Abstract
Discovery or shotgun proteomics has emerged as the most powerful technique to comprehensively map out a proteome. Reconstruction of protein identities from the raw mass spectrometric data constitutes a cornerstone of any shotgun proteomics workflow. The inherent uncertainty of mass spectrometric data and the complexity of a proteome render protein inference and the statistical validation of protein identifications a non-trivial task, still being a subject of ongoing research. This review aims to survey the different conceptual approaches to the different tasks of inferring and statistically validating protein identifications and to discuss their implications on the scope of proteome exploration.
CONTEXT
Protein Inference in Shotgun Proteomics
The shotgun proteomics approach enables biologists to identify thousands of proteins in mass spectrometric measurements of a single sample. This approach borrows from its namesake, the genome shotgun sequencing approach that reconstructs whole genomes from sequencing random DNA fragments (1). The shotgun proteomics approach operates at the level of protein fragments, i.e. peptides to reconstruct the ensemble of proteins present in a biological sample (2) Both approaches implement a divide-and-conquer strategy commonly encountered in computer science, i.e. to solve a difficult task by breaking it down to many related easy tasks (3). The reconstruction of the difficult task's solution from those of the easy tasks is typically nontrivial. The convenient physico-chemical properties of peptides render the acquisition of informative data about short protein fragments an “easy” task. The destructive nature of the shotgun proteomics approach though shifts the challenge to the computational reconstruction of protein identities from this data.
Shotgun proteomics workflows comprise three main steps. First, proteins are biochemically extracted from a biological sample and then, they are enzymatically digested to yield a complex ensemble of peptides. Protein and/or peptide ensembles are optionally further fractionated according to physical/chemical properties. Second, tandem mass spectrometry is used to sample and identify individual peptide species present in the resulting ensembles and to finally recover the set of proteins initially present in the biological sample. Mass spectrometric analysis of complex protein or respectively peptide mixtures comprises a two step scanning procedure that first registers the m/z ratios of all peptide species of a mixture, then selects, isolates and fragments one of these species and records the resulting fragment ion spectrum (4–6). Third, peptide fragment ion spectra define the data to perform inference, i.e. to infer the proteins initially present in the biological sample. Inference traditionally involves two steps, peptide spectrum matching and protein inference (7). Peptide spectrum matching refers to assigning each fragment ion spectrum a peptide sequence that best explains its signals. Protein inference reconstructs the protein composition from the peptide spectrum matches obtained in the first step. Recent less widely used approaches blur the two step setup, by either reconstructing proteins directly from the mass spectrometrical data without generating peptide spectrum matches or by simultaneously matching peptides to spectra and inferring protein identities (8).
Peptide spectrum matching is a task that admits a fragment ion spectrum as input and that consists of finding the peptide sequence best matching to the input according to a suitable objective function (score) (9). The objective function encodes our understanding of the relation between a peptide and its fragment ion spectrum and is supposed to discriminate the peptide that gave rise to the input spectrum from all other peptides. It is nontrivial to find a good objective function because the fragmentation of peptides is only partially understood (10) and, furthermore, fragment ion spectra generated from complex peptide mixtures are noisy, i.e. the fragment signals are subject to statistical fluctuation (11) and convoluted with signals from moieties other than the enriched target peptide (12). Some work recently adopted objective functions that additionally account for peptide detectability. These extensions are based on expectations to observe a specific peptide in the biological sample considering prior knowledge about protein abundance distributions and peptide ionization properties (13, 14). Most of the peptide spectrum matching approaches independently process each fragment ion spectrum. In a first step, a set of suitable candidate peptides is generated de novo (15–18) or from a sequence database (9, 19). Each candidate is scored against the fragment ion spectrum. The top scoring candidate peptide in conjunction with the fragment ion spectrum is reported as peptide spectrum match. Peptide spectrum matching has been extensively studied and reviewed in the past. For a more comprehensive overview please refer to e.g. (20).
SOLUTIONS
Protein Inference Approaches
Protein inference constitutes the second step after peptide spectrum matching and, in simple terms, typically takes the peptide spectrum matches as input and compiles a set of protein identifications that best represent the identified peptides. The protein inference task is specific to the shotgun proteomics setup (7). Enzymatic digestion of the proteins into peptides facilitates sample handling and dramatically enhance throughput. These benefits come at the cost of loosing the information which proteins gave rise to which of the identified peptides. For complex proteomes or mixtures of proteomes originating from various organisms (i.e. infectious diseases, microbial communities) peptide spectrum matches can map ambiguously to several protein entries, e.g. protein splice variants or highly conserved sequence stretches in orthologous proteins. Protein inference approaches aim to disambiguate these matches and have been implemented in various ways.
