Abstract
Proteomic studies based on abundance, activity, or interactions have been used to investigate protein functions in normal and pathological processes, but their combinatory approach has not been attempted. We present an integrative proteomic profiling method to measure protein activity and interaction using fluorescence-based protein arrays. We used an on-chip assay to simultaneously monitor the transamidating activity and binding affinity of transglutaminase 2 (TG2) for 16 TG2-related proteins. The results of this assay were compared with confidential scores provided by the STRING database to analyze the functional interactions of TG2 with these proteins. We further created a quantitative activity-interaction map of TG2 with these 16 proteins, categorizing them into seven groups based upon TG2 activity and interaction. This integrative proteomic profiling method can be applied to quantitative validation of previously known protein interactions, and in understanding the functions and regulation of target proteins in biological processes of interest.
Proteomics is the large-scale analysis of whole proteins and their role in biological systems. Abundance-based proteomics assigns protein functions in normal and pathological processes by quantification of global differences in protein expression levels (1). This classic approach identifies functional biomarkers by comparing samples from healthy individuals and patients. However, this abundance-based approach provides only indirect information about protein function (2). The abundance of a protein is not necessarily correlated with its activity because protein activities are predominantly regulated by a series of post-translational modifications (1, 2). Activity-based proteomics (activity-based protein profiling) is therefore considered an alternative approach to assigning protein functions in biological processes of interest (3). In this approach, specific activity-based probes using fluorescent, radioactive, and affinity tags are usually designed for detection of protein activity (2, 4–6). Activity-based proteomics identifies markers by comparative analyses of activity profiles between healthy and diseased cells and tissues (3, 7, 8). This approach is also used for profiling enzyme inhibitors, for developing therapeutic reagents, and for diagnosis (2, 5). Another functional proteomic approach using large-scale analysis is interactomics or interaction proteomics, which is a useful method for understanding the regulation of proteins in biological systems (9). To elucidate bioactive protein interactions with proteins or ligands, a number of technologies are currently used including the yeast two-hybrid system, affinity purification and mass spectrometry, the protein fragment complementation assay, the luminescence-based mammalian interactome, and protein arrays (9–13). Global differences in the dynamics of the interactome between healthy and diseased individuals provide new insights into causes of disease and can be used for biomarker identification and drug discovery (14–16). Thus, combinatory analyses of abundance, activity, and interaction have great potential in revealing regulation mechanisms and functions of proteins, although such an integrated proteomic approach has not been widely used.
These proteomic methods have been coupled with various detection methods including one- or two-dimensional gel electrophoresis, one- or two-dimensional liquid chromatography and tandem mass spectrometry, surface plasmon resonance, and fluorometric assays for analyses of the proteome (9, 11). In combination with specific probes, colorimetric and fluorometric assays using multiwell plates have been extensively used for the determination of the abundance and activity of various proteins. Although often limited by the amount of sample, these methods nonetheless facilitate real-time measurement of changes in protein activity and high-throughput analyses of protein abundances and activities (17). Surface plasmon resonance, a method that does not necessitate labeling of proteins, has also been used for analysis of protein abundance, activity, and binding affinity (18–20). Using only very small amounts of sample, the microarray combined with fluorometric probes is a promising technology for the rapid analysis of a wide variety of biomolecular interactions, protein abundances, and activities. This approach has been used for serodiagnosis and identification of biomarkers by abundance-based protein profiling in human sera (21–25). It has also been used for kinetic studies of carbohydrate-protein (17) and peptide-protein interactions (26, 27). In addition, this technology has been used for the rapid determination of enzyme activities and for the identification of enzyme substrates and inhibitors (24, 28–33). However, combinatory profiling of protein activities and interactions based on array technology has yet to be reported.
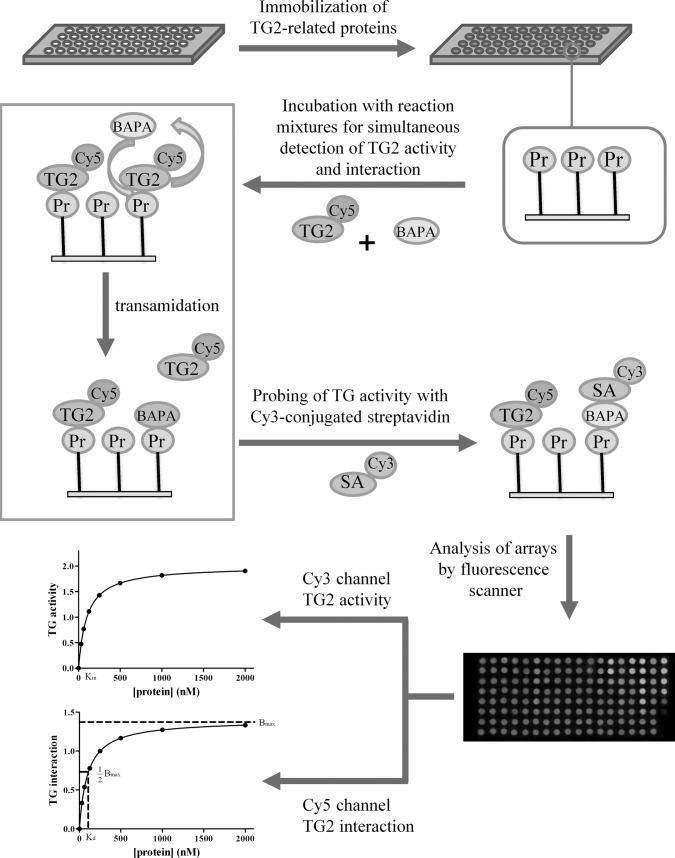
Using protein arrays, we propose as a model system an integrative proteomic approach for simultaneous profiling of the transamidating activity and interactions of transglutaminase 2 (TG2) with TG2-related proteins. TG2, known as tissue transglutaminase, is a member of the calcium-dependent transglutaminase family. Its activity and interactions are associated with a wide variety of diseases and cellular events (34). TG2 is implicated in the pathogenesis of a wide variety of diseases including inflammatory diseases such as celiac sprue, neurodegenerative disorders such as Huntington's, Alzheimer's, and Parkinson's disease, as well as cancers, cardiovascular diseases, and diabetes (34–36). TG2 is also involved in various cellular events including cell growth, cell differentiation, cell adhesion, extracellular matrix crosslinking, and apoptosis (34, 37, 38). In the present study, the transamidating activity and binding affinity of TG2 for 16 proteins were simultaneously monitored using Cy5-conjugated TG2 and protein arrays (Fig. 1). Using this large-scale analysis, we constructed a quantitative activity-interaction (AI)1 map to describe the quantitative interaction of TG2 with its related proteins. Thus, this integrative proteomic approach can be used to characterize functions and regulation mechanisms of a target protein in many biological processes of interest.
