Abstract
Inflammasomes are cytoplasmic receptors that can recognize intracellular pathogens or danger signals and are critical for interleukin 1β production. Although several key components of inflammasome activation have been identified, there has not been a systematic analysis of the protein components found in the stimulated complex. In this study, we used the isobaric tags for relative and absolute quantification approach to systemically analyze the interactomes of the NLRP3, AIM2, and RIG-I inflammasomes in nasopharyngeal carcinoma cells treated with specific stimuli of these interactomes (H2O2, poly (dA:dT), and EBV noncoding RNA, respectively). We identified a number of proteins that appeared to be involved in the interactomes and also could be precipitated with anti-apoptosis-associated speck-like protein containing caspase activation and recruitment domain antibodies after stimulation. Among them, end binding protein 1 was an interacting component in all three interactomes. Silencing of end binding protein 1 expression by small interfering RNA inhibited the activation of the three inflammasomes, as indicated by reduced levels of interleukin 1β secretion. We confirmed that end binding protein 1 directly interacted with AIM2 and ASC in vitro and in vivo. Most importantly, fluorescence confocal microscopy showed that end binding protein 1 was required for formation of the speck-like particles that represent activation of the AIM2 inflammasome. In nasopharyngeal carcinoma tissues, immunohistochemical staining showed that end binding protein 1 expression was elevated and significantly correlated with AIM2 and ASC expression in nasopharyngeal carcinoma tumor cells. In sum, we profiled the interactome components of three inflammasomes and show for the first time that end binding protein 1 is crucial for the speck-like particle formation that represents activated inflammasomes.
Nasopharyngeal carcinoma (NPC)1 is a malignancy of the head and the neck that is highly prevalent in Southern China and Southeast Asia (1). Both environmental and genetic risk factors are considered to be important for the development of NPC (2, 3); among them, Epstein-Barr virus (EBV) infection of the epithelium is the most important known factor (1). In addition to the EBV-encoded oncoprotein-mediated blockade of intracellular mechanisms in EBV-associated tumors (1), chronic inflammation is considered to be an important oncogenic factor in NPC (4). Interleukin 1 beta (IL-1β), which is an inflammatory cytokine that has oncogenic effects in many tumors (5), can be detected in NPC tumor tissues (6, 7). IL-1β secretion is mediated by cytosolic protein complexes called inflammasomes, which induce IL-1β secretion by activating catalytic caspase 1 (8). However, no previous study has examined inflammasome components in NPC tumor cells or the mechanisms of inflammasome regulation in NPC.
Inflammasomes are cytoplasmic receptors that act in innate immunity to recognize intracellular pathogen-associated molecular pattern (PAMP) or danger signal-associated molecular pattern (DAMP). A number of inflammasomes have been identified in recent years (9), and they can be classified into different subgroups according to their recognizing PAMP. These groups include the NOD-like receptors (NLR), which sense intracellular pathogens (e.g. bacteria, fungi, and parasites) and activate pro-caspase 1 with or without an adaptive protein called apoptosis-associated speck-like protein containing caspase activation and recruitment domain (ASC) (10). Activated caspase 1 then induces IL-1β secretion through direct cleavage of pro-IL-1β (8). Among the NLR family members, the NLRP3 inflammasome recognizes both pathogens and danger signals such as ATP or reactive oxygen species (ROS) generation (11, 12). Members of the two other subgroups, absence in melanoma 2 (AIM2) and retinoic acid-inducible gene I (RIG-I), sense cytoplasmic double-strand DNA and 5′-triphoshphate RNA, respectively, and then recruit ASC to activate pro-caspase 1 (13, 14). Although inflammasomes are important for pathogen defense in immune cells, recent studies have shown that inflammasomes also participate in tumorigenesis in colon cancer and melanoma (15–17). A previous report showed that EBV noncoding RNAs (EBERs) are recognized by RIG-I and activate signaling to induce type I IFN in EBV-infected B lymphocytes (18). This report is consistent with our recent unpublished observation that RIG-I is activated by EBERs in NPC cells. We additionally show that NLRP3 is triggered by tumor microenvironmental factors, such as ATP and ROS, and the clinical drug cisplatin; AIM2 recognizes EBV genomic DNA and is activated by irradiation in NPC cells. Although these inflammasomes play important role in NPC, the regulation and the interactome of these inflammasome complexes are not fully understood.
On activation by PAMP or DAMP, the activated inflammasomes tend to aggregate in the cytosol as speck-like particles (13). Biochemical and cell biological data have indicated that the core components of the inflammasome comprise the receptor, ASC, and pro-caspase 1, but an increasing number of proteins have been identified as interacting with these complexes. For example, heat-shock protein 90 (HSP90) is essential for the function of the NLRP3 and RIG-I inflammasomes (19, 20). NLRC5, another member of the NLR family, is involved in the NLRP3 inflammasome and is required for its activity (21). Rac1, a small Rho GTPase family member, is reportedly required for NLRP3 inflammasome activation during C. pneumoniae infection (22). The S. Typhimurium effector, SopE, activates caspase 1 through Rac1 activity (23), whereas Yersinia bacteria prevent caspase 1 activation by inhibiting Rac1 activity via the effector protein, YopE (24). Notably, Rac1 regulates cytoskeletal rearrangement (25), suggesting that cytoskeletal components may participate in inflammasome activation.
End-binding protein 1 (EB1), an adenomatous polyposis coli (APC)-binding protein, regulates microtubule polymerization by recruiting the plus-end tracking protein (+TIP) complex to the plus end of microtubules (26). The interaction of EB1 and the +TIP complex depends on the C-terminal (CT) domain of EB1, whereas the calponin homology (CH) domain of EB1 binds to the microtubule (26). Many studies have shown that EB1 participates in different biological processes, including mitosis, migration and signal transduction (27–29), and also that it plays an oncogenic role in cancer by affecting cell growth or migration (30, 31). However, although EB1 is known to be a cytoskeletal component that is regulated by the small GTPase, RhoA (28), its role in inflammasome activation has not yet been explored.
Here, we used the isobaric tags for relative and absolute quantification (iTRAQ) approach to systemically analyze the interactomes of the NLRP3, AIM2, and RIG-I inflammasomes in NPC cell lines treated with their specific stimuli, H2O2, poly (dA:dT), and EBER, respectively. We characterized the interactomes of the NLRP3, AIM2, and RIG-I inflammasomes in NPC cells by proteomic analysis, and report for the first time that EB1 can directly bind to the AIM2 inflammasome and is essential for speck-like particle formation in NPC cells. Finally, we suggest some possible mechanisms for EB1-associated AIM2 inflammasome activation via microtubule polymerization and RhoA activity.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The anti-pro-caspase 1, anti-EB1 for IHC, anti-RhoA, anti-tubulin, anti-GST and anti-His antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-Flag antibody was purchased from Sigma-Aldrich (St. Louis, MO). The anti-ASC antibody for immunoprecipitation and Western blot analysis was purchased from Calbiochem (Darmstadt, Germany). The anti-ASC for immunohistochemistry (IHC) staining and anti-caspase 1 (p20) were from Merck Millipore (Billerica, MA). The anti-AIM2 antibody for Western blotting was purchased from Abnova (Taipei City, Taiwan), whereas that for IHC staining was from Deciphergen Biotechnology (Cheshire, CT). The anti-RIG-I antibody was from ENZO Life Sciences (Plymouth Meeting, PA). The anti-Rac1 and anti-cdc42 antibodies for Western blotting and the anti-EB1 antibody for Western blotting and immunofluorescence were purchased from BD Transduction Laboratories (BD Biosciences, San Jose, CA). The anti-FLAG M2 affinity gel, poly (dA:dT), LPS, PMA and nocodazole were purchased from Sigma-Aldrich (St Louis, MO), the H2O2 was purchased from Calbiochem (San Diego, CA), and the paclitaxel was purchased from Bristol-Myers Squibb Co. (New York, NY). Protein G Sepharose 4 Fast Flow, Glutathione Sepharose 4B, and Ni Sepharose 6 Fast Flow were all purchased from GE Healthcare. Cy3-labeled plasmid DNA was purchased from Mirus Bio LLC (Madison, WI).
Plasmid Construction
The wild-type EB1 construct, domain-only EB1 constructs, and the wild-type AIM2 construct were PCR amplified from HK1 cDNA and cloned into HindIII/BamHI-treated pFlag-CMV2 vectors (Clontech Laboratories, Inc.). Wild-type ASC was PCR amplified from HK1 cDNA and cloned into HindIII/BamHI-treated pEGFP-N1 vectors (Clontech Laboratories, Inc.). Wild-type AIM2 was cloned into HindIII/BamHI treated pDsRed-Monomer-C1 (a kind gift from Dr. S.J. Lo; Chung Gung University, Taiwan). For GST pull-down assays, wild type and domain-only EB1 were cloned into BamHI/NotI-treated pGEX-4T2 (BD Biosciences). The wild type and domain-only AIM2 and wild-type ASC were respectively cloned into NdeI/BamHI- and NdeI/XhoI- treated pET-15b vectors (a kind gift from Dr. S.C. Hsu; Chung Gung University, Taiwan). For AIM2 and PYD domain of AIM2-expressing HK1 cells, the AIM2 and PYD domain of AIM2 were cloned into NheI/AscI treated pLKO-AS2-neo empty vectors (RNAi Core, Taiwan). For ASC-expressing HK1 cells, the ASC was cloned into AscI/EcoRI treated pLKO-AS2-neo empty vectors (RNAi Core, Taiwan).
Cell Culture
NPC TW02 and HK1 cells were cultured as previously described (32, 33) and THP-1 cells were culture in Roswell Park Memorial Institute medium (33). Lentiviruses were established following the protocol of the RNAi Core of Taiwan (http://rnail.genmed.sinica.edu.tw), and used to generate HK1 cells expressing human ASC, wild-type AIM2, and the PYD domain of AIM2. The cells were maintained with 500 μg/ml G418 (Invitrogen, Carlsbad, CA). For transfection assays, cells were transfected with poly (dA:dT) and EBER using Lipofectamine 2000 (Invitrogen).
Immunoprecipitation of ASC Complexes
HK1/ASC cells were treated with H2O2 (10 μm) for 20 h, poly (dA:dT) for 6 h, or EBER for 6 h. Cell extracts (10 mg) were collected by radioimmunoprecipitation assay (RIPA) buffer (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 1 mm ethylenediamine tetraacetate, pH 8.0, 1% Nonidet P-40, 100 mm sodium fluoride, 1 mm phenylmethylenesulfonyl fluoride) and immunoprecipitated with anti-ASC antibodies (10 μg) for 24 h. The samples were then precipitated with Protein G Sepharose 4 Fast Flow for 2 h at 4 °C, and the immunoprecipitated products were collected for iTRAQ analysis.
Preparation of Cell Extracts and Digestion of Immunoprecipitated Products
Cell extracts were prepared as previously described (34). Briefly, NPC cells were washed three times with 10 ml of PBS and lysed in hypotonic buffer (10 mm Tris, pH 7.4, 1 mm EDTA, 1 mm EGTA, 50 mm NaCl, 50 mm NaF, 20 mm Na4P2O7, 1 mm Na3VO4, 1 mm PMSF, 1 mm benzamidine, 0.5 μg/ml leupeptin, and 1% Triton-X100) on ice for 15 min. The cell lysates of four samples were collected in parallel and sonicated on ice, followed by centrifugation at 10,000 × g for 25 min at 4 °C. The resulting supernatants were used as the cell extracts. Protein concentrations were determined using the bicinchoninic acid protein assay reagent from Pierce (Rockford, IL). For tryptic in-solution digestion, the immunoprecipitated products were denatured with 8 m urea containing 50 mm triethylammonium bicarbonate (TEABC, Sigma-Aldrich), reduced with 10 mm Tris (2-carboxyethyl)-phosphine (TCEP, Sigma-Aldrich) at 37 °C for 90 min, and then alkylated with 10 mm methyl methanethiosulfonate (MMTS, Sigma-Aldrich) at room temperature for 20 min. After desalting, the protein mixtures were in-solution digested overnight at 37 °C with modified sequencing grade trypsin (Promega, Madison, WI).
