Abstract
The Venus flytrap (Dionaea muscipula) is one of the most well-known carnivorous plants because of its unique ability to capture small animals, usually insects or spiders, through a unique snap-trapping mechanism. The animals are subsequently killed and digested so that the plants can assimilate nutrients, as they grow in mineral-deficient soils. We deep sequenced the cDNA from Dionaea traps to obtain transcript libraries, which were used in the mass spectrometry-based identification of the proteins secreted during digestion. The identified proteins consisted of peroxidases, nucleases, phosphatases, phospholipases, a glucanase, chitinases, and proteolytic enzymes, including four cysteine proteases, two aspartic proteases, and a serine carboxypeptidase. The majority of the most abundant proteins were categorized as pathogenesis-related proteins, suggesting that the plant's digestive system evolved from defense-related processes. This in-depth characterization of a highly specialized secreted fluid from a carnivorous plant provides new information about the plant's prey digestion mechanism and the evolutionary processes driving its defense pathways and nutrient acquisition.
Carnivorous plants capture, digest, and “eat” animals using four different types of trapping strategies: (i) flypaper or adhesive traps (e.g. Drosera, also known as sundews, and Pinguicula, also known as butterworts), (ii) sucking bladder traps (e.g. Utricularia, also known as bladderworts), (iii) pitfall traps (e.g. Nepenthes), and (iv) snap traps (e.g. Dionaea muscipula, also known as the Venus flytrap). These plants fascinated Charles Darwin. The Venus flytrap, in particular, attracted his attention, and he described the plant as “one of the most wonderful in the world” (1). The snap trap most likely evolved from the adhesive trap, because its ability to capture larger prey than the adhesive traps gives it an evolutionary advantage (2).
The trapping motion of Dionaea muscipula is among the fastest movements in the plant kingdom, and its mechanism has been described in detail, starting with Charles Darwin's work from ∼150 years ago (3–6). The plant's leaves employ turgor pressure and hydrodynamic flow to close the trap (3). The closing is initiated by the mechanical stimulation of trigger hairs, eliciting an action potential to close the trap, which seals the fate of the animal inside (1). Then “touch” hormones such as 12-oxophytodienoic acid, which is a precursor of the phytohormone jasmonic acid, probably induce the secretion of digestive fluid (7). Touch hormones are likely to be released in response to the continuous mechanical stimulation of the trigger hairs by the prey as it struggles to escape (7). The trap may also be closed artificially by direct electrical stimulation or by the application of the bacterial phytotoxine coronatine (5, 7, 8).
The largest classes of Venus flytrap prey are spiders and flies. Highly active fliers, such as bees and wasps, are rarely caught (9). The trapped animal faces a slow death, and experiments with ants demonstrate that the prey are alive and capable of stimulating the trigger hairs up to 8 h after being caught (10). The nutrients obtained from the digestion of the different prey are important for the Venus flytrap. Among carnivorous plants in their natural habitats, the Venus flytrap appears to be the most dependent on the nitrogen obtained from its digested prey (11). The nutrients from insects and spiders give the plants a competitive advantage in their natural low-nutrient soil habitats (12).
In contrast to its trapping mechanism, only a few studies have focused on the digestion process of the Venus flytrap, and none of the involved enzymes has been purified. However, the pH during Venus flytrap digestion has been studied. The pH of the digestive fluid is 4.3, and during the secretion phase the external “stomach” is further acidified to pH 3.4 (7, 13). The optimum pH for protease activity in the fluid has been analyzed in different studies, and the resulting values range from pH 3.0 to pH 7.0 (13–16). This discrepancy is likely due to differences in the assay conditions and the substrates used as targets during the analyses (13, 16).
In our work, we have determined the protein composition of the digestive fluid of the Venus flytrap. The protein identifications were based on a two-step approach involving (i) deep sequencing of the cDNA from stimulated leaves (RNA-seq) and (ii) subsequent mass spectrometric (MS)1 analyses of the proteins in the collected digestive fluids (Fig. 1). Both the RNA-seq analyses and the digestion fluid proteomics were performed on independent samples using complementary approaches. The obtained mass spectra were searched against the two generated transcriptome databases, and the identified proteins in the secretome were abundance-ranked based on their intensity sums. Our results provide insights into the complex composition of the Venus flytrap's digestive fluid, which has vital functions in defense and nutrient digestion.
Fig. 1.
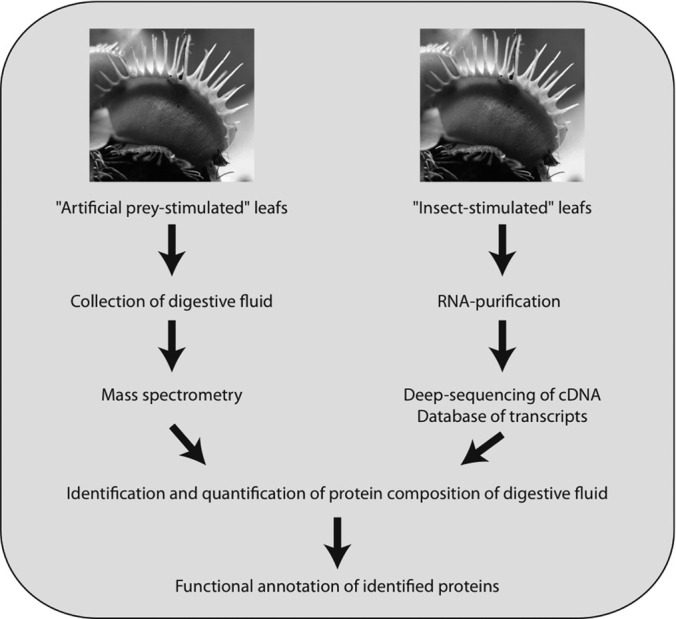
Workflow of the Venus flytrap digestive fluid analysis.
EXPERIMENTAL PROCEDURES
Plant Material for 454 Sequencing and Sampling by Filter Paper Stimulation
Dionaea muscipula plants were purchased from CRESCO Carnivora (De Kwakel, The Netherlands) and grown in plastic pots at 22 °C in a 16:8 h light:dark photoperiod. Three stimulation methods were used for the transcriptomic approach: (i) the plants were fed with ants, and the traps were collected after 24 h; (ii) the plants were sprayed with 100 μm coronatine, and the traps were harvested after 24 h; and (iii) the plants were stimulated by the placement of filter paper soaked with either 30 mm urea, 30 mm chitin, or water into the trap, and trap tissue was collected 1 and 8 h after stimulation. The material for the transcriptome analyses was harvested as follows: traps and excised trigger hairs were frozen in liquid nitrogen. Additionally, secretory cells were isolated from the inner trap surface by gently abrading the gland complexes using a razor blade. RNA was separately isolated from each sample, and for cDNA synthesis, the RNA from different tissues was pooled.
To stimulate fluid secretion for the protein analyses, the closure of healthy mature traps was initiated by tickling the trigger hairs within the trap. Because secretion does not begin without further stimulation of the trigger hairs while the trap is in the closed state, a fine piece of filter paper soaked with water was trapped in the closing snap trap, allowing for the induced movement of the trigger hairs by slight movements of the filter paper. Secreted liquid was then carefully sampled from the closed trap using a pipette tip that was inserted between the closed trap lobes.
Plant Material for Illumina Sequencing and Sampling by Magnet-based Stimulation
Plants were purchased at the Lammehave nursery (Ringe, Denmark) and grown in a walk-in plant growth chamber at 26 °C in a 12:12 h light:dark photoperiod. All of the experiments were performed on healthy mature plants. For the transcriptomics analyses, the digestion process was initiated by feeding the plants yellow mealworm beetles (Tenebrio molitor). After 3, 8, 24, 48, and 72 h, the leaves were harvested, rinsed with water to remove the partially digested beetle and beetle fragments, snap frozen in liquid nitrogen, and stored at −80 °C. For each time point, two stimulated traps were harvested.
For the protein analyses, magnet-based stimulation of the leaf was used to induce the secretion of the digestive fluid. A small stick-magnet was positioned between the trap leaves, and 100 μl of the cysteine protease inhibitor trans-epoxysuccinyl-l-leucylamido-(4-guanidino) butane (E-64; 50 μm) was added to the trap to reduce the level of adventitious proteolysis (14, 16). Then a larger magnet was applied to move the smaller magnet inside the trap, stimulating the trigger hairs and resulting in complete closure. After 48 and/or 72 h, the secreted fluid (up to 200 μl) was aspirated using a pipette that was forced in between the leaf lobes of the trap. The collected material was centrifuged, and the supernatant was used for further analyses. If not used immediately, the fluid was stored at −20 °C.
