Abstract
The accurate estimation of the number and size of cells provides relevant information on the kinetics of growth and the physiological status of a given tissue or organ. Here, we present Adiposoft, a fully automated open-source software for the analysis of white adipose tissue cellularity in histological sections. First, we describe the sequence of image analysis routines implemented by the program. Then, we evaluate our software by comparing it with other adipose tissue quantification methods, namely, with the manual analysis of cells in histological sections (used as gold standard) and with the automated analysis of cells in suspension, the most commonly used method. Our results show significant concordance between Adiposoft and the other two methods. We also demonstrate the ability of the proposed method to distinguish the cellular composition of three different rat fat depots. Moreover, we found high correlation and low disagreement between Adiposoft and the manual delineation of cells. We conclude that Adiposoft provides accurate results while considerably reducing the amount of time and effort required for the analysis.
Keywords: fat cell diameter, fat tissue cellularity, adipocyte, quantification, image analysis, software validation, obesity
Obesity is a disease associated with increased white adipose cellular mass, caused either by increased number of adipocytes (hyperplasia) or by adipocyte enlargement due to cytoplasmic lipid accumulation (hypertrophy). Therefore, the quantification of the amount and size of adipocytes in animal models is critical to properly characterize the differential response of individuals to dietary treatments (1), to test the efficacy of different food ingredients and bioactive compounds in models of diet-induced obesity, and to relate changes in cellularity with pathophysiological processes affected by obesity, such as oxidative stress, insulin resistance, and lipidemias (2–4). In summary, determining what causes the change in tissue cellularity, along with the specific location of the growth of adipose tissue, is essential to study the mechanisms driving obesity and metabolic diseases in animal models.
Noninvasive imaging techniques, such as magnetic resonance imaging or X-ray computed tomography, are commonly used for the quantification of white adipose tissue mass in vivo. Unfortunately, these techniques do not provide information about adipose cellularity and thus are unable to distinguish between hyperplasia- and hypertrophy-driven obesity. For that purpose, several high-resolution ex vivo techniques exist, such as the analysis of cells in suspension, adipocyte fractionation, flow cytometry, and the histological analysis of cellularity in stained sections using optical or electron microscopy.
Among all these methods, the analysis of cells in suspension is the most used technique, and some automated image analysis tools have been developed for the quantification of these samples (5, 6). Unfortunately though, the preparation of cells in suspension is complex because it requires the isolation of cells using collagenase. Moreover, the required tissue disaggregation causes loss of the microscopic histopathological context.
The microscopic analysis of adipocyte cellularity in hematoxylin and eosin (H&E)-stained histological sections provides a reasonable cost-benefit ratio compared with the other methods. In this type of analysis, histological images are captured using a wide-field optical microscope, printed or screened, and then manually analyzed to count the number and measure the diameter of every cell. From the diameter, the average adipocyte volume and lipid content can be mathematically derived (7, 8). Although manual delineation is considered the most accurate method, it is extremely time consuming and cannot be used in practice, except as a gold standard to validate other methods. To simplify the otherwise time-consuming analysis of these histological samples, semi-automated computer tools can be used, such as the one proposed by Chen et al (9) based on two independent, proprietary software packages, which reduce, but do not eliminate, human interaction.
In this article, we present and validate Adiposoft, open-source software that provides fast and accurate quantitative measurements of adipose tissue cellularity in histological sections. The validation is done by comparing the software with the manual analysis of histological samples and with the analysis of cells in suspension. The three methods are compared in terms of speed, sample preparation, and imaging burden, as well as by the accuracy of the analysis in different fat tissue depots.
MATERIALS AND METHODS
Animal work
Six male Wistar rats (weight 337.9 ± 14 g) were euthanized, and tissue blocks were extracted from three fat depots: retroperitoneal (RP), located in the abdominal cavity behind the peritoneum; mesenteric (MES), from the double layer of peritoneum that suspends the jejunum and ileum from the posterior wall of the abdomen; and subcutaneous (SC). Fat areas were identified, manually separated, and excised to avoid mixing white fat from different depots (10, 11). Tissue blocks from all depots were labeled either for histology or for the analysis of cells in suspension. All animal handling was done following both national and institutional guidelines of the Animal Care and Use Committee of the University of Navarra.
Sample preparation and imaging
Sample preparation for histological analysis using Adiposoft and manual validation.
Tissues were fixed in paraformaldehyde and embedded in paraffin. Two 4 μm sections per block were then stained with H&E and imaged at 20× (Plan-Neofluar objective with 0.50 NA) magnification with an AxioImager.M1 microscope (Zeiss, Germany). A total of 1,078 high-resolution images were acquired for the analysis. All images were stored in uncompressed 24-bit color TIFF format.
