Abstract
Parasitic liver flukes of the family Fasciolidae are responsible for major socioeconomic losses worldwide. However, at present, knowledge of the fundamental molecular biology of these organisms is scant. Here, we characterize, for the first time, the transcriptome and secreted proteome of the adult stage of the “giant liver fluke,” Fascioloides magna, using Illumina sequencing technology and one-dimensional SDS-PAGE and OFFGEL protein electrophoresis, respectively. A total of ∼54,000,000 reads were generated and assembled into ∼39,000 contiguous sequences (contigs); ∼20,000 peptides were predicted and classified based on homology searches, protein motifs, gene ontology, and biological pathway mapping. From the predicted proteome, 48.1% of proteins could be assigned to 384 biological pathway terms, including “spliceosome,” “RNA transport,” and “endocytosis.” Putative proteins involved in amino acid degradation were most abundant. Of the 835 secreted proteins predicted from the transcriptome of F. magna, 80 were identified in the excretory/secretory products from this parasite. Highly represented were antioxidant proteins, followed by peptidases (particularly cathepsins) and proteins involved in carbohydrate metabolism. The integration of transcriptomic and proteomic datasets generated herein sets the scene for future studies aimed at exploring the potential role(s) that molecules might play at the host–parasite interface and for establishing novel strategies for the treatment or control of parasitic fluke infections.
Parasitic liver flukes (Platyhelminthes: Trematoda) of livestock, such as Fasciola hepatica and Fasciola gigantica, are responsible for major economic losses worldwide, estimated at USD ∼3 billion, due to morbidity, mortality, and decreased productivity (1–3). The giant liver fluke, Fascioloides magna, infects a range of wild and domestic ruminants (e.g. cervids and bovids), primarily in North America and Europe (4). The life cycle of F. magna is indirect; eggs are released by mature flukes and excreted in the feces of the mammalian host. In aerated water, miracidia hatch from the eggs and penetrate the body of a susceptible, aquatic intermediate snail host (e.g. Fossaria parva or Fo. modicella in North America; Galba truncatula in Europe) within ∼2 h (5). In the intermediate host, the parasite develops through the stages of sporocysts, rediae, and cercariae; the latter larval stage emerges from the snail within ∼40–58 days (4). The cercariae encyst as metacercariae on submerged or emergent vegetation and are then ingested by a mammalian host. Once in the host, the metacercariae excyst and the juvenile flukes penetrate the intestinal walls and migrate to the liver, where they are encapsulated (in pairs) in pseudocysts formed by the hepatic parenchyma and then develop into mature flukes (4). The migration of the immature stages through the hepatic tissues, together with the large number of pseudocysts and the size of mature flukes (up to 8 cm in length (4)), can result in liver fibrosis. Clinical signs associated with infection by F. magna include lethargy, anorexia, depression, and weight loss, with sudden death occurring in heavily infected animals (4).
In livestock, the control of liver fluke infections has relied predominantly on treatment with anthelmintic drugs, such as closantel, oxyclozanide, and triclabendazole (6). Triclabendazole is considered the drug of choice against both juvenile and adult stages of liver flukes in the definitive (mammalian) host, whereas other compounds affect only the adult stage (7, 8). Thus, triclabendazole is widely and often excessively used for the treatment of trematodiases in livestock (6, 9), and this leads to a significant risk that drug resistance will develop. Indeed, there are recently published reports of triclabendazole resistance in Fa. hepatica populations in Australia (10) and in Western European countries (11–15). In addition, despite major efforts in studies aimed at developing novel intervention strategies against liver flukes (16–23), there is still a paucity of information on host–parasite interactions at the molecular level. Recent studies (3, 24, 25) have provided the first insights into the molecular biology of fasciolids by exploring the transcriptomes of the adult stages of both Fa. hepatica and Fa. gigantica (3, 25) and of the composition of the excretory/secretory (ES)1 products from both the juvenile and adult stages of Fa. hepatica (24). In these studies, proteolytic enzymes (e.g. cathepsins, asparaginyl endopeptidase cysteine proteases, and trypsin-like serine proteases) were identified as key molecules in both the transcriptome and ES products, which are likely to play key roles in parasite migration through tissues and in the modulation of immune responses in the mammalian host. Given the potential of proteolytic enzymes as vaccine candidates against trematodiases (18–20, 26), comparative analyses of the transcriptomes and ES products of liver flukes is of critical importance for an improved understanding of their molecular biology, as well as for developing new treatment and control strategies against them.
Advances in “-omic” and computational technologies for the preprocessing, assembly, and annotation of sequence data (27–29) are substantially assisting studies of the transcriptomes and proteomes of parasitic helminths (3, 28–32). In the present study we explored, for the first time on a large scale, the transcriptome of adult F. magna, using Illumina-based sequencing technology and bioinformatic analyses of sequence data, and we characterized the protein components of the ES products from this developmental stage. This insight into the molecular biology of F. magna offers unprecedented opportunities for comparative investigations of various economically important liver flukes and the design of new interventions against these parasites.
EXPERIMENTAL PROCEDURES
Procurement of Parasite Material, RNA Isolation, and Illumina Sequencing
Adult F. magna were collected from naturally infected livers of red deer (Cervus elaphus) (from the Brdy mountains, Czech Republic) and washed in 0.1 m phosphate-buffered saline (PBS), pH 7.2, at 37 °C. Total RNA was extracted from three individual adults of F. magna using TriPure reagent (Roche, Switzerland) and DNase I-treated (25). RNA amounts were estimated spectrophotometrically (NanoDrop Technologies, USA), and RNA integrity was verified using a 2100 BioAnalyser (Agilent, Santa Clara, CA). Polyadenylated (poly(A)+) RNA was purified from 10 μg of total RNA using Sera-mag oligo(dT) beads, fragmented to a length of 100 to 500 bp, reverse- transcribed using random hexamers, and end-repaired and adaptor-ligated according to the manufacturer's protocol (Illumina). Ligated products of ∼300 bp were excised from agarose and PCR-amplified (15 cycles) (25). Products were cleaned using a MinElute column (Qiagen, Netherlands) and paired-end sequenced on a Genome Analyzer II (Illumina) (33), according to the manufacturer's protocols.
Bioinformatic Analyses of Transcriptomic Sequence Data
The 100 bp single-read sequences generated from the non-normalized cDNA library representing the adult stage of F. magna were assembled using the program Oases v0.1.22 (http://www.ebi.ac.uk/∼zerbino/oases/; (34)). Adapter sequences and sequences with suboptimal read quality (i.e. PHRED score < 32.0) were eliminated. The remaining sequences (99%) were used to construct a de Bruijn graph using a k-mer value of 43 bp. Because de novo transcriptome assemblies of short-read sequence libraries can potentially lead to an underrepresentation of members of large families of molecules characterized by a high degree of sequence similarity, such as the cathepsins (25), reads with a high degree of sequence homology (98% nucleotide identity) to the conserved cathepsin propeptide inhibitor domain (I29; 177 nucleotides, nt) of a cathepsin L sequence from F. magna (GenBank protein accession number ACG50798; nucleotide EU877764.1) were used to generate contigs from the paired-end sequence datasets employing the PRICE program (http://derisilab.ucsf.edu). Eight cycles of contig extension were performed, with each new cycle using contigs (>500 nt in length) generated from the previous assembly as a template. The identity of the assembled contigs was then verified via BLASTn and BLASTx comparisons with nucleotide and amino acid sequences of trematode cathepsins, respectively, available from public databases (e.g. www.ncbi.nlm.nih.gov). The raw sequence reads from the cDNA library were then mapped to the nonredundant sequence data using the program SOAP2 (35). In brief, raw reads were aligned to the assembled, nonredundant transcriptomic data, such that each read was mapped to a unique transcript. Reads that mapped to more than one transcript (called “multi-reads”) were randomly allocated to a unique transcript, such that they were recorded only once. To provide a relative assessment of transcript abundance, the numbers of raw reads that mapped to individual contigs were normalized for sequence length (i.e. reads per kilobase per million reads (36)).
The nonredundant transcriptomic dataset for F. magna was then analyzed using an established approach (25, 37). Briefly, assembled contigs were compared (using BLASTn and BLASTx algorithms) with (i) sequences available in public databases, including that of NCBI (www.ncbi.nlm.nih.gov) and the EMBL-EBI Parasite Genome Blast Server (www.ebi.ac.uk); (ii) transcriptomic sequence data available for the related liver flukes Fa. hepatica, Fa. gigantica (3, 25), Clonorchis sinensis, and Opisthorchis viverrini (32) (http://www.gasserlab.org/); and (iii) the entire genome sequences of the blood flukes Schistosoma hematobium (38), S. japonicum (39), and S. mansoni (40) (http://www.genedb.org/; e-value cut-off: <10−5).
Proteins were conceptually translated from the predicted coding domains of individual transcriptomic sequences and their functions were predicted using InterProScan (41), employing the default search parameters. Based on their homology to conserved domains and protein families, proteins predicted for F. magna were assigned parental (i.e. level 2) gene ontology (GO) terms (i.e. “biological process,” “cellular component,” and “molecular function”) (http://www.geneontology.org/) (42) and displayed using the WEGO tool (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) (43). Inferred proteins with homologues in other organisms were mapped to conserved biological pathways utilizing the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology-Based Annotation System v.2 (44). Signal peptides were also predicted using the program SignalP 3.0, employing the neural network and hidden Markov models (45), and transmembrane domains were inferred using the program TMHMM (www.cbs.dtu.dk/services/TMHMM/) (46–48). ES peptides were predicted based on the presence of a signal peptide and sequence homology to one or more known proteins listed in the Secreted Protein (http://spd.cbi.pku.edu.cn/) (49) and/or Signal Peptide (http://proline.bic.nus.edu.sg/spdb/index.html) (50) databases.
