Abstract
Metastasis is a major obstacle that must be overcome for the successful treatment of lung cancer. Proteins secreted by cancer cells may facilitate the progression of metastasis, particularly within the phases of migration and invasion. To discover metastasis-promoting secretory proteins within cancer cells, we used the label-free quantitative proteomics approach and compared the secretomes from the lung adenocarcinoma cell lines CL1-0 and CL1-5, which exhibit low and high metastatic properties, respectively. By employing quantitative analyses, we identified 660 proteins, 68 of which were considered to be expressed at different levels between the two cell lines. High levels of A1AT were secreted by CL1-5, and the roles of A1AT in the influence of lung adenocarcinoma metastasis were investigated. Molecular and pathological confirmation demonstrated that altered expression of A1AT correlates with the metastatic potential of lung adenocarcinoma. The migration and invasion properties of CL1-5 cells were significantly diminished by reducing the expression and secretion of their A1AT proteins. Conversely, the migration and invasion properties of CL1-0 cells were significantly increased through the overexpression and secretion of A1AT proteins. Furthermore, the assembly levels of the metastasis-promoting pericellular fibronectin (FN1), which facilitates colonization of lung capillary endothelia by adhering to the cell surface receptor dipeptidyl peptidase IV (DPP IV), were higher on the surfaces of suspended CL1-5 cells than on those of the CL1-0 cells. This discovery reflects previous findings in breast cancer. In line with this finding, FN1 assembly and the lung colonization of suspended CL1-5 cells were inhibited when endogenous A1AT protein was knocked down using siRNA. The major thrust of this study is to demonstrate the effects of coupling the label-free proteomics strategy with the secretomes of cancer cells that differentially exhibit invasive and metastatic properties. This provides a new opportunity for the effective identification of metastasis-associated proteins that are secreted by cancer cells and promote experimental metastasis.
Lung cancer is the leading cause of cancer death, and ∼90% of all lung cancer deaths are attributed to metastases (1). Approximately 95% of lung cancer patients are not diagnosed until they develop symptoms, and 85% of the newly diagnosed lung cancer patients are already in the advanced stages of the disease (2, 3). Once the tumor cells have metastasized and spread throughout the lungs, the cancer is considerably more difficult to treat. Invasiveness and metastasis are major threats to successful treatment. Cancer metastasis is an intricate, multi-step process in which the tumor cells must gain both migratory and invasive properties (4). In metastasis research, there are two common in vivo models, spontaneous and experimental metastasis (5–7). In brief, spontaneous metastasis refers to primary tumor cells that are able to dissociate from the primary tumor and metastasize to the secondary organ via the circulatory system. In contrast, experimental metastasis refers to the injection of tumor cells directly into the systemic circulation. Many researchers have attempted to determine the molecular basis of these transitions in hopes of developing target-specific drugs or biomarkers for the prevention and diagnosis of metastasis. Although there have been many discoveries regarding a particular protein's influence on metastasis, the contribution of many protein targets to the metastatic process remains poorly defined.
The term “secretome” was originally coined to refer to the secretory proteins from the entire genome of Bacillus subtilis (8). The word secretome has developed a broader meaning and now refers to the proteins released by a cell, tissue, or organism through various mechanisms, which include classical secretion, nonclassical secretion, membrane protein shedding, and secretion via exosomes (9–11). Each step involved in tumor metastasis, including migration and invasion, requires specific molecular interactions by both the tumor cells and the surrounding extracellular matrix (12). Some interactions are mediated by secretory factors that function as catalytic agents or by specific recognitions. For example, cathepsins, a family of lysosomal cysteine and aspartic proteases, plays a role in breaking down the connective barriers in the extracellular matrix and basement membranes, effectively enhancing the metastasis of tumor cells (13). These unique functions correlate with invasive activity and are otherwise known as the promigratory and pro-invasive effects on cells (14, 15). With respect to cancer progression, chronic changes or abnormal secretions of certain proteins may indicate a pathologic condition and, therefore, provide suitable targets for therapeutic and biomarker discoveries (16).
Proteomic tools have been proposed as a new platform for studying complex biological functions, which entail large numbers and networks of proteins (17). Moving beyond the imposing burden of providing lists of proteins identified in certain samples, the field of quantitative proteomics yields information that specifically recognizes the differences between samples and has emerged as a very important area of research in the study of cancer. These proteomics approaches have been extensively applied to cell secretome analyses for the elucidation of disease mechanisms, diagnoses, and new drug developments (16, 18–23). To comprehensively understand the roles of the secretion-related regulations in metastatic progression, the label-free quantitative proteomics approach was used to identify metastatic-associated proteins secreted by lung adenocarcinoma cells. Comparative secretome analysis was conducted in lung adenocarcinoma cells with differing levels of migration and invasiveness (CL1-0 versus CL1-5) (24). The CL1 cell lines have been used in previous metastasis research as novel protein targets associated with lung cancer metastasis discovery (25–28). The characterized protein, A1AT, was validated for its association and functions involved with lung adenocarcinoma metastasis by subjecting the cells to experimental metastasis assays in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Cell Culture and Conditioned Medium Harvest
The human lung adenocarcinoma cell line CL1 was obtained from a 64-year-old man with a poorly differentiated adenocarcinoma (24). The CL1-0 and CL1-5 cells were provided by Dr. P.-C. Yang (Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan, Republic of China) and were cultured at 37 °C with 5% CO2 in RPMI 1640 media that was supplemented with 10% fetal bovine serum (FBS)1 (Invitrogen, Gaithersburg, MD) and 2 g/liter NaHCO3. The CL1-0 and CL1-5 cancer cells were grown in serum- containing media until the cell density reached 70–80% confluence in the culture dish. The cells were washed with serum-free media (SFM) four times before they were incubated in SFM for 24 h. After incubation, the total number of viable and dead cells in the cancer cell lines were determined using the trypan blue dye exclusion assay. The ratio of total viable cells compared with the total number of viable and dead cells represents the percentage of cell viability. The cell viability was determined to be > 98%, and the conditioned media (CM) was harvested and centrifuged to eliminate the intact cells. The harvested CM was concentrated by centrifugation in Amicon Ultra-15 tubes (molecular weight cut-off 3 kDa; Millipore, Billerica, MA). Finally, the Bradford assay (Bio-Rad, CA, USA) was used to determine the total protein concentration in the CM samples. Western blot analysis was used to determine the rate of cell lysis by considering the known cell number and the corresponding amount of the cytoskeletal protein tubulin, as the standard, when evaluating the number of lysed cells in the CM (29). From the CM sample collection to proteomics technology analyses, every cell line was performed in biological triplicate.
Sample Purification and Digestion
Each protein sample (10 μg), containing sample dye and 0.5 m dithiothreitol, was spiked with 0.2 pmol bovine serum albumin (BSA) as the internal standard and then boiled for 10 min at 95 °C and loaded onto the gel. The stacking gel-aided purification method was performed to refine the secretome samples according to the previous study (30). All of the bands were excised and digested in-gel with trypsin. The gel pieces were reduced with 80 mm dithiothreitol at 56 °C and alkylated with 150 mm iodoacetamide at room temperature, with each step requiring 1 h. Modified trypsin (1:20 (w/w) (trypsin/protein) was added to the gel pieces, and they were incubated overnight at 37 °C to allow for complete digestion. All remaining reagents from the in-gel digestion procedure and from the digested peptide samples were removed using a C18 tip (VARIAN, Palo Alto, CA).
NanoLC-MS/MS Analysis
Each peptide mixture was dissolved in 6 μl of solvent A (0.1% formic acid in water) and analyzed by online nanoflow liquid chromatography tandem mass spectrometry (LC-MS/MS) on a nanoAcquity system (Waters, Milford, MA) coupled with an LTQ-Orbitrap Velos hybrid mass spectrometer (Thermo Scientific, Bremen, Germany) that was equipped with a PicoView nanospray interface (New Objective, Woburn, MA). Peptide mixtures were loaded onto a 75-μm × 250-mm nanoACQUITY UPLC BEH130 column packed with C18 resin (Waters, Waters, Milford, MA) and were separated at a flow rate of 300 nL/min using a linear gradient of 5 to 50% solvent B (95% acetonitrile with 0.1% formic acid) for 160 min followed by a sharp increase to 85% B within 1 min and held at 85% B for an additional 15 min. Two blank washes were performed following each sample run. The LTQ Orbitrap Velos instrument was operated in data-dependent mode to automatically switch between full scan MS and MS/MS acquisition. For the collision-induced dissociation (CID)-MS/MS top20 method, full scan MS spectra (m/z 350–1600) were acquired in the Orbitrap analyzer after accumulation to a target value of 1e6 in the linear ion trap. Resolution in the Orbitrap system was set to r = 60,000 (all Orbitrap system resolution values are given at m/z 400). The 20 highest intensity peptide ions with charge states ≥2 were sequentially isolated to a target value of 5000 and fragmented in the high-pressure linear ion trap by low-energy CID with a normalized collision energy of 35%. The resulting fragment ions were scanned out in the low-pressure ion trap at the normal scan rate and recorded with the secondary electron multipliers. The ion selection threshold was 500 counts for MS/MS, and the maximum allowed ion accumulation times were 500 ms for full scans and 100 ms for CID-MS/MS measurements in the LTQ. An activation of q = 0.25 and activation time of 10 ms were used. Standard mass spectrometric conditions for all experiments were as follows: spray voltage, 1.8 kV; no sheath and auxiliary gas flow; heated capillary temperature, 200 °C; predictive automatic gain control enabled;, and an S-lens RF level of 50%.
Peptide/Protein Quantification and Identification
Progenesis software (version 4.0; Nonlinear Dynamics, Newcastle, UK) was used for intensity-based label-free quantification. On selection of a sample reference, the retention times of all eluting precursor m/z values of all other samples were thus aligned, which provided a long list of features that represented the same peptide in each sample. Only features that had a charge greater than or equal to 2 were incorporated in further analyses. Those with less were excluded. Samples from each cell line were grouped together after the alignment, and feature filtering was then complete. The raw abundances of BSA peptides that had an ion score greater than 40 were then normalized to create a scaling factor that would remove experimental variation (i.e. differing protein quantities loaded into the instrument). Multiple MS/MS spectra were frequently collected across all samples for the precursor ion; therefore, the precursor intensities could be ranked accordingly. The five highest intensity precursors for each feature were selected, and their MS/MS spectra were transformed into peak lists and exported to create their Mascot generic files. The Mascot generic files were searched against the Swiss-Prot (release date, 11/16/11; 533,049 entries; human taxonomy) and contamination database using the Mascot Daemon 2.2 server (Matrix Science, London, UK). The protein list for the contamination database was derived from The Global Proteome Machine Organization (http://www.thegpm.org/crap/index.html). When setting the search parameters, the MS and MS/MS mass tolerances were set at ± 10 ppm and ± 0.6 Da, respectively, representing two missed cleavages from the trypsin digest and variable modifications of carbamidomethylation (Cys), deamination (Asn, Gln), oxidation (Met), and a decoy search was enabled. Search results for spectrum to peptide matches were exported in .xml format and then imported into Progenesis software, enabling both peptide quantification and identification. Peptides with mascot ion scores < 33 (p < 0.01 identity threshold) were filtered out, leaving only unique peptides for corresponding proteins to be used for identification and quantification. At this point, proteins with single peptide identification and quantification were removed. The identified proteins that belong to the contamination database such as common trypsin autolysis peaks and keratin contamination were excluded. During protein quantification, any features with conflicting peptide identifications were excluded from the measurements for the affected proteins. A protein's feature intensities of all unique peptides were summed up. One-way analysis of variance (ANOVA) was used to calculate the protein p value according to the sum of the normalized abundances across all runs.
Western Blot Analysis
The fifty μg of secretory proteins from the CM were separated on a 12% SDS-PAGE gel, and iBlot (Invitrogen, Carlsbad, CA) was used to transfer the proteins to a polyvinylidene difluoride membrane in accordance with the manufacturer's protocol. The membranes were kept in a 5% nonfat milk solution for 1 h at room temperature before they were separately probed with the relevant antibodies at 4 °C overnight. The membranes were washed with Tris-buffered saline and tween 20 (TBST) three times and incubated with horseradish peroxidase-conjugated secondary antibodies at a dilution of 1:10,000 at room temperature for 1 h. The membranes were washed with TBST five times before developing them with enhanced chemiluminescence detection.
Immunohistochemistry (IHC)
Immunohistochemistry in tissue microarrays (TMAs) were required to analyze the protein levels. All of the tissue samples were obtained from patients diagnosed with primary lung adenocarcinoma and who had already undergone surgical resection. A total of 83 primary formalin-fixed and paraffin-embedded samples were provided by the Department of Pathology, National Cheng Kung University Hospital. All of the patients who provided tumor tissue specimens gave written informed consent in accordance with the rules and requirements of National Cheng Kung University Hospital's ethics committee. The hospital's tissue bank also provided TMAs generated from these samples by sampling tumor tissue cores (1.0 mm in diameter) from each paraffin-embedded tissue block in replicate. There were ∼50–60 cores that could be formed a TMA block. The TMAs were only collected to profile protein expression in lung adenocarcinoma patients. The IHC method was used to stain A1AT expression levels on the TMA blocks. IHC was then performed on 4-μm-thick formalin-fixed paraffin-embedded sections with Bond-Max Automated IHC stainer (Leica Biosystems Newcastle Ltd, Australia). Tissues were deparaffinized with xylene and pretreated with Epitope Retrieval Solution 1 (citrate buffer, pH 8.5) at 100 °C for 20 min. Subsequently, hydrogen peroxide blocking was performed for 5 min using the Bond Polymer Refine Detection Kit (Leica Biosystems Newcastle Ltd, United Kingdom). The primary antibody was incubated at room temperature for 30 min., and tissue samples were incubated with polymer at room temperature for 8 min before being developed with 3,3′- diaminobenzidine chromogen for 10 min. Counterstaining was performed with hematoxylin. Protein expression was evaluated based on the percentage of immunopositive stained cells (0, 0–4% cells; 1, 5–25% cells; 2, 26–50% cells, 3, 51–75%, and 4, 76–100%) and the intensity of the cell staining (3 (strong), 2 (moderate), 1 (weak), or 0 (no staining)). The final score is their summation. A final score greater than 2 was defined as positive staining (31).