Different data input types and analysis procedures have been proposed for protein inference. Many approaches start off from a static list of peptide spectrum matches obtained from a database search (21–26). Probabilistic approaches revisit the peptide spectrum matches and rescore these based on presence or absence of sibling matches pointing to the same protein (27–30). Other approaches perform inference in a single step by jointly fitting a probabilistic model to establish peptide spectrum matches and protein identifications at the same time (8). To benefit from multiple database search engines, a recently proposed method performs protein inference from a list of nonredundant peptides (31). Spectral alignment approaches take a special position and start off from the raw mass spectrometrical data and de novo assemble (partial) protein sequences by aligning fragment ion spectra of overlapping peptides without resorting to sequence databases (32).
The main challenge in protein inference consists of dealing with peptide spectrum matches ambiguously mapping to several protein entries in the protein database. Each approach addresses this issue by defining different notions of a protein identification. A first class of protein inference approaches maps peptide spectrum matches back to a set of ambiguous protein entries that are either defined by a priori grouping protein isoforms or reporting one representative variant for each set of isoforms (21–25). This a priori grouping effectively disambiguates the protein database and therefore allows for unambiguously mapping peptide spectrum matches to the respective groups. This approach circumvents possible ambiguities related to isoform discrimination at the cost of not resolving these ambiguities even in case of sufficiently informative data. A second class of protein inference approaches defines protein groups a posteriori, i.e. groups that take into account the acquired spectral data. Specifically, each peptide identification is associated to its supported group of protein entries. The goal of these approaches is to summarize this list into a parsimonious, i.e. minimal list of protein groups that explains all peptide identifications (7). Probabilistic approaches assign each peptide identification to a protein entry (or group of indistinguishable proteins) with highest posterior probability (27, 33, 34). On the basis of predicted peptide detectabilities (35), Alves et al. have augmented this approach by scoring protein identifications with respect to expected though unobserved peptides (34, 36). Other approaches formulate the parsimony constraint as a set cover problem (37, 38), or as bipartite graph analysis (39). These approaches represent each protein as a set of peptides that they can give rise in a shotgun proteomics experiment and then seek to find a minimal list of proteins whose peptide sets comprise (cover) all peptides supported by the spectral data. A recent approach furthermore defines protein groups with richer hierarchical structure to better guide the user in disambiguating degenerate protein identifications (37). Given sufficiently discriminative data, this class of approaches is able to resolve apparent ambiguities related to proteins with shared peptide identifications. In addition to the application of one of the above protein inference approaches, it is common practice to exclude possibly unreliable protein identifications, such as e.g. single hit protein identifications. There has been considerable debate about whether such post-processing enhances protein inference (40, 41). Latter approaches might miss protein identifications that are falsely discarded by the a priori grouping scheme or the parsimony constraint. Instead of disambiguating ambiguous peptide identifications, Farrah et al. report all proteins consistent with the spectral data (42). To be able to make statements about the occurrence of proteins in the biological sample, the authors of this study introduced the CEDAR scheme for protein identifications. This scheme defines a hierarchy of five protein identification types that are characterized by the ambiguity of their supporting peptide identifications. This approach allows the user to exploit a shotgun proteomic dataset while explicitly accounting for all protein identification ambiguities.
For the experimentalist it is difficult to choose an appropriate protein inference approach for his/her applications, given the many available protein inference variants. Although the criteria for this decision generally depend on the specific application scenario, a typical goal is to maximize the number of true protein identifications while keeping the number of spurious protein identifications low. Many of the developments discussed above aim at and provide empirical support for improving on this goal. However, general conclusions on protein inference performance are difficult due to the plethora of application scenarios. Ideally, the choice of a protein inference approach is guided by an application specific benchmark of a set of competing approaches with respect to their ability to achieve the designated goal (43). The following sections will address this issue by reviewing methods to count spurious protein identifications, factors influencing this count, and concluding remarks on how to report protein identifications in the light of these findings.
VALIDATION
False Discovery Rates for Protein Identifications
Protein identifications are not perfect. This observation is mainly related to the occurrence of spurious peptide spectrum matches. False positive peptide spectrum matches arise when the top-scoring candidate is not the source of the respective fragment ion spectrum. These events can mostly be attributed to flaws in the score related to the approximate encoding for the peptide fragmentation process and the lack of information in the fragment ion spectrum, e.g. in terms of lacking fragment ions.
