Abstract
The CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated genes) immune system of bacteria and archaea provides acquired resistance against viruses and plasmids, by a strategy analogous to RNA-interference. Key components of the defense system are ribonucleoprotein complexes, the composition of which appears highly variable in different CRISPR/Cas subtypes. Previous studies combined mass spectrometry, electron microscopy, and small angle x-ray scattering to demonstrate that the E. coli Cascade complex (405 kDa) and the P. aeruginosa Csy-complex (350 kDa) are similar in that they share a central spiral-shaped hexameric structure, flanked by associating proteins and one CRISPR RNA. Recently, a cryo-electron microscopy structure of Cascade revealed that the CRISPR RNA molecule resides in a groove of the hexameric backbone. For both complexes we here describe the use of native mass spectrometry in combination with ion mobility mass spectrometry to assign a stable core surrounded by more loosely associated modules. Via computational modeling subcomplex structures were proposed that relate to the experimental IMMS data. Despite the absence of obvious sequence homology between several subunits, detailed analysis of sub-complexes strongly suggests analogy between subunits of the two complexes. Probing the specific association of E. coli Cascade/crRNA to its complementary DNA target reveals a conformational change. All together these findings provide relevant new information about the potential assembly process of the two CRISPR-associated complexes.
Prokaryotes can protect themselves against invading elements by clustered regularly interspaced short palindromic repeats (CRISPR)1 together with various associated cas genes. The functioning of this CRISPR associated immune system shares some functional analogy with RNA interference (1–6). Upon phage encounter, bacteria can integrate small fragments of phage DNA into their genomic CRISPR loci (7–9). These virus-derived sequences, known as spacers, are separated from each other by host-derived palindromic repeat sequences. Transcription of the CRISPR loci results in long precursor CRISPR RNA (pre-crRNA), that is subsequently cleaved into short crRNA (∼60 nt) flanked by portions of the repeat sequence (10–14).
In Escherichia coli (E. coli) a complex of Cas proteins called Cascade cleaves the CRISPR transcript (pre-crRNA) in the repeat sequence to yield short CRISPR RNA (crRNA) (15). Cascade was demonstrated to be composed of five Cas proteins; CasA (Cse1), CasB (Cse2), CasC (Cas7), CasD (Cas5), and CasE (Cas6e) and belongs to the Cse-subtype (type I-E) of CRISPR/Cas (16, 17). Although each of the Cas proteins is essential for proper functioning of the Cascade, Cas6e by itself is sufficient for pre-crRNA cleavage into crRNAs (15). These crRNAs, comprise a single spacer flanked on either side by part of the repeat sequence, and are retained by Cascade. After that the Cascade complex is guided to the invading virus DNA in a sequence specific manner. Aided by the predicted nuclease and helicase Cas3, virus proliferation is prevented (15, 18, 19).
Previously, a low resolution structural model for E. coli K12 Cascade was presented in a study combining biochemical expression and purification methods with mass spectrometry (MS), electron microscopy (EM) and small angle x-ray scattering (18). This was followed shortly by a higher resolution structure determined by cryo-electron microscopy (20). Cascade was shown to contain one copy of Cse1, Cas5, Cas6e and crRNA, a Cse2 dimer and six Cas7 subunits. The structure, revealing an unusual seahorse shape complex, is a first step toward understanding CRISPR mediated interference activity. Phylogenetic analyses have specified several CRISPR-complex families, each including a set of cas genes (4, 16, 17, 21). Using an integrated structural biology approach, including native mass spectrometry and electron microscopy, we also generated a structural model for the Pseudomonas aeruginosa CRISPR-associated immune complex (Csy-complex). As opposed to Cascade that contains five different Cas proteins and crRNA, the Csy complex is assembled from only four Csy proteins (Csy1–3 and Csy4, also called Csy6f) and crRNA (22). At the amino acid sequence level the homology between both CRISPR assemblies is nearly absent (3), however, their stoichiometric composition and structural models are strikingly similar, a showcase for a powerful link between structural analogy and functional homology (22).
Here we investigate the topological arrangement, stability and structural similarities of Cascade and the Csy-complex. Based on the existing models the homology between the protein constituents is not directly evident, apart from the hexameric Cas7 and Csy3. Previous studies have determined that within the Csy-complex Csy4 has endoribonuclease activity and produces crRNA, analogous to Cas6e in Cascade. A high-resolution crystal structure of dimeric Csy4-crRNA indicates their selective sequence and structure specific interaction (12, 23). Another related endoribonuclease from Thermus thermophilus, Cse3, also shows tight interaction with crRNA (13).
We used native tandem and ion mobility mass spectrometry (IMMS) to create subunit connectivity models of both the Cascade and Csy CRISPR immune systems. IMMS of large biological assemblies is a rather new tool in structural biology. It can be used to measure the collision cross-section (Ω) of proteins and protein complexes, providing direct information about their shape and conformation (24–31). Here we compared experimentally measured Ω's of Cascade sub-modules with values calculated using molecular modeling on the basis of the cryo-EM structure of Cascade. Identification and structural characterization of incomplete Cascade and Csy modules is used to depict a minimum stable core complex that likely forms early in the assembly process of CRISPR complexes. In perfect agreement with the existing Cascade structure (20), our mass spectrometric approach reveals a tight interaction between Cas7, Cas5, Cas6e, and crRNA. Our connectivity diagrams provide the first experimental indications of the homology between all Cas and Csy proteins, suggesting the lack of a Cas5 homolog.
To extend our knowledge on the Cascade structure further we also investigated the effect of cognate DNA binding on the stability and structure of Cascade. In vivo the crRNA molecule bound to Cascade guides the complex to double stranded DNA target sequences and base pairs with the complementary DNA strand (18, 19). Our mass spectrometric study reveals that the cognate DNA interaction induces a conformational change, decreasing the compactness of Cascade, while at the same time stabilizing the ribonucleoprotein complex.
EXPERIMENTAL PROCEDURES
Mass Spectrometry of Cascade and Csy Complex
Mass spectrometry measurements were performed in positive ion mode either using an LCT electrospray time-of-flight, a modified quadrupole time-of-flight instrument, or a Synapt HDMS (all Waters, UK). Needles were made from borosilicate glass capillaries (Kwik-Fil, World Precision Instruments, Sarasota, FL) on a P97 puller (Sutter Instruments, Novato, CA), coated with a thin gold layer by using an Edwards Scancoat six pirani 501 sputter coater (Edwards Laboratories, Milpitas, USA). The instruments were adjusted for optimal performance in high mass detection (32).
Instrument settings were as follows; needle voltage around 1200 V, cone voltage around 175 V, source pressure 8.5 mbar. For tandem mass spectrometric analysis xenon was used as the collision gas at a pressure of 1.5 10−2 mbar on the modified Q-ToF1 (32). The collision voltage varied between 10–200V to monitor the sequential loss of several Cse/Cas or Csy proteins from the intact Cascade or Csy machinery respectively, or its subcomplexes. For the ion mobility measurements, on the Synapt HDMS, we used argon in the trap and the transfer at a pressure of 3 × 10−2 mbar. The trap was operated at 5V and the transfer at 12V. The ion mobility cell was filled with nitrogen at a pressure of 0.65 mbar, and we used a ramped wave height of 7–25 V and a wave velocity of 250 m/s. The frequency of the pusher was set at 180 μs. The pressure in the ToF was 1.9 × 10−6 mbar. Calibration of the ion mobility cell, essential to calculate collision cross sections, was performed as described previously (25, 29, 33). The standard deviations given for the masses are calculated from at least three independent measurements.