Knockdown and Overexpression of Proteins and Functional Assays In Vitro
Gene Knockdown and Overexpression
The siRNA transfection reagent was used to achieve gene silencing of A1AT by transfection with the siRNA, which was performed according to the manufacturer's instructions (Santa Cruz Biotechnology, Santa Cruz, CA). Half a microgram of siRNA or half a microgram of scrambled control siRNA with 6 μl of siRNA transfection reagent (3 × 105 cells/well) was used for each siRNA transfection. A1AT gene overexpression was achieved through transfection with a commercial plasmid (A1AT: BC011991, Thermo Fisher Scientific Inc., Huntsville, AL) using the TurboFect transfection reagent according to the manufacturer's instructions (OriGene Technologies, Inc., Rockville, MD). For each well of transfection, 4 μg of plasmid DNA or empty vector control with 12 μl of TurboFect transfection reagent was added to 100 μl of serum-free medium. The solution was mixed gently and overlaid onto the cells in complete medium supplemented with 10% FBS for further study.
MTT Assays
When evaluating the direct effect of siRNA knockdown and plasmid DNA overexpression on cancer cell growth in vitro, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (thiazolyl blue, MTT) proliferation analyses were performed. In the MTT assay, a 96-well plate was used, and the cells were seeded at a concentration of 3 × 103 cells per well. The cells were incubated for 24 h before being treated with the siRNA and plasmid DNA. After 24 h, 2 μl of MTT (10 mg/ml) was added to each well and incubated for 3 h at 37 °C. The cells were lysed with 100 μl of dimethyl sulfoxide. Microplate photometers measured the absorbance at 595 nm (Thermo Fisher Scientific Inc., Waltham, MA). The cell growth of each group was monitored every 24 h for 72 h total.
Bromodeoxyuridine (BrdU) Assay
A BrdU cell proliferation assay was performed according to the manufacturer's instructions (Millipore, Billerica, MA). A total of 2 × 103 cells were seeded in each well of a 96-well plate, and cell growth within each group was monitored at 24 and 48 h.
Cell Wound Healing Assay
Cell migration was examined with the commercial ibidi Culture-Insert (Applied BioPhysics, Inc., Troy, New York, USA). Cells were seeded on the insert for 12 h, and the inserts were removed. Photographs were taken at 0 h and 24 h at the same position in the cell-free gap insert with 100× magnification.
Transwell Migration and Matrigel Invasion Assay
Cell migration was examined in a membrane migration culture system with BD Falcon multiwell insert systems with 8-μm pores (BD Biosciences, San Jose, California). The upper wells contained 1 × 105 cells seeded in serum-free medium, and the lower chambers were filled with complete medium supplemented with 10% FBS to induce cell migration. After the cells were incubated at 37 °C for 24 h, the membranes were fixed with methanol, the cells were stained with a Giemsa stain and the number of cells that migrated through the membrane to the lower side was determined. The cells that managed to traverse the filter to the lower chamber were counted at 200× magnification in six different fields per filter. In preparation for the invasion assay, BD Falcon multiwell insert systems containing 8-μm pores were coated with 2 mg/ml BD Matrigel basement membrane matrix (BD Biosciences, San Jose, CA). The following steps were the same as those of the transwell migration assay steps.
Flow Cytometry
Fluorescence-activated cell sorting was performed to quantify FN1 expression on the cell surfaces (32, 33). The tumor cells were trypsinized and incubated in suspension for 2 h in 20% FBS media and were washed once with phosphate-buffered saline (PBS). Cells were incubated with a rabbit anti-FN1 antibody (diluted 1:600 in PBS with 1% BSA, Sigma) for 1 h at 4 °C before they were stained with a fluorescein isothiocyanate-conjugated donkey anti-rabbit antibody in PBS containing 1% BSA for 1 h at 4 °C and fixed in 2% paraformaldehyde in PBS. Fluorescence-activated cell sorting analysis was performed on a Coulter Epics Profile (FACSCalibur, BD Biosciences, San Jose, CA). The nonspecific fluorescence was accounted for by incubating the tumor cells with nonimmune serum rather than the primary antibody.
Lung Colony Assay
The transfected cells were incubated in suspension for 2 h in 20% FBS media. Likewise, CL1-5 cells were incubated in suspension for 2 h in 20% FBS media. Then, the cells were treated with myelin basic protein (MBP) and soluble, truncated DPP IV for 20 min (33). A total of five mice in each group were used to perform the lung colony assay and discern the levels of lung colonization. Eight-week-old nude mice were injected in the lateral tail vein with a single-cell suspension that contained 2 × 106 cells in 0.2 ml RPMI 1640 base medium. The mice were sacrificed after 8 weeks, and the lungs were removed and fixed in a 3.7% formalin fixative. The representative lung tumors were removed, fixed, and immediately embedded in paraffin. The paraffin-embedded tumors were sectioned into 4-mm layers and stained with hematoxylin and eosin (H&E) for histologic analysis.
Statistical Analysis
Statistical analyses were performed using SPSS 18.0 (SPSS Inc., Chicago, IL). A statistically significant p value less than 0.05 was required for the two-tailed tests. All experiments were performed in biological triplicate, and the results are shown as the mean ± S.D. The nonparametric Mann-Whitney U test and ANOVA were employed to analyze the all differences between groups.
RESULTS
Cell Conditioned Media Influences Lung Adenocarcinoma Cell Migration and Invasion
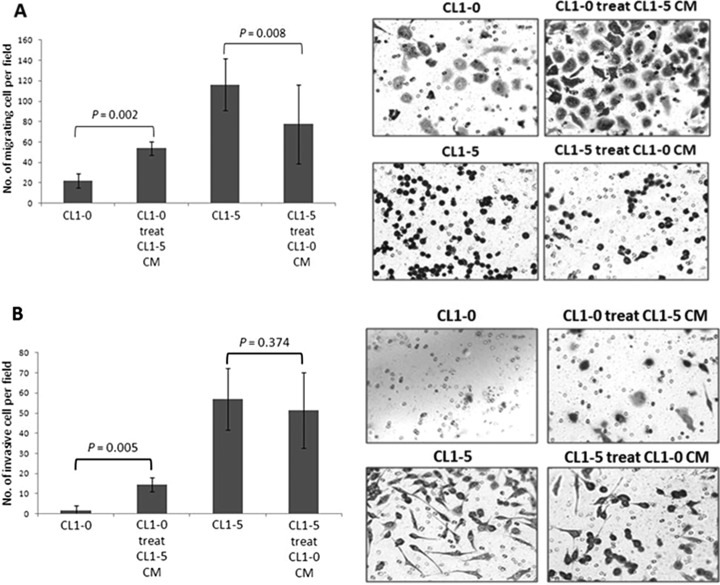
CL1-0 and CL1-5, lung adenocarcinoma cells derived from the same parental cell line with low and high invasive abilities, respectively, provide ideal model to investigate the cells' metastatic behavior in previous studies (25–28). To examine whether the secreted proteins of CL1-0 and CL1-5 cells could determinately alter the migratory and invasive abilities of cancer cells, CL1-0 and CL1-5 cells were incubated in the cell CM of CL1-5 and CL1-0, respectively, for 48 h. Thereafter, transwell migration and Matrigel invasion assays were performed to investigate the potentially altered level of cell migration and invasion. After conditioning in CL1-5 CM, the migration and invasion levels of CL1-0 cells were increased. The migration ability of CL1-5 cells was reduced after incubation in CL1-0 CM, and the invasion level of CL1-5 was slightly decreased, but the difference was not significant (Figs. 1A and 1B). Collectively, these results support the hypothesis that particular factors produced or secreted by the cells could alter the migration or invasion properties of lung adenocarcinoma cells.
Fig. 1.
The influence of cell conditioned media on the migration and invasion of lung adenocarcinoma cells was examined. CL1-0 and CL1-5 cell supernatants were collected after 48 h and then incubated with CL1-5 and CL1-0 cells, respectively, for 48 h. After 48 h, the altered migration and invasion of the cells were investigated using transwell migration and Matrigel invasion assays. Two independent experiments were performed, and each experiment was performed three times. The data are presented as the mean ± S.D., where p < 0.05 was considered statistically significant when analyzed by the Mann-Whitney U test. After conditioning the CL1-5 CM, the migration and invasive ability of CL1-0 cells increased. Conversely, CL1-5 cells were not affected by the CL1-0 CM. The results provide evidence that particular factors produced by the cells could change the migration and invasiveness of lung adenocarcinoma cells. A, Transwell migration assay. A total of 50,000∼100,000 cells were seeded into the upper wells in serum-free medium, and the lower chambers were filled with complete medium, supplemented with 10% FBS, to induce cell migration. After 24 h, the cells that had traversed the filter to the lower chamber were stained with Giemsa stain and counted microscopically (200×) in 10 different fields per filter. B, Matrigel invasion assay. The Matrigel coated the upper wells. Apart from this, the set-up was the same as the migration assay. These results indicated that particular factors produced or secreted by the cells exhibit enhanced or reduced migration or invasion properties.
Monitoring the Quality of CM Samples
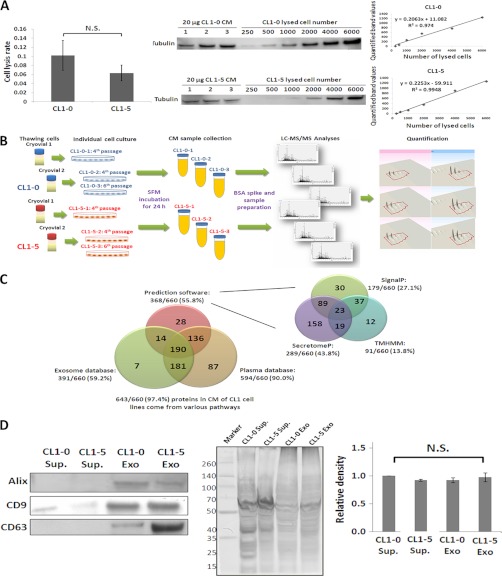
To avoid the contamination of serum constituents from the culture medium while analyzing the secretory proteins, the cells were washed four times with SFM and then incubated in SFM for the secretome sample collection. The CL1 cells were incubated in SFM for 24, 36, and 48 h to optimize the incubation time for the CM collection. This method was used to greatly deplete serum proteins and prevent intracellular contamination. The average cell viability (mean ± S.D.) of CL1-0 cells at 24, 36, and 48 h was 98.6% ± 0.6, 98.7% ± 0.3, and 94.7% ± 0.7, respectively. The average cell viability of CL1-5 cells at 24, 36, and 48 h was 98.8% ± 0.5, 96.7% ± 0.3, and 93.8% ± 0.7. The 24 h incubation period had the highest cell viability for both the CL1-0 and CL1-5 cells and was used as the CM collection time. This collection time should minimize intracellular protein contamination caused by cell lysis. Additionally, cell viability before and after SFM incubation exhibited no significant difference, and the cell viability for both cell lines was over 98% (16, 18, 29). The cell lysis rates were determined by Western blot analysis using the known cell number and the corresponding amount of the cytoskeleton protein tubulin as a standard for evaluating the number of lysed cells in the CM. The average rate of cell lysis of CL1-0 and CL1-5 cells in the CM was under 0.1% (Fig. 2A). There was no statistically significant difference between the cell lysis rates of CL1-0 and CL1-5, but cell lysis was more pronounced in CL1-0. These observations collectively indicate that cell death did not influence the recovery of proteins in CM.
Fig. 2.
Characterization of secretome samples from CL1-0 and CL1-5 cell lines. A, The distribution of the cytosolic protein tubulin from an exact cell number was evaluated to determine whether the CM contained proteins resulting from cell lysis. The rate of cell lysis was under 0.1% in both CL1 cell lines. B, The schematic representation of the label-free proteomics analysis procedure. The biological triplicates and BSA-spike were performed for the label-free quantitative approach. C, 97.4% (643/660) of the proteins were present in the CM of CL1 cells through multiple routes. These proteins were identified by the following: 1) SignalP and SecretomeP prediction software to predict classical and nonclassical secretion pathways, respectively, as well as TMHMM to predict membrane proteins; 2) the ExoCarta database, which contains results from the exosome human database; and 3) the human plasma proteome database. D, The presence of CD9, CD63, and ALG-2-interacting protein X proteins in the CL1-0 and CL1-5 CM samples proves the existence of exosomes in the CM. The abbreviation, Sup., refers to the supernatant after removing the exosomes and is the negative control. Exo indicates the exosomes were purified from CM samples via the commercial kit. An equal amount of each sample was run on SDS-PAGE and further processed with silver staining in biological triplicate as the loading controls. Each lane was quantified and statistical analyses were performed to confirm if the loading amount was equal.