Transcriptome Sequencing and Assembly
The leaves from the stimulated traps were pooled and homogenized before RNA was extracted using a previously described hot borate buffer protocol (17). Poly-A transcripts were enriched from 3.5 μg of total RNA, and the transcripts were fractionated in the presence of Zn2+. Subsequently, double-stranded cDNA synthesis was performed using random primers and RNase H. After end repair and purification, the fragments were ligated with bar-coded paired-end adapters, and fragments with insert sizes of ∼150 to 250 bp were isolated from an agarose gel. Half of the library was normalized, and the other half was amplified via PCR and purified from a gel. The library quality was assessed using capillary sequencing of randomly selected clones. The high-throughput sequencing of the cDNA samples from the beetle-stimulated traps was performed on an Illumina HiSeq 2000 instrument using a paired-end run with 2 × 50 bp. The cDNA samples isolated from the traps from the other combined stimulation approach were sequenced using a 454 GS FLX Titanium platform. The filtered reads from the two transcriptomic datasets were assembled using Oases software (56) on top of Velvet and Mira, respectively. The minimum size for the assembly was set to either 50 or 100 bases. Several parameter sets (e.g. Burrows-Wheeler Alignment) were tested to optimize the assemblies.
Sampling Procedures for Proteomics
For the filter paper stimulation method, the secreted fluid was collected into three independent pools from 15 to 20 traps stimulated for 68 h. The protein was precipitated using ice-cold acetone. Protein pellets were resuspended in 6 m urea and 2 m thiourea (pH 8). After the reduction of disulfide bonds with dithiothreitol (DTT), free cysteine residues were alkylated using iodacetamide. Proteins were predigested with Lys-C for 3 h before the dilution of the sample with four volumes of 10 mm Tris-HCl (pH 8). Trypsin was added (1 μg trypsin per 50 μg protein), and the digestions were performed at room temperature for 16 h. After acidification, the peptides were desalted over a C18 matrix prior to the MS analysis.
For the magnet-based method, digestive fluid was collected 48 h after the stimulation of 18 traps. Subsequently, three pools were prepared using the digestive fluid from five to eight plants. From these samples, 35 μl were withdrawn, and the pH was adjusted to 8.5. Subsequently, DTT was added, the samples were boiled for 5 min, and iodoacetamide was then added to alkylate free cysteine residues. After 15 min of incubation, trypsin was added (1:20 ratio), and the digestions were performed at 37 °C for 16 h.
Gel-free Proteomics of the Digestive Fluid (from Both Sampling Procedures)
The resulting peptides from each of the digests were separated using an EasyLC nanoflow HPLC system (Proxeon Biosystems, Odense, Denmark) connected to an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with a nanoESI ion source (Proxeon Biosystems, Odense, Denmark). The chromatographic separation was performed on a 15-cm fused silica emitter (100 μm i.d.) that was in-house packed with RP ReproSil-Pur C18-AQ 3 μm resin (Dr. Marisch GmbH, Ammerbuch-Entringen, Germany). The peptides were eluted using an acidic acetonitrile gradient at a flow rate of 250 nl min−1, as described elsewhere (18, 19).
MS scans (300–1800 m/z) were recorded using an Orbitrap mass analyzer at a resolution of 60,000 at 400 m/z, with 1 × 106 automatic gain control target ions and a 500-ms maximum ion injection time. The MS scans were followed by data-dependent collision-induced dissociation MS/MS scans of the five most intense multiply charged ions in the mass spectrometer at a 15,000 signal threshold, 30,000 automatic gain control target, 300-ms maximum ion injection time, 2.5-m/z isolation width, 30-ms activation time at 35 normalized collision energy, and dynamic exclusion enabled for 30 s with a repeat count of 1. Peak picking was performed using either MaxQuant 1.114 (Max Planck Institute of Biochemistry, Martinsried, Germany) or Xcalibur 2.0 (Thermo Fisher Scientific Inc., Waltham, MA). The raw data files of the in-solution digestion of the Venus flytrap secretion fluid from both sampling methods have been deposited at the Tranche database (proteomecommons.org) under the following hash key: tzwFTnv4Y04ujdmGC1tVaULYKS3OQ/0i3pmQ2P0vvvdHe6+5vX09E6zW4OzKILOJJDZ9OTXzOB8N66+5czMOCv2MORAAAAAAAAAE/A==.
Identification and Quantification of the Secretome Proteins Using the 454 Transcriptome
The acquired raw data files were searched against the 6-frame translation of the 454 transcriptome (in total, 227,604 protein entries) using MaxQuant 1.114 (20). The carbamidomethylation of cysteine residues was set as a fixed modification, and the oxidation of methionine residues was set as a variable modification. Two missed cleavages were allowed. The mass tolerance for the first search was set to 10 ppm, and the fragment mass tolerance was set to 0.5 Da. The peptide and protein false discovery rates were set to 0.01, and the identified peptides were required to have a minimum length of six amino acid residues. The assignment of the identified peptides to translated proteins was primarily based on proteotypic peptides. Peptides with more than one protein match were assigned to protein groups consisting of all of the proteins with their respective peptide matches. The retention time alignment of the precursor ions was used to extract intensity information from the peaks with matching m/z values from samples in which these peaks were not selected for data-dependent fragmentation.
Identification and Quantification of the Secretome Proteins Using the Illumina Transcriptome
For protein identification, the raw data from both sampling procedures were searched against the 6-frame translation of the Illumina transcriptome (in total, 97,728 protein entries) using Mascot 2.3.02 (Matrix Science, London, UK) (21). The searches were performed with up to one missed cleavage allowed, carbamidomethyl (C) as a fixed modification, methionine oxidation as a variable modification, a peptide mass tolerance offset of 10 ppm, a fragment mass tolerance of 0.5 Dalton, and an ion score cutoff at 20. Peptide identification was defined as peptides with scores above Mascot's homology threshold and at a significance threshold (p) of 0.01. Peptide assignments to proteins were performed according to the default Mascot settings, i.e. each redundant peptide was primarily assigned to the highest scoring protein. The described settings resulted in an average false discovery rate of 3.3%. However, additional stringent criteria for protein and peptide acceptances were applied (see below). Proteins that did not meet the quantitative thresholds and that were not identified and quantified in at least two of the six samples were rejected. To extract the quantitative information, Mascot Distiller 2.4.2.0 (Matrix science) was applied using fraction and correlation thresholds of 0.7 and 0.9, respectively. The data were parsed using MS Data Miner 1.0, which is in-house-developed software (57).
Gel-based Proteomics of the Digestive Fluid
A total of 100 μl of digestive fluid from the magnet-stimulated traps was lyophilized and subsequently dissolved in SDS sample buffer containing 30 mm DTT. The proteins were resolved in 5% to 15% acrylamide gradient gels (23). Subsequently, the gel was silver-stained, and all of the visible bands were excised and digested with trypsin (24, 25). Tryptic peptides were purified using a C18 stage tip (Proxeon Biosystem A/S part of Thermo Fisher Scientific Inc., Odense, Denmark) and were subsequently analyzed via liquid chromatography (LC)-MS/MS using an EASY-nLC (Proxeon Biosystems) connected to a Q-TOF Ultima API (Micromass/Waters, Milford, MA) mass spectrometer. As described above, the obtained mass spectra were analyzed using Mascot; however, the significant threshold value (p value) was set at 0.05 because of the lower complexity of the samples and the lower sensitivity of the instrument. In addition, the peptide mass tolerance was set at 1 Dalton because of the lower accuracy of this instrument. If the identification was based on a single peptide, then it was accepted only if the protein was also observed during the gel-free analyses using the more sensitive Orbitrap mass spectrometers.
Verification of the Peptide and Protein Assignments
We manually verified that the peptide hits corresponding to the same transcript corresponded to the same reading frame. If the peptides were identified from different reading frames, then we evaluated whether the different reading frames were likely explained by missing regions in the sequence of interest, which could have led to a frameshift. If this was the case, then the reading frames that resulted in the identification of peptides were merged in the part of the transcript that was missing (see “X” in the sequences). If the identification was based on one unique peptide, then the MS/MS data were manually validated using the following criteria: the assignment of major peaks and the occurrence of uninterrupted y- or b-ion series of at least three amino acid residues. The full list of identified peptides and proteins can be found in supplemental Tables S1 (454 transcriptome) and S2 (Illumina transcriptome). The spectra for proteins with single-peptide identifications can be found in supplemental Figs. S2 (454 transcriptome, in-solution analysis), S3 (Illumina transcriptome, in-solution analysis), and S4 (Illumina transcriptome, gel analysis). The majority of the proteins were identified and quantified by means of both the Mascot/Illumina transcriptome approach and the MaxQuant/454 transcriptome approach, which strengthens our data.
