PCR), Western blotting, immunohistochemistry (IHC), and ELISAs were performed to verify selected markers. Our integrated analyses identified 90 putative protein biomarkers that were secreted or shed to the extracellular space and over-expressed in HNSCC cell lines, relative to controls. Subsequently, the over-expression of five markers was verified in vitro at the transcriptional and translational levels using qRT-PCR and Western blotting, respectively. IHC-based validation conducted in two independent cohorts comprising of 40 and 39 HNSCC biopsies revealed that high tumor expression of PLAU, IGFBP7, MMP14 and THBS1 were associated with inferior disease-free survival, and increased risk of disease progression or relapse. Furthermore, as demonstrated using ELISAs, circulating levels of PLAU and IGFBP7 were significantly higher in the plasma of HNSCC patients compared with healthy individuals.
Head and Neck Squamous Cell Carcinoma (HNSCC)1 is the sixth most common cancer worldwide, with ∼600,000 new cases diagnosed every year (1). HNSCCs include squamous cell carcinomas (SCCs) of the oral cavity, pharynx (nasopharynx, oropharynx, and hypopharynx), and larynx. Despite improvements in therapeutic approaches, the overall 5-year survival rate of HNSCC patients has not improved over the past three decades (1, 2).
Clinical management of HNSCC is faced by several challenges, including early detection of the primary tumor, and site-specific control of the disease (3, 4). Early diagnosis is often hampered by the asymptomatic nature of HNSCC development and the inadequacy of current diagnostic methods to identify HNSCCs with sufficient specificity and sensitivity. As a result, over 60% of HNSCC patients present with advanced stages III or IV, harboring lymph node metastases (3). The outcome of HNSCC patients is reduced by the development of local recurrences, which can develop in ∼10–30% of cases, even when surgical margins are pathologically tumor-free (5). Current protocols fail to adequately stratify patients with high risk of developing tumor recurrence in order to allow for appropriate modifications in treatment.
Several potentially prognostic HNSCC biomarkers have been described including TP53 mutations, EGFR over-expression, and presence of human papillomavirus (HPV) or its surrogate marker, p16 (1, 6–8). Targeting EGFR has impacted on HNSCC therapy; however, its benefit in survival for locally advanced tumors remains modest (9). There are no other biomarkers which have influenced management of HNSCC.
Considerable efforts in the identification of secreted biomarkers for various cancers in serum or plasma have met with limited success, primarily because of the immense sample complexity, and large dynamic range, which greatly hamper the discovery of biomarkers in such fluids (10). Cancer cell line secretomes offer a valuable alternative to serum or plasma for the discovery of potential biomarkers, and have been used as a source for discovery of secreted biomarkers for prostate, ovarian, lung, and other tumors (3, 11–16). The cancer cell secretome is composed of proteins secreted or shed by tumor cells in culture and detected in the serum-free conditioned media (CM). Proteins secreted or shed by cancer cells have a chance of being released into circulation and thus have the potential to be detected in patient-derived blood in a clinical setting for disease diagnosis or prognosis (11). In vitro secretome studies typically involve growing cells to semi-confluence in normal growth media followed by incubation in serum-free media for periods of 24–48 h. The CM is then harvested for proteomic analysis, and biomarker candidates are selected, and then validated using techniques such as enzyme-linked immunosorbent assays (ELISAs) or immunohistochemistry (IHC). One hurdle associated with biomarker discovery using this approach is the selection of truly secreted proteins for validation, because intracellular contaminants can be present as a result of cell lysis. Hence, cell culture conditions must be carefully optimized to reduce cell death under serum starvation. Furthermore, parallel proteomic profiling of whole cell lysates (WCLs) can be used as a background to filter out intracellular contaminants and enrich for secreted proteins, with the assumption that the relative protein abundance of a secreted protein would be higher in the CM compared with the WCL (17–20).
In the current study, we focused on multidimensional protein identification technology (MudPIT) analyses of secretomes and whole cell lysate proteomes of three head and neck cancer (HNC) cell lines: FaDu, UTSCC8, and UTSCC42a, coupled with parallel gene expression analyses, as a platform for discovery of putative secreted HNSCC markers (21, 22). These cell lines were derived from two anatomically related subsites of the head and neck: the hypopharynx (FaDu cells) and the larynx (UTSCC8, and UTSCC42a cells). Based on protein and transcript expression data, candidates with increased abundance in the CM versus the WCL, and increased mRNA expression levels, as compared with the normal oral epithelial (NOE) cells, were selected and prioritized for validation using publicly available resources. Five candidates were selected for in vitro validation at the protein level through Western blotting against the CM of cancer cell lines and control NOE cells, and at the mRNA level using quantitative real-time PCR (qRT-PCR). In addition, using immunohistochemistry, we demonstrated that four of these candidates have higher protein expression in recurrent compared with nonrecurrent HNSCCs, and that their elevated expressions were associated with lower disease-free survival (DFS), and increased risk of disease progression. Finally, two of the candidate proteins, PLAU (urokinase) and IGFBP7 (insulin-like growth factor binding protein 7), were significantly elevated in the plasma of HNSCC patients compared with healthy controls, which in turn appeared to associate with an increased risk of death in the cancer patients.
EXPERIMENTAL PROCEDURES
Cell Culture
FaDu (human hypopharyngeal squamous cell cancer cell line), UTSCC8 and UTSCC42a (human laryngeal squamous cell cancer cell lines, kind gifts from R. Grénman, Turku University Hospital, Turku, Finland) and normal NOE cells were cultured under conditions described previously (23). FaDu cells were obtained from TCGA (Bethesda, MD), and NOEs from Celprogen (San Pedro, CA). For secretome analyses, cell lines were cultured under serum-free, phenol-red free conditions. The serum-free media were purchased from Invitrogen and included Minimum Essential Medium (GIBCO 51200) for FaDu and NOE cells, and Dulbecco's modified Eagle's medium (GIBCO 31053) for UTSCC8 and UTSCC42a cells. Cell viability was assessed with trypan blue staining (Invitrogen), and exceeded 95% following serum starvation. The cells were authenticated at the Centre for Applied Genomics (Hospital for Sick Children, Toronto, Canada) using the AmpF/STR Identifier PCR Amplification Kit (Applied Biosystems), and routinely tested for mycoplasma contamination using the Mycoalert detection kit (Lonza Group Ltd).
Protein Sample Preparation and Trypsin Digestion
Materials Ultrapure grade urea, ammonium bicarbonate, ammonium acetate, calcium chloride, HEPES and TRIS were obtained from BioShop Canada, Inc. (Burlington, ON). Ultrapure grade iodoacetamide, dithiothreitol and formic acid were obtained from Sigma. HPLC grade solvents (methanol, acetonitrile and water) were obtained from Fisher Scientific, Canada.
HNSCC cell lines were grown in normal growth media until ∼ 80% confluence in T175 cm2 culture flasks. Then, the cells were washed with phosphate buffered saline (PBS) and incubated in 30 ml of serum-free media for 48 h. Subsequently, the cells and the media were harvested. The cells were lysed by incubation in hypotonic lysis buffer (10 mm HEPES, pH 7.4) for 30 min on ice, then briefly sonicated. Triton-X-100 was then added to a final concentration of 1.5%; the cell lysates were incubated on ice for 30 min, then centrifuged at 14,000 rpm for 30 min at 4 °C. The supernatant was subsequently collected for proteomic analysis. The conditioned serum-free media was concentrated to ∼500 μl using Amicon Ultra-15 centrifugal filter tubes with a 3 kDa membrane filter (Millipore, Billerica, MA). Protein concentration was determined by Bradford assay, and 150 μg of total protein from each sample was precipitated overnight at −20 °C with five volumes of ice-cold acetone, followed by centrifugation at 21,000 × g for 15 min. The protein pellet was solubilized in 8 m urea, 2 mm dithiothreitol, 50 mm Tris-HCl, pH 8.5 at 37 °C for 1 h, followed by carboxyamidomethylation with 10 mm iodoacetamide for 1 h at 37 °C in the dark. The samples were then diluted with 50 mm ammonium bicarbonate, pH 8.5 to 1.5 m urea. Calcium chloride was added to a final concentration of 1 mm and samples were digested with 3 μg of recombinant, proteomics-grade trypsin (Promega, Madison, WI) at 37 °C overnight. The resulting peptide mixtures were solid phase-extracted with C18 spin-columns (The Nest Group Inc., Southborough, MA) according to the manufacturer's instructions, and stored at −80 °C until further use.