Identification of the Metastasis-associated Proteins From the Secretomes of Lung Adenocarcinoma Cells
To discover the secretory proteins related to cancer migration/invasion, the label-free quantitative proteomics technique was used to discern the differential expression of proteins between the low (CL1-0) and high (CL1-5) invasive/metastatic lung adenocarcinoma cell lines. Biological triplicates of CM samples were used for label-free quantitative analyses. The CM samples were collected from different cryovials and passages to ensure that the differences between the two cell lines were indeed the result of each cell line's properties (Fig. 2B). Each sample was mixed with an internal standard protein (BSA) and subjected to the stacking gel-aided method, which was recently established for cell secretome sample purification (30), followed by in-gel digestion. With each CM sample spiked with BSA, an internal standard was created, allowing for the monitoring of experimental variation during sample digestion and LC-MS measurements (34). Moreover, BSA peptides with an ion score greater than 40 were considered to have high confidence, and their features were applied for raw abundance normalization. The BSA protein ratios between two cell lines before and after normalization were 1.121 and 1.005, respectively. All features could then be normalized to determine a global scaling factor in Progenesis LC-MS software. In total, 12,219 precursor ion features were assigned to successful peptide identifications and corresponded to 660 unique proteins that contained at least two uniquely identified and quantified peptides (supplemental Table S1). Among the 660 proteins, 469 (71.1%) proteins were simultaneously identified in both CL1-0 and CL1-5 CM samples. There were 89 (13.4%) and 102 (15.5%) proteins identified exclusively in the CL1-0 and CL1-5 CM samples, respectively. Intensity-based label-free quantification requires an intensity response directly proportional to the amount of analyte. Quantification using MS intensity data proved to be a consistent method of sample processing and a stable LC-MS system to minimize analytical bias. The coefficient variation of peptide retention time (RT) along with abundances from each run can serve as indicators to observe the performance of the LC-MS system. The data indicated that the distribution of retention time and abundances were minimal. The CVs (mean ± S.D.) of RT for the 4479 human peptides and the 75 BSA peptides were 0.48% ± 0.28 and 0.54% ± 0.30, respectively. The CVs (mean ± S.D.) of normalized abundances for peptides within each group, CL1-0 and CL1-5, were 2.55% ± 4.51 and 4.34% ± 6.77%, respectively. Additionally, the coefficient of variation of normalized BSA peptide abundances in all runs was 2.96% ± 4.70 (supplemental Figs. S1 and S2). This indicates that the LC-MS conditions were stable and suitable for quantification purposes.
A total of 660 proteins were considered successful identifications and fit the following criteria: (1) peptide ion score > 32 (p < 0.01) and FDR < 1%; (2) had at least two uniquely identified peptides; and (3) had at least two quantified peptides (supplemental Table S2). The ratio distributions of peptides and proteins are shown in supplemental Fig. S3. Each protein differentially expressed between CL1-0 and CL1-5 cell lines in biological triplicates was examined via ANOVA. Further, the p values were set for multiple testing as a means of controlling the family-wise error rate (α = 0.05) using the algorithm of sequential Bonferroni (SB) (35). These p values allowed us to identify the differentially expressed proteins. Finally, 68 proteins were identified based on their differences between the two cell lines and chosen as the metastasis-associated proteins (supplemental Table S3).
Profiling of the Identified Proteins
The proteins were detected in the CM based on various pathways, such as signal peptide predominance, membrane protein shedding, exocytosis, exosome delivery or intracellular protein contamination (cell lysis). According to the Gene Ontology Database, 46.06% (304/660) of the proteins were classified as extracellular and/or membrane-related proteins (supplemental Table S2). In addition, the bioinformatics programs SignalP 4.0, which characterizes the presence of a signal peptide (36), SecretomeP 2.0, which predict proteins secreted by a nonsignal peptide trigger (37), and TMHMM, which predicts the transmembrane helices in proteins (38), were used to predict proteins released into the CM through classic secretion pathways, nonclassic secretion pathways, and/or protein shedding. Altogether, 55.76% (368/660) of the identified proteins could be released into the CM through these three pathways (Fig. 2C and supplemental Table S2). The ExoCarta database (http://exocarta.ludwig.edu.au/) was used to investigate the proportion of the 660 identified proteins that could be secreted by the cells via exosomes (39, 40). There are 2065 proteins present in the human exosome database. Of the 660 identified proteins, 59.24% (391/660) of the identified proteins exist in the human exosome database (Fig. 2C and supplemental Table S2). To confirm the existence of the exosome in the CM sample, the commercial exosome precipitation kit, ExoQuick (System Biosciences, Mountain View, CA, USA), was performed to purify the exosome from the CM sample. Three exosome markers (CD63, CD9, and ALG-2-interacting protein X) were detected in the CM samples via Western blot analyses (Fig. 2D). Among the 660 identified proteins, 84.24% (556/660) of the proteins could be released into the extracellular space via diverse mechanisms according to the prediction software and the ExoCarta database. Moreover, it is highly possible that the proteins present in human blood are released from cells and have further medical value as clinical biomarkers (41). Among the 660 identified proteins, 90.00% (594/660) were found in the plasma proteome database (http://www.plasmaproteomedatabase.org/) (Fig. 2C and supplemental Table S2) (42). Collectively, these analyses suggest that 97.42% (643/660) of the identified proteins were released into the CM via different mechanisms.
The five most significant molecular functions of the 660 identified proteins were linked to protein binding, catalytic activity and hydrolase activity according to Metacore software (GeneGo, Joseph, MI) (supplemental Fig. S4). There are extracellular and membrane proteins that transmit signals from cell-to-cell, or even act as receptors. Their primary functions are to bind signaling molecules, such as connective tissue growth factor and PLAU, and induce biochemical responses within cells. Catalytic and hydrolase activities play critical roles in cancer metastasis. In order for cancer cells to metastasize and invade a new organ, the cells must first produce hydrolytic enzymes, such as LOXL2, MMP2, TIPM1, and A1AT, and then activate/deactivate the catalytic activities of proteins to facilitate the breakdown of proteins within the extracellular matrix. By doing so, they allow tumor cells to pass into the blood and lymphatic vessels (43–45).
Confirmation of A1AT Expression Levels in CM and Clinical Specimens of Lung Adenocarcinoma Cells and Patients
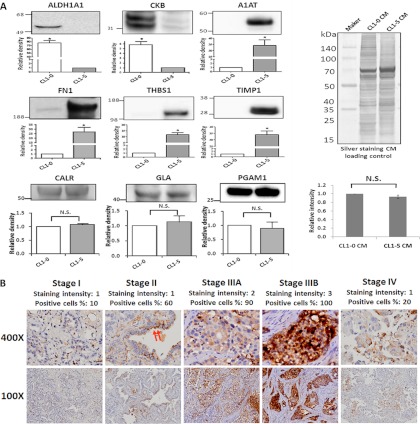
Among the 68 proteins with differentially expressed levels between the CL1-0 and CL1-5 cell lines, we focused on proteins that have at least two pieces of evidence according to the database and/or prediction software. The selected proteins also must not exhibit metastatic functions within the lung adenocarcinoma CL1 cell lines and must exhibit a significantly greater fold change (top 30) (Table I). Furthermore, proteins that lack metastasis-related studies in lung adenocarcinoma but for which evidence that they could influence cancer metastasis by function and/or clinical expression were given high priority for further investigation. Proteins that were overexpressed in the CL1-0 cells include brain-type creatine kinase and retinal dehydrogenase 1. Among the CL1-5 cells, A1AT, FN1, THBS1, and TIMP1 were selected for further confirmation via Western blot analyses (Fig. 3A). The three proteins, CALR, GLA, and PGAM1, were selected as loading controls because they had no statistically significant differences between the cell lines and because all had ratios close to 1. The findings from the Western blot analysis and those from the mass spectrometry data exhibited the same trends of protein expression. The full-length blot of A1AT and FN1 are shown in supplemental Fig. S5. We focused on up-regulated proteins in the metastatic cell line because they represent potential serum marker candidates for predicting lung adenocarcinoma metastasis, drug targets for inhibiting metastasis, and allow for a deeper understanding of the mechanism of metastasis. Further emphasis was placed on proteins not known to be involved with lung adenocarcinoma metastasis and on those that have only been studied within a few types of cancer metastasis. For these reasons, A1AT represents an ideal choice among the proteins and was further examined for its expression levels in clinical tissue specimens of distinct pathological stages.
Table I. A total of 30 identified proteins with differentially expressed levels between CL1–0 and CL1–5 cells.
| Swiss-Prot accession No. | Gene name | Protein name | Protein coverage (%) | Unique peptide no. | Quantified peptide no. | Confidence score | Fold change | SignalPa | SecretomePb | TMHMMc | Plasmad | Exosomee | Subcellular locationsf |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proteins with high levels in CL1-0 | |||||||||||||
| PLTP_HUMAN | PLTP | Phospholipid transfer protein | 16.4 | 6 | 6 | 299.33 | 56.67 | Y | Y | N | Y | Y | Secreted |
| SH3L1_HUMAN | SH3BGRL | SH3 domain-binding glutamic acid-rich-like protein | 27.2 | 2 | 2 | 109.74 | 54.38 | N | Y | N | Y | Y | Cytoplasm, nucleus |
| CFAH_HUMAN | CFH | Complement factor H | 17.6 | 14 | 14 | 696.36 | 39.13 | Y | N | N | Y | Y | Secreted |
| KCRU_HUMAN | CKMT1A | Creatine kinase U-type, mitochondrial | 11.3 | 3 | 3 | 190.41 | 36.92 | N | Y | N | Y | N | Mitochondrion inner membrane, peripheral membrane protein, intermembrane side |
| CO5A2_HUMAN | COL5A2 | Collagen alpha-2(V) chain | 12.9 | 13 | 13 | 623.58 | 29.16 | Y | N | N | Y | N | Secreted, extracellular |
| AL1A1_HUMAN | ALDH1A1 | Retinal dehydrogenase 1 | 1.2 | 19 | 18 | 1081.9 | 26.38 | N | Y | N | Y | Y | Cytoplasm |
| PCOC1_HUMAN | PCOLCE | Procollagen C-endopeptidase enhancer 1 | 22.7 | 8 | 8 | 536.54 | 19.08 | Y | Y | N | Y | N | Secreted |
| KCRB_HUMAN | CKB | Creatine kinase B-type | 19.4 | 4 | 4 | 272.68 | 14.01 | N | N | N | Y | Y | Cytoplasm |
| Proteins with high levels in CL1-5 | |||||||||||||
| LIF_HUMAN | LIF | Leukemia inhibitory factor | 22.3 | 4 | 4 | 229.63 | 202.48 | Y | Y | N | Y | N | Secreted |
| JAG1_HUMAN | JAG1 | Protein jagged-1 | 15.8 | 12 | 12 | 766.09 | 122.62 | Y | N | Y | Y | N | Membrane, single-pass type I membrane protein |
| IBP4_HUMAN | IGFBP4 | Insulin-like growth factor-binding protein 4 | 41.5 | 7 | 7 | 357.6 | 80.22 | Y | Y | N | Y | N | Secreted |
| MMP2_HUMAN | MMP2 | 72 kDa type IV collagenase | 13 | 6 | 6 | 305.92 | 70.81 | Y | Y | N | Y | N | Secreted, extracellular, membrane, nucleus |
| A1AT_HUMAN | A1AT | Alpha-1-antitrypsin | 42.1 | 16 | 16 | 1173.83 | 56.27 | Y | Y | N | Y | Y | Secreted |
| NRCAM_HUMAN | NRCAM | Neuronal cell adhesion molecule | 17 | 18 | 18 | 1134.53 | 46.98 | Y | N | Y | Y | N | Cell membrane, single-pass type I membrane protein |
| TENA_HUMAN | TNC | Tenascin | 2.3 | 4 | 4 | 175.97 | 32.8 | Y | N | Y | Y | N | Secreted, extracellular |
| CERU_HUMAN | CP | Ceruloplasmin | 19.3 | 16 | 16 | 999.39 | 28.79 | Y | Y | N | Y | Y | Secreted |
| CATB_HUMAN | CTSB | Cathepsin B | 29.5 | 9 | 9 | 518.08 | 27.04 | Y | Y | N | Y | Y | Lysosome, melanosome. |
| FUCO2_HUMAN | FUCA2 | Plasma alpha-l-fucosidase | 15.2 | 6 | 6 | 261.98 | 27.04 | Y | Y | N | Y | N | Secreted |
| SEM4B_HUMAN | SEMA4B | Semaphorin-4B | 5.3 | 4 | 4 | 219.72 | 26.96 | Y | N | Y | Y | N | Membrane, single-pass type I membrane protein |
| IL6_HUMAN | IL6 | Interleukin-6 | 25 | 5 | 5 | 265.91 | 26.65 | Y | Y | N | Y | N | Secreted |
| TIMP1_HUMAN | TIMP1 | Metalloproteinase inhibitor 1 | 36.7 | 6 | 6 | 425.88 | 26.39 | Y | Y | N | Y | N | Secreted |
| FINC_HUMAN | FN1 | Fibronectin | 36.5 | 70 | 70 | 4926.4 | 24.23 | Y | N | N | Y | Y | Secreted, extracellular |
| LAMA3_HUMAN | LAMA3 | Laminin subunit alpha-3 | 2.7 | 5 | 5 | 275.5 | 23.4 | Y | N | N | Y | Y | Secreted, extracellular, basement membrane |
| UROK_HUMAN | PLAU | Urokinase-type plasminogen activator | 33.9 | 15 | 15 | 891.44 | 21.19 | Y | Y | N | Y | Y | Secreted |
| PLBL1_HUMAN | PLBD1 | Phospholipase B-like 1 | 6.7 | 3 | 3 | 145.53 | 21.1 | Y | Y | N | N | N | Cytoplasmic granule, secreted |
| TSP1_HUMAN | THBS1 | Thrombospondin-1 | 42.9 | 38 | 37 | 2283.69 | 19.95 | Y | N | N | Y | Y | Plasma membrane, extracellular matrix |
| CSF1_HUMAN | CSF1 | Macrophage colony-stimulating factor 1 | 8.3 | 3 | 3 | 198.45 | 19.14 | Y | N | Y | Y | N | Cell membrane, single-pass membrane protein, secreted, extracellular space |
| NPTX1_HUMAN | NPTX1 | Neuronal pentraxin-1 | 14.8 | 6 | 6 | 427.84 | 18.91 | Y | Y | N | Y | N | Cytoplasmic vesicle, secretory vesicle |
| COCH_HUMAN | COCH | Cochlin | 15.3 | 7 | 7 | 414.23 | 15.53 | Y | Y | N | Y | N | Secreted, extracellular |
| CATL1_HUMAN | CTSL1 | Cathepsin L1 | 17.7 | 5 | 5 | 238.15 | 15.31 | Y | Y | N | Y | N | Lysosome |
a Results of SignalP software prediction. Y represented signal peptide probability ≧0.9, and N represented < 0.9.
b Results of SecretomeP software prediction. Y represented NN-score ≧0.5, and N represented < 0.5.
c Results of TMHMM software prediction. Y represented sequence number of predicted TMHs ≧1, and N represented 0.
d Plasma proteome database. Y represented identified protein exists in database and N represented not.
e Human exosome database of ExoCarta. Y represented identified protein exists in database and N represented not.
f Subcellular locations of identified proteins classified by Gene Ontology Database.