Isolation of ES Products
Adult F. magna (n = 15) were washed three times in large volumes of PBS and incubated for 6 h at 37 °C in sterile RPMI 1640 medium (Sigma) containing 100 IU/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, and 250 μg/ml amphotericin B (Sigma-Aldrich). Following incubation, the supernatant was centrifuged twice at 4500 g for 20 min at 2 °C, and the ES products were concentrated using Amicon Ultra-15 filters (10 kDa cut-off; Millipore, USA). Protein amounts were measured using the Quant-iT Protein Assay Kit (Invitrogen); 1 ml of ES product (concentration = 1 mg/ml) was lyophilized and stored at −20 °C until analysis.
Electrophoresis and In-gel Digestion
Two 10 μl aliquots of F. magna ES products (9 mg/ml) were incubated for 2 h at 37 °C with an equal volume of sample buffer (51). The samples were then applied to a 0.75-mm-thick, 4% stacking, and 12.5% resolving gel (prepared using the Bio-Rad PROTEAN 3) for SDS-PAGE (51). Electrophoresis was carried out using maxima of 40 mA/gel and 300 V. Proteins were stained using Coomassie Brilliant Blue and destained in 50:8.75:41.25 methanol:acetic acid:water (v:v:v). Lanes containing ES proteins were finely sliced. The gel slices were then destained twice in 200 μl of 50% acetonitrile and 25 mm NH4HCO3 for 15 min at 37 °C, desiccated using a vacuum centrifuge, resuspended in 20 mm dithiothreitol (DTT), and reduced for 1 h at 65 °C. The DTT was then removed, and the samples were alkylated by adding 1 m iodoacetamide (IAA) to a final concentration of 50 mm and incubated at 22 °C in darkness for 40 min. Gel slices were washed three times for 45 min in 25 mm NH4HCO3 and then desiccated. Individual dried slices were then allowed to swell in 20 μl of 40 mm NH4HCO3 and 10% acetonitrile containing 20 μg/ml trypsin (Sigma) for 1 h at 22 °C. An additional 50 μl of the same solution was added to the samples, and they were then incubated overnight at 37 °C. The supernatant was then removed from the gel slices, and residual peptides were washed from the slices by incubating them three times in 0.1% trifluoroacetic acid for 45 min at 37 °C. The original supernatant and extracts were combined and reduced to 10 μl in a vacuum centrifuge before mass spectral analysis.
OFFGEL Electrophoresis
For peptide separation, based on the isoelectric point, 240 μg of ES products were reduced by adding DTT to 20 mm and 10% SDS to 2% (w/v) and incubating at 65 °C for 1 h. Alkylation was then achieved by adding IAA to 50 mm and incubating the solution for 40 min in darkness at 22 °C. The sample was then co-precipitated with 1 μl of a 1 μg/μl solution of trypsin by adding nine volumes of methanol at −20 °C. After being incubated overnight at −20 °C, the sample was resuspended in 25 mm NH4HCO3 and incubated at 37 °C for 5 h with the addition of 1 μg of trypsin after 3 h. The 3100 OFFGEL fractionator and OFFGEL kit (pH 3–10; 24-well format) (Agilent Technologies) were prepared according to the manufacturer's protocols. The ES product digests and undigested proteins were diluted in the peptide- and protein-focusing buffers, respectively, to a final volume of 3.6 ml, and 150 μl of sample was loaded into individual wells. The samples were focused in a maximum current of 50 μA until 50 kilovolt hours were reached. Peptide fractions were harvested and desiccated using a vacuum centrifuge, resuspended in 25 mm NH4HCO3, and desiccated again before mass spectral analysis.
Protein Identification Using LC-MS/MS-MS
OFF-GEL electrophoresis (OGE) fractions and tryptic fragments from in-gel digests were separated chromatographically via HPLC (Dionex Ultimate 3000) using a Zorbax 300SB-C18 column (3.5 μm, 150 mm × 75 μm; Agilent Technologies) and a linear gradient of 0% to 80% solvent B for 60 min. A preconcentration step (3 min) was performed employing a Dionex μ-Precolumn cartridge (C18, 5 μm, 300 μm × 5 mm) before commencement of the gradient. A flow rate of 300 nl/min was used for all experiments. The mobile phase consisted of solvent A (0.1% formic acid (aq)) and solvent B (80/20 acetonitrile/0.1% formic acid (aq)). Eluates from the RP-HPLC column were directly introduced into the NanoSpray II ionization source of a QSTAR Elite Hybrid MS/MS System (Applied Biosystems, USA) operated in positive ion electro-spray mode. All analyses were performed using information dependant acquisition. Analyst 2.0 (Applied Biosystems) was used for data analysis. Briefly, the acquisition protocol consisted of the use of an enhanced mass spectrum scan. The three most abundant ions detected (above background) were subjected to examination using an enhanced resolution scan to confirm the charge state of the multiply charged ions. The ions with a charge state of +2 or +3 and those with an unknown charge were then subjected to collision-induced dissociation using a rolling collision energy, dependent upon the m/z and the charge state of the ion. Enhanced product ion scans were acquired, resulting in full product ion spectra for each of the selected precursors, which were then used for subsequent database searches.
Bioinformatic Analyses of Proteomic Sequence Data
Protein sequences from mass spectrometric analyses were compared to peptide sequences predicted from transcriptomic data and to proteins available from the SwissProt database (May 2011) using Mascot v.2.3.02 (http://www.matrixscience.com), employing the following search parameters: enzyme = trypsin; precursor ion mass tolerance = ±0.1; fixed modifications = methionine oxidation; variable modifications = carbamidomethylation; number of missed cleavages allowed = 2; charge states = +2 and +3. The results from the Mascot searches were validated using the program Scaffold v.3.00.06 (Proteome Software Inc., USA) (52). Peptides and proteins were identified using the Peptide Prophet algorithm (53), using a probability cutoff of 95.0% (peptides) or 99.0% probability (proteins), and contained at least two identified peptides (proteins) (54). Proteins containing similar peptides that could not be differentiated based on MS/MS analysis were grouped to satisfy the principles of parsimony. A false discovery rate of 0.1% was calculated using protein identifications validated using the Scaffold program. Relative protein abundance was determined by means of normalized spectral counting (52), and proteins encoded by transcripts were annotated by BLASTx comparisons (bit-score cutoff: >30) with data available in the nonredundant protein databases (www.ncbi.nlm.nih.gov). Proteins were also classified according to GO categories using the program InterProScan (41), and putative signal peptides and transmembrane domain(s) were predicted using the programs SignalP (55) and TMHMM (47). Putative mannose 6-phosphate glycosylation sites were identified using the NetNGlyc server (56).
RESULTS
The Transcriptome of F. magna
A total of 54,004,008 Illumina reads were produced for the adult stage of F. magna (GenBank Sequence Read Archive accession number SRA052828) (Table I). The assembly of the raw reads, together with the PRICE-assembly of cathepsin-encoding contigs (n = 9), yielded 38,934 assembled contigs (843 ± 1005 bases in length; range: 101–13,180) (Table I). A total of 26,616,266 reads (∼49.3% of all raw reads) could be remapped to the assembled contigs, with a mean depth of coverage of 847.19 ± 30,910.17 reads per sequence. The remaining 27,387,742 reads (i.e. singletons and reads of suboptimal quality) were excluded from any further analysis. The numbers of assembled F. magna contigs matching known Fa. hepatica and Fa. gigantica homologues and of contigs matching C. sinensis, O. viverrini, S. hematobium, S. japonicum, and S. mansoni homologues are listed in Table I. In total, 20,140 peptides were predicted from the transcriptome of F. magna (Table I); 11,166 of those (55.4%) had the highest homology to proteins predicted for Fa. hepatica, followed by those from Fa. gigantica (n = 10,854; 53.9%), O. viverrini (n = 9739; 48.3%), and S. mansoni (n = 9376; 46.5%) (Table I).
Table I. Summary of the nucleotide sequence data for adult Fascioloides magna prior and following assembly, and detailed bioinformatic annotation and analyses.
