Abstract
The major histocompatibility complex (MHC) class II-associated Invariant chain (Ii) is present in professional antigen presenting cells where it regulates peptide loading onto MHC class II molecules and the peptidome presented to CD4+ T lymphocytes. Because Ii prevents peptide loading in neutral subcellular compartments, we reasoned that Ii− cells may present peptides not presented by Ii+ cells. Based on the hypothesis that patients are tolerant to MHC II-restricted tumor peptides presented by Ii+ cells, but will not be tolerant to novel peptides presented by Ii− cells, we generated MHC II vaccines to activate cancer patients' T cells. The vaccines are Ii− tumor cells expressing syngeneic HLA-DR and the costimulatory molecule CD80. We used liquid chromatography coupled with mass spectrometry to sequence MHC II-restricted peptides from Ii+ and Ii− MCF10 human breast cancer cells transfected with HLA-DR7 or the MHC Class II transactivator CIITA to determine if Ii− cells present novel peptides. Ii expression was induced in the HLA-DR7 transfectants by transfection of Ii, and inhibited in the CIITA transfectants by RNA interference. Peptides were analyzed and binding affinity predicted by artificial neural net analysis. HLA-DR7-restricted peptides from Ii− and Ii+ cells do not differ in size or in subcellular location of their source proteins; however, a subset of HLA-DR7-restricted peptides of Ii− cells are not presented by Ii+ cells, and are derived from source proteins not used by Ii+ cells. Peptides from Ii− cells with the highest predicted HLA-DR7 binding affinity were synthesized, and activated tumor-specific HLA-DR7+ human T cells from healthy donors and breast cancer patients, demonstrating that the MS-identified peptides are bonafide tumor antigens. These results demonstrate that Ii regulates the repertoire of tumor peptides presented by MHC class II+ breast cancer cells and identify novel immunogenic MHC II-restricted peptides that are potential therapeutic reagents for cancer patients.
Cancer vaccines are a promising tool for cancer treatment and prevention because of their potential for inducing tumor-specific responses in conjunction with minimal toxicity for healthy cells. Cancer vaccines are based on the concept that tumor cells synthesize multiple peptides that are potential immunogens, and that with the appropriate vaccine protocol, these peptides will activate an efficacious antitumor response in the patient. Much effort has been invested in identifying and testing tumor-encoded peptides, particularly peptides presented by major histocompatibility complex (MHC)1 class I, molecules capable of activating CD8+ T-cells that directly kill tumor cells (1, 2). Fewer studies have been devoted to identifying MHC class II-restricted peptides for the activation of tumor-reactive CD4+ T-cells despite compelling evidence that Type 1 CD4+ T helper cells facilitate the optimal activation of CD8+ T-cells and the generation of immune memory, which is likely to be essential for protection from metastatic disease.
Activation of CD4+ T cells requires delivery of a costimulatory signal plus an antigen-specific signal consisting of peptide bound to an MHC II molecule. Most cells do not express MHC II or costimulatory molecules, so CD4+ T cells are typically activated by professional antigen presenting cells (APC), which endocytose exogenously synthesized antigen and process and present it in the context of their own MHC II molecules. This processing and presentation process requires Invariant chain (Ii), a molecule that is coordinately synthesized with MHC II molecules and prevents the binding and presentation of APC-encoded endogenous peptides (3, 4). As a result, tumor-reactive CD4+ T cells are activated to tumor peptides generated by the antigen processing machinery of professional APC, rather than peptides generated by the tumor cells. Because of the potential discrepancy in peptide generation between professional APC and tumor cells, and the critical role of Ii in preventing the presentation of endogenous peptides, we have generated “MHC II cancer vaccines” that consist of Ii− tumor cells transfected with syngeneic MHC class II and CD80 genes. We reasoned that MHC II+Ii−CD80+ tumor cells may present a novel repertoire of MHC II-restricted tumor peptides that are not presented by professional APC, and therefore may be highly immunogenic. Once activated, CD4+ T cells produce IFNγ and provide help to CD8+ T cells and do not need to react with native tumor cells. Therefore, the MHC II vaccines have the potential to activate CD4+ Th1 cells that facilitate antitumor immunity. In vitro (5) and in vivo (5–7) studies with mice support this conclusion. In vitro studies with human MHC II vaccines further demonstrate that the absence of Ii facilitates the activation of MHC II-restricted tumor-specific CD4+ type 1 T cells of HLA-DR-syngeneic healthy donors and cancer patients, and that the vaccines activate CD4+ T cells with a distinct repertoire of T cell receptors (8–12). A critical negative role for Ii is also supported by studies of human acute myelogenous leukemia (AML). High levels of class II-associated invariant chain peptide (CLIP), a degradation product of Ii, by leukemic blasts is associated with poor patient prognosis (13, 14), whereas down-modulation of CLIP on AML cells increases the activation of tumor-reactive human CD4+ T cells (14, 15).
We have now used mass spectrometry to identify MHC II-restricted peptides from MHC II+Ii− and MHC II+Ii+ human breast cancer cells to test the concept that the absence of Ii facilitates the presentation of unique immunogenic MHC II-restricted peptides. We report here that a subset of MHC II-restricted peptides from HLA-DR7+ breast cancer cells are unique to Ii− cells and are derived from source proteins not used by Ii+ cells. Ii− peptides have high binding affinity for HLA-DR7 and activate tumor-specific T-cells from the peripheral blood of healthy donors and breast cancer patients. This is the first study to compare the human tumor cell MHC II peptidome in the absence or presence of Ii and to demonstrate that MHC II+Ii− tumor cells present novel immunogenic MHC II-restricted peptides that are potential therapeutic reagents for cancer patients.
EXPERIMENTAL PROCEDURES
Cell Lines, Transductants, PBMC
Human breast cancer cell line MCF10CA1 (hereafter called MCF10), its nonmalignant counterpart MCF10A (16), MCF10 transductants (MCF10/DR7/CD80, MCF10/DR7/CD80/Ii, MCF10/CIITA/CD80, and MCF10/CIITA/CD80/Ii siRNA32; hereafter called /DR7, /DR7/Ii, /CIITA, and/CIITA/Ii siRNA) were cultured and/or generated as described (11, 12). Cells were expanded to ∼1 × 109 cells/line using Hyperflask tissue culture flasks (Corning, Corning, NY). MCF10 cells are: HLA-DRβ1*0401, DRβ1*0701. Peripheral blood mononuclear cells (PBMC) from healthy human donors and from breast cancer patients were obtained from the University of Maryland Medical School. Healthy donor BC100206 is HLA-DRβ1*0401and *0701; healthy donors BC100306, BC061505, and BC051505 are DRβ1*0701. Breast cancer patients 3 and 10 are HLA-DRβ1*0701. Breast cancer patient 3 is stage III, ER+/PR+, HER-2/neu−; patient 10 is stage II, ER+/PR+, HER-2/neu−. Patients were bled into ACD (citrate) tubes. Within 24 h of collection PBMC were isolated on Ficoll gradients, immediately cryopreserved at a controlled freeze rate of 1 °C/min, and stored in the vapor phase of liquid nitrogen until used. PBMC were >90% viable upon thawing. Use of human materials was approved by the UMBC IRB.
Reagents, Antibodies, and Flow Cytometry
Chemicals were purchased form Sigma Aldrich unless otherwise noted. Monoclonal antibodies L243-FITC (HLA-DR-specific), CD80-PE, Ii, CD3-FITC, CD8-PE, CD45RO-allophycocyanin, CD25-PE-Cy7, and CD56-PE-Cy7 were purchased from BD Pharmingen. HLA-A,B,C-PE-Cy5, CD4-Pacific Blue and CD4-eFluor 460 were purchased from Biolegend (San Diego, CA). Cell surface and intracellular staining for flow cytometry was performed as described (9, 12). Stained cells were analyzed using a Cyan ADP flow cytometer and Summit analysis software, v2.1 (Beckman/Coulter).