Fig. 3.
Validation of the differential expressions of the 6 selected proteins. A, Fifty micrograms of protein samples were separated on SDS-PAGE gels, transferred to polyvinylidene difluoride membranes, and probed with the indicated antibodies. Every experiment was performed in triplicate. There were 2 and 4 CM proteins with high expression in CL1-0 and CL1-5, respectively. CALR, GLA, and PGAM1 are shown as CM loading controls. Equal amounts of each cell line with silver staining were used as CM loading controls as well. B, Immunohistochemical staining of A1AT revealed a trend of increasing stain intensity and a positive association with the TNM stage of lung adenocarcinoma tissues. In a well-differentiated case, the A1AT exhibits cytoplasmic staining, especially in the apical surface of the tumor cells (red arrow).
Table II displays the A1AT IHC results concerning the correlation of several clinical pathologic factors with the expression level of A1AT among 83 tissue specimens. The tissue specimens are all from primary diagnoses of lung adenocarcinoma from 2004 to 2010, including 49 early stage (TNM stage I and II) and 34 late stage (TNM stage III and IV) specimens. The expression levels of A1AT were significantly correlated with the early and late overall TNM stage and lymph node metastasis (Fig. 3B and supplemental Fig. S6). No significant correlations were found among age, sex, or the T or the M stage.
Table II. A1AT IHC results. The correlations between clinicalpathologic factors and the expression levels of A1AT.
| Variable | No. of patients (%) | A1AT Expressiona |
||
|---|---|---|---|---|
| Negative (%) | Positive (%) | p valueb | ||
| Age | ||||
| 21–49 | 7 (8) | 4 (57) | 3 (43) | 0.797 |
| 50–59 | 28 (34) | 13 (46) | 15 (54) | |
| 60–69 | 32 (39) | 13 (41) | 19 (59) | |
| 70–79 | 16 (19) | 8 (50) | 8 (50) | |
| Sex | ||||
| Male | 33 (40) | 13 (39) | 20 (61) | 0.376 |
| Female | 50 (60) | 25 (50) | 25 (50) | |
| Overall TNM stage | ||||
| I+II | 49 (59) | 28 (57) | 21 (43) | 0.015 |
| III+IV | 34 (41) | 10 (29) | 24 (71) | |
| T stage | ||||
| T1+T2 | 66 (80) | 31 (47) | 35 (53) | 0.787 |
| T3+T4 | 17 (20) | 7 (41) | 10 (59) | |
| N stage | ||||
| N0+N1 | 55 (66) | 33 (60) | 22 (40) | 0.0004 |
| N2+N3 | 28 (34) | 5 (18) | 23 (82) | |
| M stage | ||||
| M0 | 75 (90) | 32 (43) | 43 (57) | 0.134 |
| M1 | 8 (10) | 6 (75) | 2 (25) | |
a The A1AT IHC results were evaluated by the percentage of immunopositive stained cells (score 0–4) and the intensity of the cell staining (score 0–3). The score is their summation. A final score greater than 2 was defined as a positive staining.
b Chi-square test was employed to analyze all differences between groups. p < 0.05 was considered to have a significant difference.
A1AT Does Not Influences Lung Adenocarcinoma Cancer Cell Proliferation
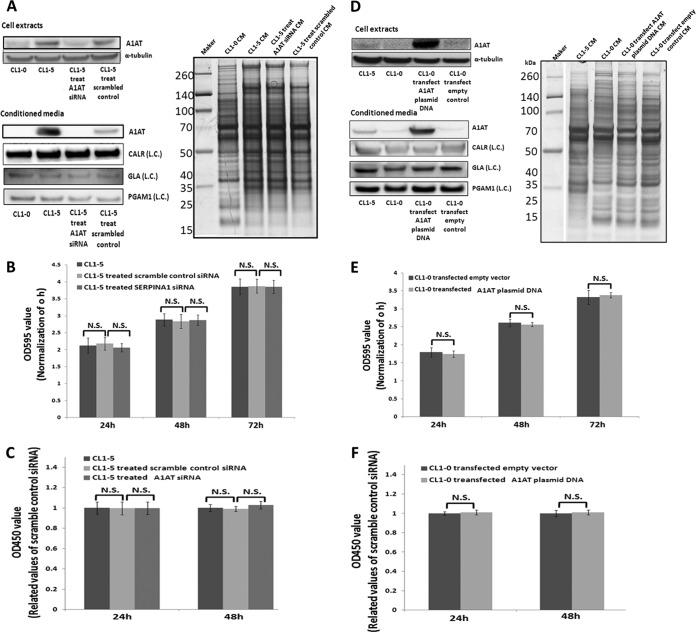
The initial step of metastasis requires proliferation of the primary tumor cells (46). To investigate A1AT's effect on tumor cell proliferation, A1AT was knocked down (Fig. 4A) and overexpressed (Fig. 4B) in CL1-5 and CL1-0 cells, respectively, and MTT (Figs. 4B and 4E) and BrdU assays (Figs. 4C and 4F) were performed to assess the effect of cell proliferation on CL1-0 (low invasive) and CL1-5 (high invasive) cells. Neither the reduction of A1AT in CL1-5 (Figs. 4B and 4C) nor the increase in the synthesis and secretion of A1AT in CL1-0 (Figs. 4E and 4F) had any influence on cancer cell proliferation from 24 to 72 h.
Fig. 4.
A1AT has no effect on lung adenocarcinoma cell proliferation. A, Western blot analysis indicated that the expression of A1AT was reduced using siRNA in the cell extract and the CM of the CL1-5 cell line. CALR, GLA, PGAM1, and SDS-PAGE sliver staining were the CM loading controls. (L.C.) indicates loading control. B, Cells (3 × 103 cells/well) were seeded onto 96-well plates. An MTT assay was performed at specific time points to evaluate cell proliferation after siRNA treatment. The results indicated no difference between the scrambled siRNA control group and the siRNA-treated A1AT group. C, BrdU assay. The results regarding the influences of cell proliferation after reduction of A1AT expression were reassessed. A1AT was overexpressed in CL1-0 cells to discern the influence on proliferation. D, The Western blot indicated that expression and secretion of A1AT was increased in CL1-0 cells via plasmid DNA transfection. E, and F, are the MTT and BrdU assays, respectively. The results reconfirmed the finding that A1AT did not regulate lung adenocarcinoma cell proliferation. N.S. indicates no significance.
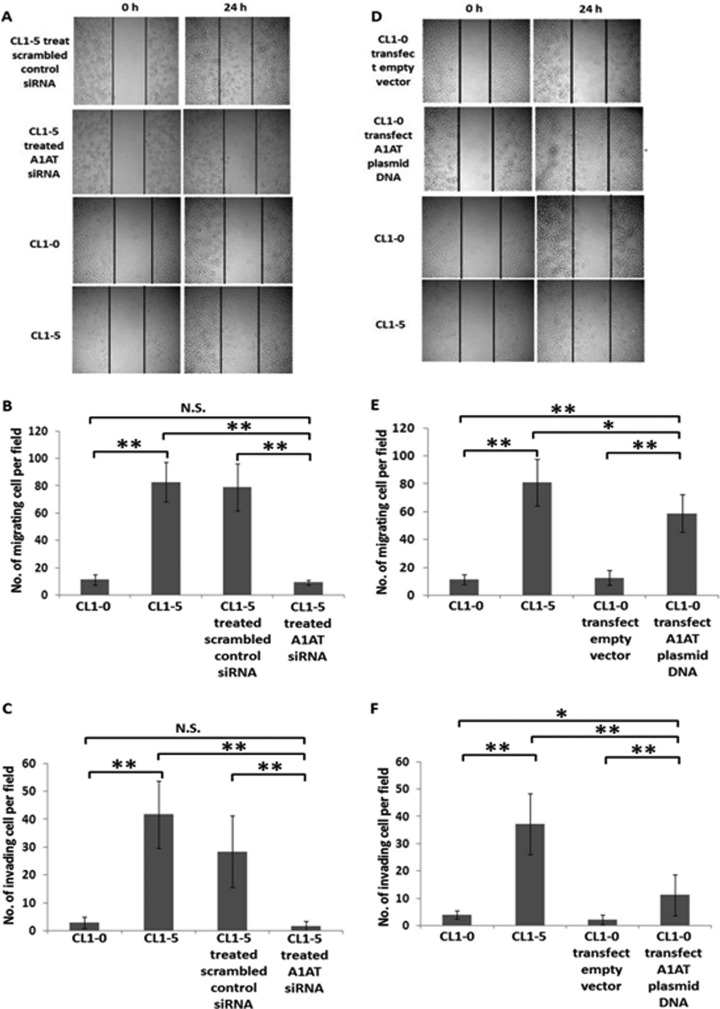
A1AT Promotes Cancer Migration and Invasion
Cancer cell migration and invasion play critical roles in metastasis and assist cancer cells as they disseminate from the primary sites (47). To further explore the possibility that the increased secretion of A1AT in the CL1-5 cells endowed them with increased migration and invasion abilities, we used siRNA to knock down the A1AT protein expression in CL1-5. We demonstrated that the reduced expression and secretion of A1AT in CL1-5 impeded both their migration and their invasion (Figs. 5A–5C). Consistently, when A1AT was overexpressed in CL1-0 cells, the migration and invasiveness of the cells were promoted (Figs. 5D–5F). The CL1-0 and CL1-5 cells, without any siRNA treatment or cDNA plasmid transfection, were used as the negative and positive controls, respectively.
Fig. 5.
A1AT prompts migration and invasiveness of lung adenocarcinoma cells. A description of A1AT knockdown and overexpression is provided in Figs. 4A and 4D, respectively. When A1AT was knocked down in CL1-5 cells, the migration and invasiveness of CL1-5 cells was reduced (A, B, and C). A, Cell wound healing assay. The extent of closure was photographed at 0 h and 24 h after treatment at 100× magnification. B, Transwell migration assay at 200× magnification. Migration was quantified by counting cells in six random fields per membrane. C, Matrigel invasion assay at 200× magnification. Invasion was quantified by counting cells in six random fields per membrane. Columns represent the average number of cells per field of at least eighteen fields from three independent experiments. Bars indicate standard deviation. In addition, A1AT was overexpressed in CL1-0 cells to investigate migration and invasiveness. Indeed, the migration and invasiveness of CL1-0 cells was increased in (D) the cell wound healing assay, (E) the transwell migration assay, and (F) the Matrigel invasion assay. All of the data indicate that A1AT protein affects the migration and invasion of lung adenocarcinoma cells. Each assay was performed in independent biological triplicates.
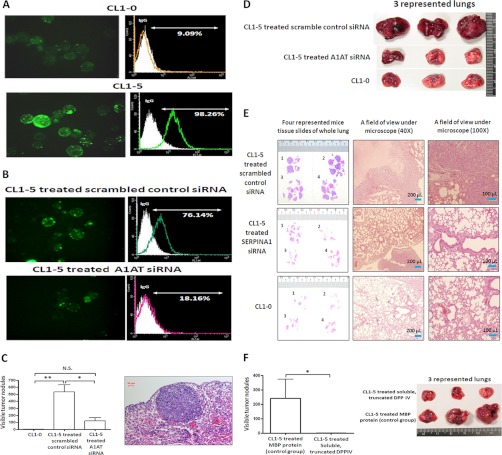
A1AT Influences the Assembly of Pericellular Polymeric FN on Suspended CL1-5 Lung Adenocarcinoma Cells and Promotes Their Abilities to Colonize the Lung
To further understand the functions or mechanisms of metastasis in which A1AT may be involved, protein-protein interactions were analyzed among the 30 differentially expressed proteins with high fold changes via the STRING 9.0 database (supplemental Fig. S7). The results indicated that FN1 had direct and/or indirect associations with A1AT. We previously demonstrated that pericellular polymeric FN (polyFN) is required for suspended breast cancer cells to colonize the lungs by adhering to DPP IV on the cell surface of lung capillary endothelia after intravenous injection (32, 33). To test whether the highly metastatic lung adenocarcinoma cell line CL1-5 is capable of assembling more pericellular polyFN than CL1-0, we performed cell surface immunofluorescence staining against FN1. We consistently found that CL1-5 cells had a significantly higher ability to assemble polyFN than did CL1-0 cells (Fig. 6A). To examine whether higher expression of A1AT in CL1-5 cells contributes to their increased ability to assemble pericellular polyFN and to validate the requirement of pericellular polyFN in promoting CL1-5 cell colonization in the lungs, we determined the effect of knocking down A1AT in CL1-5 cells on pericellular polyFN assembly and colonization ability. We demonstrated that the pericellular polyFN assembly of CL1-5 cells was markedly inhibited by transfecting siRNA against A1AT, whereas the control scramble siRNA failed to do so (Fig. 6B). We examined the effect on the lung colonization of CL1-5 cells by knocking down A1AT in immunocompromised nude mice. A1AT siRNA treatment significantly inhibited lung colonization. The number of colonies within the lungs revealed a similar trend (Figs. 6C and 6D). H&E staining of mouse lungs confirmed that the lung surface nodules were indeed colonized cancer tissue (Fig. 6E). Additionally, significant inhibition of lung colonization occurred when CL1-5 cells were treated with soluble, truncated DPP IV, suggesting that the CL1-5 cells colonize the lungs via their pericellular polyFN and bind to endothelia DPP IV (Fig. 6F).