MudPIT Analysis of HNSCC Cell Lines
A fully automated 5-cycle two-dimensional chromatography sequence was set up as previously described (22). Peptides were loaded on a 7 cm precolumn (150 μm i.d.) containing a Kasil frit packed with 3.5 cm 5 μm Magic C18 100 Å reversed-phase material (Michrom Bioresources Inc., Auburn, CA) followed by 3.5 cm Luna 5 μm SCX 100 Å strong cation exchange resin (Phenomenex, Torrance, CA). Samples were automatically loaded from a 96-well microplate auto-sampler using the EASY-nLC system (Proxeon Biosystems, Odense, Denmark) at 3 μl/min. The pre-column was connected to an 8 cm fused silica analytical column (75 μm i.d.) via a microsplitter tee (Proxeon Biosystems) to which a distal 2.2 kV spray voltage was applied. The analytical column was pulled to a fine electrospray emitter using a laser puller. For peptide separation on the analytical column, a water/acetonitrile gradient was applied at an effective flow rate of 400 nL/min, controlled by the EASY-nLC (Proxeon Biosystems). Ammonium acetate salt bumps (8 μl) were applied at concentrations of 100, 150, 200, and 500 mm followed by a water/acetonitrile gradient as described previously (22). Sample analysis was performed on a LTQ Orbitrap XL (Thermo Fisher Scientific, San Jose, CA) using previously described instrument settings (22). The MS functions and the HPLC solvent gradients were controlled by the Xcalibur data system (Thermo Fisher Scientific).
Protein Identification and Data Analysis
Raw data were converted to m/z XML using ReAdW (version 1.1) and searched against the Human IPI database (version 3.54, 75426 human sequences, (http://www.ebi.ac.uk/IPI)) using the X!Tandem search algorithm (version 2008.02.01.1). X!Tandem was searched with a fragment ion mass tolerance of 0.40 Da and a parent ion tolerance of ±10 ppm. Complete tryptic digestion was assumed; one missed cleavage site was allowed. Cysteine carbamidomethylation and methionine oxidation were specified as fixed and variable modifications, respectively. To minimize protein inference, we developed a database grouping scheme and only reported proteins with substantial peptide information, as described previously (24). Target/decoy search was performed to experimentally estimate the peptide false discovery rate (FDR), and 12 decoy sequences were identified (FDR<1%). Protein identifications with at least two unique tryptic peptides were considered, as reported previously (24, 25).
Protein Quantification by MS and Normalization
Spectral counting (SpC) was used as a measure of protein abundance. The SpCs for peptides corresponding to a protein were normalized against the total number of spectra for a given MudPIT sequence, averaged over the triplicates. To avoid division by 0, SpC values of 0 were replaced by 0.01. The relative abundance of each protein in the conditioned media versus the cell lysate was calculated for each cell line by calculating the ratio of averaged normalized SpCs in the conditioned medium versus the cell lysate.
Gene Expression Microarray
Gene expression profiling was conducted on the HNSCC cell lines cultured under normal growth conditions using a Whole Human Genome Oligo Microarray (Agilent Technologies) at the UHN Microarray Centre. The NOE cell line was used as a normal control. The fold change (FC) was calculated for each probe as compared with the expression in the NOE cells. In addition, standard statistical methods were used to calculate the minimum and maximum FC for each probe in order to provide error estimates in fold changes.
Data Mining
Human Protein Atlas (HPA) (proteinatlas.org/) was used for qualitative comparison of IHC staining of HNC tissue with normal oral mucosa. To achieve this, the strongest staining cancer and normal IHC image was given a value of 3, 2, 1, or 0 representing strong, moderate, weak, and negative staining, respectively, as reported by the HPA; the difference in the scores from cancer and normal was then calculated. For proteins with multiple antibodies, the scores were averaged. SignalP 3.0 and SecretomeP 2.0 (cbs.dtu.dk/services/) were used for prediction of classically and nonclassically secreted proteins, respectively (26). The gene products, which were not predicted to be secreted by either of these mechanisms, were searched against Exocarta (exocarta.ludwig.edu.au/) to identify whether they are present in exosome fractions (27). Comparison against published HNSCC gene expression microarrays was accomplished by linking both data sets by gene accessions using our in-house database.
Quantitative Reverse Transcriptase Real-time PCR
Primer pairs were designed for several genes (supplemental Table S1) using the Primer3 software (primer3.sourceforge.net). RNA was extracted using the Total RNA Extraction kit (Norgen, Thorold, ON) and reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) as specified by the manufacturer. Quantitative real-time PCR was performed using SYBR Green (Applied Biosystems) and an ABI Prism 7900 HT Sequence Detection System (PE Biosystems). The mean fold change in mRNA expression was calculated using the 2-ΔΔCt method, as described previously (28). The mean fold change from three independent experiments was then averaged.
Western Blotting
Proteins (20 μg) were resolved on an 8–12% SDS-PAGE gel and electro-transferred onto a PVDF membrane (Bio-Rad). Membranes were blocked for 1 h in 4% skim milk TBST (0.5% Tween 20 in TBS) buffer, and probed with the primary antibody overnight at 4 °C. Antibodies (Abcam) were used at the following concentrations: MMP14 1:2000, PLAU, TGFBI, IGFBP7, and THBS1 at 1 μg/ml. After incubation with the primary antibody, membranes were washed and incubated in the appropriate secondary HRP-conjugated antibody (Anti-rabbit IgG A9169 or anti-mouse IgG A9917, both 1:30,000 dilution, Sigma) for 2 h. Following an additional round of washing in TBST, immunoreactive protein bands were visualized with ECL Western blotting Substrate (Pierce, Rockford, IL) on an x-ray film (Bio-Flex). Membranes were stained with 0.1% (w/v) Ponceau S in 5% acetic acid to ensure equal sample loading.
Clinical Specimens
Institutional Research Ethics Board approval was obtained for this study. Primary diagnostic formalin-fixed and paraffin-embedded (FFPE) laryngeal and hypopharyngeal carcinoma biopsies from 79 patients were evaluated in 5 μm sections. The clinical characteristics of the 79 patients are shown in Table I. The specimens were divided into two cohorts: training (n = 40) and validation (n = 39) (supplemental Tables S2 and S3). The specimens were selected such that half of the patients experienced recurrences; half did not. The median follow-up times were 3 years for the training cohort; 1.85 years for the validation cohort.
Table I. Clinopathological characteristics of patients for tissue IHC studies.
| Total (n = 79) | Nonrecurrence (n = 39) | Recurrence (n = 40) | p value | |
|---|---|---|---|---|
| Age (median, range) | 65 (44–92) | 68 (44–92) | 62 (47–88) | 0.15 |
| Gender (M/F) | 66/13 | 33/6 | 33/7 | 0.80 |
| Sub-site | ||||
| Hypopharynx | 23 | 9 | 14 | 0.24 |
| Larynx | 56 | 30 | 26 | |
| Stage | ||||
| I/II/III | 36 | 18 | 18 | 0.92 |
| IVA/IVB | 43 | 21 | 22 | |
| Treatment | ||||
| Radiotherapy alone | 53 | 27 | 26 | 0.80 |
| Chemoradiotherapy | 26 | 12 | 14 |
Plasma samples were collected at diagnosis from 27 HNC patients, and 14 healthy volunteers in heparin-containing phlebotomy tubes. The clinical characteristics of these patients are shown in Tables IIA and IIB. The median follow-up time for this group of 27 patients was 2.1 years. Plasma was obtained following the phlebotomy by centrifugation at 3000 × g for 30 min at 4 °C. The clear plasma supernatant was stored at −80 °C until use. Fifteen of the plasma samples were obtained from the same patients as in the IHC validation cohort; thus these samples had matched tumor tissues for IHC analyses.