It is important to control the quality of peptide spectrum matches for both the compilation of identified peptides and their inferred proteins. Various statistical approaches have been devised to control different measures of peptide spectrum match uncertainty, the false discovery rate being the most useful one because it accounts for multiple testing (44, 45). In the context of peptide spectrum matching, the false discovery rate corresponds to the expected fraction of false positive matches. Three routes can be pursued to estimate the false discovery rate for a set of peptide spectrum matches. The false discovery rate can be derived from p values associated to each peptide spectrum match that is considered significant (44, 45). E-value calibration methods for score normalization allow us to apply this approach to data sets that have been analyzed with multiple search engines (46). This approach to false discovery rate estimation is valid as long as p values can be accurately computed (47). This requirement is though rarely met (48). The false discovery rate can be estimated from the score distributions of true and false positive peptide spectrum matches (49). This mixture distribution has to be learned in an unsupervised scenario because the information whether a match is true or false positive is not known for any match. This task has been successfully implemented in e.g. PeptideProphet (49) by resorting to Expectation Maximization (50). Recently, the target-decoy strategy became very popular to estimate the peptide spectrum match false discovery rate (51). A decoy database with nonsense protein sequences is searched in addition to the (target) protein database of the studied organism. The number of peptide spectrum matches mapping to the decoy database serves as an estimate of the number of false positive matches. If the decoy database is designed similar to the target database, then we expect the false positive matches to uniformly distribute across the target and decoy database. Elias et al. have shown that reversed, pseudo-reversed as well as scrambled databases serve equally well as decoy databases, particularly ensuring uniform distribution of false positive matches (52). Its simplicity and generic applicability make the target-decoy strategy an appealing alternative to estimate false discovery rate of peptide spectrum matches.
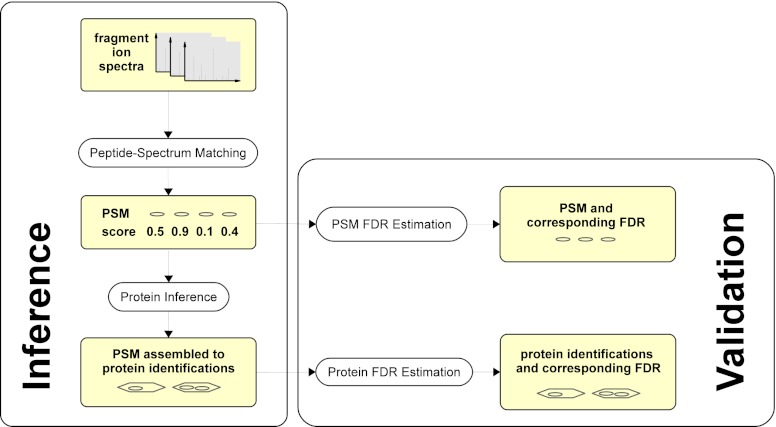
Typically, protein identifications, instead of peptide spectrum matches, are the biologically relevant outcome of a shotgun proteomics study. Therefore it is highly desirable to control the quality of a shotgun proteomics study at the level of protein identifications. Statistical validation of protein identifications has long falsely been equated with statistical validation of peptide spectrum matches (Fig. 1). It turns out, however, that errors at the level of peptide spectrum matches propagate in a nontrivial fashion to the level of protein identifications (53). Therefore, the estimation of false discovery rates for protein identifications requires appropriate approaches differing from those for validation of peptide spectrum matches and is still a topic of ongoing research.
Fig. 1.
Overview of data analysis tasks in shotgun proteomics. The inference tasks consist of assigning peptide sequences to fragment ion spectra (peptide spectrum matching) and assembly of peptide spectrum matches to protein identities (protein inference). The validation tasks consist of estimating confidence measures like false discovery rate (FDR) to the set of peptide spectrum matches and, separately, to the set of protein identifications. Solution of these tasks requires different task specific methods. Particularly, FDR estimation procedures for protein identifications differ from those for peptide spectrum matches.
Several attempts have been made to control protein identification error rates. Many approaches estimate probabilities for a protein identification to be wrong from the respective probabilities of its constituting peptide spectrum matches (27, 28, 30, 54). It turns out, however, that this kind of estimate is sensitive to the accuracy of the probability estimates for the individual peptide spectrum matches. Because these estimates are particularly difficult for peptide spectrum matches giving rise to single hit wonders in large data sets these approaches do not scale well with data set size (53) Another approach estimates the number of incorrect protein identifications assuming that false positive peptide spectrum matches distribute according to a Poisson distribution across the protein database (25, 29). Depending on the choice of different assumptions for single hit protein identifications, this strategy gives either more or less optimistic estimates for protein error rates. Naive target-decoy approaches estimate protein identification false discovery rates as described for peptide spectrum matches, i.e. by estimating the number of false positive protein identifications with the number of decoy identifications (26, 40, 54, 55) It turns out that the number of decoy protein identifications is an estimate for “mixed” protein identifications, i.e. identifications that are both supported by correct as well as incorrect peptide spectrum matches. Because a single correct supporting peptide spectrum match renders a protein identification true, the number of “mixed” protein identifications cannot generally be equated with the number of false positive protein identifications. In fact, the number of false protein identifications is likely to be smaller than the number of “mixed” protein identifications. Consequently, naive target-decoy approaches turn out to achieve too pessimistic error rates (53). The Mayu approach adapts the target-decoy strategy to the protein inference task by means of a hypergeometric model that also accounts for the occurrence of “mixed” protein identifications. The hypergeometric model formalizes and takes advantage of the observation that the statistics of the number of “mixed” protein identifications is analogous to a draw from an urn with two types of balls (e.g. black and white). In this analogy the first type of balls represents the protein entries for which there is correct support and the other type represents all other entries of the underlying protein database. Mayu has shown to achieve accurate, independently validated protein identification false discovery rates for a range of diverse datasets differing in size, underlying proteome and experimental setting (53) and been added as additional feature to PeptideAtlas (42).
