Abstract
Chromatin target of Prmt1 (Chtop) is a vertebrate-specific chromatin-bound protein that plays an important role in transcriptional regulation. As its mechanism of action remains unclear, we identified Chtop-interacting proteins using a biotinylation-proteomics approach. Here we describe the identification and initial characterization of Five Friends of Methylated Chtop (5FMC). 5FMC is a nuclear complex that can only be recruited by Chtop when the latter is arginine-methylated by Prmt1. It consists of the co-activator Pelp1, the Sumo-specific protease Senp3, Wdr18, Tex10, and Las1L. Pelp1 functions as the core of 5FMC, as the other components become unstable in the absence of Pelp1. We show that recruitment of 5FMC to Zbp-89, a zinc-finger transcription factor, affects its sumoylation status and transactivation potential. Collectively, our data provide a mechanistic link between arginine methylation and (de)sumoylation in the control of transcriptional activity.
Transcription factor activity is often controlled by post-translational modifications such as acetylation, phosphorylation, methylation, and sumoylation. Some modifications are associated with both gene activation and repression, whereas others appear to be more exclusive: asymmetrical dimethylation of arginine residues is restricted to transcriptional activation, whereas modification by sumoylation correlates with inhibition of transcription (1).
Arginine methylation occurs frequently within glycine-arginine-rich (GAR)1 regions and is catalyzed by members of the protein arginine methyltransferase (Prmt) family. These enzymes are subdivided in two major classes: type I enzymes catalyze the formation of asymmetrically dimethylated arginines (aDMA), whereas type II enzymes form symmetrically dimethylated arginines (sDMA) (2). Prmt1 and Prmt4/Carm1 (Coactivator-associated arginine methyltransferase 1) are the major type I enzymes and both are critical for mammalian development (3–4). Their substrates include RNA-binding proteins, nuclear matrix proteins, cytokines, and transcriptional regulators (2). Prmt1 methylates transcription factors such as Runx1 and STAT1 thereby promoting their transcriptional activity (5–6). Furthermore, Prmt1 and Prmt4 are recruited by nuclear hormone receptors and other transcription factors including YY1, p53, and NF-κB (7–10), resulting in the methylation of additional coactivators and histones. Prmt4 methylates histone H3 at arginine 17 and 26, whereas Prmt1 targets histone H4 at arginine 3 for methylation promoting subsequent acetylation of histone H3 at lysine 9 and histone H3 at lysine 14 (11) and further activating events (12).
Small ubiquitin-like modifier (SUMO) has an important regulatory function in several cellular processes, including DNA repair, cell cycle progression, signal transduction, chromatin structure and transcriptional regulation (13). Mammalian cells express four SUMO paralogs (SUMO-1 to SUMO-4). SUMO-1 differs in sequence by about 50% from SUMO-2 and 3, whereas SUMO-2 and SUMO-3 are 97% identical to each other. Conjugation of SUMO to target proteins occurs by a series of reactions conducted by the E1 activating enzyme, E2 conjugating enzyme, and an E3 SUMO ligase (14). The reverse desumoylation process is mediated by the isopeptidase activity of SUMO-specific proteases (Senps). In mammals, six members of Senps have been reported, known as Senp1–3 and Senp5–7. Sumoylation of multiple transcription factors, including Sp3, Sox6, Zeb1, and Zbp-89, has a negative effect on their transactivation potential, as it promotes the recruitment of repressive complexes (15–17). Many components of the repressor complexes CoREST1, NuRD, PRC1, Setdb1, and MEC themselves are also sumoylated, or have SUMO interacting motifs (SIMs). This suggests that sumoylation plays an important role in the formation and/or stabilization of these complexes (18).
We previously identified Friend of Prmt1 (Fop), also known as Small protein rich in arginine and glycine (SRAG), encoded by the mouse 2500003M10Rik and human C1orf77 genes, respectively (19–20). Recently, the name Chromatin Target of Prmt1 (Chtop) has been assigned to this gene/protein by the HUGO Gene Nomenclature Committee (HGNC). For reasons of clarity, we will also use the name Chtop for the murine homolog. Chtop is a chromatin-associated protein that plays an important role in the ligand-dependent activation of estrogen target genes such as TFF1 (pS2) in breast cancer cells (19). In addition, it is a critical regulator of γ-globin gene expression (21). However, little is known about the molecular mechanism of transcriptional control mediated by Chtop.
Chtop contains a central GAR region that is recognized and methylated by Prmt1. Because arginine methylation controls protein-protein interactions, we used a biotinylation-proteomics approach to identify proteins that bind Chtop in the presence and absence of Prmt1. In this study we identified and characterized a protein complex that binds specifically to methylated Chtop. As this nuclear complex consists of five proteins—SUMO1/sentrin/SMT3 specific peptidase 3 (Senp3), proline-glutamate and leucine rich protein 1 (Pelp1), LAS1-like protein (Las1L), Testis expressed 10 protein (Tex10), and WD repeat domain 18 protein (Wdr18)—we call it Five Friends of Methylated Chtop (5FMC). We show that Pelp1 is critical for the integrity of 5FMC and that Chtop and 5FMC are recruited by Zinc finger binding protein-89 (Zbp-89), thereby regulating both (de)sumoylation of, and transactivation by, Zbp-89.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Cells
The following plasmids have been described previously: GST-PELP1 deletions (22), T7-Pelp1 (23), pMT2_HA_Chtop, HA_mPRMT1 WT, and HA_mPRMT1 EQ (19). The cDNA of human SUMO-2 (hSMT3b) was kindly provided by Dr. Guntram Suske (Philipps-Universität Marburg, Germany). The cDNA of full-length LAS1L was obtained from Open Biosystems (Clone ID 3140243; Huntsville, AL, USA), full-length cDNA of Wdr18 was obtained from RZPD/imaGenes (clone IRAVp968G04150D6; Berlin, Germany), and full-length cDNA of Senp3 was obtained from RZPD/imaGenes (clone IRAVp968B0184D6; Berlin, Germany). The cDNA of PELP1 was subcloned from T7-PELP1. After the introduction of the 23-amino acid (aa) biotinylation tag into the pMT2_HA (24) and pMT2_HA_Chtop, LAS1L, Senp3, and PELP1 were cloned to pMT2_bio_HA using SalI and NotI, Wdr18 was cloned using SalI, and SUMO-2 was cloned using SalI and EcoRI. Bio_HA_LAS1L, Bio_HA_Senp3, Bio_HA_PELP1, Bio_HA_Wdr18, Bio_HA_SUMO-2, and Bio_HA_Chtop were subcloned into the erythroid expression vector pEV-neo (25) and electroporated into mouse erythroleukemia (MEL) cells expressing the BirA biotin ligase (26). To make the Gateway pSG513_myc destination plasmid, the Attr1-CmR-ccdb-Attr2 fragment was subcloned from pDEST17 (Invitrogen) to a modified pSG5 (Stratagene, La Jolla, CA) using HindIII, downstream the myc-tag sequence that was introduced to the pSG513 plasmid using EcoRI and BamHI. cDNAs for Senp3, Wdr18, and LAS1L were cloned into pDONR221 (Invitrogen), from which they were cloned by Gateway LR reaction to pSG513_myc. Internal deletion mutants of LAS1L and Wdr18 were generated using the QuikChange site-directed mutagenesis kit (Stratagene). MEL and 293T cells were grown in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen) supplemented with 10% fetal calf serum (FCS).