Fig. 1.
Schematic diagram for the simultaneous analysis of transamidating activity and interaction of TG2 with TG2-related proteins. BAPA, 5-(biotinamido)pentylamine; Pr, protein; SA, streptavidin; TG2, transglutaminase 2.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
3-Aminopropyltrimethoxysilane, dithiothreitol, thrombin, Cy3-conjugated streptavidin, human serum albumin (HSA), fibrinogen and fibronectin (from human plasma), α-synuclein (human recombinant), retinoblastoma (human recombinant), E-cadherin (human recombinant), and G-actin were obtained from Sigma. 5-(biotinamido)pentylamine (BAPA) was obtained from Pierce Science (Rockford, IL). TG2 from guinea pig liver was purchased from Innovative Research (Novi, Michigan). Human recombinant aldolase A, grancalcin, S100 calcium binding protein A7 (S100A7), osteopontin, B-cell lymphoma 2 (Bcl-2), Sma- and Mad-related protein 2 (SMAD2), and annexin A1 were purchased from ATGen (Seongnam, Korea). Cy5 NHS ester and Bio-spin columns were purchased from Amersham Biosciences (Piscataway, NJ) and Bio-Rad (Hercules, CA), respectively. Human recombinant GST and Rac1 (GST-free) were prepared as previously reported (22).
Preparation of Amine Arrays
Amine-modified glass slides were fabricated according to the procedures of Jung et al. (22). Briefly, glass slides were cleaned with a solution of NH4OH:H2O2:H2O (1:1:5, v/v/v) at 70 °C for 10 min. The slides were immersed in 1.5% 3-aminopropyltrimethoxysilane solution (v/v) in 95% ethanol for 2 h, dried under air, and baked at 110 °C. Teflon tape containing arrayed holes of 1.5 mm diameter were attached to the modified glass slides to prepare well-type amine arrays.
Conjugation of TG2 and Its Related Proteins with Cy5 NHS Ester and Characterization of the Cy5-conjugates
TG2 was labeled with Cy5 NHS ester as previously reported (22). TG2 (100 μl, 100 μg/ml) in 100 mm sodium bicarbonate buffer (pH 8.3) was mixed with 2 μl of 1 mg/ml Cy5 mono NHS-ester in 10% dimethyl sulfoxide and incubated for 2 h on ice. To quench the reactions, 5 μl of 1 m Tris-HCl (pH 8.0) was added to the reaction solution. Reaction mixtures were loaded onto 1.5 ml Sephadex G-25 columns, and Cy5-conjugated TG2 was eluted by centrifugation for 3 min at 1050 × g. To calculate the molar concentrations of Cy5 and TG2 in Cy5-conjugated TG2, molar extinction coefficients of pure TG2 at 280 nm and Cy5 at 280 and 650 nm were determined using the Nanodrop® ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE). TG2 concentrations of Cy5-conjugates were determined using equation (1),
 |
where A280 is the absorbance of Cy5-conjugates at 280 nm, a is the molar extinction coefficient of Cy5 at 280 nm, b is the molar extinction coefficient of Cy5 at 650 nm, and c is the molar extinction coefficient of TG2 at 280 nm.
TG2-related proteins were also labeled with Cy5 NHS ester and absorbance of Cy5-conjugates was determined using the Nanodrop® ND-1000 UV-Vis spectrophotometer. Protein concentrations of Cy5-conjugates were determined using equation (1), where c is the molar extinction coefficient of the proteins at 280 nm.
Determination of Protein Concentration Bound to the Array Surface
We used dry-off measurements to determine the concentrations of proteins bound to the surface of well-type amine arrays, as previously described (39). Briefly, various concentrations of Cy5-conjugated proteins in 1 μl of 9.3 mm phosphate buffer (pH 7.4) were applied in duplicate sets to amine arrays for 1 h at 37 °C. One set of array spots (washed set) was washed with 0.1% Tween 20 in PBS and with milli-Q-purified water whereas another set of array spots (dried set) was dried under ambient conditions. The resulting arrays were scanned with a fluorescence scanner (ScanArray Express, PerkinElmer Life and Analytical Sciences). Protein concentrations bound to the array surface were calculated from fluorescence intensities obtained from washed sets using standard curves generated from dried sets of array spots.
Simultaneous Transamidating Activity and Binding Assay of TG2 Against TG2-related Proteins
To simultaneously determine transamidating activity and interaction of TG2 with TG2-related proteins, we selected 16 commercially available proteins (fibrinogen, fibronectin, α-synuclein, retinoblastoma, aldolase A, grancalcin, S100A7, osteopontin, Bcl-2, SMAD2, annexin A1, Rac1, GST, E-cadherin, G-actin, and HSA) from the list of predicted functional partners on the STRING database, one of the widely used resources for prediction of functional interactions between two proteins, and from the literature (29, 40, 41). For each protein pair, STRING provides a probabilistic confidence score, representing the probability of a functional linkage between two proteins (42). The scores of TG2 with TG2-related proteins are shown in Table I. Sixteen proteins with the indicated concentrations were immobilized on the surface of well-type amine arrays for 1 h at 37 °C and blocked with 2 μm BSA containing 0.1% (v/v) Tween 20 in phosphate buffered saline (PBS) (8.1 mm Na2HPO4, 1.2 mm KH2PO4, 2.7 mm KCl, and 138 mm NaCl, pH 7.4) for 30 min at 37 °C. The resulting protein arrays were washed with 0.1% Tween 20 in PBS and with milli-Q-purified water. Reaction mixtures were prepared in 30 μl of solution containing 40 mm Tris-HCl (pH 7.5), 140 mm NaCl, 2 mm CaCl2, 50 mm dithiothreitol, 5 mm BAPA, 0.01% (v/v) Triton X-100, and 10 μg/ml Cy5-conjugated TG2. Then, 0.7 μl of reaction mixture was applied to array wells and incubated at 37 °C for 30 min. After washing with 0.1% (v/v) Tween-20 in PBS and with milli-Q-purified water, BAPA incorporation catalyzed by TG2 was probed with 10 μg/ml Cy3-conjugated streptavidin at 37 °C for 30 min. Following washing with 0.1% (v/v) Tween-20 in PBS and with milli-Q-purified water, the arrays were dried under compressed air and scanned with a fluorescence scanner using 543 nm and 633 nm lasers (PerkinElmer Life and Analytical Sciences). Fluorescence intensities (FI) of array spots were analyzed using the ScanArray Express program (PerkinElmer Life and Analytical Sciences). Fluorescence intensities at Cy3 and Cy5 channels represent TG2 activities and interactions, respectively, with TG2-related proteins.