| Raw reads (paired-end) | 54,004,008 |
| Contigs (average length ± S.D.; min–max length) | 38,934 (843 ± 1005; 101–13,180) |
| GC content (%) | 44.04 |
| Raw reads mapped to contigs (%) | 26,616,266 (49.3) |
| Containing an open reading frame (%) | 20,140 (52) |
| With homologues in Fasciola hepatica (%) | 25,405 (65.2) |
| Fasciola gigantica | 23,025 (59.1) |
| Clonorchis sinensis | 5488 (14.1) |
| Opisthorchis viverrini | 7533 (19.3) |
| Schistosoma mansoni | 4147 (10.6) |
| Schistosoma japonicum | 3966 (10.2) |
| Schistosoma haematobium | 3941 (10.1) |
| Returning InterProScan results (%) | 15,414 (76.5) |
| Number of InterPro terms | 2965 |
| Gene ontology (%) | 7127 (35.4) |
| Number of biological process terms (level 2) | 379 |
| cellular component | 140 |
| molecular function | 533 |
| Returning a KOBAS result (%) | 7811 (38.8) |
| Number of predicted biological pathways (KEGG) | 235 |
| Predicted proteins with signal peptides (%) | 835 (4.1) |
| transmembrane domains (%) | 2854 (14.2) |
| Homologous to proteins in the SPD database (%) | 4087 (20.3) |
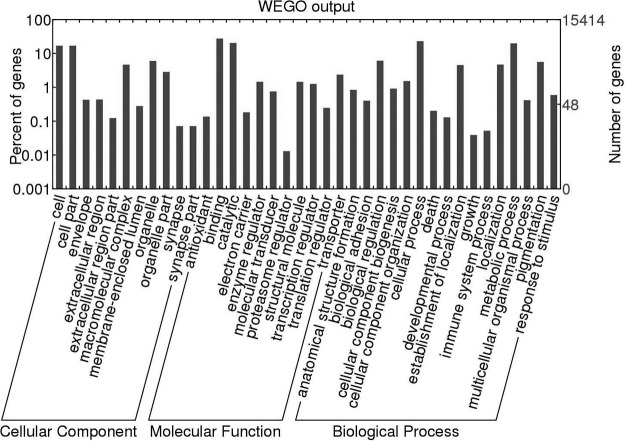
Proteins inferred from the transcriptome of F. magna were then categorized according to the presence of conserved (Pfam and/or InterPro) domains/signatures; 15,414 (76.5%) sequences could be mapped to known proteins, characterized by 2965 different conserved domains (Table I). Of the 20,140 proteins predicted for F. magna, 35.4% could be assigned to 379 “biological process,” 140 “cellular component,” and 533 “molecular function” parental (level 2) terms (Table I). The predominant terms were “cellular process” and “metabolic process” for “biological process” (15.7% and 10.1%, respectively), “cell” and “cell part” for “cellular component” (28.6% and 13.3%, respectively), and “ATP binding” and “catalytic binding” for “molecular function” (36.4% and 34.8%, respectively) (Fig. 1). From the proteome predicted for F. magna, 9690 (48.1%) proteins mapped to homologous proteins in the KEGG database, which were assigned to 384 biological pathway terms (Table I), including “spliceosome” (n = 105 molecules), “RNA transport” (86), and “endocytosis” (76) (supplemental Table S1). Putative proteins involved in amino acid degradation (i.e. cathepsins, n = 15) were most abundant (Fig. 2), in accordance with previous studies of the transcriptomes of other parasitic trematodes, including Fa. hepatica (3, 24), C. sinensis (see Refs. 57–60), O. viverrini,2 and Paragonimus westermani (61). A total of 835 (4.1%) ES proteins were predicted from the transcriptome of F. magna (Table I), 4% of which were inferred to be peptidases (n = 32), including cathepsin B and L cysteine proteases (n = 7) (supplemental Table S2).
Fig. 1.
Gene ontology. Bar graph displaying the gene ontology terms (according to the categories “cellular component,” “molecular function,” and “biological process”) assigned to peptides inferred from transcriptomic data for Fascioloides magna and plotted using the WEGO tool (43).
Fig. 2.
Transcription profiling. Bar graph showing the 25 most abundant proteins encoded in the transcriptome of Fascioloides magna.
Proteomic Analysis of ES Proteins
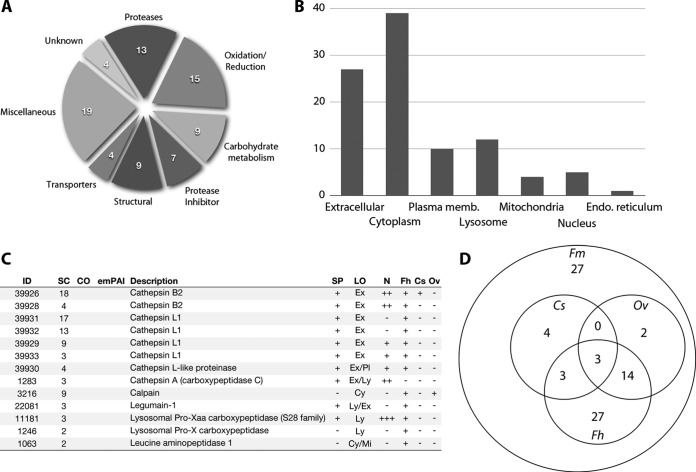
F. magna ES proteins were subjected to SDS-PAGE, and the protein constituents were identified using tandem mass spectroscopy. More than 37,000 spectra from in-gel digests and OGE analysis were used for Mascot searches of the transcriptome. Using Scaffold, 80 proteins were identified by at least two peptides at a 99.0% probability and with an estimated false discovery rate of 0.1% (supplemental Tables S3 and S4). Analysis of SDS-PAGE gels yielded 79 protein identifications, with OGE contributing to one additional identification (supplemental Table S4). The proteins identified were assigned to functional categories on the basis of InterPro domains and/or GO categories. The largest functional group represented antioxidant proteins, followed by peptidases and proteins linked to carbohydrate metabolism; approximately one-quarter of the proteins identified could not be assigned to a major functional group, and they were thus grouped into a “miscellaneous” group (Fig. 3A). In accordance with respective functional categories, GO terms highly represented among the proteins included “cysteine-type peptidase activity,” “oxidoreductase activity,” “serine-type endopeptidase inhibitor activity,” and “carbohydrate metabolic process.” Using the spectral counting method included in Scaffold, the cysteine protease inhibitor cystatin was found to be the most abundant protein in the ES products, followed by two cathepsin L1 proteases, cathepsin B, calpain, and ferritin. Cathepsin L was the most abundant proteinase identified in the ES products, with four different isoforms identified, as well as a cathepsin L-like protease. Similarly, multiple isoforms of cathepsin B2 and fatty-acid-binding protein were identified. In all cases, isoforms were reported only if at least one unique peptide (which scored significantly) was assigned to the isoform (see Fig. 4). Based on SignalP and TMHMM analyses (47, 55), ∼30% of the identified proteins were inferred to contain a classical secretory signal peptide and represented mainly proteases and proteins associated with carbohydrate metabolism. Six proteins were predicted to contain more than two transmembrane helices, indicative of a membrane-bound protein. Protein localization was predicted using PSORT (62). The majority of ES products were predicted to be either extracellular (= 27) or cytoplasmic (= 39), with smaller numbers inferred to be localized or co-localized to the nucleus, plasma membrane, and/or mitochondria (Fig. 3B). Twelve proteins were assigned a lysosomal localization; to further validate this finding, all proteins with a predicted signal sequence were analyzed for the presence of potential mannose 6-phosphate glycosylation sites. Of the 23 proteins with predicted signal peptides, 10 proteins had a glycosylation probability of >0.75, and 6 had a probability of >0.50. Proteins inferred to be glycosylated included known lysosomal proteins, such as Pro-Xaa carboxypeptidase, alpha-mannosidase, alpha-glucosidase, and cathepsins A and B2.
Fig. 3.
Overview of proteins identified in the excretory/secretory (ES) products of Fascioloides magna. A, Functional categories of the 80 proteins identified in the ES products of F. magna. Antioxidant proteins represented the largest functional group identified, followed by proteases, proteins involved in carbohydrate metabolism, and protease inhibitors. Four proteins could not be assigned to any functional category, and ∼25% were assigned to miscellaneous functional categories. B, Putative subcellular localizations of the identified proteins. Locations were predicted using PSORT (62) and based on evidence from the peer-reviewed literature. C, Proteases identified in the ES of F. magna. Abbreviations: SC, number of unique peptides assigned to the protein; CO, percent cover; LO, the putative subcellular localization of the protein. The presence of a signal peptide (SP) or an N-linked glycosylation site (N) is denoted by “+”; the presence of the protein in the ES of adult Fasciola hepatica (Fh), Clonorchis sinensis (Cs), or Opisthorchis viverrini (Ov) is also denoted by “+.” D, Venn diagram depicting the number of similar proteins identified in the ES of adult F. magna (Fm), Fa. hepatica (Fh), O. viverrini (Ov), and C. sinensis (Cs).
Fig. 4.
Cathepsin L1. Sequence alignment of four cathepsin L1 isoforms and a cathepsin l-like protease (39930) identified in the excretory/secretory (ES) products of Fascioloides magna. Peptides (assigned using Mascot) representing each isoform or multiple isoforms are highlighted in green and red, respectively. Each amino acid sequence corresponding to the mature protein is delimited by a solid black line.
Proteases represented a significant proportion (∼16%) of the proteins identified in the ES products. Thirteen proteases were identified in these products: nine cysteine proteases, including cathepsin B and L, calpain, and legumain; three carboxypeptidases, including cathepsin A; and a leucine aminopeptidase (Fig. 3C). Cathepsins were the most represented proteins, with examples of cathepsins A, B, and L identified. Based on BLAST-comparisons, all cathepsin L proteins identified in the ES products shared significant homology with Fa. hepatica or F. magna cathepsin L1, in accordance with the results from the analysis of the F. magna transcriptome (see Fig. 2). In addition to cathepsins identified by proteomic means, cathepsins D and L3 and a cathepsin B-like protease, the corresponding transcripts of which were detected in the F. magna transcriptome, were not found in the ES proteome. The cathepsins identified correlated with the most abundantly expressed transcripts, suggesting that unidentified proteases were expressed at levels below the detection limit of the mass spectrometric method used (Table II). Analysis of the peptides detected in Mascot searches showed that only one peptide representing cathepsin L (ID: 39933) (see Fig. 4) originated from the pro-region of the protease, indicating that these proteases were predominantly present in an activated form. Conversely, three pro-region peptides were identified from one of the cathepsin B2 isoforms (ID: 39926) (supplemental Fig. S1), suggesting the presence of inactive forms of this protease.