Determination of the Proteins' Relative Abundances
To calculate the relative abundances, we initially calculated the sum of the ion intensities from the extracted ion chromatograms for all of the identified peptides in that particular analysis (i.e. the total ion intensity). Subsequently, the individual samples were normalized. Then, the relative abundance (or fraction of the total) of a particular protein (weighted by the protein's molecular weight) was derived using the following formula: (MS intensity sum for the peptides belonging to a specific protein/total ion intensity of the sample)/molecular weight of the transcript. Subsequently, the average value of the relative abundances was calculated based on samples from the same stimulation method when the protein was present. The abundance ranking performed here was similar to the emPAI calculation (26); however, we based our rankings on summed ion intensities rather than peptide counts, which is analogous to the iBAQ quantification (27). Transcript molecular weight was used for protein size normalization. If only one or two peptides were identified and quantifiable for a single protein, then those peptides were used for quantitation in any case. Proteins were excluded if they were identified and quantifiable in only one of the six LC-MS/MS samples. We focused on the identified proteases in the Results section, and we searched specifically for proteases in the obtained data. Consequently, if the identified protein was a protease, then we carefully searched the peptide spectra, and if acceptable (see supplemental Fig. S2) the protein was included in Table II even if it was identified and quantified in only one of the six samples. Based on this screening, only contig 18374 was included.
Table II. List of the identified and quantified proteins from the secreted fluid after two different stimulation methods (paper-based and magnet-based) and after matching against two different transcriptomes (454 and Illumina); the abundance ranking within each sample is indicated.
| Identifier (454-transcript) | Identifier (Illumina-transcript) | Accession (A. thaliana) | Putative function | Paper-based stimulation | Average rank in both transcripts |
|
|---|---|---|---|---|---|---|
| Magnet-based stimulation | ||||||
| Locus_8322_Transcript_1/1_Confidence_1.000 | No BLAST hit with E-value < 1 | 5 | 1 | |||
| DM_TRA02_REP_contig53074 | Locus_610_Transcript_2/2_Confidence_1.000 | AT3G12500 | Chitinase | 5 | 2 | |
| NG-5590_Gland_cleanedcontig148924 | Locus_223_Transcript_110/117_Confidence_0.063 | AT5G45890 | Cysteine protease (dionain-3) | 6 | 3 | |
| DM_TRA02_REP_contig82037 | No BLAST hit with E-value < 1 | 16 | 4 | |||
| DM_TRA02_REP_contig53027 | Locus_223_Transcript_111/117_Confidence_0.063 | AT4G33720 | PR (pathogenesis-related) protein | 5 | 5 | |
| DM_TRA02_contig15221 | Locus_369_Transcript_1/1_Confidence_1.000 | AT4G33355 | PR (pathogenesis-related) protein | 20 | 5 | |
| DM_TRA02_REP_contig50549 | Locus_270_Transcript_1/2_Confidence_1.000 | AT4G11650 | Osmotin-like protein | 29 | 6 | |
| DM_TRA02_contig105872 | AT1G11905 | Protein of unknown function | 22 | 6 | ||
| DM_TRA02_REP_contig53296 | AT4G33355 | PR (pathogenesis-related) protein | 7 | |||
| NG-5590_Gland_cleanedcontig74818& contig124417 | Locus_21_Transcript_5/6_Confidence_0.188 | AT5G45890; AT1G | Cysteine protease (dionain-2) | 54 | 9 | |
| NG-5590_Gland_cleanedcontig110078 | AT1G47128 | Cysteine proteinase (Dionain 4) | 10 | 9 | ||
| DM_TRA02_contig14398 | Locus_326_Transcript_1/1_Confidence_1.000 | AT2G38530 | Lipid transfer protein | 24 | 10 | |
| DM_TRA02_contig352 | Locus_1885_Transcript_2/2_Confidence_1.000 | AT5G06860 | Polygalacturonase inhibiting protein | 8 | 11 | |
| NG-5590_Gland_cleanedcontig146186 | Locus_2155_Transcript_2/2_Confidence_1.000 | AT3G10410 | Serine carboxypeptidase-like 49 | 13 | 11 | |
| Locus_3837_Transcript_1/1_Confidence_1.000 | AT2G02990 | Ribonuclease T2 family | 15 | 13 | ||
| DM_TRA02_contig19199 | Locus_673_Transcript_1/1_Confidence_1.000 | AT3G57260 | Beta 1,3-glucanase | 1 | 14 | |
| DM_TRA02_contig16330 | Locus_52_Transcript_2/5_Confidence_0.385 | AT1G14540 | Peroxidase superfamily protein | 2 | 15 | |
| NG-5590_Gland_cleanedcontig2880 | Locus_448_Transcript_1/1_Confidence_1.000 | AT5G54370 | Late embryogenesis abundant protein | 15 | ||
| DM_TRA02_contig33225 | No BLAST hit with E-value < 1 | 16 | ||||
| DM_TRA02_REP_contig51333 | Locus_3455_Transcript_1/1_Confidence_1.000 | AT5G59970 | Histone superfamily protein | 20 | 17 | |
| DM_TRA02_contig18767 | AT4G35790 | Protein with phospholipase D activity | 17 | |||
| DM_TRA02_REP_contig85615 | Locus_462_Transcript_4/6_Confidence_0.200 | AT1G53130 | PR (pathogenesis-related) protein | 18 | ||
| DM_TRA02_REP_contig55271 | Locus_522_Transcript_1/2_Confidence_1.000 | AT1G51060 | A histone H2A protein | 25 | 18 | |
| Locus_21_Transcript_3/6_Confidence_0.562 | AT5G45890 | Cysteine proteinase (dionain-1) | 19 | |||
| DM_TRA02_contig14593 | XP_002510033 | Branched Chain amino acid pathway | 19 | |||
| Locus_462_Transcript_2/6_Confidence_0.300 | AT4G26880 | Stigma-specific Stig1 family protein | 20 | |||
| Locus_5180_Transcript_1/1_Confidence_1.000 | AT1G79820 | Suppressor of G protein beta1 (SGB1) | 27 | 21 | ||
| DM_TRA02_contig12450 | Locus_5610_Transcript_1/1_Confidence_1.000 | AT5G05340 | Peroxidase superfamily protein | 19 | 22 | |
| DM_TRA02_REP_contig78697 | No BLAST hit with E-value < 1 | 15 | 22 | |||
| Locus_6940_Transcript_1/1_Confidence_1.000 | AT5G05340 | Peroxidase superfamily protein | 21 | 22 | ||
| DM_TRA02_contig53&contig6292 | Locus_8046_Transcript_1/1_Confidence_1.000 | AT5G50400 | Purple acid phosphatase 27 (PAP27) | 10 | 23 | |
| DM_TRA02_contig14693 | No BLAST hit with E-value < 1 | 32 | 24 | |||
| Locus_5274_Transcript_1/2_Confidence_0.800 | AT2G07698 | ATPase, F1 complex, alpha subunit protein | 33 | 25 | ||
| NG-5590_Gland_cleanedcontig104489 | AT4G35790 | Encodes a protein with phospholipase D activity | 25 | |||
| DM_TRA02_contig7401 | Locus_597_Transcript_3/3_Confidence_0.667 | AT5G14780 | NAD-dependent formate dehydrogenase | 41 | 26 | |
| DM_TRA02_contig16324 | AT3G04720 | PR (pathogenesis-related) protein | 26 | |||
| DM_TRA02_contig24716 | No BLAST hit with E-value < 1 | 18 | 27 | |||
| DM_TRA02_REP_contig50397 | Locus_864_Transcript_1/1_Confidence_1.000 | AT5G06860 | Polygalacturonase inhibiting protein | 5 | 28 | |
| DM_TRA02_contig9524 | AT2G27130 | Lipid transfer protein (LTP) family protein | 28 | |||
| DM_TRA02_REP_contig87892 | Locus_5734_Transcript_1/1_Confidence_1.000 | AT2G17120 | Lysm domain GPI-anchored protein 2 precursor | 35 | 29 | |
| DM_TRA02_REP_contig55391 | AT4G26880 | stigma-specific Stig1 family protein | 30 | |||
| DM_TRA02_contig19291 | Locus_187_Transcript_30/39_Confidence_0.191 | AT4G39330 | Cinnamyl alcohol dehydrogenase 9 | 41 | 31 | |
| DM_TRA02_REP_contig63924 | VITISV_027092 | Hypothetical protein, peroxidase-like | 4 | |||
| DM_TRA02_contig6244 | Locus_9868_Transcript_1/1_Confidence_1.000 | AT5G45960 | GDSL-like lipase | 10 | ||
| DM_TRA02_REP_contig53347 | Locus_5819_Transcript_1/1_Confidence_1.000 | AT3G52780 | Purple acid phosphatase 20 | 12 | ||
| NG-5590_Gland_cleanedcontig146154 | Locus_258_Transcript_1/1_Confidence_1.000 | AT5G48430 | Aspartyl protease family protein-like | 13 | ||
| Locus_2276_Transcript_1/1_Confidence_1.000 | AT5G09810 | Actin | 14 | |||
| Locus_2818_Transcript_1/1_Confidence_1.000 | AT1G68290 | ENDO 2 (endonuclease 2) | 17 | |||
| DM_TRA02_REP_contig79562 | AT3G50990 | Peroxidase superfamily protein | 17 | |||