Sample Preparation for Mass Spectrometry
Cascade (Cse11Cse22Cas76Cas51Cas6e1/crRNA1), Cse22Cas76Cas51Cas6e1/crRNA1, lacking Cse1, Cas76Cas51Cas6e1/crRNA1, lacking Cse1 and Cse2, and the Csy complex were purified as described previously (15, 18, 22). For mass spectrometric analysis of these assemblies the buffer was exchanged to 150 mm ammonium acetate pH 8.0, using centrifugal filter units with a cut-off of 10 kDa (Millipore, England). Buffer exchange was completed after five sequential steps at 4 °C, and concentrated CRISPR associated complex solutions were obtained. All protein complexes were used to perform denaturing, native and in solution dissociation mass spectrometric experiments. Unless mentioned otherwise the sprayed concentrations were 5 μm (intact complex).
Optimal conditions we evaluated to generate subcomplexes for Cascade were found to be 5% iso-propanol added to the spray solution, whereas 15% acetonitrile was used to optimally create subassemblies for the Csy system. To perform exact mass measurements of the individual Cse/Cas/Csy proteins, CRISPR complexes were analyzed under denaturing conditions (50% acetonitrile, 50% deionized water (MilliQ), 0.1% formic acid). When appropriate, the ssDNA-probe was added to the Cascade complexes in a twofold excess. All samples were buffer exchanged as described previously. The sequence of the 5′ biotin-TEG-labeled single stranded target DNA was 5′-CTGTTGGCAAGCCAGGATCTGAACAATACCGT-3′ (Biolegio, Nijmegen, Netherlands). The sequence is complementary to the spacer region of the crRNA, thus only base pairing the core of the crRNA and not the flanking (repeating) regions on crRNA.
Molecular Modeling
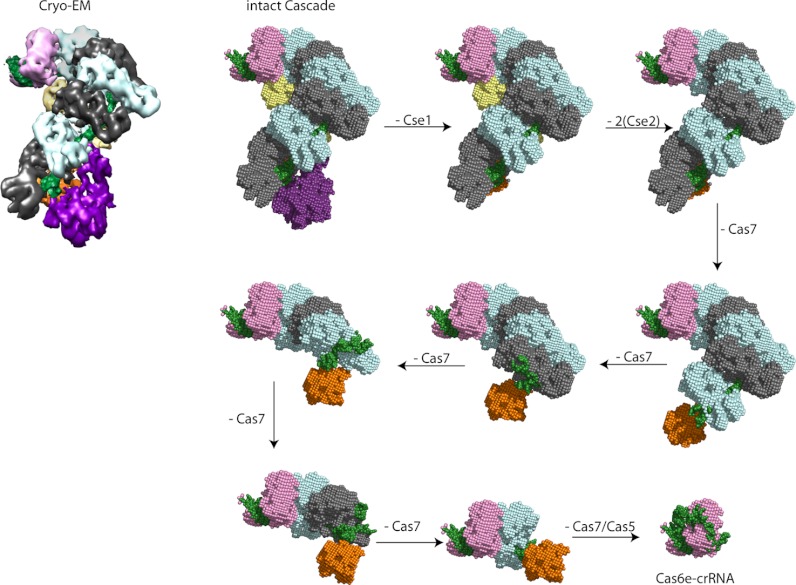
The theoretical Ω's of the ribonucleoprotein (sub)complexes were calculated using molecular modeling. The projection approximation (PA) algorithm implemented in Masslynx (Waters) was used for these calculations (34), as it provides for many systems Ω's in good agreement with experimental data (26, 27, 29, 34, 35). We used the cryo-EM map of the Cascade complex and its individual subunits (20). The segmented volumes for each of the components were extracted from the cryo-EM structure. These segmented volumes were used to generate structural models of all (sub)complexes measured with IMMS. First we used Astas, to fill the cryo-EM map of Cascade and its individual subunits with dummy C-alpha atoms in order to create PDB-like files of the structures. Recommended contour levels were used, varying between 2 and 2.5. The individual subunits were fitted in the cryo-EM map of Cascade, using Chimera software. The structure of the modeled subcomplexes were optimized using Pymol, whereby we allowed the crRNA to tightly interact (adopting a more collapsed conformation) with its neighboring subunits in the small subcomplexes, extending toward its expelled state upon the formation of the larger complexes. To create a homologous model for the Csy (sub)complexes, the crystal structures of the known subunit Csy4 (PDB: 2XLI) and a distant homolog of Csy3, Csa2 from Sulfolobus solfataricus (PDB: 3PS0) were fitted in the cryo-EM map of the Cascade complex (12, 36). The crystal structure of Csa2, is the only available structure of a protein from the Cas7 superfamily, known to form crescent shaped oligomeric structures (36). It is important to realize that the chosen Csa2 structure only provides a model to correlate our IMMS data. At the sequence level these proteins are not alike, however, we expect them to have a similar functional role, as both form an extended structure that support the crRNA over its entire length (36). Csa2 contains an RNA binding domain (RNA recognition motif, RRM), a ferredoxin like fold that serves in RNA recognition. However, Csa2 is not classified as a typical RRM containing protein, of which many structures have been deposited in the PDB. Although only part of the Csa2 structure might originate from the RRM, the rest of the structure differs (36).
Like for Cascade, the modeled Csy modeled subcomplexes were optimized in Pymol (The PyMOL Molecular Graphics System, Version 1.1 Schrödinger, LLC).
RESULTS AND DISCUSSION
Mass Spectrometric Analysis of E. coli Cascade
Native mass spectra of Cascade revealed a large ribonucleoprotein complex with a measured mass of 405 kDa as the most dominant species, for which we deduced a stoichiometry of Cse11Cse2Cas76Cas51Cas6e1/crRNA1 (Fig. 1A) (18). However, under our experimental MS conditions Cse1 easily dissociates from the Cascade complexes as indicated by the presence of the 349 kDa species (Cse22Cas76Cas51Cas6e1/crRNA1). In addition, a distribution corresponding to the Cse2 dimer (42.5 kDa) was observed low in the m/z region (inset Fig. 1A). Table I provides an overview of all measured masses throughout the work presented here.
Fig. 1.
A, Native mass spectrum of intact Cascade, showing a dominant charge state distribution that is consistent with a species of 405,365 ± 135 Da. Two additional subcomplexes were identified having molecular masses of 349,399 ± 84 Da (Cascade lacking Cse1) and 42,524 ± 8 Da (dimeric Cse2, inset). B, Mass spectrum of Cascade sprayed from a solution containing 5% iso-propanol, triggering in solution dissociation of the complex. Several additional charge state distributions that correspond to Cascade subcomplexes were identified. All complexes annotated in (A) and (B) are indicated by colors shown in the inset on the right. C, Native mass spectrum of the Csy system of Pseudomonas aeruginosa, showing a dominant charge state distribution that is consistent with a the intact complex of 350,414 ± 22 Da. Satellite peaks originate from incomplete removal of the purification tag of Csy4, one of the complex components, from the complex resulting into a complex mass of 352,494 ± 16. In addition two subcomplexes were identified having molecular masses of 265,307 ± 9 Da, lacking both Csy1 and Csy2, and a dimeric subcomplex of the endoribonuclease Csy4 and crRNA (40.867 Da), see inset. Adapted from (22).