Transient Transfection, Immunoprecipitation, GST Pull-down Assay and Western Blot Analysis
Transient transfections in 293T cells, immunoprecipitations and Western blot analysis were performed as described previously (27). For immunoprecipitations combined with Benzonase (Novagen) incubation, 250 units of Benzonase were used followed by 3 h incubation at 4 °C. Nitrocellulose membranes were blocked in 1% bovine serum albumin (BSA), probed with the appropriate primary antibodies and analyzed using the Odyssey Infrared Imaging System (Li-Cor Biosciences, Cambridge, UK). GST pull-down assays were performed as described previously (22). Western blots were probed with the following primary antibodies: Prmt1 (07-404), Asym24 (07-414), and Cbx4 (09-029) were from Upstate, Charlottesville, VA; Actin (clone I-19; sc-1616), Taf1β (clone H-120; sc-25564), Lamin B (sc-6216), HA (monoclonal F7; sc-7392), HA (polyclonal Y11; sc-805), Myc (monoclonal 9E10; sc-40), Myc (polyclonal A-14; sc-789), and Pol II (polyclonal N-20; sc-899) were from Santa Cruz Biotechnology, Santa Cruz, CA; Tex10 (17372-1-AP), Las1L (16010-1-AP), Senp3 (17659-1-AP) and Wdr18 (15165-1-AP) were from Proteintech Group; Zbp-89 (ab69933) was from Abcam, Cambridge, MA, SUMO 2/3 (clone 1E7; M114-3) was from MBL, Woburn, MA; Pelp1 (A300-876A) was from Bethyl Laboratories, Montgomery, TX; T7 (69522-3) was from Novagen, Madison, WI; Taf1α (B100-56353) was from Novus Biologicals, Littleton, CO and Chtop (KT64) was from Absea Biotechnology. Ring1B antibody was kindly provided from Dr. Miguel Vidal (Madrid, Spain).
Cell Lysates and Mass Spectrometry (MS)
Preparation of nuclear and whole cell extracts from MEL and 293T cells were carried out as described previously (27). Purification of biotinylated proteins, digestion with trypsin (Promega, sequencing grade) on paramagnetic streptavidin beads and liquid chromatography-tandem MS (LC-MS/MS) were performed as described previously (19, 26). Nanoflow LC-MS/MS was performed on an 1100 series capillary LC system (Agilent Technologies, Santa Clara, CA) coupled to an LTQ-Orbitrap mass spectrometer (Thermo), operating in positive mode and equipped with a nanospray source. Peptide mixtures were trapped on a ReproSil C18 reversed phase column (Dr Maisch GmbH; column dimensions 1.5 cm × 100 μm, packed in-house) at a flow rate of 8 μl/min. Peptide separation was performed on ReproSil C18 reversed phase column (Dr Maisch GmbH; column dimensions 15 cm × 50 μm, packed in-house) using a linear gradient from 0 to 80% B (A = 0.1% formic acid; B = 80% (v/v) acetonitrile, 0.1% formic acid) in 70 min and at a constant flow rate of 200 nl/min using a splitter. The column eluent was directly sprayed into the electrospray ionization (ESI) source of the mass spectrometer. Mass spectra were acquired in continuum mode; fragmentation of the peptides was performed in data-dependent mode. Peak lists were automatically created from raw data files using the Mascot Distiller software (version 2.3; MatrixScience). The Mascot search algorithm (version 2.2, MatrixScience) was used for searching against the IPI_mouse database (version 3.83, containing 60,010 sequences and 27,475,843 residues). The peptide tolerance was set to 10 ppm and the fragment ion tolerance was set to 0.8 Da. A maximum number of two missed cleavages by trypsin were allowed and carbamidomethylated cysteine and oxidized methionine were set as fixed and variable modifications, respectively. Search results were parsed into a home-built database system for further analysis. Entries were parsed if they had a minimum peptide Mascot score of 25, and a significance threshold of p < 0.05; the option “require red bold” was also selected. Using these parameters yields an estimated peptide false discovery rate (FDR) of 3–5% against a target decoy database. The Mascot data have been uploaded to the PRIDE database (28) (www.ebi.ac.uk/pride) under accession numbers 21750–21767. The data was converted using PRIDE Converter (29) (http://pride-converter.googlecode.com).
Lentivirus Mediated Knockdown
The Chtop and Prmt1 shRNA lentiviral vectors were described previously (19, 21). For mouse Pelp1, Senp3, and human PELP1, SENP3, WDR18, LAS1L, clones from the TRC Mission shRNA library ((30); Sigma Aldrich, St. Louis, MO, USA) were used for knockdown experiments in MEL and 293T cells respectively, including a non-targeting shRNA control virus (SHC002). Lentivirus was produced by transient transfection of 293T cells as described before (31). The following clones were used from the TRC shRNA library: TRCN0000177043 (shPelp1 #1), TRCN0000178252 (shPelp1 #2), TRCN0000031014 (shSenp3 #1), TRCN0000031017 (shSenp3 #2), TRCN0000159617 (shPELP1 #2), TRCN0000159673 (shPELP1 #3), TRCN0000004106 (shSENP3 #1), TRCN0000004107 (shSENP3 #2), TRCN0000078088 (shWDR18 #1), TRCN0000078089 (shWDR18 #2), TRCN0000121835 (shLAS1L #2), TRCN0000142144 (shLAS1L #4), TRCN0000035931 (sh hPRMT1).
Size-exclusion Chromatography and Subcellular Fractionation
Nuclear extracts from MEL cells expressing BirA biotin ligase enzyme, were chromatographed over a Superose 6 column (Amersham Biosciences) using an AKTA fast-performance liquid chromatography apparatus. Fractions were collected and precipitated with trichloroacetic acid and analyzed by Western blotting. Subcellular fractionation was performed as described previously (19).
RT, QPCR, ChIP Assay, and Statistical Analysis
Reverse transcription (RT), RT-quantitative PCR (RT-QPCR), and ChIP were performed as described previously (32). Primers used for RT-QPCR and ChIP-QPCR are summarized in supplemental Table S1. ANOVA statistical analysis was performed by GraphPad Prism 5.02.
RESULTS
Methylation Dependent and Independent Interactions of Chtop
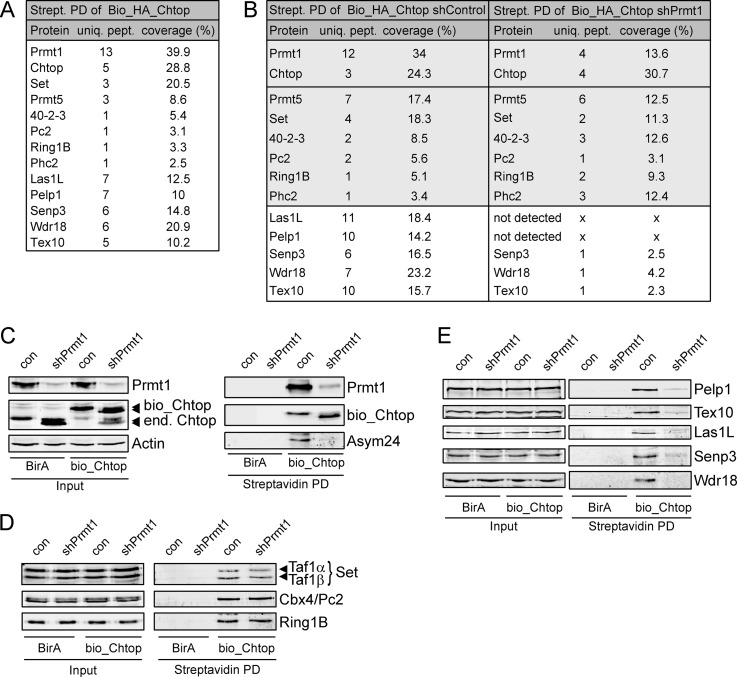
To identify interaction partners of Chtop, an N-terminal double-tagged version of Chtop protein (Bio_HA_Chtop) was expressed in MEL cells. These cells also stably expressed BirA, a bacterial biotin ligase which efficiently biotinylates the Bio-tag (26). Note that tagged Chtop was not overexpressed, as endogenous levels were reduced in Bio_HA-Chtop transfected cells (Fig. 1C). Protein complexes from nuclear lysates were recovered by streptavidin pull down followed by nanoflow liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS) and were compared with samples from cells expressing BirA alone (Fig. 1A, supplemental Table S2). This confirmed the association of Chtop with Prmt1 and Prmt5, factors that we have previously identified as Chtop binding proteins (19). MS analysis also revealed the binding of three members of the polycomb repressor complex 1 (PRC1; Pc2, Ring1B, and Phc2) (33–35), the SET nuclear oncoprotein (36), and the mRNA export protein 40-2-3 (37). Furthermore, high MASCOT scores were obtained for the proteins Pelp1, Las1L, Tex10, Senp3, and Wdr18, four of which were previously copurified with the MLL complex and were recently described as regulators of ribosome biogenesis (38–40). Pelp1 is a coactivator involved in nuclear hormone signaling (41), whereas Senp3 is a SUMO-specific protease (42). The WD-repeat protein Wdr18, Tex10, and Las1L had not been characterized previously. To investigate whether arginine methylation of Chtop played a role in these interactions, we performed lentiviral-mediated knockdown of Prmt1 in Bio_HA_Chtop expressing MEL_BirA cells and used a similar purification approach. When compared with cells transduced with a control lentivirus, no major differences were observed in the binding of Chtop to Prmt5, Pc2, Ring1B, Phc2, 40-42-3, and Set (Fig. 1B, supplemental Table S2). Interestingly, copurification of Las1L, Pelp1, Tex10, Senp3, and Wdr18 was strongly reduced or absent when Chtop was hypomethylated. Next, we confirmed the methylation (in)dependent interactions by streptavidin affinity purification, followed by Western blot analysis. Chtop is hypomethylated in the absence of Prmt1, as indicated by its faster migration pattern and by staining with an antibody that specifically recognizes aDMA (Asym24; Fig. 1C). Endogenous Chtop interactors were efficiently recovered in Bio_HA_Chtop pull downs, whereas no background staining was observed in MEL_BirA cells (Figs. 1D, 1E). In addition, no association was observed with Wdr18 in the absence of Prmt1, whereas the interactions with Pelp1, Las1L, Tex10 and Senp3 were strongly reduced. These associations do not depend on the presence if Prmt1, as only wildtype but not enzymatic inactive Prmt1 could rescue the knockdown (supplemental Fig. S1). Together, these results validate the interactions identified by MS and show that methylation of Chtop is required for the recruitment of Pelp1, Las1L, Tex10, Senp3, and Wdr18.