| DM_TRA02_contig126504 | AT2G42840 | PDF1 (protodermal factor 1) | 21 | |||
| DM_TRA02_contig120869 | Locus_1790_Transcript_1/2_Confidence_1.000 | AT5G03240; AT3G | Ubiquitin extension protein | 23 | ||
| DM_TRA02_contig14293 | Locus_1849_Transcript_1/1_Confidence_1.000 | AT1G71695 | Peroxidase superfamily protein | 23 | ||
| DM_TRA02_REP_contig72380 | XP_003536264 | Peroxidase superfamily protein | 23 | |||
| DM_TRA02_REP_contig49581 | Locus_468_Transcript_1/1_Confidence_1.000 | AT4G28390 | AAC3 (ADP/ATP carrier 3) | 28 | ||
| DM_TRA02_REP_contig137982 | AT1G71695 | Peroxidase superfamily protein | 28 | |||
| DM_TRA02_contig14688 | Locus_1267_Transcript_1/1_Confidence_1.000 | AT5G08680 | ATP synthase beta-subunit | 31 | ||
| NG-5590_Gland_cleanedcontig79407 | Locus_3385_Transcript_1/2_Confidence_1.000 | AT5G22850 | Aspartyl protease family protein (dionaeasin-2) | 31 | ||
| DM_TRA02_REP_contig50840 | Locus_175_Transcript_1/1_Confidence_1.000 | AT1G78300 | G-box binding factor GF14 omega | 34 | ||
| Locus_5247_Transcript_1/1_Confidence_1.000 | AT3G12580 | Heat shock protein 70 | 34 | |||
| DM_TRA02_REP_contig60010 | AT3G54420 | ATEP3; chitinase | 34 | |||
| DM_TRA02_REP_contig56545 | XP_003380422 | superoxide dismutase [Trichinella spiralis] | 35 | |||
| DM_TRA02_contig7075 | AT5G08680 | ATP synthase beta-subunit | 36 | |||
| DM_TRA02_REP_contig48999 | AT5G60390 | elongation factor 1-alpha/EF-1-alpha | 37 | |||
| DM_TRA02_contig12608 | AT1G68290 | ENDO 2 (endonuclease 2) | 38 | |||
| DM_TRA02_contig1301 | AT1G13440 | Glyceraldehyde-3-phosphate dehydrogenase C2 (GAPC2) | 39 | |||
| DM_TRA02_contig16247 | Locus_1944_Transcript_1/1_Confidence_1.000 | AT4G37530 | Peroxidase superfamily protein | 40 | ||
| DM_TRA02_contig13240 | AT3G54420 | ATEP3; chitinase | 41 | |||
| DM_TRA02_contig347 | Locus_4642_Transcript_1/1_Confidence_1.000 | AT5G67130 | Phosphodiesterase superfamily protein | 42 | ||
| NG-5590_Gland_cleanedcontig105066 | AT1G03220 | Aspartyl protease family protein-like | 42 | |||
| DM_TRA02_REP_contig50509 | AT5G02500 | HSC70–1 (heat shock cognate protein 70–1) | 44 | |||
| DM_TRA02_contig28745 | No BLAST hit with E-value < 1 | 45 | ||||
| DM_TRA02_REP_contig129322 | AT5G41210 | Glutathione transferase | 48 | |||
| DM_TRA02_contig16780 | AT2G36460 | Fructose-bisphosphate aldolase, putative | 49 | |||
| DM_TRA02_REP_contig48954 | AT2G44490 | Thioglucosidase | 50 | |||
| DM_TRA02_contig14440 | AT5G17920 | Methionine synthase | 54 | |||
| DM_TRA02_contig18374 | AT2G03200 | Eukaryotic aspartyl protease family protein (dionaeasin-1) | 55 | |||
Functional Annotation and Alignment Analyses
Each identified protein was functionally annotated via comparison with Arabidopsis thaliana using the TAIR BLAST 2.2.8 tool with the default settings (www.arabidopsis.org). The highest scoring hit was used for the annotation. If a homologous protein was not identified using this approach, a comparison with the NCBInr database was performed using the NCBI BLAST tool. The MEROPS peptidase database (merops.sanger.ac.uk) and the Biology Workbench software from the San Diego Supercomputer Center (workbench.sdsc.edu) were used for sequence analyses and alignments of the identified proteolytic enzymes (28).
RESULTS
Deep Sequencing of the cDNA from Stimulated Venus Flytrap Snap Trap Leaf Lobes
In the present study, we used transcriptomics-generated databases to facilitate the subsequent proteomics-based identification and quantification of the proteins in Venus flytrap digestive fluid (Fig. 1). To increase the likelihood of including all of the relevant transcripts, we employed two different deep sequencing RNA-seq technologies to generate two comprehensive cDNA databases. After the raw data were cleaned, the 454 transcriptome was assembled from 3.5 million reads. In total, 37,934 contigs with matching homologies in the plant kingdom were assembled with an average length of 550 (Table I). In the Illumina transcriptome, ∼41 and 120 million paired-end reads of 50 bp lengths were obtained for the non-normalized sample and the normalized sample, respectively. The data from these two sequencing runs were combined and used for contig and transcript assemblies. A hash value of 41 was selected for the contig and transcript assembly, and 16,288 transcripts were assembled with an average length of 747 (Table I).
Table I. Transcriptome properties.
| Illumina | 454 | |
|---|---|---|
| Number of contigs | 16,288 | 37,934 |
| Sum of the contig lengths | 12,177,595 | 21,138,288 |
| Cut-off contig length | 100 | 50 |
| Average length of the contigs | 747 | 550 |
The Basic Pattern of the Protein Composition of the Venus Flytrap Digestive Fluid
If the Venus flytrap plants had been stimulated to secrete digestive fluid by a natural prey, then the peptides derived from “prey proteins” would have influenced the mass spectrometry analyses, likely affecting the sensitivity of the analyses, as well as the quantification and identification of the secreted Venus flytrap proteins. A standardized protocol for digestive fluid harvesting would have been difficult to implement if natural prey had been used. Instead, we developed two methods to stimulate the secretion of the digestive fluid without adding real prey.
Upon magnet-based stimulation, the secretion process was initially monitored over 3 days. After 24 h, moisture was detected in the traps, but the amounts were too small for sampling. Up to 200 μl of digestive fluid was collected 48 h (10 replicates) and 72 h (three replicates) after stimulation. The traps that were emptied after 48 h were allowed to continue to secrete for an additional 24 h after stimulation (three replicates). The proteins in the collected samples were separated by SDS-PAGE (supplemental Fig. S1), resulting in the same pattern of protein bands. This suggested that the variation over time and from plant to plant in the overall protein composition of the digestive fluid was low under the conditions employed.
Identification and Abundance Ranking of the Proteins in the Venus Flytrap Digestive Fluid
The secreted proteins from the magnet-based and filter-paper-based stimulations were analyzed by LC-MS/MS. Six datasets were searched against both of the generated transcriptomes and analyzed using MaxQuant and Mascot Distiller. Using this approach, we identified and quantified 76 proteins in the digestive fluid (Table II, supplemental Table S3), with 32 proteins detected in both transcripts. Furthermore, 34 were present in only the 454 transcriptome (supplemental Tables S1 and S4), and 10 proteins were present in only the Illumina transcriptome (supplemental Tables S2 and S5). In total, 66 proteins were identified in the secreted fluid upon filter-paper-based stimulation, and 42 proteins were identified in the secretion fluid after magnet-based stimulation (Table II). Of these 42 proteins, 30 proteins were also identified upon filter-paper-based stimulation, demonstrating large overlap (71%) between the sampling procedures (Table II). However, the abundance ranking displayed differences between the two sampling procedures, emphasizing the importance of using complementary methods. The rankings based on MaxQuant and Mascot Distiller were generally in good agreement (supplemental Table S3). The 32 proteins identified from both transcriptome databases displayed significant identical overlaps over long, continuous stretches. Frequently, the shorter contig from one transcript was completely contained within another, longer transcript (supplemental Table S6). This finding clearly supports the value of the complementary approach utilized in this analysis of the Venus flytrap secretome.