Current approaches to statistical validation of protein identifications assume wrong peptide spectrum matches as the single source of erroneous protein identifications. This assumption does not hold true in the context of complex proteomes featuring protein entries with overlapping sequences as for instance protein isoforms or splice variants. Protein inference approaches that assign ambiguous peptide spectrum matches to a single protein might suffer from events where correct peptide spectrum matches are associated to an incorrect protein identity. These events constitute an additional source of errors in the course of protein inference. To the best of our knowledge, there is still no published method to estimate the frequency of these subtle errors, thereby constituting a relevant and interesting target for future research. In the light of emerging targeted proteomics approaches like selected reaction monitoring (56) it is furthermore conceivable that reliable disambiguation of protein identities will be tackled by specifically providing additional informative experimental data.
PROTEIN INFERENCE IN PRACTICE
Data Set and Database Size Matter
The size of the database used for peptide spectrum matching and protein inference influences protein identification false discovery rates. At the level of peptide spectrum matching and for invariant filter criteria, larger protein databases contribute more confounding peptide sequences that lead to a larger amount of false positive peptide spectrum matches. More stringent filter criteria are required counteract this trend and to achieve an acceptable confidence level. More stringent filter criteria though come at the cost of increased false negative rates, i.e. increased number of correct peptide spectrum matches achieving below threshold scores. Besides this effect, the size of protein databases additionally affects protein inference performance by another mechanism. This phenomenon can be seen by considering the behavior of an incorrect peptide spectrum match, randomly mapping to some entry of the protein database. The more entries the database comprises the more likely the incorrect peptide spectrum match will map to a new, so far unsupported protein entry and thereby give rise to a false positive protein identification (Fig. 2). These trends taken together strongly advocate to prefer small protein databases that in particular exclude exceedingly rare protein entries.
Fig. 2.
Error relation between peptide spectrum matches and protein identifications. Impact of dataset and database size on discrepancy between false discovery rate of peptide spectrum matches and protein identifications. Protein database entries are represented as colored circles. true/false peptide spectrum matches (PSM) are depicted as green/red discs. True protein identifications (PID) are supported by at least one correct peptide spectrum match and tagged with a checkmark. The larger the data set or database size, the more pronounced the discrepancy of false discovery rates (FDR) at the level of peptide spectrum matches (PSM) and protein identifications (PID). For large datasets the apparent proteome coverage can deviate significantly from the coverage of true positive (tP) protein identifications.
Successful deep sequencing projects for various model organisms have achieved substantial proteome coverage by resorting to well curated protein databases featuring low redundancy (22–24, 57). These studies cover around 50% of the respective sequence databases, indicating a reasonable tradeoff between constraining the size of the protein database while retaining sufficient diversity for comprehensive discovery. These considerations are more intricate in proteogenomic projects that aim at genome annotation and discovery of novel gene models from shotgun proteomic data (58, 59). The nature of these projects entails the use of large sequence databases that account for all possible protein coding regions of a genome. Proteogenomic studies for various model organisms resorted to six frame translated genomic databases and expressed sequence tag (EST)1 databases to achieve this goal (60–64). The number of peptides in such databases is in the order of billions and further grows by two orders of magnitude if single amino acid mutations are considered, too (58). Several strategies have been pursued to faithfully compress these databases. A simple heuristic consists of only considering open reading frames of at least average exon length. More sophisticated lossless compression approaches involve the use of exon database graphs (65) and Bruijn graph representation of EST databases (66). Two pass database search approaches combine the benefits of achieving low error rates (and computational efficiency) and comprehensive discovery by first confidently identifying data supported genomic regions and secondly mapping the fragment ion spectra to a subdatabase that comprises an enumeration of ab initio predicted gene models for this subset of regions (67, 68). The applicability of EST databases for protein inference is further complicated because a single gene product can map to several sequence tags and the mapping in general is nontrivial (68, 69). The choice of databases and compression strategies can be guided by a benchmark with respect to a useful optimality criterion, e.g. the number of protein identifications or gene model discoveries at a user defined protein false discovery rate (43).
Data-set size has an important influence on protein identification false discovery rates. This influence is related to the different behavior of true and false positive peptide spectrum matches. Typically, only a small fraction of proteins represented in the protein database are actually present, or at least present at a level that is within the dynamic range of the mass spectrometer, in the studied biological sample. Therefore, true peptide spectrum matches start to redundantly map to the same protein entries with growing dataset size. The rate of true new protein discoveries slows down with data-set size. False positive peptide spectrum matches do not feature this redundant behavior (or at least to a significantly lower magnitude) and thereby contribute to a constant rate of false new protein discoveries over a wide range of dataset sizes. These observations lead to the trend of protein false discovery rates growing with data-set size while keeping the false discovery rate for peptide spectrum matches fixed (Fig. 2). For large data sets acquired to map out complete proteomes twenty fold differences between these two types of false discovery have been observed (53). Because of this significant effect it is advisable to control the quality of a larger shotgun proteomics experiment at the level of protein identifications.
