Abstract
Background:
Hepatitis C is a disease with significant global impact. The distribution of hepatitis C virus (HCV) genotypes in Mashhad (the Northeast and the biggest city after the capital of Iran) is unknown. The purpose of this study was to determine the prevalence of HCV genotypes among HCV seropositive patients, and to study the relationship between types, virologic and demographic features of patients in Mashhad.
Methods:
Three hundred and eighty-two clinical specimens obtained from HCV-infected patients referred to Ghaem Hospital in Mashhad during a period of 2009 to 2010 were selected. HCV genotype was determined by Nested PCR amplification of HCV core gene using genotype specific primers.
Results:
Totally, 299 patients were male (79.9%). The most common HCV genotype was genotype 3a, with 150 (40%) of subjects. Genotype 1a was the other frequent genotype, with 147(39.2%) subjects. Frequency of genotypes for 1b, 5 and 2 was 41(10.9%), 13(3.4%) and 9(2.4%), respectively. Mix genotype including 1a+1b in 4 (1.04%), 1a+3a in 3 (0.8%) was found in 7 patients. Four percent out of these samples had an undetermined genotype. Among the hemophilia patient, there were 13(48.1%) genotypes as 1a, 3(11.1%) 1b and 10(37%) 3a, respectively.
Conclusion:
The dominant HCV genotype among patients living in Mashhad was 3a. This study gives added evidence of the predominant HCV genotypes in Iran.
Keywords: HCV, Genotyping, Nested PCR, Iran
Introduction
Hepatitis C infection is known as a serious health problem. It is one of the main causes of chronic viral hepatitis, and also cirrhosis and hepatocellular carcinoma in the world (1). The worldwide prevalence of hepatitis C virus (HCV) infection is estimated as 2% to 3% of the global population with approximately 200 million infected people (2). Prevention of HCV depends on an evaluation of global HCV infection distribution, assessment of its risk factors, and determination of accelerating factors in disease progression. Several host factors such as male sex, older age at infection, long disease duration, and various viral factors such as genotype and peripheral viral load have been described to effect on disease progression. However, due to the lack of a vaccine or some form of post-exposure prophylaxis, an accurate epidemiological assessment to plan primary prevention actions in any given population is essential (1).
HCV is classified into six major genotypes, several subtypes, and about 100 different strains based on variations in HCV RNA (3,4). Different HCV isolates worldwide show substantial nucleotide sequence variability throughout the viral genome. These viral types and subtypes differ in their geographical distribution (5–8). HCV genotype information is one of the most important factors in order to prediction of the medical treatment response, treatment duration and the dose of ribavirin (9,10). In addition, HCV genotyping is a useful tool to determine its molecular epidemiology, as they are indicative of transmission route of infection. HCV genotyping also has been used in the development of reliable diagnostic tests and vaccines (11).
Several studies on HCV genotype have been performed in Iran. No such studies on the HCV genotyping are available from this region of Iran. Therefore, this study was conducted to determine the genotype distribution of HCV in a population of patients in Mashhad, the Northeast of Iran.
Materials and Methods
Clinical samples
Three hundred and eighty-two clinical specimens obtained from HCV-infected patients living in Mashhad were selected for this study. Patients had well-documented hepatitis C infection, as confirmed by clinical and laboratory findings. Serum samples were obtained from all patients during a period of October 2009 to October 2010 and stored at −80°C until genotyping assay was carried out. The diagnosis of hepatitis C infection was made on the basis of the presence of anti-HCV antibodies in sera detected by third-generation commercially available enzyme-linked immunosurbent assay (ELISA) kits (DiaSorin, Spain).
RNA extraction and cDNA synthesis
Viral RNA was extracted from HCV positive serum samples according to the protocol provided by the commercial High Pure Viral RNA Kit (Boehringer, Germany). Briefly, 200 μl of plasma were treated with 400 μl of guanidine hydrochloride, supplemented by poly (A) carrier RNA. After vortexing, nucleic acids specifically bind to the surface of glass fibers precast into a column. Non-specifically bound material was removed by washing solution of the kit. Finally, nucleic acids were eluted in a volume of 50 μl of RNase-free water.