HLA-DR Induction
Cells were incubated at 37 °C for 48 h with 200 units IFNγ/ml. Washed cells were stained with mAbs to HLA-DR to ascertain HLA-DR expression.
MHC II Peptide Isolation, LC-MS/MS Analysis
Approximately 1 × 109 cultured cells per cell line were harvested and lysed on ice for 1 h in lysis buffer (20 mm Tris-HCl, pH 8.0; 150 mm NaCl,1% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate; (17) containing a Complete Mini Protease Inhibitor Mixture tablet (Roche). Lysates were ultra centrifuged at 129,888 × g for 1 h at 4 °C in a SW40Ti swinging bucket rotor and the supernatants harvested and stored at −80 °C. MHC II peptides were obtained by HPLC (Biologic HR, BioRad, Hercules, CA) as follows: Thawed supernatants were pre-cleared on a 5 ml Protein G Sepharose column equilibrated with lysis buffer and then applied to a 2 ml in-house generated L243 mAb (pan HLA-DR) Sepharose column (9) equilibrated with two column volumes of lysis buffer. The loaded column was sequentially washed with 20 column volumes of 20 mm Tris-HCl, 150 mm NaCl, pH 8.0; 20 mm Tris-HCl, 1 m NaCl, pH 8.0, followed by 20 column volumes of 20 mm Tris-HCl, pH 8.0. MHC II-peptide complexes were eluted with five column volumes of 0.2 N acetic acid, and eluates were lyophilized and stored at −20 °C. Lyophilized material was resuspended in 50 μl water and acidified to ∼pH3 with glacial acetic acid to dissociate peptides from MHC II molecules. The resulting material was analyzed on a LTQ XL (ThermoFisher) mass spectrometer interfaced with a UltiMate 3000 nanoLC (Dionex). Peptides were concentrated on FACET biocompatible strong cation exchange (SCX) trapping columns (Protea Biosciences, Morgantown, WV) and separated on Acclaim PepMap 100 columns (C18,5 μm, 100 Å, 75 μm i.d. × 15 cm, Dionex) at 300 nl/min using a 70 min. gradient program. Mobile phases were: 0.1% formic acid in water (A), 80% acetonitrile, 20% water with addition of 0.1% formic acid (B), and 2 m ammonium bicarbonate in water (C). Peptide samples were loaded onto the SCX trapping column and washed with 4% B, 96% A for 0.75 min, then eluted onto an analytical column over 10 min at 15% C, 4% B. Peptides were separated by a gradient of mobile phase B, which was 15, 45, 100, and 4% at min. 20, 40, 50, and 55, respectively. Eluted peptides were ionized on a Thermo Scientific (nanospray ionization) NSI source, equipped with a dynamic NSI probe. The electrospray capillary was at 200 °C temperature and under 1.7 kV voltage. For each cell line two affinity purifications were performed, and for each affinity purification two LC-MS/MS runs were conducted. Mass spectrometric analysis was performed using unattended data-dependent acquisition mode, in which the mass spectrometer automatically switched between acquiring a survey mass spectrum (full MS) and consecutive CID of up to four most abundant ions (MS/MS). To facilitate identification of a broad range of peptides, dynamic exclusion for MS/MS was used. Individual precursor ions were selected no more than twice over the duration of 20 s, and were then placed in the exclusion list for 120 s. The m/z tolerance window for dynamic exclusion was 1.5 Da.
Peptide Identification
Spectra were searched against the International Protein Index (IPI) human database, (version 3.26, 67665 entries) on BioWorks 3.3.1 SP1 platform (ThermoFisher), using SEQUEST and the following search parameters: peptide mass tolerance: 2 Da; fragment mass tolerance: 1 Da, no enzyme specificity. Results of the SEQUEST searches (.out files) were converted into mzXML files and analyzed on Peptide Prophet using Trans-Proteomic Pipeline version 4.3 revision 1 (18). Peptides that were identified in both affinity purifications and both LC-MS/MS runs were considered as reliable identifications. These peptides had a minimal Xcorr score of 1.5 for the charge state of 1, a minimal Xcorr score of 2.0 for the charge state of 2, a minimal Xcorr score of 2.5 for the charge state of 3, and a minimal Peptide Prophet probability of 0.07. Peptides with Peptide Prophet probability greater than 0.05 were subjected to Artificial Neural Network (ANN) analysis. We purposefully included all peptides identified with a more relaxed filtering criterion to facilitate MHC II peptide binding prediction accuracy. Subcellular localization of source proteins for peptides was determined using LOCATE subcellular localization database (http://locate.imb.uq.edu.au/) and WOLF Psort (http://wolfpsort.org/).
Artificial Neural Network Analysis
Two hundred and sixty seven published HLA-DR7-restricted peptides of 9–25 amino acids in length (MHCBN database, version 4.0; http://www.imtech.res.in/raghava/mhcbn/index.html) were used to train an ANN. The network was a three layer back propagation network, trained to produce four output classes, representing highest to lowest binding affinities. The amino acid strings were initially coded by amino acid names. A “1” in a specific position of the 20 node input corresponded to the particular amino acid in the ordered list of 20 amino acids; the other positions being set to “0.” The binding region was assumed to be nine amino acids in length. Therefore, larger peptides were broken into (n-9 + 1) 9-mers prior to coding. This produced 1318 9-mer sequences. A 9-mer was coded by 9 lines of 20 input nodes and 4 output nodes representing the affinity levels. Members of the smaller affinity classes were duplicated as necessary to provide roughly equal representation in the training set. Training was stopped when the root mean square error reached 0.30. Nine-mers whose highest score was in the expected affinity group were chosen for continued study. The network was thus used to index relevant 9-mers within the larger peptide sequence and produced 122, 106, 12, and 27 peptides in high (H), medium (M), low, and nonbinding groups, respectively. A new neural network was then trained using members from each binding affinity group. Each amino acid was represented by its Borstnick-Hofacker (B-H) distance (19) to each of the other 20 amino acids. These distances were divided into 16 bins, spanning the range of B-H distances from 0.1 to 1.6 units. Thus each amino acid was represented by 20 lines of 16 elements each. The order of the 20 lines was Leu, Ser, Arg, Ala, Val, Pro, Thr, Gly, Ile, Tyr, Asn, Lys, Asp, Glu, Cys, Phe, Trp, and Met. If Leu was coded, the first line would have all zeros, and a “1” somewhere representing the B-H distance between Leu and the particular amino acid. For example, val for line 5 had a “1” in the first bin on the right for a 0.1 B-H distance (rounded from 0.14). Coding a nine amino acid sequence therefore required 9 blocks of 20 lines of 16 elements each, or 2709 zeros and 171 ones. Coding the sequences in this way revealed patterns involving near-neighbor substitutions in forming classifications. The network had 2880 input neurons, 10 hidden neurons, and four output neurons. It was trained on 174 examples drawn from the four affinity classes and tested on 33 and 53 additional H and M class examples. A test example was judged correct if its highest score occurred in the expected class and the score was 0.5 or higher. The network correctly identified 29 of 32 high affinity peptides as such and generated a single false positive in 103 test peptides. This network was then used to test the peptides isolated from HLA-DR7 molecules of Ii+ and Ii− MCF10 cell lines. Peptides >9 amino acids were analyzed for all possible contiguous 9-mer sequences in the larger peptide sequence. The resultant 9-mer sequences were then coded in the same manner as those used for training the network. Five peptides unique to Ii− cells and two peptides present in both Ii− and Ii+ cells scored >0.92 (H binding class).