Table I. Comparison of predicted protein association scores with experimental values of TG2 activity (normalized FI) and interaction (Kd).
Confidence scores were obtained from the STRING database. TG2 activities of the TG2-related proteins were normalized by median-centering. N.D., nondetectable; AI grade, activity-interaction grade; A, activity; H, high; L, low; I, interaction; S, strong; W, weak; N, non-detectable.
| Protein | Confidence score | Integrative proteomic profiling |
||
|---|---|---|---|---|
| TG2 activity (normalized) | TG2 interaction Kd (nM) | AI grade | ||
| Fibronectin | 0.996 | 4.199 | 6.44 | AHIS |
| Annexin A1 | 0.747 | 0.396 | 6.99 | ALIS |
| Grancalcin | 0.708 | 0.453 | 3.82 | ALIS |
| G-actina | 0.688 | 4.012 | 25.42 | AHIW |
| E-cadherin | 0.659 | 5.653 | N.D. | AHIN |
| Retinoblastoma | 0.649 | 2.881 | 7.33 | AHIS |
| S100A7 | 0.638 | 0.565 | 2.82 | ALIS |
| Aldolase A | 0.619 | 1.074 | 43.21 | ALIW |
| α-synuclein | 0.619 | 0.926 | 44.93 | ALIW |
| Osteopontin | 0.593 | 3.230 | N.D. | AHIN |
| GST* | 0.530 | 0.415 | 469.5 | ALIW |
| Bcl-2 | 0.372 | 0.708 | 26.83 | ALIW |
| Rac1 | 0.218 | N.D. | 15.41 | ANIW |
| SMAD2 | 0.196 | 0.232 | N.D. | ALIN |
| Fibrinogen | No information | 3.175 | 1.15 | AHIS |
| HSA | No information | 1.356 | N.D. | ALIN |
a β-actin and GSTP1 were used to determine the confidence scores of G-actin and GST from the STRING database.
Determination of Dissociation Constants (Kd) Between TG2 and TG2-related Proteins
We quantitatively analyzed binding affinities of TG2 with 16 TG2-related proteins by determining the dissociation constants (Kd). Kd values were calculated by following the modified Langmuir isotherm using GraphPad PRISM (GraphPad Software; San Diego, CA) as follows:
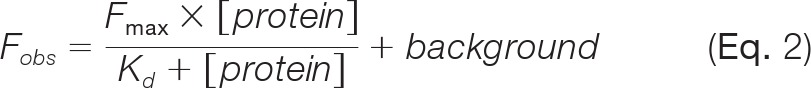 |
where Fobs is the fluorescence intensity of triplicate spots, Fmax is the maximum fluorescence at saturation, [protein] is the protein concentration on the surface, and Kd is the apparent equilibrium dissociation constant.
Data Analysis
ScanArray Express software was used for quantitation of fluorescence intensities and extraction of data. TG2 activities were normalized by the median-centering.
RESULTS
Characterization of Cy5-conjugated TG2
To better characterize the regulation and functions of proteins in biological systems, we developed an integrative proteomic profiling method to simultaneously monitor the activities and interactions of target proteins with their related proteins in a high-throughput assay. We used TG2 and 16 related proteins as a model system for investigating this proteomic profiling. TG2 was conjugated with the Cy5 NHS ester to visualize TG2 interaction with the proteins, and BAPA and Cy3-conjugated streptavidin were used for probing TG2 activity for the proteins, as illustrated in Fig. 1.
To calculate molar concentrations of Cy5 and TG2 in Cy5-conjugated TG2, we determined molar extinction coefficients of TG2 at 280 nm and Cy5 at 280 nm and 650 nm using the Nanodrop® spectrophotometer. The molar extinction coefficient of Cy5 at 650 nm was 219,500 M−1cm−1, whereas that of TG2 at 280 nm was 109,600 M−1cm−1. However, the molar extinction coefficient of Cy5 at 280 nm was very low (3600 M−1cm−1, ∼2% of that of Cy5 at 650 nm) (Fig. 2A). We then reacted 1.27 pmole of TG2 with 1-, 5-, 10-, 20-, and 40-fold molar excesses of Cy5 NHS ester and determined Cy5 and TG2 concentrations in Cy5-conjugated TG2 using the extinction coefficient of Cy5 and equation (1), respectively. The concentration of Cy5 in the Cy5-conjugates increased in proportion to the molar excess of the Cy5 NHS ester, and the molar ratio of Cy5 to TG2 increased from 0.65 to 7.59 (Fig. 2B). However, the TG2 concentration in Cy-5 conjugates was in a similar range from 0.78 μm to 0.84 μm, demonstrating that up to a 40-fold molar excess, the efficiency of TG2 conjugation with Cy5 NHS ester was not significantly affected.
Fig. 2.