ITRAQ Reagent Labeling and Fractionation by Strong Cation Exchange Chromatography
The peptides were labeled with the iTRAQ reagent (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. Briefly, one unit of label (defined as the amount of reagent required to label 100 μg of protein) was thawed and reconstituted in 70 μl ethanol. The peptide mixtures were reconstituted with 25 μl iTRAQ dissolution buffer. Aliquots of iTRAQ 114, 115, 116, and 117 were combined with peptide mixtures from the immunoprecipitated products of untreated HK1 cells, HK1 cells treated with EBER, H2O2, and poly (dA:dT), respectively, and incubated at room temperature for 1 h. The peptide mixtures were then pooled and dried by vacuum centrifugation. Each dried peptide mixture was reconstituted and acidified with 0.5 ml buffer A (0.1% formic acid and 25% acetonitrile, pH 2.5) and subjected to fractionation by SCX chromatography using the Ettan MDLC system (GE Healthcare, Taipei, Taiwan). The iTRAQ-labeled peptides were loaded onto a 2.1 mm × 250 mm BioBasic SCX column with a 5-μm particle size and a 300-μm pore size (Thermo Electron, San Jose, CA). The peptides were eluted at a flow rate of 100 μl/min with the following gradient: 0–10% buffer B (300 mm NH4Cl, 0.1% formic acid and 25% acetonitrile, pH 2.5) for 20 min, 10–20% buffer B for 35 min, 20–50% buffer B for 15 min, and 50–100% buffer B for 10 min. The elution was monitored by absorbance at 220 nm, and fractions were collected every 1 min. Each fraction was vacuum dried, resuspended in 0.1% formic acid (20 μl), and subjected to desalting and concentration using a Zip-tip packed in-house with C18 resin (5–20 μm, LiChroprep RP-18; Merck).
Liquid Chromatography-electrospray Ionization-tandem MS (LC-ESI MS/MS) Analysis by LTQ-OrbitrapPQD
For analysis of the iTRAQ-labeled peptide mixtures, each peptide fraction was reconstituted in HPLC buffer A (0.1% formic acid), loaded across a trap column (Zorbax 300SB-C18, 0.3 × 5 mm; Agilent Technologies, Wilmington, DE) at a flow rate of 0.2 μl/min in HPLC buffer A, and separated on a resolving 10-cm analytical C18 column (inner diameter, 75 μm; tip, 15-μm; New Objective, Woburn, MA). The peptides were eluted at a flow rate of 0.25 μl/min using the following linear gradient: 2–30% HPLC buffer B (99.9% acetonitrile containing 0.1% formic acid) for 63 min, 30–45% buffer B for 5 min, and 45–95% buffer B for 2 min. The LC setup was coupled on-line to an LTQ-Orbitrap Discovery system (Thermo Fisher, San Jose, CA) operated using the Xcalibur 2.0 software (Thermo Fisher). Intact peptides were detected in the Orbitrap at a resolution of 30,000. Internal calibration was performed using the ion signal of (Si(CH3)2O)6H+ at m/z 445.120025 as a lock mass (35). Peptides were selected for MS/MS using the PQD operating mode with a normalized collision energy setting of 27%, and fragment ions were detected in the LTQ system (36, 37). We used a data-dependent procedure that alternated between one MS scan followed by three MS/MS scans for the three most abundant precursor ions in the MS survey scan. The m/z values selected for MS/MS were dynamically excluded for 180 s. The electrospray voltage applied was 1.8 kV. Both MS and MS/MS spectra were acquired using the 4 microscan setting with a maximum fill-time of 1000 and 100 ms for MS and MS/MS, respectively. Automatic gain control was used to prevent over-filling of the ion trap, and 5 × 104 ions were accumulated in the ion trap for generation of the PQD spectra. For MS scans, the m/z scan range was set at 350 to 2000 Da.
Database Searching and Data Analysis
All MS/MS samples were analyzed using the MASCOT search algorithm (Version 2.1, Matrix Science, London, UK). Mascot was used to search the SwissProt database (released Jun 15, 2010, selected for Homo sapiens, 20367 entries), assuming trypsin as the digestion enzyme. For protein identification, the following settings were used: 10 ppm mass tolerance was permitted for intact peptide masses; 0.5 Da was allowed for PQD fragment ions; one missed cleavage was allowed from the trypsin digest; oxidized methionine (+16 Da) was accepted as a potential variable modification; and iTRAQ (Nterminal, +144 Da), iTRAQ (K, +144 Da) and MMTS (C, +46 Da) were taken as the fixed modifications.
The MASCOT search results for each SCX elution were further processed using the Trans-Proteomic Pipeline (TPP, version 4.0), which includes the programs PeptideProphet, ProteinProphet, and Libra (38–40). PeptideProphet is a peptide probability score program that aids in the assignment of peptide MS spectra (41); it assigns peptides to a unique protein or protein family, allows filtering of large-scale data sets, and predicts the sensitivity and false-positive identification error rates (42). We used a ProteinProphet probability score of ≥ 0.90 to ensure an overall false positive rate below 1%, and excluded proteins identified with only a single peptide hit. Protein quantification was achieved with the Libra program (38, 39, 43), using the default setting. A weighted average of the peptide iTRAQ ratios per protein was used to quantify the protein. The following were excluded: peptides with iTRAQ reporter ion intensities lower than 30, peptides with an iTRAQ ratio beyond twofold deviation from the mean ratio, and proteins with ≤ 2 spectra. Proteins with iTRAQ ratios below the low range (mean ratio - 0.2) were considered to be decreased in the ASC-associated protein complexes, whereas those above the high range (mean ratio + 0.2) were considered to be increased in the protein complexes. Information on PeptideProphet, ProteinProphet, and Libra can be found on the website for the Institute for Systems Biology of the Seattle Proteome Center (http://www.proteomecenter.org/).
RNA Interference
SMARTpool reagents, including four 21-bp RNA duplexes targeting each of RIG-I, RhoA and cdc42 were purchased from Dharmacon (Lafayette, CO). Stealth RNAi reagents, including three 25-bp RNA duplexes targeting each of NLRP3, AIM2, EB1, and Rac1 were purchased from Invitrogen (Carlsbad, CA). The negative-control siRNA was synthesized by Research Biolabs (Singapore). Cells were transfected with 75 pmol dsRNA duplexes using Lipofectamine 2000 according to the manufacturer's instructions. After 6 h incubation, the dsRNA complexes were removed and the cells were re-plated in 5 ml fresh medium. The oligonucleotide sequences of the dsRNA duplexes are presented in supplementary Table S3.
In Vitro Synthesis of EBER1
In this study, we used EBER1 as the EBER stimulation. EBER1 cDNAs were generated from C666–1 NPC cells by reverse transcription (primer: 5′-CCCTTTACATGTTGTGGGTGCAAAACTAGCCA) and cloned downstream of the T7 promoter in the pGEMTeasy vector (Promega) to generate pGEMT-EBER1. T7 promoter-fused EBER1 was amplified by PCR (primers: EBER1, 5′-TGTAATACGACTCACTATAGGGAGGACCTACGCTGCCCTAGAGGTT and 5′-AAAACATGCGGACCACCAGCTGGTACTTG), and 1 μg of purified PCR product was used for in vitro transcription, which was performed for 4 h according to the manufacturer's instructions (Epicenter, Madison, WI). Purified EBER1 (3 μg) treated with or without RNase A was checked on a 6% denaturing polyacrylamide gel and by RT-PCR.
IL-1β ELISA
Cell culture supernatants were assayed for human IL-1β using the Human IL-1β ELISA Ready-SET-Go Kit (eBioscience, San Diego, CA) according to the manufacturer's instructions.
Quantitative RT-PCR
Quantitative RT-PCR analysis was performed as previously described (34). The primer sequences for NLRP3 were 5′ TCTGTGTGTGGGACTGAAGCA (sense) and 5′ TACTGATGCAAGATCCTGACAACA (antisense). The gene expression results were normalized with regard to the expression of GADPH.
GST Pull-down Assays
All recombinant proteins were expressed in BL21 (DE3) E. coli. Glutathione-Sepharose beads complexed with GST, wild-type GST-EB1 and domain-only GST-EB1 were respectively incubated with purified wild-type His-AIM2 or wild-type His-ASC in RIPA buffer for 24 h at 4 °C. The Ni-Sepharose beads complexed with His, wild-type His-AIM2 and domain-only His-AIM2 were respectively incubated with purified wild-type GST-EB1 in RIPA buffer for 24 h at 4 °C. After six washes in RIPA buffer, all samples were collected for Western blot analysis.
Western Blot Analysis
Whole-cell extracts were homogenized and lysed in a buffer containing 50 mm Tris-Cl (pH 7.5), 150 mm NaCl, 10 mm MgCl2, 1 mm ethylenediamine tetraacetate (pH 8.0), 1% Nonidet P-40, 100 mm sodium fluoride, 1 mm phenylmethylenesulfonyl fluoride, and 2 μl/ml protease inhibitor mixture (Sigma-Aldrich). Protein concentrations were determined using a protein assay reagent (Bio-Rad), and equal amounts of proteins (30–50 μg/lane) were resolved on a 12% SDS-polyacrylamide gel. The proteins were then electro-transferred onto nitrocellulose (NC) membranes (Amersham Biosciences). The blots were blocked with 5% nonfat powdered milk in TBS, incubated overnight with the respective primary antibodies at 4 °C, and then incubated with HRP-conjugated rabbit/goat/mouse anti-IgG (Invitrogen) for 1 h at room temperature. Protein bands were detected by enhanced chemiluminescence (Pierce ECL, Thermo Scientific) on Fuji SuperRx films.
Immunofluorescence Microscopy
Cells grown on polylysine-coated coverslips were fixed with 3.7% formaldehyde, permeabilized, and blocked with 0.1% saponin containing 1% BSA for 20 min. The coverslips were incubated with the indicated primary antibodies for 2 h, and then with the appropriate fluorophore-conjugated secondary antibodies for 1 h at room temperature. Nuclei were stained with 4′-6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). All coverslides were mounted with the VECTASHEILD reagent (Vector Laboratories Inc., CA, USA) and visualized by confocal microscopy using a ZEISS LSM510 META laser scanning microscope (Carl Zeiss, Germany) with a 63 × 1.32 NA oil immersion objective.
Clinical Specimens
The retrospective cohort comprised 95 NPC patients who were admitted to Chang Gung Memorial Hospital (CGMH) at Lin-Kou from 1990 to 2000. Clinical stage was defined according to the 2002 cancer staging system revised by the American Joint Committee on Cancer. These included 32 stage-I 26 stage-II, 20 stage-III, and 17 stage-IV patients comprising 70 men and 25 women ranging from 14 to 78 years of age (mean age 46). Histological typing was done according to the World Health Organization (WHO) classification criteria (44). This study was reviewed and approved by the institutional review board and ethics committee of CGMH. Informed consent was obtained from all patients.
Immunohistochemistry
Samples of NPC and adjacent normal nasopharyngeal tissues were obtained from patients undergoing surgery, and were frozen immediately after surgical resection. Immunohistochemistry was performed according to the previously described procedures (45, 46). Staining for AIM2, ASC, and EB1 was carried out using the Envision-kit (DAKO, Carpinteria, CA). Briefly, the sections were deparaffinized with xylene, dehydrated with ethanol, and then heated in 0.01 m citrate buffer (pH 6.0). Endogenous peroxidase activities were inactivated in 3% H2O2 for 10 min at room temperature, and the sections were blocked with 3% normal goat serum in 0.2 m PBS (pH 7.4). Samples were then incubated with specific antibody for 2 h at room temperature. Secondary antibody-coated polymer peroxidase complexes (DAKO) were then applied for 30 min at room temperature, followed by treatment with substrate/chromogen (DAKO) and a further incubation for 5–10 min at room temperature. The slides were counterstained with hematoxylin, and protein expression was assessed by quantitative scoring of the staining intensity and the proportion of positively stained cells. The staining intensity was graded as 0, 1, 2, or 3 to indicate undetectable, weak, moderate, and strong staining, respectively. These scores were multiplied by the percentage of cells that showed positive staining. The resulting scores, which were taken as reflecting protein expression, were used to classify the specimens/patients into two groups: “high-level” expression (ASC scores > 120, AIM2 scores > 140, EB1 scores > 60) and “low-level” expression (ASC scores ≤ 120, AIM2 scores ≤ 140, EB1 scores ≤ 60). ASC-, AIM2-, EB1-positive tumor cells in representative microscopic fields were scored independently by two experienced pathologists.