PBMC Activation
Seven MS-identified peptides and Her2/neu peptide 776 (20, 21) were synthesized in the University of Maryland, Baltimore biopolymer facility. Peptides were assessed for their ability to activate T cells as previously described (9, 11, 12). Briefly, PBMC from HLA-DR7+ donors were primed with peptide (2 μg/ml), expanded with IL-15, and boosted with Ii− MCF10 vaccine cells, MHC II+Ii+ transductants, or nonmalignant MCF10A cells. T-cell activation was assessed by measuring IFNγ production by ELISA. Percent T-cell activation = (100% × (pg IFNγ for experimental peptide/pg IFNγ for Her2 p776)).
Peptide Binding Assay
The procedures of (22) and (23) were modified as follows: DR7/CD80 cells (6 × 105 cells/well of 96 well plates) were washed twice with excess ice-cold PBS and cells were resuspended in 100 μl of 0.1315 m citric acid in PBS (pH∼3) for 60 s. Solution was neutralized by addition of 150 μl Iscoves Modified Dulbecco's medium with 2% FCS (IMDM-2% fetal calf serum), cells washed twice with PBS-2% fetal calf serum, and 0 to 350 μg/ml of peptide in PBS-2% fetal calf serum (250 μl/well) or PBS-2% fetal calf serum added to each well. Following 16–24 h incubation at 4 °C, cells were washed, stained with L243-FITC mAb and analyzed by flow cytometry. Delta mean channel fluorescence (ΔMCF) = (MCF of cells + peptide) − (MCF of cells + PBS-2% fetal calf serum).
Statistical and Bioinformatics Analysis
Statistical analyses of IFNγ production and comparison of T-cell activation by tumor-derived peptides versus Her2 p776 peptide was determined by two-tailed equal and/or unequal variance t test (Microsoft Excel 2007). p values < 0.05 were considered significant. Error bars represent the standard error of the mean.
RESULTS
MCF10 Transductants Express MHC II and Do Not Express Ii
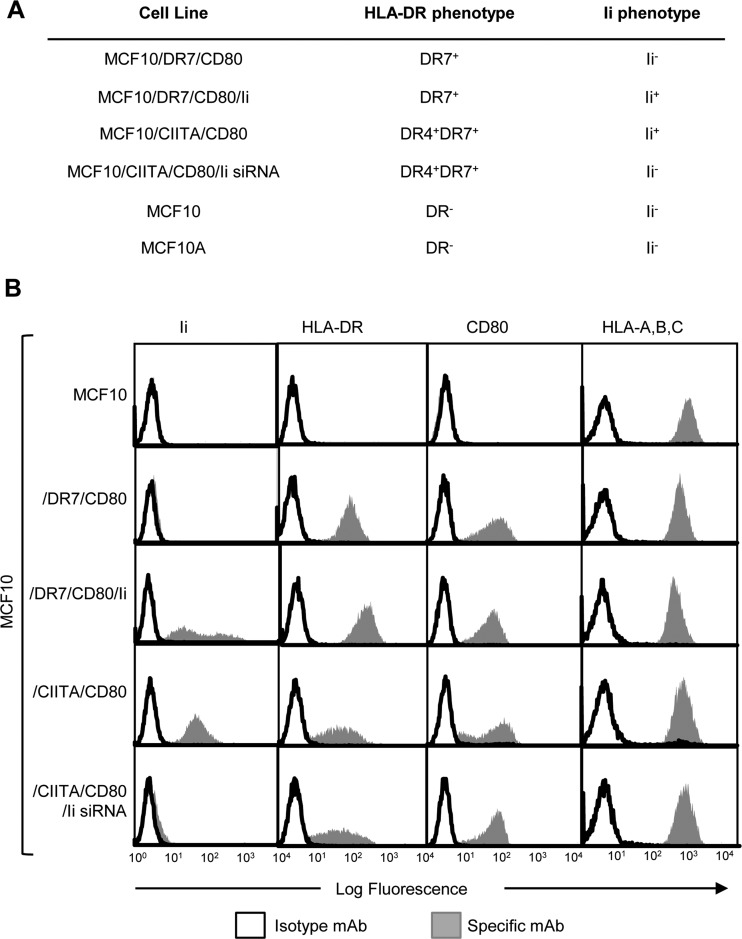
We have used human mammary adenocarcinoma MCF10 cells (16) (genotype HLA-DR4, -DR7) that constitutively express MHC I and do not constitutively express MHC II, Ii, or CD80 (Fig. 1A, 1B). Two types of transfectants have been generated: (1) Stable transfectants that are HLA-DR7+CD80+ and differentially express Ii (MCF10/DR7/CD80 and MCF10/DR7/CD80/Ii; hereafter called /DR7 and /DR7/Ii) were generated by transfection with a bicistronic vector encoding the α and β chains of HLA-DR7, a vector encoding the CD80 costimulatory molecule, ± a vector encoding Ii. These transfectants were used for analyzing peptides bound to the HLA-DR7 allele. (2) MCF10 cells stably expressing multiple MHC II alleles were generated by transfection with the MHC class II transactivator (CIITA), a transcription factor that coordinately up-regulates all MHC II alleles, Ii, and MHC II-associated proteins (24). Ii expression in CIITA transfectants was extinguished by cotransfection with siRNA for Ii. Ii+ and Ii− CIITA transfectants (MCF10/CIITA/CD80 and MCF10/CIITA/CD80/Ii siRNA; hereafter called /CIITA and /CIITA/Ii siRNA) were used to obtain a broader pool of MHC II-restricted peptides because these cells express multiple HLA-DR alleles. All transfectants were generated as described (11, 12), and stably expressed their transgenes as assessed by flow cytometry (Fig. 1B).
Fig. 1.
A, HLA-DR and Ii phenotype for parental cells, vaccines, and transfectants. HLA-DR genotype of MCF10 and MCF10A is DR4, DR7. Untransfected cells do not express plasma membrane HLA-DR. B, MCF10 vaccine cells express MHC I, MHC II and CD80, and do not express Invariant chain (Ii). MCF10 parental cells, vaccine cells, and Ii+ MHC II+ positive MCF10 transfectants were stained with antibodies to Ii, HLA-DR, CD80, and HLA-ABC. Data are from 1 of 25 independent experiments.