Sample preparation for the analysis based on cells in suspension.
White fat areas were cut into small pieces. Each fragment was digested at 37°C with collagenase I (2.5 mg ml−1) (Worthington Biochemical, Lakewood, NJ) in Krebs-Ringer bicarbonate buffer (KRBA) containing albumin (3.5 g 100 ml−1) and glucose (6 mM) at 7.4 pH. We used 2 ml of digestion solution per gram of adipose tissue. After 30 min of incubation under continuous vigorous shaking (60 cycles min−1), fat cells were filtered through a nylon mesh (400 µm) and washed three times with KRBA to eliminate the stroma-vascular fraction and collagenase. The filtered fat cells were diluted 1:1 in KRBA and loaded in a neubauer chamber. A total of 738 brightfield, wide-field images were captured using a CKX31 Compact inverted microscope equipped with a 40× magnification objective lens and a C5060WZ Olympus compact camera. All images were stored as JPG files.
Analysis
Automated analysis of histological sections using Adiposoft.
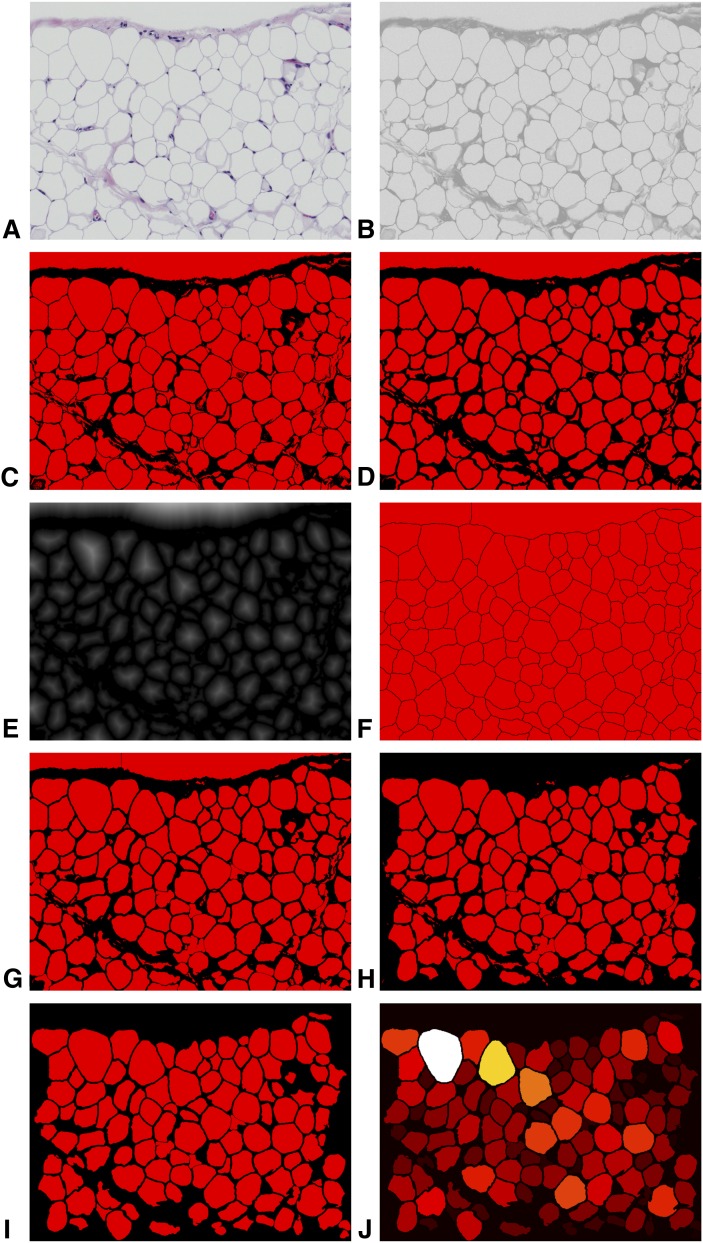
The sequence of image analysis steps automatically implemented by Adiposoft is shown in Fig. 1. This sequence was executed in batch mode on the entire set of images stored in a predefined directory. Briefly, each input color image (Fig. 1A) is split into its three-color channels. The red channel (Fig. 1B) is binarized using Otsu's automatic thresholding method (12) that extracts the bright fat areas from dark cell nuclei and membranes (Fig. 1C). Because cells often merge in areas that contain thin cellular walls, we detect the cell boundaries in those regions using a morphological filter. Namely, we apply a morphological Opening followed by a Median filter (13) to remove small objects and reinforce cell boundaries (Fig. 1D). Next, we apply a seeded Watershed algorithm (14, 15) to the filtered image using as seeds the peaks of the distance transform of the previously computed image (Fig. 1E). Next, we use a logical AND operation to combine the result of the Watershed (Fig. 1F) with the output of the initial thresholding (Fig. 1C), thus reinforcing the cellular boundaries and forcing the separation of clustered adipocytes (Fig. 1G). Objects touching the image borders are deleted (Fig. 1H). The remaining objects are measured (number, size, and Feret diameter), and those with a diameter below a predefined threshold (25 μm) are removed from the image (Fig. 1I). All the objects left in the image are then labeled according to their sizes (Fig. 1J), and the output data is stored in an Excel file. From the diameters, the mean fat-cell volume and lipid content are derived mathematically using the method described in Bourgeois et al. (16).