Fig. 6.
A1AT triggers the assembly of pericellular polyFN on lung adenocarcinoma cells to promote lung colonization. A, CL1-0 and CL1-5 cells were trypsinized and placed in a suspension culture for 2 h, stained with rabbit anti-FN antibodies followed by a GFP-conjugated donkey anti-rabbit IgG secondary antibody, observed under a fluorescent microscope, and detected using flow cytometry. Both IF and flow cytometry was used to demonstrate the different levels of FN1 assembly on the cell surfaces of CL1-0 and CL1-5 cells. The assembly levels of pericellular polyFN of CL1-5 cells were higher than those of CL1-0. The Olympus FB1000 microscope was used, and the fluorescent filter set for 488 excitation at 600× magnification. B, Evaluation of polyFN1 assembly on the cell surface after scrambled siRNA control and A1AT siRNA transfection was performed using IF and flow cytometry. The pericellular polyFN assembly of CL1-5 cells was inhibited when the expression of A1AT was reduced by transfecting siRNA. C, In the nude mice model, a tail vein injection was performed using the CL1-5 cells treated with the scrambled siRNA control and A1AT siRNA or CL1-0 cells, using five mice for each treatment. After 8 weeks, the mice were sacrificed and the tumor nodules of their lungs were measured. A tumor size of 0.5 mm in diameter was counted as one nodule. A significant inhibition of lung experimental metastasis was observed in mice that were injected with A1AT siRNA CL1-5 cells. Three of five lungs are shown in (D). E, Represented tumor sections of these three groups were stained with H&E. F, CL1-5 cells were incubated in suspension for 2 h in 20% FBS media. The cells were subsequently treated with myelin basic protein (MBP) and soluble, truncated dipeptidyl peptidase IV (DPP IV) for 20 min. The five mice in each group were sacrificed after 8 weeks, and the lungs were removed and fixed in a 3.7% formalin fixative. On injection of the soluble, truncated DPP IV-treated CL1-5 cells, lung colonization among the nude mice was completely inhibited. There are three representative lungs shown in each group.
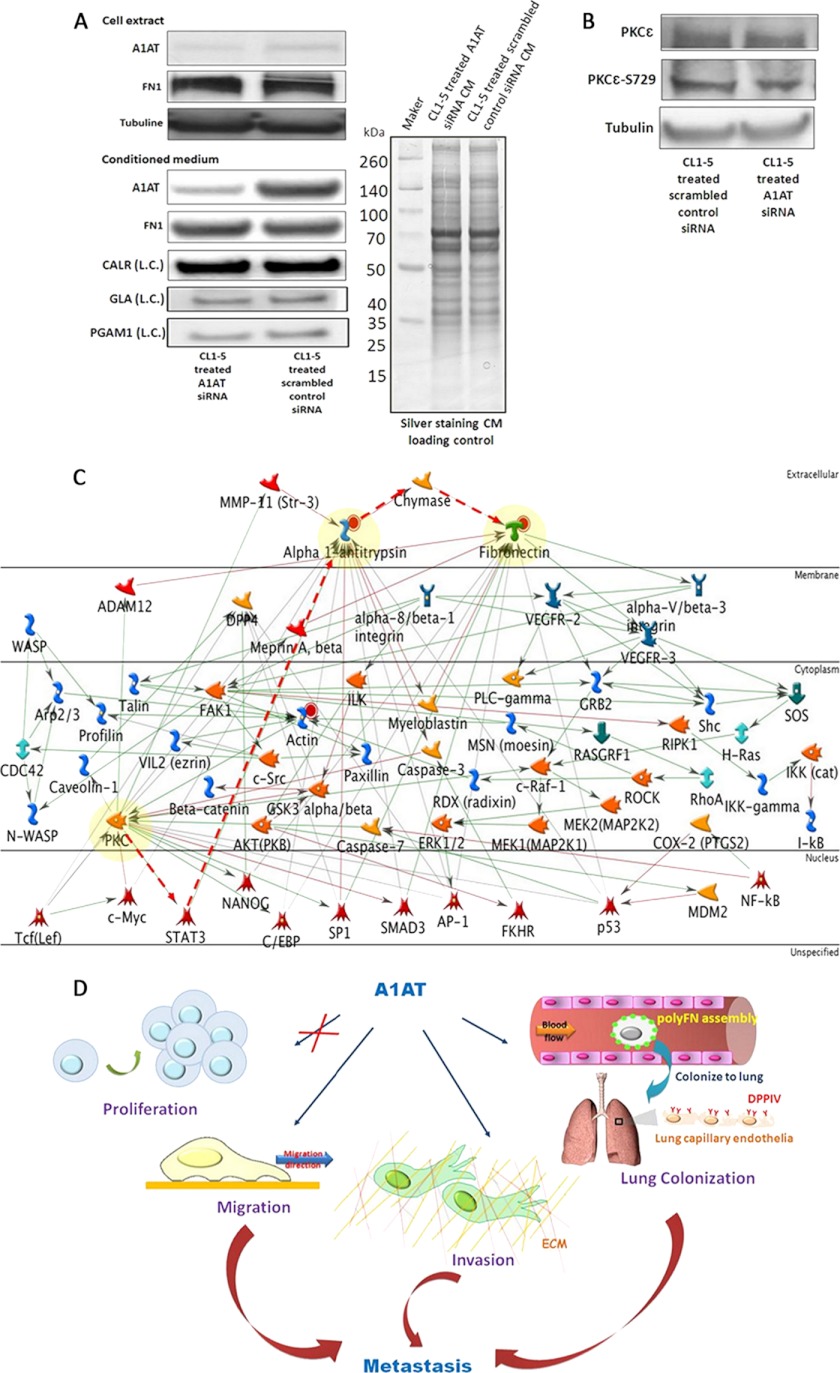
The Possible Mechanisms of A1AT That Influence the Assembly of Pericellular Polymeric FN
Knockdown of A1AT successfully impeded polyFN assembly on CL1-5 cell surfaces (Fig. 6B), and this could be achieved without influencing their FN1 synthesis or secretion (Fig. 7A). Phosphorylated PKC epsilon (PKCε) promotes lung colonization by triggering polyFN assembly (48), and there are three possibilities by which PKCε directs lung colonization in lung adenocarcinoma cells via pericellular polyFN assembly: 1) A1AT regulates PKCε activity; 2) phosphorylated PKCε regulates A1AT functions; or 3) A1AT is independent of PKCε in regulating polyFN assembly. The first possibility was inspected via siRNA reduction of A1AT expression levels followed by Western blot. The data indicate no variation among the phosphorylated PKCε as these proteins were knocked down (Fig. 7B). This would suggest that A1AT may be regulated by either phosphorylated PKCε or independent regulation of polyFN assembly, the two remaining possibilities. Unfortunately, there is lack of specific kinase inhibitors of PKCε to investigate the second possibility. Thereby, to deduce the relationship between PKCε and A1AT regarding the regulation of migration/invasion and polyFN assembly in lung colonization, we employed the Metacore interactome prediction software (GeneGo, Joseph, MI) (Fig. 7C).
Fig. 7.
The proposed mechanisms of A1AT for modulating experimental metastasis in lung adenocarcinoma.
A, A1AT influenced polyFN1 assembly but not FN1 synthesis and secretion, as determined by siRNA combined with Western blot analysis. CALR, GLA, PGAM1, and SDS-PAGE silver staining served as the CM loading controls. The abbreviation (L.C.) indicates the loading control. B, The phosphorylation level of PKCε was not affected upon A1AT knockdown in the CL1-5 cells. C, The proposed mechanism of A1AT-modulated polyFN1 assembly on cell surfaces via phosphorylated PKCε regulates A1AT. The Metacore interactome prediction software was used to predict the interaction with PKCε, A1AT, and FN1. The symbols and links shown on the network illustration include the following: transcription factor ( ), generic kinase (
), generic kinase ( ), metalloprotease (
), metalloprotease ( ), generic protease (
), generic protease ( ), generic binding protein (
), generic binding protein ( ), lipid phosphatase (
), lipid phosphatase ( ), generic receptor (
), generic receptor ( ), receptor ligand (
), receptor ligand ( ), positive effect (
), positive effect ( ), negative effect (
), negative effect ( ), unspecified effect (
), unspecified effect ( ), the proposed possible pathway (
), the proposed possible pathway ( ). D, This scheme summarizes the functions in which A1AT is involved when facilitating lung adenocarcinoma experimental metastasis. A1AT mediates migration and invasion abilities but not the proliferation of cancer cells. Additionally, it regulates polyFN assembly on lung adenocarcinoma cell surfaces to colonize on the lung via adhering to DPP IV on lung capillary endothelia.
). D, This scheme summarizes the functions in which A1AT is involved when facilitating lung adenocarcinoma experimental metastasis. A1AT mediates migration and invasion abilities but not the proliferation of cancer cells. Additionally, it regulates polyFN assembly on lung adenocarcinoma cell surfaces to colonize on the lung via adhering to DPP IV on lung capillary endothelia.
DISCUSSION
Cancer cell metastasis is a major cause of cancer death. Unfortunately, the underlying molecular mechanisms remain enigmatic, which results in a lack of efficient therapy and prevention. Nevertheless, proteins secreted by cancer cells are known to play key roles in the promotion of tumor transformation and progression (49, 50). In this study, we hypothesized that cells can produce factors that promote or suppress cancer cell metastasis-related functions, such as migration and invasiveness. Indeed, the data indicated that CL1-0 and CL1-5 cell migration (Fig. 1A) and invasiveness (Fig. 1B) were influenced when conditioned with the other's CM. Interestingly, the results reveal that the large variation and the invasiveness of CL1-5 cells exhibited no effect after CL1-0 CM conditioning. This may be caused by the cells continually producing their own original factors. A total of 660 CM proteins secreted by the lung adenocarcinoma cell lines CL1-0 (low metastatic) and CL1-5 (highly metastatic) were characterized, and the proteins within the two cells lines that passed multiple hypothesis tests were considered differentially expressed. Sixty-eight unique proteins were expressed at high levels in the CL1-0 and CL1-5 cell lines (supplemental Table S3). Previous studies have indicated that two of the proteins among these 68, connective tissue growth factor and COL6A1, have been known to take on their roles as suppressor and promoter in metastasis, respectively, in the CL1-0 and CL1-5 cell lines (30, 51). The related publications of the 68 metastasis-related proteins are listed in supplemental Table S4. These findings provide credibility to our quantified results, generated by secretomic analysis along with the label-free quantitative approach, regarding the discovery of metastasis-associated proteins. Among the 68 proteins, 30 proteins were considered high confidence as metastasis-associated secretory proteins. These 30 were selected because they have at least two pieces of evidence for their role as metastasis-associated secretory proteins as suggested by the database and/or prediction software, did not exhibit functions of metastasis in the lung adenocarcinoma CL1 cell line, and have significantly greater fold change (Table I).
The CM should predominantly contain extracellular and membrane-bound proteins (9). Using subcellular localization and prediction software, the secretion of these proteins was observed. Collectively, 55.82% of the proteins can be released into the extracellular space by signal peptide-directed pathways, non-signal peptide-directed pathways, and/or membrane protein shedding. A small portion of intracellular proteins in the secretome analysis has been associated with the nonspecific liberation of cytoplasmic proteins as a consequence of cell lysis (Fig. 2A). The presence of many intracellular proteins in the CM may also be associated with an active release of proteins through nonclassical secretion mechanisms or unknown pathways (52–54). Recent evidence suggests that various mechanisms, such as cargo protein delivery, targeting motif interactions, membrane flip-flop, distinct transport systems, and intracellular vesicles release, e.g. exosomes, may contribute to the presence of intracellular proteins in the CM (55).
Recently, studies of exosomes have emerged as an important research topic, particularly in the fields of immunology and oncology (10, 11, 56–59). Exosome proteins include cytoplasmic, nuclear, and membrane proteins (39, 40). According to the human exosome database in ExoCarta, 59.24% of the 391 identified proteins in our study are generally thought to be exosomal proteins (Fig. 2C). Western blot analysis confirmed the existence of exosomes in our CM samples (Fig. 2D). The presence of high levels of cytoplasmic and nuclear proteins in the secretomes of cancer cells may indicate that a non-classical exosome secretion pathway is highly activated. In the data from previous research, evidence has been provided to suggest that hypoxic tumor cells actually secrete exosomes, which are capable of modulating their microenvironment and successfully facilitate angiogenesis and metastasis potential (59). Therefore, this observation suggests that further studies are necessary to determine whether the exosomes affect cancer metastasis.