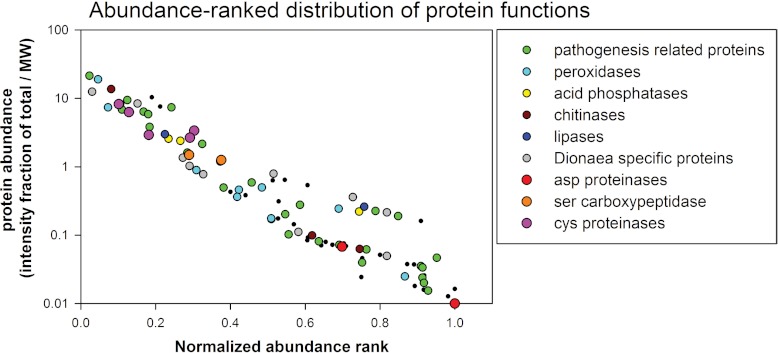
In order to annotate the identified proteins based on function, the corresponding translated transcripts were homology searched against the well-annotated Arabidopsis thaliana proteome (supplemental Tables S4 and S5). Among the most abundant proteins in the Venus flytrap secretome were homologs of proteases, chitinases, osmotin-like protein, pathogenesis-related proteins, lipid transfer proteins, peroxidases, and beta-1,3-glucanase, which all belong to families of pathogenesis-related proteins (29). The presence of defense-related proteins has also been observed in the digestive fluid of Nepenthes (30), indicating that the digestive process in these two plants is functionally similar, although the plants belong to two different families, namely, Droseraceae and Nepenthaceae. Nine of the transcripts were homologous to different proteases, showing that the proteases are one of the two largest protein families in the secretome. This result correlates with the degrading function of the fluid. The sequence of one of the identified proteases was identical to the peptide sequence of a Venus flytrap digestive fluid cysteine protease from a previous study, except for one amino acid residue (16). In that particular study, the protease was termed “dionain.” Here, we adhered to this nomenclature and named the cysteine proteases dionain-1, dionain-2, dionain-3, and dionain-4. Dionain-1 is the protease that contains the previously sequenced peptides (Table II). Nine peroxidase homologs, including one of the top-ranking proteins from the filter-paper-based sampling procedure, were identified. These data indicate that peroxidases have an important role in the digestive fluid. For each identified protein, we plotted the summed ion intensity fraction of the total against the normalized abundance rank in the two transcriptomes. The normalized abundance ranks were calculated as the abundance rank divided by the total number of proteins identified in each sample. When performing this ranking, it became apparent that proteins with antimicrobial/defense functions were among the most abundant proteins in the Venus flytrap secretion fluid (Fig. 2). Cysteine proteinases were consistently the most abundant proteases, followed by serine carboxypeptidases and aspartic proteases.
Fig. 2.
The summed ion intensity fraction of the totals for the proteins identified in the two transcriptome versions plotted against the normalized abundance rank within the respective sample. Different colors indicate proteins of a similar function.
To strengthen our findings on the in-solution-based abundance ranking and composition analysis of the Venus flytrap digestive fluid, we conducted a complementary gel study (supplemental Fig. S1). Digestive fluid was collected from magnet-stimulated traps, and the proteins were separated on a silver-stained polyacrylamide gel prior to analysis via LC-MS/MS. The obtained spectra were queried against the Illumina transcriptome (supplemental Table S7). The majority of the proteins identified in the gel-based analysis were among the average top-10 ranked proteins present in the in-solution analysis of the magnet-based stimulation. Although there is a general relationship between the summed peptide intensities and the amount of protein present (27), outliers with very few tryptic peptides compatible with an MS analysis can also occur. This is likely to be the case for the relatively low ranking of the dionain-1 protein. This protein has been described previously as the major protein in the digestive fluid (16), and we identified this protein in four of the bands from the gel, which indicates the prominence of this protease. However, this protein was quantified by only one peptide, so its ranking is relatively low (supplemental Table S7).
The Proteolytic Enzymes of the Venus Flytrap Digestive Fluid
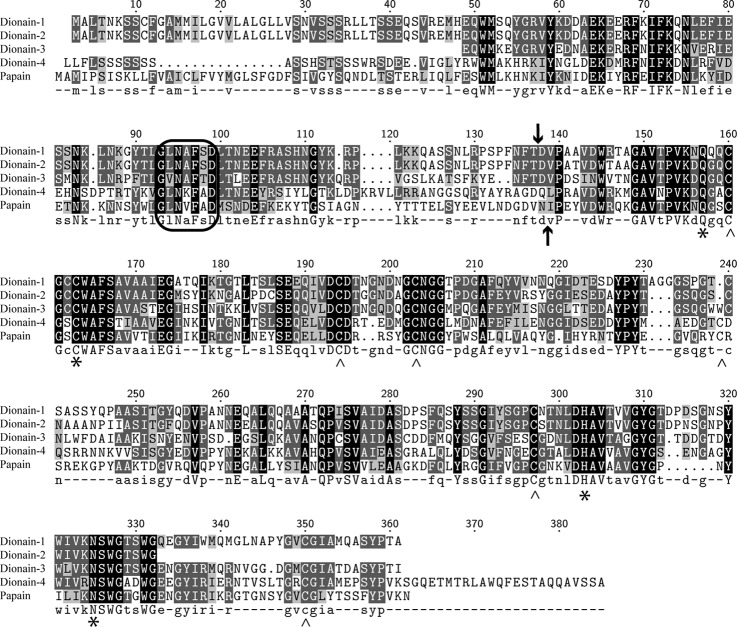
The protein sequences of four identified cysteine proteases were compared with those of other proteases using the MEROPS BLAST service (28). Dionains 1–3 and dionain-4 most resembled the cysteine proteases SPG31-like peptidase (31) and pseudotzain (32), respectively. All four proteases belong to the subfamily C1A, represented by the cysteine protease papain and to which the identified sequences were aligned (Fig. 3). The alignment illustrated that all of the reactive site residues and disulfide bridge-forming cysteines in papain were conserved in all of the dionains. The similarity to papain was also apparent because the pH optima of dionains are expected to be acidic, which is also the case for papain (33). In addition, the alignment showed that an evolutionarily conserved motif (Gly-Xxx-Asn-Xxx-Phe-Xxx-Asp), pivotal for the pH-dependent autoactivation of cysteine proteases, was present in the pro-peptide of dionains (34, 35). These results suggest that autoactivation is a part of the dionain activation mechanism. Indeed, SDS-PAGE of the digestive fluid proteins (collected in the absence of E-64) followed by the Edman degradation of dionain-1 (data not shown; also Ref. 16) revealed that activation occurred due to the proteolysis of a peptide bond within the same region observed for papain (Fig. 3).
Fig. 3.
The alignment of the open reading frames of the Venus flytrap cysteine proteases with papain. The encircled area represents a conserved motif that is essential for the conversion from zymogen to the mature protease in papain and cathepsin L. The upward-pointing arrow (↑) indicates the cleavage site between the activation peptide and the mature protease in papain, and the downward-pointing arrow (↓) indicates the cleavage site between the activation peptide and the mature protease in dionain-1. The asterisks (*) represent the active site residues in papain, and the carets (∧) represent the cysteine residues involved in the disulfide bridges. The alignment strongly suggests that the active site residues and disulfide bridges are conserved between the five different plant cysteine proteases.
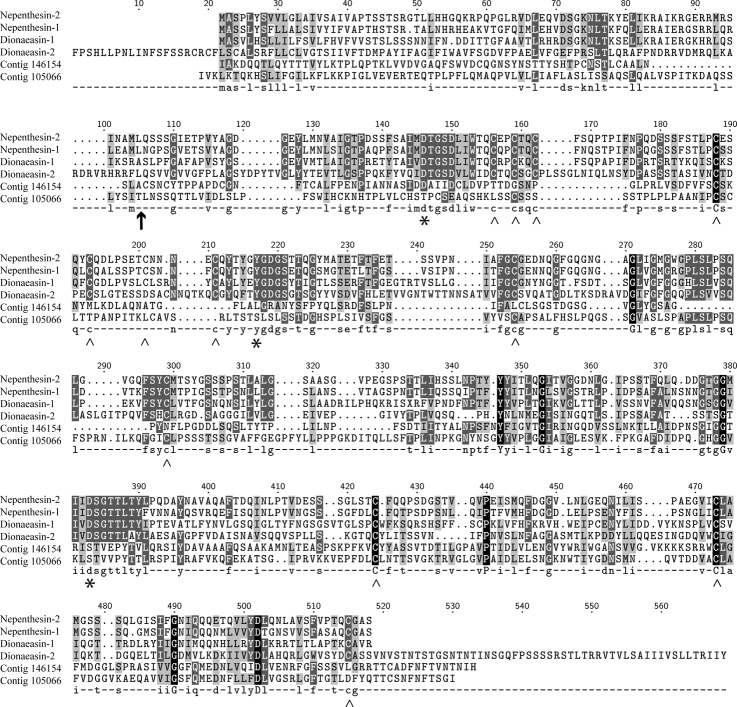
The other large group of proteolytic enzymes in the secretome was the aspartic proteases. To characterize these proteases, the sequences were analyzed using BLAST against the MEROPS database, which demonstrated that these proteases belong to subfamily A1B. This subfamily is represented by the nepenthesin from Nepenthes gracilis (36). The other large aspartic protease subfamily is A1A and is represented by pepsin, which is important for digestion processes in vertebrates. The main parts of the proteases in subfamilies A1A and A1B are most active at acidic pHs, but A1B differs from A1A in that it normally has six disulfide bridges, which are likely responsible for the remarkable stability of these proteins (37). Two of the identified aspartic proteases (contig 146154 and contig 105066 when using the 454 transcriptome naming) did not contain the active site residues found in the nepenthesins (Fig. 4). In addition, the cysteine patterns were different. These data indicate that although these proteases belong to the A1B subfamily, they are unable to display catalytic activities. An NCBI-BLAST search of these sequences indicated that the two proteins might be xylanase and endoglucanase inhibitor proteins, which are involved in plant defense (38). Taken together, two of the putative aspartic proteases are likely not involved in the degradation of prey proteins in the Venus flytrap.