Table I. Overview of mass spectrometric characterization of Cascade and Cascade modules, including ion mobility collision cross section values.
n.d., not determined.
| Complex | Theoretical mass (kDa) | Experimental mass (kDa) | Ω (nm2) | Ω (nm2) iso-propanol | Modeled Ω (nm2) |
|---|---|---|---|---|---|
| Cse11Cse22Cas76Cas51Cas6e1/crRNA1 | 405,095 | 405,365 ± 135 | 151.3 ± 1.5 | 151.4 ± 1.5 | 157.9 |
| Cse22Cas76Cas51Cas6e1/crRNA1 | 349,051 | 349,399 ± 84 | 134.8 ± 1.9 | 134.8 ± 1.9 | 139.9 |
| Cas76Cas51Cas6e1/crRNA1 | 306,602 | 306,957 ± 88 | 129.4 ± 1.7 | 129.2 ± 1.8 | 137.9 |
| Cas75Cas51Cas6e1/crRNA1 | 266,707 | 267,013 ± 67 | n.d. | 116.6 ± 1.7 | 123.1 |
| Cas74Cas51Cas6e1/crRNA1 | 226,813 | 227,156 ± 128 | n.d. | 99.9 ± 1.8 | 104.8 |
| Cas73Cas51Cas6e1/crRNA1 | 186,918 | 187,254 ± 78 | n.d. | 85.3 ± 1.7 | 90.5 |
| Cas72Cas51Cas6e1/crRNA1 | 147,024 | 147,155 ± 63 | n.d. | 70.8 ± 1.4 | 74.9 |
| Cas71Cas51Cas6e1/crRNA1 | 107,130 | 107,340 ± 58 | n.d. | 52.9 ± 0.9 | 58.7 |
| Cse22 | 42,521 | 42,524 ± 8 | n.d. | n.d. | 26.6 |
| Cas6e1/crRNA1 | 42,026 | 42,071 ± 46 | n.d. | 25.6 ± 0.7 | 27.2 |
| Cse22Cas76Cas51Cas6e1/crRNA1 | |||||
| Cse22Cas76Cas51Cas6e1/crRNA1 | 349,050 | 349,407 ± 43 | 133.0 ± 3.4 | 133.5 ± 2.0 | 139.9 |
| Cas76Cas51Cas6e1/crRNA1 | 306,529 | 306,838 ± 93 | 128.0 ± 2.0 | 127.1 ± 2.4 | 137.9 |
| Cas75Cas51Cas6e1/crRNA1 | 266,635 | 266,870 ± 58 | n.d. | 115.2 ± 2.0 | 123.1 |
| Cas73Cas51Cas6e1/crRNA1 | 186,845 | 186,960 ± 22 | n.d. | 85.1 ± 1.9 | 90.5 |
| Cas72Cas51Cas6e1/crRNA1 | 146,952 | 146,833 ± 173 | n.d. | 68.4 ± 1.7 | 74.9 |
| Cas76Cas51Cas6e1/crRNA1 | |||||
| Cas76Cas51Cas6e1/crRNA1 | 324,085 | 324,724 ± 14 | 135.6 ± 2.2 | 135.4 ± 1.9 | 137.9 |
| Cas75Cas51Cas6e1/crRNA1 | 281,289 | 281,189 ± 17 | n.d. | 119.1 ± 1.6 | 123.1 |
| Cas73Cas51Cas6e1/crRNA1 | 195,695 | 195,896 ± 91 | n.d. | n.d. | 90.5 |
| Cas72Cas51Cas6e1/crRNA1 | 152,898 | 152,999 ± 31 | n.d. | n.d. | 74.9 |
| Cas71Cas51Cas6e1/crRNA1 | 110,102 | 110,021 ± 64 | n.d. | n.d. | 58.7 |
| In the presence of ssDNA | |||||
| Cse11Cse22Cas76Cas51 Cas6e1/crRNA1/ssDNA1 | 415,455 | 415,850 ± 51 | 160.9 ± 0.4 | n.d. | 164.2 |
| Cas76Cas51Cas6e1/crRNA1/ssDNA1 | 316,962 | 317,657 ± 149 | 136.6 ± 1.6 | n.d. | n.d. |
The occurrence of subassemblies provides valuable information about the architecture of Cascade from its individual building blocks. A common trend observed in native MS is that only proteins located at the periphery of a complex, having weak contacts with the other subunits tend to easily dissociate (37–39). Therefore, our data indicate that Cse1 is likely positioned at the periphery of the Cascade complex, in agreement with the known structure (18, 20, 40).
To further probe the topology of Cascade we performed both gas-phase and in-solution dissociation experiments, using collision induced dissociation (tandem MS) and in-solution disruption of the protein-protein interactions, respectively. In native MS, protein complexes are typically sprayed from an aqueous ammonium acetate solution at physiological pH. Addition of a low percentage of organic modifiers to the electrospray solution, generates Cascade intermediates that may be used as indicators of the complex interaction network or topology (38, 41, 42).
In solution dissociation of Cascade, using iso-propanol, results in a series of intermediate modules. Each of the complexes was subjected to tandem mass spectrometry for unambiguous identification of its build-up. The intermediates ranged from a complex of Cas6e with crRNA only, up to Cascade lacking only Cse1 (Fig. 1B and Table I), but intact Cascade was also still detected. In solution, the facile dissociation of Cse1 was followed by the loss of dimeric Cse2 (i.e. no monomeric Cse2) (see supplemental Fig. S1), validating the strong interaction between the Cse2 subunits. A series of six consecutive Cascade subcomplexes were identified exhibiting mass differences of ∼40 kDa, representing the successive loss of Cas7 subunits. In tandem MS these intermediates, ranging in mass from 107 kDa up to 307 kDa, all expelled a Cas7 subunit. In each Cas6e the Cas5 subunit could dissociate from the complex. All subcomplexes contained crRNA and Cas6e strongly suggesting that Cas6e tightly interacts with the crRNA. In line with that, we detected the hetero-dimeric Cas6e-crRNA module (42,071 ± 46 Da, Table I). The Cas6e-crRNA module proved to be very resistant to dissociation, whereby even at high collision energies no disruption of the hetero-dimer was observed. Instead, the Cas6e subunit fragmented by loss of peptide fragments (supplemental Fig. S2). The stability of this small complex explains why it turned out to be a suitable candidate for structure determination by x-ray crystallography. High-resolution co-crystal structures for the Cse3-cRNA from Thermus thermophilus and homologous Csy4-crRNA dimer are already available (12, 13, 43).