The CM proteins with high expression levels in the cells that have high invasive/metastatic properties are potential serum marker candidates for predicting lung adenocarcinoma metastasis, drug targets for inhibiting metastasis, and important prometastatic factors for a more detailed understanding of metastasis mechanisms. Of the 30 proteins that were expressed at higher levels in the CL1-5 cells, certain proteins are known to play key roles in lung adenocarcinoma metastasis (supplemental Table S4).
A1AT was selected as a potentially important metastasis-promoting secretory protein that is not only novel in lung adenocarcinoma but also less recognized in other types of cancer. A protease inhibitor among the serpin family, alpha-1-antitrypsin (A1AT) is more commonly known as a serum trypsin inhibitor (60). A common disease caused by A1AT is alpha-1 antitrypsin deficiency. This autosomal recessive genetic disorder is caused by the defective production of A1AT, which causes decreased A1AT activity in the blood and, eventually, the lungs. With this decrease, neutrophil elastase will start to break down elastin, compromising the elasticity of the lungs, resulting in serious respiratory complications. The list of further health risks includes emphysema and chronic obstructive pulmonary disease (61, 62). Previous studies have also determined that A1AT deficiency increases the risk of lung cancer (63). However, the definitive role of A1AT in cancer progression is still unknown. Previous studies indicate that A1AT has a different molecular weight in its intracellular (∼50 kDa) and extracellular forms (mature form, ∼55 kDa) because of differential glycosylation (64, 65). Additionally, there are various biosynthesis productions of A1AT because of polymerization, post-translational modification and genetic variants (66–69). This would provide several bands that can be detected in the cell lysate via Western blot (supplemental Fig. S5).
Clinical research has been revealed that the serum levels of A1AT are higher in cancer patients than in healthy individuals (70–73). Additionally, A1AT is involved in the distant metastasis of various cancer types in clinical expression, including ovarian, cervical, colorectal, breast, and lung adenocarcinomas (74–78). The tissue specimens of primary site were used to perform the TMAs followed by IHC for investigation of A1AT expression levels in lung adenocarcinoma patients. Protein expression levels within the primary site of tissues may reveal information regarding the mechanisms by which cancer cells leave the primary site and spread to distant sites. In previous research, cancer cells revealed different properties in the primary and distant sites, providing clues about the nature of this process. For instance, epithelial-to-mesenchymal transition (EMT) is a critical step in which cancer cells gain the ability to leave the primary site (79–81). As another example, mesenchymal-epithelial transition (MET) involves the transition from spindle-shaped mesenchymal cells to planar arrays of polarized epithelia cells. This major shift plays a role in the establishment and stabilization of distant metastases in that it allows cancerous cells to attain epithelial properties and integrate into distant organs (81, 82). Although the mechanisms that allow cells to locate themselves on distant sites are very important, our research focuses on the mechanisms that allow cancer cells to disseminate from the primary site. In our A1AT IHC results, A1AT exhibits significant correlations with overall TNM stages and lymph node metastasis. This would raise the accuracy of clinical stage determination and assist doctors when deciding the proper treatments for their patients. In addition, an increase in the level of A1AT may be a prognostic factor for monitoring patients' progression after treatment (83). However, the exact clinical usage of A1AT would require a greater sample size. There was no significant connection to distant metastasis (M stage), which may have been the result of a sample size only consisting of eight patients in the M1 stage (with distant metastasis). Notably, among the eight patients in the M1 stage, three patients were in the early N stage (N0-N1), and the A1AT expressions in all cases was negative. Interestingly, among the five M1 stage patients in the late N stage (N2-N3), A1AT expression among three patients were also negative. In total, among the 28 patients within the late N stage, the A1AT expression of only five patients was negative, three of which belonged to patients in the M1 stage. The results may hint that when cancer cells locate themselves in the primary site, they secrete A1AT to facilitate migration and invasiveness in lung adenocarcinoma. However, when cancer cells prepare to enter the processes of distant metastasis, various pathways may be triggered to reduce A1AT secretion from the cancer cells at the primary site. Previous research has demonstrated that proteins can express multiple functions to mediate different stages of cancer development (84–86). For instance, the protein Box 1 has different functions at the primary site and in distant organs (87). Another possibility is that different metastatic mechanisms produce different A1AT levels, including lymphatic or hematogenous dissemination (88). Here, we show for the first time that the colony formation of the lung adenocarcinoma cells is regulated by the expression of A1AT.
The complex process of metastasis is a multi-stage process in which cancer cells proliferate, migrate, invade surrounding microenvironments, intravasate, transit in the blood or lymph, and extravasate and form a growth at a new site (89). Within lung cancer, common sites of metastasis include the brain, pleura, contralateral lung, etc (90). However, it is still unclear how the lung adenocarcinoma cells metastasize to other sites. For this reason, it was necessary to find at exactly which step A1AT participates in facilitating metastasis. The results indicated that A1AT regulates lung adenocarcinoma cell migration and invasion but not proliferation (Figs. 4 and 5). A1AT may regulate migration/invasion via mediation of chymase activity. Chymase is widely studied as it is expressed on mast cells, as only mast cells appear capable of accumulating chymases in secretory granules. According to previous studies, chymase can activate angiotensin II, MMP2, and MMP9 to promote tumor cell angiogenesis, migration, and invasion or cleave and inactivate TIMP1 (91–94). In our proteomics data, TIMP1, MMP2, and A1AT are highly expressed in CL1-5 cells. In the body's microenvironment, lung adenocarcinoma cells may release high levels of A1AT and inhibit the chymase activity of surrounding mast cells. The situation may further trigger tumor cells to release TIMP1 proteins, thereby reducing MMP2. This will trigger other mechanisms to increase MMP2 and begin facilitating cancer metastasis, such as TNF-α (95). The association of chymase activity and A1AT is certainly worthy of further research. Although migration and invasion are essential cellular events within the solid tissues in the facilitation of cancer metastasis (96), once tumor cells are traveling in the circulation, they are in a suspended state and require additional cellular activities to enable their colonization in distant organs (97). Hence, bioinformatics was utilized to uncover possible mechanisms of metastasis in which A1AT is involved. The protein-protein interaction database (STRING) provided a feasible association between A1AT and FN1.
PolyFN assembly on suspended cancer cell surfaces is required for rat breast cancer cells to colonize the lungs by adhering to dipeptidyl peptidase IV (DPP IV) expressed on lung capillary endothelia. The specificity of DPP IV/polyFN adhesion in lung colonization was confirmed by reduced colonization ability when the tumor cells were treated with soluble, truncated DPP IV or when the tumor cells were injected into Fischer 344/CRJ rats, a rat sub-strain with a significantly decreased lung endothelial DPP IV expression (32, 33, 98). The supposition was made that polyFN assembly may be a significant factor in the migration of lung adenocarcinoma cells toward the lungs. To be more precise, the same mechanism by which lung adenocarcinoma cells metalize to the lung was validated in CL1-5 cells, which exhibited a high invasive ability (Fig. 6A). We not only found a novel mechanism in the experimental metastasis of lung adenocarcinoma cells but also discovered a novel molecule, A1AT, that regulates this mechanism. Additionally, we observed that knocking down A1AT impeded polyFN assembly on CL1-5 cell surfaces (Fig. 6B) without influence their FN1 synthesis and secretion (Fig. 7A). Remarkably, after A1AT was overexpressed in the CL1-0 cell line, the assembly level of polyFN was uninfluenced after 2 h in suspended condition (data not shown). Trypsinization of adherent cells may cleave cell surface proteins and receptors for the assembly of polyFN on the cell surface, such as integrin α5β1 (99–101). Therefore, the time of suspension culture for CL1-0 cells with overexpression of A1AT was extended for 4, 6, 12, and 24 h to regenerate their receptors for optimal polyFN assembly. However, the levels of polyFN assembly were unchanged in all of these conditions. The results proved to be the equivalent of our previous findings in breast cancer (32). This outcome implies that more than one molecule is required to trigger polyFN assembly on lung adenocarcinoma cell surfaces. Furthermore, cell migration and invasion can be promoted by the over-expression of A1AT. However, this is not the case for pericellular polyFN assembly. These results indicate that A1AT may have specific functions within different processes of experimental metastasis, and the association between migration/invasion and pericellular polyFN assemblies may be independent of each other. LAMA3, which was identified as a high-expression protein in CL1-5 cells, might be one of the possible molecules involved in polyFN assembly (102, 103). The findings are worthy of further investigation.
Phosphorylated PKC epsilon (PKCε) is known to promote lung colonization via polyFN assembly (48). The data refuted the possibility of A1AT regulates PKCε activity (Fig. 7B). There are insufficient specific kinase inhibitors of PKCε to investigate the possibility that phosphorylated PKCε regulates A1AT functions. To predict the relationship between PKCε and A1AT in the regulation of polyFN assembly in lung colonization, Metacore interactome prediction software (GeneGo, Joseph, MI, USA) was used (Fig. 7C). Various putative pathways were thus revealed in connecting the signaling flows between PKCε and A1AT. For example, Stat3 may serve as a downstream effecter of PKCε in regulating A1AT (104, 105). Furthermore, A1AT may possibly prevent cell surface-fibronectin connection disruption by inhibiting chymase activity for polyFN assembly mediation (106, 107). If the hypothesis that PKCε regulates A1AT to direct lung colonization of lung adenocarcinoma turn out to be false, the possibility that there may be independent regulatory mechanisms for PKCε and A1AT may then represent the means by which they both promote lung colonization of lung adenocarcinoma. The discovery of these possible mechanisms merits further investigation for the greater understanding of metastasis.
In summary, we used a proteomics approach to gain a new perspective on the metastasis-associated secretory proteins from lung adenocarcinoma cell lines. The protein A1AT exhibited high expression levels in CL1-5 cells and was selected according to its essentiality, novelty and association with migration/invasion in the promotion of metastasis. The metastasis-promoting activities of migration/invasion and pericellular polyFN assembly were higher in CL1-5 than in CL1-0 cells. A1AT significantly regulated the migratory and invasive properties, the polyFN assembly, and the lung colonization potential of the CL1-0 and/or CL1-5 cells (Fig. 7D). The quantitative proteomics analysis of the CL1-0/CL1-5 secretomes provided a pivotal tool to unveil the biological significance of A1AT in human lung adenocarcinoma experimental metastasis, which may further contribute to the underlying mechanisms of lung cancer metastasis and to the identification of new therapeutic targets for the effective treatment of lung cancer patients.
Supplementary Material
Acknowledgments
We thank the National Cheng-Kung University Proteomics Research Core Laboratory for assistance in mass spectrometry analysis for protein identification and Mr. Corbett Hart Moy from Graduate Institute of Teaching Chinese as a Second Language (at the National Taiwan Normal University) for help in English editing. Proteomic mass spectrometry analyses were performed by the Core Facilities for Protein Structural Analysis located at the Institute of Biological Chemistry, Academia Sinica, supported by a National Science Council grant (NSC100-2325-B-001-029) and the Academia Sinica. Computational analyses and data mining were performed using the system provided by the Bioinformatics Core at the National Cheng Kung University, supported by the National Science Council, Taiwan. We are grateful for the support from the Tissue Bank, Research Center of Clinical Medicine, National Cheng Kung University Hospital.
Footnotes
* This study was supported by Grant NSC-99-2923-M-006-001-MY3, 101-2325-B-006-003, and Grant 100-2113-M-006-002-MY3 (Liao, PC) from the National Science Council; Landmark Project of National Cheng Kung University and the National Cheng Kung University Project of Promoting Academic Excellence & Developing World Class Research Centers (Liao, PC) from the Ministry of Education of Taiwan; and Grants NSC95-2320-B-006-075-MY3, NSC99-2320-B-006-028-MY3, and NSC99-2627-B-006-023 (Cheng, HC) from the National Science Council. It was also partially supported by Sustainable Environment Research Center (Liao, PC).
 This article contains supplemental Figs. S1 to S7 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S7 and Tables S1 to S3.
1 The abbreviations used are:
- FBS
- Fetal bovine serum
- A1AT
- Alpha 1-antitrypsin
- ANOVA
- Analysis of variance
- BrdU
- Bromodeoxyuridine (5-bromo-2′-deoxyuridine
- BSA
- Bovine serum albumin
- CID
- Collision-induced dissociation
- CM
- Conditioned medium
- Da
- Dalton
- DPP IV
- dipeptidyl peptidase IV
- DTT
- Dithiothreitol
- Exo
- Exosome
- FDR
- False-discovery rate
- FN1
- Fibronectin
- H&E
- Hematoxylin and eosin stain
- IF
- Immunofluorescence
- IHC
- Immunohistochemistry
- LC
- Liquid chromatography
- MBP
- Myelin basic protein
- MMP2
- 72 kDa type IV collagenase
- MS/MS
- Tandem mass spectrometry
- MTT
- 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
- PBS
- Phosphate buffered saline
- PKCε
- Protein kinase C epsilon
- polyFN
- Polymeric fibronectin
- ppm
- Parts per million
- SB
- sequential Bonferroni
- SD
- Standard deviation
- SFM
- Serum-free medium
- siRNA
- Small interfering RNA
- Stat3
- Signal transducer and activator of transcription 3
- TMA
- Tissue microarray
- TBST
- Tris-buffered saline and tween 20
- THBS1
- Thrombospondin 1
- TIMP1
- Metalloproteinase inhibitor 1
- TMHMM
- Transmembrane hidden Markov model
- TNF-α
- Tumor necrosis factor-alpha
- TNM
- The TNM classification of malignant tumors.