Statistical Analysis
Statistical analyses were performed using the SPSS 13.0 statistical software package. Expression correlations were evaluated using the Chi-Square test. The cut-off values to define high intensity of IHC staining in tumor cells were determined from Receiver Operating Characteristics curve analysis.
RESULTS
Identification of Protein Components Involved in Inflammasome Complexes of NPC Cells by iTRAQ-LC-MS/MS Analysis
Many reports have shown that inflammasomes are involved in tumor development (15, 47, 48). In NPC, we found that NLRP3, AIM2, and RIG-I inflammasomes were up-regulated in an NPC Affymetrix microarray database (supplemental Fig. S1). These three inflammasomes require the adaptive protein, ASC, to activate pro-caspase 1. Here, in an effort to identify novel protein components of activated inflammasome complexes in NPC cells, we conducted immunoprecipitation of ASC complexes from ASC-expressing HK1 (HK1/ASC) cells untreated or treated with EBER, H2O2, and poly (dA:dT) using total cell extract, followed by iTRAQ-based quantitative proteomic analysis (Fig. 1A) and two-dimensional liquid chromatography-mass spectrometry (LC-MS/MS). The two-dimensional fractionation of the labeled peptides involved an offline SCX-based separation followed by an online reverse phase fractionation (Fig. 1A). The resulting MS/MS spectra were analyzed using the SwissProt database (released Jun 15, 2010, selected for Homo sapiens, 20367 entries) with the MASCOT algorithm. The search results were further evaluated using the open-source TPP software (version 4.0) with stringent criteria regarding protein probability (≥0.90) and at least two peptide hits for each identified protein. The false discovery rate (FDR) of protein detection was empirically determined by searching the dataset against a random SwissProt database using the same search parameters and TPP cutoffs. The estimated FDR of 0.9% was calculated as the number of reverse proteins divided by the number of forward proteins.
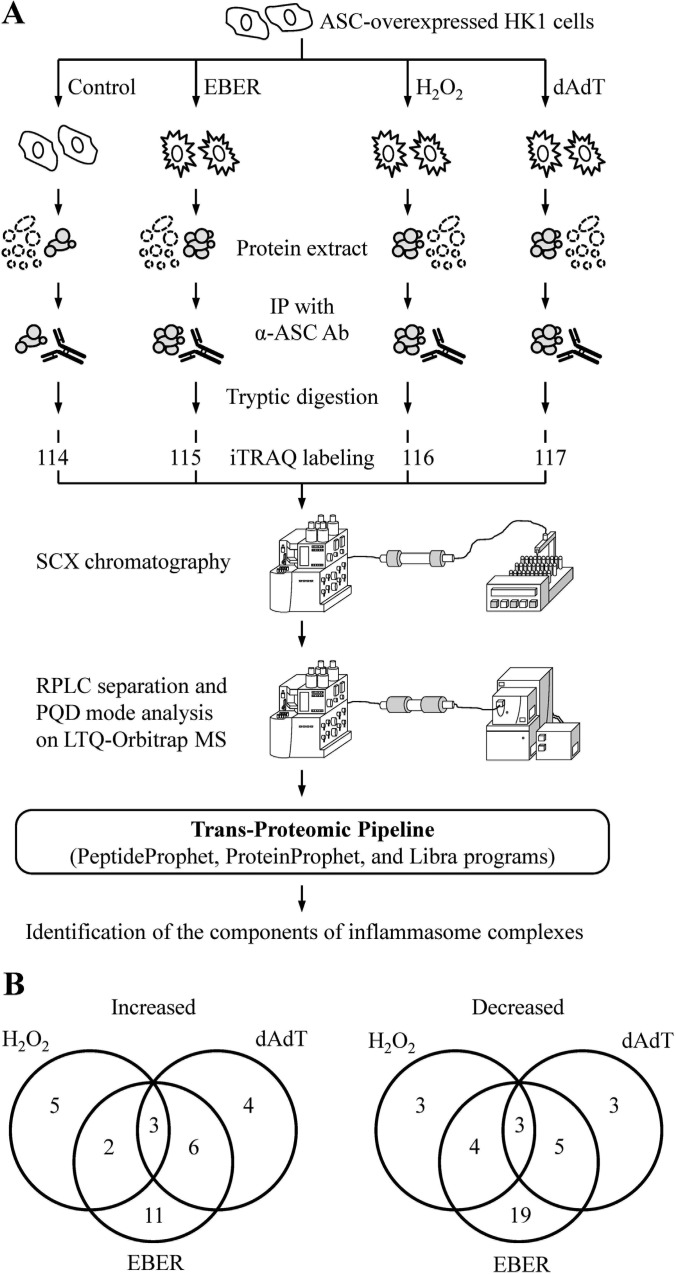
Fig. 1.
Differentially expressed proteins in the activated inflammasome complexes of HK1/ASC cells, as assessed by iTRAQ. A, A flow chart showing our iTRAQ-based proteomic analysis of inflammasome complexes. B, The numbers of proteins found to be elevated or decreased after H2O2, poly (dA:dT) or EBER treatment, as assessed by iTRAQ.
Using this approach, we identified 172 nonredundant proteins in the inflammasome complexes of HK1/ASC cells (supplementary Table S1). After the removal of proteins with fewer than three spectra, we obtained 131 proteins that we considered quantifiable complex components (supplementary Table S2). Among them, comparing with untreated control, we detected 8, 13, and 22 proteins with increase levels and 10, 11, and 31 proteins with decrease levels in inflammasome complexes from HK1/ASC cells treated with H2O2, poly (dA:dT), and EBER, respectively (Table I; Fig. 1B). Among the 22 proteins associated with EBER treatment (i.e. RIG-I inflammasome activation), six belong to the heterogeneous nuclear ribonucleoprotein family. Consistent with previous reports (19, 20), we also identified heat shock protein 90 (HSP90) (supplementary Table S1) as being involved in the NLRP3 and RIG-I inflammasomes, respectively. Notably, we found three proteins that were elevated after all treatments: (1) dihydrolipoamide S-succinyltransferase (DLST), which catalyzes the overall conversion of 2-oxoglutarate to succinyl-CoA and CO (49); (2) end-binding protein 1 (EB1), which is involved in microtubule polymerization, mitosis, and migration (26); (3) and the 60S ribosomal protein, L34 (RL34), which participates in translation (50). Our findings suggest that these proteins may be recruited to the inflammasome complexes upon stimulation. Three other proteins were found to be down-regulated after all treatments: (1) structural maintenance of chromosomes protein 3 (SMC3), which is involved in mitosis (51); (2) thrombospondin-1 (TSB1), which participates in tumor development (52); (3) and titin (TTN), which regulates the function of muscle cells (53). These proteins may play negative regulatory roles in inflammasome activation. To investigate the components of the activated inflammasome, we herein focused on the up-regulated proteins identified in our iTRAQ assay.
Table I. The list of level changed proteins in ASC-pulled down complexes after treatments.
“UN” means untreatment; “-” means no difference after treatment.
| Gene symbol | Accession No. | Protein name | Protein probability | No. of identified peptides | Present coverage % | iTRAQ ratio |
Peptide No. for quantification | Quantitative result |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H2O2/UN (116/114) | S.D. | dAdT/UN (117/114) | S.D. | EBER/UN (115/114) | S.D. | H2O2 | dAdT | EBER | |||||||
| ALB | P02768 | Serum albumin | 1.00 | 8 | 11.7 | 1.21 | 0.40 | 1.06 | 0.27 | 1.04 | 0.41 | 29 | Up | - | - |
| ALS2CR4 | Q96Q45 | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein | 1.00 | 4 | 15.3 | 1.31 | 0.23 | 1.06 | 0.23 | 1.11 | 0.24 | 11 | Up | - | - |
| CAV1 | Q03135 | Caveolin-1 | 1.00 | 2 | 9.6 | 0.73 | 0.15 | 0.84 | 0.08 | 0.80 | 0.10 | 7 | Down | - | Down |
| CCT3 | P49368 | T-complex protein 1 subunit gamma | 1.00 | 3 | 6.2 | 0.88 | 0.15 | 0.95 | 0.14 | 0.76 | 0.11 | 6 | - | - | Down |
| CCT6A | P40227 | T-complex protein 1 subunit zeta | 1.00 | 5 | 18.5 | 0.98 | 0.16 | 1.33 | 0.15 | 0.95 | 0.21 | 5 | - | Up | - |
| DHX9 | Q08211 | ATP-dependent RNA helicase A | 1.00 | 3 | 2.0 | 1.07 | 0.09 | 0.93 | 0.06 | 1.43 | 0.20 | 7 | - | - | Up |
| DLST | P36957 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | 1.00 | 2 | 4.6 | 1.66 | 0.70 | 2.42 | 1.46 | 2.19 | 1.19 | 3 | Up | Up | Up |
| DYNC1H1 | Q14204 | Cytoplasmic dynein 1 heavy chain 1 | 1.00 | 3 | 0.9 | 0.92 | 0.11 | 0.94 | 0.16 | 0.79 | 0.13 | 5 | - | - | Down |
| EB1 (MAPRE1) | Q15691 | End binding protein 1 | 1.00 | 4 | 17.5 | 1.70 | 0.29 | 1.47 | 0.20 | 1.49 | 0.27 | 32 | Up | Up | Up |
| FASN | P49327 | Fatty acid synthase | 1.00 | 9 | 5.7 | 0.74 | 0.19 | 0.97 | 0.15 | 0.95 | 0.17 | 10 | Down | - | - |
| FN1 | P02751 | Fibronectin | 1.00 | 2 | 0.8 | 1.21 | 0.19 | 1.06 | 0.16 | 1.49 | 0.58 | 8 | Up | - | Up |
| GPHN | Q9NQX3 | Gephyrin | 1.00 | 9 | 14.9 | 1.00 | 0.29 | 0.98 | 0.27 | 1.31 | 0.48 | 13 | - | - | Up |
| GRN | P28799 | Granulins | 1.00 | 7 | 16.5 | 1.05 | 0.48 | 0.90 | 0.29 | 0.74 | 0.55 | 27 | - | - | Down |
| H1FX | Q92522 | Histone H1x | 1.00 | 2 | 14.6 | 1.07 | 0.15 | 0.67 | 0.33 | 0.74 | 0.05 | 5 | - | Down | Down |
| HIST1H2AB | P04908 | Histone H2A type 1-B/E | 1.00 | 2 | 21.7 | 1.07 | 0.13 | 1.26 | 0.18 | 1.54 | 0.29 | 4 | - | Up | Up |
| HNRNPA1 | P09651 | Heterogeneous nuclear ribonucleoprotein A1 | 1.00 | 6 | 30.9 | 1.00 | 0.12 | 1.01 | 0.16 | 1.82 | 0.17 | 11 | - | - | Up |
| HNRNPA2B1 | P22626 | Heterogeneous nuclear ribonucleoprotein A2/B1 | 1.00 | 8 | 25.5 | 1.07 | 0.21 | 0.98 | 0.12 | 1.84 | 0.33 | 25 | - | - | Up |
| HNRNPA3 | P51991 | Heterogeneous nuclear ribonucleoprotein A3 | 1.00 | 3 | 16.7 | 0.94 | 0.20 | 0.96 | 0.10 | 1.47 | 0.38 | 6 | - | - | Up |