Fig. 1.
Identification of Chtop-interacting proteins. A, List of Chtop interacting proteins indentified by mass spectrometry (MS). Protein complexes from nuclear lysates of MEL_BirA cells ectopically expressing Bio_HA_Chtop were recovered by MS. Proteins, unique peptides (uniq. pept.) and percent coverage (coverage %) are indicated. B, List of Chtop interacting proteins in MEL_BirA cells treated with control lentivirus (shControl) and lentivirus expressing shRNA against Prmt1 (shPrmt1) identified by MS. Interactions with major differences between shControl and shPrmt1 treated cells are shown in a white box. C, Chtop is hypomethylated in the absence of Prmt1. Whole cell lysates (Input) and streptavidin pull downs (Streptavidin PD) were analyzed for Prmt1, Chtop and asymmetric dimethyl arginine (Asym24) residues. Actin staining serves as a loading control. Ectopically expressed (bio_Chtop) and endogenous Chtop (end. Chtop) are indicated. D, Chtop methylation-independent interactions. Whole cell lysates (Input) and streptavidin pull-downs (Streptavidin PD) were analyzed with the antibodies indicated. E, Chtop methylation-dependent interactions. Whole-cell lysates (Input) and streptavidin pull-downs (Streptavidin PD) were analyzed with the antibodies indicated.
5FMC is a Nuclear Complex
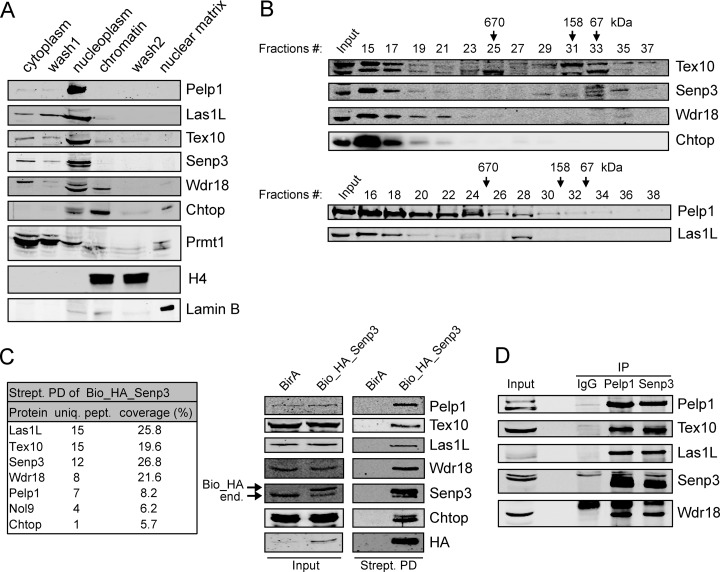
We have previously shown that the majority of Chtop is bound to chromatin (19). To elucidate where the newly identified methylation-specific partners of Chtop localize in the cell, we performed biochemical fractionation of MEL cells. This revealed that all five proteins were mainly found in the nucleoplasm, whereas low levels were also detected in the cytoplasmic and chromatin fractions (Fig. 2A). To examine whether the five proteins form a complex, we first performed size-exclusion chromatography of MEL nuclear extracts. The elution patterns of Pelp1, Las1L, Tex10, Senp3 and Wdr18, as well as Chtop, overlapped substantially (Fig. 2B). The molecular mass of the positive fractions was >1MDa, indicating that the factors were present in a high molecular weight protein complex or were bound to chromatin. Similar experiments in human 293T cells revealed comparable results, although larger proportions of LAS1L, TEX10, and WDR18 were detected in fractions corresponding to lower molecular mass (supplemental Fig. S2). Next, doubled tagged (Bio_HA) Pelp1, Las1L, Senp3, and Wdr18 were stably expressed in MEL_BirA cells. Of note, we were not able to exogenously express the Tex10 protein, probably because of protein stability issues. Associated proteins were identified by streptavidin pull down in nuclear lysates followed by nanoLC-MS/MS and were compared with samples from MEL_BirA only cells. In all four experiments, the associated proteins with the highest MASCOT score were Pelp1, Las1L, Tex10, Senp3, and Wdr18 (Fig. 2C (Bio_HA_Senp3) and supplemental Fig. S3 (Pelp1, Wdr18, and Las1L)). Moreover, in the MS analysis of Bio_HA_Senp3, Bio_HA_Wdr18 and Bio_HA_Pelp1 we observed an association with the Nol9 protein. The Nol9 ortholog in S. pombe (Grc3) was shown to associate with Las1 and the yeast IPI complex that consist of Rix1, Ipi1 and Crb3 (43). These proteins share homologous regions with Pelp1, Tex10 and Wdr18, respectively. Moreover, it was recently shown in human cells that NOL9 interacts with PELP1, TEX10, WDR18, LAS1L, and SENP3 (40). Chtop was only detected in the Bio_HA_Senp3 purification (Fig. 2C, left panel). The MS results were confirmed by immunoblot analysis of the streptavidin pull downs of tagged proteins (Fig. 2C, right panel and supplemental Fig. S3), and immunoprecipitations of endogenous proteins from both mouse and human cells (Fig. 2D and supplemental Fig. S4, respectively). Taken together, these results show that Pelp1, Las1L, Tex10, Senp3, and Wdr18 form a nuclear multi-protein complex. As this complex binds selectively to methylated Chtop, we named it Five Friend of Methylated Chtop, or 5FMC.
Fig. 2.
