Abstract
We describe a method that combines an optimized titanium dioxide protocol and hydrophilic interaction liquid chromatography to simultaneously enrich, identify and quantify phosphopeptides and formerly N-linked sialylated glycopeptides to monitor changes associated with cell signaling during mouse brain development. We initially applied the method to enriched membrane fractions from HeLa cells, which allowed the identification of 4468 unique phosphopeptides and 1809 formerly N-linked sialylated glycopeptides. We subsequently combined the method with isobaric tagging for relative quantification to compare changes in phosphopeptide and formerly N-linked sialylated glycopeptide abundance in the developing mouse brain. A total of 7682 unique phosphopeptide sequences and 3246 unique formerly sialylated glycopeptides were identified. Moreover 669 phosphopeptides and 300 formerly N-sialylated glycopeptides differentially regulated during mouse brain development were detected. This strategy allowed us to reveal extensive changes in post-translational modifications from postnatal mice from day 0 until maturity at day 80. The results of this study confirm the role of sialylation in organ development and provide the first extensive global view of dynamic changes between N-linked sialylation and phosphorylation.
The development of novel methods to simultaneously monitor multiple protein post-translational modifications (PTMs)1 is an attractive tool for researchers. There is increasing evidence that both phosphorylation and glycosylation play important roles in cellular signaling networks during development and transformation of cells. Development of the mammalian brain is initiated during the embryonic stage and continues until adulthood. The brain originates through the proliferation of the telencephalon, the anterior part of the neural tube. Following differentiation, cells begin to migrate and associate into different brain structures. The brain structures are reorganized with the extension of axons and dendrites to communicate via synaptic terminal interactions (1, 2). These molecular interactions are governed by cell surface receptors that are often post-translationally modified with both N-linked glycans and phosphate groups, and studies have suggested that extracellular glycans play vital roles in the regulation of signal transduction pathways (3). For example, the myelin-associated glycoprotein (MAG) binds to cell surface glyco-conjugates GD1a, GT1b and Nogo receptors to form signaling complexes that inhibit axon outgrowth, whereas inhibition of Rho kinase reverses this process in a number of nerve cell types (4). There is growing evidence that both the differentiation and migration of neurons and the guidance of axons are regulated by sialic acid-containing glycoconjugates (5–7). Dietary supplementation of sialic acid leads to increases in sialic acid-containing glycoproteins in the frontal cortex and is associated with faster learning and memory in piglets (8). The nervous system contains an abundant array of sialylated molecules and it is therefore not surprising that changes in the sialiome (the content of sialylated glycoproteins (9)) of a neuron can regulate activity. Removal of sialic acids from membrane proteins by NEU3 in primary neurons leads to actin depolymerization and axonal growth through TrkA-mediated signaling (10). Moreover, the modulation of phosphorylation events because of changes in cell membrane sialylation has been described in cancer (11, 12). Tumors induced in sialyltransferase-deficient animals show altered expression of genes associated with focal adhesion signaling and display decreased phosphorylation of focal adhesion kinase, a target of β1-integrins (13). Sialylated glycoconjugates include N-linked glycans (attached to asparagine residues), O-linked glycans (attached to hydroxylated residues) and glycolipids. N-linked and O-linked glycans are predominantly processed through the endoplasmic reticulum and Golgi, and their protein targets are generally membrane associated, cell-surface or found in extracellular environments. Additional glycoconjugates include single sugar modifications such as O-linked N-acetylglucosamine, glycosaminoglycans, large lipopolysaccharides, and peptidoglycans.
The ability to identify and quantify PTM in proteins using mass spectrometry (MS) relies on specific enrichment techniques to purify modified peptides-of-interest from among a complex mixture. As modified peptides are normally present in sub-stoichiometric levels compared with nonmodified peptides, they are generally not detected by MS without such specific enrichment. Many methods are available to enrich for single PTM, including phosphorylation and glycosylation. Titanium dioxide (TiO2) chromatography was originally described for enrichment of phophopeptides from peptide mixtures using similar peptide loading conditions as used for immobilized metal affinity chromatography (14–16). However, using this procedure resulted in significant co-enrichment of nonphosphorylated peptides. Later we demonstrated that TiO2 was able to selectively purify phosphorylated peptides and sialic acid-containing N-glycopeptides (9, 17) if peptide samples are loaded onto the TiO2 resin in a buffer containing high organic solvent, very low pH and a multifunctional acid, such as 2,5- dihydroxybenzoic acid or glycolic acid.
A recent study demonstrated the first simultaneous enrichment of N-glycopeptides and phosphopeptides from a complex peptide mixture (18). Peptides from mouse brain were separated using electrostatic repulsion hydrophilic interaction chromatography to identify 738 unique glycosylation sites representing 446 glycoproteins, and 915 unique phosphorylation sites from 382 phosphoproteins. This method however, required 3 mg of starting material and did not demonstrate the ability to selectively enrich sialylated glycopeptides from glycopeptides displaying neutral glycans. Furthermore, only a comparatively low number of phosphopeptides could be identified considering the generous protein load investigated. The method was also unable to separate deglycosylated peptides from phosphopeptides and no quantitative capabilities were shown.
Here we report a novel multidimensional strategy that employs TiO2 chromatography to enrich for sialylated glycopeptides and phosphopeptides followed by PNGase F treatment of the eluent and μHPLC hydrophilic interaction liquid chromatography (HILIC) to fractionate and separate formerly N-linked sialylated glycopeptides and phosphopeptides from complex membrane protein preparations of a variety of biological samples. The development of a quantitative N-linked sialiomics and phosphoproteomic strategy that is able to simultaneously monitor cell-(extracellular)-cell interactions and receptor signaling will be a valuable tool to study tissue development and cell stimulation.
EXPERIMENTAL PROCEDURES
HeLa Cell Membrane Preparation
HeLa (human cervix epithelial adenocarcinoma) cells were grown in Dulbecco's modified Eagle's medium (GIBCO BRL Invitrogen, Carlsbad, CA) until 80% confluence (107 cells as starting material). The cells were washed once in ice-cold PBS buffer containing protease inhibitors (Roche). Membrane protein fractionation was performed essentially as previously described (19, 20). Briefly, the cells were lysed in 100 mm Na2CO3, pH 11 containing protease inhibitors and phosphatase inhibitors (Roche) by sonication using a tip-probe sonicator. The lysate was incubated at 4 °C with gentle rotation for 1.5 h followed by ultracentrifugation at 100,000 × g for 1.5 h. After ultracentrifugation, the pellet was recovered and washed with 20 mm triethylammonium bicarbonate (TEAB). The membrane pellet was then re-dissolved in 30 μl of 6 m urea, 2 m thiourea and endoproteinase Lys-C (Wako Chemicals, Japan) was added to the solution at 1:100 enzyme/protein and incubated for 3 h at room temperature. After incubation, the solution was diluted to 1 m urea with 20 mm TEAB and a total of 5 μg of modified trypsin (Promega) added. The solution was incubated overnight at room temperature. After digestion, formic acid (final concentration 1%) was added to the peptide solution in order to precipitate lipids that would interfere with the downstream purification (17) and the solution was centrifuged for 10 min at 14,000 × g. The supernatant was stored at −80 °C for further analysis.
Mouse Brain Membrane Preparation
Postnatal 0, 8, 2,1 and 80 days old mice (C57BL/6, males) were euthanized by decapitation and total brain matter removed and immediately snap-frozen in liquid nitrogen. Experiments were performed according to the ethics guidelines of the Society of Laboratory Animal Science. Frozen tissue (3 brains from each time point) was homogenized in lysis buffer containing 100 mm Na2CO3, protease inhibitor mixture (Roche), phosphatase inhibitor mixture (Roche), pH 11, by dounce homogenization and tip-probe sonication at 4 °C. The lysate was shaken at 4 °C for 1 h and centrifuged at 150,000 × g for 1.5 h at 4 °C. After centrifugation, the supernatant was removed and the membrane pellet washed with 50 mm TEAB.