Ten microliters of the eluted RNA were used to carry out reverse transcription. cDNA synthesis was done with AMV reverse transcriptase (Promega, USA) and random hexamer. Two microliters of this cDNA were submitted for the first round of PCR reaction. Before performing of the genotyping reactions, in order to evaluation of the extracted RNA quality, after cDNA synthesis, PCR with beta-globin primers was done for all of the samples. Nucleotid sequences of these primers were: BGF: GAAGAGCCAAGGACAGGTAC and BGR: GGAAAATAGACCAATAGGCAG. The expected size for beta globin bond was 407 bp.
HCV genotyping assay
HCV genotype was determined by using the type-specific detection system of Ohno et al. (11). According to the protocol, reverse transcription was followed by two rounds of nucleic acid amplification. 2.5μM of S2 and A2 primers (which are the sense and the anti-sense primers for the core region, respectively), 0.2 mM dNTPs and 2 units of Taq DNA polymerase (Promega, USA) were utilized in the first round of amplification. Thermal cycles of the first PCR were as follows: initial denaturation at 94°C for 5 min; preliminary 20 cycles of amplification in three steps: 94°C for 1 min, 45°C for 1 min, 72°C for 1 min followed by 20 additional cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min.
In the second round, two different primer mixtures were prepared. Mix1 included the S7, S2a, G1b, G2a, G2b, and G3b primers and Mix2 contained the S7, G1a, G3a, G4, G5a, and G6a primers (Table 1).
Table 1:
oligonucleotide primers used for genotyping of Hepatitis C
| Primer | Nucleotide position | Sequence (5-3) |
|---|---|---|
| S2 | core protein | GGGAGGTCTCGTAGACCGTGCACCATG |
| A2 | core protein | GAG(AC)GG(GT)AT(AG)TACCCCATGAG(AG)TCGGC |
| S7 | core protein | AGACCGTGCACCATGAGCAC |
| S2a | core protein | AACACTAACCGTCGCCCACAA |
| G1a | core protein | GGATAGGCTGACGTCTACCT |
| G1b | core protein | CCTGCCCTCGGGTTGGCTA(AG) |
| G2a | core protein | CACGTGGCTGGGATCGCTCC |
| G2b | core protein | GGCCCCAATTAGGACGAGAC |
| G3a | core protein | GCCCAGGACCGGCCTTCGCT |
| G3b | core protein | CGCTCGGAAGTCTTACGTAC |
| G4 | core protein | CCCGGGAACTTAACGTCCAT |
| G5a | core protein | GAACCTCGGGGGGAGAGCAA |
| G6a | core protein | GGTCATTGGGGCCCCAATGT |
Then, 0.5 μl of first-round PCR product was used and two PCR reactions were carried out for each sample, one with primer mix 1 and the other with mix 2. Thermal cycles of second PCR were as follows: 30 cycles of (94°C for 1 minute, 62°C for 45 seconds and 72°C for 1 minute). All reactions were performed with negative sera as a negative control.
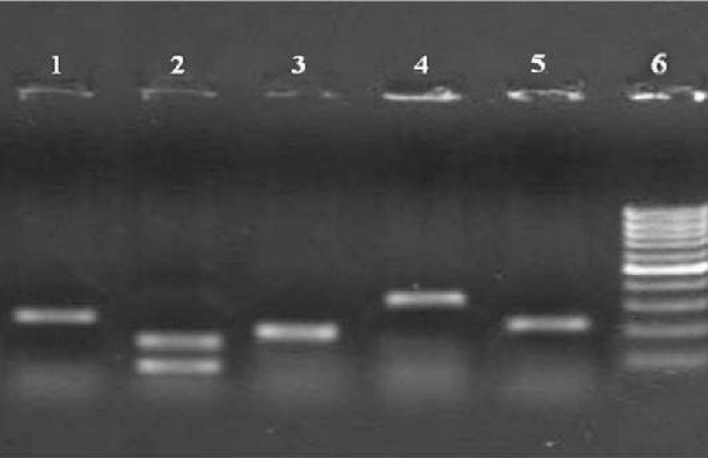
After DNA amplification, data analysis was done based on the presence or absence of specific bands of amplified DNA in agarose gel (2%). The HCV genotype for each sample was determined by identifying the genotype specific DNA bands, as showed in Fig. 1.
Fig. 1:
Electrophoresis patterns of PCR products from different HCV genotypes. Line 1: genotype 1b, 234 bp ; line 2: genotype 2a, 139 and 190 bp; line 3: genotype 1a, 208 bp; line 4: genotype 5a, 320 bp; line 5: genotype 3a, 232 bp; line 6: 100bp Ladder
In this study HCV RNA viral load was detected using Cobas Amplicor Test (Version 2, Roche Molecular Systems).