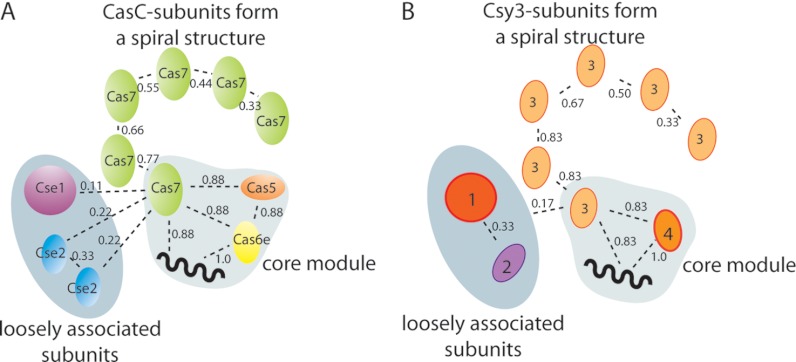
To define a model of subunit connectivity for Cascade we first looked for a minimum stable core to which other Cas proteins may associate. Considering the ten Cascade subcomplexes identified (Fig. 1B and Table I) we see that Cas6e and crRNA were present in nearly all identified intermediates. Next, a hetero-tetrameric assembly of unit stoichiometry, namely Cas71Cas51Cas6e1/crRNA1, is most often present; observed in eight out of the ten complexes. The frequency of all subunits appearing throughout the intermediates was weighted and incorporated into one connectivity diagram (Fig. 2A). Such an analysis indicates that the Cas71Cas51Cas6e1/crRNA1 forms a putative core module, to which the remaining five Cas7 subunits bind in a successive fashion. Likely, hereby the crRNA serves as a string along which the protein subunits bind in a helical arrangement (20). The data furthermore show that both a single copy of Cas7 and Cas5 tightly interact with the heterodimeric complex Cas6e-crRNA. The association of only Cas7 to form trimeric Cas7-Cas6e-crRNA, or the binding of Cas5 to the heterodimer to generate Cas5-Cas6e-crRNA was not observed. Dimeric Cse2 and Cse1 interact weaker with the proposed core module, illustrated by their lower connectivity score. Our connectivity data also validate that the homodimeric interaction between the Cse2 subunits is stronger than their interaction with the core module.
Fig. 2.
Subunit connectivity diagram of Cascade, indicating a stable core composed of Cas71Cas51Cas6e1/crRNA1. For each protein-protein or protein-crRNA interaction a connectivity score is given, which is based on the observed frequency of that specific contact. Two loosely associated modules were identified, Cse1 and dimeric Cse2, both have low scores.
Mass Spectrometric Analysis of P. aeruginosa Csy-complex
The analysis of the intact assembly of P. aeruginosa revealed a 350 kDa complex with a stoichiometry alike the Cas proteins forming Cascade, namely Csy11Csy21Csy36Csy41/crRNA (Fig. 1C) (22). Besides the intact complex, also a complex lacking Csy1 and Csy2 was present under physiological conditions, indicating a peripheral location for these subunits. As for Cascade we subsequently triggered the formation of intermediate modules by in solution dissociation. Table II lists the resulting subassemblies and their identities were confirmed by subsequent tandem mass spectrometric analysis. Two stable hetero-dimeric modules were observed, Csy4-crRNA and Csy1-Csy2 (supplemental Fig. S3). The identification of this Csy1-Csy2 dimer is in line with their facile dissociation from the intact Csy-system.
Table II. Overview of Csy complex modules identified by native mass spectrometry.
n.o., not observed; n.d., not determined; n.a., not available.
| Complex | Theoretical mass (kDa) | Experimental mass (kDa) | Ω (nm2) | Modeled Ω (nm2) |
|---|---|---|---|---|
| Csy11213641/crRNA1 | 350,323 | 350,414 ± 22 | 133.7 ± 0.5 | n.a. |
| Csy3641/crRNA1 | 265,255 | 265,307 ± 9 | 108.9 ± 1.8 | 110.6 |
| Csy3541/crRNA1 | 227,856 | 227,907 ± 13 | 92.4 ± 0.6 | 96.2 |
| Csy3441/crRNA1 | 190,457 | 190,568 ± 21 | n.d. | 84.4 |
| Csy3341/crRNA1 | 153,058 | 153,139a | 71.5 ± 2.4 | 71.3 |
| Csy3241/crRNA1 | 115,660 | 114,038 ± 27 | 57.3 ± 2.4 | 55.9 |
| Csy3141/crRNA1 | 78,260 | n.o. | n.d. | 44.8 |
| Csy41/crRNA1 | 40,860 | 40,867 ± 1 | 25.6 ± 0.7 | 26.6 |
| Csy1121 | 85,068 | 85,616 ± 2 | 51.3 ± 0.3 | n.a. |
| Csy21 | 36,070 | 36,122 ± 10 | 23.9 ± 1.1 | n.a. |
a Only one mass measurements was included.
Overall, the intermediate modules generated for the Csy-complex resemble those observed for Cascade and allow us to speculate about possible protein homologs based on the similarities in their connectivity diagrams (Fig. 2). For both CRISPR associated assemblies a core containing Cas7/Csy3 and Cas6e/Csy4-crRNA can be defined. In addition to these three components the core of Cascade comprises a fourth subunit, Cas5 that is lacking in the Csy-system. Although only four Csy proteins, instead of five Cascade subunits, assemble into an intact functional complex our data suggest that the Csy-complex does not contain a Cas5 counterpart. Besides the similarities in core composition Cascade and the Csy-complex are both surrounded by two loosely associated subunits, Cse1 and dimeric Cse2, and Csy 1 and Csy2 respectively. Our current model does not allow us to assign how these two Cas and Csy proteins correlate but their similar behavior and location in the intact CRISPR associated complexes further strengthens our hypothesis of their homologous function.