Table II. Numbers of cathepsin-encoding transcripts and corresponding proteins identified in the transcriptome and excretory/secretory products of Fascioloides magna, respectively.
| Transcripts | Proteins | |
|---|---|---|
| Cathepsin L1 | 18 | 4 |
| Cathepsin L-like protease | 2 | 1 |
| Cathepsin L3 | 3 | 0 |
| Cathepsin B2 | 18 | 2 |
| Cathepsin B-like protease | 1 | 0 |
| Cathepsin A | 9 | 1 |
| Cathepsin D | 1 | 0 |
| Total | 70 | 8 |
The ES products of F. magna were compared with those characterized in previous studies of Fa. hepatica (24, 63–66), C. sinensis (67), and O. viverrini (68). Approximately 65% of the proteins identified here were also detected in at least one of these flukes, but only enolase, actin, and triose-phosphate isomerase were identified in all three species. The ES proteomes of F. magna and Fa. hepatica were most similar, with 47 of 80 proteins identified here found in both species; a major difference between these proteomes was the identification of five isoforms of cathepsin L1 in F. magna and ten in Fa. hepatica. The 27 proteins unique to F. magna were less abundant and membrane-bound and/or structural, although eight of them contained a secretory signal peptide. Apart from the difference in cathepsin L1 isoform numbers, the protease profile of F. magna differed from that of Fa. hepatica only by the presence of cathepsin A. However, the protease profiles of both F. magna and Fa. hepatica (Fasciolidae) were markedly different from those of C. sinsensis and O. viverrini (Opisthorchiidae) (67, 68), with only cathepsin B2 and legumain detected in the ES products of both families (Figs. 3C and 3D).
DISCUSSION
The present study provides a comprehensive snapshot of the transcriptome and the ES proteome of the adult stage of F. magna and represents an invaluable resource for fundamental investigations of the molecular biology of liver flukes that is of both veterinary and public health importance. Almost two-thirds of F. magna transcripts sequenced in the present study were similar to molecules identified in the transcriptomes of Fa. hepatica and Fa. gigantica, respectively (3, 25), which is indicative of the biological similarities among members of the family Fasciolidae. Most abundant in the transcriptome of F. magna were molecules containing a predicted signal peptide; this finding is in accordance with the results of a previous study of the transcriptome of Fa. gigantica (25) and likely reflects the crucial role(s) that secreted proteins play in the biology of these organisms (69). Of the 835 predicted proteins with a signal peptide in F. magna, 80 were identified in the ES products from this organism at a confidence level of 99%, in accordance with previous proteomic analyses of the ES products of other liver flukes (i.e. 20 to 90 proteins identified) (24, 66–68). This finding suggests that, in addition to peptides that were undetectable because of their low molecular weight and concentration and/or possible “endogenous” secretion, most proteins in ES products of F. magna were identified. In a previous study of the transcriptome and secreted proteome of Fa. hepatica, Robinson et al. (24) also observed a discrepancy between the number of secreted proteins predicted from transcriptomic data and that of proteins identified in the ES products from this species. In the present study, a total of 42 transcripts encoding cathepsins (e.g. cathepsins B and L) were detected in the transcriptome of F. magna, of which 7 were predicted to contain a signal peptide indicative of secretion (see supplemental Table S2), and 8 were identified in the ES products (see Fig. 3C). A possible explanation for the absence of a predicted signal peptide in transcripts encoding proteins identified in the ES products is that some members of the cathepsin family might be excreted/secreted via a “nonclassical” pathway that does not involve signal peptide cleavage (70). Conversely, the difference between the number of cathepsin-encoding transcripts predicted from transcriptomic data and the number of cathepsins identified in the ES products supports the hypothesis that the transcriptome of fasciolids encodes “endogenous” cathepsins that function in key biological pathways, such as egg production, protein turnover, and molecular remodelling (71), and “exogenous” cathepsins with roles that appear to relate to the digestion of host molecules (72, 73).
In parasitic trematodes, some cathepsins (e.g. cathepsins B and L) are known to be encoded by multigene families (74), which poses a challenge for both the de novo assembly of transcripts encoding different, particularly closely related isoforms and the identification of such isoforms using mass spectroscopy. The accurate characterization of isoforms via spectrometry is highly dependent on their sequence similarity and their abundance in the matrix subjected to analysis (in this case, ES products). Thus, it is possible that the high sequence similarity of the cathepsin L proteins identified in the transcriptome impaired mass spectral identification of low-abundant cathepsins L, which, in some cases, relied on the specific detection of variation of only two or three peptides. The presence of multiple isoforms of secreted cathepsin L has been reported previously for both Fa. hepatica (24, 66) and Fa. gigantica (25). In Fa. hepatica, these proteins have been shown to be crucial for parasite survival, mediating essential processes such as the digestion of host macromolecules and the suppression of the host immune response (20). In Fa. hepatica, 24 different cathepsin L isoforms have been identified and classified into five clades (designated Clades 1–5; FhCL1–5) (65); members of Clades 1, 2, and 5 were shown to be present among ES products from adult worms, whereas members of Clades 3 and 4 were detected exclusively in ES products from juvenile worms, suggesting that different isoforms play distinct roles in molecular mechanisms linked to the invasion of and survival in the vertebrate host (65). The expansion of the cathepsin family of peptidases in Fa. hepatica is the result of a series of gene duplication events (65, 75). The sequence diversity displayed by members of the cathepsin protein family, together with their broad range of substrate specificities, has been hypothesized to play a role in the ability of parasitic trematodes to infect a wide range of mammalian hosts (20, 75, 76). In the present study, transcripts encoding cathepsins with significant homology to FhCL3 were identified in the transcriptome of adult F. magna; however, the corresponding proteins were not detected in the ES products of this trematode by proteomic means. This finding contrasts with a previous analysis of Fa. hepatica, in which molecules encoding FhCL3 were not detected in the transcriptome of the adult worm (65). Five distinct cathepsin L isoforms were identified in the ES proteome of F. magna, including four isoforms and a cathepsin L-like protease, all significant homologues of FhCL1. However, some S2 active site residues of the F. magna cathepsin L (see Fig. 4) peptidases were similar to those of cathepsins expressed specifically by the juvenile stages of Fa. hepatica (65). The developmental regulation of the expression of cathepsin L is likely to represent a potential adaptation of different developmental stages of the parasite to the diverse environmental conditions encountered throughout its life cycle. Based on these observations, it is tempting to speculate that if developmental regulation of cathepsin L peptidases occurs in F. magna, it will differ from that in Fa. hepatica. Given the potential of cathepsins as vaccine targets against fascioliasis (20), further studies aimed at elucidating the expression profiles of cathepsin-encoding transcripts in different stages of the F. magna life cycle, as well as the presence of members of this protein family in the ES products of the juvenile stages, will be crucial.
In addition to the cathepsin L isoforms, six other proteases were detected in the ES products from F. magna, namely, two isoforms of cathepsin B2, a cathepsin A, a legumain, two isoforms of lysosomal Pro-Xaa carboxypeptidase and a leucine aminopeptidase (see Fig. 3C). Like cathepsin L, cathepsin B is thought to be expressed as an inactive zymogen and is activated following the removal of a pre-pro-region by an asparaginyl endopeptidase (legumain) (77), also present in the ES products from F. magna. In the present study, predominantly activated forms of cathepsin L were identified, as evidenced by the lack of peptides assigned to the pre-pro-region. In a single case, the amino acid sequence of one of the cathepsin B isoforms identified included three pre-pro-region peptides, typical of the inactive form of this protein; however, each of the peptides from this region was represented by only one spectrum, whereas spectral counts of peptides from the mature sequence were higher (e.g. up to 23 for one of the peptides). Therefore, it is likely that inactivated proteases constituted a small proportion of the total number of cathepsins B detected in the ES products. In addition, for the second isoform of cathepsin B2 identified herein (Table III), the Asn in the region of the amino acid sequence, which precedes the first residue of the mature protein (typical of all cathepsin L peptidases from Fa. hepatica and proposed to act as a substrate for possible activation of these proteins by legumain proteases) (77), was substituted for by a Glu residue. The same substitution was observed in three of the cathepsins L of F. magna (Fig. 4). Several of the proteases identified in the F. magna ES products were of lysosomal origin, including cathepsin A, the two Pro-Xaa carboxypeptidases, and, possibly, legumain (see Fig. 3C). On the basis of PSORT and the presence of mannose 6-phosphate glycosylation sites, 12 lysosomal proteins were identified in the F. magna ES products, which constituted ∼15% of the total identifications; after cystatin and the cathepsin proteases, lysosomal proteins were most abundant in the ES products. Recent proteomic studies of S. mansoni (blood fluke) and Fa. hepatica vomitus also identified a number of lysosomal proteins as enzymes putatively involved in digestive processes (66, 78). In S. mansoni, this observation led to the hypothesis that lysosomal proteases might be actively secreted into the gut lumen, the pH of which facilitates the activity of these proteins, in order to enable the digestion of plasma components as well as hemoglobin (78). A legumain, a pro-X carboxylpeptidase, a beta-glucosidase, several isoforms of ferritin, and the Niemann-Pick C2 protein were also identified here as constituents of the ES products from F. magna. The presence of these proteins in the F. magna ES products, as well as serpin and kunitz-type protease inhibitors, is likely a consequence of the fact that, in vitro, F. magna readily regurgitates the contents of its digestive tract into the culture medium, as previously observed for Fa. hepatica (66). Indeed, the same proteins were also identified in vomitus from both of the latter trematodes and S. mansoni (66, 78).