REFERENCES
- 1. Hanahan D., Weinberg R. A. (2000) The hallmarks of cancer. Cell. 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 2. Sugiura H., Yamada K., Sugiura T., Hida T., Mitsudomi T. (2008) Predictors of survival in patients with bone metastasis of lung cancer. Clin. Orthop. Relat. Res. 466, 729–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D'Addario G., Früh M., Reck M., Baumann P., Klepetko W., Felip E., and ESMO Guidelines Working Group (2010) Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 21, 116–119 [DOI] [PubMed] [Google Scholar]
- 4. Woodhouse E. C., Chuaqui R. F., Liotta L. A. (1997) General mechanisms of metastasis. Cancer 80, 1529–1537 [DOI] [PubMed] [Google Scholar]
- 5. Box G. M., Eccles S. A. (2011) Simple experimental and spontaneous metastasis assays in mice. Methods Mol Biol. 769, 311–329 [DOI] [PubMed] [Google Scholar]
- 6. Khanna C., Hunter K. (2005) Modeling metastasis in vivo. Carcinogenesis 26, 513–523 [DOI] [PubMed] [Google Scholar]
- 7. Fidler I. J., (2006) Models for spontaneous metastasis. Cancer Res. 66, 9787. [DOI] [PubMed] [Google Scholar]
- 8. Tjalsma H., Bolhuis A., Jongbloed J. D., Bron S., van Dijl J. M. (2000) Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64, 515–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Volmer M. W., Stühler K., Zapatka M., Schöneck A., Klein-Scory S., Schmiegel W., Meyer H. E., Schwarte-Waldhoff I. (2005) Differential proteome analysis of conditioned media to detect Smad4 regulated secreted biomarkers in colon cancer. Proteomics. 5, 2587–2601 [DOI] [PubMed] [Google Scholar]
- 10. Simpson R. J., Lim J. W., Moritz R. L., Mathivanan S. (2009) Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics 6, 267–283 [DOI] [PubMed] [Google Scholar]
- 11. Mathivanan S., Lim J. W., Tauro B. J., Ji H., Moritz R. L., Simpson R. J. (2010) Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell. Proteomics 9, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liotta L. A., Kohn E. C. (2001) The microenvironment of the tumour-host interface. Nature 411, 375–379 [DOI] [PubMed] [Google Scholar]
- 13. Nomura T., Katunuma N. (2005) Involvement of cathepsins in the invasion, metastasis and proliferation of cancer cells. J. Med. Invest. 52, 1–9 [DOI] [PubMed] [Google Scholar]
- 14. Lane T. F., Sage E. H. (1994) The biology of SPARC, a protein that modulates cell-matrix interactions. FASEB J. 8, 163–173 [PubMed] [Google Scholar]
- 15. De S., Chen J., Narizhneva N. V., Heston W., Brainard J., Sage E. H., Byzova T. V. (2003) Molecular pathway for cancer metastasis to bone. J. Biol. Chem. 278, 39044–39050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang Y. H., Wu C. C., Chang K. P., Yu J. S., Chang Y. C., Liao P. C. (2009) Cell secretome analysis using hollow fiber culture system leads to the discovery of CLIC1 protein as a novel plasma marker for nasopharyngeal carcinoma. J. Proteome Res. 8, 5465–5474 [DOI] [PubMed] [Google Scholar]
- 17. Grant S. G., Blackstock W. P. (2001) Proteomics in neuroscience: from protein to network. J. Neurosci. 21, 8315–8318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu C. C., Hsu C. W., Chen C. D., Yu C. J., Chang K. P., Tai D. I., Liu H. P., Su W. H., Chang Y. S., Yu J. S. (2010) Candidate serological biomarkers for cancer identified from the secretomes of 23 cancer cell lines and the human protein atlas. Mol. Cell. Proteomics 9, 1100–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grønborg M., Kristiansen T. Z., Iwahori A., Chang R., Reddy R., Sato N., Molina H., Jensen O. N., Hruban R. H., Goggins M. G., Maitra A., Pandey A. (2006) Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol. Cell. Proteomics 5, 157–171 [DOI] [PubMed] [Google Scholar]
- 20. Xue H., Lü B., Zhang J., Wu M., Huang Q., Wu Q., Sheng H., Wu D., Hu J., Lai M. (2010) Identification of serum biomarkers for colorectal cancer metastasis using a differential secretome approach. J. Proteome Res. 9, 545–555 [DOI] [PubMed] [Google Scholar]
- 21. Sintiprungrat K., Singhto N., Sinchaikul S., Chen S. T., Thongboonkerd V. (2010) Alterations in cellular proteome and secretome upon differentiation from monocyte to macrophage by treatment with phorbol myristate acetate: insights into biological processes. J. Proteomics 73, 602–618 [DOI] [PubMed] [Google Scholar]
- 22. Keller M., Rüegg A., Werner S., Beer H. D. (2008) Active caspase-1 is a regulator of unconventional protein secretion. Cell 132, 818–831 [DOI] [PubMed] [Google Scholar]
- 23. West G. M., Tucker C. L., Xu T., Park S. K., Han X., Yates J. R., 3rd, Fitzgerald M. C. (2010) Quantitative proteomics approach for identifying protein-drug interactions in complex mixtures using protein stability measurements. Proc. Natl. Acad. Sci. U.S.A. 107, 9078–9082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang P. C., Luh K. T., Wu R., Wu C. W. (1992) Characterization of the mucin differentiation in human lung adenocarcinoma cell lines. Am. J. Respir. Cell Mol. Biol. 7, 161–171 [DOI] [PubMed] [Google Scholar]
- 25. Kao Y. R., Shih J. Y., Wen W. C., Ko Y. P., Chen B. M., Chan Y. L., Chu Y. W., Yang P. C., Wu C. W., Roffler S. R. (2003) Tumor-associated antigen L6 and the invasion of human lung cancer cells. Clin. Cancer Res. 9, 2807–2816 [PubMed] [Google Scholar]
- 26. Shih J. Y., Yang S. C., Hong T. M., Yuan A., Chen J. J., Yu C. J., Chang Y. L., Lee Y. C., Peck K., Wu C. W., Yang P. C. (2001) Collapsin response mediator protein-1 and the invasion and metastasis of cancer cells. J. Natl. Cancer Inst. 93, 1392–1400 [DOI] [PubMed] [Google Scholar]
- 27. Wang C. C., Tsai M. F., Hong T. M., Chang G. C., Chen C. Y., Yang W. M., Chen J. J., Yang P. C. (2005) The transcriptional factor YY1 upregulates the novel invasion suppressor HLJ1 expression and inhibits cancer cell invasion. Oncogene 24, 4081–4093 [DOI] [PubMed] [Google Scholar]
- 28. Kuo J. C., Wang W. J., Yao C. C., Wu P. R., Chen R. H. (2006) The tumor suppressor DAPK inhibits cell motility by blocking the integrin-mediated polarity pathway. J. Cell Biol. 172, 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu H. Y., Chang Y. H., Chang Y. C., Liao P. C. (2009) Proteomics analysis of nasopharyngeal carcinoma cell secretome using a hollow fiber culture system and mass spectrometry. J. Proteome Res. 8, 380–389 [DOI] [PubMed] [Google Scholar]
- 30. Chiu K. H., Chang Y. H., Wu Y. S., Lee S. H., Liao P. C. (2011) Quantitative secretome analysis reveals that COL6A1 is a metastasis-associated protein using stacking gel-aided purification combined with iTRAQ labeling. J. Proteome Res. 10, 1110–1125 [DOI] [PubMed] [Google Scholar]
- 31. Goldstein N. S., Ferkowicz M., Odish E., Mani A., Hastah F. (2003) Minimum formalin fixation time for consistent estrogen receptor immunohistochemical staining of invasive breast carcinoma. Am. J. Clin. Pathol. 120, 86–92 [DOI] [PubMed] [Google Scholar]
- 32. Cheng H. C., Abdel-Ghany M., Elble R. C., Pauli B. U. (1998) Lung endothelial dipeptidyl peptidase IV promotes adhesion and metastasis of rat breast cancer cells via tumor cell surface-associated fibronectin. J. Biol. Chem. 273, 24207–24215 [DOI] [PubMed] [Google Scholar]
- 33. Cheng H. C., Abdel-Ghany M., Pauli B. U. (2003) A novel consensus motif in fibronectin mediates dipeptidyl peptidase IV adhesion and metastasis. J. Biol. Chem. 278, 24600–24607 [DOI] [PubMed] [Google Scholar]
- 34. Han C. L., Chen J. S., Chan E. C., Wu C. P., Yu K. H., Chen K. T., Tsou C. C., Tsai C. F., Chien C. W., Kuo Y. B., Lin P. Y., Yu J. S., Hsueh C., Chen M. C., Chan C. C., Chang Y. S., Chen Y. J. (2011) An informatics-assisted label-free approach for personalized tissue membrane proteomics: case study on colorectal cancer. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.003087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Holm S. (1979) A simple sequentially rejective multiple test procedure. Scand. J. Statist. 6, 65–70 [Google Scholar]
- 36. Thomas N. P., Søren B., Gunnar V. H., Henrik N. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 37. Bendtsen J. D., Jensen L. J., Blom N., Von Heijne G., Brunak S. (2004) Feature based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356 [DOI] [PubMed] [Google Scholar]
- 38. Möller S., Croning M. D., Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. (2001) Bioinformatics 17, 646–653 [DOI] [PubMed] [Google Scholar]
- 39. Mathivanan S., Simpson R. J. (2009) ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000 [DOI] [PubMed] [Google Scholar]
- 40. Mathivanan S., Ji H., Simpson R. J. (2010) Exosomes: extracellular organelles important in intercellular communication. J. Proteomics 73, 1907–1920 [DOI] [PubMed] [Google Scholar]
- 41. Anderson N. L., Polanski M., Pieper R., Gatlin T., Tirumalai R. S., Conrads T. P., Veenstra T. D., Adkins J. N., Pounds J. G., Fagan R., Lobley A. (2004) The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol. Cell. Proteomics 3, 311–326 [DOI] [PubMed] [Google Scholar]
- 42. Muthusamy B., Hanumanthu G., Reshmi R., Sriranjini S., Suresh S., Rekha B., Srinivas D., Karthick L., Vrushabendra B. M., Sharma S., Mishra G., Rashmi B. P., Kadekar S., Chatterjee P., Mangala K. S., Shivashankar H. N., Chandrika K. N., Deshpande N., Suresh M., Kannabiran N., Niranjan V., Nalli A., Prasad T. S. K., Arun K. S., Reddy R., Chandran S., Jadhav T., Julie D., Mahesh M., John S. L., Palvankar K., Sudhir D., Bala P., Jithesh M. K., Rashmi N. S., Vishnupriya G., Dhar K., Reshma S., Chaerkady R., Prasad C. K., Gandhi T. K. B., Harsha H. C., Mohan S. S., Deshpande K. S., Sarker M., Pandey A. (2005) Plasma proteome database as a resource for proteomics research. Proteomics 5, 3531–3536 [DOI] [PubMed] [Google Scholar]
- 43. Lee M., Fridman R., Mobashery S. (2004) Extracellular proteases as targets for treatment of cancer metastases. Chem. Soc. Rev. 33, 401–409 [DOI] [PubMed] [Google Scholar]
- 44. Jee B. K., Park K. M., Surendran S., Lee W. K., Han C. W., Kim Y. S., Lim Y. (2006) KAI1/CD82 suppresses tumor invasion by MMP9 inactivation via TIMP1 up-regulation in the H1299 human lung carcinoma cell line. Biochem. Biophys. Res. Commun. 342, 655–661 [DOI] [PubMed] [Google Scholar]
- 45. Deryugina E. I., Quigley J. P. (2006) Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 25, 9–34 [DOI] [PubMed] [Google Scholar]
- 46. Hunter K. W., Crawford N. P., Alsarraj J. (2008) Mechanisms of metastasis. Breast Cancer Research 10, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Friedl P., Wolf K. (2003) Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer 3, 362–374 [DOI] [PubMed] [Google Scholar]
- 48. Huang L., Cheng H. C., Isom R., Chen C. S., Levine R. A., Pauli B. U. (2008) Protein kinase Cepsilon mediates polymeric fibronectin assembly on the surface of blood-borne rat breast cancer cells to promote pulmonary metastasis. J. Biol. Chem. 283, 7616–7627 [DOI] [PubMed] [Google Scholar]
- 49. Wajapeyee N., Serra R. W., Zhu X., Mahalingam M., Green M. R. (2008) Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 132, 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Samani A. A., Yakar S., LeRoith D., Brodt P. (2007) The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr. Rev. 28, 20–47 [DOI] [PubMed] [Google Scholar]
- 51. Chang C. C., Shih J. Y., Jeng Y. M., Su J. L., Lin B. Z., Chen S. T., Chau Y. P., Yang P. C., Kuo M. L. (2004) Connective tissue growth factor and its role in lung adenocarcinoma invasion and metastasis. J. Natl. Cancer Inst. 96, 364–375 [DOI] [PubMed] [Google Scholar]
- 52. Böttger A., Strasser D., Alexandrova O., Levin A., Fischer S., Lasi M., Rudd S., David C. N. (2006) Genetic screen for signal peptides in Hydra reveals novel secreted proteins and evidence for non-classical protein secretion. Eur. J. Cell Biol. 85, 1107–1117 [DOI] [PubMed] [Google Scholar]
- 53. Bendtsen J. D., Jensen L. J., Blom N., Von Heijne G., Brunak S. (2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356 [DOI] [PubMed] [Google Scholar]
- 54. Mor-Vaknin N., Punturieri A., Sitwala K., Markovitz D. M. (2003) Vimentin is secreted by activated macrophages. Nat. Cell Biol. 5, 59–63 [DOI] [PubMed] [Google Scholar]