Reduction, Alkylation, Protein Digestion, and iTRAQ Labeling
The membrane pellets were resuspended in 6 m urea, 2 m thiourea, 50 mm TEAB, pH 8.0 and an aliquot removed for protein concentration determination by amino acid analysis and Qubit fluorescent measurement (Invitrogen). Proteins were reduced with 10 mm dithiolthreitol for 1 h at 30 °C followed by alkylation with 50 mm iodoacetamide for 1 h at 30 °C in the dark. Samples were diluted 1:10 with 50 mm TEAB, pH 8.0 and digested with trypsin at an enzyme to substrate ratio of 1:50 for 12 h at room temperature. The samples were acidified to a final concentration of 2% formic acid and 0.1% trifluoroacetic acid (TFA) and centrifuged at 20000 × g for 30 mins to precipitate the lipids (17). The supernatant from the HeLa cell membrane protein preparation was saved for TiO2 enrichment. The peptides derived from the mouse brain membrane proteins were desalted prior to isobaric tagging for relative quantification (iTRAQ) labeling using a Hydrophilic-Lipophilic-Balance solid phase extraction (HLB-SPE) (Waters, Bedford, MA) cartridge according to the manufacturer's instructions. The peptide mixtures were eluted from the HLB-SPE column with 70% acetonitrile (ACN), 0.1% TFA and dried by vacuum centrifugation. Peptides were resuspended in 200 mm TEAB, pH 8.0. A total of 100 μg for each time point was labeled with 4-plex iTRAQTM (Applied Biosystems, Foster City, CA) as described by the manufacturer (0 days, iTRAQ-114; 8 days, iTRAQ-115; 21 days, iTRAQ-116; 80 days, iTRAQ-117). After labeling, the samples were mixed 1:1:1:1 and dried by vacuum centrifugation to ∼100 μl. Analysis was performed in biological triplicate using three pooled mouse brains for each replicate equating to a total of 9 mouse brains for each time point.
Enrichment of Sialic Acid Containing Glycopeptides and Phosphopeptides by TiO2 Chromatography
Samples were made up to 1 ml loading buffer (1 m glycolic acid, 80% ACN, 5% TFA) by adding 100% TFA and ACN. The samples were incubated with TiO2 beads (GL Sciences, Japan, 5 μm; using a total of 0.6 mg TiO2 beads per 100 μg of peptides) and shaken at room temperature for 15 min in batch format. The suspension was centrifuged at 1000 × g for 1 min and the supernatant loaded onto a second batch of TiO2 (containing half the amount of TiO2 as initially used) and shaken at room temperature for 15 mins. The two batches of TiO2 were washed with 100 μl of loading buffer and centrifuged at 1000 × g for 1 min. The supernatant was removed and the beads washed with 100 μl washing buffer 1 (80% ACN, 1% TFA) and centrifuged at 1000 × g for 1 min. The supernatant was removed and the beads were washed with 100 μl washing buffer 2 (20% ACN, 0.2% TFA) and centrifuged at 1000 × g for 1 min. The supernatant was removed and the beads were dried in a vacuum centrifuge for 5 mins. The bound peptides were eluted with 100 μl of 1% ammonium hydroxide by vortexing for 15 mins and then centrifuged at 1000 × g for 1 min. The eluted peptides were dried by vacuum centrifugation to produce the enriched phosphopeptide/sialylated glycopeptide fraction. For the mouse brain development study, unbound peptides and subsequent washes were combined and dried by vacuum centrifugation to produce the nonmodified peptide fraction. The nonmodified peptide fraction was resuspended in 0.1% TFA and purified by HLB-SPE and the eluted peptides dried by vacuum centrifugation.
PNGase F Deglycosylation and Desalting
The enriched phosphopeptide/sialylated glycopeptide fraction was resuspended in 50 μl of 50 mm TEAB, pH 8.0 and deglycosylated with 500 U of PNGase F (New England Biolabs, Ipswich, MA) and 0.1 U Sialidase A (Prozyme, Hayward, CA) for 12 h at 37 °C. After incubation, the peptide mixture was diluted 1:1 with 1% TFA and purified on an Oligo R3 reversed phase micro-column as described previously (17) and the eluted peptides were dried by vacuum centrifugation. An aliquot of the enriched phospho- and N-linked sialylated glyco-peptides without PNGase F treatment was analyzed directly by nano-LC-MS/MS to identify chemically deamidated peptides occurring within the sequon for N-glycosylation (NxS/T/C) that could be interpreted as false positives after PNGase F treatment (21, 22).
TSKgel Amide-80 Hydrophilic Interaction Liquid Chromatography (HILIC) μHPLC
The phosphopeptide and formerly N-linked sialylated glycopeptide mixtures were resuspended in 90% ACN, 0.1% TFA and injected onto an in-house packed TSKgel Amide-80 HILIC (Tosoh, 5 μm) 320 μm × 170 mm μHPLC column using an Agilent 1200 HPLC system (17). The phosphopeptides and formerly N-sialylated glycopeptides were eluted using a gradient from 90% ACN, 0.1% TFA to 60% ACN, 0.1% TFA over 35 mins at a flow rate of 6 μl/min. Fractions were automatically collected at 1 min intervals after UV detection at 210 nm and the fractions were combined to a total of 12–14 fractions according to UV detection. All fractions were dried by vacuum centrifugation.
Reversed Phase Nano-liquid Chromatography Tandem Mass Spectrometry (nano-LC-MS/MS)
Each HILIC fraction was resuspended in 0.3 μl 100% formic acid and diluted to 5 μl in 0.05% TFA. The peptides were separated by nano-LC-MS/MS on an in-house packed 17 cm x 100 μm Reprosil-Pur C18-AQ column (3 μm; Dr. Maisch GmbH, Germany) using an Easy-LC nano-HPLC (Proxeon, Odense, Denmark). The HPLC gradient was 0–34% solvent B (A = 0.1% formic acid; B = 90% ACN, 0.1% formic acid) in 120 mins at a flow of 250 nL/min. Mass spectrometric analysis was performed using an LTQ Orbitrap XL or an LTQ Orbitrap Velos (Thermo Scientific, Bremen, Germany). An MS scan (400–2000 m/z) was recorded in the Orbitrap at a resolution of 30,000 at 400 m/z for a target of 1e6 ions. For analysis of HeLa peptides, data-dependent collision-induced dissociation (CID) MS/MS analysis of the top seven most intense ions was performed on an LTQ Orbitrap XL. Parameters for acquiring CID were as follows; activation time = 15 ms, normalized energy = 35, Q-activation = 0.25, dynamic exclusion = enabled with repeat count 1, exclusion duration = 30 s and, intensity threshold = 30,000, target ions = 2e4. The iTRAQ labeled mouse brain peptides were analyzed on an LTQ Orbitrap XL and an LTQ Orbitrap Velos. For Orbitrap XL analysis, the 2 most intense ions were fragmented by CID (multistage activation (MSA)) in the LTQ followed by high-energy collision induced dissociation (HCD) MS/MS of the corresponding ions with detection in the Orbitrap analyzer, in order to obtain abundant iTRAQ reporter ions. Parameters for acquiring MSA (activation of neutral losses of 49, 32.6, 24.5) were as follows; activation time = 15 ms, normalized energy = 35, Q-activation = 0.25, dynamic exclusion = enabled with repeat count 1, exclusion duration = 30 s and, intensity threshold = 30,000, target ions 2e4. Parameters for acquiring HCD were as follows; activation time = 5 ms, normalized energy = 55, dynamic exclusion = enabled with repeat count 1, exclusion duration = 30 s. For Orbitrap Velos analysis, the top seven most intense ions were fragmented by HCD MS/MS. Parameters for acquiring HCD were as follows; activation time = 0.1 ms, normalized energy = 48, dynamic exclusion enabled with repeat count 1, exclusion duration = 30 s, intensity threshold = 5000, target ions = 2e5.