Table III. Fascioloides magna excretory/secretory proteins identified using tandem mass spectrscopy.
| ID | SC | CO | Description | SP | TM | LO | N | Fh | Cs | Ov |
|---|---|---|---|---|---|---|---|---|---|---|
| Proteases | ||||||||||
| 39926 | 18 | 38 | Cathepsin B2 | + | - | Ex | ++ | + | + | - |
| 39928 | 4 | 11 | Cathepsin B2 | + | - | Ex | ++ | + | - | - |
| 39931 | 17 | 44 | Cathepsin L1 | + | - | Ex | - | + | - | - |
| 39932 | 13 | 37 | Cathepsin L1 | + | - | Ex | - | + | - | - |
| 39929 | 9 | 29 | Cathepsin L1 | + | - | Ex | + | + | - | - |
| 39933 | 3 | 27 | Cathepsin L1 | + | - | Ex | + | + | - | - |
| 39930 | 4 | 30 | Cathepsin L-like proteinase | + | - | Ex/Pl | + | + | - | - |
| 1283 | 3 | 7 | Cathepsin A (carboxypeptidase C) | + | - | Ex/Ly | ++ | - | - | - |
| 3216 | 9 | 15 | Calpain | - | - | Cy | - | + | - | + |
| 22081 | 3 | 29 | Legumain-1 | + | - | Ly/Ex | - | + | - | - |
| 11181 | 3 | 7 | Lysosomal Pro-Xaa carboxypeptidase (S28 family) | + | - | Ly | +++ | + | - | - |
| 1246 | 2 | 11 | Lysosomal Pro-X carboxypeptidase | - | - | Ly | - | + | - | - |
| 1063 | 2 | 5 | Leucine aminopeptidase 1 | - | - | Cy/Mi | - | + | - | - |
| Oxidation/reduction | ||||||||||
| 2257 | 11 | 25 | Ferritin-like protein | - | - | Cy | - | + | - | + |
| 24226 | 7 | 55 | Yolk ferritin | - | - | Cy | - | + | - | + |
| 6813 | 4 | 17 | Yolk ferritin | - | - | Cy | - | + | - | + |
| 3565 | 4 | 18 | Yolk ferritin | - | - | Ex | - | + | - | + |
| 3449 | 5 | 16 | Ferritin heavy chain | - | - | Cy | - | + | - | + |
| 1228 | 7 | 22 | Glutathione S-transferase | - | - | Cy | - | + | - | + |
| 23322 | 2 | 65 | Mu-glutathione transferase | - | - | Cy/Ex | - | + | - | + |
| 1482 | 4 | 10 | Dihydrolipoamide dehydrogenase | - | - | Mi | - | - | + | - |
| 22000 | 4 | 54 | Peroxiredoxin | - | - | Ex | - | + | - | - |
| 21268 | 2 | 21 | Thioredoxin | - | - | Cy/Ex | - | + | - | + |
| 4552 | 3 | 37 | Fatty-acid-binding protein Fh15 | - | - | Cy | - | + | - | + |
| 22680 | 3 | 36 | Fatty-acid-binding protein Fh15 | - | - | Cy | - | + | - | + |
| 21349 | 2 | 19 | Fatty-acid-binding protein type 3 | - | - | Cy/Ex | - | + | - | + |
| 21954 | 2 | 21 | Fatty-acid-binding protein type 3 | - | - | Cy/Ex/Nu | - | + | - | + |
| 5430 | 2 | 9 | Short chain dehydrogenase | - | - | Cy | - | + | + | - |
| Carbohydrate metabolism | ||||||||||
| 1498 | 6 | 16 | Hexokinase | - | - | Ex | - | + | - | - |
| 11097 | 6 | 20 | Alpha-glucosidase | - | - | Ly | + | - | - | - |
| 11549 | 6 | 26 | Lysosomal alpha-mannosidase | + | - | Ly | +++ | - | - | - |
| 10957 | 6 | 15 | Lysosomal alpha-mannosidase | + | - | Ly | - | - | - | - |
| 11712 | 4 | 10 | Beta-galactosidase | + | - | Ly | ++ | - | - | - |
| 1121 | 4 | 10 | Alpha-galactosidase | + | - | Ly | + | - | - | - |
| 12521 | 4 | 17 | Lysosomal alpha-glucosidase | + | - | Ly | +++ | - | - | - |
| 1772 | 4 | 7 | Beta-hexosaminidase B | + | - | Ly | +++ | - | - | - |
| 11649 | 2 | 7 | Enolase | - | - | Cy | - | + | + | + |
| Protease inhibitor | ||||||||||
| 8767 | 38 | 32 | Cystatin | - | - | Cy | - | + | - | + |
| 2249 | 8 | 18 | Leukocyte elastase inhibitor | - | - | Cy | - | - | - | - |
| 7623 | 2 | 9 | Leukocyte elastase inhibitor | - | - | Cy | - | - | - | - |
| 2998 | 6 | 21 | Serpin | - | - | Mi | - | + | - | - |
| 4671 | 2 | 7 | Serpin B | - | - | Pl | - | + | - | - |
| 22726 | 4 | 30 | Kunitz-type proteinase inhibitor | + | - | Ex | - | + | - | - |
| 22727 | 2 | 17 | Kunitz-type protease inhibitor–like protein | + | - | Ex | - | + | - | - |
| Structural | ||||||||||
| 1276 | 8 | 17 | Plastin-2 | - | - | Cy | - | - | - | - |
| 4303 | 5 | 4 | Myoferlin | - | 7 | Pl | - | - | - | + |
| 5968 | 4 | 34 | Ribosomal protein L40-like isoform 1 | - | - | Cy/Nu | - | - | - | - |
| 2802 | 3 | 11 | Tegumental protein 20.8 kDa (dynein light chain) | - | - | Cy | - | - | - | + |
| 9449 | 2 | 20 | Transgelin | - | - | Cy | - | - | - | - |
| 1108 | 2 | 23 | Cyclophilin A | - | - | Ex | - | - | - | - |
| 12338 | 2 | 16 | CD63 antigen (Tetraspanin) | - | 3 | Pl | - | + | - | - |
| 21424 | 2 | 20 | Tetraspanin-CD63 receptor | - | 3 | Pl | - | + | - | - |
| 6044 | 2 | 10 | Actin | - | - | Cy | - | + | + | + |
| Transporters | ||||||||||
| 1261 | 4 | 4 | Multidrug resistance protein 1, 2, 3 | - | - | Pl | - | - | - | |
| 11051 | 3 | 8 | Glucose transporter | + | 10 | Pl | - | + | - | |
| 11222 | 3 | 7 | Solute carrier family 22 member 5 | - | 5 | Pl | - | + | - | |
| 2147 | 2 | 5 | ATP synthase, H+ transporting | - | - | Pl | + | + | - | |
| Miscellaneous | ||||||||||
| 11672 | 10 | 29 | Annexin A7 | - | - | Cy | - | + | - | - |
| 2360 | 5 | 13 | Annexin A6 | - | - | Cy | - | + | - | - |
| 3162 | 4 | 10 | Annexin A7 | - | - | Cy | - | + | - | - |
| 3411 | 7 | 27 | Triose-phosphate isomerase | - | - | Ex | - | + | + | + |
| 2363 | 6 | 37 | Cytidine deaminase | - | - | Cy/Ex | - | - | - | - |
| 2295 | 4 | 23 | Heat shock protein 70 | - | - | Nu/Cy | - | - | + | - |
| 8898 | 5 | 3 | Basement membrane-specific heparan sulfate proteoglycan core protein | - | - | Nu | - | - | - | - |
| 14949 | 4 | 29 | Hemoglobin F2 | - | - | Cy | - | + | - | - |
| 10852 | 4 | 20 | Cubilin | - | - | Cy/Nu | - | - | - | - |
| 9902 | 3 | 2 | GPI-anchored surface glycoprotein | + | 3 | Pl | +++ | + | - | - |
| 11521 | 3 | 7 | LAMA-like protein 2 precursor | - | - | Er/Ex | - | + | - | - |
| 21652 | 2 | 51 | LAMA-like protein 2 precursor | - | - | Cy/Ex | - | + | - | - |
| 11828 | 3 | 27 | SH3-containing protein SH3GLB1-like | - | - | Cy | - | - | - | - |
| 11926 | 2 | 10 | Ornithine aminotransferase | - | - | Cy/Ex | - | - | - | - |
| 1810 | 2 | 6 | Carbonic anhydrase | + | - | Pl | + | + | - | - |
| 2270 | 2 | 14 | Niemann-Pick C2 protein | + | - | Ly | ++ | + | - | - |
| 22436 | 2 | 64 | Acid sphingomyelinase-like phosphodiesterase 3a | + | - | Ex/Ly | + | - | - | - |
| 12409 | 2 | 10 | Proteophosphoglycan ppg4 | - | - | Cy/Ex | - | - | - | - |
| 4722 | 2 | 18 | Translation initiation inhibitor | - | - | Cy | - | - | - | - |
| Unknown | ||||||||||
| 11997 | 4 | 15 | Hypothetical protein CLF_104189 [Clonorchis sinensis] | - | - | Mi/Cy | - | - | - | - |
| 22473 | 4 | 5 | Unknown | - | - | Cy | - | - | - | - |
| 24853 | 2 | 11 | Unknown | - | - | Cy | - | - | - | - |
| 21118 | 2 | 32 | Unknown | - | - | Cy/Ex | - | - | - | - |
SC, number of unique peptides assigned to the protein; CO, percent cover; LO, putative subcellular localization of the protein; TM, number of transmembrane domains. The presence of a signal peptide (SP) or an N-linked glycosylation site (N) is denoted by “+”; the presence of the protein in the ES of adult Fasciola hepatica (Fh), Clonorchis sinensis (Cs), or Opisthorchis viverrini (Ov) is also denoted by “+.”