Ion Mobility MS and Computational Modeling on Cascade and Csy (sub)Complexes
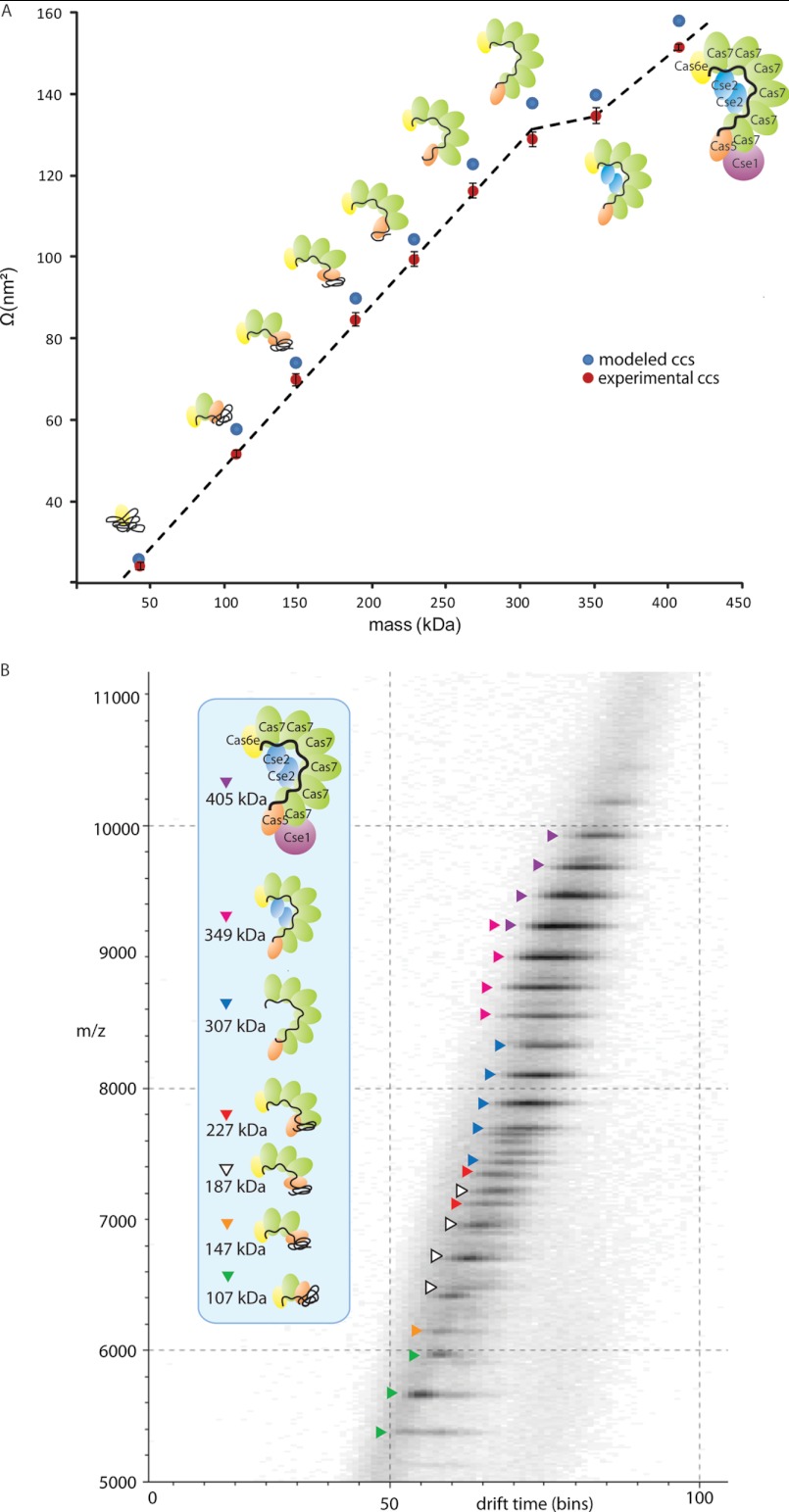
Using IMMS we further investigated the quaternary structure of the core-module Cas71D1E1-crRNA, as well as of all other stable Cascade sub-complexes. IMMS is gaining momentum in structural biology (27), yet only a limited number of studies have applied this technique to macromolecular protein complexes. These reported studies have been quite consistent in their findings that solution phase structures of larger protein assemblies can (at least partly) be retained in the gas phase (25–27, 29–31, 44, 45). In IMMS gas-phase ions are separated in a drift tube filled with inert gas molecules under the influence of a weak electric field. For larger ions the time required to traverse through the ion mobility cell is longer than for smaller ions, simply because the friction that the small ions encounter in the gas-filled chamber is lower. The measured drift time of the ions can be converted into a collision cross section (Ω) or averaged projected area, which is a direct measure for the ions' shape (46). We measured the Ω 's of intact Cascade and of its submodules. To ensure that a low percentage of iso-propanol does not seriously affect the overall structure of the assembly, we included several control experiments. We determined the Ω of Cascade sprayed from electrospray solutions with and without iso-propanol (Table I, Fig. 3). These data did not reveal any differences in the Ω 's determined, suggesting that the overall conformation of Cascade and intermediate modules, is not significantly altered upon the addition of iso-propanol. As a final control, we included two Cascade subcomplexes in the IMMS studies, purified from strains engineered to lack either the Cse1 or both Cse1 and Cse2, termed here Cse2Cas76Cas51Cas6e1/crRNA1 and Cas76Cas51Cas6e1/crRNA1 respectively (18). The two complexes mimic sub-complex formation, without the need for organic solvents. Mass spectrometric analysis of Cse2Cas76Cas51Cas6e1/crRNA1 showed that its mass was 349,406 ± 38 Da, in agreement with the theoretical mass of 349,051 Da (Fig. 4A and Table I), and that its stoichiometry was consistent with intact Cascade. Two other subcomplexes with molecular masses of 186,960 ± 22 Da and 146,833 ± 174 Da were also detected, originating from species lacking dimeric Cse2 and either three or four Cas7 proteins, respectively. The stoichiometry of the Cas76Cas51Cas6e1/crRNA1 complex was also in line with Cascade. However, Cas76Cas51Cas6e1/crRNA1 seems to be less stable than Cascade under physiological conditions (Fig. 4B). All the identified subcomplexes can be related to those observed after Cascade destabilization, and provide evidence that structural rearrangements in the (sub)complexes, because of propanol addition, can be neglected. The exact masses of the individual Cse/Cas proteins that are present in Cse2Cas76Cas51Cas6e1/crRNA1 and Cas76Cas51Cas6e1/crRNA1 are listed in supplemental Table S1. Table I shows that the Ω's determined for protein assemblies with equal stoichiometry, in the presence or absence of iso-propanol, all are in good agreement. These results validate the use of Ω values of all Cascade modules. The Ω 's for Cascade are plotted versus the mass of each (sub)assembly (Fig. 3). As previously mentioned, Cas6e and crRNA represent the components of the Cascade core-module, together with a single subunit of Cas5 and Cas7. It is evident that association of Cas7 subunits to this tetrameric entity follows a near linear increase in Ω (data point 2–7 in Fig. 3A). This indicates that the six Cas7 subunits do not form a closely packed (e.g. ring-like) structure within Cascade, but instead suggests the formation of a more string-like open structure of all Cas7 subunits. In contrast, the binding of the Cse2 homodimer results in only a marginal increase in Ω, suggesting that the Cse2 in good agreement with the cryo-EM structure (20), that reveals that the Cse2 dimer is partly located in a cavity inside the Cascade complex. With the addition of Cse1 Cascade is completed. Cse1 association results in a significant increase in Ω, in correspondence with its peripheral location within the complex. These data seem to agree very well with the available structural model of Cascade (18, 20), and therefore we used the cryo-EM image and molecular modeling calculations to calculate theoretical Ω 's. To enable such a calculation the EM surface map of Cascade was filled with dummy carbon-alpha atoms from which a PDB-like file was generated. To generate the structures of the smaller Cascade subcomplexes we modeled the crRNA to become compact contacting the surface of the remaining subunits. Using these constraints, the Ω's of the modeled structure for Cas6e-crRNA compares well with the experimental value, 27.2 and 25.6 ± 0.6 nm2, respectively. Likewise subcomplexes were modeled to compact structure, including a compact state for crRNA and Cas7 and Cas5 to be in close proximity to Cas6e. For each subcomplex structure (Fig. 5) a PDB-like file was generated and used for Ω's calculations. The modeled Ω's values are also plotted in Fig. 3A and show very favorable agreement with our experimental data (Table I). Similarly we determined the experimental Ω's for the Csy system and its subcomplexes by IMMS (Table II). Although high-resolution structural models for the Csy system are lacking, apart for Csy4 bound to crRNA, we modeled intermediate assembly structures using Csa2, a very distant homolog of Csy3 (supplemental Fig. S3). So far, Csa2 is the only available crystal structure of the Cas7 superfamily. It plays a central role in the CRISPR/cas immunity complex of Sulfolobus solfataricus, and reveals a crescent-shaped structure (36). Our IMMS data support that Csy3, the component with a copy number of six in the intact assembly, forms into an open spiral shaped structure. Eventually this is complemented by the peripheral subunits Csy1 and Csy2.