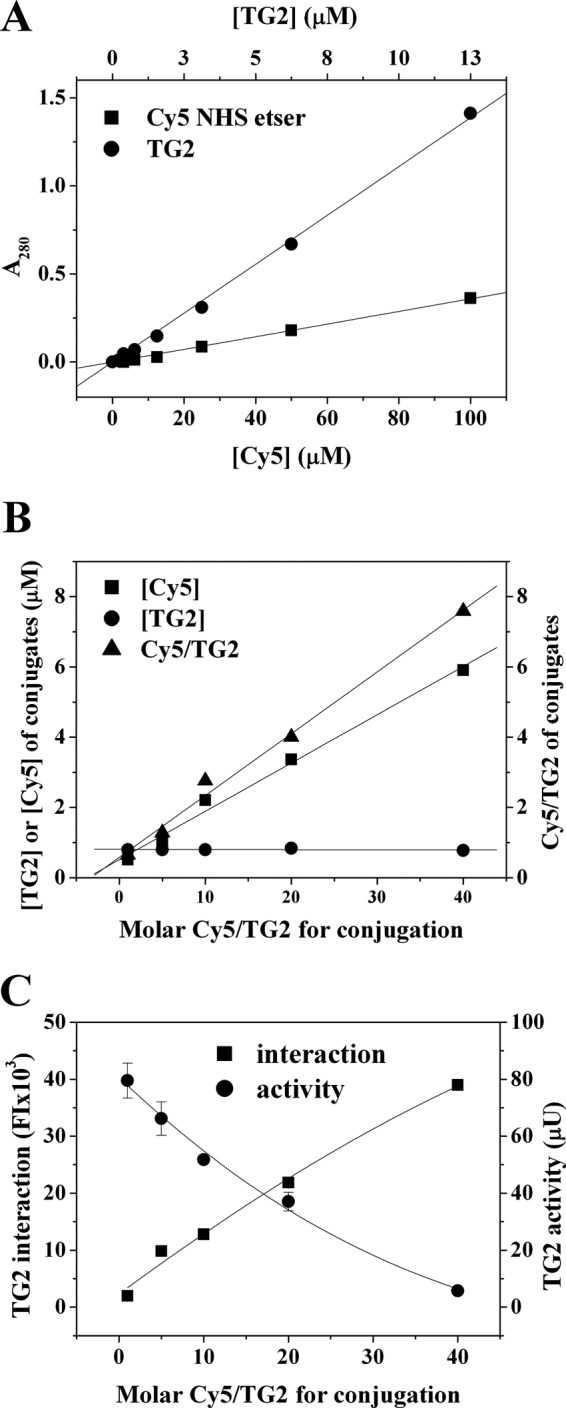
Characterization of Cy5-conjugated TG2. A, Molar extinction coefficients of TG2 and Cy5 NHS ester at 280 nm. B, Concentrations of TG2 and Cy5 and molar ratio of Cy5/TG2 in Cy5-congugated TG2. TG2 was conjugated with 1- to 40-fold molar excess of Cy5 mono NHS-ester, and concentrations of Cy5 and TG2 in the conjugates were determined as described in Methods. C, Reaction mixtures containing 10 μg/ml Cy5-conjugated TG2 were applied to fibrinogen arrays and incubated for 30 min. Transamidating activity and interaction of Cy5-conjugated TG2 with fibronectin were simultaneously determined as described in Methods. The results are expressed as means ± S.D. from three separate experiments.
Because covalent conjugation of an enzyme with a fluorophore may affect the activity of the enzyme, we determined the optimal molar ratio of Cy5 NHS ester to TG2 for TG2 conjugation by measuring transamidating activity and interaction of Cy5-conjugated TG2 using fibronectin arrays (Fig. 2C). The transamidating activity of Cy5-conjugated TG2 was dependent on the molar ratio of Cy5 NHS ester to TG2. The transamidating activity of Cy5-conjugated TG2 at the ratio of 1:1 (Cy5 NHS ester:TG2) decreased to about 61% of unconjugated TG2, and continuously decreased in proportion to an increase in the concentration of Cy5 NHS ester, with a minimal activity at a ratio of 40:1. We then investigated the effect of Cy5-conjugation on TG2 interaction using fibronectin arrays. Interaction of Cy5-conjugated TG2 with fibronectin increased with increased molar ratio of Cy5 NHS ester and TG2 (Fig. 2C), which was opposite to the effect of Cy5-conjugation on the transamidating activity of the conjugated TG2. Thus, for conjugation of TG2 with the Cy5 NHS ester, we chose the molar ratio of 10:1 to minimize the loss of transamidating activity and to maximize the interaction of Cy5-conjugated TG2 with fibronectin.
Determination of the Transamidating Activity of TG2 with TG2-related Proteins
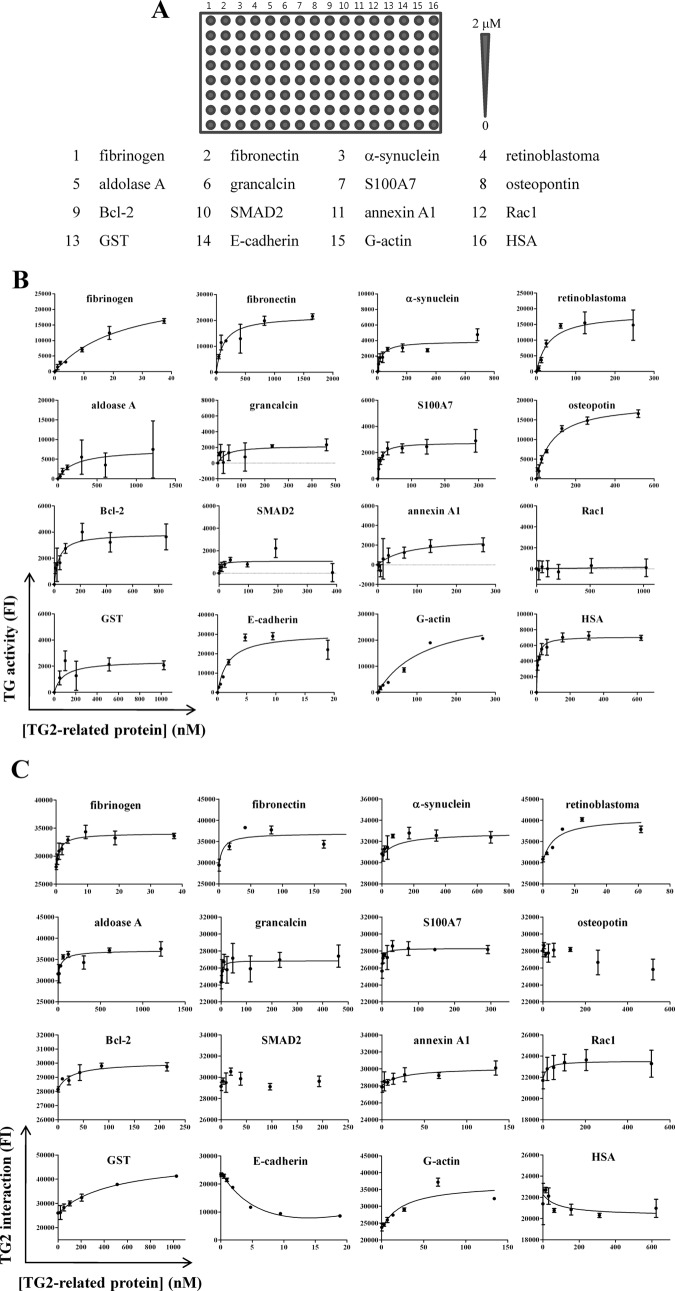
For quantitative analysis of the transamidating activity and interaction of TG2 for TG2-related proteins, we fabricated protein arrays by immobilizing 16 proteins, with indicated concentrations, to the amine surface of well-type arrays, then incubated with reaction mixtures containing 10 μg/ml of Cy5-conjugated TG2 (Fig. 3A). The concentrations of the proteins bound to the array surface were determined by dry-off measurements using solid-phase standard curves. Fluorescence intensities obtained with Cy3 and Cy5 channels represent the transamidating activity and interaction of TG2 with its related proteins, respectively.