The vast majority of F. magna ES products represented a complex mixture of proteins, predominantly of extracellular or cytoplasmic origin. In particular, a significant proportion of nonclassically secreted ES proteins, including a number of proteins predicted to be membrane bound, such as a tetraspanin and a glucose transporter, were identified. Unlike ES products from gastrointestinal parasitic nematodes, such as Ancylostoma caninum (31), in which classically secreted proteins are very abundant, nonclassically secreted proteins have been consistently shown to represent a significant proportion of ES products from trematode parasites, including S. japonicum (79), C. sinensis (67), O. viverrini (68), and Fa. hepatica (66). For the latter trematode, it has been proposed that the presence of these proteins in the ES products is a result of stress-induced shedding of the tegument during culture (64). However, morphological studies of this parasite have noted rapid turnover of its tegument, facilitated by subtegumental cells, leading to the hypothesis that the shedding of the tegument and associated proteins occurring in vivo might represent an immuno-defensive strategy (24, 80). Two phospholipases and an ABC transporter were also identified among the ES products of F. magna. The presence of ABC transporters in the tegumental membrane of Fa. hepatica has prompted comparisons to the ER/Golgi-independent secretion of IL-1β and caspase-1 in mammalian cells (24). This process involves the formation of plasma membrane “blebs,” mediated by ABC transporters, which are subsequently released as microvesicles after phospholipase-mediated fusion with the plasma membrane (81); the presence of both phospholipases and ABC transporters in the ES products of F. magna suggests that a similar process might occur in this trematode. Although some ES products identified in the present study might relate to gut content regurgitated by the parasite (66), 15 antioxidant proteins were identified, none of which possessed a signal peptide indicative of secretion. A similar profile of ES antioxidant proteins, including fatty-acid-binding proteins, glutathione S-transferase, peroxiredoxin, and thioredoxin, has been described for Fa. hepatica (24). In the latter trematode, these proteins are proposed to play an important role in the evasion of host immune responses, which likely relates to the protection of the parasite from reactive oxygen species released by host immune cells (82–85), an inhibition of the proliferative potential of spleen cells (e.g. in rats) (86), and/or the recruitment and alternative activation of macrophages (86–88). All antioxidant proteins identified here, with the exception of dihydrolipoamide dehydrogenase, were also detected in the ES products from adult Fa. hepatica, suggesting that these proteins play a role in immune evasion (82–84, 86–88). In Fa. hepatica, a variable antioxidant profile was observed in ES products from different developmental stages (24), which led to the hypothesis that these nonclassically secreted antioxidant proteins were transported through an alternative, transtegumental, secretory pathway (24). However, the existence of such a pathway in fasciolid trematodes remains to be demonstrated.
The availability of entire genome sequences of related species of liver flukes, such as C. sinensis (89) and blood flukes (38–40), now provides unprecedented opportunities to (i) conduct comparative proteomic comparisons; (ii) elucidate the structures and functions of key molecules (e.g. “endogenous” and “exogenous” cathepsins); (iii) explore novel biological pathways (e.g. transtegumental, secretory pathways); and (iv) establish relationships among genes, transcripts, and proteins involved specifically in the parasite's invasion of, establishment in, and interactions with the host. Advancing these areas will provide a basis for studies aimed at exploring the potential of these molecules as targets for the development of novel strategies for the control of trematodiases.
Supplementary Material
Footnotes
* Currently, the Gasser Lab is supported by funding from the Australian Research Council (ARC), Melbourne Water Corporation, and the National Health and Medical Research Council (NHMRC) (R. B. G.). Support from the Victorian Life Sciences Computation Initiative (VLSCI) and IBM Collaboratory is also acknowledged (R. B. G.). The Hofmann and Loukas Labs are also supported by grants from the NHMRC and ARC. This study was also supported by the Linkage Infrastructure, Equipment and Facilities (LIEF) scheme from ARC (J. M.), the Czech Science Foundation (grant nos. P502/10/P248 and 206/09/H026), the Czech Ministry of Education (grant nos. MSM LC06009 and MSM 0021620820), and (partially) UNCE of the Charles University (grant no. 204017) (M. K.). J. M. holds a Career Development Award (CDA1), N. D. Y. holds an Early Career Researcher (ECR) Fellowship, and A. L. holds a Principal Research Fellowship from NHMRC.
 This article contains supplemental Figure S1 and Tables S1 to S4.
This article contains supplemental Figure S1 and Tables S1 to S4.
2 Young et al., unpublished data.
1 The abbreviations used are:
- BLAST
- Basic Local Alignment Search Tool
- ES
- excretory/secretory
- GO
- gene ontology
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- DTT
- dithiothreitol
- IAA
- iodoacetamide
- NCBI
- National Center for Biotechnology Information
- OGE
- OFF-GEL electrophoresis
- PBS
- phosphate-buffered saline
- SDS-PAGE
- sodium dodecyl-sulfate polyacrylamide gel electrophoresis.
REFERENCES
- 1. Boray J. C. (1997) Chemotherapy of infections with Fasciolidae. In Immunology, Pathobiology and Control of Fasciolosis (Boray J. C., ed.), pp. 83–97, MSD AGVET, Rahway, NJ [Google Scholar]
- 2. Kaplan R. M. (2001) Fasciola hepatica: a review of the economic impact in cattle and considerations for control. Vet. Ther. 2, 40–50 [PubMed] [Google Scholar]
- 3. Young N. D., Hall R. S., Jex A. R., Cantacessi C., Gasser R. B. (2010) Elucidating the transcriptome of Fasciola hepatica——a key to fundamental and biotechnological discoveries for a neglected parasite. Biotechnol. Adv. 28, 222–231 [DOI] [PubMed] [Google Scholar]
- 4. Pybus M. J. (2001). Liver flukes. In Parasitic Diseases in Wild Mammals (Samuel W. M., Pybus M. J., Kocan A. A., eds.), pp. 121–149, Iowa State Press, Iowa City, IA [Google Scholar]
- 5. Coil W. H. (1977) The penetration of Fascioloides magna miracidia into the snail host Fossaria bulimoides. A scanning electron microscope study. Zeitschrift für Parasitenkunde 52, 53–59 [DOI] [PubMed] [Google Scholar]
- 6. Fairweather I. (2005) Triclabendazole: new skills to unravel an old(ish) enigma. J. Helminthol. 79, 227–234 [DOI] [PubMed] [Google Scholar]
- 7. Maes L., Vanparijs O., Lauwers H., Deckers W. (1990) Comparative efficacy of closantel and triclabendazole against Fasciola hepatica in experimentally infected sheep. Vet. Rec. 127, 450–452 [PubMed] [Google Scholar]
- 8. Keiser J., Utzinger J. (2004) Chemotherapy for major food-borne trematodes: a review. Expert Opin. Pharmacother. 5, 1711–1726 [DOI] [PubMed] [Google Scholar]
- 9. Flynn R. J., Mulcahy G., Elsheikha H. M. (2010) Coordinating innate and adaptive immunity in Fasciola hepatica infection: implications for control. Vet. Parasitol. 169, 235–240 [DOI] [PubMed] [Google Scholar]
- 10. Overend D. J., Bowen F. L. (1995) Resistance of Fasciola hepatica to triclabendazole. Aust. Vet. J. 72, 275–276 [DOI] [PubMed] [Google Scholar]
- 11. Coles G. C., Rhodes A. C., Stafford K. A. (2000) Activity of closantel against adult triclabendazole-resistant Fasciola hepatica. Vet. Rec. 146, 504. [DOI] [PubMed] [Google Scholar]
- 12. Moll L., Gaasenbeek C. P. H., Vellema P., Borgsteede F. H. M. (2000) Resistance of Fasciola hepatica against triclabendazole in cattle and sheep in the Netherlands. Vet. Parasitol. 91, 153–158 [DOI] [PubMed] [Google Scholar]
- 13. Thomas I., Coles G. C., Duffus K. (2000) Triclabendazole-resistant Fasciola hepatica in southwest Wales. Vet. Rec. 146, 200. [PubMed] [Google Scholar]
- 14. Fairweather I. (2009) Triclabendazole progress report, 2005–2009: an advancement of learning? J. Helminthol. 83, 139–150 [DOI] [PubMed] [Google Scholar]
- 15. Gaasenbeek C. P. H., Moll L., Cornelissen J., Vellema P., Borgsteede F. H. M. (2001) An experimental study on triclabendazole resistance of Fasciola hepatica in sheep. Vet. Parasitol. 95, 37–43 [DOI] [PubMed] [Google Scholar]
- 16. Spithill T. W., Dalton J. P. (1998) Progress in development of liver fluke vaccines. Parasitol. Today 14, 224–228 [DOI] [PubMed] [Google Scholar]
- 17. Piacenza L., Acosta D., Basmadjian I., Dalton J. P., Carmona C. (1999) Vaccination with cathepsin L proteinases and with leucine aminopeptidase induces high levels of protection against fascioliasis in sheep. Infect. Immun. 67, 1954–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dalton J. P., Mulcahy G. (2001) Parasite vaccines—a reality? Vet. Parasitol. 98, 149–167 [DOI] [PubMed] [Google Scholar]