Fig. 3.
A, Plot of the Ω versus the mass of Cascade sub(complexes). The experimental values are plotted in orange, the theoretical Ω 's calculated from the modeled structures are depicted in purple. A near linear trend line is observed upon the dissociation of each Cas7 subunit from the complex, suggesting the formation of an open string-like scaffold by the six Cas7 proteins. The linear correlation between Ω and mass is not valid when dimeric Cse2 disassembles from the complex, likely because it occupies a cavity of the complex (boxed). The release Cse1 considerably decreases the Ω of Cascade, confirming the peripheral location of this subunit within Cascade. B, Drift time chromatogram, showing the separation of Cascade and the subcomplexes, based on drift time (shown in bins, 100 bins equals 18 ms) and m/z. Subcomplexes are color-coded. The inset identifies the various species.
Fig. 4.
A, Native mass spectrum of Cse22Cas76Cas51Cas6e1/crRNA1. The dominant species present corresponds to a mass of 349,407 ± 43 Da, in good agreement with the expected stoichiometry (theoretical mass 349,051 Da). B, Native mass spectrum of Cas76Cas51Cas6e1/crRNA1, with a mass of 324,546 ± 96 Da. The expression of the complex without the presence of Cse1 and Cse2 destabilizes Cascade, as shown by the increased abundance of several subcomplexes. All subcomplexes were previously observed after the in solution dissociation of complete Cascade by adding 5% iso-propanol.
Fig. 5.
Modeled structures for Cascade subcomplexes based on the cryo-EM density map (20). A dummy-atom filled model was generated from the EM data of the intact complex. Models of the smaller modules were based on the structure of the intact complex, lacking specific subunits, whereby the crRNA (in green) was allowed to rearrange to contact tightly on to the remaining proteins. The other Cas subunits are depicted in the following colors: Cse1, purple; Cse2, yellow; Cas7, gray and blue (for clarity the consecutive Cas7 subunits alternate in color); Cas5, orange; and Cas6e, pink.
Conformation and Stability of Cascade on Cognate DNA Binding
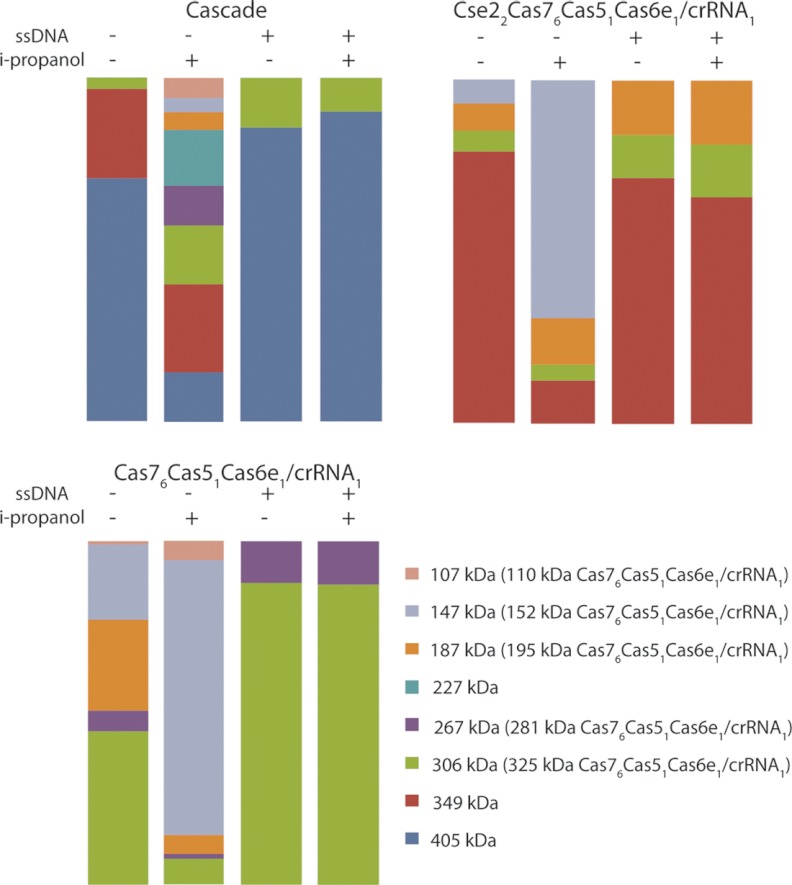
Last, we studied the effect of ssDNA binding to Cascade on the overall shape and stability of the complex. In vivo Cascade and Csy complexes bind specifically to cognate DNA via their crRNA ligand (18, 19, 36). This step is essential to block viral infection or plasmid conjugation. Our MS data revealed that only one ssDNA molecule binds to Cascade, Cse2Cas76Cas51Cas6e1/crRNA1 or Cas76Cas51Cas6e1/crRNA1 evidenced by the increase in the mass of the used ssDNA construct (∼10 kDa, supplemental Fig. S4). Interestingly, the native MS data indicate that all complexes become more stable upon association with ssDNA, as evidenced by the significant reduction of in-solution disassembly (e.g. a decrease in Cse1 dissociation for intact Cascade). This finding was further substantiated by subsequent addition of iso-propanol to these solutions. Even under these circumstances the ssDNA bound Cascade complexes displayed a significant resistance to dissociation, even at elevated concentrations of iso-propanol (Fig. 6).
Fig. 6.
Bar diagrams summarizing the summed intensities of all complexes present for (A) Cascade, (B) Cse22Cas76Cas51Cas6e1/crRNA1, and (C) Cas76Cas51Cas6e1/crRNA1 under varying conditions (with and without i-propanol or ssDNA). The overall conclusion from these experiments is that ssDNA binding significantly increases the stability of Cascade, Cse22Cas76Cas51Cas6e1/crRNA1 or Cas76Cas51Cas6e1/crRNA1. Clearly a reduction in intermediate complexes formed is observed, after triggering in solution dissociation of the complexes by the addition of iso-propanol to the electrospray solution.
By IMMS we observed a small but significant structural change for Cascade upon its association to ssDNA as evidenced by the measured Ω's. Solely based on the increase in mass because of ssDNA association we expected an increase in Ω of ∼3 nm2. Our data revealed a significantly higher increase in Ω of ∼9 nm2, which can only be explained by an altered conformation of the complex (Table I). Our IMMS data are in agreement with EM data indicating that Cascade undergoes conformational changes upon target DNA binding and target RNA binding (18, 20). The theoretically calculated Ω for Cascade containing a double stranded nucleotide helix is 164.2 nm2, versus a Ω of 157.9 nm2 for a modeled Cascade, corresponding to an increase of >6 nm2.