Table IIA. Clinical characteristics of healthy volunteer and HNSCC patient plasma used for ELISAs.
| Factors | Healthy volunteers (n = 14) | HNSCC (n = 27) | p value (Controls vs. HNSCC) |
|---|---|---|---|
| Age (median, range) | 52 (22–64) | 71 (48–94) | 0.009 |
| Gender (M/F) | 8/6 | 23/4 | 0.05 |
| Sub-site | |||
| Hypopharynx | 7 | ||
| Larynx | 20 | ||
| Treatment | |||
| Radiotherapy alone | 19 | ||
| Chemoradiotherapy | 8 |
Table IIB. Clinical characteristics of HNSCC patient plasma used for ELISAs stratified by recurrence status.
| Factors | HNSCC (n = 27) | Nonrecurrence (n = 15) | Recurrence (n = 12) | p value (nonrecurrence vs. recurrence) |
|---|---|---|---|---|
| Age (median, range) | 71 (48–94) | 65 (44–94) | 68 (47–82) | 0.98 |
| Gender (M/F) | 23/4 | 13/2 | 10/2 | 0.81 |
| Sub-site | ||||
| Hypopharynx | 7 | 4 | 3 | 0.92 |
| Larynx | 20 | 11 | 9 | |
| Treatment | ||||
| Radiotherapy alone | 19 | 12 | 7 | 0.40 |
| Chemoradiotherapy | 8 | 3 | 5 |
Immunohistochemistry
Protein expression was evaluated on formalin-fixed and paraffin-embedded tumor sections using microwave antigen retrieval with 0.01 m citric acid (pH 6.0) in combination with LSAB+ System-HRP (Dako). Rabbit polyclonal anti-PLAU (1:75 dilution, Abcam), rabbit polyclonal anti-IGFBP7 (1:500 dilution, Abcam), rabbit polyclonal anti-MMP14 (1:300 dilution, Abcam), mouse monoclonal anti-THBS1 (1:50 dilution, Abcam), and rabbit polyclonal anti-TGFBI (1:300 dilution, Sigma) antibodies were utilized. Primary antibody was omitted as a negative control. Tumor cytoplasmic staining levels were scored according to intensity of immuno-expression, with 0, 1, 2, and 3 indicating negative, weak, moderate, and strong staining of tumor cells, respectively. IHC scoring was performed without prior knowledge of any clinopathological parameters; each tissue section was scored twice and the result was then averaged.
Statistical Analysis
The Wilcoxon rank-sum test was used to compare IHC straining intensities and plasma protein concentrations between different groups of patients. Overall survival (OS) was calculated from the date of diagnosis to death or the last follow-up time. Disease-free survival was calculated from the date of diagnosis to the date of death or date of relapse or the date of last follow-up. OS and DFS were estimated by the Kaplan-Meier (KM) method. The comparison of the KM curves between the high (above-median) and low (below-median) protein expression groups was based on the log-rank test. Cox proportional-hazard model was used to estimate hazard ratios (HRs) and corresponding 95% confidence intervals (CIs). Two sided test was used. For the training set, we compared the IHC expression between the recurrence and nonrecurrence groups. The IHC biomarkers with p ≤ 0.05 were considered of nominal significance and were further assessed by an independent validation set. Assuming similar effect size as in the IHC training set, power analysis was applied for the validation set. Based on 20 recurrent and 19 nonrecurrent patients in the validation set, and a significance level α of 0.01 to adjust for multiple testing, the validation set had at least 0.86 power to detect statistically significant differences of biomarker expression between the recurrent versus nonrecurrent groups. Power calculations were based on a two-sided test using Power and Sample Size 2008 (PASS) software package (NCSS, Kaysville, Utah). SAS 9.2 software was used for statistical analysis.
ELISA
The concentration of four candidate proteins was measured by ELISA in plasma samples of patients with or without HNSCC. The concentration of PLAU (ELISA kit from MyBiosource), IGFBP7 (ELISA kit from Antibodies-Online), MMP14 (ELISA kit from MyBiosource), and THBS1 (ELISA kit from R&D systems) were determined according to manufacturer's instructions.
Data Sets
Microarray data of HNSCC and NOE cells have been submitted to Gene Expression Omnibus with the accession number GSE40185. The proteomic data associated with this manuscript may be downloaded from the Proteome Commons (http://www.proteomecommons.org) Tranche network using the following hash code: 2by4ZjbUxu+7AQhgVoTOHgFQ5kjXqZweYzUq6ySwJ8dUZ7CLK3TVP6FF9u4ukxMDbyy9vivP8kkMxDwQfOrbPu7ruOoAAAAAAAAoeg==.
RESULTS
Secretome and Transcriptome Analysis of HNSCC Cell Lines
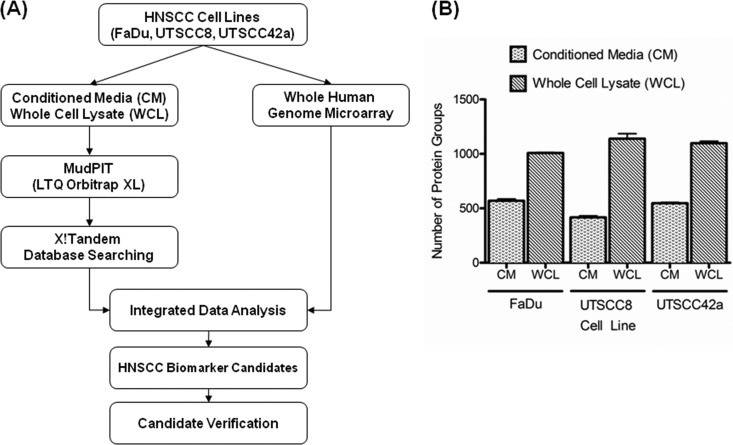
The workflow and experimental design of this study are shown in Fig. 1A. As a result of MudPIT-based proteomic analyses of conditioned media and the whole cell lysates of the three HNC cell lines, 1850 protein groups were identified with high confidence (i.e. gene products). The complete lists of protein and peptide identifications are presented in supplemental Tables S4 and S5. In total, 809 gene products were identified in the CM of the HNC cell lines, with 626, 464, and 626 gene products identified in the media of FaDu, UTSCC8 and UTSCC42a cells, respectively (Fig. 1B). Furthermore, 1632 gene products were identified in the whole cell lysates of HNC cell lines, with 1093, 1182, 1267 gene products identified in FaDu, UTSCC8, and UTSCC42a cells, respectively (Fig. 1B).
Fig. 1.
Proteomic analysis of HNSCC cell line models. A, Outline of the experimental workflow for identification of secreted proteins for HNSCC. B, Average number of protein groups identified in the conditioned media and whole cell lysates of FaDu, UTSCC8 and UTSCC42a cells. Each sample was analyzed in triplicate MudPIT runs.