5FMC is a nuclear complex. A, Chtop methylation dependent interaction proteins are localized mainly in the nucleoplasm. MEL_BirA cells were biochemically fractionated as described under “Experimental Procedures”. Cytoplasmic, nucleoplasmic, chromatin and nuclear matrix were tested using Pelp1, Las1L, Tex10, Senp3, Wdr18 antibodies against endogenous proteins. Chtop, Prmt1, H4, and Lamin B served as controls for individual fractions. B, MEL_BirA cell nuclear extracts were analyzed by sized-exclusion chromatography on a Superose 6 column. Proteins eluted from the indicated fractions were blotted with the indicated antibodies. Molecular mass markers are indicated at the top. C, Senp3 interactions in MEL cells. Whole cell lysates (Input) and streptavidin pull downs (Strept. PD) from MEL_BirA (BirA) and MEL_BirA cells expressing biotinylated Senp3 (Bio_HA_Senp3) were analyzed by MS (table) and Western blotting. Immunoblot probed with the indicated antibodies. Arrows indicate endogenous (end.) and biotinylated (Bio_HA) Senp3. D, 5FMC is a nuclear complex. Endogenous association between the 5FMC components. MEL_BirA cell nuclear lysates were analyzed by immunoprecipitation (IP) and Western blotting with the antibodies indicated.
Pelp1 is the Core Subunit and Critical for 5FMC Stability
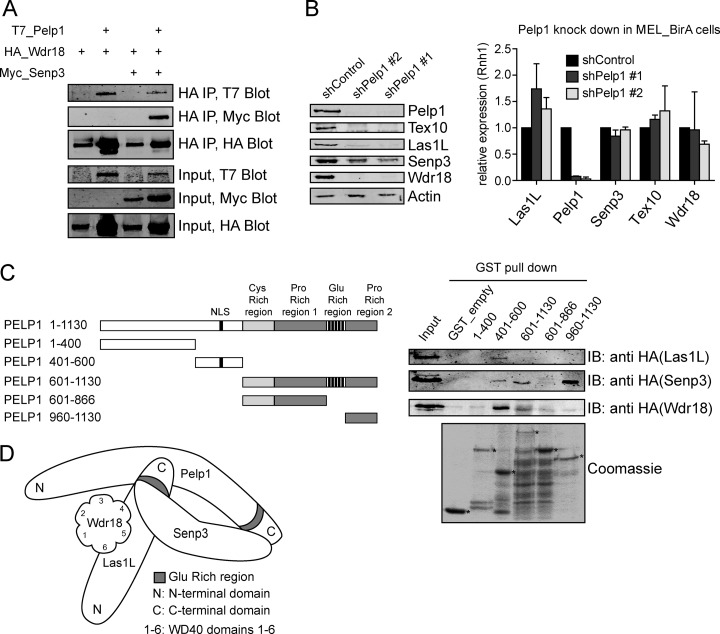
To further study the composition of the 5FMC complex, we transiently cotransfected tagged 5FMC components in 293T cells, followed by coimmunoprecipitation (co-IP). We found that T7_Pelp1 is efficiently recovered in HA_Wdr18 IPs, whereas Myc_Senp3 interacts with HA_Wdr18 (Fig. 3A). Moreover, Myc_Senp3 co-purifies with HA_Wdr18 only when T7_Pelp1 is cotransfected (Fig. 3A, lanes 3 and 4). In addition, we observed that cotransfection of T7_Pelp1 resulted in higher protein levels of HA_Wdr18 and Myc_Senp3 (Fig. 3A, lanes 2 and 4). Identical results were obtained when Myc_Senp3 was immunoprecipitated (data not shown). No conclusive results could be obtained with ectopically expressed Tex10 and Las1L, probably because of stability issues. These results show that Pelp1 is required for the interaction between Wdr18 and Senp3 and that Pelp1 might be required for the stability of these proteins. To further study the potential central role of Pelp1 within the 5FMC complex, we depleted endogenous mouse Pelp1 in MEL_BirA cells and endogenous human PELP1 in 293T cells by lentiviral-mediated knockdown using two different shRNAs (Fig. 3B and supplemental Fig. S5A). Interestingly, the protein levels of Senp3 and Las1L were dramatically decreased, whereas Wdr18 and Tex10 could not be detected in the absence of endogenous Pelp1 (Fig. 3B, left panel). Quantitative RT-PCR showed that the reduced protein levels were caused by protein stability rather than reduced mRNA levels (Fig. 3B, right panel). The individual knockdown of LAS1L, SENP3, or WDR18 had no significant effect on the protein levels of the other 5FMC subunits. Furthermore, partial complexes exist upon depletion of LAS1L, SENP3, and WDR18 (supplemental Fig. S5). These results indicated that Pelp1 is the central component of the 5FMC complex and that the integrity of 5FMC is essential for the stability of its components. Analysis of the primary sequence of Pelp1 showed that Pelp1 contains a cysteine-rich region, two proline-rich regions and a C-terminal glutamine-rich region (41). To map the interactions between Pelp1 and other 5FMC components, we tested the ability of HA-tagged Las1L, Wdr18, and Senp3 proteins to bind to various domains of Pelp1 fused to GST. HA_Las1L and HA_Wdr18 interacted with the GST_Pelp1 fusion containing amino acids (aa) 401–600, whereas HA_Senp3 mainly interacted with domain 960–1130. Interactions were further studied using a series of deletion mutants of Las1L. These experiments showed that the C-terminal part of Las1L (aa 552–734) mediated the interaction with Pelp1, whereas the central domain (aa 370–552) mediated the interaction with Wdr18. The binding to Senp3 could not be mapped in detail: any deletion between aa 188 and 734 disrupted the interaction (supplemental Fig. S6A). Similar experiments with Wdr18 deletion constructs revealed that the region containing WD40 domains 4–6 were required for binding to Las1L, whereas deletion of any WD40 domain disrupted the binding to Pelp1 (supplemental Fig. S6B). These initial domain-mapping experiments suggest complex multi-intermolecular interactions and are in line with the proposed model shown in Fig. 3D.
Fig. 3.
Pelp1 is the core component of the 5FMC complex. A, Pelp1 is required for interaction between Senp3 and Wdr18. 293T cells were transiently transfected with expression vectors encoding T7_Pelp1, HA_Wdr18 and Myc_Senp3. Cell lysates were analyzed by IP and Western blotting with the antibodies indicated. B, Pelp1 is needed for the stability of the 5FMC complex. MEL_BirA cells were treated with the indicated shRNAs. Nuclear lysates were analyzed by Western blotting with the indicated antibodies. Actin staining serves as a loading control. Total RNA was extracted from MEL_BirA cells transduced with the indicated shRNA and analyzed by RT-QPCR for Las1L, Pelp1, Senp3, Tex10, and Wdr18. Error bars: S.D. of triplicate experiment. C, Mapping the interaction regions of Pelp1. Schematic representation of Pelp1 deletion constructs. GST fused Pelp1 domains and GST alone (GST_empty) were immobilized onto glutathione beads (lower panel stained with Coomassie) and used to pull down nuclear cell lysates from 293T cells expressing HA_Las1L, HA_Senp3 and HA_Wdr18. Asterisks indicate GST fusion proteins. D, Schematic representation of interaction between Pelp1, Las1L, Senp3, and Wdr18 protein domains.