CONCLUSIONS
CRISPR defense systems have the remarkable ability to constantly adapt their composition at the genomic level (4, 47). Although the proteins present within each complex form a constant element, the crRNA component that is part of the assembly through CRISPR adaptation is varied. Targeting of previously encountered invader DNA is achieved by base pairing between the crRNA and the target sequence, thereby specifically interfering with viral proliferation (1, 2).
Despite the lack of sequence homology between various CRISPR systems, the two protein complexes we investigated showed a striking similarity at the structural and topology level (18, 22). Because the activity of protein complexes is often determined by their three-dimensional and quaternary organization, this could explain the general function of CRISPR mediated defense systems.
We studied the topology of E. coli Cascade and the P. aeruginosa Csy-system by a combination of tandem and ion mobility mass spectrometric experiments to identify the stable core complex of the two CRISPR systems. Both cores contain crRNA, an endoribonuclease (Cas6e or Csy4), and one copy of the hexameric protein crescent, either Cas7 or Csy3. In addition to these three components the stable core of Cascade also contains Cas5, however, a fourth member of the core is missing in the Csy-assembly. These data indicate that the Csy-complex does not contain a Cas5 homolog. This may be explained by the fact that the Cascade is composed of five different Cse/Cas sub-units, whereas the Csy complex only contains four. Recently, Makarova et al. (47), showed that Cascade subunits Cas5, Cas6e and Cas7 all contain a RNA recognition motif. Furthermore, in the cryo-EM model structure Cas5 most likely interacts with the 5′ handle of the crRNA (20). In the absence of Cas5, another protein subunit within the Csy complex should take over this role, possibly Csy3. The remaining Cse and Csy proteins, Cse1, Cse2, Csy1, and Csy2 take on a more peripheral location and weakly associate to the CRISPR associated complexes.
A recent computational approach performed by Makarova et al., agrees, albeit only in part, with our mass spectrometry data. Based on a detailed sequence analysis they hypothesized a homologous function for Cse1 and Csy1 and Cas5 and Csy2 (47). The facile dissociation of Csy1 and Cse1 from the Csy-system or Cascade complex respectively, would confirm their homology. However, unlike the suggested finding that Cas5 and Csy2 have some resemblance, we speculate that the Csy-complex lacks this Cas5-like protein component. Cascade is able to interact with DNA in a sequence specific fashion via Cse1 (20), whereas the CRISPR complex from P. aeruginosa has not shown to possess this special binding feature, leading to the conclusion that the Csy-complex may lack a Cse1 homolog. These three different approaches clarify that it is not trivial to assign the homology at the individual protein level of the extremely divergent CRISPR components, and more experimental evidence is required to conclusively classify all proteins. Our mass spectrometry approach provides a complementary method to reveal the similarities in structural composition providing hints about protein functionality in analogous CRISPR complexes.
The structural information we obtained for all intermediate assemblies as well as intact Cascade by ion mobility mass spectrometry are in agreement with the existing structural models. Our data provided further evidence that the Cas7 sub-units form an open string like structure, whereby Cse2 is buried as a tight-dimer in a cavity within this string like structure. Finally, we examined the interaction of Cascade with target ssDNA. By using IMMS we did observe a small structural difference in Cascade in its DNA paired state, consistent with earlier indications provided by electron microscopy and small angle x-ray scattering (18, 20). Evidently, when crRNA base pairs with a single strand of DNA (or RNA) a series of short helical segments is formed that together comprises a more firm structural unit within Cascade. Possibly, this limits the flexibility of Cascade and increases its overall stability. As limited conformational flexibility is a prerequisite for successful electron microscopy and x-ray crystallography studies we suggest that stable Cascade-ssDNA bound complexes may be suitable candidates to further optimize structural studies on this system.
Supplementary Material
Footnotes
* This work was supported by the Netherlands Organization for Scientific Research (NWO) (VENI 700.58.402) to E.v.D., and (VENI 863.08.014) to S.J.J.B. We thank The Netherlands Proteomics Centre for financial support.
 This article contains supplemental Figs. S1 to S4 and Table S1.
This article contains supplemental Figs. S1 to S4 and Table S1.
1 The abbreviations used are:
- CRISPR
- Clustered regularly interspaced short palindromic repeats
- Cas
- CRISPR associated genes/proteins
- Cascade
- CRISPR associated complex for antiviral defense
- crRNA
- CRISPR RNA
- Csy
- CRISPR system yersinia
- EM
- Electron microscopy
- IMMS
- Ion mobility mass spectrometry
- Ω
- collision cross-section.
REFERENCES
- 1. Horvath P., Barrangou R. (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170 [DOI] [PubMed] [Google Scholar]
- 2. Karginov F. V., Hannon G. J. (2010) The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell 37, 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Makarova K. S., Grishin N. V., Shabalina S. A., Wolf Y. I., Koonin E. V. (2006) A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van der Oost J., Jore M. M., Westra E. R., Lundgren M., Brouns S. J. (2009) CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 34, 401–407 [DOI] [PubMed] [Google Scholar]
- 5. Deveau H., Garneau J. E., Moineau S. (2010) CRISPR/Cas system and its role in phage-bacteria interactions. Ann. Rev. Microbiol. 64, 475–493 [DOI] [PubMed] [Google Scholar]
- 6. Marraffini L. A., Sontheimer E. J. (2010) CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., Horvath P. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 [DOI] [PubMed] [Google Scholar]
- 8. Yosef I., Goren M. G., Qimron U. (2012) Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Swarts D. C., Mosterd C., van Passel M. W., Brouns S. J. (2012) CRISPR Interference Directs Strand Specific Spacer Acquisition. PLoS One 7, e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carte J., Pfister N. T., Compton M. M., Terns R. M., Terns M. P. (2010) Binding and cleavage of CRISPR RNA by Cas6. RNA 16, 2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deltcheva E., Chylinski K., Sharma C. M., Gonzales K., Chao Y., Pirzada Z. A., Eckert M. R., Vogel J., Charpentier E. (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haurwitz R. E., Jinek M., Wiedenheft B., Zhou K., Doudna J. A. (2010) Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329, 1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sashital D. G., Jinek M., Doudna J. A. (2011) An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat. Structural Mol. Biol. 18, 680–687 [DOI] [PubMed] [Google Scholar]