In the context of large shotgun proteomics projects aiming at extensive proteome coverage it is desirable to minimize the number of experiments not only to save resources but also to keep dataset size small and thereby enhance protein inference. Experiment design aims at minimizing the data-set size by identifying experiments that are expected to produce the most informative data, i.e. to most effectively explore a proteome. Four routes have been pursued to suggest informative experiments: (1) A priori simulation of shotgun proteomics experiments have been carried out to benchmark various fractionation schemes. Shotgun proteomics experiments have been modeled as consecutive fractionation steps at the protein and peptide level that uniformly distribute species into fractions with random omissions to account for sample losses along the course of the experiment. These simulations suggested that separation at the protein level results in more significant gains in proteome coverage than fractionation at the peptide level (70). (2) Directed mass spectrometry approaches exploit a small number of initial shotgun proteomics experiments to, first, identify informative MS1 precursor signals and, second, to perform targeted experiments that specifically generate fragment ion spectra for the selected precursors (71–73). (3) A posteriori analysis of protein identification statistics has been exploited to design experiments that specifically enrich for underrepresented identification types, as for instance for short and basic proteins in the context of a Drosophila sequencing project (22). (4) Finally, proteome coverage prediction approaches lend themselves to determine which experiments to carry out how many times in a multidimensional shotgun proteomics scenario to optimally improve proteome coverage (74, 75). Application of these methods to design shotgun proteomics studies renders them more efficient and, as delineated above, also more informative and reliable.
GUIDELINES
Reporting Protein Identifications
Shotgun proteomics projects typically aim at comprehensively and precisely reconstructing the protein composition of the studied biological sample. Ideally, the list of reported protein identifications should be exempt from spurious identification and exactly reflect the sample proteins. This goal is probably not achievable. Fixing the protein false discovery rate at a reasonably low level (e.g. 1%) and asking for the maximal number of protein identifications constitutes a reasonable alternative goal.
There has been substantial debate on guidelines for reporting protein identifications. Rigid guidelines like the general exclusion of single hit wonders are recurring suggestions in this context. These rigid guidelines predominantly aim at ensuring high quality of the reported identifications and at avoiding the inflation of identification lists with erroneous entries. However, these suggestions neglect the second part of the delineated aim, i.e. the aim of maximizing the number of identifications at a desired quality. In fact, recent studies show evidence that retaining single hit wonders instead is advantageous since these still comprise many correct identifications (40, 43, 53). Besides these results on the specific rule of single hit wonder exclusion, focusing on error avoidance by means of rigid guidelines is generally prone to missing out on sophisticated protein inference approaches that internally deal with e.g. unreliable single hits and yet recover more protein identifications at the same quality, i.e. protein false discovery rate. These conceptual considerations motivate guidelines that simply require reporting the protein false discovery rate of a protein identification list and thereby leave the choice of protein inference approach to the experimentalist.
CONCLUSION
Protein inference is a task arising in shotgun proteomics that aims at mapping back peptide spectrum matches to entries in the underlying protein database. Because of its conceptual simplicity, breadth and depth, shotgun proteomics is likely to keep on playing a pivotal role in exploratory stages of proteomics projects. Protein inference will therefore keep proteomics researchers busy for a while, either as consumers or developers that tackle some of the still open and intricate validation issues. It will be furthermore be interesting to see how similar tasks will arise in new emerging peptide centric mass spectrometry based proteomics technologies and to what extent we will be able to transfer the lessons learned in the shotgun proteomics scenario.
Acknowledgments
We thank Ruedi Aebersold and Moritze Treeck for helpful comments and discussions on the topics discussed in this review.
Footnotes
1 The abbreviation used is:
- EST
- expressed sequence tag.