Chtop Recruits 5FMC to Zbp-89
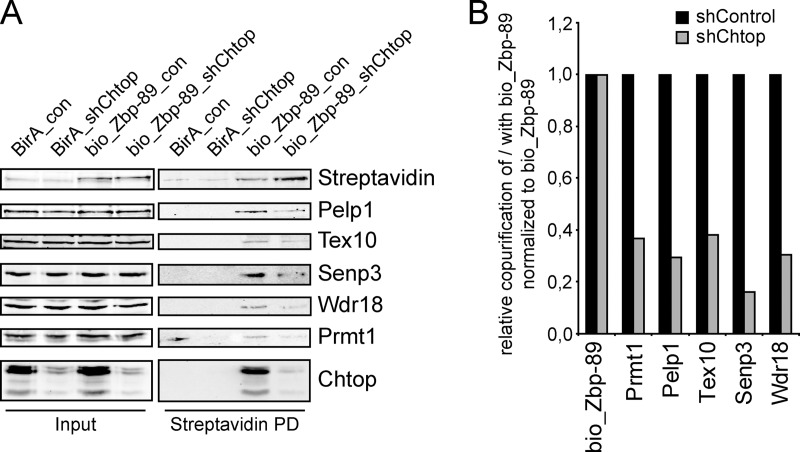
MS analysis of the Zbp-89 interactome revealed Chtop and several Chtop-associated factors as potential interaction partners of Zbp-89 in MEL cells (manuscript in preparation). To further explore the possibility that 5FMC interacts with Zbp-89, we performed bio_Zbp-89 pull downs in MEL_BirA cells followed by immunoblotting. We observed that Chtop, Prmt1 and 5FMC components associated with Zbp-89 (supplemental Fig. S7). These interactions were DNA independent, as degradation of DNA by Benzonase treatment did not affect the efficiency of co-purification. To investigate whether Chtop is required for the interaction between Zbp-89 and 5FMC, we examined the association of Zbp-89 and 5FMC complex components in Chtop knockdown cells. Bio_Zbp-89 was precipitated more efficiently when Chtop protein levels were reduced, suggesting that bio_Zbp-89 was more accessible in the absence of Chtop (Fig. 4A). Indeed, co-purification of 5FMC complex components was reduced to ∼30% of control samples (Figs. 4A and 4B). These results indicate that Chtop is required for the association of Zbp-89 with the 5FMC complex.
Fig. 4.
Chtop recruits 5FMC to Zbp-89. A, Zbp-89 is associated with Chtop and 5FMC complex. Nuclear cell lysates (Input) of MEL_BirA cells (BirA) and MEL_BirA cells expressing biotinylated Zbp-89 (bio_Zbp-89) treated with lentivirus expressing shRNA against Chtop (shChtop) and control lentivirus (shControl) analyzed by streptavidin pull down (Streptavidin PD) and Western blotting with the antibodies indicated. B, Quantification of protein levels using the Odyssey Infrared Imaging System.
Senp3 Regulates the Sumoylation Status of Zbp-89
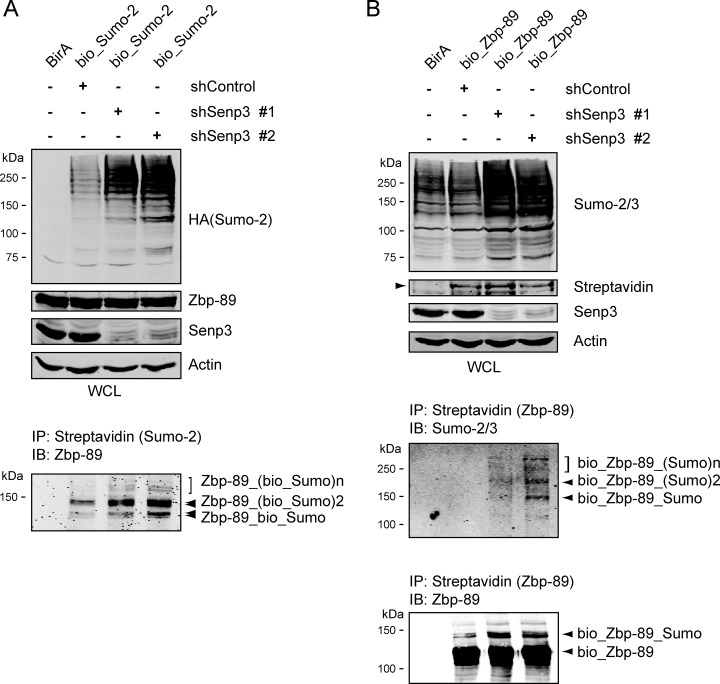
It has been reported that Zbp-89 can be post-translationally modified by SUMO in transient overexpression experiments (44). To investigate whether endogenous Zbp-89 could be sumoylated, we first performed streptavidin pull downs from bio_HA_SUMO-2 expressing MEL_BirA nuclear extracts. Staining with an antibody recognizing Zbp-89 detects multiple sumoylated Zbp-89 species (Fig. 5A, lower panel), in line with the observation that Zbp-89 contains at least two domains that can be sumoylated (44). Knockdown of Senp3, using two different shRNAs, in these cells led to a significant increase of SUMO-2 detection in whole cell lysates, as well as of sumoylated Zbp-89. It should be noted that the reduction of Senp3 expression affected cell growth and survival, thereby limiting the effect of the knockdown. Next, we performed similar experiments in MEL cells expressing bio_Zbp-89. Streptavidin pull downs probed with an anti-SUMO 2/3 antibody showed that Senp3 depletion resulted in an increase of the levels of SUMO modified bio_Zbp-89 and the appearance of a slower mobility form of bio_Zbp-89 when probed with anti-Zbp-89 antibody (Fig. 5B). The upper band is consistent with SUMO modification. Taken together, these results show that Zbp-89 is sumoylated in vivo, and that Senp3 plays a role in this process.
Fig. 5.
Senp3 regulates Zbp-89 sumoylation. A–B, Senp3 plays a role in Zbp-89 sumoylation. (A) MEL_BirA (BirA) and MEL_BirA cells expressing bio_HA_Sumo2 (bio_Sumo2) were treated with lentiviruses expressing two different shRNAs against Senp3 (shSenp3 #1, shSenp3 #2) and control lentivirus (shControl). Whole cell lysates (WCL) analyzed by Western blotting with the indicated antibodies. Actin staining serves as a loading control. Nuclear extracts were pull down using magnetic streptavidin beads and analyzed by Western blotting with anti-Zbp-89 antibody. B, MEL_BirA (BirA) and MEL_BirA cells expressing biotinylated Zbp-89 (bio_Zbp-89) were treated with lentiviruses expressing two different shRNAs against Senp3 (shSenp3 #1, shSenp3 #2) and control lentivirus (shControl). Whole cell lysates (WCL) analyzed by Western blotting with the antibodies indicated. Actin staining serves as a loading control. Nuclear extracts were pulled down using magnetic streptavidin beads and analyzed by Western blotting with anti-Zbp-89 and anti Sumo-2/3 antibodies.