Fig. 4.
The alignment of the Venus flytrap aspartate proteases with nepenthesin 1 and nepenthesin 2 identified from Nepenthes gracilis. The upward-pointing arrow (↑) indicates the cleavage site between the activation peptide and the mature nepenthesin proteases. The asterisks (*) represent the active site residues in the nepenthesins, and the carets (∧) indicate the positions of the cysteine residues involved in the disulfide bridges in the nepenthesins. The alignment strongly suggests that two of the Venus flytrap aspartic proteases are catalytically active proteases (referred to as dionaeasins), in contrast to the two other identified proteins (contigs 146154 and 105066), which are likely not catalytically active proteases.
In contrast, the other two Venus flytrap aspartic proteases contained the active site residues and nearly all of the disulfide bridge-forming cysteine residues at the same positions as in the nepenthesins. Consequently, we named these sequences dionaeasin-1 (contig 18374) and dionaeasin-2 (contig 79407), which is analogous to the nepenthesins from Nepenthes. These dionaeasins are likely involved in prey digestion. However, these two proteases were less abundant in the digestive fluid than the cysteine proteases (Table II and Fig. 2), a finding also emphasized by the fact that dionaeasin-1 was found in only one of the six LC-MS/MS analyses. These results suggest that aspartic proteases have a minor role in the degradation process. This is in contrast to Nepenthes, in which the nepenthesins are the most prominent proteolytic enzymes.
Regardless of the sampling procedure, the identified serine carboxypeptidase was relatively abundant (Table II), and it was identified using the less sensitive gel-based approach (supplemental Table S7). These results suggest that this protease has an important role in the digestion process. Sequence analyses revealed that it belongs to the S10 family of serine proteases and to the plant serine carboxypeptidase III group (MEROPS ID S10.009) (39). The S10 family, represented by carboxypeptidase Y, is active only at acidic pHs, which makes it different from all of the other serine protease families (except for the S53 family). This activity at a low pH correlates with the finding of this protease in the acidic Venus flytrap digestive fluid.
DISCUSSION
Dionaea muscipula, the Venus flytrap plant, is not eaten by animals and is rarely infected by microbes due to its high content of defense metabolites and proteins (40). Instead, it actively traps, kills, and consumes animals. To gain insight into its digestive processes, we analyzed the composition of the fluid secreted by the plant. Most previous studies of the Venus flytrap's digestive fluid (13–15, 41) used an enzyme activity-based approach to characterize the composition of the fluid. In contrast to these indirect methods, we used a combination of transcriptomics and proteomics to identify the major proteins present in the secreted fluid. Using the enzyme activity-based approach, protease, chitinase, peroxidase, phosphatase, and nuclease activities were detected (14, 15). With regard to protease activity, it has been suggested that the major protease in Venus flytrap digestive fluid is a cysteine protease and that carboxypeptidase activity is also present (14). With the present proteome study, we can explain these different enzymatic activities and correlate them with specific protein sequences.
Transcriptomics-facilitated Proteomics
A direct MS-based identification of the secreted proteins in Venus flytrap digestive fluid was complicated by the lack of the Venus flytrap genome. Recently, deep sequencing technologies have revolutionized the transcriptomics field (42). These methods are largely unbiased and high-throughput, and they provide a large dynamic range (43). To facilitate MS-based protein identifications and to obtain more sequence information, in contrast to a de novo MS approach, we used deep sequencing methods to produce a cDNA library of the transcribed mRNA sequences in stimulated Venus flytrap leaves (Fig. 1). The use of the assembled transcriptome as a database for protein identification was significantly more comprehensive and faster than de novo peptide sequencing. The rapid development of the “next-generation sequencing” field and the decreasing costs of this technology suggest that the approach applied here is generally applicable for proteome-based studies of organisms and systems for which genome sequence information is limited (44).
Secreted Proteins are Actively Synthesized in the Traps
The gel-based analysis of the secreted fluid confirmed that all of the major proteins (i.e. those visible on the silver-stained gel) in the digestive fluid were identified based on the transcriptome derived from the trap tissue. This analysis demonstrates that the cDNA sequences of the major secreted proteins are present in the transcriptome database. Indirectly, it shows that the mRNA coding for these proteins is present in the stimulated traps. Therefore, our results reveal that the Venus flytrap does not exclusively secrete proteins into the digestive fluid from preformed vesicles. Instead, the presence of mRNA indicates that the plant also synthesizes these proteins in the trap during the digestion process. These results are substantiated by the finding that only a limited amount of digestive fluid was present in the traps even after 24 h. If the proteins to be secreted had been present in storage vesicles and ready to be released, then we would have expected the digestive fluid at an earlier time point. These results are supported by previous findings showing that protein synthesis occurs during the secretory phase and that some of the synthesized protein is directly secreted (41).
Abundance Ranking of the Secreted Proteins
In total, 71% of the proteins identified upon magnet-based stimulation were also identified by filter-paper-based stimulation, demonstrating a large overlap between the two stimulation procedures. However, the abundance ranking displayed some differences between the two procedures, at least when the integer ranks were compared. Because more proteins were identified using the filter-paper-based method, these rank numbers contained larger values than those from the magnet-based sampling. When the rankings were standardized for the number of identified proteins (Fig. 2), the abundances were more similar.
The low number of identified peptides for some of the proteins and the rather low abundance ranks likely can be explained by mispredicted nucleotides in some of the transcripts, which is consistent with previous findings demonstrating that contigs obtained from Illumina RNA-seq contain ∼5% mispredicted nucleotides (45). Furthermore, the complete open reading frames were not sequenced for all of the transcripts, which can be partly explained by the low amounts of the respective mRNAs. In addition, the assembly programs used, Velvet and Mira, do not perform perfectly during redundancy reduction even at an error rate of 1% in the transcriptome (46).
The cysteine proteases were difficult to quantify and rank properly. These proteases contain a large activation peptide, and the arginine and lysine contents of the mature proteases are low; as a result, only a few tryptic peptides are applicable for MS analyses. These proteins also contain several glycosylation sites, which hamper MS-based identification. In addition, many of the peptides contain cysteine residues, and in our work these were more difficult to detect even though the proteins were alkylated. Thus, the ranking of e.g. dionain-1, the cysteine protease that was previously found to be a major component of the digestive fluid (16), was likely too low relative to its actual abundance. The high numbers of cysteine residues and putative disulfide bridges (Figs. 3 and 4) indicated that the identified proteins were compact and stable. Consequently, these proteins are likely to be relatively resistant to the promiscuous protease degradations taking place during digestion. Similar to other plant proteases (47), potential N-linked glycosylation sites indicate that the identified proteases could be glycosylated, stabilizing the proteins with respect to proteolytic degradation.
The Composition of the Digestive Environment Sheds Light on the Prey Digestion Mechanism
The low pH of the Venus flytrap digestive fluid is similar to that of other carnivorous plants (e.g. Nepenthes) and to the digestive fluid pHs among vertebrates. However, in vertebrates and Nepenthes, the proteolytically active enzymes are predominantly aspartic proteases (36, 48). In contrast, our findings suggest that cysteine proteases are the most abundant class of proteases in the digestive fluid of the Venus flytrap, followed by a serine carboxypeptidase and aspartic proteases. This composition and diversity of proteases has not been observed in other digestive fluids. The enzyme composition that resembles our findings the most is the intestinal protein digestion cascade employed by some invertebrates (49–51), which similarly includes aspartic proteases and cysteine proteases from the same protease families (the pepsin family (A1) and the papain family (C1)) observed in the Venus flytrap. In general, cysteine proteases have a neutral pH optimum. However, adaptations to acidic pH optima have been observed among the lysosomal cathepsins, which are primarily involved in unspecific bulk protein degradation (52). The protease composition of the Venus flytrap's digestive fluid, with three classes of peptidases, is likely a potent digestion system, emphasizing the strong dependence of Dionaea on the nutrients supplied through prey capture and digestion (11). Particularly as the pH of the digestive fluid changes over time (7), the different enzymes might reach their maximum activities at different digestion stages after the prey is captured. As previously mentioned, the natural habitat of Venus flytrap plants is low-nutrient soils, and the plants depend on nutrients obtained by digesting trapped prey. These identified proteases are likely involved in the release of nitrogen from the prey proteins. In addition to proteases, a number of other hydrolytic enzymes are present in the digestive fluid, and the fact that nucleases, phosphatases, and phospholipases were identified indicates that phosphate is similarly obtained from the prey's nucleic acids, proteins, and cell membranes.