REFERENCES
- 1. Fleischmann R. D., Adams M. D., White O., Clayton R. A., Kirkness E. F., Kerlavage A. R., Bult C. J., Tomb J. F., Dougherty B. A., Merrick J. M., McKenney K., Sutton G., FitzHugh W., Fields C., Gocayne J. D., Scott J., Shirley R., Liu L.-l., Glodek A., Kelley J. M., Weidman J. F., Phillips C. A., Spriggs T., Hedblom E., Cotton M. D., Utterback T. R., Hanna M. C., Nguyen D. T., Saudek D. M., Brandon R. C., Fine L. D., Fritchman J. L., Fuhrmann J. L., Geoghagen N. S. M., Gnehm C. L., McDonald L. A., Small K. V., Fraser C. M., Smith H. O., Venter J. C. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512 [DOI] [PubMed] [Google Scholar]
- 2. Hunt D. F., Yates J. R., 3rd, Shabanowitz J., Winston S., Hauer C. R. (1986) Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 83, 6233–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cormen T. H. (2009) Introduction to algorithms. 3rd ed., MIT Press [Google Scholar]
- 4. McLafferty F. W. (1981) Tandem mass spectrometry. Science 214, 280–287 [DOI] [PubMed] [Google Scholar]
- 5. Zubarev R. A., Horn D. M., Fridriksson E. K., Kelleher N. L., Kruger N. A., Lewis M. A., Carpenter B. K., McLafferty F. W. (2000) Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 72, 563–573 [DOI] [PubMed] [Google Scholar]
- 6. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nesvizhskii A. I., Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics 4, 1419–1440 [DOI] [PubMed] [Google Scholar]
- 8. Shen C., Wang Z., Shankar G., Zhang X., Li L. (2008) A hierarchical statistical model to assess the confidence of peptides and proteins inferred from tandem mass spectrometry. Bioinformatics 24, 202–208 [DOI] [PubMed] [Google Scholar]
- 9. Eng J. K., McCormack A. L., Yates J. R., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 10. Wysocki V. H., Tsaprailis G., Smith L. L., Breci L. A. (2000) Mobile and localized protons: a framework for understanding peptide dissociation. J. Mass Spectrom. 35, 1399–1406 [DOI] [PubMed] [Google Scholar]
- 11. Tabb D. L., Smith L. L., Breci L. A., Wysocki V. H., Lin D., Yates J. R., 3rd (2003) Statistical characterization of ion trap tandem mass spectra from doubly charged tryptic peptides. Anal. Chem. 75, 1155–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Michalski A., Cox J., Mann M. (2011) More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 10, 1785–1793 [DOI] [PubMed] [Google Scholar]
- 13. Mallick P., Schirle M., Chen S. S., Flory M. R., Lee H., Martin D., Ranish J., Raught B., Schmitt R., Werner T., Kuster B., Aebersold R. (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25, 125–131 [DOI] [PubMed] [Google Scholar]
- 14. Li Y. F., Arnold R. J., Tang H., Radivojac P. (2010) The importance of peptide detectability for protein identification, quantification, and experiment design in MS/MS proteomics. J. Proteome Res. 9, 6288–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Taylor J. A., Johnson R. S. (2001) Implementation and uses of automated de novo peptide sequencing by tandem mass spectrometry. Anal. Chem. 73, 2594–2604 [DOI] [PubMed] [Google Scholar]
- 16. Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., Lajoie G. (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 2337–2342 [DOI] [PubMed] [Google Scholar]
- 17. Frank A., Pevzner P. (2005) PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem. 77, 964–973 [DOI] [PubMed] [Google Scholar]
- 18. Fischer B., Roth V., Roos F., Grossmann J., Baginsky S., Widmayer P., Gruissem W., Buhmann J. M. (2005) NovoHMM: a hidden Markov model for de novo peptide sequencing. Anal. Chem. 77, 7265–7273 [DOI] [PubMed] [Google Scholar]
- 19. Mann M., Wilm M. (1994) Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 66, 4390–4399 [DOI] [PubMed] [Google Scholar]
- 20. Nesvizhskii A. I. (2010) A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteomics 73, 2092–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martens L., Hermjakob H., Jones P., Adamski M., Taylor C., States D., Gevaert K., Vandekerckhove J., Apweiler R. (2005) PRIDE: the proteomics identifications database. Proteomics 5, 3537–3545 [DOI] [PubMed] [Google Scholar]
- 22. Brunner E., Ahrens C. H., Mohanty S., Baetschmann H., Loevenich S., Potthast F., Deutsch E. W., Panse C., de Lichtenberg U., Rinner O., Lee H., Pedrioli P. G., Malmstrom J., Koehler K., Schrimpf S., Krijgsveld J., Kregenow F., Heck A. J., Hafen E., Schlapbach R., Aebersold R. (2007) A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25, 576–583 [DOI] [PubMed] [Google Scholar]
- 23. Baerenfaller K., Grossmann J., Grobei M. A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320, 938–941 [DOI] [PubMed] [Google Scholar]
- 24. Schrimpf S. P., Weiss M., Reiter L., Ahrens C. H., Jovanovic M., Malmström J., Brunner E., Mohanty S., Lercher M. J., Hunziker P. E., Aebersold R., von Mering C., Hengartner M. O. (2009) Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 7, e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. States D. J., Omenn G. S., Blackwell T. W., Fermin D., Eng J., Speicher D. W., Hanash S. M. (2006) Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat. Biotechnol. 24, 333–338 [DOI] [PubMed] [Google Scholar]