Statistical analysis
Statistical analysis was performed using SPSS version 11.5 software. Descriptive statistics were used to calculate the mean and standard deviation. All information was compared with clinical data including age, sex, and HCV viral load. Differences in virologic, biochemical and demographic features between groups were analyzed using chi-squared test and Student’s t-test. The significance level was set at a P-value of ≤ 0.05.
Results
Patients
In the present study all the 382 cases with hepatitis C infection were evaluated. 299 patients of this study were male (79.7%) and 76(30.3%) of them were female with a mean age of 40 (range 13–76 years; 33.9±11.83). Among them, there were 26 (6.8%) with hemophilia.
Distribution of HCV genotypes
In this study, genotype 3a showed the highest frequency with 150 (40%) of subjects. Genotype 1a was the other frequent genotype, with 147 (39.2%) subjects. Frequency of Genotype of 1b, 5 and 2a were 41(10.9%), 13(3.4%) and 9(2.4%), respectively. As shown in Fig. 1, the expected size of the genotype-specific bands in primer mixture (1) was 234 bp for genotype (1b), 139 bp and 190 bp for genotype (2a). For HCV genotype 2a, the antisense primer G2a potentially can be annealed to a nucleotide sequences in genotype 4. So a specific sense primer for genotype 2 (S2a), was added, resulting in two bands is visible. As shown in Fig.2, the expected size of the genotype-specific bands in primer mixture (2) was 208 bp for genotype (1a), 232 bp for genotype (3a), 320 bp for genotype (5a) and 336 bp for genotype (6a).
Furthermore, 7(1.82%) patients were infected simultaneously by two genotypes; 4(1.04%) by the 1a+1b genotypes, and 3(0.8%) by the 1a+3a genotypes. 15(4%) out of these samples had an undetermined genotype. It should be noted that among the hemophilia patients, there were 13(48.1%) genotypes 1a, 3(11.1%) 1b and 10(37%) 3a, respectively.
HCV RNA viral load in 212 patients was measured. The patients with genotype 3a had high levels of HCV viremia (Mean = 4.9 million copies/ml). In addition, in patients with genotype 1a, the average of HCV RNA was 2.1 million copies/ml.
Discussion
HCV is now recognized as an important public health problem globally. More than 50% of the infected individuals develop severe chronic hepatitis with liver cirrhosis and hepatocellular carcinoma. Indeed, among all types of viral hepatitis diagnosed in the local community, HCV is responsible for up to 56% of total cases (2,12).
Regional differences in the prevalence of HCV are focused by many studies (13,14). The most countries have prevalence rates from 1 to 2%, meanwhile, higher rates have been reported in Asian countries, including Egypt (15%), Pakistan (4.7%) and Taiwan (4.4%) (15). The last study on HCV prevalence in Iran was on healthy blood donors in 2010, and showed a seroprevalence of 0.5% (2).
The HCV genotypes should be identified before starting any treatment for HCV infection, as it affect in selection of an appropriate antiviral therapy. Available evidence in various regions of the world shows the differences in distribution of HCV genotypes. Genotype 1a is most frequently found in Northern Europe and North America, whereas lb is the most common genotype in Japan, Southern and Eastern Europe. Genotype 2 is present in most developed countries, but is less common than type 1. Genotype 3 is endemic in South-East Asia and India. Type 4 is the most prevalent genotype in the Middle East, Egypt, and Central Africa, type 5 is almost exclusively found in South Africa, and finally, type 6 is present in Hong Kong, Macau and Vietnam (16–19).
In Iran, located in the Middle East, there have been regional reports on the hepatitis genotyping among infected patients. The goal of present study was to identify the prevalence of HCV genotypes in Mashhad (the northeast of Iran). The results showed that the HCV genotype 3a is predominant type in Mashhad. Present results are nearly similar to those studies which have already been conducted in Isfahan and Shiraz, Iran (13,14). Our investigation was inconsistent to earlier studies from Tehran that reported genotype 1a was the most prevalent genotype in Iran (7,20). Comparing to the studies of Iran's neighbor countries, it can be understood that the most common genotype of Pakistan is type 3a. No study is available on geographic variation in the prevalence of various HCV genotypes from Afghanistan. However, external displacement can be consider, particularly refugees to neighboring countries including Afghanistan and Pakistan which harbor higher infected population of HCV (15).