- 14. Wang R., Preamplume G., Terns M. P., Terns R. M., Li H. (2011) Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure 19, 257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brouns S. J., Jore M. M., Lundgren M., Westra E. R., Slijkhuis R. J., Snijders A. P., Dickman M. J., Makarova K. S., Koonin E. V., van der Oost J. (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haft D. H., Selengut J., Mongodin E. F., Nelson K. E. (2005) A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1, e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Makarova K. S., Haft D. H., Barrangou R., Brouns S. J., Charpentier E., Horvath P., Moineau S., Mojica F. J., Wolf Y. I., Yakunin A. F., van der Oost J., Koonin E. V. (2011) Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jore M. M., Lundgren M., van Duijn E., Bultema J. B., Westra E. R., Waghmare S. P., Wiedenheft B., Pul U., Wurm R., Wagner R., Beijer M. R., Barendregt A., Zhou K., Snijders A. P., Dickman M. J., Doudna J. A., Boekema E. J., Heck A. J., van der Oost J., Brouns S. J. (2011) Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Structural Mol. Biol. 18, 529–536 [DOI] [PubMed] [Google Scholar]
- 19. Westra E. R., van Erp P. B., Kunne T., Wong S. P., Staals R. H., Seegers L. C., Bollen S., Jore M. M., Semenova E., Severinov K., de Vos W. M., Dame R. T., de Vries R., Brouns S. J., Van der Oost J. (2012) CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell 46, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wiedenheft B., Lander G. C., Zhou K., Jore M. M., Brouns S. J., van der Oost J., Doudna J. A., Nogales E. (2011) Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477, 486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kunin V., Sorek R., Hugenholtz P. (2007) Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8, R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wiedenheft B., van Duijn E., Bultema J., Waghmare S., Zhou K., Barendregt A., Westphal W., Heck A., Boekema E., Dickman M., Doudna J. A. (2011) RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. U. S. A. 108, 10092–10097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sternberg S. H., Haurwitz R. E., Doudna J. A. (2012) Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA 18, 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bush M. F., Hall Z., Giles K., Hoyes J., Robinson C. V., Ruotolo B. T. (2010) Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal. Chem. 82, 9557–9565 [DOI] [PubMed] [Google Scholar]
- 25. Ruotolo B. T., Benesch J. L., Sandercock A. M., Hyung S. J., Robinson C. V. (2008) Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protocols 81, 1139–1152 [DOI] [PubMed] [Google Scholar]
- 26. Ruotolo B. T., Giles K., Campuzano I., Sandercock A. M., Bateman R. H., Robinson C. V. (2005) Evidence for macromolecular protein rings in the absence of bulk water. Science 310, 1658–1661 [DOI] [PubMed] [Google Scholar]
- 27. Uetrecht C., Rose R. J., van Duijn E., Lorenzen K., Heck A. J. (2010) Ion mobility mass spectrometry of proteins and protein assemblies. Chem. Soc. Rev. 39, 1633–1655 [DOI] [PubMed] [Google Scholar]
- 28. von Helden G., Wyttenbach T., Bowers M. T. (1995) Conformation of macromolecules in the gas phase: use of matrix-assisted laser desorption methods in ion chromatography. Science 267, 1483–1485 [DOI] [PubMed] [Google Scholar]
- 29. Uetrecht C., Barbu I. M., Shoemaker G. K., van Duijn E., Heck A. J. (2011) Interrogating viral capsid assembly with ion mobility mass spectrometry: from sheet to sheath-like structures. Nat. Chem. 3, 126–132 [DOI] [PubMed] [Google Scholar]
- 30. Pukala T. L., Ruotolo B. T., Zhou M., Politis A., Stefanescu R., Leary J. A., Robinson C. V. (2009) Subunit architecture of multiprotein assemblies determined using restraints from gas-phase measurements. Structure 17, 1235–1243 [DOI] [PubMed] [Google Scholar]
- 31. Smith D. P., Radford S. E., Ashcroft A. E. (2010) Elongated oligomers in beta2-microglobulin amyloid assembly revealed by ion mobility spectrometry-mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 107, 6794–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van den Heuvel R. H., van Duijn E., Mazon H., Synowsky S. A., Lorenzen K., Versluis C., Brouns S. J., Langridge D., van der Oost J., Hoyes J., Heck A. J. (2006) Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Anal. Chem. 78, 7473–7483 [DOI] [PubMed] [Google Scholar]
- 33. van Duijn E., Barendregt A., Synowsky S., Versluis C., Heck A. J. (2009) Chaperonin complexes monitored by ion mobility mass spectrometry. J. Am. Chem. Soc. 131, 1452–1459 [DOI] [PubMed] [Google Scholar]
- 34. Giles K., Williams J. P., Campuzano I. (2011) Enhancements in travelling wave ion mobility resolution. Rapid Commun. Mass Spectrom. 25, 1559–1566 [DOI] [PubMed] [Google Scholar]
- 35. Smith D. P., Knapman T. W., Campuzano I., Malham R. W., Berryman J. T., Radford S. E., Ashcroft A. E. (2009) Deciphering drift time measurements from travelling wave ion mobility spectrometry-mass spectrometry studies. in Eur. J. Mass Spectrom. 15, 113–130 [DOI] [PubMed] [Google Scholar]
- 36. Lintner N. G., Kerou M., Brumfield S. K., Graham S., Liu H., Naismith J. H., Sdano M., Peng N., She Q., Copié V., Young M. J., White M. F., Lawrence C. M. (2011) Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J. Biol. Chem. 286, 21643–21656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lorenzen K., Vannini A., Cramer P., Heck A. J. (2007) Structural biology of RNA polymerase III: mass spectrometry elucidates subcomplex architecture. Structure 15, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 38. Synowsky S. A., van den Heuvel R. H., Mohammed S., Pijnappel P. W., Heck A. J. (2006) Probing genuine strong interactions and post-translational modifications in the heterogeneous yeast exosome protein complex. Mol. Cell. Proteomics 5, 1581–1592 [DOI] [PubMed] [Google Scholar]
- 39. Koschubs T., Lorenzen K., Baumli S., Sandström S., Heck A. J., Cramer P. (2010) Preparation and topology of the Mediator middle module. Nucleic Acids Res. 38, 3186–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sashital D. G., Wiedenheft B., Doudna J. A. (2012) Mechanism of Foreign DNA Selection in a Bacterial Adaptive Immune System. Mol. Cell 46, 606–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hernández H., Dziembowski A., Taverner T., Séraphin B., Robinson C. V. (2006) Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep. 7, 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lorenzen K., van Duijn E. (2010) Native mass spectrometry as a tool in structural biology. Curr. Prot. Protein Sci. 62, 17.12.11–17.12.17 [DOI] [PubMed] [Google Scholar]
- 43. Haurwitz R. E., Sternberg S. H., Doudna J. A. (2012) Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 31, 2824–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shoemaker G. K., van Duijn E., Crawford S. E., Uetrecht C., Baclayon M., Roos W. H., Wuite G. J., Estes M. K., Prasad B. V., Heck A. J. (2010) Norwalk virus assembly and stability monitored by mass spectrometry. Mol. Cell. Proteomics 9, 1742–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Uetrecht C., Versluis C., Watts N. R., Roos W. H., Wuite G. J., Wingfield P. T., Steven A. C., Heck A. J. (2008) High resolution mass spectrometry of viral assemblies: Molecular composition and stability of dimorphic hepatitis B virus capsids. Proc. Natl. Acad. Sci. U. S. A. 105, 9216–9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pringle S. D., Giles K., Wildgoose J. L., Williams J. P., Slade S. E., Thalassinos K., Bateman R. H., Bowers M. T., Scrivens J. H. (2007) An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-TOF instrument. Int. J. Mass Spectrom. 261, 1–12 [Google Scholar]
- 47. Makarova K. S., Aravind L., Wolf Y. I., Koonin E. V. (2011) Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct 6, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.