- 26. Zhang B., Chambers M. C., Tabb D. L. (2007) Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 28. Eriksson J., Fenyö D. (2003) Probity: a protein identification algorithm with accurate assignment of the statistical significance of the results. J. Proteome Res. 3, 32–36 [DOI] [PubMed] [Google Scholar]
- 29. Serang O., MacCoss M. J., Noble W. S. (2010) Efficient marginalization to compute protein posterior probabilities from shotgun mass spectrometry data. J. Proteome Res. 9, 5346–5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sadygov R. G., Liu H., Yates J. R. (2004) Statistical models for protein validation using tandem mass spectral data and protein amino acid sequence databases. Anal. Chem. 76, 1664–1671 [DOI] [PubMed] [Google Scholar]
- 31. Shteynberg D., Deutsch E. W., Lam H., Eng J. K., Sun Z., Tasman N., Mendoza L., Moritz R. L., Aebersold R., Nesvizhskii A. I. (2011) iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, 10.1074/mcp.M111.007690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bandeira N., Clauser K. R., Pevzner P. A. (2007) Shotgun protein sequencing: assembly of peptide tandem mass spectra from mixtures of modified proteins. Mol. Cell. Proteomics 6, 1123–1134 [DOI] [PubMed] [Google Scholar]
- 33. Gerster S., Qeli E., Ahrens C. H., Buhlmann P. (2010) Protein and gene model inference based on statistical modeling in k-partite graphs. Proc. Natl. Acad. Sci. U. S. A. 107, 12101–12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Y. F., Arnold R.J., Li Y., Radivojac P., Sheng Q., Tang H. (2009) A Bayesian approach to protein inference problem in shotgun proteomics. J. Computational Biol. 16, 1183–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang H., Arnold R. J., Alves P., Xun Z., Clemmer D. E., Novotny M. V., Reilly J. P., Radivojac P. (2006) A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics 22, e481–488 [DOI] [PubMed] [Google Scholar]
- 36. Alves P., Arnold R. J., Novotny M. V., Radivojac P., Reilly J. P., Tang H. (2007) Advancement in protein inference from shotgun proteomics using peptide detectability. Pacific Symposium on Biocomputing 12, 409–470 [PubMed] [Google Scholar]
- 37. Koskinen V. R., Emery P. A., Creasy D. M., Cottrell J. S. (2011) Hierarchical clustering of shotgun proteomics data. Mol. Cell. Proteomics 10, M110 003822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang X., Dondeti V., Dezube R., Maynard D. M., Geer L. Y., Epstein J., Chen X., Markey S. P., Kowalak J. A. (2004) DBParser: web-based software for shotgun proteomic data analyses. J. Proteome Res. 3, 1002–1008 [DOI] [PubMed] [Google Scholar]
- 39. Ma Z. Q., Dasari S., Chambers M. C., Litton M. D., Sobecki S. M., Zimmerman L. J., Halvey P. J., Schilling B., Drake P. M., Gibson B. W., Tabb D. L. (2009) IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 8, 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gupta N., Pevzner P. A. (2009) False discovery rates of protein identifications: a strike against the two-peptide rule. J. Proteome Res. 8, 4173–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grobei M. A., Qeli E., Brunner E., Rehrauer H., Zhang R., Roschitzki B., Basler K., Ahrens C. H., Grossniklaus U. (2009) Deterministic protein inference for shotgun proteomics data provides new insights into Arabidopsis pollen development and function. Genome Res. 19, 1786–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Farrah T., Deutsch E. W., Omenn G. S., Campbell D. S., Sun Z., Bletz J. A., Mallick P., Katz J. E., Malmström J., Ossola R., Watts J. D., Lin B., Zhang H., Moritz R. L., Aebersold R. (2011) A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.006353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Claassen M., Reiter L., Hengartner M. O., Buhmann J. M., Aebersold R. (2012) Generic comparison of protein inference engines. Mol. Cell. Proteomics 11, 10.1074/mcp.O110.007088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statistical Soc. 57, 289–300 [Google Scholar]
- 45. Storey J. D., Tibshirani R. (2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alves G., Ogurtsov A. Y., Wu W. W., Wang G., Shen R. F., Yu Y. K. (2007) Calibrating E-values for MS2 database search methods. Biol. Direct 2, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gupta N., Bandeira N., Keich U., Pevzner P. A. (2011) Target-decoy approach and false discovery rate: when things may go wrong. J. Am. Soc. Mass Spectrom. 22, 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim S., Gupta N., Pevzner P. A. (2008) Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J. Proteome Res. 7, 3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 50. Dempster A. P., Laird N. M., Rubin D. B. (1977) Maximum likelihood from incomplete data via the EM algorithm. J. Roy. Statistical Soc. 39, 1–38 [Google Scholar]
- 51. Moore R. E., Young M. K., Lee T. D. (2002) Qscore: an algorithm for evaluating SEQUEST database search results. J. Am. Soc. Mass Spectrom. 13, 378–386 [DOI] [PubMed] [Google Scholar]