Epidemiological differences in age distribution of the major HCV genotype have investigated (17). European studies show genotype 1a and 3a are relatively more common in young individuals with a history of intravenous drug use. In some studies, type 1b infected patients were significantly older than those infected by other genotypes and a significant proportion of younger patients were infected with type 3 (21). In this study, type 3a infection was found predominantly in younger patients (42.9% in age 30 or less) and type 1a was common in those aged 50 or more.
Several studies have indicated that infection with certain HCV genotypes is influenced by the route of its transmission. Genotype la was the major genotype in patients receiving multiple transfusions, while genotype 3a was detected primarily in intravenous drug users (22). In this study, data regarding route of transmission were not available, so it was not possible to associate HCV genotype with route of transmission. It would be interesting to explore such relationships in the population of Mashhad.
HCV viral load and viral genotype are two key predictive markers, knowledge of which might be valuable in the treatment decisions. Viral load seems to be a useful prognostic marker for the outcome of antiviral therapy. In fact, patients with high level viremia show a poor response to interferon therapy than those with low viral load. However, the correlation between HCV viral load and genotypes remains controversial (23, 24).
In the present study, HCV RNA in the number of patient (n= 212) was measured. Analyzed data showed that the majority of patients with HCV type 3a infections had high levels of viremia (N=89, Mean = 4.9 million copies/ml). HCV RNA levels of type 3a infected patients in this study were significantly higher than those in the HCV type 1 of infected patients (4.9 and 2.1 million copies/ml, respectively). This was in contrast with studies that have shown a random distribution of HCV RNA levels among patients infected with different genotypes (24, 25).
Finally, according to recent studies, genotype analysis should be part of treatment algorithms at some point in the process. This study highlighted that the dominant HCV genotype among patients living in Mashhad is 3a. This study gives added evidence of the predominant HCV genotypes in Iran.
Ethical considerations
Ethical issues (Including plagiarism, Informed Consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc) have been completely observed by the authors.
Acknowledgments
Funding for this study was provided by the Research Assistant in Mashhad University of Medical Sciences, grant No. 64707 and the authors are grateful for their financial assistance. The study received approval from the Research Ethics Board in Mashhad University of Medical Sciences. Finally, the authors acknowledge Farnood Rajabzadeh for his collaboration in this study. We have no competing interests to declare.
References
- 1.Martins T, Narciso-Schiavon JL, Schiavon Lde L. Epidemiology of hepatitis C virus infection. Rev Assoc Med Bras. 2011;57(1):107–12. doi: 10.1590/s0104-42302011000100024. [DOI] [PubMed] [Google Scholar]
- 2.Merat S, Rezvan H, Nouraie M, Jafari E, Abolghasemi H, Radmard AR, et al. Seroprevalence of hepatitis C virus: the first population-based study from Iran. Int J Infect Dis. 2010;14s:e113–e116. doi: 10.1016/j.ijid.2009.11.032. [DOI] [PubMed] [Google Scholar]
- 3.Duarte CAB, Foti L, Nakatani SM, Riedigerc IN, Poersch CO, Pavoni DP, et al. A Novel Hepatitis C Virus Genotyping Method Based on Liquid Microarray. PloS One. 2010;5:e12822. doi: 10.1371/journal.pone.0012822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu P, Cai XC, Ding W, Zhang Q, Norris ED, Greene JR. HCV genotyping using statistical classification approach. J Biomed Sci. 2009;16:62. doi: 10.1186/1423-0127-16-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavalheiro NP. Hepatitis C: Genotyping. Braz J Infect Dis. 2007;11(5):25–27. Suppl. 1. [Google Scholar]
- 6.Omrani MD, Khadem Ansari MH. Hepatitis C virus genotyping by Melting curve analysis in West Azarbaijan, Northwest of Iran. Hepat Mon. 2009;9:133–36. [Google Scholar]