MHC II+Ii− Tumor Cells Present MHC II-restricted Peptides Not Presented by MHC II+Ii+ Cells
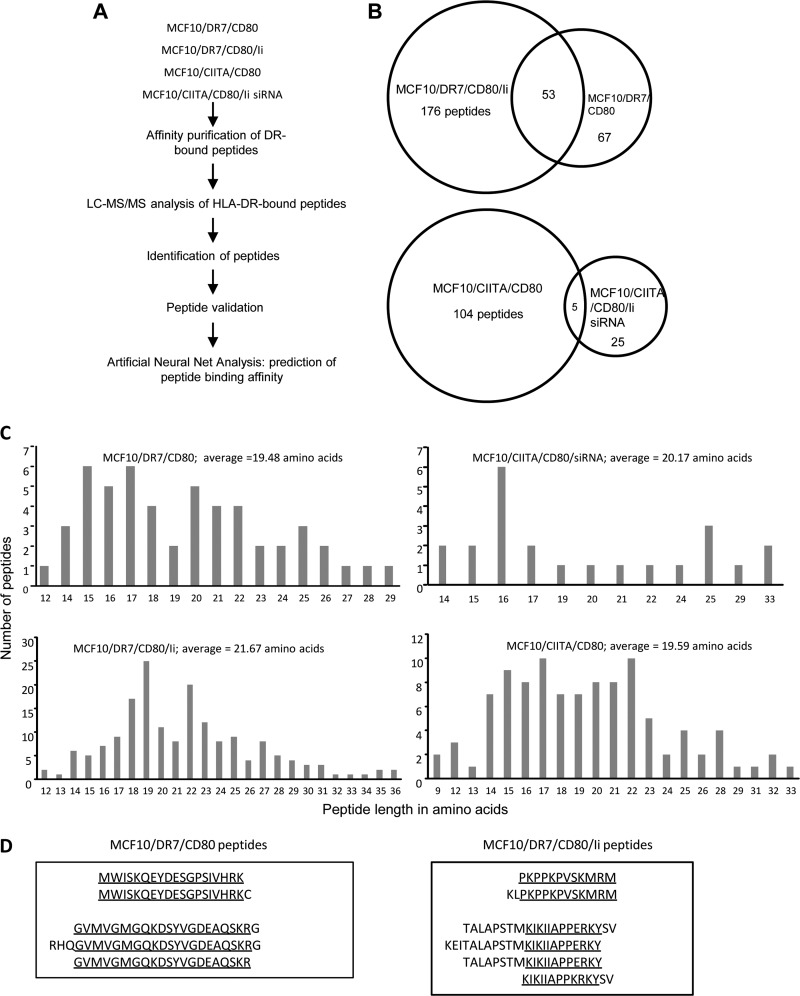
To determine if the presence of Ii impacts the repertoire of MHC II-restricted peptides, two independent batches of Ii+ and Ii− cells were expanded in culture and their HLA-DR-bound peptides were affinity purified, sequenced by mass spectrometry, and identified by SEQUEST database analysis (Fig. 2A). Two hundred and ninety-six peptides were identified from /DR7 and /DR7/Ii cells, and 134 peptides were identified from /CIITA and /CIITA/Ii siRNA cells that were present in both preparations and both LC-MS/MS runs (Fig. 2B and supplemental Table S1 (/DR7 and /DR7/Ii peptides) and supplemental Table S2 (/CIITA and /CIITA/Ii siRNA peptides). For the HLA-DR7 transduced lines, 22.6% of peptides were unique to /DR7, 59.4% of peptides were unique to /DR7/Ii cells, and 17.9% of peptides were shared by /DR7 and /DR7/Ii cells. For the CIITA transductants, 18.6% of peptides were unique to /CIITA/Ii siRNA cells, 77.6% of peptides were unique to /CIITA cells, and 3.7% of peptides were shared by /CIITA and /CIITA/Ii siRNA cells. These results demonstrate that the absence of Ii enables the presentation of MHC II-restricted peptides that are not presented by Ii+ cells.
Fig. 2.
A, Experimental design for isolation and identification of HLA-DR-bound peptides. HLA-DR bound peptides from two independent preparations of DR+Ii− MCF10 vaccine cells and from DR+Ii+ MCF10 transfectants were purified using affinity chromatography, sequenced using LC-MS/MS, assigned to proteins using SEQUEST, and analyzed using ANN. Peptides with the highest predicted HLA-DR7 binding affinity were selected for further study. B, HLA-DR+ MCF10 vaccines and Ii+ transfectants present distinct and partially overlapping repertoires of peptides. Values are the number of peptides that are unique or shared between DR+Ii− MCF10 vaccine cells and DR+Ii+ MCF10 transfectants. C, Peptides isolated from HLA-DR7-transfected and CIITA-transfected Ii− and Ii+ MCF10 cells are similar in length. D, Peptides isolated from HLA-DR7-transfected Ii− and Ii+ MCF10 cells contain nested sequences. Data are from two independent analysis of each preparation of cells.
Peptides from Ii− (/DR7 and /CIITA/Ii siRNA) and Ii+ (/DR7/Ii and /CIITA) cells ranged in length from 9 to 36 amino acids, with an average length of 19.48, 20.17, 21.67, and 19.59, respectively (Fig. 2C). Many of the peptides were derived from the same protein cleaved at different locations, giving rise to sets of nested peptides. /DR7/Ii unique peptides contained eight nested sets, whereas there were no nested sets among /DR7 unique peptides. Seven and four nested sets were identified for /CIITA and /CIITA/Ii siRNA cells, respectively. Fig. 2D shows examples of nested sets. Nested sets of peptides ranging in length from 13–23 are characteristic of MHC II-restricted peptides (25, 26) and demonstrate that the peptides presented by tumor cells share physical characteristics with MHC II-restricted peptides presented by professional APC.
Ii− Cells Present Peptides Derived from Source Proteins Not Used by Ii+ Cells
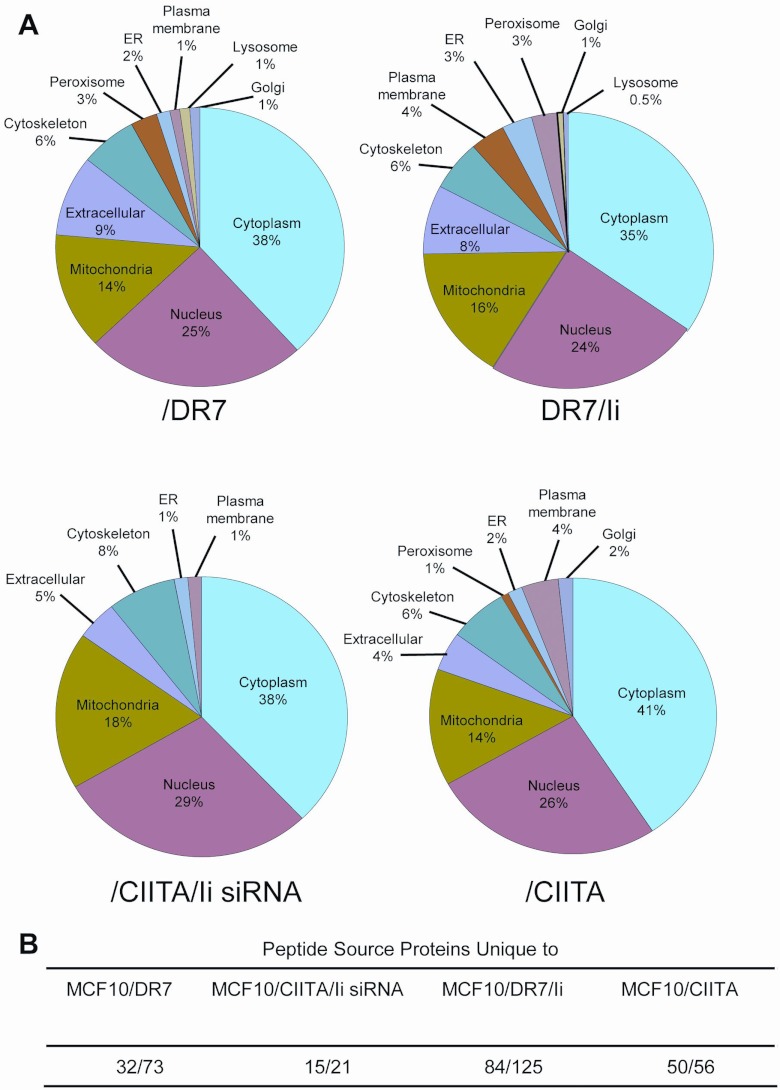
Protein Prophet was used to identify the source proteins for the peptides, which are shown in supplemental Tables S3 and S4. Annotated spectra for the proteins identified by a single peptide per protein are shown in supplemental Figs. S1–S3. The source proteins were analyzed using the LOCATE subcellular localization database and WOLF Psort to identify the cellular source and determine if the peptides presented by Ii− and Ii+ cells were derived from proteins residing in similar locations (Fig. 3A). The source proteins for all three categories of peptides (unique to Ii− or Ii+ cells; shared by Ii− and Ii+ cells) and for both DR7 and CIITA transductants originated in similar compartments, with most proteins residing in the cytoplasm, nuclei, and mitochondria, and fewer in the endoplasmic reticulum, cytoskeleton, peroxisome, plasma membrane, lysosome, and Golgi.