Data Analysis
Raw files were analyzed using Proteome Discoverer v1.3 beta (Thermo Scientific). MS/MS spectra were converted to .mgf files and searched against the UniProt Human database (June 2011, 51,661 entries) or SwissProt Mouse database (March 2012; 16,513 entries) using Mascot v2.3.02. Database searches were performed with the following fixed parameters: precursor mass tolerance 10 ppm; MS/MS mass tolerance 0.6 Da (CID data) or 0.02 Da (HCD data); 1 possible missed cleavage, N-terminal and lysine iTRAQ, cysteine carbamidomethylation and full trypsin cleavage. Variable modifications included: methionine oxidation; serine, threonine, tyrosine phosphorylation; asparagine and glutamine deamidation. Additional searches were performed with iTRAQ as a variable modification to ensure labeling efficiency was >90% based on the number of peptides observed with and without iTRAQ. Shared peptide sequences were reported as protein grouped accessions. False discovery rates were obtained using Percolator (23) selecting identification with a q-value equal or less than 0.01. Peptides with a Mascot ion score <18 (HCD data) or <26 (LTQ-CID data) were removed. PhosphoRS score was used to determine the phosphosite localization probability (24). iTRAQ quantification was performed using Proteome Discoverer with reporter ion area integration within a 50 ppm window. Ratios were normalized against the median peptide ratio and only peak areas of >1000 were used. When more than one spectra of an identified PTM was present in a single replicate, the ratios were averaged. Comparison of total protein versus PTM quantification values was performed manually for the significantly regulated values. Raw mass spectrometer output data files are available for download at the Proteome Commons Tranche data repository (https://proteomecommons.org/group.jsp?i=395) under the project name “Simultaneous enrichment of phosphorylated and sialylated glycopeptides.”
Bioinformatic Analysis
The results from the database search were introduced into ProteinCenter (Proxeon A/S, Odense, Denmark) in order to retrieve biological and functional information. The quantitative data were analyzed using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, www.ingenuity.com) software to find biological networks regulated during brain development. Each protein identifier with the quantitative information was uploaded and mapped to its corresponding object in Ingenuity's Knowledge Base to compute molecular networks. ProteinCenter was used to map the cellular location of the identified proteins using Gene Ontology and transmembrane domain predictions using TMAP (25). Cluster analysis was performed with the fuzzy c-means algorithm (26) with optimal parameters obtained using (27). Motif analysis was performed using MotifX (28). Statistical analysis included analysis of variance (ANOVA) and limma testing (29).
RESULTS AND DISCUSSION
Simultaneous Identification of Phosphopeptides and Formerly N-linked Sialylated Glycopeptides from HeLa Cells
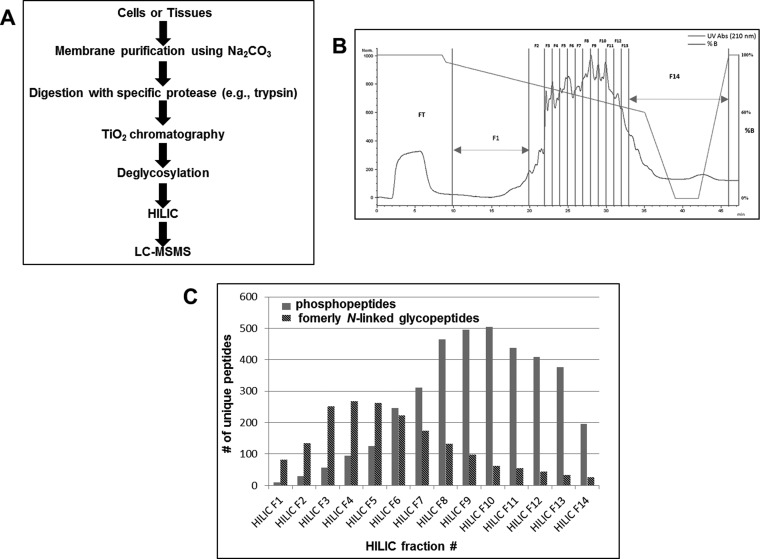
The strategy (Fig. 1A) was developed using HeLa cell membrane fractions that were isolated by Na2CO3 treatment combined with ultracentrifugation (20). After isolation of an enriched membrane fraction, the proteins were cleaved by a specific protease (e.g. trypsin) and peptides (200 μg) subjected to TiO2 chromatography. The enriched phosphopeptides and sialylated glycopeptides were then eluted from the TiO2 and treated with Sialidase A/PNGase F to remove N-linked glycan structures from the sialylated glycopeptides, resulting in the enzymatic conversion of Asn to Asp (mass increase of +0.9840 Da) at the former site of N-linked glycan attachment and found in the N-linked glycosylation consensus motif N-X-S/T/C (X cannot be proline). The phosphopeptides and formerly N-linked sialylated glycopeptides were then separated by HILIC into 14 fractions (Fig. 1B) prior to MS/MS. The deglycosylated peptides were on average more hydrophobic than the phosphopeptides and thus the two species are partially separated from each other using μHILIC separation (Fig. 1C). A total of 4,468 unique phosphopeptide sequences (excluding oxidation, deamidation and the position of the phosphate group(s)) representing 2076 phosphoprotein groups were identified with 3588 phosphorylation sites localized with >90% probability (supplemental Table S1). Furthermore, 1809 unique peptides containing a deamidated asparagine within the motif for N-linked glycosylation (NXS/T/C) (excluding oxidation) i.e. formerly N-sialylated glycopeptides, representing 843 glycoprotein groups were also identified from membrane protein-enriched fractions derived from HeLa cells (supplemental Table S2). The utility of μHPLC HILIC enables the separation of formerly sialylated glycopeptides and phosphopeptides after an optimized TiO2 enrichment. In addition to this, the relatively low amount of starting material (200 μg) makes this approach compatible with multiplexed isotopic labeling strategies such as iTRAQ and is therefore an attractive approach for studies involving limited sample amount. The applicability of modification-based proteomics to the clinical setting often involves limited sample amount. In this sense, choosing the most efficient method is vital. In an attempt to compare different phosphoproteomic enrichment methods, Ficarro and coworkers compared the analytical efficiencies of various methods expressed as the number of unique phosphopeptides identified per microgram of peptide starting material (30). The majority of phosphoproteomic studies have previously identified ∼1–10 identifications (ID)/μg. Hennrich and coworkers used SCX-WAX-RP to identify 11010 phosphopeptides from 500 μg (22 ID/μg) (31), Ficarro and co-workers utilized RP-SCX-RP to identify more than 6000 phosphopeptides from 100 μg (>60 ID/μg) (32) (obtained by analyzing more than 60 individual fractions), and Masuda and coworkers used hydroxyl acid-modified metal oxide chromatography (HAMMOC) to identify 995 phosphopeptides from ∼1 μg (∼995 ID/μg) (32). The current study identified 4468 phosphopeptides from 200 μg (22 ID/μg). It should be noted that MS instrument performance and analysis time is also a major factor influencing the number of identifications. Furthermore, comparison of robustness and reproducibility of the techniques have not been evaluated. In addition, the sample quantity in the present study was determined using amino acid composition analysis, which ensures an accurate (± 5%) quantitation of sample material. Although we employed membrane protein-enriched fractions to perform the HeLa study, it is equally applicable to whole cell/tissue lysates, or indeed any isolated subcellular organelles. Phosphoproteomic studies are often conducted on whole cell fractions or on cell lysates split into soluble and insoluble (membrane-associated) fractions. N-linked glycoproteins are predominantly membrane-associated or extracellular in vivo, and glycoproteomic strategies have been described as the ideal approach to capture the membrane subproteome (33). Subcellular localization analysis of the 2076 phosphoprotein groups revealed that 51% of the identified proteins had a Gene Ontology subcellular location described as membrane-associated, with 50% containing more than one predicted transmembrane domain (supplemental Fig. S1). This predominance of membrane proteins likely explains the relatively lower number of identified phosphopeptides observed in this study compared with others, where whole cell lysates and larger amounts of starting cells are generally employed (34). Furthermore, 81% of the identified formerly sialylated glycoproteins were predicted to be membrane localized according to Gene Ontology, with 70% containing more than one predicted transmembane domain. Fifty-six percent of the identified formerly sialylated glycoproteins (only 13% of identified phosphoproteins) contained a predicted signal sequence confirming previous reports that a glycoproteomic approach is able to very successfully enrich for such proteins in proteomic studies (supplemental Fig. S1).