| HNRNPD | Q14103 | Heterogeneous nuclear ribonucleoprotein D0 | 1.00 | 3 | 12.5 | 1.07 | 0.44 | 1.03 | 0.23 | 1.84 | 1.23 | 11 | - | - | Up |
| HNRNPH1 | P31943 | Heterogeneous nuclear ribonucleoprotein H | 1.00 | 3 | 6.2 | 0.96 | 0.17 | 0.88 | 0.07 | 1.67 | 0.14 | 4 | - | - | Up |
| HNRNPU | Q00839 | Heterogeneous nuclear ribonucleoprotein U | 1.00 | 5 | 7.5 | 1.04 | 0.12 | 1.44 | 0.35 | 1.54 | 0.28 | 4 | - | Up | Up |
| HTRA2 | O43464 | Serine protease HTRA2, mitochondrial | 1.00 | 6 | 21.8 | 1.21 | 0.30 | 0.98 | 0.22 | 1.11 | 0.30 | 10 | Up | - | - |
| IFFO2 | Q5TF58 | Intermediate filament family orphan 2 | 1.00 | 3 | 17.1 | 0.83 | 0.09 | 1.99 | 0.37 | 2.05 | 0.29 | 10 | - | Up | Up |
| KARS | Q15046 | Lysyl-tRNA synthetase | 1.00 | 2 | 2.5 | 0.78 | 0.07 | 0.94 | 0.05 | 0.83 | 0.07 | 10 | Down | - | Down |
| KCTD3 | Q9Y597 | BTB/POX domain-containing protein KCTD3 | 1.00 | 4 | 8.3 | 0.93 | 0.11 | 0.78 | 0.11 | 0.77 | 0.15 | 8 | - | Down | Down |
| LMNA | P02545 | Lamin-A/C | 1.00 | 2 | 3.9 | 0.81 | 0.32 | 0.75 | 0.23 | 0.72 | 0.26 | 3 | - | Down | Down |
| MTHFD1 | P11586 | C-1-tetrahydrofolate synthase, cytoplasmic | 1.00 | 2 | 3.7 | 0.80 | 0.09 | 0.79 | 0.13 | 0.64 | 0.18 | 7 | - | Down | Down |
| MVP | Q14764 | Major vault protein | 1.00 | 11 | 21.2 | 0.86 | 0.24 | 0.72 | 0.24 | 1.56 | 0.68 | 18 | - | Down | Up |
| MYH9 | P35579 | Myosin-9 | 1.00 | 3 | 2.6 | 1.42 | 0.27 | 1.00 | 0.17 | 1.53 | 0.56 | 3 | Up | - | Up |
| MYO1B | O43795 | Myosin-Ib | 1.00 | 3 | 3.3 | 0.78 | 0.13 | 0.84 | 0.11 | 0.70 | 0.22 | 11 | Down | - | Down |
| NCL | P19338 | Nucleolin | 1.00 | 7 | 8.6 | 0.83 | 0.13 | 1.05 | 0.21 | 1.35 | 0.22 | 10 | - | - | Up |
| NIF3L1 | Q9GZT8 | NIF3-like protein 1 | 1.00 | 2 | 4.8 | 1.21 | 0.16 | 1.02 | 0.07 | 1.02 | 0.10 | 7 | Up | - | - |
| NOLC1 | Q14978 | Nucleolar and coiled-body phosphoprotein 1 | 1.00 | 3 | 6.3 | 0.84 | 0.09 | 1.27 | 0.13 | 1.35 | 0.21 | 3 | - | Up | Up |
| NPM1 | P06748 | Nucleophosmin | 1.00 | 3 | 16.1 | 1.04 | 0.20 | 1.25 | 0.17 | 1.88 | 0.29 | 8 | - | Up | Up |
| PCNA | P12004 | Proliferating cell nuclear antigen | 1.00 | 3 | 12.3 | 1.04 | 0.30 | 0.93 | 0.07 | 0.84 | 0.44 | 3 | - | - | Down |
| PHB | P35232 | Prohibitin | 1.00 | 2 | 7.0 | 1.05 | 0.08 | 0.97 | 0.11 | 0.87 | 0.11 | 3 | - | - | Down |
| PHB2 | Q99623 | Prohibitin-2 | 1.00 | 7 | 26.8 | 1.01 | 0.22 | 0.91 | 0.20 | 0.83 | 0.17 | 18 | - | - | Down |
| PKM2 | P14618 | Pyruvate kinase isozymes M1/M2 | 1.00 | 13 | 41.1 | 0.77 | 0.17 | 0.84 | 0.18 | 0.61 | 0.21 | 29 | Down | - | Down |
| PRKDC | P78527 | DNA-dependent protein kinase catalytic subunit | 1.00 | 7 | 3.1 | 0.92 | 0.29 | 0.95 | 0.14 | 0.85 | 0.25 | 13 | - | - | Down |
| PSMC1 | P62191 | 26S protease regulatory subunit 4 | 1.00 | 2 | 7.3 | 0.93 | 0.11 | 0.86 | 0.06 | 0.73 | 0.13 | 4 | - | - | Down |
| RAN | P62826 | GTP-binding nuclear protein Ran | 1.00 | 4 | 25.5 | 0.90 | 0.15 | 0.62 | 0.06 | 0.64 | 0.14 | 9 | - | Down | Down |
| RPL10 | P27635 | 60S ribosomal protein L10 | 1.00 | 3 | 18.7 | 0.86 | 0.06 | 1.28 | 0.13 | 0.94 | 0.05 | 7 | - | Up | - |
| RPL13 | P26373 | 60S ribosomal protein L13 | 1.00 | 3 | 14.2 | 1.14 | 0.08 | 1.29 | 0.17 | 1.28 | 0.24 | 7 | - | Up | - |
| RPL17 | P18621 | 60S ribosomal protein L17 | 1.00 | 3 | 13.6 | 0.72 | 0.11 | 1.15 | 0.25 | 0.97 | 0.16 | 5 | Down | - | - |
| RPL18 | Q07020 | 60S ribosomal protein L18 | 1.00 | 3 | 25.5 | 0.82 | 0.15 | 0.78 | 0.23 | 1.00 | 0.23 | 9 | - | Down | - |
| RPL34 | P49207 | 60S ribosomal protein L34 | 1.00 | 2 | 13.7 | 1.22 | 0.26 | 1.29 | 0.26 | 1.58 | 0.36 | 6 | Up | Up | Up |
| RPL5 | P46777 | 60S ribosomal protein L5 | 1.00 | 4 | 10.8 | 0.95 | 0.07 | 1.03 | 0.22 | 1.29 | 0.17 | 6 | - | - | Up |
| RPS11 | P62280 | 40S ribosomal protein S11 | 1.00 | 5 | 37.3 | 0.90 | 0.13 | 1.01 | 0.13 | 0.82 | 0.12 | 10 | - | - | Down |
| RPS23 | P62266 | 40S ribosomal protein S23 | 1.00 | 2 | 16.1 | 1.17 | 0.19 | 1.26 | 0.14 | 1.22 | 0.19 | 5 | - | Up | - |
| RPS27A | P62988 | Ubiquitin | 1.00 | 2 | 15.8 | 0.86 | 0.04 | 0.88 | 0.16 | 0.78 | 0.07 | 5 | - | - | Down |
| RPS4X | P62701 | 40S ribosomal protein S4, X isoform | 1.00 | 3 | 8.0 | 1.12 | 0.17 | 1.62 | 0.22 | 1.82 | 0.26 | 7 | - | Up | Up |
| RPS7 | P62081 | 40S ribosomal protein S7 | 1.00 | 3 | 19.6 | 0.77 | 0.30 | 0.98 | 0.39 | 0.90 | 0.17 | 3 | Down | - | - |
| RPS9 | P46781 | 40S ribosomal protein S9 | 1.00 | 6 | 25.4 | 0.93 | 0.14 | 1.03 | 0.15 | 0.82 | 0.12 | 8 | - | - | Down |
| SFPQ | P23246 | Splicing factor, proline- and glutamine-rich | 1.00 | 6 | 11.2 | 1.00 | 0.15 | 1.10 | 0.14 | 1.32 | 0.25 | 14 | - | - | Up |
| SLC25A3 | Q00325 | Phosphate carrier protein, mitochondrial | 1.00 | 2 | 5.3 | 1.10 | 0.21 | 1.02 | 0.32 | 0.84 | 0.24 | 5 | - | - | Down |
| SLC25A5 | P05141 | ADP/ATP translocase 2 | 1.00 | 6 | 18.8 | 0.94 | 0.32 | 0.99 | 0.25 | 0.84 | 0.37 | 14 | - | - | Down |
| SMC3 | Q9UQE7 | Structural maintenance of chromosome protein 3 | 1.00 | 2 | 4.6 | 0.74 | 0.15 | 0.83 | 0.11 | 0.83 | 0.03 | 4 | Down | Down | Down |
| TCOF1 | Q13428 | Treacle protein | 1.00 | 2 | 2.1 | 0.99 | 0.06 | 1.19 | 0.17 | 0.86 | 0.08 | 3 | - | - | Down |
| TCP1 | P17987 | T-complex protein 1 subunit alpha | 1.00 | 3 | 5.8 | 1.03 | 0.05 | 0.93 | 0.17 | 0.78 | 0.17 | 5 | - | - | Down |
| TECR | Q9NZ01 | Trans-2,3-enoyl-CoA reductase | 1.00 | 2 | 5.5 | 0.92 | 0.08 | 0.88 | 0.18 | 0.70 | 0.08 | 3 | - | - | Down |
| THBS1 | P07996 | Thrombospondin-1 | 1.00 | 2 | 2.2 | 0.71 | 0.21 | 0.69 | 0.13 | 0.70 | 0.21 | 3 | Down | Down | Down |
| TRIM21 | P19474 | 52 kDa Ro protein | 1.00 | 25 | 57.5 | 1.35 | 0.35 | 1.02 | 0.26 | 1.27 | 0.59 | 113 | UP | - | - |
| TSC1 | Q92574 | Hamartin | 1.00 | 3 | 2.9 | 1.02 | 0.21 | 1.02 | 0.36 | 0.62 | 0.14 | 3 | - | - | Down |
| TTN | Q8WZ42 | Titin | 1.00 | 4 | 0.2 | 0.62 | 0.06 | 0.82 | 0.09 | 0.61 | 0.06 | 180 | Down | Down | Down |
| TUBB | P07437 | Tubulin beta chain | 1.00 | 15 | 45.3 | 0.95 | 0.29 | 0.97 | 0.29 | 0.82 | 0.22 | 42 | - | - | Down |
| TUFM | P49411 | Elongation factor Tu, mitochondrial | 1.00 | 2 | 3.8 | 0.86 | 0.14 | 0.92 | 0.11 | 0.85 | 0.18 | 4 | - | - | Down |
| UQCRC2 | P22695 | Cytochrome b-c1 complex subunit 2, mitochondrial | 1.00 | 3 | 9.1 | 0.92 | 0.34 | 0.81 | 0.24 | 0.88 | 0.53 | 6 | - | Down | - |
Biological Process Network Analysis of iTRAQ Candidates by Metacore
According to previous reports, cytoskeletal rearrangement may be involved in inflammasome activation (22, 54). To evaluate the candidates identified by our iTRAQ analysis, we used the Metacore software to analyze the process networks that use these candidates. Our results indicated that proteins significantly involved in the translation-related, cytoskeleton-related and protein folding categories were differentially present in the 131 quantified proteins (Table II). Furthermore, Metacore analysis of the 67 level-changed candidates revealed the potential involvement of mitosis, microtubule regulation, cell adhesion regulation and mRNA processing (Table II). Among the candidates that were preferentially present in all of threes inflammasomes, EB1 (encoded by MAPRE1), which participates in microtubule regulation and mitosis (Table II; supplemental Fig. S2) and was up-regulated in response to all three treatments, was selected for further study.