- 19. Dalton J. P., Brindley P. J., Knox D. P., Brady C. P., Hotez P. J., Donnelly S., O'Neill S. M., Mulcahy G., Loukas A. (2003) Helminth vaccines: from mining genomic information for vaccine targets to systems used for protein expression. Int. J. Parasitol. 33, 621–640 [DOI] [PubMed] [Google Scholar]
- 20. Dalton J. P., O'Neill S., Stack C., Collins P., Walshe A., Sekiya M., Doyle S., Mulcahy G., Hoyle D., Khaznadji E., Moire N., Brennan G., Mousley A., Kreshchenko N., Maule A. G., Donnelly S. M. (2003) Fasciola hepatica cathepsin L-like proteases: biology, function, and potential in the development of first generation liver fluke vaccines. Int. J. Parasitol. 33, 1173–1181 [DOI] [PubMed] [Google Scholar]
- 21. McManus D. P., Dalton J. P. (2006) Vaccines against the zoonotic trematodes Schistosoma japonicum, Fasciola hepatica and Fasciola gigantica. Parasitology 133, S43–S61 [DOI] [PubMed] [Google Scholar]
- 22. Acosta D., Cancela M., Piacenza L., Roche L., Carmona C., Tort J. F. (2008) Fasciola hepatica leucine aminopeptidase, a promising candidate for vaccination against ruminant fasciolosis. Mol. Biochem. Parasitol. 158, 52–64 [DOI] [PubMed] [Google Scholar]
- 23. Jayaraj R., Piedrafita D., Dynon K., Grams R., Spithill T. W., Smooker P. M. (2009) Vaccination against fasciolosis by a multivalent vaccine of stage-specific antigens. Vet. Parasitol. 160, 230–236 [DOI] [PubMed] [Google Scholar]
- 24. Robinson M. W., Menon R., Donnelly S. M., Dalton J. P., Ranganathan S. (2009) An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica: proteins associated with invasion and infection of the mammalian host. Mol. Cell. Proteomics 8, 1891–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Young N. D., Jex A. R., Cantacessi C., Hall R. S., Campbell B. E., Spithill T. W., Tangkawattana S., Tangkawattana P., Laha T., Gasser R. B. (2011) A portrait of the transcriptome of the neglected trematode, Fasciola gigantica—biological and biotechnological implications. PLoS Negl. Trop. Dis. 5, e1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kasny M., Mikes L., Hampl V., Dvorak J., Caffrey C. R., Dalton J. P., Horak P. (2009) Chapter 4. Peptidases of trematodes. Adv. Parasitol. 69, 205–297 [DOI] [PubMed] [Google Scholar]
- 27. DeMarco R., Verjovski-Almeida S. (2009) Schistosomes—proteomics studies for potential novel vaccines and drug targets. Drug Discov. Today 14, 472–478 [DOI] [PubMed] [Google Scholar]
- 28. Chuan J., Feng Z., Brindley P. J., McManus D. P., Han Z., Jianxin P., Hu W. (2010) Our wormy world: genomics, proteomics and transcriptomics in East and southeast Asia. Adv. Parasitol. 73, 327–371 [DOI] [PubMed] [Google Scholar]
- 29. Cantacessi C., Campbell B. E., Gasser R. B. (2012) Key strongylid nematodes of animals—impact of next-generation transcriptomics on systems biology and biotechnology. Biotechnol. Adv. 30, 469–488 [DOI] [PubMed] [Google Scholar]
- 30. Verjovski-Almeida S., DeMarco R. (2008) Current developments in Schistosoma proteomics. Acta Trop. 108, 183–185 [DOI] [PubMed] [Google Scholar]
- 31. Mulvenna J., Hamilton B., Nagaraj S. H., Smyth D., Loukas A., Gorman J. J. (2009) Proteomics analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Mol. Cell. Proteomics 8, 109–121 [DOI] [PubMed] [Google Scholar]
- 32. Young N. D., Campbell B. E., Hall R. S., Jex A. R., Cantacessi C., Laha T., Sohn W. M., Sripa B., Loukas A., Brindley P. J., Gasser R. B. (2010) Unlocking the transcriptomes of two carcinogenic parasites, Clonorchis sinensis and Opisthorchis viverrini. PLoS Negl. Trop. Dis. 4, e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bentley D. R., Balasubramanian S., Swerdlow H. P., George D., Gietzen K. J., Goddard C. P., Golda G. S., Granieri P. A., Green D. E., Gustafson D. L., Hansen N. F., Harnish K., Haudenschild C. D., Heyer N. I., Hims M. M., Ho J. T., Horgan A. M., Hoschler K., Hurwitz S., Ivanov D. V., Johnson M. Q., James T., Huw Jones T. A., Kang G. D., Kerelska T. H., Kersey A. D., Khrebtukova I., Kindwall A. P., Kingsbury Z., Kokko-Gonzales P. I., Kumar A., Laurent M. A., Lawley C. T., Lee S. E., Lee X., Liao A. K., Loch J. A., Lok M., Luo S., Mammen R. M., Martin J. W., McCauley P. G., McNitt P., Mehta P., Moon K. W., Mullens J.W., Newington T., Ning Z., Ling Ng B., Novo S. M., O'Neill M. J., Osborne M. A., Osnowski A., Ostadan O., Paraschos L. L., Pickering L., Pike A. C., Pike A. C., Chris Pinkard D., Pliskin D. P., Podhasky J., Quijano V. J., Raczy C., Rae V. H., Rawlings S. R., Chiva Rodriguez A., Roe P. M., Rogers J., Rogert Bacigalupo M. C., Romanov N., Romieu A., Roth R. K., Rourke N. J., Ruediger S. T., Rusman E., Sanches-Kuiper R. M., Schenker M. R., Seoane J. M., Shaw R. J., Shiver M. K., Short S. W., Sizto N. L., Sluis J. P., Smith M. A., Ernest Sohna Sohna J., Spence E. J., Stevens K., Sutton N., Szajkowski L., Tregidgo C. L., Turcatti G., Vandevondele S., Verhovsky Y., Virk S. M., Wakelin S., Walcott G. C., Wang J., Worsley G. J., Yan J., Yau L., Zuerlein M., Rogers J., Mullikin J. C., Hurles M. E., McCooke N. J., West J. S., Oaks F. L., Lundberg P. L., Klenerman D., Durbin R., Smith A. J. (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schulz M. H., Zerbino D. R., Vingron M., Birney E. (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28, 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li R., Yu C., Li Y., Lam T. W., Yiu S. M., Kristiansen K., Wang J. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 [DOI] [PubMed] [Google Scholar]
- 36. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 37. Cantacessi C., Jex A. R., Hall R. S., Young N. D., Campbell B. E., Joachim A., Nolan M. J., Abubucker S., Sternberg P. W., Ranganathan S., Mitreva M., Gasser R. B. (2010) A practical, bioinformatic workflow system for large data sets generated by next-generation sequencing. Nucleic Acids Res. 38, e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Young N. D., Jex A. R., Li B., Liu S., Yang L., Xiong Z., Li Y., Cantacessi C., Hall R. S., Xu X., Chen F., Wu X., Zerlotini A., Oliveira G., Hofmann A., Zhang G., Fang X., Kang Y., Campbell B. E., Loukas A., Ranganathan S., Rollinson D., Rinaldi G., Brindley P. J., Yang H., Wang J., Wang J., Gasser R. B. (2012) Whole-genome sequence of Schistosoma haematobium. Nat. Genet. 44, 221–225 [DOI] [PubMed] [Google Scholar]
- 39. The Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium (2009) The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature 460, 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Berriman M., Haas B. J., LoVerde P. T., Wilson R. A., Dillon G. P., Cerqueira G. C., Mashiyama S. T., Al-Lazikani B., Andrade L. F., Ashton P. D., Aslett M. A., Bartholomeu D. C., Blandin G., Caffrey C. R., Coghlan A., Coulson R., Day T. A., Delcher A., DeMarco R., Djikeng A., Eyre T., Gamble J. A., Ghedin E., Gu Y., Hertz-Fowler C., Hirai H., Hirai Y., Houston R., Ivens A., Johnston D. A., Lacerda D., Macedo C. D., McVeigh P., Ning Z., Oliveira G., Overington J. P., Parkhill J., Pertea M., Pierce R. J., Protasio A. V., Quail M. A., Rajandream M. A., Rogers J., Sajid M., Salzberg S. L., Stanke M., Tivey A. R., White O., Williams D. L., Wortman J., Wu W., Zamanian M., Zerlotini A., Fraser-Liggett C. M., Barrell B. G., El-Sayed N. M. (2009) The genome of the blood fluke Schistosoma mansoni. Nature 460, 352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hunter S., Apweiler R., Attwood T. K., Bairoch A., Bateman A., Binns D., Bork P., Das U., Daugherty L., Duquenne L., Finn R. D., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Laugraud A., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., Mistry J., Mitchell A., Mulder N., Natale D., Orengo C., Quinn A. F., Selengut J. D., Sigrist C. J., Thimma M., Thomas P. D., Valentin F., Wilson D., Wu C. H., Yeats C. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res. 37, D211-D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L., Wang J. (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34, W293-W297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., Kong L., Gao G., Li C. Y., Wei L. (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316-W322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 46. Sonnhammer E. L. L., von Heijne G., Krogh A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, pp. 175–182, AAAI, Menlo Park, CA: [PubMed] [Google Scholar]
- 47. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 48. Moller S., Croning M. D. R., Apweiler R. (2001) Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17, 646–653 [DOI] [PubMed] [Google Scholar]