5FMC is Critical for Zbp-89 Dependent Gene Regulation
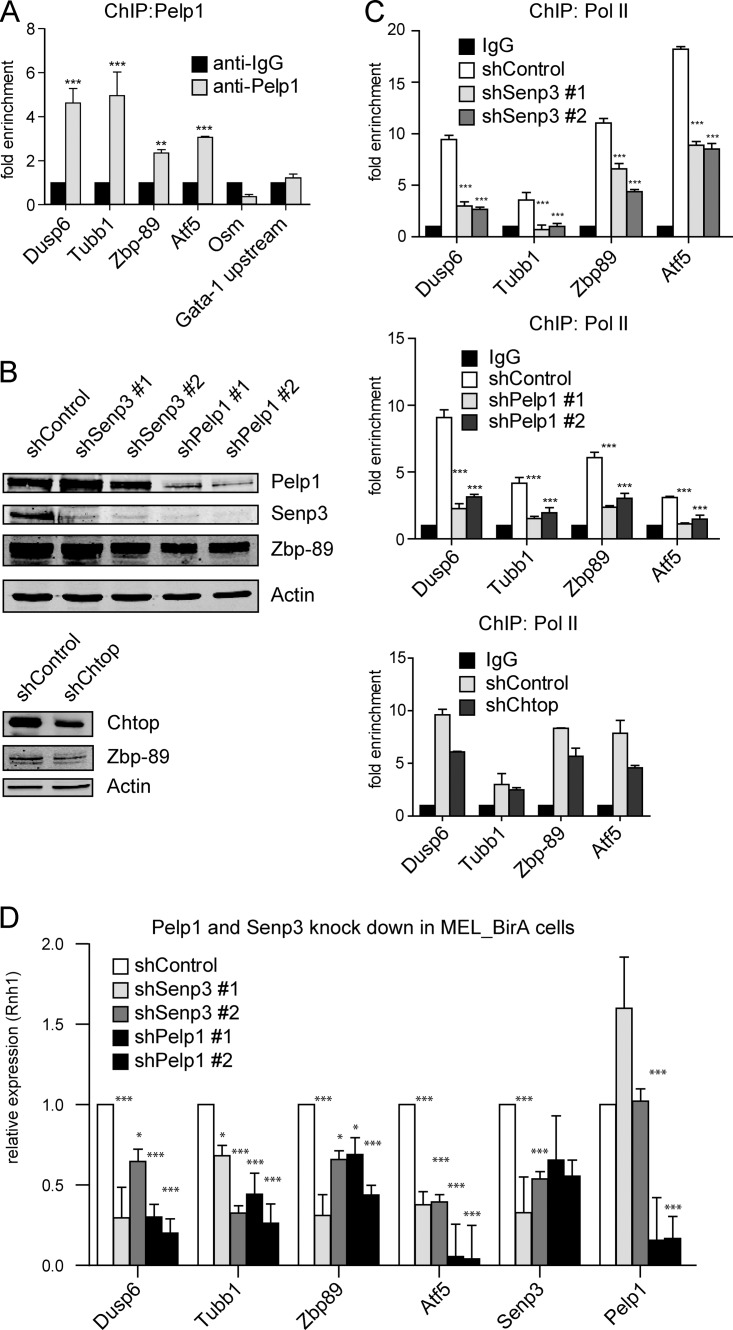
The observations that 5FMC is a desumoylating complex that is recruited by Zbp-89 suggest that it is involved in transcriptional regulation. To examine whether 5FMC is recruited to Zbp-89 target genes we performed chromatin immunoprecipitation (ChIP) experiments for Pelp1, the core component of the complex. We used MEL_BirA cells that ectopically expressed bio_Pelp1, as this increased sensitivity (not shown). After chromatin precipitation, the promoter regions of three (Dusp6, Zbp-89, Atf5) and the coding region of one (Tubb1) Zbp-89 target genes, that were identified by ChIP-sequencing as binding sites of Zbp-89 (manuscript in preparation) and confirmed by ChIP (supplemental Fig. S8A), were analyzed with the corresponding primers. Occupancy by Pelp1 was indeed observed for these Zbp-89 target genes (Fig. 6A). To further investigate the potential role of 5FMC in transcription regulation, we depleted Pelp1, Senp3 and Chtop in MEL_BirA cells using two different shRNAs (Fig. 6B) and performed ChIP using an antibody against RNA polymerase II (Pol II). Reduced levels of Pelp1, Senp3 and Chtop resulted in reduced Pol II occupancy at the promoter regions of Dusp6, Zbp-89, Atf5 and the coding region of Tubb1 when compared with cells treated with scrambled control shRNA (Fig. 6C). To validate the Pol II ChIP observations we performed quantitative RT-PCR analysis for the Zbp-89 target genes after depletion of Senp3 and Pelp1. As expected, Dusp6, Tubb1, Zbp-89, and Atf5 mRNA levels were decreased (Fig. 6D). To exclude that the observed differences in occupancy by Pol II were not caused by altered occupancy by Zbp-89 we performed ChIP using an antibody against Zbp-89. Binding of Zbp-89 to the regions of its target genes was unaffected on depletion of Senp3 and Pelp1, whereas binding of Zbp-89 was reduced when Chtop was depleted (supplemental Fig. S8B). The reduction of Zbp-89 binding upon depletion of Chtop was because of reduced protein levels of Zbp-89 (Fig. 6B, lower panel). The same regions were also tested for changes in the histone modifications H3K4 and H3K27, but no changes were detected (not shown). Collectively, our data indicate that 5FMC is recruited to Zbp-89 target genes and that it is involved in their transcriptional activation by Zbp-89. This most likely involves desumoylation of Zbp-89 and possibly of other factors.
Fig. 6.
5FMC is involved in the regulation of Zbp-89 target genes. A, Pelp1 is recruited at the promoter regions of Dusp6, Zbp-89, Atf5 and the coding region of Tubb1. MEL_BirA cells that ectopically expressed bio_Pelp1 analyzed by ChIP using Pelp1 antibody for the indicated gene promoter or coding regions. The promoter region of the Osm gene and the region upstream of Gata-1 promoter (Gata-1 upstream) were used as negative controls. *** indicates p < 0.001, ** indicates p < 0.01. Error bars: S.D. of triplicate experiment. B, Knockdown of Pelp1, Senp3 and Chtop in MEL_BirA cells. MEL_BirA cells were treated with the indicated shRNAs. Cell lysates were analyzed by Western blotting with the indicated antibodies. Actin staining serves as a loading control. C, Pelp1, Senp3 and Chtop knockdowns reduced RNA polymerase II (Pol II) occupancy at the promoter regions of Dusp6, Zbp-89, Atf5 and the coding region of Tubb1. MEL_BirA cells were treated as in (B). ChIP analysis at the indicated regions was performed using Poll II antibody. Error bars: S.D. of triplicate experiment. D, Regulation of Zbp-89 target genes by Senp3 and Pelp1. Total RNA was extracted from MEL_BirA cells treated as in (B) and analyzed by RT-QPCR for Dusp6, Tubb1, Zbp-89, Atf5, Senp3 and Pelp1. *** indicates p < 0.001, ** indicates p < 0.01, * indicates p < 0.05. Error bars: S.D. of triplicate experiment.
DISCUSSION
In the present study, we have identified 5FMC, a desumoylating protein complex that exclusively binds to arginine-methylated Chtop. It acts as a key regulator of Zbp-89 sumoylation and is required for full transcriptional activation of Zbp-89 dependent genes. To our knowledge, this is the first description of a mechanism that adds specificity to desumoylation processes.
The 5FMC complex is composed of five proteins: Pelp1, Las1L, Tex10, Senp3, and Wdr18. These factors are among the most abundant proteins present in Chtop purifications, but only when Chtop is methylated by Prmt1. Little is known about most of the components of the 5FMC complex. Pelp1 is a coactivator involved in nuclear hormone signaling (41), Senp3 is a SUMO-specific protease (42), whereas Wdr18, Las1L, and Tex10 have not been characterized yet. In yeast, the proteins Rix1, Ipi1, and Ipi3 (S. cerevisiae)/Crb3 (S. pombe) share conserved regions with Pelp1, Tex10, and Wdr18, respectively. They form the IPI complex and have been shown to function in ribosomal RNA processing (45). It was recently shown that Las1, the yeast ortholog of Las1L, is also associated with the IPI complex (43). Furthermore, PELP1, TEX10, LAS1L, SENP3, and WDR18 were recently linked to ribosome biogenesis in human cells, indicating an evolutionary conserved complex with a role in ribosomal RNA processing. In human cells, the complex was localized mainly in the nucleoplasm with a subfraction present in the nucleolus (39–40). In this study, we observed a similar distribution in mouse cells.
Additionally, several studies suggested a role for 5FMC components in transcriptional regulation and (de)sumoylation events, although this had not been explored further. Doseff and Arndt proposed in their initial identification of Las1 in S. cerevisiae that it functions as a transcription factor (46), whereas it was recently shown to localize to heterochromatic regions (43). In human cells, components of the 5FMC complex were first detected in the MS analysis of the MLL1-WDR5 complex, a complex that regulates transcription activation by H3K4 methylation (38). The 5FMC complex was also found in a recent study on human coregulator protein complex networks obtained from integrative mass spectrometry-based analysis of 3290 antibody-based affinity purifications (47). Thus, the data obtained with this approach independently support the composition of the 5FMC complex reported here. Furthermore, PELP1, LAS1L, TEX10, and SENP3 were also detected together with components of the CoREST1/HDAC1 corepressor complex in a MS study for proteins that interact with SUMO-2 (48). Sumoylation is important for stability and recruitment of repressive complexes such as CoREST, NuRD, and SetDB1 (18), indicating that desumoylation of transcription factors and corepressors is required for derepression.