- 7.Keyvani H, Alizadeh AH, Alavian SM, Ranjbar M, Hatami S. Distribution frequency of hepatitis C virus genotypes in 2231 patients in Iran. Hepatol Res. 2007;37(2):101–3. doi: 10.1111/j.1872-034X.2007.00015.x. [DOI] [PubMed] [Google Scholar]
- 8.Ramia S, Eid-Fares J. Distribution of hepatitis C virus genotypes in the Middle East. Int J Infect Dis. 2006;10:272–77. doi: 10.1016/j.ijid.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Zarkesh-Esfahani S, Kardi MT, Edalati M. Hepatitis C virus genotype frequency in Isfahan province of Iran: a descriptive cross-sectional study. Virol J. 2010;7:69. doi: 10.1186/1743-422X-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao H, Zhang H, Zhang H, Zhao J, Lu Z, Jin G, et al. Clinical evaluation of a colorimetric oligonucleotide chip for genotyping hepatitis C virus. Clin Biochem. 2010;43:214–19. doi: 10.1016/j.clinbiochem.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Ohno T, Mizokami M, Wu RR, Saleh MG, Ohba KI, Orito E, et al. New Hepatitis C Virus (HCV) Genotyping System That Allows for Identification of HCV Genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microb. 1997;35(1):201–207. doi: 10.1128/jcm.35.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wasley A, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin Liver Dis. 2000;20:1–16. doi: 10.1055/s-2000-9506. [DOI] [PubMed] [Google Scholar]
- 13.Zarkesh-Esfahani SH, Kardi MT, Edalati M. Hepatitis C virus genotype frequency in Isfahan province of Iran: a descriptive cross-sectional study. Virol J. 2010;7:69. doi: 10.1186/1743-422X-7-69.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziyaeyan M, Alborzi A, Badiee P, Moeini M. Prevalence of hepatitis C virus genotypes in chronic infected patients, southern Iran. J J M. 2011;4(3):141–146. [Google Scholar]
- 15.Sievert W, Altraif I, Razavi HA, Abdo A, Ali Ahmed E, AlOmair A, et al. A systematic review of hepatitis C virus epidemiology in Asia, Australia and Egypt. Liver Int. 2011:61–80. doi: 10.1111/j.1478-3231.2011.02540.x. [DOI] [PubMed] [Google Scholar]
- 16.Kabir A, Alavian SM, Keyvani H. Distribution of hepatitis C virus genotypes in patients infected by different sources and its correlation with clinical and virological parameters: a preliminary study. Comp Hepatol. 2006;5:4–9. doi: 10.1186/1476-5926-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoofnagle JH, Wahed AS, Brown RS, Jr, Howell CD, Belle SH. Early changes in hepatitis C virus (HCV) levels in response to peginterferon and ribavirin treatment in patients with chronic HCV genotype 1 infection. J Infect Dis. 2009;199:1112–20. doi: 10.1086/597384. [DOI] [PubMed] [Google Scholar]
- 18.Madhavi C, Thippavuzzula R, Rao R, Aejaz MH, Habibullah CMa, Narasu L, et al. Genotyping of Hepatitis C virus (HCV) in infected patients from South India. Infect Genet Evol. 2007;7:724–30. doi: 10.1016/j.meegid.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Park JC, Kim JM, Kwon OJ, Lee KR, Chai YG, Oh HB. Development and clinical evaluation of a microarray for hepatitis C virus Genotyping. J Virol Methods. 2010;163:269–75. doi: 10.1016/j.jviromet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Safieh Amini S, Mahmoodi Farahani Majd Abadi M, Alavian SM, Joulaie M, Ahmadipour MH. Distribution of Hepatitis C Virus Genotypes in Iran: A Population-Based Study. Hepat Mon. 2009;9:95–102. [Google Scholar]
- 21.Meffre C, Delarocque-Astagneau E, Pioche C, Dubois F, Roudot-Thoraval F, et al. Hepatitis C testing, care and treatment: evolution over ten years in France. Hepatology. 2007;46:625A. [Google Scholar]
- 22.Pawlotsky JM, Chevaliez S. Preventing hepatitis C virus recurrence in liver transplant recipients: a role for adoptive immunotherapy. Hepatology. 2010;51(3):1072–6. doi: 10.1002/hep.23579. [DOI] [PubMed] [Google Scholar]
- 23.Anderson JC, Simonetti J, Fisher DG, Williams J, Yamamura Y, Rodriguez N, et al. Comparison of different HCV viral load and genotyping assays. J Clin Virol. 2003;28(1):27–37. doi: 10.1016/s1386-6532(02)00235-4. [DOI] [PubMed] [Google Scholar]
- 24.Chakravarti A, Dogra G, Verma V, Srivastava AP. Distribution pattern of HCV genotypes and its association with viral load. Indian J Med Res. 2011;133:326–331. [PMC free article] [PubMed] [Google Scholar]