- 55. Nickel W. (2003) The mystery of nonclassical protein secretion. A current view on cargo proteins and potential export routes. Eur. J. Biochem. 270, 2109–2119 [DOI] [PubMed] [Google Scholar]
- 56. Théry C., Ostrowski M., Segura E. (2009) Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 57. Clayton A., Mason M. D. (2009) Exosomes in tumour immunity. Curr. Oncol. 16, 46–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Andre F., Schartz N. E., Movassagh M., Flament C., Pautier P., Morice P., Pomel C., Lhomme C., Escudier B., Le Chevalier T., Tursz T., Amigorena S., Raposo G., Angevin E., Zitvogel L. (2002) Malignant effusions and immunogenic tumour-derived exosomes. Lancet 360, 295–305 [DOI] [PubMed] [Google Scholar]
- 59. Park J. E., Tan H. S., Datta A., Lai R. C., Zhang H., Meng W., Lim S. K., Sze S. K. (2010) Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteomics 9, 1085–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gettins P. G. (2002) Serpin structure, mechanism, and function. Chem. Rev. 102, 4751–4804 [DOI] [PubMed] [Google Scholar]
- 61. Kumar V., Abbas A. K., Fausto N. (2005) Robbin and Cotran Pathological Basis of Disease (7th ed.) Elsevier/Saunders, 911–912 ISBN 978-0-7216-0187-3 [Google Scholar]
- 62. Stoller J. K., Aboussouan L. S. (2005) Alpha1-antitrypsin deficiency. Lancet 365, 2225–2236 [DOI] [PubMed] [Google Scholar]
- 63. Yang P., Sun Z., Krowka M. J., Aubry M. C., Bamlet W. R., Wampfler J. A., Thibodeau S. N., Katzmann J. A., Allen M. S., Midthun D. E., Marks R.S., de Andrade M. (2008) Alpha1-antitrypsin deficiency carriers, tobacco smoke, chronic obstructive pulmonary disease, and lung cancer risk. Arch. Intern. Med. 168, 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gross V., Geiger T., Tran-Thi T. A., Gauthier F., Heinrich P. C. (1982) Biosynthesis and secretion of alpha 1-antitrypsin in primary cultures of rat hepatocytes. Characterization of differently glycosylated intracellular and extracellular forms. Eur J Biochem. 129, 317–323 [DOI] [PubMed] [Google Scholar]
- 65. Bolmer S., Kleinerman J. (1986) Isolation and characterization of alpha 1-antitrypsin in PAS-positive hepatic granules from rats with experimental alpha 1-antitrypsin deficiency. Am J Pathol. 123, 377–389 [PMC free article] [PubMed] [Google Scholar]
- 66. Alam S., Li Z., Janciauskiene S., Mahadeva R. (2011) Oxidation of Z α1-antitrypsin by cigarette smoke induces polymerization: a novel mechanism of early-onset emphysema. Am. J. Respir. Cell Mol. Biol. 245, 261–269 [DOI] [PubMed] [Google Scholar]
- 67. Purkayastha P., Klemke J. W., Lavender S., Oyola R., Cooperman B. S., Gai F. (2005) Alpha 1-antitrypsin polymerization: a fluorescence correlation spectroscopic study. Biochemistry 44, 2642–2649 [DOI] [PubMed] [Google Scholar]
- 68. Salahuddin P. (2010) Genetic variants of alpha1-antitrypsin. Curr Protein Pept Sci. 11, 101–117 [DOI] [PubMed] [Google Scholar]
- 69. Gooptu B., Miranda E., Nobeli I., Mallya M., Purkiss A., Brown S. C., Summers C., Phillips R. L., Lomas D. A., Barrett T. E. (2009) Crystallographic and cellular characterisation of two mechanisms stabilising the native fold of alpha1-antitrypsin: implications for disease and drug design. J. Mol. Biol. 387, 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. El-Akawi Z. J., Al-Hindawi F. K., Bashir N. A. (2008) Alpha-1 antitrypsin (alpha1-AT) plasma levels in lung, prostate and breast cancer patients. Neuroendocrinol. Lett. 29, 482–484 [PubMed] [Google Scholar]
- 71. El-Akawi Z. J., Abu-Awad A. M., Sharara A. M., Khader Y. (2010) The importance of alpha-1 antitrypsin (alpha1-AT) and neopterin serum levels in the evaluation of non-small cell lung and prostate cancer patients. Neuroendocrinol. Lett. 31, 113–116 [PubMed] [Google Scholar]
- 72. Comunale M. A., Rodemich-Betesh L., Hafner J., Wang M., Norton P., Di Bisceglie A. M., Block T., Mehta A. (2010) Linkage specific fucosylation of alpha-1-antitrypsin in liver cirrhosis and cancer patients: implications for a biomarker of hepatocellular carcinoma. PLoS One. 5, e12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Al Bugami M. M., Farahat K. L., Al-Ashgar H. I., Hainau B. (2004) Fibrolamellar hepatocellular carcinoma with alpha-one antitrypsin liver disease. Saudi J. Gastroenterol. 10, 92–95 [PubMed] [Google Scholar]
- 74. Normandin K., Péant B., Le Page C., de Ladurantaye M., Ouellet V., Tonin P. N., Provencher D. M., Mes-Masson A. M. (2010) Protease inhibitor SERPINA1 expression in epithelial ovarian cancer. Clin. Exp. Metastasis 27, 55–69 [DOI] [PubMed] [Google Scholar]
- 75. Woźniak B., Mila-Kierzenkowska C., WoŸniak A., Drewa G., Sopońska M., Drewa T., Krzyzyńska-Malinowska E., Makarewicz R., Kowalski T., Szmytkowska K. (2008) The effect of combined therapy on activity of cathepsin D and alpha-1-antitrypsin in the blood serum of women with cervical cancer. Eur. J. Gynaecol. Oncol. 29, 617–619 [PubMed] [Google Scholar]
- 76. Scamuffa N., Siegfried G., Bontemps Y., Ma L., Basak A., Cherel G., Calvo F., Seidah N. G., Khatib A. M. (2008) Selective inhibition of proprotein convertases represses the metastatic potential of human colorectal tumor cells. J. Clin. Invest. 118, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mbeunkui F., Metge B. J., Shevde L. A., Pannell L. K. (2007) Identification of differentially secreted biomarkers using LC-MS/MS in isogenic cell lines representing a progression of breast cancer. Proteome Res. 6, 2993–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zelvyte I., Wallmark A., Piitulainen E., Westin U., Janciauskiene S. (2004) Increased plasma levels of serine proteinase inhibitors in lung cancer patients. Anticancer Res. 24, 241–247 [PubMed] [Google Scholar]
- 79. Yang M. H., Wu K. J. (2008) TWIST activation by hypoxia inducible factor-1 (HIF-1): implications in metastasis and development. Cell Cycle. 7, 2090–2096 [DOI] [PubMed] [Google Scholar]
- 80. Garber K. (2008) Epithelial-to-mesenchymal transition is important to metastasis, but questions remain. J. Natl. Cancer Inst. 100, 232–239 [DOI] [PubMed] [Google Scholar]
- 81. Yang J., Weinberg R.A. (2008) Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 14, 818–829 [DOI] [PubMed] [Google Scholar]
- 82. Thompson E. W., Williams E. D. (2008) EMT and MET in carcinoma–clinical observations, regulatory pathways and new models. Clin. Exp. Metastasis 25, 591–592 [DOI] [PubMed] [Google Scholar]
- 83. Sung H. J., Cho J. Y. (2008) Biomarkers for the lung cancer diagnosis and their advances in proteomics. BMB Rep. 41, 615–625 [DOI] [PubMed] [Google Scholar]
- 84. Mancini M., Toker A. (2009) NFAT proteins: emerging roles in cancer progression. Nat. Rev. Cancer 9, 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Egeblad M., Werb Z. (2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 86. Koivunen J., Aaltonen V., Peltonen J. (2006) Protein kinase C (PKC) family in cancer progression. Cancer Lett. 235, 1–10 [DOI] [PubMed] [Google Scholar]
- 87. Nguyen D. X., Bos P. D., Massagué J. (2009) Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer 9, 274–284 [DOI] [PubMed] [Google Scholar]
- 88. Wong S. Y., Hynes R. O. (2006) Lymphatic or hematogenous dissemination: how does a metastatic tumor cell decide? Cell Cycle 5, 812–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chambers A. F., Groom A. C., MacDonald I. C. (2002) Dissemination and growth of cancer cells in metastatic sites. Nature Rev. Cancer 2, 563–572 [DOI] [PubMed] [Google Scholar]
- 90. Stenbygaard L. E., Sørensen J. B. (1999) Small bowel metastases in non-small cell lung cancer. Lung Cancer 26, 95–101 [DOI] [PubMed] [Google Scholar]
- 91. Kondo K., Muramatsu M., Okamoto Y., Jin D., Takai S., Tanigawa N., Miyazaki M. (2006) Expression of chymase-positive cells in gastric cancer and its correlation with the angiogenesis. J. Surg. Oncol. 93, 36–42 [DOI] [PubMed] [Google Scholar]
- 92. He S. H., Chen P., Chen H. Q. (2003) Modulation of enzymatic activity of human mast cell tryptase and chymase by protease inhibitors. Acta Pharmacol. Sin. 24, 923–929 [PubMed] [Google Scholar]
- 93. Frank B. T., Rossall J. C., Caughey G. H., Fang K. C. (2001) Mast cell tissue inhibitor of metalloproteinase-1 is cleaved and inactivated extracellularly by agr-chymase. J. Immunol. 166, 2783–2792 [DOI] [PubMed] [Google Scholar]
- 94. Tchougounova E., Lundequist A., Fajardo I., Winberg J. O., Abrink M., Pejler G. (2005) A key role for mast cell chymase in the activation of pro-matrix metalloprotease-9 and pro-matrix metalloprotease-2. J. Biol. Chem. 280, 9291–9296 [DOI] [PubMed] [Google Scholar]
- 95. Eichler W., Friedrichs U., Thies A., Tratz C., Wiedemann P. (2002) Modulation of matrix metalloproteinase and TIMP-1 expression by cytokines in human RPE cells. Invest. Ophthalmol. Vis. Sci. 43, 2767–2773 [PubMed] [Google Scholar]
- 96. Zeidman I. (1957) Metastasis: a review of recent advances. Cancer Res. 17, 157–162 [PubMed] [Google Scholar]
- 97. Tarin D., Price J. E. (1981) Influence of microenvironment and vascular anatomy on “metastatic” colonization potential of mammary tumors. Cancer Res. 41, 3604–3609 [PubMed] [Google Scholar]
- 98. Hung T. T., Wu J. Y., Liu J. F., Cheng H. C. (2009) Epitope analysis of the rat dipeptidyl peptidase IV monoclonal antibody 6A3 that blocks pericellular fibronectin-mediated cancer cell adhesion. FEBS J. 276, 6548–6559 [DOI] [PubMed] [Google Scholar]
- 99. Colombi M., Zoppi N., De Petro G., Marchina E., Gardella R., Tavian D., Ferraboli S., Barlati S. (2003) Matrix assembly induction and cell migration and invasion inhibition by a 13-amino acid fibronectin peptide. J. Biol. Chem. 278, 14346–14355 [DOI] [PubMed] [Google Scholar]
- 100. McKeown-Longo P. J., Mosher D. F. (1985) Interaction of the 70,000-mol-wt amino-terminal fragment of fibronectin with the matrix-assembly receptor of fibroblasts. J. Cell Biol. 100, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Sechler J. L., Schwarzbauer J. E. (1998) Control of cell cycle progression by fibronectin matrix architecture. J. Biol. Chem. 273, 25533–25536 [DOI] [PubMed] [Google Scholar]
- 102. Xu J., Bae E., Zhang Q., Annis D. S., Erickson H. P., Mosher D. F. (2009) Display of cell surface sites for fibronectin assembly is modulated by cell adherence to (1)F3 and C-terminal modules of fibronectin. PLoS One. 4, e4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Henry M. D., Satz J. S., Brakebusch C., Costell M., Gustafsson E., Fässler R., Campbell K. P. (2001) Distinct roles for dystroglycan, beta1 integrin and perlecan in cell surface laminin organization. J. Cell Sci. 114, 1137–1144 [DOI] [PubMed] [Google Scholar]
- 104. Aziz M. H., Hafeez B. B., Sand J. M., Pierce D. B., Aziz S. W., Dreckschmidt N. E., Verma A. K. (2010) Protein kinase Cvarepsilon mediates Stat3Ser727 phosphorylation, Stat3-regulated gene expression, and cell invasion in various human cancer cell lines through integration with MAPK cascade (RAF-1, MEK1/2, and ERK1/2). Oncogene 29, 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Morgan K., Marsters P., Morley S., van Gent D., Hejazi A., Backx M., Thorpe E. R., Kalsheker N. (2002) Oncostatin M induced alpha1-antitrypsin (AAT) gene expression in Hep G2 cells is mediated by a 3′ enhancer. Biochem. J. 365, 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. He S. H., Chen P., Chen H. Q. (2003) Modulation of enzymatic activity of human mast cell tryptase and chymase by protease inhibitors. Acta Pharmacol. Sin. 24, 923–929 [PubMed] [Google Scholar]
- 107. Pat B., Chen Y., Killingsworth C., Gladden J. D., Shi K., Zheng J., Powell P. C., Walcott G., Ahmed M. I., Gupta H., Desai R., Wei C. C., Hase N., Kobayashi T., Sabri A., Granzier H., Denney T., Tillson M., Dillon A. R., Husain A., Dell'italia L. J. (2010) Chymase inhibition prevents fibronectin and myofibrillar loss and improves cardiomyocyte function and LV torsion angle in dogs with isolated mitral regurgitation. Circulation 122, 1488–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.