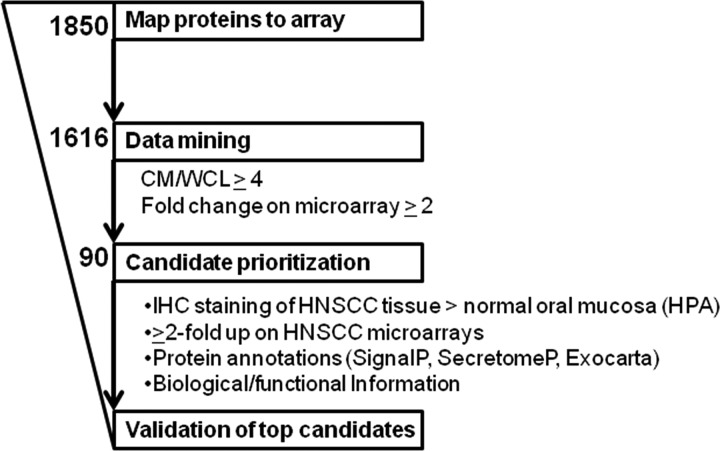
To identify genes that were potentially de-regulated in HNSCC, gene expression profiling was conducted on the HNC cell lines cultured under normal growth conditions. The integration of the protein expression data obtained by MudPIT with the gene expression microarray profiles resulted in 1616 (87%) gene products, which could be linked to probes on the chip (i.e. protein-mRNA pairs), and which were used for further evaluation (Fig. 2).
Fig. 2.
Data mining scheme used for candidate prioritization.
Integrative Data Mining for Marker Prioritization
To obtain a list of promising candidates for potential validation, gene products with at least four-fold increased mean spectral counts in the CM versus the WCL, and a minimum 2-fold up-regulation in HNSCC cell lines according to mRNA expression relative to the NOE cells, were included for further consideration. To be included in the shortlist, these criteria had to be fulfilled in at least two of the three cell lines at both the protein and mRNA level. Ninety putative HNSCC biomarkers fulfilled these criteria (Fig. 2). These two filters were chosen primarily to enrich for markers that were preferentially secreted, and therefore, presumably more abundant in the CM as opposed to the WCL, and up-regulated in the HNSCC cell lines, compared with the NOEs, using the gene expression profiles generated in parallel. In addition, selection for gene products with higher abundance in the CM versus the WCL was used to deplete for potentially contaminating intracellular proteins present because of cell lysis.
As validation of all potential candidates was not feasible, several publicly available resources were used to prioritize these 90 proteins, and then systematically select candidates for downstream verification (Fig. 2 and supplemental Table S6). First, the candidates were prioritized based on differences in IHC staining intensity of HNC versus normal oral mucosal tissues, as per Human Protein Atlas database. Thirty-nine candidates appeared to have stronger staining in HNC tissue as compared with normal tissues; thus were prioritized for validation. Nineteen had no difference in staining between tumor and normal tissue, 25 had no IHC information available, and the remaining seven proteins had stronger staining in normal as compared with HNC tissues. The IHC staining data from HPA provided evidence for differential protein expression between tumor and normal tissues from the human head and neck region, thereby corroborating the protein and gene expression patterns observed in the HNSCC in vitro models. Secondly, expressions of the 90 markers were mapped to two previously published microarray expression data sets for primary human HNC samples (29, 30). The former study determined the expression profiles of 22 paired HNSCC and adjacent nontumor tissue samples from the same patients (including four laryngeal and 1 hypopharyngeal squamous cell carcinomas) (29). The latter report analyzed the expression profiles of 34 hypopharyngeal squamous cell carcinomas and four normal tissues (30). Thirty candidates mapped to at least one of these microarrays; 25 of which were up-regulated at least two-fold in HNSCCs versus controls; thus prioritized for validation. The gene expression profiles provided further evidence for enrichment of several candidates in HNSCCs as compared with nontumor tissue. Third, we investigated whether these proteins were secreted via the canonical secretion pathway, noncanonical secretion pathway, or released via exosomes using SignalP 3.0, SecretomeP 2.0 and Exocarta, respectively (26, 27). Using these tools, 78 of the 90 candidates were highly likely to be secreted by one of these mechanisms, and prioritized for validation by giving preference to candidates that target to the extracellular space. Fourth, a literature search was performed to identify biomarker candidates that may have biologically relevant roles in cancer development, particularly in HNSCC. Lastly, candidates with available antibodies for IHC and ELISAs were prioritized for validation.
In Vitro Marker Validation
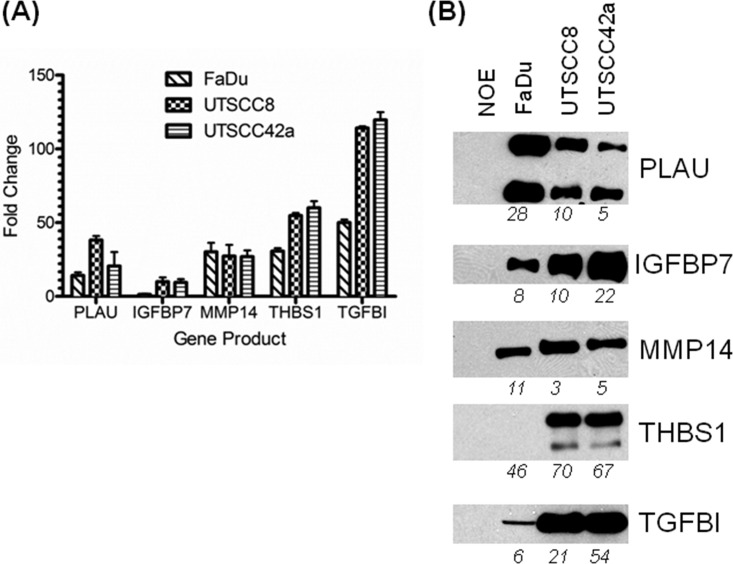
The top 5 ranked candidates were selected and validated in vitro using qRT-PCR and Western blotting against the CM of HNSCC and NOE cells (Fig. 3). PLAU, IGFBP7, MMP14 (matrix metalloproteinase 14), THBS1 (thrombospondin-1), and TGFBI (transforming growth factor, beta-induced) were verified as up-regulated at both the mRNA (Fig. 3A) and protein levels (Fig. 3B) in HNSCC cells and their respective CMs, as compared with the NOEs. Equal protein loading was verified through Ponceau S membrane staining (supplemental Fig. S1). Although THBS1 was confirmed to be up-regulated in the HNSCC cell lines at the transcript level, THBS1 protein was not detected in the CM of FaDu cells possibly because of a modification at the epitope site (Fig. 3B); this protein was nonetheless readily detectable by MS with high confidence. In general, the relative transcript and protein expression levels of these markers determined by qRT-PCR and Western blotting exhibited a similar trend to the levels identified through global gene expression profiling and semiquantitative spectral counting, respectively. As the next step, we attempted to validate the expression of these markers in a three-dimensional tumor model of HNSCC using IHC in FaDu xenograft tumor tissue sections, and corroborated that all five proteins were indeed expressed in these tumors (supplemental Fig. S2).
Fig. 3.
In vitro verification of selected markers. A, Quantitative real-time PCR results show up-regulation of the selected genes in HNSCC cell lines in comparison to a normal oral epithelial cell line. B, Immunoblotting against the selected markers in serum-free conditioned media following 48-hour incubation. Membranes were stained with 0.1% (w/v) Ponceau S in 5% acetic acid to ensure equal sample loading (supplemental Fig. S1). Total number of spectra identified for selected candidates by mass spectrometry shown on the bottom of each blot. The upper band for THBS1 may be a protein isoform.