Fig. 3.
Simultaneous transamidating activity and binding assays of TG2 with TG2-related proteins. A, Protein arrays were fabricated by immobilizing 16 TG2-related proteins, with indicated concentrations, onto well-type amine arrays. B, C, Reaction mixtures (1 μl) containing 10 μg/ml Cy5-conjugated TG2 were applied to protein arrays for 30 min. Transamidating (TG) activities and interactions of TG2 with 16 proteins were simultaneously determined as described in Methods. The results are expressed as means of fluorescence intensities (FI) ± S.D. from three separate experiments.
For the analysis of transamidating activity, fluorescence intensities at the Cy3 channel were plotted against the concentrations of 16 TG2-related proteins. Most of the proteins, except Rac1, showed a concentration-dependent increase of fluorescence with saturation at 1000 nm (Fig. 3B). TG2 activity was not detectable with Rac1, indicating that there was no nonspecific binding of BAPA to Rac1. Consistently, fluorescence was not detectable when reaction mixtures without TG2 were applied to arrays of fibrinogen and G-actin, with concentrations ranging from 20 nm to 2 μm (applied concentrations; data not shown), demonstrating that the transamidating activity was mostly contributed by TG2-mediated biotinylation of proteins on arrays. To analyze suitability of the proteins as substrates for TG2, we monitored maximal fluorescence intensities for 15 proteins (Table I). The resulting transamidating activities were normalized using the median centering, then used to construct an activity-interaction (AI) map (Fig. 4). The 16 proteins were assigned to three groups based upon the mean value (1.830) of the normalized transamidating activities: high activity (greater than the mean, AH), low activity (lower than the mean, AL), and nondetectable activity (AN) (Table I). Six proteins (fibronectin, G-actin, E-cadherin, retinoblastoma, osteopontin, and fibrinogen) were grouped into the AH group. Nine proteins—annexin A1, grancalcin, S100 calcium binding protein A7 (S100A7), aldolase A, α-synuclein, GST, Bcl-2, SMAD2, and HSA—belonged to the ALgroup. Rac1 was a member of the AN group. Thus, by determining transamidating activity using protein arrays, we successfully applied the integrative proteomic profiling approach to ascertain the suitability of the 16 proteins as TG2 substrates.
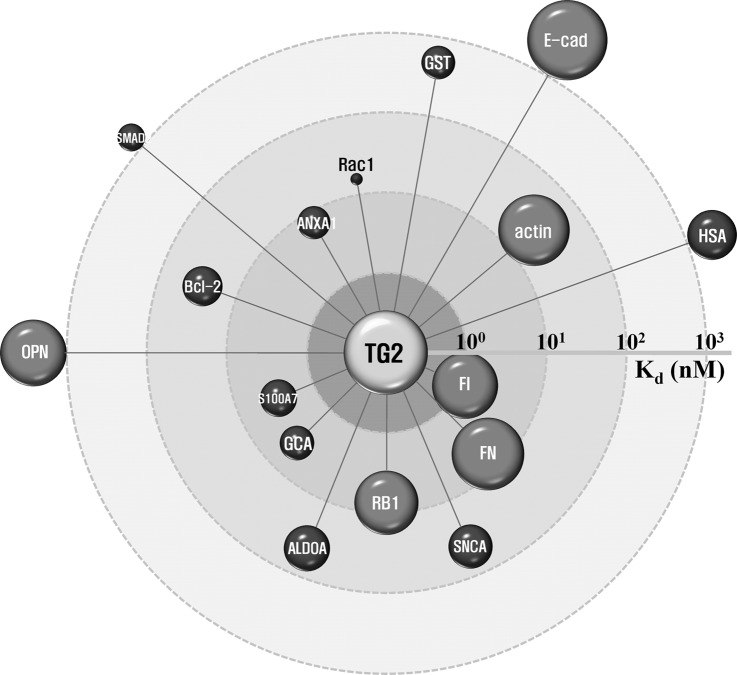
Fig. 4.
Quantitative AI map of TG2 with its related proteins. Normalized transamidating activities and dissociation constants (Kd) of TG2 with 16 proteins were used to construct an integrative proteomic map. In this map, the node volume represents the transamidating activity and the distance between the node and TG2 indicates the Kd value. ALDOA, aldolase A; ANXA, annexin A1; Bcl2, B-cell lymphoma 2; E-cad, E-cadherin; FI, fibrinogen; FN, fibronectin; GCA, grancalcin; GST, gluatathione S-transferase; HSA, human serum albumin; OPN, osteopontin; RB1, retinoblastoma; S100A7, S100 calcium binding protein A7; SMAD2, Sma- and Mad-related protein 2; SNCA, α-synuclein.
Determination of TG2 Affinity with TG2-related Proteins
For quantitative analysis of the interaction of TG2 with its related proteins, fluorescence intensities obtained at the Cy5 channel from the simultaneous activity and binding assay of TG2 (Fig. 3A), were plotted against the concentrations of 16 TG2-related proteins and fitted using equation (2) (Fig. 3C). With the exception of osteopontin, SMAD2, E-cadherin, and HSA, binding of TG2 increased in proportion to the concentration of protein. The binding affinity (Kd) was determined using equation (2) and ranged from 1.15 nm to 469.50 nm (Table I), but was not detectable for the four proteins. The Kd values, together with transamidating activities, were used to construct a quantitative AI map (Fig. 4).
The 16 proteins were assigned to three groups using the log Kd values (logKd); strong (logKd < 1, IS), weak (logKd > 1, IW), and nondetectable (IN) interaction groups (Table I). Six proteins (fibronectin, annexin A1, grancalcin, retinoblastoma, S100A7, and fibrinogen) showed a strong interaction with TG2 (logKd < 1). The log Kd values of another six proteins (G-actin, aldolase A, α-synuclein, GST, Bcl-2, and Rac1) were higher than 1 (logKd > 1), suggesting a weak interaction with TG2. The Kd values for E-cadherin, osteopontin, SMAD2, and HSA were undetectable, indicating that these proteins did not interact with TG2. Thus, we could simultaneously and quantitatively analyze the transamidating activity and binding affinity of TG2 for 16 TG2-related proteins by the use of integrative proteomic profiling in conjunction with protein arrays.