Three chitinases were also identified, including one of the proteins found to be most abundant in the digestive fluid regardless of the stimulation method. These chitinases would be expected to degrade the exoskeletons of captured insects or spiders and thereby facilitate enzymatic access to the inner part of the prey. Furthermore, chitinases are pathogenesis-related proteins that might prevent microbial growth on the trapped prey during the digestion process.
It has been suggested that prey proteins in the Venus flytrap are initially oxidized in order to facilitate their subsequent proteolysis (53), and it has been demonstrated that Nepenthes gracilis uses free radicals during the digestion process (54). Plumbagin, a low-molecular-weight compound present in Venus flytrap digestive fluid, likely facilitates this oxidation (40). The identified peroxidases from the present study are likely involved in these oxidative processes. Thus, our findings support the hypothesis that the oxidation of prey molecules facilitates the digestion mechanisms of the Venus flytrap.
The functions of the hydrolytic enzymes in the digestive fluid are intuitively easy to envision. These enzymes are likely directly involved in prey digestion. The functions of some of the other proteins present in the fluid are more challenging to elucidate, and a functional annotation based on the name of the best match in a homology search (Table II) does not necessarily shed light on the in vivo role of the protein. The roles of these proteins in the digestion mechanism remain to be investigated.
The Digestive Fluid Proteome Suggests a Shift from Defense-related Processes to Digestion-related Processes among the Carnivorous Plants
The only previously characterized digestive fluid proteome from a carnivorous plant was derived from Nepenthes (30). The depth of that de novo sequencing-based study was lower than in the present study; however, aspartic proteases (nepenthesin I and II), a chitinase, a glucanase, a xylosidase, and a thaumatin-like protein were identified. These protein classes were, with the exception of the xylosidase, also identified in the present analysis of the Venus flytrap, indicating the conserved functions of the digestive fluid among carnivorous plant species. Similar to our results, the Nepenthes digestive proteins are also predominantly pathogenesis-related proteins. Higher plants express pathogenesis-related proteins as a response to an attack by pathogens, and consequently, many of these proteins possess hydrolytic activities that are potentially applicable to prey digestion in carnivorous plants. The identification of several defense-related proteins suggests that carnivorous plants have exploited the hydrolytic properties of these pathogenesis-related proteins (55). Many pathogenesis-related proteins are resistant to low pHs and to proteolytic degradation (29), making them functional in digestive fluids. During the evolution of carnivory in plants, there has likely been a shift from a pathogen-related response to a prey-related response and a shift from the hydrolysis and destruction of the pathogens to the hydrolysis and digestion of the prey. The defense-related proteins in digestive fluid likely still display antibacterial and antifungal effects, as in e.g. poplar extrafloral nectaries (22), in order to avoid pathogenic attacks during the digestion process.
CONCLUSION
The present characterization of Venus flytrap digestive fluid employed deep sequencing of the transcriptome followed by its assembly and subsequent use as a database during the proteomic analyses. This study demonstrates the use of high-throughput technologies in expanding molecular analyses to organisms for which the genome sequence is unknown. The Venus flytrap secretome reveals a unique diversity of hydrolytic enzymes, and the results shed light on the purpose and mechanisms of digestion. Furthermore, the Dionaea secretome contains a high proportion of pathogenesis-related proteins, suggesting that the capability of carnivorous plants to digest prey evolved from a plant defense system.
Supplementary Material
Acknowledgments
We thank Katharina Markmann (Aarhus, Denmark) and Tania A. Nielsen (Aarhus, Denmark) for help with RNA purification; Tom A. Mortensen (Aarhus, Denmark) and the nursery Lammehave (Denmark) for assistance and helpful suggestions regarding the cultivation of Venus flytrap plants; Fasteris SA (Switzerland) for library preparation, Illumina sequencing, and Illumina transcriptome assembly; Kerstin Zander (Golm) for help in sample preparation for mass spectrometry; and Brigitte Neumann for excellent technical assistance (Würzburg).
Footnotes
* This work was supported by a grant from the Danish Research Council for Strategic Research to J.J.E. and an ERC Advanced Grant to R.H.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- DTT
- dithiothreitol
- LC
- liquid chromatography
- MS
- mass spectrometric.
REFERENCES
- 1. Darwin C. (1875) Insectivorous Plants, Murray, London [Google Scholar]
- 2. Gibson T. C., Waller D. M. (2009) Evolving Darwin's ‘most wonderful’ plant: ecological steps to a snap-trap. New Phytol. 183, 575–587 [DOI] [PubMed] [Google Scholar]
- 3. Forterre Y., Skotheim J. M., Dumais J., Mahadevan L. (2005) How the Venus flytrap snaps. Nature 433, 421–425 [DOI] [PubMed] [Google Scholar]
- 4. Markin V. S., Volkov A. G., Jovanov E. (2008) Active movements in plants: mechanism of trap closure by Dionaea muscipula Ellis. Plant Signal Behav. 3, 778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ueda M., Tokunaga T., Okada M., Nakamura Y., Takada N., Suzuki R., Kondo K. (2010) Trap-closing chemical factors of the Venus flytrap (Dionaea muscipulla Ellis). Chembiochem. 11, 2378–2383 [DOI] [PubMed] [Google Scholar]
- 6. Williams S. E., Bennett A. B. (1982) Leaf closure in the Venus flytrap: an acid growth response. Science 218, 1120–1122 [DOI] [PubMed] [Google Scholar]
- 7. Escalante-Perez M., Krol E., Stange A., Geiger D., Al-Rasheid K. A., Hause B., Neher E., Hedrich R. (2011) A special pair of phytohormones controls excitability, slow closure, and external stomach formation in the Venus flytrap. Proc. Natl. Acad. Sci. U.S.A. 108, 15492–15497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Volkov A. G., Carrell H., Baldwin A., Markin V. S. (2009) Electrical memory in Venus flytrap. Bioelectrochemistry 75, 142–147 [DOI] [PubMed] [Google Scholar]
- 9. Griggs R. F. (1935) Victims of the Venus flytrap. Science 81, 7–8 [Google Scholar]
- 10. Lichtner F. T., Williams S. E. (1977) Prey capture and factors controlling trap narrowing in Dionaea (Droseraceae). Am. J. Bot. 64, 881–886 [Google Scholar]
- 11. Schulze W., Schulze E. D., Schulze I., Oren R. (2001) Quantification of insect nitrogen utilization by the Venus fly trap Dionaea muscipula catching prey with highly variable isotope signatures. J. Exp. Bot. 52, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 12. Adamec L. (1997) Mineral nutrition of carnivorous plants: a review. Bot. Rev. 63, 273–299 [Google Scholar]
- 13. Takahashi K., Matsumoto K., Nishi W., Muramatsu M., Kubota K. (2009) Comparative studies on the acid proteinase activiteis in the digestive fluids of NEPENTHES, CEPHALOTOUS, DIONAEA, and DROSERA. Carnivorous Plant Newsletter 38, 75–82 [Google Scholar]
- 14. Robins R. I., Juniper B. E. (1980) The secretory cycle of Dionaea-muscipula ellis. IV. The enzymology of the secretion. New Phytol. 86, 401–412 [Google Scholar]
- 15. Scala J., Iott K., Schwab D. W., Semersky F. E. (1969) Digestive secretion of Dionaea muscipula (Venus's-flytrap). Plant Physiol. 44, 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takahashi K., Suzuki T., Nishii W., Kubota K., Shibata C., Isobe T., Dohmae N. (2011) A cysteine endopeptidase (“dionain”) is involved in the digestive fluid of Dionaea muscipula (Venus's fly-trap). Biosci. Biotech. Bioch. 75, 346–348 [DOI] [PubMed] [Google Scholar]
- 17. Hogslund N., Radutoiu S., Krusell L., Voroshilova V., Hannah M. A., Goffard N., Sanchez D. H., Lippold F., Ott T., Sato S., Tabata S., Liboriussen P., Lohmann G. V., Schauser L., Weiller G. F., Udvardi M. K., Stougaard J. (2009) Dissection of symbiosis and organ development by integrated transcriptome analysis of lotus japonicus mutant and wild-type plants. PLoS One 4, e6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Engelsberger W. R., Schulze W. X. (2012) Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J. 69, 978–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rasmussen M. I., Refsgaard J. C., Peng L., Houen G., Hojrup P. (2011) CrossWork: software-assisted identification of cross-linked peptides. J. Proteomics 74, 1871–1883 [DOI] [PubMed] [Google Scholar]