Clinical Validation of Five Proteins in Primary HNSCC Tissues
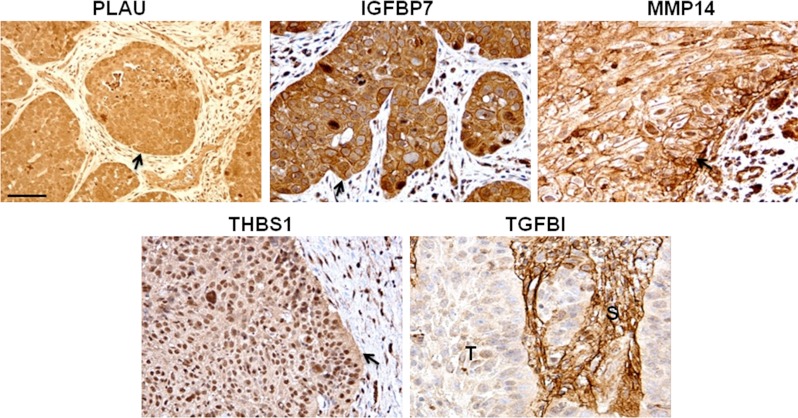
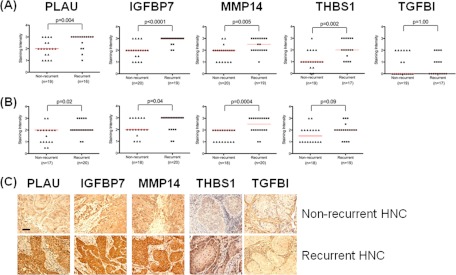
We next sought to determine whether these proteins were also expressed in primary human HNSCC tissues. IHC evaluations confirmed that all five proteins were indeed expressed in primary HNSCC biopsies (Fig. 4), albeit TGFBI expression appeared to be limited to the surrounding stroma, and not the cancer tissues. Nonetheless, the potential prognostic value of these five proteins was assessed. To that end, IHC for these five proteins were evaluated first on a training set (n = 40), followed by a validation cohort (n = 39), wherein half of these selected patients had relapsed, and were group matched for known clinical parameters including age, gender, stage at presentation, and treatment (Table I, supplemental Tables S2 and S3). For a few samples, the IHC staining could not be evaluated because of poor quality of the tissue or staining morphology, hence, accounting for the variations in the number of samples. IHC expression levels of PLAU, IGFBP7, MMP14, and THBS1 were significantly higher in recurrent versus nonrecurrent HNSCCs in the training set (p = 0.004, p < 0.0001, p = 0.005, p = 0.002, respectively) (Fig. 5A). Only TGFBI failed to be differentially-expressed between relapsed versus nonrelapsed samples; hence it was no longer evaluated in the validation cohort. Nonetheless, it was gratifying to observe that a similar pattern of IHC expression was corroborated in the second independent validation set (n = 40), (p = 0.02 for PLAU; p = 0.04 for IGFBP7; p = 0.004 for MMP14; and p = 0.09 for THBS1) (Fig. 5B).
Fig. 4.
Verification of selected markers in HNSCCs by immunohistochemistry. Representative images shown. Scale bar 50 μm. Arrows and (T) indicate tumor tissue; (S) indicates stroma. All markers except TGFBI were localized in the cytoplasm of tumor cells and had some degree of expression at the plasma membrane. In contrast to the other four markers, TGFBI was predominantly expressed in stromal cells and had no or weak expression in the majority of tumor cells.
Fig. 5.
Immunohistochemistry on pretreatment human HNSCC biopsies. A, Training set. B, Validation set. Wilcoxon rank-sum test was used to compare the median immunoexpression levels of biomarker candidates in nonrecurrent to recurrent HNSCCs (p values shown). C, Representative IHC staining images of nonrecurrent and recurrent HNSCCs. Scale bar 50 μm.
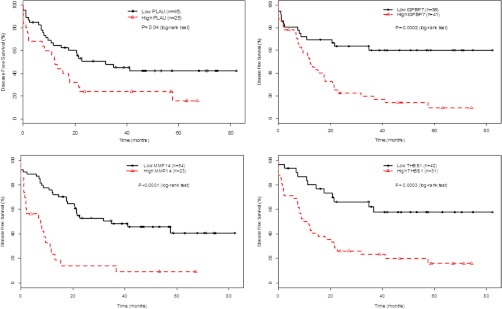
Next, the prognostic value of PLAU, IGFBP7, MMP14 and THBS1 was evaluated through estimation of OS and DFS using the Kaplan-Meier method (Fig. 6, supplemental Table S7). The IHC expression data were combined for both the training and validation cohorts in order to better gauge the potential prognostic value of these biomarkers. The patients were divided into two categories based on the median IHC staining intensity scores for each marker: high versus low expression. On multivariate analysis, patients' tumors with high expression level of any of these four proteins experienced a significantly worse DFS compared with those with low expression (p = 0.04, p = 0002, p = 0.0001, p = 0.0003 for PLAU, IGFBP7, MMP14, and THBS1, respectively). Furthermore, tumors with high IGFBP7 and THBS1 expression were also associated with lower OS rates (p = 0.005, p = 0.002 for IGFBP7 and THBS1, respectively) (supplemental Fig. 3). Accordingly, the respective hazard ratios for disease progression or death were also significantly higher for patients' whose tumors had high expression of these proteins (supplemental Table 7). Specifically, elevated IGFBP7 (HR = 3.17, p = 0.0007); MMP14 (HR = 3.95, p < 0.0001), or THBS1 (HR = 3.07, p = 0.0011) were associated with increased risk of disease progression. Similar associations were observed for increased risk of death for IGFBP7 (HR = 3.46; p = 0.004), and THBS1 (HR = 3.37; p = 0.01).
Fig. 6.
Kaplan-Meier estimates of disease-free survival among patients with HNSCC according to PLAU, THBS1, IGFBP7 and MMP14 expression. The patients were divided into high versus low expression based on the median IHC staining intensity scores for each marker. The comparison of the KM curves between the high (above-median) and low (below-median) protein expression groups was based on the log-rank test (p values shown).
Preliminary Validation in Patient-derived Plasma Samples
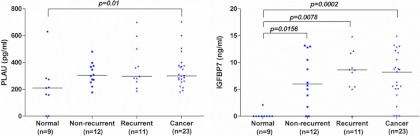
Given that these biomarkers were identified from the “secretome,” we next asked if any of these proteins could be measured in the plasma of HNSCC patients. Neither MMP14 nor THBS1 were differentially secreted between cancer patients and healthy volunteers (supplemental Fig. S4). However, PLAU, and IGFBP7 were both significantly over-expressed in HNC patients, compared with healthy volunteers (respective p values of p = 0.01 and p = 0.0002) (Fig. 7, Table IIA, supplemental Table S8). In particular, the median level of IGFBP7 was ∼35-fold higher in HNC patients, compared with almost undetectable levels in healthy volunteers (absent in eight out of nine controls), raising the intriguing possibility of IGFBP7 being a potential diagnostic marker for HNC.
Fig. 7.
Verification of PLAU and IGFBP7 in plasma of HNSCC patients and healthy controls. Wilcoxon rank-sum test was used to compare plasma levels (significant p values shown).
Among the 23 HNC patients tested for plasma IGFBP7 levels, 11 had already experienced a recurrence; 12 did not; there was a trend toward a higher plasma level of IGFBP7 in relapsed patients, although this did not reach statistical significance. When patients were stratified into high and low plasma PLAU or IGFBP7 expression based on the median concentration, however, patients with higher PLAU or IGFBP7 levels had a two-fold increased risk of death (HR = 2.03, p = 0.3 for PLAU and HR-2.00, p = 0.33 for IGBFP7) (supplemental Table S9).
DISCUSSION
Using a shotgun proteomics approach, starting with three HNSCC cell lines of laryngeal and hypopharyngeal origin, PLAU, IGFBP7, MMP14, and THBS1 were identified as four potential prognostic tumor tissue markers for clinical outcome. Furthermore, for the first time, PLAU and IGFBP7 were determined to be elevated in the plasma of HNC patients, with a trend toward higher risk of death from disease for those with higher circulating levels at time of initial diagnosis, validating the utility of such approaches in identifying potentially novel and useful biomarkers for HNC.