Fig. 1.
A, Illustration of the strategy for simultaneous isolation and analysis of phosphorylated peptides and formerly N-linked sialylated glycopeptides using TiO2 in combination with micro-HILIC separation and LC-MS/MS. B, HILIC chromatogram showing the UV trace of eluting phosphopeptides and formerly N-linked sialylated glycopeptides. The fractions selected for LC-MS/MS analysis are illustrated by F1-F14. The red line illustrates the elution gradient. C, The number of unique phosphopeptides (blue) and formerly N-linked sialylated glycopeptides (red) identified in each of the fractions from the HILIC separation shown in B.
Identification and Quantification of Phosphopeptides and Formerly N-linked Sialylated Glycopeptides During Mouse Brain Development
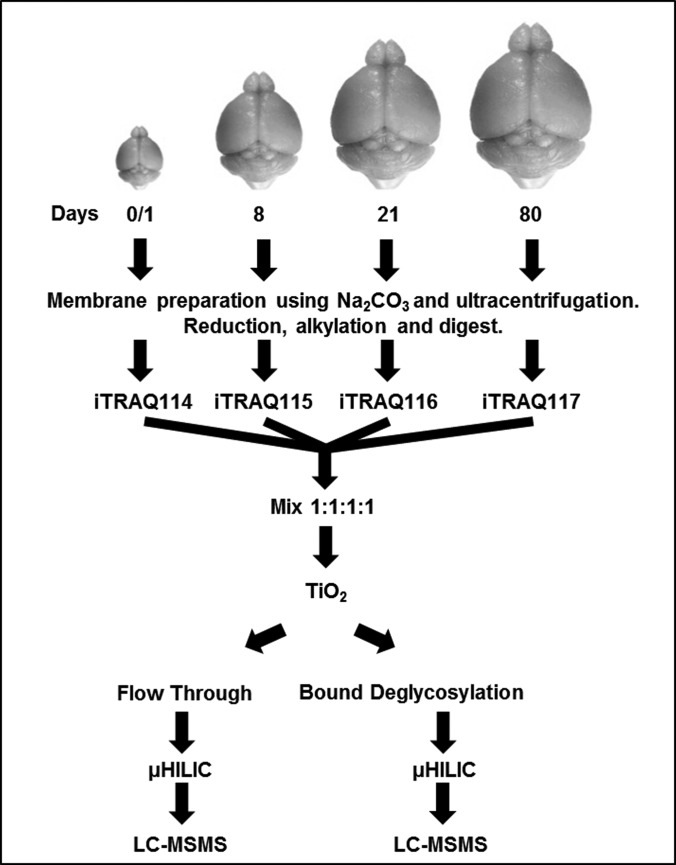
To further illustrate the strength of a simultaneous enrichment of phosphopeptides and N-linked sialylated glycopeptides, the method was applied to tryptic digests of membrane protein-enriched fractions derived from mouse brain tissue (Fig. 2). Phosphopeptides and formerly sialylated glycopeptides were quantitatively compared in peptide mixtures from membrane-associated proteins originating from the developing mouse brain (postnatal day 0/1, 8, 21, and 80) using the iTRAQ quantification strategy. Membrane proteins from each developmental time point (three mouse brains for each time point) were pooled and digested with trypsin followed by four-plex iTRAQTM labeling. A total of 400 μg of labeled peptides (4 × 100 μg from each condition) were subjected to our strategy. To determine whether changes identified in modified peptides were associated with a concomitant change in protein abundance, we also fractionated the peptides that did not bind to TiO2 (“nonmodified flow-through”) using HILIC (12 fractions) and analyzed the fractions by nano-LC-MS/MS. The entire protocol was performed in biological triplicate using three pooled mouse brains for each replicate with a total of nine mouse brains for each time point (three brains × three biological replicates). A total of 7682 unique phosphopeptide sequences (ignoring oxidation, deamidation and the position of the phosphate group(s)) representing 2835 phosphoprotein groups were identified (supplemental Table S3). Localization of the phosphorylation sites identified >11000 unique phosphopeptides, of which 7915 unique phosphorylation sites were identified with a localization probability greater than >90%. Data analysis revealed 17% (1306/7682) of the unique phosphopeptide sequences were identified in all three replicates, 34% (2608/7682) of the unique phosphopeptide sequences were identified in two of three replicates, and 49% (3769/7682) of the unique phosphopeptide sequences were confidently identified in one replicate (supplemental Table S3). Phosphopeptide grouping between the replicates was achieved by assigning phosphosite localization to either <10%, 25%, 33%, 50%, or >90% bins. Only phosphopeptide sequences with identical localizations between the replicates were grouped and used for average and standard deviation calculations. Average biological coefficients of variance (CVs) for phosphopeptides quantified in all three replicates were 23%, 29%, and 40% for day 8/day 0, day 21/day 0, and day 80/day 0, respectively. Evaluation of phosphopeptides regulated during development revealed 669 that were significantly altered in abundance (p < 0.05; ANOVA observed in three biological replicates). A total of 3246 unique formerly sialylated glycopeptides (defined as those containing a deamidated asparagine characteristic of PNGase F treatment and within the motif for N-linked glycosylation (NXS/T/C)); moreover 1152 glycoprotein groups were identified (supplemental Table S4). Of these, 747 unique formerly N-sialylated glycopeptides were identified in all three replicates, 857 unique formerly N-sialylated glycopeptides were identified in two of three replicates, and 1638 unique formerly N-sialylated glycopeptides were identified in one replicate. Average biological CVs of formerly N-sialylated glycopeptides quantified in all three replicates were 21%, 26%, and 35% for day 8/day 0, day 21/day 0, and day 80/day 0, respectively. Evaluation of formerly N-sialylated glycopeptides regulated during development revealed 300 that were significantly altered in abundance (p < 0.05; ANOVA observed in three biological replicates). In total, 502 protein groups contain at least one phosphorylation site and a former N-sialylated glycosylation site.
Fig. 2.
Illustration of the strategy for quantitative iTRAQ analysis of phosphopeptides, formerly N-linked sialylated glycopeptides and nonmodified peptides derived from mouse brain membrane proteins through early postnatal brain development (day 0, 8, 16, and 80).
In an attempt to determine whether the changes we observed in post-translationally modified peptides were as a consequence of a change in overall protein abundance or to the level of the PTM itself, we next analyzed the TiO2 flow-through (nonmodified peptides) by HILIC fractionation and LC-MS/MS. A total of 4641 protein groups were identified with 1742 protein groups quantified in all three replicates (supplemental Table S5). Average biological CVs were 21%, 24%, and 39% for day 8/day 0, day 21/day 0, and day 80/day 0, respectively. Median ratios prior to normalization were 1.02, 0.93, and 0.92 for day 8/day 0, day 21/day 0, and day 80/day 0, respectively.