Fig. 3.
HLA-DR7-restricted peptides are derived from proteins residing in multiple subcellular compartments. A, Subcellular localization of source proteins for peptides was determined using LOCATE subcellular localization database and WOLF Psort. B, Peptide source proteins unique to each cell line. Unique source proteins/total source proteins per cell line.
Although source proteins for peptides from Ii− and Ii+ cells originated in the same subcellular compartments, a subset of peptides in Ii− cells were derived from proteins not used by Ii+ cells (Fig. 3B). Specifically, 29 and 19% of source proteins used by Ii− /DR7 and CIITA/Ii siRNA cells were not represented in the protein pool from /DR7/Ii and /CIITA Ii+ cells, respectively. Likewise, 53.5% and 74.6% of source proteins used by Ii+ /DR7/Ii and /CIITA cells were not used by Ii− /DR7 and /CIITA/Ii siRNA cells, respectively. These results suggest that a subset of peptides presented by Ii− and Ii+ cells are derived from a different pool of source proteins.
Peptides Presented by Ii− and Ii+ Cells Have Similar Binding Affinity for MHC II and Contain Redundant Binding Motifs
To determine if Ii expression affects the binding affinity of MHC II-restricted peptides, all of the peptides identified by LC-MS/MS from /DR7 and /DR7/Ii cells (1385 and 2219 peptides, respectively) were subjected to Artificial Neural Net (ANN) analysis. The peptides of /CIITA transductants were not included in this analysis because we could not determine which of these peptides were restricted to HLA-DR7 versus HLA-DR4. The ANN was trained on the MHCBN database of 267 peptides with known high, medium, or low binding affinity for HLA-DR7 (27, 28). Of the peptides in Ii− /DR7 cells, 5.1% scored >0.92, placing them at the top of the high affinity class, and 8.7% scored >0.62 placing them lower in the high affinity class. The remaining peptides scored <0.62 and were in medium and low affinity classes. Approximately the same percentage of peptides from Ii+ /DR7/Ii cells were in each class (5.4% highest affinity, 8.2% next highest affinity). Surprisingly, ANN analysis identified a previously unpublished binding motif in the peptides isolated from /DR7 and /DR7/Ii cells (Fig. 4A). Most of the peptides from both Ii− and Ii+ cells in the high and medium binding affinity classes contained multiple binding cores indicating that these peptides could bind to HLA-DR7 in multiple registers. Fig. 4B shows a representative peptide that contains four potentially high or medium binding regions. These data demonstrate that Ii does not impact the binding affinity of MHC II-restricted peptides, and suggest that strong binding could be because of multiple binding sites within a given peptide.
Fig. 4.
MS-identified peptides isolated from MCF10/DR7/CD80 and MCF10/DR7/CD80/Ii cells bind peptides in multiple registers and have predicted high affinity binding to HLA-DR7. A, Consensus HLA-DR7 binding motif for MS-identified peptides as determined by ANN, and published HLA-DR7 binding motif from SYFPEITHI database. B, MS-identified peptides contain multiple overlapping binding motifs for HLA-DR7. Example of one peptide is shown. C, Seven MS-identified peptides with high predicted binding affinity used in the subsequent PBMC activation studies of Fig. 5 and supplemental Fig. S4.
Peptides From Ii− Cells Bind to HLA-DR7 and Activate Tumor-specific T Cells from Healthy Donors and Breast Cancer Patients
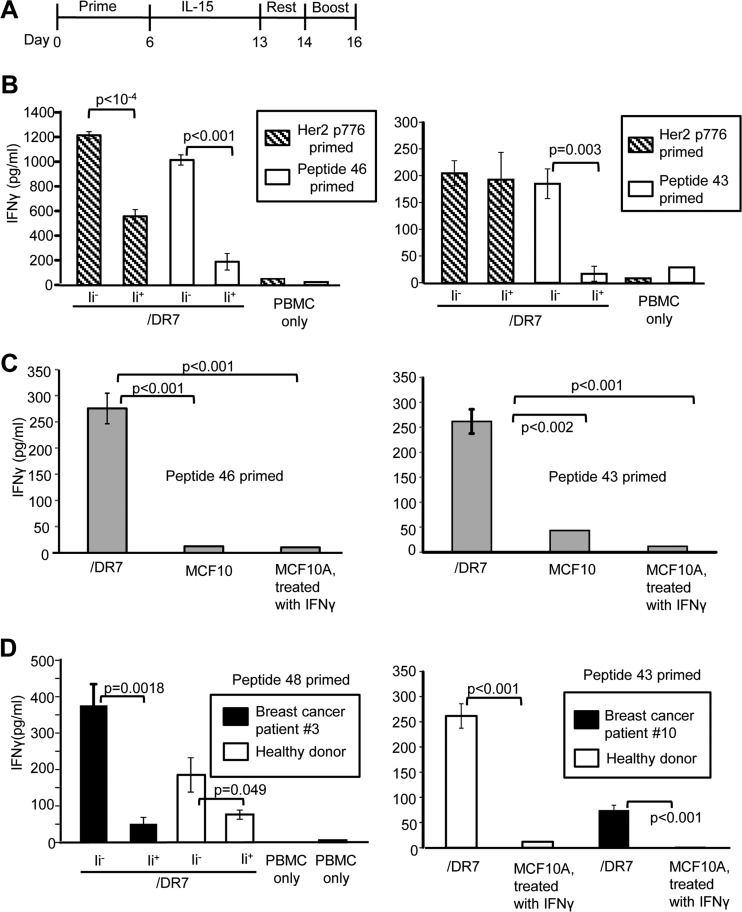
To determine if the peptides identified by MS were bonafide tumor antigens, five peptides unique to Ii− vaccines and two peptides shared by Ii+ and Ii− cells from the highest predicted binding class and isolated from HLA-DR7-transfectants were selected for further study (Fig. 4C). To ascertain the relative immunogenicity of the MS-identified peptides, T cell activation by these peptides was compared with T-cell activation by the immunogenic Her2/neu peptide p776–790 (Her2 p776) (21). HLA-DR7+ healthy donor PBMC were primed with peptides, rested, expanded in IL-15, and boosted with /DR7 or /DR7/Ii cells (Fig. 5A). Priming with peptides derived from Ii− vaccine cells (peptides 46 and 43) and boosting with the corresponding Ii− vaccine cells gave maximum activation as measured by IFNγ production (Fig 5B). Similar results were obtained for the three other peptides unique to Ii− cells (peptides 45, 47, and 48; supplemental Fig. S4A) and for the two peptides shared by Ii− and Ii+ cells (peptides 42 and 44; supplemental Fig. S4B). An irrelevant peptide (HEL 46–61) gave only background levels of IFNγ (supplemental Fig. S4C). Tumor-derived peptides activated T cells because >90% of the cells after priming and before boosting were CD3+, and depletion of CD3+ cells before boosting eliminated IFNγ production (75.5 ± 17.6 pg/ml undepleted versus 0 pg/ml CD3-depleted). Table I compares the relative activation by the MS-identified peptides as compared with activation by Her2 p776. Peptides 43, 47, and 48 (present only in Ii− cells) and peptides 42 and 44 (shared by Ii− and Ii+ cells) were as effective as Her2 p776 peptide in activating PBMC, and Ii− peptide 45 was significantly more effective. These results demonstrate that the peptides identified by mass spectrometry are authentic antigens expressed by MCF10 tumor cells, and that peptides generated in the absence of Ii are immunogenic.