Analogous shotgun proteomics-driven approaches for identification and verification of secreted cancer biomarkers for various in vitro tumor models have been previously employed by others with numerous variations across groups in experimental design including the mode of sample preparation, mass spectrometry techniques, biomarker candidate selection and validation methods (15). Large scale validations examining the potential diagnostic or prognostic value of protein candidates utilizing clinically annotated patient samples have been limited and are one of the primary bottlenecks of secretome-based biomarker discovery, partly owing to limited access to patient samples and lack of commercially available ELISAs. Nonetheless, some groups have validated circulating biomarkers discovered with this approach to harbor promising clinical value (31, 32). Taken together, secretome studies demonstrated in principle that the cancer cell line secretome could be a valuable source of biomarkers, hence, we extended this approach to the discovery of laryngeal and hypopharyngeal carcinoma biomarkers.
The following protein candidates were verified to potentially harbor prognostic or diagnostic significance: PLAU, MMP14, IGFBP7, and THBS1. PLAU and MMP14 are proteases which promote tumor invasiveness and metastasis through their involvement in the degradation of the extracellular matrix. They have been previously implicated in HNC as well as other tumor types, and their inhibition has been explored for therapeutic purposes (17, 33–47). IGFBP7 is a tumor suppressor protein shown to be down-regulated in solid tumors of the liver, lung, breast, prostate, colon, and skin (48–50). There is evidence that it can also be up-regulated during specific stages of tumor development, e.g. in early-stage thyroid cancers; furthermore, its expression appears to be tissue-specific (51). To the best of our knowledge, the role or expression of IGFBP7 has not been previously investigated in HNC; however, in other solid tumor models, it has been shown to mediate cell growth and adhesion (50). Last, THBS1 is a secreted glycoprotein involved in tumor progression through regulation of extracellular matrix remodeling and angiogenesis; however, its role in HNC also remains to be fully elucidated, although THBS1 has been shown to promote tumor invasion via the urokinase receptor (52). These proteins appear to have highly diverse yet inter-related functions as they are all part of a complex extracellular matrix remodeling network. Furthermore, as suggested by the significant correlation found between the tumor immunoexpression of these markers (data not shown), they are likely promoting tumor progression co-operatively through extracellular matrix degradation, cell growth and angiogenesis. These are key biological processes occurring at the cell surface or the extracellular vicinity of tumor cells; thus, it serves as no surprise that proteins involved in these diverse processes were identified in this study.
Ralhan et al. reported the only secretome profiling of a similar HNC subsite, examining the CM of the human laryngeal SCC cell line (SCC38), in addition to oral SCC cell lines, which has resulted in fewer protein identifications, likely because of a different proteomics analysis strategy (53). We further extended our results by systematic validation in well-annotated patient tissue and plasma samples. Notably, nine proteins were commonly identified between the study by Ralhan et al. and the current study, including PLAU, IGFBP7, MMP14, and TGFBI, thereby supporting the findings presented here.
A significant effort was undertaken to validate our secretome-based results, by: (1) candidate up-regulation in vitro; (2) protein expression in FaDu xenografts; (3) IHC verification in two independent sets of primary HNSCC tissues of the same tumor types (laryngeal and hypopharyngeal SCCs); (4) evaluation of the prognostic value of these biomarkers; and (5) measurement of the secreted markers in plasma of HNSCC patients and controls. As a result, in a multivariate survival analysis, we observed that high tumor PLAU, IGFBP7, MMP14, and THBS1 expression correlated with inferior DFS, and increased risk of death and disease progression. Furthermore, plasma levels of PLAU and IGFBP7 may be used to distinguish subjects with and without cancer. Taken together, these data indicate that PLAU and IGFBP7 could be potentially utilized as diagnostic or prognostic biomarkers for squamous cell carcinomas of the larynx and the hypopharynx. Elevated plasma PLAU and IGFBP7 levels in HNSCC patients could also be an indication of increased risk of death; however, this requires validation in a larger cohort of HNSCC patient plasma samples in the future. No association was observed between plasma IGFBP7 and PLAU levels with disease progression or recurrence possibly because of the small sample size examined in each group. In addition, although all proteins were detected in plasma (pg/ml to ng/ml range), only PLAU and IGFBP7 demonstrated a significantly higher expression in the HNSCC patients. A similar differential expression pattern was not observed for MMP14 and THBS1, possibly because they are also secreted by additional cell types (e.g. stromal cells) thereby potentially masking the signal emanating from the tumor itself (37, 40).
Nevertheless, there remain limitations to this study, in that despite group-matching the clinical parameters to our best ability, there was a slight age bias with a younger population in the group of patients with recurrent HNSCC in the IHC training cohort. Similarly, although the training cohort included only stage III/IV patient tissues, the validation set also included stages I and II patient samples. The primary reason behind this imperfect matching is based on limited sample availability. Similarly, the plasma samples obtained from healthy subjects versus HNSCC patients were not age or gender-matched, with the latter being significantly older than the former and composed of higher proportion of males. These study limitations, along with inherent patient-to-patient, intra-tumoral heterogeneity, and modest patient sample sizes, are potential caveats to the current study; hence further validation using larger cohorts will be required before these biomarkers can be further advanced to guide clinical management (54). Nonetheless, considering the small cohort of plasma samples and the initial discovery of putative markers emanating from an in vitro cell culture model, these results are highly encouraging for future evaluations.
In conclusion, the cancer cell line secretome provides a rich source of proteins for biomarker discovery using a simplified model system. In the current study, this system enabled the identification of several novel biomarker candidates for HNSCC. Further validation efforts would be required to confirm the diagnostic and prognostic value of these proposed markers in plasma, and to evaluate their specificity and sensitivity in relation to additional clinical parameters. Finally, through a prospective longitudinal study, it would be important to determine whether PLAU and IGFBP7 might serve as biomarkers to monitor disease response, or serve as early indicators of disease relapse.
Supplementary Material
Footnotes
* This study was funded by the Canadian Institutes of Health Research (CIHR) to T.K. and FF.L. (MOP-114896). L.S. was supported by the CIHR Canada Graduate Scholarship (CGS) Master's Award and the Terry Fox Foundation Strategic Training Initiative for Excellence in Radiation Research of the 21st Century (EIRR21) at the Canadian Institutes of Health Research (CIHR). T.K. is supported by the Canada Research Chair Program. FF.L. is the Dr. Mariano Elia Chair for Head & Neck Oncology. We gratefully acknowledge the philanthropic support from the Wharton Family, Joe's Team, and Gordon Tozer. Support is also provided from the Campbell Family Institute for Cancer Research and the Ministry of Health and Long-term Planning.
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S9.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S9.
1 The abbreviations used are:
- HNSCC
- head and neck squamous cell carcinoma
- SCC
- squamous cell carcinoma
- CM
- conditioned media
- IHC
- immunohistochemistry
- WCL
- whole cell lysate
- MudPIT
- multidimensional protein identification technology
- HNC
- head and neck cancer
- NOE
- normal oral epithelial
- qRT-PCR
- quantitative real-time PCR
- DFS
- disease-free survival
- PLAU
- urokinase
- IGFBP7
- insulin-like growth factor binding protein 7
- FDR
- false discovery rate
- SpC
- spectral counting
- FC
- fold change
- HPA
- human protein atlas
- OS
- overall survival
- KM
- Kaplan–Meier
- HR
- hazard ratio
- CI
- confidence interval
- MMP14
- matrix metalloproteinase 14
- THBS1
- thrombospondin-1
- TGFBI
- transforming growth factor, beta-induced, 68kDa.