- 20. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 21. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 22. Escalante-Perez M., Jaborsky M., Lautner S., Fromm J., Muller T., Dittrich M., Kunert M., Boland W., Hedrich R., Ache P. (2012) Poplar extrafloral nectaries: two types, two strategies of indirect defenses against herbivores. Plant Physiol. 159, 1176–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bury A. F. (1981) Analysis of protein and peptide mixtures—evaluation of three sodium dodecyl sulfate-polyacrylamide gel-electrophoresis buffer systems. J. Chromatogr 213, 491–500 [Google Scholar]
- 24. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 25. Sanggaard K. W., Karring H., Valnickova Z., Thogersen I. B., Enghild J. J. (2005) The TSG-6 and IalphaI interaction promotes a transesterification cleaving the protein-glycosaminoglycan-protein (PGP) cross-link. J. Biol. Chem. 280, 11936–11942 [DOI] [PubMed] [Google Scholar]
- 26. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially Modified Protein Abundance Index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 27. Schwannhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–473 [DOI] [PubMed] [Google Scholar]
- 28. Rawlings N. D., Barrett A. J., Bateman A. (2010) MEROPS: the peptidase database. Nucleic Acids Res. 38, D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Loon L. C., Rep M., Pieterse C. M. (2006) Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162 [DOI] [PubMed] [Google Scholar]
- 30. Hatano N., Hamada T. (2008) Proteome analysis of pitcher fluid of the carnivorous plant Nepenthes alata. J. Proteome Res. 7, 809–816 [DOI] [PubMed] [Google Scholar]
- 31. Chen G. H., Huang L. T., Yap M. N., Lee R. H., Huang Y. J., Cheng M. C., Chen S. C. (2002) Molecular characterization of a senescence-associated gene encoding cysteine proteinase and its gene expression during leaf senescence in sweet potato. Plant Cell Physiol. 43, 984–991 [DOI] [PubMed] [Google Scholar]
- 32. Tranbarger T. J., Misra S. (1996) Structure and expression of a developmentally regulated cDNA encoding a cysteine protease (pseudotzain) from Douglas fir. Gene 172, 221–226 [DOI] [PubMed] [Google Scholar]
- 33. Hoover S. R., Kokes E. L. (1947) Effect of pH upon proteolysis by papain. J. Biol. Chem. 167, 199–207 [PubMed] [Google Scholar]
- 34. Vernet T., Berti P. J., de Montigny C., Musil R., Tessier D. C., Menard R., Magny M. C., Storer A. C., Thomas D. Y. (1995) Processing of the papain precursor. The ionization state of a conserved amino acid motif within the Pro region participates in the regulation of intramolecular processing. J. Biol. Chem. 270, 10838–10846 [DOI] [PubMed] [Google Scholar]
- 35. Collins P. R., Stack C. M., O'Neill S. M., Doyle S., Ryan T., Brennan G. P., Mousley A., Stewart M., Maule A. G., Dalton J. P., Donnelly S. (2004) Cathepsin L1, the major protease involved in liver fluke (Fasciola hepatica) virulence: propetide cleavage sites and autoactivation of the zymogen secreted from gastrodermal cells. J. Biol. Chem. 279, 17038–17046 [DOI] [PubMed] [Google Scholar]
- 36. Athauda S. P. B., Matsumoto K., Rajapakshe S., Kuribayashi M., Kojima M., Kubomura-Yoshida N., Iwamatsu A., Shibata C., Inoue H., Takahashi K. (2004) Enzymic and structural characterization of nepenthesin, a unique member of a novel subfamily of aspartic proteinases. Biochem. J. 381, 295–306; Erratum (2004) Biochem. J.382, 1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takahashi K., Athauda S. B. P., Matsumoto K., Rajapakshe S., Kuribayashi M., Kojima M., Kubomura-Yoshida N., Iwamatsu A., Shibata C., Inoue H. (2005) Nepenthesin, a unique member of a novel subfamily of aspartic proteinases: enzymatic and structural characteristics. Curr. Protein Pept. Sci. 6, 513–525 [DOI] [PubMed] [Google Scholar]
- 38. Sansen S., De Ranter C. J., Gebruers K., Brijs K., Courtin C. M., Delcour J. A., Rabijns A. (2004) Structural basis for inhibition of Aspergillus niger xylanase by triticum aestivum xylanase inhibitor-I. J. Biol. Chem. 279, 36022–36028 [DOI] [PubMed] [Google Scholar]
- 39. Drzymala A., Bielawski W. (2009) Isolation and characterization of carboxypeptidase III from germinating triticale grains. Acta Biochim. Biophys. Sin. 41, 69–78 [DOI] [PubMed] [Google Scholar]
- 40. Tokunaga T., Takada N., Ueda M. (2004) Mechanism of antifeedant activity of plumbagin, a compound concerning the chemical defense in carnivorous plant. Tetrahedron Lett. 45, 7115–7119 [Google Scholar]
- 41. Robins R. J., Juniper B. E. (1980) The secretory cycle of Dionaea-muscipula ellis. II. Storage and synthesis of the secretory proteins. New Phytol. 86, 297–311 [Google Scholar]
- 42. Shendure J., Ji H. (2008) Next-generation DNA sequencing. Nat. Biotechnol. 26, 1134–1145 [DOI] [PubMed] [Google Scholar]
- 43. Wang Z., Gerstein M., Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brautigam A., Shrestha R. P., Whitten D., Wilkerson C. G., Carr K. M., Froehlich J. E., Weber A. P. (2008) Low-coverage massively parallel pyrosequencing of cDNAs enables proteomics in non-model species: comparison of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. J. Biotechnol. 136, 44–53 [DOI] [PubMed] [Google Scholar]
- 45. Wall C. E., Cozza S., Riquelme C. A., McCombie W. R., Heimiller J. K., Marr T. G., Leinwand L. A. (2011) Whole transcriptome analysis of the fasting and fed Burmese python heart: insights into extreme physiological cardiac adaptation. Physiol. Genomics 43, 69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bräutigam A., Mullick T., Schliesky S., Weber A. P. (2011) Critical assessment of assembly strategies for non-model species mRNA-Seq data and application of next-generation sequencing to the comparison of C(3) and C(4) species. J. Exp. Bot. 62, 3093–3102 [DOI] [PubMed] [Google Scholar]
- 47. Choi K. H., Laursen R. A. (2000) Amino-acid sequence and glycan structures of cysteine proteases with proline specificity from ginger rhizome Zingiber officinale. Eur. J. Biochem. 267, 1516–1526 [DOI] [PubMed] [Google Scholar]
- 48. Kageyama T. (2002) Pepsinogens, progastricsins, and prochymosins: structure, function, evolution, and development. Cell Mol. Life Sci. 59, 288–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Williamson A. L., Brindley P. J., Knox D. P., Hotez P. J., Loukas A. (2003) Digestive proteases of blood-feeding nematodes. Trends Parasitol. 19, 417–423 [DOI] [PubMed] [Google Scholar]
- 50. Delcroix M., Sajid M., Caffrey C. R., Lim K. C., Dvorak J., Hsieh I., Bahgat M., Dissous C., McKerrow J. H. (2006) A multienzyme network functions in intestinal protein digestion by a platyhelminth parasite. J. Biol. Chem. 281, 39316–39329 [DOI] [PubMed] [Google Scholar]
- 51. Knox D. (2011) Proteases in blood-feeding nematodes and their potential as vaccine candidates. Adv. Exp. Med. Biol. 712, 155–176 [DOI] [PubMed] [Google Scholar]
- 52. Barrett A. J. (1992) Cellular proteolysis. An overview. Ann. N.Y. Acad. Sci. 674, 1–15 [DOI] [PubMed] [Google Scholar]
- 53. Galek H., Osswald W. F., Elstner E. F. (1990) Oxidative protein modification as predigestive mechanism of the carnivorous plant Dionaea muscipula: an hypothesis based on in vitro experiments. Free Radic. Biol. Med. 9, 427–434 [DOI] [PubMed] [Google Scholar]
- 54. Chia T. F., Aung H. H., Osipov A. N., Goh N. K., Chia L. S. (2004) Carnivorous pitcher plant uses free radicals in the digestion of prey. Redox Rep. 9, 255–261 [DOI] [PubMed] [Google Scholar]
- 55. Mithofer A. (2011) Carnivorous pitcher plants: insights in an old topic. Phytochemistry 72, 1678–1682 [DOI] [PubMed] [Google Scholar]
- 56. Schulz M. H., Zerbino D. R., Vingron M., Birney E. (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics DOI: 10.1093/bioinformatics/bts094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dyrlund T. F., Poulsen E. T., Scavenius C., Sanggaard K. W., Enghild J. J. (2012) MS Data Miner: a web-based software tool to analyze, compare and share mass spectrometry protein identifications. Proteomics, in press, DOI: 10.1002/pmic.201200109 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.