Fig. 5.
MS-identified HLA-DR7-restricted peptides from MCF10/DR7/CD80 vaccine cells activate tumor-specific PBMC from HLA-DR7+ healthy donors and breast cancer patients. A, PBMC activation scheme: HLA-DR7+ PBMC were primed with peptide for 6 days, expanded with IL-15 for 7 days, rested for 24 h, boosted for 48 h with Ii− (MCF10/DR7/CD80) or Ii+ (MCF10/DR7/CD80/Ii) cells, and IFNγ production measured by ELISA. B, PBMC were primed with Her2 p776 and either peptide 46 (healthy donor PBMC BC100206) or peptide 43 (healthy donor PBMC BC100306), and boosted with the indicated cells shown on the x axis. C, Peptide-activated PBMC do not react with nonmalignant breast tissue cells. PBMC from healthy donor BC061005 were primed with either peptide 46 or 43 and boosted with MCF10/DR7/CD80 or nonmalignant HLA-DR+ MCF10A cells. D, MS-identified peptides activate breast cancer patients' PBMC. HLA-DR7+ PBMC from healthy donor BC051005 and breast cancer patient 3 (left panel) were primed with peptide 48 (left panel) or healthy donor BC061005 and breast cancer patient 10 were primed with peptide 43 (right panel) and boosted with MCF10/DR7/CD80 or nonmalignant MCF10A cells.
Table I. The seven peptides with predicted highest binding affinity to HLA-DR7 efficiently activated PBMC from HLA-DR7+ healthy donors.
PBMC activation was assessed as in Fig. 5A–5D. Representative individual experiments from a total of 12 independent experiments. Percent PBMC activation = 100% × (pg IFNγ for experimental peptide/pg IFNγ for Her2 p776). Response >100% indicates priming with the indicated peptide yielded more IFNγ than priming with Her2 p776. * indicates p < 0.05 for experimental peptide vs. p776 as assessed by t test.
| ID# | Source | Peptide sequence, ID#, and cell source | Percent PBMC activation relative to Her2 p776 |
|---|---|---|---|
| 43 | Ii− | KVAPAPAVVKKQEAKK | 90% |
| 45 | Ii− | RTYAGGTASATKVSASSGATSKS | 333%* |
| 46 | Ii− | SESAAAPAFASSSSEVNPAPKFHW | 83%* |
| 47 | Ii− | RRMRLTHCGLQEKHL | 52% |
| 48 | Ii− | GKFYLVIEELSQLFRSLVPIQL | 120% |
| 42 | Ii− and Ii+ | RQTVAVGVIKAVDKKA | 96% |
| 44 | Ii− and Ii+ | ELEEALANASDAELCDIAAILGMHTLM | 107% |
To determine if the peptides activate T cells that react only with malignant cells, healthy donor PBMC were primed with peptide 46 or 43 and boosted with malignant cells (MCF10/DR7/CD80) or with nonmalignant HLA-DR7+ MCF10A cells (Fig. 5C). MCF10A cells were rendered HLA-DR7+ by treatment with IFNγ (supplemental Fig. S5). Boosting with MCF10A cells yielded minimal IFNγ, demonstrating that the peptides activate tumor-specific PBMC that do not react with nonmalignant cells.
Peptide binding affinity for HLA-DR was determined by assessing the ability of the peptides to stabilize HLA-DR expression. α and β chains of HLA-DR heterodimers are stabilized by bound peptide. Mild acid treatment releases peptide and causes dissociation of the chains. Neutralization in the presence of peptide restores HLA-DR conformation. The mAb L243 recognizes HLA-DR heterodimers and not dissociated α and β chains (29, 30), allowing for the assessment of peptide binding to renatured molecules. DR7/CD80 cells were acid treated, neutralized, and then incubated with peptide 43, 45, or 46. Her2 776 peptide, which binds to HLA-DR7 with high affinity (Kd = 190 nm) (21) served as a positive control, and an irrelevant peptide (HEL 46–61) served as a negative control. HLA-DR levels were assessed by flow cytometry after labeling with L243 mAb. Peptides 43 and 45 restored HLA-DR levels slightly better than Her2 peptide, whereas peptide 46 was slightly less effective that Her2 peptide. Irrelevant HEL peptide did not increase HLA-DR7 levels (supplemental Fig. S6). Peptides that bind to MHC II with a Kd<1500 nm are considered to have high binding affinity (21). Therefore, these results demonstrate that peptides 43, 45, and 46 bind to HLA-DR7 with high affinity.
To determine if the peptides have the capacity to activate PBMC from breast cancer patients, PBMC from breast cancer patient 3 and from a healthy donor were primed with peptide 48 and boosted with either /DR7 or /DR7/Ii cells (Fig. 5D). Peptide 48 activated PBMC from the breast cancer patient and from the healthy donor, and consistent with the results of Figs. 5B and 5C, Ii− peptides preferentially activated T cells specific for Ii− tumor cells. Similar results were obtained for peptide 43 and PBMC from breast cancer patient 10. These results further confirm that peptides 43 and 48 are bonafide tumor peptides and demonstrate that these peptides activate PBMC from breast cancer patients.
DISCUSSION
Mass spectrometry is a powerful tool for identifying novel peptide antigens with speed and high throughput, and is currently unsurpassed by other peptide sequencing methods. However, the analysis of complex peptide mixtures by mass spectrometry based proteomics has potential drawbacks such as under-sampling and limited dynamic range (31). Under-sampling is a phenomenon in which not all peptides present in a sample are fragmented because of limitations in chromatographic separation, ionization artifacts, and data-dependent sampling of peptides by the mass spectrometer. If a peptide is not fragmented, then its sequence is not determined and not all peptides in a sample are identified. Complex biological peptide mixtures can have a dynamic range of concentration that is orders of magnitude larger than can be detected by modern mass spectrometers. Therefore, it is difficult to detect peptides that are of low abundance, and a complete HLA-DR7 peptidome may not be obtained.
In the present study we have identified the HLA-DR7-restricted peptide repertoire of a breast cancer cell-based vaccine and have shown that the absence if Invariant chain produces a distinct peptide repertoire. The peptides we identified possess classic attributes of MHC II peptides, such as nested sets and average length of ∼20 amino acids. However, the HLA-DR7 peptidome of Ii− cells contains novel peptides that are not presented by Ii+ cells, and Ii− cells process source proteins that are not used by Ii+ cells. The novel peptides are immunogenic as demonstrated by in vitro T-cell activation assays using healthy donor and breast cancer patients' PBMC, and they do not activate PBMC that react with nonmalignant cells. It has been predicted that the diversity of MHC II ligands can reach many thousands of peptides (32), so only a subset of the HLA-DR7 peptidome has been identified in the present study. However, the novel peptides we have identified are functional, immunogenic and tumor-specific, making them potential agents for therapeutic cancer vaccines.
Surprisingly, ANN, the bioinformatics approach we used to identify MHC II-restricted peptides and predict their binding affinity, yielded a binding motif that was not identified by previous analyses (33, 34). The validity of ANN was confirmed in that six of the seven peptides that were identified by ANN as being high affinity binders were strongly immunogenic for four healthy donors and two breast cancer patients. ANN analysis has the advantage of not only identifying key anchor residues, but also evaluates the contributions of neighboring residues and therefore may give a more accurate assessment of affinity.