REFERENCES
- 1. Leemans C. R., Braakhuis B. J., Brakenhoff R. H. (2011) The molecular biology of head and neck cancer. Nat. Rev. Cancer 11, 9–22 [DOI] [PubMed] [Google Scholar]
- 2. Matta A., Ralhan R., DeSouza L. V., Siu K. W. (2010) Mass spectrometry-based clinical proteomics: head-and-neck cancer biomarkers and drug-targets discovery. Mass Spectrom. Rev. 29, 945–961 [DOI] [PubMed] [Google Scholar]
- 3. Schaaij-Visser T. B., Brakenhoff R. H., Leemans C. R., Heck A. J., Slijper M. (2010) Protein biomarker discovery for head and neck cancer. J. Proteomics 73, 1790–1803 [DOI] [PubMed] [Google Scholar]
- 4. de Jong M. C., Pramana J., Knegjens J. L., Balm A. J., van den Brekel M. W., Hauptmann M., Begg A. C., Rasch C. R. (2010) HPV and high-risk gene expression profiles predict response to chemoradiotherapy in head and neck cancer, independent of clinical factors. Radiother. Oncol. 95, 365–370 [DOI] [PubMed] [Google Scholar]
- 5. Tabor M. P., Brakenhoff R. H., Ruijter-Schippers H. J., Kummer J. A., Leemans C. R., Braakhuis B. J. (2004) Genetically altered fields as origin of locally recurrent head and neck cancer: a retrospective study. Clin. Cancer Res. 10, 3607–3613 [DOI] [PubMed] [Google Scholar]
- 6. Poeta M. L., Manola J., Goldwasser M. A., Forastiere A., Benoit N., Califano J. A., Ridge J. A., Goodwin J., Kenady D., Saunders J., Westra W., Sidransky D., Koch W. M. (2007) TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 357, 2552–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ang K. K., Harris J., Wheeler R., Weber R., Rosenthal D. I., Nguyen-Tân P. F., Westra W. H., Chung C. H., Jordan R. C., Lu C., Kim H., Axelrod R., Silverman C. C., Redmond K. P., Gillison M. L. (2010) Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 363, 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shi W., Kato H., Perez-Ordonez B., Pintilie M., Huang S., Hui A., O'Sullivan B., Waldron J., Cummings B., Kim J., Ringash J., Dawson L. A., Gullane P., Siu L., Gillison M., Liu F. F. (2009) Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J. Clin. Oncol. 27, 6213–6221 [DOI] [PubMed] [Google Scholar]
- 9. Bonner J. A., Harari P. M., Giralt J., Cohen R. B., Jones C. U., Sur R. K., Raben D., Baselga J., Spencer S. A., Zhu J., Youssoufian H., Rowinsky E. K., Ang K. K. (2010) Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 11, 21–28 [DOI] [PubMed] [Google Scholar]
- 10. Tonack S., Aspinall-O'Dea M., Jenkins R. E., Elliot V., Murray S., Lane C. S., Kitteringham N. R., Neoptolemos J. P., Costello E. (2009) A technically detailed and pragmatic protocol for quantitative serum proteomics using iTRAQ. J. Proteomics 73, 352–356 [DOI] [PubMed] [Google Scholar]
- 11. Kulasingam V., Diamandis E. P. (2007) Proteomics analysis of conditioned media from three breast cancer cell lines: a mine for biomarkers and therapeutic targets. Mol. Cell. Proteomics 6, 1997–2011 [DOI] [PubMed] [Google Scholar]
- 12. Wu C. C., Hsu C. W., Chen C. D., Yu C. J., Chang K. P., Tai D. I., Liu H. P., Su W. H., Chang Y. S., Yu J. S. (2010) Candidate serological biomarkers for cancer identified from the secretomes of 23 cancer cell lines and the human protein atlas. Mol. Cell. Proteomics 9, 1100–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sardana G., Jung K., Stephan C., Diamandis E. P. (2008) Proteomic analysis of conditioned media from the PC3, LNCaP, and 22Rv1 prostate cancer cell lines: discovery and validation of candidate prostate cancer biomarkers. J. Proteome Res. 7, 3329–3338 [DOI] [PubMed] [Google Scholar]
- 14. Gunawardana C. G., Kuk C., Smith C. R., Batruch I., Soosaipillai A., Diamandis E. P. (2009) Comprehensive analysis of conditioned media from ovarian cancer cell lines identifies novel candidate markers of epithelial ovarian cancer. J. Proteome Res. 8, 4705–4713 [DOI] [PubMed] [Google Scholar]
- 15. Makridakis M., Vlahou A. (2010) Secretome proteomics for discovery of cancer biomarkers. J. Proteomics 73, 2291–2305 [DOI] [PubMed] [Google Scholar]
- 16. Planque C., Kulasingam V., Smith C. R., Reckamp K., Goodglick L., Diamandis E. P. (2009) Identification of five candidate lung cancer biomarkers by proteomics analysis of conditioned media of four lung cancer cell lines. Mol. Cell. Proteomics 8, 2746–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Du B., Wang P., Guo X., Du B. (1999) Expression of membrane type 1-matrix metalloproteinase in laryngeal carcinoma. Pathol. Oncol. Res. 5, 214–217 [DOI] [PubMed] [Google Scholar]
- 18. Faça V. M., Ventura A. P., Fitzgibbon M. P., Pereira-Faca S. R., Pitteri S. J., Green A. E., Ireton R. C., Zhang Q., Wang H., O'Briant K. C., Drescher C. W., Schummer M., McIntosh M. W., Knudsen B. S., Hanash S. M. (2008) Proteomic analysis of ovarian cancer cells reveals dynamic processes of protein secretion and shedding of extra-cellular domains. PloS One 3, e2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Faça V. M., Hanash S. M. (2009) In-depth proteomics to define the cell surface and secretome of ovarian cancer cells and processes of protein shedding. Cancer Res. 69, 728–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Loei H., Tan H. T., Lim T. K., Lim K. H., So J. B., Yeoh K. G., Chung M. C. (2012) Mining the Gastric Cancer Secretome: Identification of GRN as a Potential Diagnostic Marker for Early Gastric Cancer. J. Proteome Res. 11, 1759–1772 [DOI] [PubMed] [Google Scholar]
- 21. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 22. Taylor P., Nielsen P. A., Trelle M. B., Hørning O. B., Andersen M. B., Vorm O., Moran M. F., Kislinger T. (2009) Automated 2D peptide separation on a 1D nano-LC-MS system. J. Proteome Res. 8, 1610–1616 [DOI] [PubMed] [Google Scholar]
- 23. Hui A. B., Lenarduzzi M., Krushel T., Waldron L., Pintilie M., Shi W., Perez-Ordonez B., Jurisica I., O'Sullivan B., Waldron J., Gullane P., Cummings B., Liu F. F. (2010) Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin. Cancer Res. 16, 1129–1139 [DOI] [PubMed] [Google Scholar]
- 24. Gortzak-Uzan L., Ignatchenko A., Evangelou A. I., Agochiya M., Brown K. A., St Onge P., Kireeva I., Schmitt-Ulms G., Brown T. J., Murphy J., Rosen B., Shaw P., Jurisica I., Kislinger T. (2008) A proteome resource of ovarian cancer ascites: integrated proteomic and bioinformatic analyses to identify putative biomarkers. J. Proteome Res. 7, 339–351 [DOI] [PubMed] [Google Scholar]
- 25. Sodek K. L., Evangelou A. I., Ignatchenko A., Agochiya M., Brown T. J., Ringuette M. J., Jurisica I., Kislinger T. (2008) Identification of pathways associated with invasive behavior by ovarian cancer cells using multidimensional protein identification technology (MudPIT). Mol. Biosyst. 4, 762–773 [DOI] [PubMed] [Google Scholar]
- 26. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protocols 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 27. Mathivanan S., Simpson R. J. (2009) ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000 [DOI] [PubMed] [Google Scholar]