Nonenzymatic deamidation has previously been observed during the extended incubations with either trypsin or PNGase F digestions (35). We have previously demonstrated that false positive glycosylations may occur in large-scale datasets by mis-annotation of chemically deamidated Asn residues within an N-linked sequon (21, 22). To control for this, we employed the method set out in (21). Analysis of an aliquot of the enriched phospho- and N-linked sialylated glycopeptides was undertaken without PNGase F treatment and interrogation of the TiO2 flow-through resulted in the identification of 202 unique deamidated peptides with 39 containing a deamidation within the motif for N-linked glycosylation (NxS/T/C). This gives rise to an approximate 1.2% glycosite FDR (39/3246) (supplemental Table S6). A further improvement to these negative control experiments could include the combination of an optimized trypsin and PNGase F protocol described in (35) with or without incubation of the tryptic peptides at increased pH and temperature to observe the formation of deamdiated peptides.
Functional annotation of identified glyco- and phosphoproteins revealed that 62% of the glycoproteins and 22% of the phosphoproteins have a predicted signal sequence. Moreover, 78% of the formerly N-linked sialylated glycoproteins and 59% of the phosphoproteins were predicted to contain more than one transmembrane domain (supplemental Fig. S2), indicating a high enrichment of integral membrane and membrane-bound proteins using this approach. This is consistent with the Gene Ontology subcellular localization of the identified glycoproteins and phosphoproteins, with the highest abundance group consisting of membrane-associated proteins (92 and 71% for glycoproteins and phosphoproteins, respectively). It should be noted that the small percentage of N-linked glycoproteins assigned to a cytoplasmic localization are most likely associated with the ER and/or Golgi apparatus, or furthermore could exist in an extracellular form. To assess whether the enrichment strategy is potentially biased for or against a specific class of phosphopeptide, we performed motif analysis using MotifX on phosphopeptides with a PhosphoRS probability of >0.9 (supplemental Fig. S3). Enriched motifs associated with mouse brain development included: (1) proline at the +1 position after the phosphorylation site (p + 1) combined with lysine at p + 3, p + 4 or p + 5; (2) arginine at P-2 or P-3; and (3) aspartic acid and glutamic acid at p + 2. These results indicated that the strategy is able to enrich for both acidic and basic phosphopeptides.
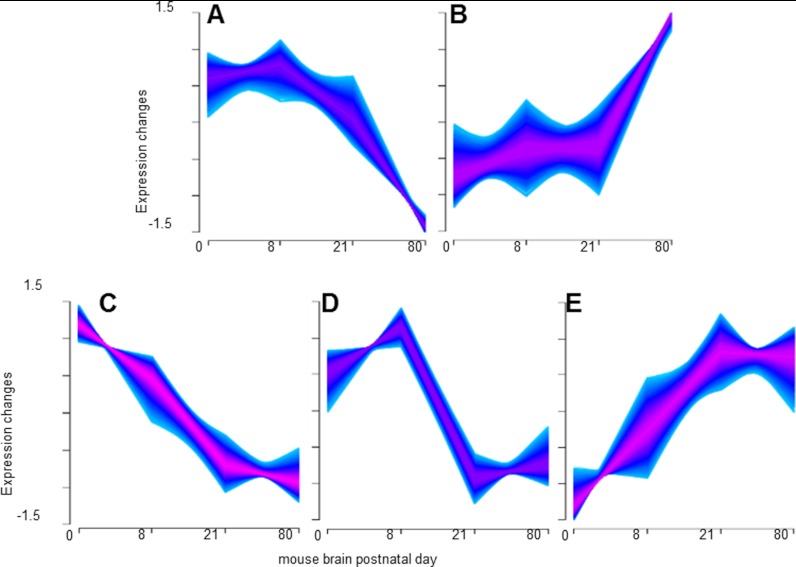
The compatibility of the method with low abundance starting material enabled us to investigate the temporal changes of phosphorylation, sialylation and membrane proteome in the postnatal brain. Figs. 3A–3E shows the results of fuzzy c-means clustering of the total combined quantified phosphopeptides, formerly sialylated glycopeptides and nonmodified peptides. Clusters in Figs. 3B and 3E contain many peptides from proteins involved in long-term potentiation consistent with the increase of synaptic strength between two neurons. Examples included increased abundance of ionotropic glutamate receptors GRIA2 and 3, GRIN2A and 2B, and metabotropic glutamate receptors GRM2, 3, and 7. This was correlated with a general increase in Ca2+ signaling, for example; increases in total CAMK2A and 2B, and total PRKCG during brain development. Clusters in Figs. 3A, 3C, and 3D contain many proteins involved in axon growth and elongation consistent with a general decrease in axonal guidance and cellular movement, including netrin receptor DCC and nerve growth factor receptor.
Fig. 3.
A–E, Fuzzy c-means clustering of the total combined quantified phosphopeptides, formerly sialylated N-linked glycopeptides, and nonmodified peptides for peptides/proteins observed in 1, 2, and 3 replicates.
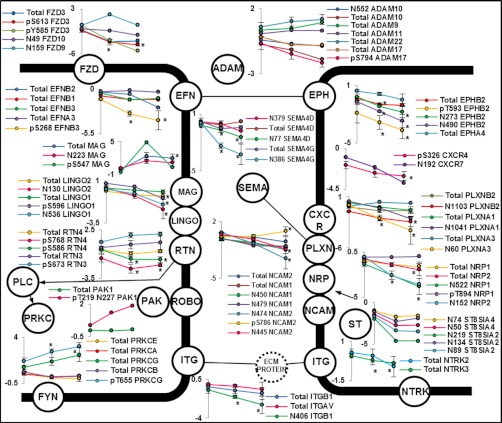
Isoform-specific and site-specific quantification of PTM are critical for a full understanding of the developing brain. Altered quantification of several isoform-specific PTM sites from a number of receptor tyrosine kinases, including the ephrin receptors and neurotrophic tyrosine kinases were observed during development (Fig. 4). Ephrin receptors and their ephrin ligands (Eph-ephrin) exhibit bidirectional signaling during cell-cell interactions (36–38). Very little is known about the developmental regulation of glycosylation on ephrin signaling. We observed several formerly sialylated glycopeptides on ephrin receptors and ephrin ligands that were decreased in abundance during brain development. Quantification of a number of nonmodified peptides derived from these ephrin receptors and ligands showed no significant differences in abundance during development, confirming that the changes we observed were associated with altered glycosylation during development rather than altered genetic expression and protein abundance. The impact of glycosylation on tyrosine kinase activation is currently unknown however; we hypothesize that the glycosylation status is likely to have a significant impact on bidirectional signaling through the alteration of protein-protein interactions. An additional role of glycosylation on receptor signaling is the ability of glycans to alter proteolytic processing (39). We observed alterations in the abundance of formerly N-linked sialylated glycopeptides from a number of A disintegrin and metalloproteases (ADAMs). The glycosylation state of ADAM10 has been correlated with enzyme activity (40) and ADAMs are known to cleave a number of ephrin-A receptors to enable contact repulsion (41). ADAM isoforms have also been implicated in brain development (42); however, the effects of glycosylation or phosphorylation on enzyme function is currently unknown.
Fig. 4.
Regulated PTMs and protein levels associated with axon guidance signaling. Expressed as log2(ratio to day 0/1) * p < 0.05 observed in all three replicates.