In professional APC MHC class II molecules are loaded with peptides in MIIC endosomal compartments when the acidic pH causes the release of Ii from the MHC II molecule. The peptides are derived from either exogenous antigens that are endocytosed and degraded in the endocytic vesicles, or from endogenously synthesized nuclear and cytosolic material that accesses endosomal compartments by autophagy (35). Because the vaccine cells lack Ii, their MHC II molecules can potentially be loaded with peptide in any of the subcellular compartments preceding the MIIC. Therefore, newly synthesized and recycling MHC II molecules in the vaccines are likely to be exposed to a broader repertoire of peptides than MHC II molecules of Ii+ (i.e. professional) APC. The finding that Ii− peptides consist of fewer nested sets relative to Ii+ peptides further supports the concept that peptides are either loaded onto MHC class II molecules, or that MHC II molecules traffic through different subcellular compartments in Ii− and Ii+ cells, resulting in the novel peptides identified in this report. Surprisingly, most of the peptides identified from Ii+ cells and Ii− cells are derived from endogenous proteins, consistent with the concept that autophagy is a prominent source of MHC II-restricted peptides in Ii+ cells. The absence of Ii may facilitate MHC II trafficking through the secretory pathway rather than the endosomal pathway where they encounter a different peptide repertoire. The absence of Ii and its intrinsic endosomal trafficking sequence (36) may or may not affect MHC II trafficking because the MHC II β chain also contains an endosomal trafficking signal (37). Our previous antigen presentation studies using endosomal and proteosomal inhibitors support peptide loading in the endosomal route (38, 39); however, confocal microscopy studies indicate that vaccine cell MHC II molecules also traffic via the secretory pathway (40).
The different peptidomes of Ii− and Ii+ cells combined with the observation that some of the peptides of Ii− cells are derived from source proteins not present in the repertoire of Ii+ cells, confirm our original hypothesis that the absence of Ii facilitates the presentation of novel peptides. Whether the differences in intracellular trafficking patterns or differential use of endocytosis or autophagy are responsible for the different peptidomes requires further study. Regardless of which pathway and process are used, the absence of Ii enables the presentation of immunogenic, bonafide nonconventional tumor peptides.
Supplementary Material
Acknowledgments
We thank Lydia Grmai for help with culturing MCF10 transductants for MHC II peptide purification, Christopher Ecker for help with peptide subcellular localization analysis, Virginia Clements for technical support, Dr. Amy Fulton for the MCF10A cells, Drs. Saranya Chumsri and Dean Mann for healthy donor and breast cancer patients' PBMC, and Drs. Elizabeth Jaffee and Sheherazade Sadegh-Nasseri for advice on assessing peptide binding affinity.
Footnotes
* This work was supported by National Institutes of Health (NIH) (NIH RO1CA84232 and RO1CA115880. OC is the recipient of DOD breast cancer program pre-doctoral fellowship W81XWH-10-1-0027 and was partially supported by NIH CBI T32GM 066706.
 This article contains supplemental Figs. S1 to S6 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S6 and Tables S1 to S4.
1 The abbreviations used are:
- ANN
- artificial neural net analysis
- APCs
- Antigen presenting cells
- CIITA
- MHC class II transactivator
- /CIITA
- MCF10 cells transfected with CIITA
- /CIITA/CD80
- CIITA cells transfected with and expressing CD80
- /CIITA/Ii siRNA
- /CIITA cells down-regulated for Ii by RNA interference
- CLIP MHC
- class II-associated invariant chain peptide
- /DR7
- MCF10 cells transfected with and expressing HLA-DR7
- /DR7/CD80
- /DR7 cells transfected with and expressing CD80
- HEL
- Hen egg lysozyme
- HLA
- Human leukocyte antigen
- HLA-DR7
- MHC class II DR7 allele
- IFNγ
- interferon gamma
- Ii
- invariant chain
- mAb
- Monoclonal antibody
- MCF10
- malignant human breast cancer cells
- MCF10A
- nonmalignant human breast cancer cells
- MHC
- major histocompatibility complex
- PBMC
- peripheral blood mononuclear cells
- PE
- phycoerythrin
- PE-Cy5
- -Cy7 PE coupled to cyanin −5, −7
- Th1
- Type 1 CD4+ T helper lymphocytes.
REFERENCES
- 1. Fang L., Lonsdorf A. S., Hwang S. T. (2008) Immunotherapy for advanced melanoma. J. Invest. Dermatol. 128, 2596–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mellman I., Coukos G., Dranoff G. (2011) Cancer immunotherapy comes of age. Nature 480, 480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Neefjes J., Jongsma M..L., Paul P., Bakke O. (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 [DOI] [PubMed] [Google Scholar]
- 4. Germain R. N. (2011) Uncovering the role of invariant chain in controlling MHC class II antigen capture. J. Immunol. 187, 1073–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Armstrong T. D., Clements V. K., Martin B. K., Ting J. P., Ostrand-Rosenberg S. (1997) Major histocompatibility complex class II-transfected tumor cells present endogenous antigen and are potent inducers of tumor-specific immunity. Proc. Natl. Acad. Sci. U.S.A. 94, 6886–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Humphreys R. E., Hillman G. G., von Hofe E., Xu M. (2004) Forcing tumor cells to present their own tumor antigens to the immune system: a necessary design for an efficient tumor immunotherapy. Cell. Mol. Immunol. 1, 180–185 [PubMed] [Google Scholar]
- 7. Baskar S., Glimcher L., Nabavi N., Jones R.T., Ostrand-Rosenberg S. (1995) Major histocompatibility complex class II+B7–1+ tumor cells are potent vaccines for stimulating tumor rejection in tumor-bearing mice. J. Exp. Med. 181, 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bosch J. J., Thompson J. A., Srivastava M. K., Iheagwara U. K., Murray T. G., Lotem M., Ksander B. R., Ostrand-Rosenberg S. (2007) MHC class II-transduced tumor cells originating in the immune-privileged eye prime and boost CD4(+) T lymphocytes that cross-react with primary and metastatic uveal melanoma cells. Cancer Res. 67, 4499–4506 [DOI] [PubMed] [Google Scholar]
- 9. Dissanayake S. K., Thompson J. A., Bosch J. J., Clements V. K., Chen P. W., Ksander B. R., Ostrand-Rosenberg S. (2004) Activation of tumor-specific CD4(+) T lymphocytes by major histocompatibility complex class II tumor cell vaccines: a novel cell-based immunotherapy. Cancer Res. 64, 1867–1874 [DOI] [PubMed] [Google Scholar]
- 10. Srivastava M. K., Bosch J. J., Thompson J. A., Ksander B. R., Edelman M. J., Ostrand-Rosenberg S. (2008) Lung cancer patients' CD4(+) T cells are activated in vitro by MHC II cell-based vaccines despite the presence of myeloid-derived suppressor cells. Cancer Immunol. Immunother. 57, 1493–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thompson J. A., Dissanayake S. K., Ksander B. R., Knutson K. L., Disis M. L., Ostrand-Rosenberg S. (2006) Tumor cells transduced with the MHC class II Transactivator and CD80 activate tumor-specific CD4+ T cells whether or not they are silenced for invariant chain. Cancer Res. 66, 1147–1154 [DOI] [PubMed] [Google Scholar]