Table II. Metacore process network analysis of quantificated and level changed candidates in inflammasome complexes.
| Quantificated candidates (n = 131) | |
|---|---|
| GeneGo process network | p value |
| Translation_Translation initiation | 1.03E-32 |
| Translation_Elongation-Termination | 1.71E-31 |
| Cytoskeleton_Spindle microtubules | 1.20E-03 |
| Protein folding_Protein folding nucleus | 1.70E-03 |
| Level changed candidates (n = 67) | ||
|---|---|---|
| GeneGo process network | p value | Candidates |
| Translation_Translation initiation | 5.01E-11 | RPL10, RPL13, RPL17, RPL18, RPL34, and RPL5 |
| RPS11, RPS23, RPS27A, RPS4X, RPS7 and RPS9 | ||
| Translation_Elongation-Termination | 2.85E-10 | RPL10, RPL13, RPL17, RPL18, RPL34, and RPL5 |
| RPS11, RPS23, RPS27A, RPS4X, RPS7 and RPS9 | ||
| Cytoskeleton_Spindle microtubules | 2.56E-05 | DYNC1H1, RAN, SMC3, TCP1 and TUBB |
| Cell cycle_Mitosis | 8.90E-05 | DYNC1H1, EB1, H1FX, RAN, RPS27A, SMC3, and TUBB |
| Cell adhesion_Integrin-mediated cell-matrix adhesion | 3.06E-04 | CAV1, FN1, MYH9 and TSC1 |
| Cytoskeleton_Cytoplasmic microtubules | 3.17E-04 | DYNC1H1, EB1, TCP1, and TUBB |
| Transcription_mRNA processing | 1.80E-03 | HNRNPA1, HNRNPA2B1, HNRNPA3, HNRNPD, HNRNPH1, HNRNPU, and SFPQ |
EB1 is a Novel Component of Inflammasome Complexes and is Important for their Activation in NPC Cells
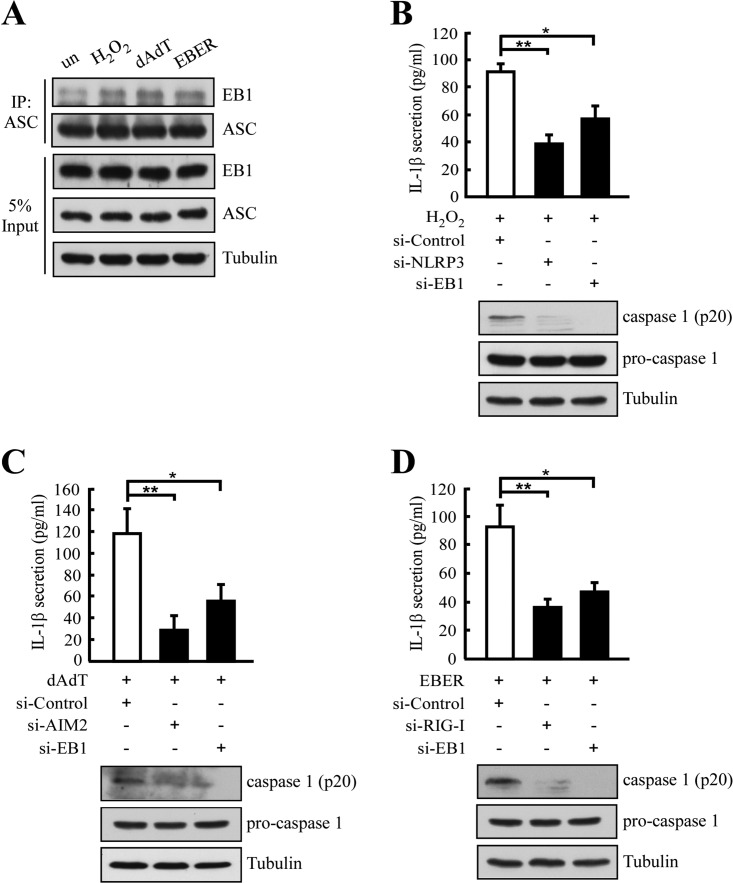
To determine if EB1 is involved in the stimulation-induced formation of the inflammasome complex, we first carried out co-immunoprecipitation assays using extracts of HK1/ASC cells treated with H2O2, poly (dA:dT) or EBER, which activate NLRP3, AIM2 and RIG-I, respectively. As shown in Fig. 2A, immunoprecipitation with an anti-ASC antibody showed that ASC-associated EB1 was elevated in cells treated with all three inflammasome-specific stimuli. We then tested whether EB1 is involved in the activation of inflammasomes by using small interfering RNA (siRNA) to decrease the expression of EB1, NLRP3, AIM2, or RIG-I (supplemental Fig. S3). The resulting knockdown cells were treated with the corresponding stimuli, and ELISA was used to measure the levels of the cytokine, IL-1β, in the culture media. As shown in Fig. 2B, the induction of IL-1β by H2O2 was significantly reduced in cells treated with si-EB1 and si-NLRP3, but not si-Control. Furthermore, the activated caspase 1 (p20), as another indicator for inflammasome activation, was also decreased after knockdown NLRP3 or EB1, comparing with si-Control. Similar inhibitions of IL-1β secretion and decrease of activated caspase 1 were detected in cells transfected with si-EB1 or si-AIM2 and subsequently treated with poly (dA:dT) (Fig. 2C), and in cells transfected with si-EB1 or si-RIG-I and subsequently treated with EBER (Fig. 2D). Together, these results suggest that EB1 is a common component present in the NLRP3, AIM2, and RIG-I inflammasome complexes, and that it is required for their activation. Among the three inflammasomes examined, the knockdown effect of si-EB1 was more profound for AIM2 inflammasome activation in response to poly (dA:dT) treatment. Thus, the following studies were performed using the AIM2 inflammasome.
Fig. 2.
EB1 is involved in inflammasome complexes and is required for their activity. A, Pull-down assays of inflammasome complexes from cells treated with NLRP3-, AIM2-, or RIG-I-specific stimuli. Complexes were immunoprecipitated from HK1/ASC cells using anti-ASC antibodies, and EB1 levels were detected with a specific antibody. The amount of ASC was used as the pull-down control. B, HK1 cells were pretreated with control, NLRP3, or EB1 siRNA for 48 h and then treated with H2O2 (10 μm). After 24 h, media were collected for IL-1β ELISA. C, HK1 cells were pretreated with control, AIM2, or EB1 siRNA for 48 h and treated with poly (dA:dT) for 2 h. After a further incubation for 10 h, media were collected for IL-1β ELISA. D, HK1 cells were prepared as described in (C), but treated with EBER instead of poly (dA:dT). Cell extracts were collected, and then indicated molecules were detected by specific antibodies. Symbols: *, p < 0.05 and **, p < 0.01. The results are presented as the means ± S.D. of three experiments.
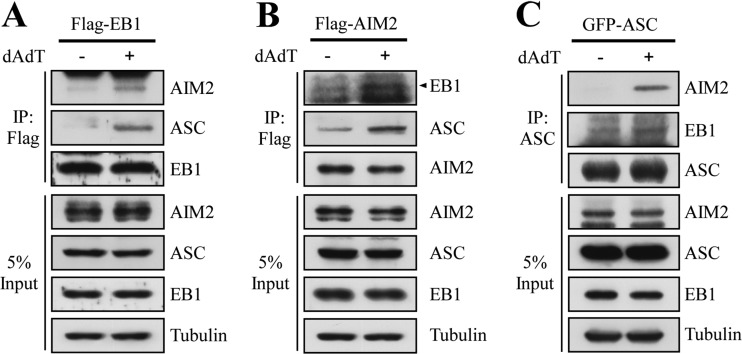
EB1 is a 30-kDa, 268-amino acid microtubule-binding protein; its amino terminus contains a calponin homology (CH) domain, which is found in many proteins involved in microtubule binding, whereas its carboxyl terminus contains a C-terminal (CT) dimerization domain that is required to recruit the +TIP complex during microtubule polymerization (25). Next, we evaluated whether EB1 physically associates with the inflammasome complex by co-immunoprecipitation assays of AIM2 and ASC. NPC TW02 cells were transfected with Flag-tagged EB1 (Flag-EB1) and stimulated with poly (dA:dT). Cell extracts were precipitated with anti-Flag antibodies, and endogenous AIM2 and ASC were detected by specific antibodies. As shown in Fig. 3A, AIM2 and ASC were detected more highly in immunoprecipitates from poly (dA:dT)-treated cell extracts compared with those from untreated cells. Similarly, more EB1 and ASC proteins were detected in cells transfected with Flag-AIM2 and treated with poly (dA:dT) compared with controls (Fig. 3B). Furthermore, AIM2 was preferentially precipitated with anti-ASC antibodies in poly (dA:dT)-treated cells, and more EB1 was detected in treated cell extracts than in untreated controls (Fig. 3C). To investigate the time course of the association of EB1 with AIM2 inflammasome, we transfected flag-AIM2 in NPC TW02 cells and then precipitated AIM2 complex by anti-flag antibodies at indicated time points. As shown in supplemental Fig. S4, the EB1 and ASC were associated with the ectopically expressed flag-AIM2 without poly (dA:dT) stimulation. An enhanced association was detected at 2 h and reached plateau at 4–6h after poly (dA:dT) treatment. Together, these results suggest that EB1 may be recruited to the AIM2 inflammasome in response to poly (dA:dT) stimulation.
Fig. 3.
EB1 associates with the AIM2 inflammasome after poly (dA:dT) treatment of NPC TW02 cells. A, NPC TW02 cells were transfected with flag-EB1 for 48 h, treated with/without poly (dA:dT) for 2 h, and then further incubated for 4 h. Cell extracts were collected and subjected to immunoprecipitation with an anti-Flag matrix, and Western blotting of the indicated proteins was performed using specific antibodies. Flag-EB1 was detected using an anti-Flag antibody. B, NPC TW02 cells were transfected with Flag-AIM2 for 48 h, treated with/without poly (dA:dT) for 2 h, and incubated for an additional 4 h. Cell extracts were collected and subjected to immunoprecipitation using an anti-Flag matrix. The indicated proteins were detected by Western blotting. Flag-AIM2 was detected using an anti-Flag antibody. C, NPC TW02 cells were transfected with GFP-ASC for 48 h, treated with/without poly (dA:dT) for 2 h, and further incubated for 4 h. Cell extracts were collected and subjected to immunoprecipitation using anti-ASC antibodies. The indicated proteins were detected by Western blotting. GFP-ASC was detected using an anti-GFP antibody.
EB1 Directly Interacts with AIM2
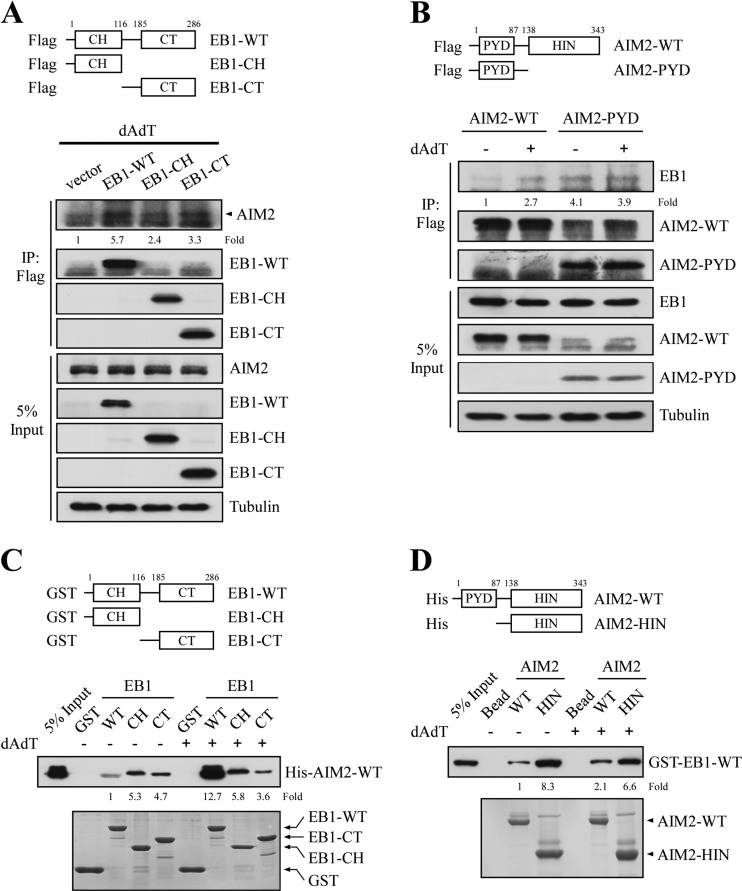
To dissect the domains of EB1 responsible for its interaction with the AIM2 inflammasome, we generated a series of Flag-tagged EB1 CH- and CT domain-specific constructs (EB1-CH and EB1-CT in Fig. 4A) and examined their associations with AIM2 in response to poly (dA:dT) treatment of NPC TW02 cells. Cells were individually transfected with the empty vector, WT and domain-specific EB1 constructs, treated with poly (dA:dT), and then subjected to co-immunoprecipitation assays. As shown in Fig. 4A, EB1-WT showed a strong association with AIM2 (5.7-fold increase), comparing with vector control. Both the CH and CT domains could interact with AIM2, although EB1-CT (3.3-fold) showed a slightly stronger interaction than EB1-CH (2.4-fold). To investigate whether AIM2 interacts with EB1 in a reciprocal manner, we conducted co-immunoprecipitation assays using an anti-Flag matrix to precipitate EB1 from lysates of HK1 cells stably expressing Flag-AIM2-WT or Flag-AIM2-PYD, with or without poly (dA:dT) treatment. The pyrin domain of AIM2 (AIM2-PYD), which interacts with ASC to activate pro-caspase 1, interacted with EB1 both with/without stimulation (3.9- and 4.1-fold), whereas for the AIM2-WT binding was enhanced (2.7-fold increase) by the treatment (Fig. 4B). We then tested the direct binding between AIM2 and EB1 using GST pull-down assays. As shown in Fig. 4C, GST-EB1-WT pulled down significantly more purified His-AIM2 following poly (dA:dT) treatment (12.7-fold increase), whereas GST-EB1-CH and GST-EB1-CT pulled down purified His-AIM2 in the absence (5.3- and 4.7-fold) and presence (5.8-fold and 3.6-fold) of poly (dA:dT). These results suggest that that the interaction of EB1 and AIM2 is affected by double-strand DNA stimulation. The direct binding of EB1 with purified His-ASC was further confirmed by using GST-EB1-WT and purified His-ASC in pull down assay, as our results showed that GST-EB1-CT specifically bound to His-ASC, whereas GST-EB1-CH did not (supplemental Fig. S5). Furthermore, reciprocal immunoprecipitation experiments also supported the direct interaction between EB1 and AIM2. As shown in Fig. 4D, His-AIM2-WT pulled down twofold more GST-EB1 after poly (dA:dT) treatment. Although the HIN domain of AIM2 (His-AIM2-HIN), which recognizes intracellular DNA, interacted strongly with GST-EB1 in the presence (6.6-fold) or absence (8.3-fold) of stimulation. Together, these results indicate that EB1 directly interacts with both AIM2 and ASC, and that these EB1-AIM2 and EB1-ASC interactions are likely to be important for AIM2 inflammasome activation.