Our results indicate that Pelp1 is the core component of 5FMC. Pelp1 has the ability to interact with nuclear receptors (NRs) and enhances transcription of their target genes (23). Pelp1 has been shown to interact with the acetyltransferases CBP and p300 (22), KDM1, a member of the CoREST1 repressor complex (49), and deacetylases, including components of the NuRD repressor complex (50). Interestingly, Pelp1 was also identified in a blind screen for SUMO-2 interacting proteins (51). This opens the possibility that Pelp1 acts, in addition to its scaffold function within 5FMC, as a Sumo-2 sensor to detect Senp3 substrates.
As Chtop is strongly associated with chromatin (19), whereas 5FMC mainly resides in the nucleoplasm (this paper), the interactions between Chtop and 5FMC are most likely transient and highly dynamic. Possibly, Chtop recruits 5FMC complex, to desumoylate its substrates, in a “hit-and-run” manner. In addition, 5FMC may very well act as a desumoylation complex outside the chromatin environment and independent of Chtop, e.g. in ribosome biogenesis (39).
In line with our previous observation that Chtop colocalizes to H3K27me3 (19), we found that Chtop interacts with the Prc1 complex. In contrast to 5FMC, this association does not depend on the methylation status of Chtop. Intriguingly, the Pc2 component of Prc1 (also known as Cbx4) has been identified as a SUMO-ligase for several transcriptional regulators (52–53). Although these studies mainly focused on SUMO-1 modification, Chtop may recruit 5FMC as an antagonist of Pc2/Cbx4.
We have shown that Chtop recruits 5FMC to Zbp-89 and that Zbp-89 is subsequently desumoylated by Senp3, resulting in higher Pol II levels on Zbp-89 target genes. We anticipated that depletion for Senp3 and Pelp1 would also affect H3K4 and/or H3K27 methylation of these regions, as 5FMC might be connected with the MLL1-WDR5 (38) and Prc1 complexes (this paper). However, no changes were observed, which is in line with the observation that overexpression of catalytically inactive Senp3 does not affect diMeH3K4 of specific promoters (48).
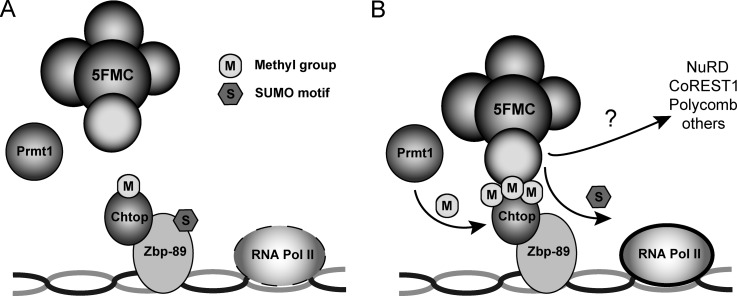
Collectively, these observations suggest a model where methylated Chtop recruits the 5FMC complex to factors like Zbp-89. Subsequently, the Senp3 protease desumoylates Zbp-89 and possibly additional components of repressor complexes, resulting in the stimulation of transcription of target genes (Fig. 7).
Fig. 7.
5FMC complex recruitment to Zbp-89 stimulates transcription of target genes. A–B, Model illustrating 5FMC complex function in Zbp-89 dependent gene expression. Methylated Chtop recruits the 5FMC complex to transcription factor Zbp-89. Upon 5FMC binding, Senp3 protease desumoylates Zbp-89 and possibly additional components of repressor complexes, resulting in the stimulation of transcription of target genes.
Supplementary Material
Acknowledgments
We thank Karel Bezstarosti and Erikjan Rijkers for technical assistance.
Footnotes
* This work was supported by the Netherlands Genomics Initiative (93518009 and 93511036), the Landsteiner Foundation for Blood Transfusion Research (1040), the Netherlands Scientific Organization (NWO DN 82-301 and ZonMW 912-07-019 and 40-00812-98-08032) and the Centre for Biomedical Genetics.
 This article contains supplemental Tables S1 and S2 and Figs. S1 to S8.
This article contains supplemental Tables S1 and S2 and Figs. S1 to S8.
AUTHOR CONTRIBUTIONS: F.G., S.P. and T.B. designed experiments. P.F., N.G., T.B., F.E., F.P. and A.A. performed experiments. P.F., T.B. analyzed results. J.D. provided expertise, analysis tools and infrastructure. J.D. analyzed data. P.F., S.P., and T.B. wrote the paper.
1 The abbreviations used are:
- GAR
- glycine-arginine-rich
- aDMA
- asymmetrically dimethylated arginines
- sDMA
- symmetrically dimethylated arginines
- SUMO
- Small ubiquitin-like modifier
- SIMs
- SUMO interacting motifs
- Chtop
- Chromatin target of Prmt1
- HGNC
- HUGO Gene Nomenclature Committee
- MEL
- mouse erythroleukemia.
REFERENCES
- 1. Verger A., Perdomo J., Crossley M. (2003) Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 4, 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bedford M. T., Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pawlak M. R., Scherer C. A., Chen J., Roshon M. J., Ruley H. E. (2000) Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol. Cell. Biol. 20, 4859–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O'Brien K. B., Alberich-Jordà M., Yadav N., Kocher O., Diruscio A., Ebralidze A., Levantini E., Sng N. J., Bhasin M., Caron T., Kim D., Steidl U., Huang G., Halmos B., Rodig S. J., Bedford M. T., Tenen D. G., Kobayashi S. (2010) CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development 137, 2147–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao X., Jankovic V., Gural A., Huang G., Pardanani A., Menendez S., Zhang J., Dunne R., Xiao A., Erdjument-Bromage H., Allis C. D., Tempst P., Nimer S. D. (2008) Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 22, 640–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mowen K. A., Tang J., Zhu W., Schurter B. T., Shuai K., Herschman H. R., David M. (2001) Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell 104, 731–741 [DOI] [PubMed] [Google Scholar]
- 7. Wang H., Huang Z. Q., Xia L., Feng Q., Erdjument-Bromage H., Strahl B. D., Briggs S. D., Allis C. D., Wong J., Tempst P., Zhang Y. (2001) Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293, 853–857 [DOI] [PubMed] [Google Scholar]
- 8. An W., Kim J., Roeder R. G. (2004) Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117, 735–748 [DOI] [PubMed] [Google Scholar]
- 9. Rezai-Zadeh N., Zhang X., Namour F., Fejer G., Wen Y. D., Yao Y. L., Gyory I., Wright K., Seto E. (2003) Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 17, 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Covic M., Hassa P. O., Saccani S., Buerki C., Meier N. I., Lombardi C., Imhof R., Bedford M. T., Natoli G., Hottiger M. O. (2005) Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. EMBO J. 24, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li X., Hu X., Patel B., Zhou Z., Liang S., Ybarra R., Qiu Y., Felsenfeld G., Bungert J., Huang S. (2010) H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood 115, 2028–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng D., Côté J., Shaaban S., Bedford M. T. (2007) The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25, 71–83 [DOI] [PubMed] [Google Scholar]
- 13. Hay R. T. (2005) SUMO: a history of modification. Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 14. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 15. Stielow B., Sapetschnig A., Wink C., Krüger I., Suske G. (2008) SUMO-modified Sp3 represses transcription by provoking local heterochromatic gene silencing. EMBO Rep. 9, 899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fernández-Lloris R., Osses N., Jaffray E., Shen L. N., Vaughan O. A., Girwood D., Bartrons R., Rosa J. L., Hay R. T., Ventura F. (2006) Repression of SOX6 transcriptional activity by SUMO modification. FEBS Lett. 580, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 17. Wang J., Scully K., Zhu X., Cai L., Zhang J., Prefontaine G. G., Krones A., Ohgi K. A., Zhu P., Garcia-Bassets I., Liu F., Taylor H., Lozach J., Jayes F. L., Korach K. S., Glass C. K., Fu X. D., Rosenfeld M. G. (2007) Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446, 882–887 [DOI] [PubMed] [Google Scholar]
- 18. Garcia-Dominguez M., Reyes J. C. (2009) SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim. Biophys. Acta 1789, 451–459 [DOI] [PubMed] [Google Scholar]