Evidence of a role for the large family of semaphorins in neuronal development and induction of signaling is mounting. The family is divided into 8 sub-families and include both attractants and repellents (43). The primary receptors for semaphorins are members of the plexin family made up of nine members (44). The diversity of these protein interactions has been extensively studied however, the complexity continues to be unraveled with observation of semaphorin-plexin heterodimers (45), semaphorin-proteoglycan interactions (46), semaphorin-integrin interactions (47) and semaphorin-plexin-RTK interactions (48). It is now well-established that semaphorin-induced signaling through phosphorylation networks is critical for development (49). We observed a general decrease in the abundance of formerly N-linked sialylated glycopeptides derived from more than five semaphorin isoforms during mouse brain development (Fig. 4). The role of glycosylation on semaphorin/plexin function is currently unknown. The observation of developmental regulation of glycosylation on these proteins indicates that further research to elucidate function is warranted. A systems-wide approach capable of simultaneously monitoring multiple isoforms and pathways will be a valuable tool to unravel the complexity of this dynamic network.
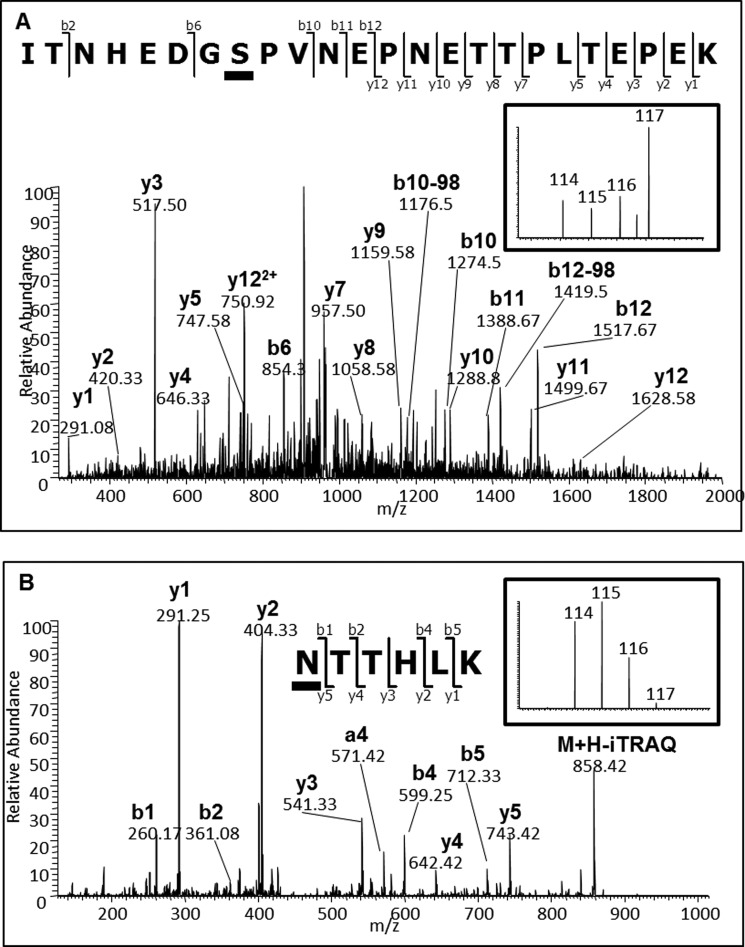
One of the most extensively studied molecules involved in brain development is the neural cell adhesion molecule (NCAM). The glycosylation state of NCAM is developmentally regulated by a polymer of α2,8-linked sialic acid (PolySA), which alters its biophysical properties and binding capabilities (50). Our study detected decreased levels of the formerly N-glycosylated tryptic peptides containing Asn-450 and Asn-479 of NCAM1 during development (Fig. 4). These sites are modified with PolySA and this PTM is lost during mouse brain development (51). These data were correlated with decreased levels of formerly glycosylated peptides containing Asn-50 and Asn-74 from CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase (ST8SIA4), one of the two enzymes responsible for PolySA synthesis associated with modification of NCAM. Interestingly, we also found decreased abundances for a number of formerly N-glycosylated peptides from NCAM2 and phosphopeptides from both NCAM1 and NCAM2. Fig. 5A shows the LTQ-CID-MS/MS spectrum of a novel phosphopeptide containing modification at Ser-786 from NCAM2, which showed reciprocal regulation in the adult brain to the formerly N-linked sialylated glycopeptide containing Asn-445 during development (Fig. 5B). The same doubly phosphorylated peptide containing Ser-786 and Thr-795 was also identified and, a doubly phosphorylated peptide containing Ser-780 and Ser-786 that was also deamidated within a glycosylation motif at Asn-792 was identified. However, all of these peptides were identified in < 2 biological replicates highlighting the complexity of NCAM PTM state. Further experiments are necessary to investigate these PTM intricacies and potential influence on function.
Fig. 5.
Annotated CID MS/MS spectra of NCAM2 derived peptides. A, Phosphopeptide ITNHEDGSPVNEPNETTPLTEPEK [1006. 1420 m/z (3+, 3.02ppm)] with the underlined serine corresponding to phosphorylated Ser-786 and; B, Formerly sialylated N-linked glycopeptide NTTHLK [501.7949 m/z (2+, 0.22ppm)] with the underlined asparagine corresponding to the formerly glycosylated Asn-445. Inserts show iTRAQ reporter regions from the corresponding HCD MS/MS spectra.
Analysis of the TiO2 flow-through fraction by μHILIC and MS/MS allowed for the assessment of changes in PTMs versus total protein (supplemental Table S7). 495 protein groups were quantified as containing significantly regulated phosphopeptide(s) and/or glycopeptide(s) of which total protein quantification was obtained for 256 (52%) in all three replicates. Of these 256 protein groups, 205 (80%) were quantified as not significantly regulated (p value > 0.05; ANOVA). As an example, data generated for Reticulon-4 shows that the quantification of nonmodified peptides does not correlate with the phosphorylation state, indicating that the modification levels are altered rather than the overall protein abundance. Total protein levels at postnatal 80 days were not significantly different from day 0 (p value > 0.05; limma), whereas the phosphorylation state of Ser-690 was significantly down-regulated by more than 2-fold (p value < 0.05; limma). A further example where former-glycopeptide quantification did not correlate with total protein levels was with observed on semaphorin-4G and -4D. Former glycopeptides were significantly down-regulated during development, whereas total protein levels were not significantly regulated. The observed change in abundance of the former glycopeptides could be linked to either the relative amount of glycosylation at that site, or a change in the composition of the attached glycan (reduced levels of terminal sialic acids). A further possibility could be that the change relates to core fucosylation, which has recently been shown to inhibit the action of trypsin on lysines or arginines close to the glycosylation site (52). It should also be noted that O-linked sialylated glycopeptides will also be enriched with this method and could, in theory, be simultaneously analyzed. The analysis of intact O-linked glycopeptides is however, particularly difficult owing to the large heterogeneity present. Furthermore, unambiguous site localization of O-linked glycopeptides would require alternative fragmentation methods than those presented in this study.
Although not specifically the focus of this study, it is noteworthy to mention the ability of this method to potentially quantify extracellular phosphorylation events. Very little is known about the kinases/phosphatases involved in this process however, preliminary evidence suggests that extracellular phosphorylation plays a role in neuronal signal transduction (53, 54) and long-term potentiation in hippocampal neurons (55). The combination of the presented method with specific analysis of extracellular extracts and/or the use of cell impermeable kinase inhibitors (56) may be a valuable approach to investigate this relatively unexplored phenomenon.