- 52. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 53. Reiter L., Claassen M., Schrimpf S. P., Jovanovic M., Schmidt A., Buhmann J. M., Hengartner M. O., Aebersold R. (2009) Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell. Proteomics 8, 2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Price T. S., Lucitt M. B., Wu W., Austin D. J., Pizarro A., Yocum A. K., Blair I. A., FitzGerald G. A., Grosser T. (2007) EBP, a program for protein identification using multiple tandem mass spectrometry datasets. Mol. Cell. Proteomics 6, 527–536 [DOI] [PubMed] [Google Scholar]
- 55. Nesvizhskii A. I., Vitek O., Aebersold R. (2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 [DOI] [PubMed] [Google Scholar]
- 56. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 58. Castellana N., Bafna V. (2010) Proteogenomics to discover the full coding content of genomes: a computational perspective. J. Proteomics 73, 2124–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ansong C., Purvine S. O., Adkins J. N., Lipton M. S., Smith R. D. (2008) Proteogenomics: needs and roles to be filled by proteomics in genome annotation. Briefings Functional Genomics Proteomics 7, 50–62 [DOI] [PubMed] [Google Scholar]
- 60. Bitton D. A., Smith D. L., Connolly Y., Scutt P. J., Miller C. J. (2010) An integrated mass-spectrometry pipeline identifies novel protein coding-regions in the human genome. PloS one 5, e8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Desiere F., Deutsch E. W., Nesvizhskii A. I., Mallick P., King N. L., Eng J. K., Aderem A., Boyle R., Brunner E., Donohoe S., Fausto N., Hafen E., Hood L., Katze M. G., Kennedy K. A., Kregenow F., Lee H., Lin B., Martin D., Ranish J. A., Rawlings D. J., Samelson L. E., Shiio Y., Watts J. D., Wollscheid B., Wright M. E., Yan W., Yang L., Yi E. C., Zhang H., Aebersold R. (2005) Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 6, R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fermin D., Allen B. B., Blackwell T. W., Menon R., Adamski M., Xu Y., Ulintz P., Omenn G. S., States D. J. (2006) Novel gene and gene model detection using a whole genome open reading frame analysis in proteomics. Genome Biol. 7, R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Loevenich S. N., Brunner E., King N. L., Deutsch E. W., Stein S. E., FlyBase Consortium, Aebersold R., Hafen E. (2009) The Drosophila melanogaster PeptideAtlas facilitates the use of peptide data for improved fly proteomics and genome annotation. BMC Bioinformatics 10, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Merrihew G. E., Davis C., Ewing B., Williams G., Käll L., Frewen B. E., Noble W. S., Green P., Thomas J. H., MacCoss M. J. (2008) Use of shotgun proteomics for the identification, confirmation, and correction of C. elegans gene annotations. Genome Res. 18, 1660–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tanner S., Shen Z., Ng J., Florea L., Guigó R., Briggs S. P., Bafna V. (2007) Improving gene annotation using peptide mass spectrometry. Genome Res. 17, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Edwards N. J. (2007) Novel peptide identification from tandem mass spectra using ESTs and sequence database compression. Mol. Syst. Biol. 3, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Roos F. F., Jacob R., Grossmann J., Fischer B., Buhmann J. M., Gruissem W., Baginsky S., Widmayer P. (2007) PepSplice: cache-efficient search algorithms for comprehensive identification of tandem mass spectra. Bioinformatics 23, 3016–3023 [DOI] [PubMed] [Google Scholar]
- 68. Kuster B., Mortensen P., Andersen J. S., Mann M. (2001) Mass spectrometry allows direct identification of proteins in large genomes. Proteomics 1, 641–650 [DOI] [PubMed] [Google Scholar]
- 69. Shafer P., Lin D. M., Yona G. (2006) EST2Prot: mapping EST sequences to proteins. BMC Genomics 7, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Eriksson J., Fenyo D. (2007) Improving the success rate of proteome analysis by modeling protein-abundance distributions and experimental designs. Nat. Biotechnol. 25, 651–655 [DOI] [PubMed] [Google Scholar]
- 71. Schmidt A., Claassen M., Aebersold R. (2009) Directed mass spectrometry: towards hypothesis-driven proteomics. Curr. Opinion Chem. Biol. 13, 510–517 [DOI] [PubMed] [Google Scholar]
- 72. Schmidt A., Gehlenborg N., Bodenmiller B., Mueller L. N., Campbell D., Mueller M., Aebersold R., Domon B. (2008) An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol. Cell. Proteomics 7, 2138–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zerck A., Nordhoff E., Resemann A., Mirgorodskaya E., Suckau D., Reinert K., Lehrach H., Gobom J. (2009) An iterative strategy for precursor ion selection for LC-MS/MS based shotgun proteomics. J. Proteome Res. 8, 3239–3251 [DOI] [PubMed] [Google Scholar]
- 74. Claassen M., Aebersold R., Buhmann J. M. (2009) Proteome coverage prediction with infinite Markov models. Bioinformatics 25, i154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Claassen M., Aebersold R., Buhmann J. M. (2011) Proteome coverage prediction for integrated proteomics datasets. J. Computational Biol. 18, 283–293 [DOI] [PubMed] [Google Scholar]