- 28. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 29. Kuriakose M. A., Chen W. T., He Z. M., Sikora A. G., Zhang P., Zhang Z. Y., Qiu W. L., Hsu D. F., McMunn-Coffran C., Brown S. M., Elango E. M., Delacure M. D., Chen F. A. (2004) Selection and validation of differentially expressed genes in head and neck cancer. Cell Mol. Life Sci. 61, 1372–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cromer A., Carles A., Millon R., Ganguli G., Chalmel F., Lemaire F., Young J., Dembélé D., Thibault C., Muller D., Poch O., Abecassis J., Wasylyk B. (2004) Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene 23, 2484–2498 [DOI] [PubMed] [Google Scholar]
- 31. Kulasingam V., Zheng Y., Soosaipillai A., Leon A. E., Gion M., Diamandis E. P. (2009) Activated leukocyte cell adhesion molecule: a novel biomarker for breast cancer. Int. J. Cancer 125, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xue H., Lü B., Zhang J., Wu M., Huang Q., Wu Q., Sheng H., Wu D., Hu J., Lai M. (2010) Identification of serum biomarkers for colorectal cancer metastasis using a differential secretome approach. J. Proteome Res. 9, 545–555 [DOI] [PubMed] [Google Scholar]
- 33. Schafer J. M., Peters D. E., Morley T., Liu S., Molinolo A. A., Leppla S. H., Bugge T. H. (2011) Efficient targeting of head and neck squamous cell carcinoma by systemic administration of a dual uPA and MMP-activated engineered anthrax toxin. PloS One 6, e20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yasuda T., Sakata Y., Kitamura K., Morita M., Ishida T. (1997) Localization of plasminogen activators and their inhibitor in squamous cell carcinomas of the head and neck. Head Neck 19, 611–616 [DOI] [PubMed] [Google Scholar]
- 35. Myoung H., Kim M. J., Hong S. D., Lee J. I., Lim C. Y., Hong S. P. (2002) Expression of membrane type I-matrix metalloproteinase in oral squamous cell carcinoma. Cancer letters 185, 201–209 [DOI] [PubMed] [Google Scholar]
- 36. Shiomi T., Okada Y. (2003) MT1-MMP and MMP-7 in invasion and metastasis of human cancers. Cancer Metastasis Rev. 22, 145–152 [DOI] [PubMed] [Google Scholar]
- 37. Zhang H., Liu M., Sun Y., Lu J. (2009) MMP-14 can serve as a prognostic marker in patients with supraglottic cancer. Eur. Arch. Otorhinolaryngol. 266, 1427–1434 [DOI] [PubMed] [Google Scholar]
- 38. Suojanen J., Salo T., Koivunen E., Sorsa T., Pirila E. (2009) A novel and selective membrane type-1 matrix metalloproteinase (MT1-MMP) inhibitor reduces cancer cell motility and tumor growth. Cancer Biol. Ther. 8, 2362–2370 [DOI] [PubMed] [Google Scholar]
- 39. de Vicente J. C., Lequerica-Fernandez P., Santamaria J., Fresno M. F. (2007) Expression of MMP-7 and MT1-MMP in oral squamous cell carcinoma as predictive indicator for tumor invasion and prognosis. J. Oral. Pathol. Med. 36, 415–424 [DOI] [PubMed] [Google Scholar]
- 40. Rosenthal E. L., Matrisian L. M. (2006) Matrix metalloproteases in head and neck cancer. Head Neck 28, 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Imanishi Y., Fujii M., Tokumaru Y., Tomita T., Kanke M., Kanzaki J., Kameyama K., Otani Y., Sato H. (2000) Clinical significance of expression of membrane type 1 matrix metalloproteinase and matrix metalloproteinase-2 in human head and neck squamous cell carcinoma. Human Pathol. 31, 895–904 [DOI] [PubMed] [Google Scholar]
- 42. Kurahara S., Shinohara M., Ikebe T., Nakamura S., Beppu M., Hiraki A., Takeuchi H., Shirasuna K. (1999) Expression of MMPS, MT-MMP, and TIMPs in squamous cell carcinoma of the oral cavity: correlations with tumor invasion and metastasis. Head Neck 21, 627–638 [DOI] [PubMed] [Google Scholar]
- 43. Parolini S., Flagiello D., Cinquetti A., Gozzi R., Cristini S., Cappiello J., Nicolai P., Rusnati M., Presta M., Tosatti M. M. (1996) Up-regulation of urokinase-type plasminogen activator in squamous cell carcinoma of human larynx. Br. J. Cancer 74, 1168–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Strojan P., Budihna M., Smid L., Vrhovec I., Skrk J. (1998) Urokinase-type plasminogen activator (uPA) and plasminogen activator inhibitor type 1 (PAI-1) in tissue and serum of head and neck squamous cell carcinoma patients. Eur. J. Cancer 34, 1193–1197 [DOI] [PubMed] [Google Scholar]
- 45. Pasini F. S., Brentani M. M., Kowalski L. P., Federico M. H. (2001) Transforming growth factor beta1, urokinase-type plasminogen activator and plasminogen activator inhibitor-1 mRNA expression in head and neck squamous carcinoma and normal adjacent mucosa. Head Neck 23, 725–732 [DOI] [PubMed] [Google Scholar]
- 46. Zhang Z., Pan J., Li L., Wang Z., Xiao W., Li N. (2011) Survey of risk factors contributed to lymphatic metastasis in patients with oral tongue cancer by immunohistochemistry. J. Oral. Pathol. Med. 40, 127–134 [DOI] [PubMed] [Google Scholar]
- 47. Dass K., Ahmad A., Azmi A. S., Sarkar S. H., Sarkar F. H. (2008) Evolving role of uPA/uPAR system in human cancers. Cancer Treatment Rev. 34, 122–136 [DOI] [PubMed] [Google Scholar]
- 48. Chen D., Yoo B. K., Santhekadur P. K., Gredler R., Bhutia S. K., Das S. K., Fuller C., Su Z. Z., Fisher P. B., Sarkar D. (2011) Insulin-like growth factor-binding protein-7 functions as a potential tumor suppressor in hepatocellular carcinoma. Clin. Cancer Res. 17, 6693–6701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shersher D. D., Vercillo M. S., Fhied C., Basu S., Rouhi O., Mahon B., Coon J. S., Warren W. H., Faber L. P., Hong E., Bonomi P., Liptay M. J., Borgia J. A. (2011) Biomarkers of the insulin-like growth factor pathway predict progression and outcome in lung cancer. Annals Thoracic Surg. 92, 1805–1811; discussion 1811 [DOI] [PubMed] [Google Scholar]
- 50. Hwa V., Oh Y., Rosenfeld R. G. (1999) Insulin-like growth factor binding proteins: a proposed superfamily. Acta Paediatr. Suppl. 88, 37–45 [DOI] [PubMed] [Google Scholar]
- 51. Vizioli M. G., Possik P. A., Tarantino E., Meissl K., Borrello M. G., Miranda C., Anania M. C., Pagliardini S., Seregni E., Pierotti M. A., Pilotti S., Peeper D. S., Greco A. (2011) Evidence of oncogene-induced senescence in thyroid carcinogenesis. Endocrine-related Cancer 18, 743–757 [DOI] [PubMed] [Google Scholar]
- 52. Albo D., Tuszynski G. P. (2004) Thrombospondin-1 up-regulates tumor cell invasion through the urokinase plasminogen activator receptor in head and neck cancer cells. J. Surg. Res. 120, 21–26 [DOI] [PubMed] [Google Scholar]
- 53. Ralhan R., Masui O., Desouza L. V., Matta A., Macha M., Siu K. W. (2011) Identification of proteins secreted by head and neck cancer cell lines using LC-MS/MS: Strategy for discovery of candidate serological biomarkers. Proteomics 11, 2363–2376 [DOI] [PubMed] [Google Scholar]
- 54. Gerlinger M., Rowan A. J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P., Varela I., Phillimore B., Begum S., McDonald N. Q., Butler A., Jones D., Raine K., Latimer C., Santos C. R., Nohadani M., Eklund A. C., Spencer-Dene B., Clark G., Pickering L., Stamp G., Gore M., Szallasi Z., Downward J., Futreal P. A., Swanton C. (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.