- 19. van Dijk T. B., Gillemans N., Stein C., Fanis P., Demmers J., van de Corput M., Essers J., Grosveld F., Bauer U. M., Philipsen S. (2010) Friend of Prmt1, a novel chromatin target of protein arginine methyltransferases. Mol. Cell. Biol. 30, 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zullo A. J., Michaud M., Zhang W., Grusby M. J. (2009) Identification of the small protein rich in arginine and glycine (SRAG): a newly identified nucleolar protein that can regulate cell proliferation. J. Biol. Chem. 284, 12504–12511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Dijk T. B., Gillemans N., Pourfarzad F., van Lom K., von Lindern M., Grosveld F., Philipsen S. (2010) Fetal globin expression is regulated by Friend of Prmt1. Blood 116, 4349–4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nair S. S., Mishra S. K., Yang Z., Balasenthil S., Kumar R., Vadlamudi R. K. (2004) Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res. 64, 6416–6423 [DOI] [PubMed] [Google Scholar]
- 23. Vadlamudi R. K., Wang R. A., Mazumdar A., Kim Y., Shin J., Sahin A., Kumar R. (2001) Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor alpha. J. Biol. Chem. 276, 38272–38279 [DOI] [PubMed] [Google Scholar]
- 24. Kaufman R. J., Davies M. V., Pathak V. K., Hershey J. W. (1989) The phosphorylation state of eucaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Mol. Cell. Biol. 9, 946–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Needham M., Gooding C., Hudson K., Antoniou M., Grosveld F., Hollis M. (1992) LCR/MEL: a versatile system for high-level expression of heterologous proteins in erythroid cells. Nucleic Acids Res. 20, 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Boer E., Rodriguez P., Bonte E., Krijgsveld J., Katsantoni E., Heck A., Grosveld F., Strouboulis J. (2003) Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 100, 7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Dijk T. B., van Den Akker E., Amelsvoort M. P., Mano H., Löwenberg B., von Lindern M. (2000) Stem cell factor induces phosphatidylinositol 3′-kinase-dependent Lyn/Tec/Dok-1 complex formation in hematopoietic cells. Blood 96, 3406–3413 [PubMed] [Google Scholar]
- 28. Vizcaino J. A., Côté R., Reisinger F., Barsnes H., Foster J. M., Rameseder J., Hermjakob H., Martens L. (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 38, D736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barsnes H., Vizcaino J. A., Eidhammer I., Martens L. (2009) PRIDE Converter: making proteomics data-sharing easy. Nat. Biotechnol. 27, 598–599 [DOI] [PubMed] [Google Scholar]
- 30. Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E. (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 31. Zufferey R., Nagy D., Mandel R. J., Naldini L., Trono D. (1997) Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15, 871–875 [DOI] [PubMed] [Google Scholar]
- 32. Esteghamat F., van Dijk T. B., Braun H., Dekker S., van der Linden R., Hou J., Fanis P., Demmers J., van Ijcken W., Ozgur Z., Horos R., Pourfarzad F., von Lindern M., Philipsen S. (2011) The DNA binding factor Hmg20b is a repressor of erythroid differentiation. Haematologica 96, 1252–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shao Z., Raible F., Mollaaghababa R., Guyon J. R., Wu C. T., Bender W., Kingston R. E. (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98, 37–46 [DOI] [PubMed] [Google Scholar]
- 34. Alkema M. J., Bronk M., Verhoeven E., Otte A., van 't Veer L. J., Berns A., van Lohuizen M. (1997) Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Dev. 11, 226–240 [DOI] [PubMed] [Google Scholar]
- 35. Levine S. S., Weiss A., Erdjument-Bromage H., Shao Z., Tempst P., Kingston R. E. (2002) The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22, 6070–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nagata K., Kawase H., Handa H., Yano K., Yamasaki M., Ishimi Y., Okuda A., Kikuchi A., Matsumoto K. (1995) Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc. Natl. Acad. Sci. U.S.A. 92, 4279–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hautbergue G. M., Hung M. L., Walsh M. J., Snijders A. P., Chang C. T., Jones R., Ponting C. P., Dickman M. J., Wilson S. A. (2009) UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr. Biol. 19, 1918–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dou Y., Milne T. A., Tackett A. J., Smith E. R., Fukuda A., Wysocka J., Allis C. D., Chait B. T., Hess J. L., Roeder R. G. (2005) Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121, 873–885 [DOI] [PubMed] [Google Scholar]
- 39. Finkbeiner E., Haindl M., Muller S. (2011) The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 30, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Castle C. D., Cassimere E. K., Denicourt C. (2012) LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol. Biol. Cell 23, 716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vadlamudi R. K., Kumar R. (2007) Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl. Recept. Signal. 5, e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nishida T., Tanaka H., Yasuda H. (2000) A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267, 6423–6427 [DOI] [PubMed] [Google Scholar]
- 43. Kitano E., Hayashi A., Kanai D., Shinmyozu K., Nakayama J. (2011) Roles of fission yeast grc3 protein in ribosomal RNA processing and heterochromatic gene silencing. J. Biol. Chem. 286, 15391–15402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chupreta S., Brevig H., Bai L., Merchant J. L., Iñiguez-Lluhi J. A. (2007) Sumoylation-dependent control of homotypic and heterotypic synergy by the Kruppel-type zinc finger protein ZBP-89. J. Biol. Chem. 282, 36155–36166 [DOI] [PubMed] [Google Scholar]
- 45. Krogan N. J., Peng W. T., Cagney G., Robinson M. D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D. P., Beattie B. K., Lalev A., Zhang W., Davierwala A. P., Mnaimneh S., Starostine A., Tikuisis A. P., Grigull J., Datta N., Bray J. E., Hughes T. R., Emili A., Greenblatt J. F. (2004) High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 13, 225–239 [DOI] [PubMed] [Google Scholar]
- 46. Doseff A. I., Arndt K. T. (1995) LAS1 is an essential nuclear protein involved in cell morphogenesis and cell surface growth. Genetics 141, 857–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Malovannaya A., Lanz R. B., Jung S. Y., Bulynko Y., Le N. T., Chan D. W., Ding C., Shi Y., Yucer N., Krenciute G., Kim B. J., Li C., Chen R., Li W., Wang Y., O'Malley B. W., Qin J. (2011) Analysis of the human endogenous coregulator complexome. Cell 145, 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ouyang J., Shi Y., Valin A., Xuan Y., Gill G. (2009) Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol. Cell. 34, 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nair S. S., Nair B. C., Cortez V., Chakravarty D., Metzger E., Schüle R., Brann D. W., Tekmal R. R., Vadlamudi R. K. (2010) PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-alpha target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 11, 438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mishra S. K., Balasenthil S., Nguyen D., Vadlamudi R. K. (2004) Cloning and functional characterization of PELP1/MNAR promoter. Gene 330, 115–122 [DOI] [PubMed] [Google Scholar]
- 51. Rosendorff A., Sakakibara S., Lu S., Kieff E., Xuan Y., DiBacco A., Shi Y., Gill G. (2006) NXP-2 association with SUMO-2 depends on lysines required for transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 103, 5308–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wotton D., Merrill J. C. (2007) Pc2 and SUMOylation. Biochem. Soc. Trans. 35, 1401–1404 [DOI] [PubMed] [Google Scholar]
- 53. Kagey M. H., Melhuish T. A., Wotton D. (2003) The polycomb protein Pc2 is a SUMO E3. Cell 113, 127–137 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.