Fig. 4.
EB1 directly interacts with AIM2 in vitro. A, NPC TW02 cells were transfected with empty vector or vectors encoding Flag-EB1-WT, Flag-EB1-CH, or Flag-EB1-CT. After 48 h, the cells were treated with poly (dA:dT) for 2 h and incubated for 4 h, and then harvested for immunoprecipitation using an anti-Flag matrix. The levels of AIM2 were assessed by Western blotting after immunoprecipitation. The fold-change numbers were obtained from three independent experiments. B, HK1 cells stably expressing Flag-AIM2-WT or Flag-AIM2-PYD (introduced by lentiviral infection) were treated with or without poly (dA:dT) and subjected to immunoprecipitation using an anti-Flag matrix. The levels of EB1 were assessed by Western blotting. C, Purified GST, GST-EB1-WT, GST-EB1-CH, and GST-EB1-CT fusion proteins were immobilized on glutathione agarose, and then separately incubated with purified His-AIM2-WT fusion proteins for 24 h. The bound proteins were washed, and then analyzed by Western blotting with an anti-His antibody. The fold-change numbers were derived from three independent experiments. The amounts of GST fusion proteins were assessed by Coomassie blue staining. D, Purified His-AIM2-WT or His-AIM2-HIN were immobilized on Ni-charged agarose, and then incubated with purified GST-EB1-WT for 24 h. The bound proteins were washed, and then analyzed by Western blotting with an anti-GST antibody. The fold-change numbers were obtained from three independent experiments. The amount of His fusion proteins were assessed by Coomassie blue staining.
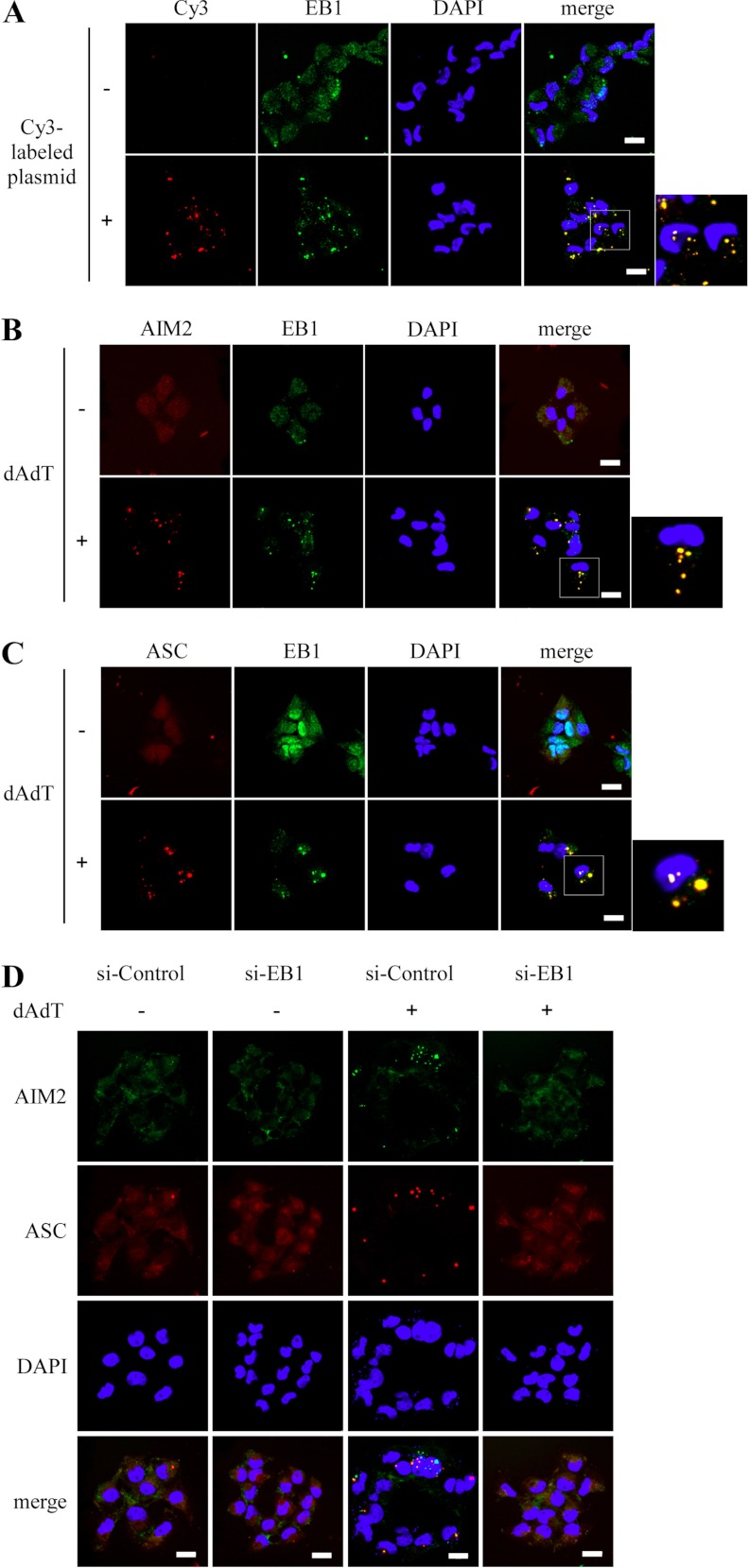
EB1 Colocalizes with AIM2 and ASC and is Required for AIM2 Speck-like Particle Formation in NPC Cells
Inflammasome activation triggers speck-like particle formation in the cytosol, where the complexes are formed (13). To confirm that the activated AIM2 inflammasome contains EB1 in addition to AIM2 and ASC, we examined if these proteins colocalized in activated speck-like particles. We transfected commercial Cy3-labeled plasmid DNA into NPC TW02 cells to activate the AIM2 inflammasome, and then investigated whether the labeled DNA and EB1 colocalized with endogenous AIM2 by fluorescence confocal microscopy. As shown in Fig. 5A, EB1 did indeed colocalize with Cy3-labeled plasmid DNA in the cytosolic speck-like particles. Similarly, EB1 colocalized with AIM2 or ASC in speck-like particles following poly (dA:dT) treatment (Fig. 5B and 5C). We then tested whether EB1 affected speck formation by using siRNA to knock down EB1 in NPC TW02 cells, transfecting the cells with GFP-AIM2 or DsRed-ASC, and using immunofluorescence confocal microscopy to examine complex formation with or without poly (dA:dT) treatment. The formation of speck-like particles of the AIM2 inflammasome was indeed disrupted when cells were treated with EB1-specific siRNA (Fig. 5D), indicating that EB1 is involved in speck-like particle formation. Collectively, these findings indicate that EB1 is required for AIM2 inflammasome complex formation, which is an important process in the activation of procaspase 1.
Fig. 5.
EB1 colocalizes with the AIM2 inflammasome and is required for speck-like particle formation in NPC TW02 cells. A, Cells were treated with or without Cy3-labeled plasmids for 2 h, and were further incubated for 4 h. Cells were fixed and were stained with an anti-EB1 antibody, and the colocalization of Cy3-labled plasmids and EB1 was observed by confocal immunofluorescence microscopy. B, and C, Cells were transfected with DsRed-AIM2 or DsRed-ASC for 48 h, treated with poly (dA:dT) for 2 h, and further incubated for 4 h. The cells were then fixed and stained with an anti-EB1 antibody, and the colocalization of EB1 with DsRed-AIM2 or DsRed-ASC was observed by confocal immunofluorescence microscopy. D, NPC TW02 cells expressing control siRNA or EB1 siRNA were cotransfected with GFP-AIM2 and DsRed-ASC for 48 h, treated with or without poly (dA:dT) for 2 h, further incubated for 4 h, and then fixed. The colocalization of GFP-AIM2 and DsRed-ASC was observed by confocal fluorescence microscopy. Scale bar = 20 μm.
The Expression of EB1 is Statistically Correlated with that of AIM2 and ASC in NPC Patients
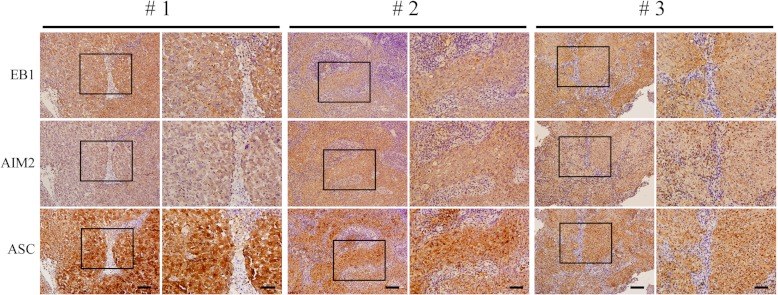
To examine whether EB1 participates in AIM2 inflammasome activation in vivo, and to assess the potential oncogenic role of the EB1 over-expression observed in various cancers (31, 55–57), we investigated the correlation of EB1 expression with AIM2 and ASC in NPC biopsy tissues. Immunohistochemical staining showed that EB1, AIM2, and ASC were highly expressed in NPC tumor tissues (Fig. 6). A correlational study carried out in 95 NPC biopsy samples showed that high expression of EB1 was statistically correlated with high expression of AIM2 (p < 0.05) and ASC (p < 0.01) (Table III). Thus, it appears likely that EB1 plays a physiologically relevant role in NPC.
Fig. 6.
Correlation of EB1 expression with AIM2 and ASC in NPC patients. Immunohistochemical staining of EB1, AIM2, and ASC were detected by specific antibodies in consecutive NPC tissue sections from three NPC patients. The results are shown at 200× magnification (left, scale bar = 200 μm) and 400× magnification (right, scale bar = 100 μm).