- 12. Thompson J. A., Srivastava M. K., Bosch J. J., Clements V. K., Ksander B. R., Ostrand-Rosenberg S. (2008) The absence of invariant chain in MHC II cancer vaccines enhances the activation of tumor-reactive type 1 CD4+ T lymphocytes. Cancer Immunol. Immunother. 57, 389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chamuleau M. E., Souwer Y., Van Ham S. M., Zevenbergen A., Westers T. M., Berkhof J., Meijer C. J., van de Loosdrecht A. A., Ossenkoppele G. J. (2004) Class II-associated invariant chain peptide expression on myeloid leukemic blasts predicts poor clinical outcome. Cancer Res. 64, 5546–5550 [DOI] [PubMed] [Google Scholar]
- 14. van Luijn M. M., Chamuleau M. E., Thompson J. A., Ostrand-Rosenberg S., Westers T. M., Souwer Y., Ossenkoppele G. J., van Ham S. M., van de Loosdrecht A. A. (2010) Class II-associated invariant chain peptide down-modulation enhances the immunogenicity of myeloid leukemic blasts resulting in increased CD4+ T-cell responses. Haematologica 95, 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Luijn M. M., van den Ancker W., Chamuleau M. E., Zevenbergen A., Westers T. M., Ossenkoppele G. J., van Ham S. M., van de Loosdrecht A. A. (2011) Absence of class II-associated invariant chain peptide on leukemic blasts of patients promotes activation of autologous leukemia-reactive CD4+ T cells. Cancer Res. 71, 2507–2517 [DOI] [PubMed] [Google Scholar]
- 16. Pauley R. J., Soule H. D., Tait L., Miller F. R., Wolman S. R., Dawson P. J., Heppner G. H. (1993) The MCF10 family of spontaneously immortalized human breast epithelial cell lines: models of neoplastic progression. Eur. J. Cancer Prev. 2 67–76 [PubMed] [Google Scholar]
- 17. Depontieu F. R., Qian J., Zarling A. L., McMiller T. L., Salay T. M., Norris A., English A. M., Shabanowitz J., Engelhard V. H., Hunt D. F., Topalian S. L. (2009) Identification of tumor-associated, MHC class II-restricted phosphopeptides as targets for immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 106, 12073–12078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., Aebersold R. A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Borstnik B., Hofacker G. L. (1985) Functional aspects of the neutral patterns in protein evolution, Guiderland: Academic Press [Google Scholar]
- 20. Sotiriadou R., Perez S. A., Gritzapis A. D., Sotiropoulou P. A., Echner H., Heinzel S., Mamalaki A., Pawelec G., Voelter W., Baxevanis C. N., Papamichail M. (2001) Peptide HER2(776–788) represents a naturally processed broad MHC class II-restricted T cell epitope. Br. J. Cancer 85, 1527–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salazar L. G., Fikes J., Southwood S., Ishioka G., Knutson K. L., Gooley T. A., Schiffman K., Disis M. L. (2003) Immunization of cancer patients with HER-2/neu-derived peptides demonstrating high-affinity binding to multiple class II alleles. Clin. Cancer Res. 9, 5559–5565 [PubMed] [Google Scholar]
- 22. Thomas A. M., Santarsiero L. M., Lutz E. R., Armstrong T. D., Chen Y. C., Huang L. Q., Laheru D. A., Goggins M., Hruban R. H., Jaffee E. M. (2004) Mesothelin-specific CD8(+) T cell responses provide evidence of in vivo cross-priming by antigen-presenting cells in vaccinated pancreatic cancer patients. J. Exp. Med. 200, 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kessler J. H., Mommaas B., Mutis T., Huijbers I., Vissers D., Benckhuijsen W. E., Schreuder G. M., Offringa R., Goulmy E., Melief C. J., van der Burg S. H., Drijfhout J. W. (2003) Competition-based cellular peptide binding assays for 13 prevalent HLA class I alleles using fluorescein-labeled synthetic peptides. Human Immunol. 64, 245–255 [DOI] [PubMed] [Google Scholar]
- 24. Steimle V., Siegrist C. A., Mottet A., Lisowska-Grospierre B., Mach B. (1994) Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science 265, 106–109 [DOI] [PubMed] [Google Scholar]
- 25. Lippolis J. D., White F. M., Marto J. A., Luckey C. J., Bullock T. N., Shabanowitz J., Hunt D. F., Engelhard V. H. (2002) Analysis of MHC class II antigen processing by quantitation of peptides that constitute nested sets. J. Immunol. 169, 5089–5097 [DOI] [PubMed] [Google Scholar]
- 26. Suri A., Lovitch S. B., Unanue E. R. (2006) The wide diversity and complexity of peptides bound to class II MHC molecules. Curr. Opin. Immunol. 18, 70–77 [DOI] [PubMed] [Google Scholar]
- 27. Bhasin M., Singh H., Raghava G. P. (2003) MHCBN: a comprehensive database of MHC binding and non-binding peptides. Bioinformatics. 19, 665–666 [DOI] [PubMed] [Google Scholar]
- 28. Lata S., Bhasin M., Raghava G. P. (2009) MHCBN 4.0: A database of MHC/TAP binding peptides and T-cell epitopes. BMC Res. Notes 2, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arimilli S., Cardoso C., Mukku P., Baichwal V., Nag B. (1995) Refolding and reconstitution of functionally active complexes of human leukocyte antigen DR2 and myelin basic protein peptide from recombinant alpha and beta polypeptide chains. J. Biol. Chem. 270, 971–977 [DOI] [PubMed] [Google Scholar]
- 30. Justesen S., Harndahl M., Lamberth K., Nielsen L. L., Buus S. (2009) Functional recombinant MHC class II molecules and high-throughput peptide-binding assays. Immunome Res. 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nilsson T., Mann M., Aebersold R., Yates J. R., 3rd, Bairoch A., Bergeron J.J. (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat. Methods 7, 681–685 [DOI] [PubMed] [Google Scholar]
- 32. Purcell A. W., Gorman J. J. (2004) Immunoproteomics: Mass spectrometry-based methods to study the targets of the immune response. Mol. Cell. Proteomics 3, 193–208 [DOI] [PubMed] [Google Scholar]
- 33. Gelder C., Davenport M., Barnardo M., Bourne T., Lamb J., Askonas B., Hill A., Welsh K. (1998) Six unrelated HLA-DR-matched adults recognize identical CD4+ T cell epitopes from influenza A haemagglutinin that are not simply peptides with high HLA-DR binding affinities. Int. Immunol. 10, 211–222 [DOI] [PubMed] [Google Scholar]
- 34. Chicz R. M., Urban R. G., Gorga J. C., Vignali D. A., Lane W. S., Strominger J. L. (1993) Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J. Exp. Med. 178, 27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crotzer V. L., Blum J. S. (2009) Autophagy and its role in MHC-mediated antigen presentation. J. Immunol. 182, 3335–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berger A. C., Roche P. A. (2009) MHC class II transport at a glance. J. Cell Sci. 122, 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lechler R., Aichinger G., Lightstone L. (1996) The endogenous pathway of MHC class II antigen presentation. Immunol. Rev. 151, 51–79 [DOI] [PubMed] [Google Scholar]
- 38. Dissanayake S. K., Tuera N., Ostrand-Rosenberg S. (2005) Presentation of endogenously synthesized MHC class II-restricted epitopes by MHC class II cancer vaccines is independent of transporter associated with Ag processing and the proteasome. J. Immunol. 174, 1811–1819 [DOI] [PubMed] [Google Scholar]
- 39. Qi L., Rojas J. M., Ostrand-Rosenberg S. (2000) Tumor cells present MHC class II-restricted nuclear and mitochondrial antigens and are the predominant antigen presenting cells in vivo. J. Immunol. 165, 5451–5461 [DOI] [PubMed] [Google Scholar]
- 40. Bosch J. J., Iheagwara U. K., Reid S., Srivastava M. K., Wolf J., Lotem M., Ksander B. R., Ostrand-Rosenberg S. (2010) Uveal melanoma cell-based vaccines express MHC II molecules that traffic via the endocytic and secretory pathways and activate CD8(+) cytotoxic, tumor-specific T cells. Cancer Immunol. Immunother. 59, 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.