Integrative Proteomic Networks of Quantitative Transamidating Activity and Interaction of TG2 with TG2-related Proteins
Visualization of protein interaction networks is the best method for presenting known and predicted protein interactions, using experimental and computational information from databases. Web-based databases, such as STRING, provide extensive information on interactomes. However, these databases provide predictive rather than quantitative information of protein activity and dissociation constant (Kd). Using data derived from large-scale analyses, we circumvented this limitation by constructing a quantitative AI map of TG2 with 16 TG2-related proteins (Fig. 4). In this map, the node size and distance between TG2 and various nodes represent the transamidating activity and binding affinity, respectively, of TG2 for different proteins. Four proteins (E-cadherin, osteopontin, SMAD2, and HSA) are located outside the Kd of 1,000, because Kd values were not detectable for these proteins. The AI map can therefore provide a systematic view of protein interactions based both on activity and binding affinity.
The 16 proteins were assigned to seven groups by an AI grading system; high activity and strong interaction (AHIS), high activity and weak interaction (AHIW), high activity and nondetectable interaction (AHIN), low activity and strong interaction (ALIS), low activity and weak interaction (ALIW), low activity and nondetectable interaction (ALIN), and nondetectable activity and weak interaction groups (ANIS) (Table I). The AI grading system is based on the combination of three activity groups (AH, AL, and AN) and three interaction groups (IS, IW, and IN). No proteins were assigned to the group for ANIS or ANIN. Thus, this integrative proteomic profiling based on the quantitative AI map and AI grading system is a potential approach to construct real functional networks of the quantitative activities and interactions of target proteins with various proteomes to understand the functions and regulation of the target proteins.
DISCUSSION
In this report, we present an integrative proteomic profiling method for the simultaneous analysis of activities and interactions of target proteins, in order to provide an overview of the functions and regulation mechanisms of target proteins in biological processes of interest. TG2 was used as a model system because this protein is involved in the pathogenesis of various diseases including celiac disease, and in a variety of cellular events (34, 36). TG2 is a member of the transglutaminase family and catalyzes the formation of an ε-(γ-glutamyl)lysine bond between glutamine and lysine in a Ca2+-dependent manner (29, 34). In the Ca2+-dependent transamidation reaction, Ca2+ activates transamidating activity of TG2 by inducing a conformational change. The side chain of glutamine forms a thioester with the active site cysteine of TG2 by transfer of an acyl acceptor amine to form an isopeptide bond (34, 43). TG2 is regulated by small molecules including Ca2+, GTP, and ATP (34). In simultaneous assays using fibrinogen and G-actin arrays, GTPγS suppressed transamidating activity and decreased binding affinity (increase in Kd) of TG2 for the proteins (data not shown). These results demonstrate that the integrative proteomic profiling can reveal regulation of TG2 activity and interaction for related proteins by small molecules. Thus, the simultaneous determination of transamidating activities and interactions of TG2 with its related proteins using this integrative proteomic approach can be used for revealing functions and regulation mechanisms of TG2, as well as for screening TG2-related proteins.
To investigate physiologically important interactions and/or enzymatic reactions, we selected 16 TG2-related proteins from the STRING database. Each protein pair in the STRING database provides a probabilistic confidence score, representing a rough estimate of how likely a given association describes a functional linkage between two proteins (42). The scores of the predicted functional associations of TG2 with the 16 proteins were compared with transamidating activities and dissociation constants obtained by simultaneous profiling (Table I). In general, confidence scores correlated well with the quantitative transamidating activities and interactions obtained by integrative proteomic profiling of the proteins. As expected from the highest confidence score among the 16 proteins, fibronectin had a strong binding affinity to TG2 and was categorized in the AHIS group by the AI grading system. Annexin A1 and grancalcin, with relatively higher confidence scores, also showed a strong interaction with TG2 and belonged to the ALIS or AHIW group. Bcl-2 and Rac1, with lower confidence scores, had a weak interaction with TG2 (the ALIW or ANIW groups). Interestingly, retinoblastoma showed a strong binding affinity with a high transamidating activity, even though the protein had a moderate confidence score. Thus, to characterize the functions and regulation of proteins in biological systems, it is necessary to evaluate interaction information provided by web-based databases, including STRING, with novel approaches such as integrative proteomic profiling based on the quantitative AI map as well as the AI grading system.
We assigned 16 proteins into seven groups using a quantitative AI map, based upon transamidating activity and binding affinity (Kd) of TG2 with the proteins. Three proteins (fibronectin, retinoblastoma, and fibrinogen) exhibited high activity and strong interaction (AHIS group), indicating that the three proteins are both good substrates and can strongly interact with TG2. Consistent with previous reports, fibronectin is one of the best-known interacting proteins and substrates for TG2. It plays a major role in cell adhesion, growth, migration, and differentiation (44). Retinoblastoma is known as a potential TG2 substrate in U937 monocytic cells undergoing apoptosis (37). Confidence score for fibrinogen is not available from the STRING database, but the protein is one of the best-known substrates used in a transamidating activity assay (29). Three proteins (annexin A1, grancalcin, and S100A7) may be closely related to TG2 by interaction rather than by transamidating activity in TG2-related cellular events (ALIS group). Actin is known as a substrate for TG2 in human leukemia cells undergoing apoptosis (45). Consistent with a previous report (45), G-actin was found to be a good substrate for transamidating activity of TG2, but bound weakly to this protein (AHIW group). Four proteins (aldolase A, α-synuclein, GST, and Bcl-2) exhibited low activity and weak interaction (ALIW group). E-cadherin and osteopontin showed a high transamidating activity with nondetectable interaction (AHIN group). Interestingly, the interactions of TG2 with E-cadherin and osteopontin decreased with increasing protein concentration, suggesting that TG2 interacts weakly with E-cadherin and osteopontin, and dissociates from those proteins upon forming new isopeptide bonds between the proteins and BAPA in the intermediate state. Rac1 showed a weak interaction with TG2 with no detectable activity (ANIW group). Finally, based upon our studies, SMAD2 and HSA might be used as substrates for transamidating with no interaction (ALIN group). Thus, the AI grading system based on a quantitative AI map derived from the integrative proteomic profiling was successfully used for annotating the activities and interactions of TG2 with its related proteins.
In conclusion, using protein arrays, we successfully performed simultaneous profiling of the transamidating activities and interactions of TG2 with 16 proteins, and constructed a quantitative AI map to provide a systematic view of protein interactions based on activity and binding affinity. We then assigned the 16 proteins into seven groups using the AI grading system. This integrative proteomic approach can therefore provide a global visualization of functions and regulation mechanisms of target proteins in biological systems. In addition, the approach can validate possible protein networks derived from web-based databases and can specify network data into functional interactions of target proteins with various proteomes. Thus, integrative proteomic profiling has the potential to provide a systemic view of protein functions in many biological processes.