Table III. The correlation of EB1 expression with AIM2 or ASC in NPC patients.
| Level | AIM2 |
p value | ASC |
p value | |||
|---|---|---|---|---|---|---|---|
| High | Low | High | Low | ||||
| EB1 | High | 40 | 13 | p < 0.05 | 32 | 21 | p < 0.001 |
| Low | 23 | 19 | 10 | 32 | |||
DISCUSSION
Inflammasome activation is believed to play a role in tumorigenesis (17, 47), but the detailed molecular basis of inflammasome activation and its role in malignancy is not yet fully understood. Here, we used iTRAQ-based proteomic analysis to characterize the interactomes of the NLRP3, AIM2, and RIG-I inflammasomes. ASC pull-down experiments demonstrated that a novel component, EB1, was preferentially recruited to the activated NLRP3, AIM2, and RIG-I inflammasomes stimulated with H2O2, poly (dA:dT) and EBER, respectively (Table I and Fig. 2A). Metacore network analysis showed that many of the differentially expressed candidates were involved in microtubule-associated processes (Table II); among them, EB1 was elevated in response to all three stimuli. We confirmed the direct interaction of EB1 with AIM2 and ASC in NPC cells and in vitro. Knockdown of EB1 expression abolished inflammasome-induced IL-1β secretion and disrupted the formation of speck-like particles. In NPC tissue samples, we further verified the expression of EB1 and its correlation with AIM2 and ASC, not only providing support for the physiological relevance of this mechanism but also linking EB1 to NPC. Thus, our findings suggest that EB1 serves as a crucial molecule for inflammasome assembly, extending our knowledge of the inflammasome complex beyond its core machinery.
The systematic iTRAQ analysis of proteins in three independent activated inflammasomes proved to be a powerful approach to search for novel components in these biological interactomes. Unlike previous studies that proteomic screened the downstream targets of activated caspase 1 (58, 59), we herein used proteomics to identify proteins that participate in the assembly of activated inflammasome complexes. We found that many nucleotide-binding proteins of the hnRNP family appear to be involved in the responses to poly (dA:dT) or EBER (Table I), suggesting that these proteins may be involved in the association of nucleic-acid binding during inflammasome activation. The hnRNP family proteins are important for mRNA processing and mRNA stability (34), and are considered to have oncogenic potential in many cancers, including NPC (44). Our findings provide evidence that these proteins may play an additional role in the activation of cytoplasmic nucleotide-sensing inflammasomes (AIM2 and RIG-I). Consistent with a previous report that many microtubule-associated proteins, including EB1, were up-regulated during macrophage activation (60), our iTRAQ analysis identified EB1 and numerous other microtubule-associated proteins that may participate in inflammasome activation (supplemental Table 1). We also identified an association between the activated NLRP3 inflammasome and HSP90, which was previously reported to stabilize the NLRP3 inflammasome prior to activation (61). Thus, this protein appears to also play an important role in the pre-activated inflammasome. Notably, most of the candidate proteins differed between H2O2-stimulated NLRP3 inflammasomes and the other two stimulated inflammasome types (Table I). Thus, it could prove interesting to re-examine the NLRP3 inflammasome using alternative approaches, such as immunoprecipitation with anti-NLRP3 antibodies followed by direct mass spectrometric identification.
Here, we showed that EB1 directly interacted with AIM2 and ASC, both in NPC cells and in vitro. It has been reported that the SxIP (Ser-x-Ile-Pro) amino acid sequence is the major EB1 binding motif in other EB1 interacting proteins, such as APC and CLASP2 (62). Using the DILIMOT algorithm sequence analysis software (62), we identified a similar amino acid sequence, Ser-x-x-Lys-Pro, in APC, AIM2, and ASC (data no shown). Further studies using site-specific mutagenesis to disrupt the interaction site could help verify this potential binding site. When an inflammasome receptor interacts with a cytoplasmic pathogen, it undergoes a conformational change that is important for its association with ASC and their downstream targets (63). Based on our observation that the interaction between EB1 and AIM2 was markedly increased following poly (dA:dT) treatment in vitro (Fig. 3B), we speculate that AIM2 may undergo a conformational change after interacting with cytoplasmic DNA, thereby exposing the EB1-binding motif and facilitating this binding ability. We found that the CT domain of EB1 is involved in the interactions with AIM2 and ASC. The CT domain has been reported to recruit the +TIP complex to stabilize the plus ends of microtubules (26). We found that the CH domain of EB1, which is the microtubule-binding domain, could directly interact with AIM2 in vitro, but this association was weaker than that of the CT domain with AIM2 in vivo (Fig. 4A). These observations suggest that the CH domain has a higher binding affinity for microtubules than for AIM2.
Although we herein show that EB1 is an AIM2 inflammasome-interacting protein that is required for speck-like particle formation, the molecular mechanism through which EB1 affects speck-like particle formation is not yet known. Because EB1 regulates microtubule polymerization and recruits the +TIP complex to the plus end of the microtubule, we speculate that EB1 may recruit the AIM2 inflammasome to form the speck-like particle on the plus end of the microtubule. EB1-mediated microtubule stabilization was reported to be regulated by the small Rho GTPase, RhoA (28), and another small Rho GTPase, Rac1, was shown to regulate inflammasome activation in macrophage (21). Our knockdown analysis of individual small GTPases in NPC cells showed that poly (dA:dT)-induced, AIM2-mediated IL-1β secretion was decreased by silencing of RhoA, but not Rac1 or cdc42 (supplemental Fig. S6A). In addition, poly (dA:dT)-induced, AIM2-mediated IL-1β secretion was significantly reduced in NPC cells treated with the microtubule polymerization inhibitor, nocodazole (supplemental Fig. S6B). In contrast, treatment of these cells with paclitaxel, which stabilizes microtubule polymerization, slightly increased AIM2-mediated IL-1β secretion (supplemental Fig. S5B). Thus, we propose that AIM2 inflammasome activation may depend on plus-end microtubule polymerization, and cytoskeletal regulation may affect inflammasome activation by mediating complex formation. Indeed, a report showed that the NLRP3 inflammasome was constitutively activated without ligand stimulation in metastatic melanoma cells, but not in primary melanoma cells (64), possibly suggesting that cytoskeletal rearrangement may participate in inflammasome activation. Collectively, our findings link microtubule stabilization to AIM2 inflammasome activation. Further investigation of AIM2 inflammasome activation in other cell types subjected to knockdown of various small Rho GTPases may clarify whether specific small GTPases are required for inflammasome activation via stabilization of EB1.
Because inflammasomes are important for defense against pathogens infection in immune cells, here, we showed that EB1 is the novel component in inflammasome complex and important for their activation in NPC cells. EB1 was reported to be up-regulated during macrophage activation in THP-1 cells (60), and knockdown of EB1 significantly reduced IL-lβ secretion (p < 0.05) and caspase 1 activation in THP-1 cells after poly (dA:dT) treatment (supplemental Fig. S7), suggesting that EB1 is also involved in AIM2 inflammasome activation in immune cells.
The elevated expression of EB1 and its strong correlation with AIM2 and ASC expression in NPC tumor cells suggest that our findings are of strong physiological relevance. We previously noted that ASC and AIM2 are independent prognostic biomarkers capable of predicting better local recurrence-free survival of patients under current treatment regimes (unpublished data). EB1 has been reported as a biomarker for many cancers (54–56), so we may presume that EB1 also serves as an NPC biomarker. Furthermore, increasing evidence suggests that inflammation and inflammasome activation are involved in cancer tumorigenesis (17, 46). Here, we show for the first time that EB1 is important for inflammasome activation via microtubule stabilization. Thus, future studies are warranted to test whether cytoskeletal integrity or cell mobility activities could be targeted for the development of new therapeutic strategies against inflammasome-derived chronic inflammatory diseases.
Supplementary Material
Footnotes
* This work was supported by grants provided by the Ministry of Education, Taiwan (to Chang Gung University), the National Science Council, Taiwan (Grants NSC 101-2320-B-182-010-MY3 to Y.-S. Chang), and the Chang Gung Memorial Hospital, Taiwan (Grant CMRPD190133 to Y.-S. Chang).
 This article contains supplemental Figs. S1 to S7 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S7 and Tables S1 to S3.
1 The abbreviations used are:
- NPC
- nasopharyngeal carcinoma
- EBV
- Epstein-Barr virus
- IL-1β
- interleukin 1 beta
- PAMP
- pathogen-associated molecular pattern
- DAMP
- danger signal-associated molecular pattern
- NLR
- NOD-like receptors
- ROS
- reactive oxygen species
- AIM2
- absence in melanoma 2
- RIG-I
- retinoic acid-inducible gene I
- ASC
- apoptosis-associated speck-like protein containing caspase activation and recruitment domain
- HSP90
- heat shock protein 90
- EBER
- EBV noncoding RNA
- EB1
- end binding protein 1
- APC
- adenomatous polyposis coli
- +TIP
- plus-end tracking protein
- CT
- C-terminal
- CH
- calponin homology
- iTRAQ
- isobaric tags for relative and absolute quantification.
REFERENCES
- 1. Chan A. T. (2010) Nasopharyngeal carcinoma. Ann. Oncol. 21 Suppl 7, vii308–312 [DOI] [PubMed] [Google Scholar]
- 2. Sriamporn S., Vatanasapt V., Pisani P., Yongchaiyudha S., Rungpitarangsri V. (1992) Environmental risk factors for nasopharyngeal carcinoma: a case-control study in northeastern Thailand. Cancer Epidemiol. Biomarkers Prev. 1, 345–348 [PubMed] [Google Scholar]
- 3. Bei J. X., Li Y., Jia W. H., Feng B. J., Zhou G., Chen L. Z., Feng Q. S., Low H. Q., Zhang H., He F., Tai E. S., Kang T., Liu E. T., Liu J., Zeng Y. X. (2010) A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat. Genet. 42, 599–603 [DOI] [PubMed] [Google Scholar]
- 4. Chang K. P., Chang Y. T., Wu C. C., Liu Y. L., Chen M. C., Tsang N. M., Hsu C. L., Chang Y. S., Yu J. S. (2011) Multiplexed immunobead-based profiling of cytokine markers for detection of nasopharyngeal carcinoma and prognosis of patient survival. Head Neck 33, 886–897 [DOI] [PubMed] [Google Scholar]
- 5. Apte R. N., Dotan S., Elkabets M., White M. R., Reich E., Carmi Y., Song X., Dvozkin T., Krelin Y., Voronov E. (2006) The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor-host interactions. Cancer Metastasis Rev. 25, 387–408 [DOI] [PubMed] [Google Scholar]
- 6. Xie L., Xu L., He Z., Zhou W., Wang L., Zhang L., Lan K., Ren C., Liu W., Yao K. (2000) Identification of differentially expressed genes in nasopharyngeal carcinoma by means of the Atlas human cancer cDNA expression array. J. Cancer Res. Clin. Oncol. 126, 400–406 [DOI] [PubMed] [Google Scholar]
- 7. Zhu Y., Xu Y., Wei Y., Liang W., Liao M., Zhang L. (2008) Association of IL-1B gene polymorphisms with nasopharyngeal carcinoma in a Chinese population. Clin. Oncol. 20, 207–211 [DOI] [PubMed] [Google Scholar]
- 8. Martinon F., Burns K., Tschopp J. (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 9. Davis B. K., Wen H., Ting J. P. (2011) The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29, 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bryant C., Fitzgerald K. A. (2009) Molecular mechanisms involved in inflammasome activation. Trends Cell Biol. 19, 455–464 [DOI] [PubMed] [Google Scholar]
- 11. Duncan J. A., Bergstralh D. T., Wang Y., Willingham S. B., Ye Z., Zimmermann A. G., Ting J. P. (2007) Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl. Acad. Sci. U.S.A. 104, 8041–8046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou R., Tardivel A., Thorens B., Choi I., Tschopp J. (2010) Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 11, 136–140 [DOI] [PubMed] [Google Scholar]