CONCLUSION
We have described a method to simultaneously identify and quantify phosphopeptides and formerly N-linked sialylated glycopeptides, and have demonstrated the applicability of this method to both cell and tissue-based biological samples. The results presented in this study were generated from relatively low amounts of starting material meaning that the method can be extended to the analysis of a variety of other biological systems where changes in cell-surface receptor signaling are likely to lead to significant functional consequences. The method combined TiO2 and salt-free fractionation with HILIC, which limits the number of de-salting steps required and therefore minimizes sample loss. The goal of proteomics is to provide a complete systems-wide view of proteins and their modifications. By combining the analysis of two highly significant mammalian PTM in a single experiment, this strategy provides a rapid and more complete view of a biological system. The growing evidence that protein PTM cross-talk could generate a higher level of functional and genetic regulation means that development of novel methods to monitor multiple PTM is required. Where samples are precious and in low amounts, such methods need to generate as much orthogonal information as is possible in a single experiment. Modification-based experiments must also consider the relative impacts of additional PTM, including acetylation, ubiquitinylation, methylation and many others. It is theoretically possible to include the analysis of more PTM into the current approach by performing additional enrichments on the ‘nonmodified’ peptides in the flow-through from TiO2.
We have also provided an extensive overview of the developmental regulation of phosphorylation and glycosylation in the murine brain and have revealed a number of novel findings regarding the complexity of the regulation of glycosylation during tissue development. The results of this study provide a framework for specific functional analysis of PTM during development and could be combined with targeted workflows to monitor specific sites. We utilized whole mouse brains for analysis meaning all morphological information is lost. Combining the current approach with dissected regions of the mouse brain will also add additional information regarding the control of brain morphology by protein PTM.
Supplementary Material
Acknowledgments
We thank colleagues in the Department of Biochemistry and Molecular Biology at the University of Southern Denmark for helpful discussion.
Footnotes
* This study was supported by the Lundbeck Foundation (M.R.L. - Junior Group Leader Fellowship), the Danish Natural Science Research Council (MRL09-06-5989) and the National Health and Medical Research Council (NHMRC) of Australia (S.J.C. NHMRC 571002; M.E.G NHMRC 531700 and 571070). Additional support was from the Center Institute, New South Wales, Australia. G.P. is supported by the Danish Medical Science Research Council (grant No. 11-107551).
 This article contains supplemental Tables S1 to S7 and Figs. S1 to S3.
This article contains supplemental Tables S1 to S7 and Figs. S1 to S3.
1 The abbreviations used are:
- PTM
- post-translational modification
- MAG
- myrlin associated glycoprotein
- TiO2
- titanium dioxide
- HILIC
- hydrophilic interaction liquid chromatography
- TEAB
- triethylammonium bicarbonate
- iTRAQ
- isobaric tracking for relative quantification
- ADAM
- A disintegrin and metalloproteases
- NCAM
- neural cell adhesion molecule.
REFERENCES
- 1. Yamamoto N., Tamada A., Murakami F. (2002) Wiring of the brain by a range of guidance cues. Prog. Neurobiol. 68, 393–407 [DOI] [PubMed] [Google Scholar]
- 2. Siddiqui T. J., Craig A. M. (2011) Synaptic organizing complexes. Curr. Opin. Neurobiol. 21, 132–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parker R. B., Kohler J. J. (2010) Regulation of intracellular signaling by extracellular glycan remodeling. ACS Chem. Biol. 5, 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mehta N. R., Lopez P. H., Vyas A. A., Schnaar R. L. (2007) Gangliosides and Nogo receptors independently mediate myelin-associated glycoprotein inhibition of neurite outgrowth in different nerve cells. J. Biol. Chem. 282, 27875–27886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quirico-Santos T., Fonseca C. O., Lagrota-Candido J. (2010) Brain sweet brain: importance of sugars for the cerebral microenvironment and tumor development. Arq. Neuropsiquiatr. 68, 799–803 [DOI] [PubMed] [Google Scholar]
- 6. Kontou M., Weidemann W., Bork K., Horstkorte R. (2009) Beyond glycosylation: sialic acid precursors act as signaling molecules and are involved in cellular control of differentiation of PC12 cells. Biol. Chem. 390, 575–579 [DOI] [PubMed] [Google Scholar]
- 7. Wang B. (2009) Sialic acid is an essential nutrient for brain development and cognition. Annu. Rev. Nutr. 29, 177–222 [DOI] [PubMed] [Google Scholar]
- 8. Wang B., Yu B., Karim M., Hu H., Sun Y., McGreevy P., Petocz P., Held S., Brand-Miller J. (2007) Dietary sialic acid supplementation improves learning and memory in piglets. Am. J. Clin. Nutr. 85, 561–569 [DOI] [PubMed] [Google Scholar]
- 9. Larsen M. R., Jensen S. S., Jakobsen L. A., Heegaard N. H. (2007) Exploring the sialiome using titanium dioxide chromatography and mass spectrometry. Mol. Cell. Proteomics 6, 1778–1787 [DOI] [PubMed] [Google Scholar]
- 10. Rodriguez J. A., Piddini E., Hasegawa T., Miyagi T., Dotti C. G. (2001) Plasma membrane ganglioside sialidase regulates axonal growth and regeneration in hippocampal neurons in culture. J. Neurosci. 21, 8387–8395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen J. Y., Tang Y. A., Huang S. M., Juan H. F., Wu L. W., Sun Y. C., Wang S. C., Wu K. W., Balraj G., Chang T. T., Li W. S., Cheng H. C., Wang Y. C. (2011) A novel sialyltransferase inhibitor suppresses FAK/paxillin signaling and cancer angiogenesis and metastasis pathways. Cancer Res. 71, 473–483 [DOI] [PubMed] [Google Scholar]
- 12. Liu Y. C., Yen H. Y., Chen C. Y., Chen C. H., Cheng P. F., Juan Y. H., Khoo K. H., Yu C. J., Yang P. C., Hsu T. L., Wong C. H. (2011) Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc. Natl. Acad. Sci. U.S.A. 108, 11332–11337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hedlund M., Ng E., Varki A., Varki N. M. (2008) Alpha 2–6-Linked sialic acids on N-glycans modulate carcinoma differentiation in vivo. Cancer Res. 68, 388–394 [DOI] [PubMed] [Google Scholar]
- 14. Kuroda I., Shintani Y., Motokawa M., Abe S., Furuno M. (2004) Phosphopeptide-selective column-switching RP-HPLC with a titania precolumn. Anal. Sci. 20, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 15. Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 16. Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jorgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 17. Palmisano G., Lendal S. E., Engholm-Keller K., Leth-Larsen R., Parker B. L., Larsen M. R. (2010) Selective enrichment of sialic acid-containing glycopeptides using titanium dioxide chromatography with analysis by HILIC and mass spectrometry. Nat. Protoc. 5, 1974–1982 [DOI] [PubMed] [Google Scholar]
- 18. Zhang H., Guo T., Li X., Datta A., Park J. E., Yang J., Lim S. K., Tam J. P., Sze S. K. (2010) Simultaneous characterization of glyco- and phosphoproteomes of mouse brain membrane proteome with electrostatic repulsion hydrophilic interaction chromatography. Mol. Cell. Proteomics 9, 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nouwens A. S., Cordwell S. J., Larsen M. R., Molloy M. P., Gillings M., Willcox M. D., Walsh B. J. (2000) Complementing genomics with proteomics: the membrane subproteome of Pseudomonas aeruginosa PAO1. Electrophoresis 21, 3797–3809 [DOI] [PubMed] [Google Scholar]