Footnotes
* This work was supported in part by the Korean Ministry of Health and Welfare through the National R&D Program for Cancer Control (1020420) and the Korea Research Foundation through the Basic Research Program (2008-05943).
1 The abbreviations used are:
- AI map
- activity-interaction map
- BAPA
- 5-(biotinamido)pentylamine
- Bcl-2
- B-cell lymphoma 2
- HSA
- human serum albumin
- Kd
- dissociation constants
- S100A7
- S100 calcium binding protein A7
- SMAD2
- Sma- and Mad-related protein 2
- TG2
- transglutaminase 2.
REFERENCES
- 1. Sadaghiani A. M., Verhelst S. H., Bogyo M. (2007) Tagging and detection strategies for activity-based proteomics. Curr. Opin. Chem. Biol. 11, 20–28 [DOI] [PubMed] [Google Scholar]
- 2. Schmidinger H., Hermetter A., Birner-Gruenberger R. (2006) Activity-based proteomics: enzymatic activity profiling in complex proteomes. Amino Acids 30, 333–350 [DOI] [PubMed] [Google Scholar]
- 3. Nomura D. K., Dix M. M., Cravatt B. F. (2010) Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer 10, 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fonović M., Bogyo M. (2008) Activity-based probes as a tool for functional proteomic analysis of proteases. Expert Rev. Proteomics 5, 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillet L. C., Namoto K., Ruchti A., Hoving S., Boesch D., Inverardi B., Mueller D., Coulot M., Schindler P., Schweigler P., Bernardi A., Gil-Parrado S. (2008) In-cell selectivity profiling of serine protease inhibitors by activity-based proteomics. Mol. Cell. Proteomics 7, 1241–1253 [DOI] [PubMed] [Google Scholar]
- 6. Goddard J. P., Reymond J. L. (2004) Enzyme assays for high-throughput screening. Curr. Opin. Biotechnol. 15, 314–322 [DOI] [PubMed] [Google Scholar]
- 7. Wiedl T., Arni S., Roschitzki B., Grossmann J., Collaud S., Soltermann A., Hillinger S., Aebersold R., Weder W. (2011) Activity-based proteomics: Identification of ABHD11 and ESD activities as potential biomarkers for human lung adenocarcinoma. J. Proteomics 74, 1884–1894 [DOI] [PubMed] [Google Scholar]
- 8. Viertler M., Schittmayer M., Birner-Gruenberger R. (2012) Activity based subcellular resolution imaging of lipases. Bioorg. Med. Chem. 20, 628–632 [DOI] [PubMed] [Google Scholar]
- 9. Kool J., Jonker N., Irth H., Niessen W. M. (2011) Studying protein-protein affinity and immobilized ligand-protein affinity interactions using MS-based methods. Anal. Bioanal. Chem. 401, 1109–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., McCartney R. R., Schmidt M. C., Rachidi N., Lee S. J., Mah A. S., Meng L., Stark M. J., Stern D. F., De Virgilio C., Tyers M., Andrews B., Gerstein M., Schweitzer B., Predki P. F., Snyder M. (2005) Global analysis of protein phosphorylation in yeast. Nature 438, 679–684 [DOI] [PubMed] [Google Scholar]
- 11. Lievens S., Eyckerman S., Lemmens I., Tavernier J. (2010) Large-scale protein interactome mapping: strategies and opportunities. Expert Rev. Proteomics 7, 679–690 [DOI] [PubMed] [Google Scholar]
- 12. Tarassov K., Messier V., Landry C. R., Radinovic S., Serna Molina M. M., Shames I., Malitskaya Y., Vogel J., Bussey H., Michnick S. W. (2008) An in vivo map of the yeast protein interactome. Science 320, 1465–1470 [DOI] [PubMed] [Google Scholar]
- 13. Barrios-Rodiles M., Brown K. R., Ozdamar B., Bose R., Liu Z., Donovan R. S., Shinjo F., Liu Y., Dembowy J., Taylor I. W., Luga V., Przulj N., Robinson M., Suzuki H., Hayashizaki Y., Jurisica I., Wrana J. L. (2005) High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307, 1621–1625 [DOI] [PubMed] [Google Scholar]
- 14. Goehler H., Lalowski M., Stelzl U., Waelter S., Stroedicke M., Worm U., Droege A., Lindenberg K. S., Knoblich M., Haenig C., Herbst M., Suopanki J., Scherzinger E., Abraham C., Bauer B., Hasenbank R., Fritzsche A., Ludewig A. H., Büssow K., Coleman S. H., Gutekunst C. A., Landwehrmeyer B. G., Lehrach H., Wanker E. E. (2004) A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell. 15, 853–865 [DOI] [PubMed] [Google Scholar]
- 15. Nibbe R. K., Markowitz S., Myeroff L., Ewing R., Chance M. R. (2009) Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol. Cell. Proteomics 8, 827–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ruffner H., Bauer A., Bouwmeester T. (2007) Human protein-protein interaction networks and the value for drug discovery. Drug Discov. Today 12, 709–716 [DOI] [PubMed] [Google Scholar]
- 17. Liang P. H., Wang S. K., Wong C. H. (2007) Quantitative analysis of carbohydrate-protein interactions using glycan microarrays: determination of surface and solution dissociation constants. J. Am. Chem. Soc. 129, 11177–11184 [DOI] [PubMed] [Google Scholar]
- 18. Jung J. W., Jung S. H., Kim H. S., Yuk J. S., Park J. B., Kim Y. M., Han J. A., Kim P. H., Ha K. S. (2006) High-throughput analysis of GST-fusion protein expression and activity-dependent protein interactions on GST-fusion protein arrays with a spectral surface plasmon resonance biosensor. Proteomics 6, 1110–1120 [DOI] [PubMed] [Google Scholar]
- 19. Jung S. H., Kong D. H., Park J. H., Lee S. T., Hyun J., Kim Y. M., Ha K. S. (2010) Rapid analysis of matrix metalloproteinase-3 activity by gelatin arrays using a spectral surface plasmon resonance biosensor. Analyst 135, 1050–1057 [DOI] [PubMed] [Google Scholar]
- 20. Mori T., Inamori K., Inoue Y., Han X., Yamanouchi G., Niidome T., Katayama Y. (2008) Evaluation of protein kinase activities of cell lysates using peptide microarrays based on surface plasmon resonance imaging. Anal. Biochem. 375, 223–231 [DOI] [PubMed] [Google Scholar]