- 13. Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., Alnemri E. S. (2009) AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poeck H., Bscheider M., Gross O., Finger K., Roth S., Rebsamen M., Hannesschläger N., Schlee M., Rothenfusser S., Barchet W., Kato H., Akira S., Inoue S., Endres S., Peschel C., Hartmann G., Hornung V., Ruland J. (2010) Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat. Immunol. 11, 63–69 [DOI] [PubMed] [Google Scholar]
- 15. Allen I. C., TeKippe E. M., Woodford R. M., Uronis J. M., Holl E. K., Rogers A. B., Herfarth H. H., Jobin C., Ting J. P. (2010) The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 207, 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zaki M. H., Vogel P., Body-Malapel M., Lamkanfi M., Kanneganti T. D. (2010) IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J. Immunol. 185, 4912–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dunn J. H., Ellis L. Z., Fujita M. (2012) Inflammasomes as molecular mediators of inflammation and cancer: potential role in melanoma. Cancer Lett. 314, 24–33 [DOI] [PubMed] [Google Scholar]
- 18. Samanta M., Iwakiri D., Kanda T., Imaizumi T., Takada K. (2006) EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 25, 4207–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mayor A., Martinon F., De Smedt T., Pétrilli V., Tschopp J. (2007) A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat. Immunol. 8, 497–503 [DOI] [PubMed] [Google Scholar]
- 20. Matsumiya T., Imaizumi T., Yoshida H., Satoh K., Topham M. K., Stafforini D. M. (2009) The levels of retinoic acid-inducible gene I are regulated by heat shock protein 90-alpha. J. Immunol. 182, 2717–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davis B. K., Roberts R. A., Huang M. T., Willingham S. B., Conti B. J., Brickey W. J., Barker B. R., Kwan M., Taxman D. J., Accavitti-Loper M. A., Duncan J. A., Ting J. P. (2011) Cutting edge: NLRC5-dependent activation of the inflammasome. J. Immunol. 186, 1333–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eitel J., Meixenberger K., van Laak C., Orlovski C., Hocke A., Schmeck B., Hippenstiel S., N′Guessan P. D., Suttorp N., Opitz B. (2012) Rac1 regulates the NLRP3 inflammasome which mediates IL-1beta production in Chlamydophila pneumoniae infected human mononuclear cells. PLoS One 7, e30379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muller A. J., Hoffmann C., Galle M., Van Den Broeke A., Heikenwalder M., Falter L., Misselwitz B., Kremer M., Beyaert R., Hardt W. D. (2009) The S. Typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe 6, 125–136 [DOI] [PubMed] [Google Scholar]
- 24. Schotte P., Denecker G., Van Den Broeke A., Vandenabeele P., Cornelis G. R., Beyaert R. (2004) Targeting Rac1 by the Yersinia effector protein YopE inhibits caspase-1-mediated maturation and release of interleukin-1beta. J. Biol. Chem. 279, 25134–25142 [DOI] [PubMed] [Google Scholar]
- 25. Ridley A. J. (2006) Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16, 522–529 [DOI] [PubMed] [Google Scholar]
- 26. Jiang K., Akhmanova A. (2011) Microtubule tip-interacting proteins: a view from both ends. Curr. Opin. Cell Biol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 27. Berrueta L., Kraeft S. K., Tirnauer J. S., Schuyler S. C., Chen L. B., Hill D. E., Pellman D., Bierer B. E. (1998) The adenomatous polyposis coli-binding protein EB1 is associated with cytoplasmic and spindle microtubules. Proc. Natl. Acad. Sci. U.S.A. 95, 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wen Y., Eng C. H., Schmoranzer J., Cabrera-Poch N., Morris E. J., Chen M., Wallar B. J., Alberts A. S., Gundersen G. G. (2004) EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6, 820–830 [DOI] [PubMed] [Google Scholar]
- 29. Morrison E. E. (2009) The APC-EB1 interaction. Adv. Exp. Med. Biol. 656, 41–50 [DOI] [PubMed] [Google Scholar]
- 30. Wang Y., Zhou X., Zhu H., Liu S., Zhou C., Zhang G., Xue L., Lu N., Quan L., Bai J., Zhan Q., Xu N. (2005) Overexpression of EB1 in human esophageal squamous cell carcinoma (ESCC) may promote cellular growth by activating beta-catenin/TCF pathway. Oncogene 24, 6637–6645 [DOI] [PubMed] [Google Scholar]
- 31. Schober J. M., Cain J. M., Komarova Y. A., Borisy G. G. (2009) Migration and actin protrusion in melanoma cells are regulated by EB1 protein. Cancer Lett. 284, 30–36 [DOI] [PubMed] [Google Scholar]
- 32. Chen L. C., Chung I. C., Hsueh C., Tsang N. M., Chi L. M., Liang Y., Chen C. C., Wang L. J., Chang Y. S. (2010) The antiapoptotic protein, FLIP, is regulated by heterogeneous nuclear ribonucleoprotein K and correlates with poor overall survival of nasopharyngeal carcinoma patients. Cell Death Differ. 17, 1463–1473 [DOI] [PubMed] [Google Scholar]
- 33. Chen L. C., Chen C. C., Liang Y., Tsang N. M., Chang Y. S., Hsueh C. (2011) A novel role for TNFAIP2: its correlation with invasion and metastasis in nasopharyngeal carcinoma. Mod. Pathol. 24, 175–184 [DOI] [PubMed] [Google Scholar]
- 34. Chen L. C., Liu H. P., Li H. P., Hsueh C., Yu J. S., Liang C. L., Chang Y. S. (2009) Thymidine phosphorylase mRNA stability and protein levels are increased through ERK-mediated cytoplasmic accumulation of hnRNP K in nasopharyngeal carcinoma cells. Oncogene 28, 1904–1915 [DOI] [PubMed] [Google Scholar]
- 35. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 36. Griffin T. J., Xie H., Bandhakavi S., Popko J., Mohan A., Carlis J. V., Higgins L. (2007) iTRAQ reagent-based quantitative proteomic analysis on a linear ion trap mass spectrometer. J. Proteome Res. 6, 4200–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bantscheff M., Boesche M., Eberhard D., Matthieson T., Sweetman G., Kuster B. (2008) Robust and sensitive iTRAQ quantification on an LTQ Orbitrap mass spectrometer. Mol. Cell. Proteomics 7, 1702–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu H. P., Wu C. C., Kao H. Y., Huang Y. C., Liang Y., Chen C. C., Yu J. S., Chang Y. S. (2011) Proteome-wide dysregulation by PRA1 depletion delineates a role of PRA1 in lipid transport and cell migration. Mol. Cell. Proteomics 10, 10.1074/mcp.M900641-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mueller L. N., Brusniak M. Y., Mani D. R., Aebersold R. (2008) An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J. Proteome Res. 7, 51–61 [DOI] [PubMed] [Google Scholar]
- 40. Nesvizhskii A. I., Vitek O., Aebersold R. (2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 [DOI] [PubMed] [Google Scholar]
- 41. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 42. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 43. Khan A. P., Poisson L. M., Bhat V. B., Fermin D., Zhao R., Kalyana-Sundaram S., Michailidis G., Nesvizhskii A. I., Omenn G. S., Chinnaiyan A. M., Sreekumar A. (2010) Quantitative proteomic profiling of prostate cancer reveals a role for miR-128 in prostate cancer. Mol. Cell. Proteomics 9, 298–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen L. C., Hsueh C., Tsang N. M., Liang Y., Chang K. P., Hao S. P., Yu J. S., Chang Y. S. (2008) Heterogeneous ribonucleoprotein k and thymidine phosphorylase are independent prognostic and therapeutic markers for nasopharyngeal carcinoma. Clin. Cancer Res. 14, 3807–3813 [DOI] [PubMed] [Google Scholar]
- 45. Yu C. J., Chang K. P., Chang Y. J., Hsu C. W., Liang Y., Yu J. S., Chi L. M., Chang Y. S., Wu C. C. (2011) Identification of guanylate-binding protein 1 as a potential oral cancer marker involved in cell invasion using omics-based analysis. J. Proteome Res. 10, 3778–3788 [DOI] [PubMed] [Google Scholar]
- 46. Chang Y. T., Wu C. C., Shyr Y. M., Chen T. C., Hwang T. L., Yeh T. S., Chang K. P., Liu H. P., Liu Y. L., Tsai M. H., Chang Y. S., Yu J. S. (2011) Secretome-based identification of ULBP2 as a novel serum marker for pancreatic cancer detection. PloS one 6, e20029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hu B., Elinav E., Flavell R. A. (2011) Inflammasome-mediated suppression of inflammation-induced colorectal cancer progression is mediated by direct regulation of epithelial cell proliferation. Cell Cycle 10, 1936–1939 [DOI] [PubMed] [Google Scholar]
- 48. Verma D., Bivik C., Farahani E., Synnerstad I., Fredrikson M., Enerback C., Rosdahl I., Soderkvist P. (2012) Inflammasome polymorphisms confer susceptibility to sporadic malignant melanoma. Pigment Cell Melanoma Res., in press [DOI] [PubMed] [Google Scholar]
- 49. McCartney R. G., Rice J. E., Sanderson S. J., Bunik V., Lindsay H., Lindsay J. G. (1998) Subunit interactions in the mammalian alpha-ketoglutarate dehydrogenase complex. Evidence for direct association of the alpha-ketoglutarate dehydrogenase and dihydrolipoamide dehydrogenase components. J. Biol. Chem. 273, 24158–24164 [DOI] [PubMed] [Google Scholar]
- 50. Panagiotidis C. A., Huang S. C., Tsirka S. A., Kyriakidis D. A., Canellakis E. S. (1988) Regulation of polyamine biosynthesis in Escherichia coli by the acidic antizyme and the ribosomal proteins S20 and L34. Adv. Exp. Med. Biol. 250, 13–24 [DOI] [PubMed] [Google Scholar]
- 51. Bessat M., Ersfeld K. (2009) Functional characterization of cohesin SMC3 and separase and their roles in the segregation of large and minichromosomes in Trypanosoma brucei. Mol. Microbiol. 71, 1371–1385 [DOI] [PubMed] [Google Scholar]
- 52. Kazerounian S., Yee K. O., Lawler J. (2008) Thrombospondins in cancer. Cell. Mol. Life Sci. 65, 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krüger M., Linke W. A. (2009) Titin-based mechanical signalling in normal and failing myocardium. J. Mol. Cell Cardiol. 46, 490–498 [DOI] [PubMed] [Google Scholar]
- 54. Broz P., Monack D. M. (2011) Molecular mechanisms of inflammasome activation during microbial infections. Immunol. Rev. 243, 174–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sahab Z. J., Hall M. D., Zhang L., Cheema A. K., Byers S. W. (2010) Tumor Suppressor RARRES1 Regulates DLG2, PP2A, VCP, EB1, and Ankrd26. J. Cancer 1, 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Orimo T., Ojima H., Hiraoka N., Saito S., Kosuge T., Kakisaka T., Yokoo H., Nakanishi K., Kamiyama T., Todo S., Hirohashi S., Kondo T. (2008) Proteomic profiling reveals the prognostic value of adenomatous polyposis coli-end-binding protein 1 in hepatocellular carcinoma. Hepatology 48, 1851–1863 [DOI] [PubMed] [Google Scholar]
- 57. Ralhan R., Desouza L. V., Matta A., Chandra Tripathi S., Ghanny S., Datta Gupta S., Bahadur S., Siu K. W. (2008) Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol. Cell. Proteomics 7, 1162–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Agard N. J., Maltby D., Wells J. A. (2010) Inflammatory stimuli regulate caspase substrate profiles. Mol. Cell. Proteomics 9, 880–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lamkanfi M., Kanneganti T. D., Van Damme P., Vanden Berghe T., Vanoverberghe I., Vandekerckhove J., Vandenabeele P., Gevaert K., Nunez G. (2008) Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell. Proteomics 7, 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Patel P. C., Fisher K. H., Yang E. C., Deane C. M., Harrison R. E. (2009) Proteomic analysis of microtubule-associated proteins during macrophage activation. Mol. Cell. Proteomics 8, 2500–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dagenais M., Skeldon A., Saleh M. (2012) The inflammasome: in memory of Dr. Jurg Tschopp. Cell Death Differ. 19, 5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Honnappa S., Gouveia S. M., Weisbrich A., Damberger F. F., Bhavesh N. S., Jawhari H., Grigoriev I., van Rijssel F. J., Buey R. M., Lawera A., Jelesarov I., Winkler F. K., Wuthrich K., Akhmanova A., Steinmetz M. O. (2009) An EB1-binding motif acts as a microtubule tip localization signal. Cell 138, 366–376 [DOI] [PubMed] [Google Scholar]
- 63. Jin T., Perry A., Jiang J., Smith P., Curry J. A., Unterholzner L., Jiang Z., Horvath G., Rathinam V. A., Johnstone R. W., Hornung V., Latz E., Bowie A. G., Fitzgerald K. A., Xiao T. S. (2012) Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor. Immunity 36, 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Okamoto M., Liu W., Luo Y., Tanaka A., Cai X., Norris D. A., Dinarello C. A., Fujita M. (2010) Constitutively active inflammasome in human melanoma cells mediating autoinflammation via caspase-1 processing and secretion of interleukin-1beta. J. Biol. Chem. 285, 6477–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.