- 20. Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parker B. L., Palmisano G., Edwards A. V., White M. Y., Engholm-Keller K., Lee A., Scott N. E., Kolarich D., Hambly B. D., Packer N. H., Larsen M. R., Cordwell S. J. (2011) Quantitative N-linked glycoproteomics of myocardial ischemia and reperfusion injury reveals early remodeling in the extracellular environment. Mol. Cell. Proteomics 10, M110.006833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Palmisano G., Melo-Braga M. N., Engholm-Keller K., Parker B. L., Larsen M. R. (2012) Chemical deamidation: a common pitfall in large-scale N-linked glycoproteomic mass spectrometry-based analyses. J. Proteome Res. 11, 1949–1957 [DOI] [PubMed] [Google Scholar]
- 23. Käll L., Canterbury J. D., Weston J., Noble W. S., MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 24. Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 25. Persson B., Argos P. (1994) Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J. Mol. Biol. 237, 182–192 [DOI] [PubMed] [Google Scholar]
- 26. Cannon R. L., Dave J. V., Bezdek J. C. (1986) Efficient Implementation of the Fuzzy c-Means Clustering Algorithms. IEEE Trans. Pattern Anal. Mach Intell. 8, 248–255 [DOI] [PubMed] [Google Scholar]
- 27. Schwämmle V., Jensen O. N. (2010) A simple and fast method to determine the parameters for fuzzy c-means cluster analysis. Bioinformatics 26, 2841–2848 [DOI] [PubMed] [Google Scholar]
- 28. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 29. Smyth G. K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 30. Ficarro S. B., Zhang Y., Carrasco-Alfonso M. J., Garg B., Adelmant G., Webber J. T., Luckey C. J., Marto J. A. (2011) Online nanoflow multidimensional fractionation for high efficiency phosphopeptide analysis. Mol. Cell. Proteomics 10, O111.011064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hennrich M. L., Groenewold V., Kops G. J., Heck A. J., Mohammed S. (2011) Improving depth in phosphoproteomics by using a strong cation exchange-weak anion exchange-reversed phase multidimensional separation approach. Anal. Chem. 83, 7137–7143 [DOI] [PubMed] [Google Scholar]
- 32. Masuda T., Sugiyama N., Tomita M., Ishihama Y. (2011) Microscale phosphoproteome analysis of 10,000 cells from human cancer cell lines. Anal. Chem. 83, 7698–7703 [DOI] [PubMed] [Google Scholar]
- 33. Cordwell S. J., Thingholm T. E. (2010) Technologies for plasma membrane proteomics. Proteomics 10, 611–627 [DOI] [PubMed] [Google Scholar]
- 34. Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3. [DOI] [PubMed] [Google Scholar]
- 35. Hao P., Ren Y., Alpert A. J., Sze S. K. (2011) Detection, evaluation and minimization of nonenzymatic deamidation in proteomic sample preparation. Mol. Cell. Proteomics 10, O111.009381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wilkinson D. G. (2001) Multiple roles of EPH receptors and ephrins in neural development. Nat. Rev. Neurosci. 2, 155–164 [DOI] [PubMed] [Google Scholar]
- 37. Chen Y., Fu A. K., Ip N. Y. (2008) Bidirectional signaling of ErbB and Eph receptors at synapses. Neuron Glia Biol. 4, 211–221 [DOI] [PubMed] [Google Scholar]
- 38. Jørgensen C., Sherman A., Chen G. I., Pasculescu A., Poliakov A., Hsiung M., Larsen B., Wilkinson D. G., Linding R., Pawson T. (2009) Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science 326, 1502–1509 [DOI] [PubMed] [Google Scholar]
- 39. Hsiao C. C., Cheng K. F., Chen H. Y., Chou Y. H., Stacey M., Chang G. W., Lin H. H. (2009) Site-specific N-glycosylation regulates the GPS auto-proteolysis of CD97. FEBS Lett. 583, 3285–3290 [DOI] [PubMed] [Google Scholar]
- 40. Escrevente C., Morais V. A., Keller S., Soares C. M., Altevogt P., Costa J. (2008) Functional role of N-glycosylation from ADAM10 in processing, localization and activity of the enzyme. Biochim. Biophys. Acta 1780, 905–913 [DOI] [PubMed] [Google Scholar]
- 41. Janes P. W., Saha N., Barton W. A., Kolev M. V., Wimmer-Kleikamp S. H., Nievergall E., Blobel C. P., Himanen J. P., Lackmann M., Nikolov D. B. (2005) Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 123, 291–304 [DOI] [PubMed] [Google Scholar]
- 42. Markus A., Yan X., Rolfs A., Luo J. (2011) Quantitative and dynamic expression profile of premature and active forms of the regional ADAM proteins during chicken brain development. Cell Mol. Biol. Lett. 16, 431–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tran T. S., Kolodkin A. L., Bharadwaj R. (2007) Semaphorin regulation of cellular morphology. Annu. Rev. Cell Dev. Biol. 23, 263–292 [DOI] [PubMed] [Google Scholar]
- 44. Takahashi T., Fournier A., Nakamura F., Wang L. H., Murakami Y., Kalb R. G., Fujisawa H., Strittmatter S. M. (1999) Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 99, 59–69 [DOI] [PubMed] [Google Scholar]
- 45. Ayoob J. C., Terman J. R., Kolodkin A. L. (2006) Drosophila Plexin B is a Sema-2a receptor required for axon guidance. Development 133, 2125–2135 [DOI] [PubMed] [Google Scholar]
- 46. Kantor D. B., Chivatakarn O., Peer K. L., Oster S. F., Inatani M., Hansen M. J., Flanagan J. G., Yamaguchi Y., Sretavan D. W., Giger R. J., Kolodkin A. L. (2004) Semaphorin 5A is a bifunctional axon guidance cue regulated by heparan and chondroitin sulfate proteoglycans. Neuron 44, 961–975 [DOI] [PubMed] [Google Scholar]
- 47. Pasterkamp R. J., Peschon J. J., Spriggs M. K., Kolodkin A. L. (2003) Semaphorin 7A promotes axon outgrowth through integrins and MAPKs. Nature 424, 398–405 [DOI] [PubMed] [Google Scholar]
- 48. Swiercz J. M., Worzfeld T., Offermanns S. (2008) ErbB-2 and met reciprocally regulate cellular signaling via plexin-B1. J. Biol. Chem. 283, 1893–1901 [DOI] [PubMed] [Google Scholar]
- 49. Franco M., Tamagnone L. (2008) Tyrosine phosphorylation in semaphorin signalling: shifting into overdrive. EMBO Rep. 9, 865–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rutishauser U. (2008) Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 9, 26–35 [DOI] [PubMed] [Google Scholar]
- 51. Edelman G. M., Chuong C. M. (1982) Embryonic to adult conversion of neural cell adhesion molecules in normal and staggerer mice. Proc. Natl. Acad. Sci. U.S.A. 79, 7036–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deshpande N., Jensen P. H., Packer N. H., Kolarich D. (2010) GlycoSpectrumScan: fishing glycopeptides from MS spectra of protease digests of human colostrum sIgA. J. Proteome Res. 9, 1063–1075 [DOI] [PubMed] [Google Scholar]
- 53. Ehrlich Y. H., Davis T. B., Bock E., Kornecki E., Lenox R. H. (1986) Ecto-protein kinase activity on the external surface of neural cells. Nature 320, 67–70 [DOI] [PubMed] [Google Scholar]
- 54. Ehrlich Y. H., Snider R. M., Kornecki E., Garfield M. G., Lenox R. H. (1988) Modulation of neuronal signal transduction systems by extracellular ATP. J. Neurochem. 50, 295–301 [DOI] [PubMed] [Google Scholar]
- 55. Chen W., Wieraszko A., Hogan M. V., Yang H. A., Kornecki E., Ehrlich Y. H. (1996) Surface protein phosphorylation by ecto-protein kinase is required for the maintenance of hippocampal long-term potentiation. Proc. Natl. Acad. Sci. U.S.A. 93, 8688–8693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nagashima K., Nakanishi S., Matsuda Y. (1991) Inhibition of nerve growth factor-induced neurite outgrowth of PC12 cells by a protein kinase inhibitor which does not permeate the cell membrane. FEBS Lett. 293, 119–123 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.