- 49. Chen Y., Zhang Y., Yin Y., Gao G., Li S., Jiang Y., Gu X., Luo J. (2005) SPD—a web based secreted protein database. Nucleic Acids Res. 33, D169-D173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Choo K. H., Tan T. W., Ranganathan S. (2005) SPdb—a signal peptide database. BMC Bioinformatics 6, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–907 [DOI] [PubMed] [Google Scholar]
- 52. Searle B. C. (2010) Scaffold: a bioinformatics tool for validating MS/MS-based proteomics studies. Proteomics 10, 1265–1269 [DOI] [PubMed] [Google Scholar]
- 53. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 54. Nesvizhsii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 55. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 56. Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649 [DOI] [PubMed] [Google Scholar]
- 57. Cho P. Y., Lee M. J., Kim T. I., Kang S. Y., Hong S. J. (2006) Expressed sequence tag analysis of adult Clonorchis sinensis, the Chinese liver fluke. Parasitol. Res. 99, 602–608 [DOI] [PubMed] [Google Scholar]
- 58. Cho P. Y., Kim T. I., Whang S. M., Hong S. J. (2008) Gene expression profile of Clonorchis sinensis metacercariae. Parasitol. Res. 102, 277–282 [DOI] [PubMed] [Google Scholar]
- 59. Kang J. M., Bahk Y. Y., Cho P. Y., Hong S. J., Kim T. S., Sohn W. M., Na B. K. (2010) A family of cathepsin F cysteine proteases of Clonorchis sinensis is the major secreted proteins that are expressed in the intestine of the parasite. Mol. Biochem. Parasitol. 170, 7–16 [DOI] [PubMed] [Google Scholar]
- 60. Yoo W. G., Kim D. W., Ju J. W., Cho P. Y., Kim T. I., Cho S. H., Choi S. H., Park H. S., Kim T. S., Hong S. J. (2011) Developmental transcriptomic features of the carcinogenic liver fluke, Clonorchis sinensis. PLoS Negl. Trop. Dis. 5, e1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim T. S., de Guzman J. V., Kong H. H., Chung D. I. (2006) Comparison of gene representation between diploid and triploid Paragonimus westermani by expressed sequence tags analyses. J. Parasitol. 92, 803–816 [DOI] [PubMed] [Google Scholar]
- 62. Horton P., Park K., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585-W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jefferies J. R., Campbell A. M., vanRossum A. J., Barrett J., Brophy P. M. (2001) Proteomic analysis of Fasciola hepatica excretory-secretory products. Proteomics 1, 1128–1132 [DOI] [PubMed] [Google Scholar]
- 64. Morphew R. M., Wright H. A., LaCourse E. J., Porter J., Barrett J., Woods D. J., Brophy P. M. (2011) Towards delineating functions within the Fasciola secreted cathepsin I protease family by integrating in vivo based sub-proteomics and phylogenetics. PLoS Negl. Trop. Dis. 5, e937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Robinson M. W., Tort J. F., Lowther J., Donnelly S. M., Wong E., Xu W., Stack C. M., Padula M., Herbert B., Dalton J. P. (2008) Proteomics and phylogenetic analysis of the cathepsin L protease family of the helminth pathogen Fasciola hepatica: expansion of a repertoire of virulence-associated factors. Mol. Cell. Proteomics 7, 1111–1123 [DOI] [PubMed] [Google Scholar]
- 66. Wilson R. A., Wright J. M., de Castro-Borges W., Parker-Manuel S. J., Dowle A. A., Ashton P. D., Young N. D., Gasser R. B., Spithill T. W. (2011) Exploring the Fasciola hepatica tegument proteome. Int. J. Parasitol. 41, 1347–1359 [DOI] [PubMed] [Google Scholar]
- 67. Zheng M., Hu K., Liu W., Hu X., Hu F., Huang L., Wang P., Hu Y., Huang Y., Li W., Liang C., Yin X., He Q., Yu X. (2011) Proteomic analysis of excretory secretory products from Clonorchis sinensis adult worms: molecular characterization and serological reactivity of excretory-secretory antigen-fructose-1,6-bisphosphatase. Parasitol. Res. 1, 1–8 [DOI] [PubMed] [Google Scholar]
- 68. Mulvenna J., Sripa B., Brindley P. J., Gorman J., Jones M. K., Colgrave M. L., Jones A., Nawaratna S., Laha T., Suttiprapa S., Smout M. J., Loukas A. (2010) The secreted and surface proteomes of the adult stage of the carcinogenic human liver fluke Opisthorchis viverrini. Proteomics 10, 1063–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Robinson M. W., Corvo I., Jones P. M., George A. M., Padula M. P., To J., Cancela M., Rinaldi G., Tort J. F., Roche L., Dalton J. P. (2011) Collagenolytic activities of the major secreted cathepsin L peptidases involved in the virulence of the helminth pathogen, Fasciola hepatica. PLoS Negl. Trop. Dis. 4, e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nickel W. (2003) The mystery of nonclassical protein secretion. A current view on cargo proteins and potential export routes. Eur. J. Biochem. 270, 2109–2119 [DOI] [PubMed] [Google Scholar]
- 71. Robinson M. W., Colhoun L. M., Fairweather I., Brennan G. P., Waite J. H. (2001) Development of the vitellaria of the liver fluke, Fasciola hepatica, in the rat host. Parasitology 123, 509–518 [DOI] [PubMed] [Google Scholar]
- 72. Chen J. M., Dando P. M., Stevens R. A., Fortunato M., Barrett A. J. (1998) Cloning and expression of mouse legumain, a lysosomal endopeptidase. Biochem. J. 335, 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Turk D., Guncar G. (2003) Lysosomal cysteine proteases (cathepsins): promising drug targets. Acta Crystallogr. D Biol. Crystallogr. 59, 203–213 [DOI] [PubMed] [Google Scholar]
- 74. Robinson M. W., Dalton J. P., Donnelly S. (2008) Helminth pathogen cathepsin proteases: it's a family affair. Trends Biochem. 33, 601–608 [DOI] [PubMed] [Google Scholar]
- 75. Irving J. A., Spithill T. W., Pike R. N., Whisstock J. C., Smooker P. M. (2003) The evolution of enzyme specificity in Fasciola spp. J. Mol. Evol. 57, 1–15 [DOI] [PubMed] [Google Scholar]
- 76. Tort J., Brindley P. J., Knox D., Wolfe K. H., Dalton J. P. (1999) Proteinases and associated genes of parasitic helminths. Adv. Parasitol. 43, 161–266 [DOI] [PubMed] [Google Scholar]
- 77. Dalton J. P., Brindley P. J., Donnelly S., Robinson M. W. (2009) The enigmatic asparaginyl endopeptidase of helminth parasites. Trends Parasitol. 25, 59–61 [DOI] [PubMed] [Google Scholar]
- 78. Hall S. L., Braschi S., Truscott M., Mathieson W., Cesari I. M., Wilson R. A. (2011) Insights into blood feeding by schistosomes from a proteomic analysis of worm vomitus. Mol. Biochem. Parasitol. 179, 18–29 [DOI] [PubMed] [Google Scholar]
- 79. Liu F., Cui S. J., Hu W., Feng Z., Wang Z. Q., Han Z. G. (2008) Excretory/secretory proteome of the adult developmental stage of human blood fluke, Schistosoma japonicum. Mol. Cell. Proteomics 8, 1236–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dalton J. P., Skelly P., Halton D. W. (2004) Role of the tegument and gut in nutrient uptake by parasitic platyhelminths. Can. J. Zool. 24, 211–232 [Google Scholar]
- 81. MacKenzie A., Wilson H. L., Kiss-Toth E., Dower S. K., North R. A., Surprenant A. (2001) Rapid secretion of interleukin-1β by microvescicle shedding. Immunity 15, 825–835 [DOI] [PubMed] [Google Scholar]
- 82. McGonigle L., Mousley A., Marks N. J., Brennan G. P., Dalton J. P., Spithill T. W., Day T. A., Maule A. G. (2008) The silencing of cysteine proteases in Fasciola hepatica newly excysted juveniles using RNA interference reduces gut penetration. Int. J. Parasitol. 38, 149–155 [DOI] [PubMed] [Google Scholar]
- 83. Salazar-Calderón M., Martin-Alonso J. M., Ruiz de Eguino A. D., Parra F. (2001) Heterologous expression and functional characterization of thioredoxin from Fasciola hepatica. Parasitol. Res. 87, 390–395 [DOI] [PubMed] [Google Scholar]
- 84. Salazar-Calderón M., Martin Alonso J. M., Castro A. M., Parra F. (2003) Cloning, heterologous expression in Escherichia coli and characterization of a protein disulfide isomerase from Fasciola hepatica. Mol. Biochem. Parasitol. 126, 15–23 [DOI] [PubMed] [Google Scholar]
- 85. Sekiya M., Mulcahy G., Irwin J. A., Stack C. M., Donnelly S. M., Xu W., Collins P., Dalton J. P. (2006) Biochemical characterisation of the recombinant peroxiredoxin (FhePrx) of the liver fluke, Fasciola hepatica. FEBS Lett. 580, 5016–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cervi L., Rossi G., Masih D. T. (1999) Potential role for excretory-secretory forms of glutathione-S-transferase (GST) in Fasciola hepatica. Parasitology 119, 627–633 [DOI] [PubMed] [Google Scholar]
- 87. Donnelly S., O'Neill S. M., Sekiya M., Mulcahy G., Dalton J. P. (2005) Thioredoxin peroxidase secreted by Fasciola hepatica induces the alternative activation of macrophages. Infect. Immun. 73, 166–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Donnelly S., Stack C. M., O'Neill S. M., Sayed A. A., Williams D. L., Dalton J. P. (2008) Helminth 2-Cys peroxiredoxin drives Th2 responses through a mechanism involving alternatively activated macrophages. FASEB J. 22, 4022–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wang X., Chen W., Huang Y., Sun J., Men J., Liu H., Luo F., Guo L., Lv Z., Deng C., Zhou C., Fan Y., Li X., Huang L., Hu Y., Liang C., Hu X., Xum J., Yum X. (2011) The draft genome of the carcinogenic human liver fluke Clonorchis sinensis. Genome Biol. 12, R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.