- 21. Han M. K., Oh Y. H., Kang J., Kim Y. P., Seo S., Kim J., Park K., Kim H. S. (2009) Protein profiling in human sera for identification of potential lung cancer biomarkers using antibody microarray. Proteomics 9, 5544–5552 [DOI] [PubMed] [Google Scholar]
- 22. Jung J. W., Jung S. H., Yoo J. O., Suh I. B., Kim Y. M., Ha K. S. (2009) Label-free and quantitative analysis of C-reactive protein in human sera by tagged-internal standard assay on antibody arrays. Biosens. Bioelectron. 24, 1469–1473 [DOI] [PubMed] [Google Scholar]
- 23. Shafer M. W., Mangold L., Partin A. W., Haab B. B. (2007) Antibody array profiling reveals serum TSP-1 as a marker to distinguish benign from malignant prostatic disease. Prostate 67, 255–267 [DOI] [PubMed] [Google Scholar]
- 24. Kwon M. H., Kong D. H., Jung S. H., Suh I. B., Kim Y. M., Ha K. S. (2011) Rapid determination of blood coagulation factor XIII activity using protein arrays for serodiagnosis of human plasma. Anal. Chem. 83, 2317–2323 [DOI] [PubMed] [Google Scholar]
- 25. AbdAlla S., Lother H., Langer A., el Faramawy Y., Quitterer U. (2004) Factor XIIIA transglutaminase crosslinks AT1 receptor dimers of monocytes at the onset of atherosclerosis. Cell 119, 343–354 [DOI] [PubMed] [Google Scholar]
- 26. Jones R. B., Gordus A., Krall J. A., MacBeath G. (2006) A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439, 168–174 [DOI] [PubMed] [Google Scholar]
- 27. Wolf-Yadlin A., Sevecka M., MacBeath G. (2009) Dissecting protein function and signaling using protein microarrays. Curr. Opin. Chem. Biol. 13, 398–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwon M. H., Jung S. H., Kim Y. M., Ha K. S. (2011) Simultaneous activity assay of two transglutaminase isozymes, blood coagulation factor XIII and transglutaminase 2, using fibrinogen arrays. Anal. Chem. 83, 8718–8724 [DOI] [PubMed] [Google Scholar]
- 29. Kwon M. H., Jung J. W., Jung S. H., Park J. Y., Kim Y. M., Ha K. S. (2009) Quantitative and rapid analysis of transglutaminase activity using protein arrays in mammalian cells. Mol. Cells 27, 337–343 [DOI] [PubMed] [Google Scholar]
- 30. Kong D.-H., Jung S.-H., Lee S.-T., Ha K.-S. (2010) On-chip assay of matrix metalloproteinase-3 activity using fluorescence-conjugated gelatin arrays. BioChip J. 4, 210–216 [Google Scholar]
- 31. Mugherli L., Burchak O. N., Balakireva L. A., Thomas A., Chatelain F., Balakirev M. Y. (2009) In situ assembly and screening of enzyme inhibitors with surface-tension microarrays. Angew Chem. Int. Ed. Engl. 48, 7639–7644 [DOI] [PubMed] [Google Scholar]
- 32. Salisbury C. M., Maly D. J., Ellman J. A. (2002) Peptide microarrays for the determination of protease substrate specificity. J. Am. Chem. Soc. 124, 14868–14870 [DOI] [PubMed] [Google Scholar]
- 33. Kong D. H., Jung S. H., Lee S. T., Kim Y. M., Ha K. S. (2012) Monitoring of proteolytic enzyme activity using phase transition-based peptide arrays. Biosens. Bioelectron. 36, 147–153 [DOI] [PubMed] [Google Scholar]
- 34. Park D., Choi S. S., Ha K. S. (2010) Transglutaminase 2: a multi-functional protein in multiple subcellular compartments. Amino Acids 39, 619–631 [DOI] [PubMed] [Google Scholar]
- 35. Reif S., Lerner A. (2004) Tissue transglutaminase–the key player in celiac disease: a review. Autoimmun Rev. 3, 40–45 [DOI] [PubMed] [Google Scholar]
- 36. Iismaa S. E., Mearns B. M., Lorand L., Graham R. M. (2009) Transglutaminases and disease: lessons from genetically engineered mouse models and inherited disorders. Physiol. Rev. 89, 991–1023 [DOI] [PubMed] [Google Scholar]
- 37. Oliverio S., Amendola A., Di Sano F., Farrace M. G., Fesus L., Nemes Z., Piredda L., Spinedi A., Piacentini M. (1997) Tissue transglutaminase-dependent posttranslational modification of the retinoblastoma gene product in promonocytic cells undergoing apoptosis. Mol. Cell. Biol. 17, 6040–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ohtake Y., Nagashima T., Suyama-Satoh S., Maruko A., Shimura N., Ohkubo Y. (2007) Transglutaminase catalyzed dissociation and association of protein-polyamine complex. Life Sci. 81, 577–584 [DOI] [PubMed] [Google Scholar]
- 39. Jung S. H., Kong D. H., Park S. W., Kim Y. M., Ha K. S. (2012) Quantitative kinetics of proteolytic enzymes determined by a surface concentration-based assay using peptide arrays. Analyst DOI: 10.1039/c2an35080g [DOI] [PubMed] [Google Scholar]
- 40. Griffin M., Casadio R., Bergamini C. M. (2002) Transglutaminases: nature's biological glues. Biochem. J. 368, 377–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Esposito C., Caputo I. (2005) Mammalian transglutaminases. Identification of substrates as a key to physiological function and physiopathological relevance. FEBS J. 272, 615–631 [DOI] [PubMed] [Google Scholar]
- 42. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., von Mering C. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fleckenstein B., Qiao S. W., Larsen M. R., Jung G., Roepstorff P., Sollid L. M. (2004) Molecular characterization of covalent complexes between tissue transglutaminase and gliadin peptides. J. Biol. Chem. 279, 17607–17616 [DOI] [PubMed] [Google Scholar]
- 44. Li B., Antonyak M. A., Druso J. E., Cheng L., Nikitin A. Y., Cerione R. A. (2010) EGF potentiated oncogenesis requires a tissue transglutaminase-dependent signaling pathway leading to Src activation. Proc. Natl. Acad. Sci. U.S.A. 107, 1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nemes Z., Jr., Adany R., Balazs M., Boross P., Fésüs L. (1997) Identification of cytoplasmic actin as an abundant glutaminyl substrate for tissue transglutaminase in HL-60 and U937 cells undergoing apoptosis. J. Biol. Chem. 272, 20577–20583 [DOI] [PubMed] [Google Scholar]