Fig. 1.
Sequence of image analysis operations performed with Adiposoft. A: Initial image. B: 8-bits red channel. C: Binarized version of the red channel. D: Result of the morphological Opening of (C). E: Distance transform of (D). F: Watershed of (D)using as seed the maxima of the distance transform (E). G: AND operation between (C) and (F). H: Deletion of objects touching the edges of the image. I: Deletion of small objects. J: Final labeled image.
Manual analysis of histological sections.
The set of histological images was also manually analyzed. The method used is not completely manual, but it requires intensive user interaction: First, all the adipocytes were manually seeded using ImageJ's multipoint selection (17). Then the adipocytes were automatically separated using the Watershed transform plugin. Interactive tools were used, whenever necessary, to correct inaccurate results. The adipocyte area, Feret diameter, and number were measured.
Automatic analysis of cells in suspension.
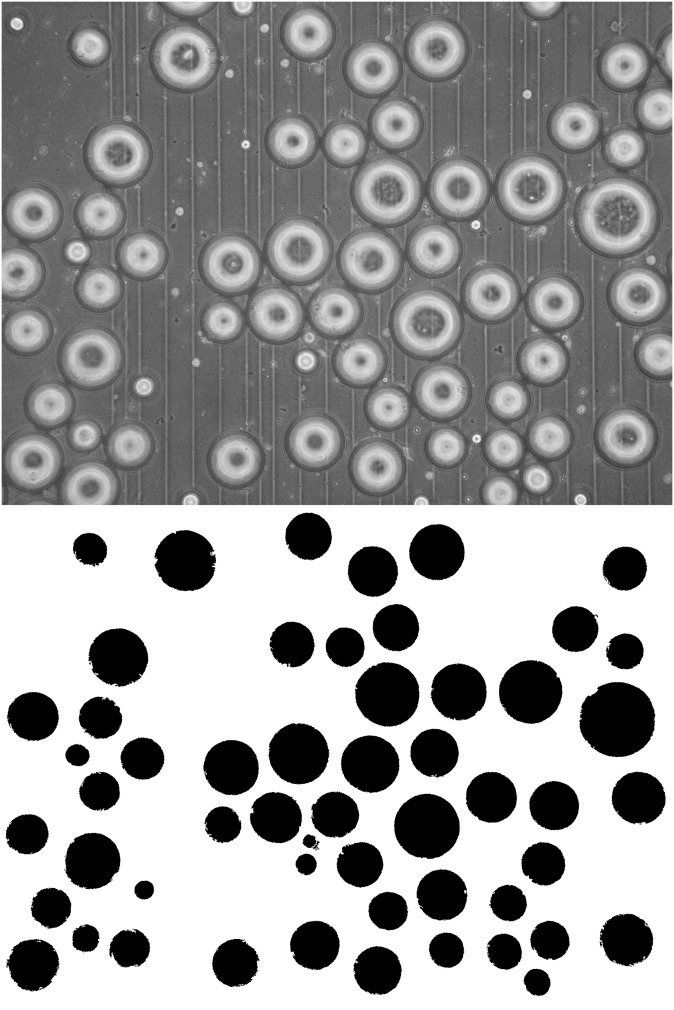
The images of cells in suspension were analyzed using a set of simple image analysis and measurement routines available in ImageJ. First, each image is split into its three-color components, and the green channel is binarized using ImageJ's embedded automated threshold. Then the area, diameter, and number of cells are calculated. Fig. 2 shows an image with cells in suspension and the output of the segmentation algorithm.
Fig. 2.
(Top) Original image cells in suspension. (Bottom) Segmentation result.
Software platform and availability.
Adiposoft was entirely developed in MATLAB v.7.11 (MathWorks, Natick, MA) using the DIPlib v2.2 C Image Processing library (18) under Linux OS Fedora 14. To run Adiposoft, these or subsequent versions of MATLAB and DIPlib for any OS are required. We also migrated Adiposoft to JAVA and packaged it as an open-source ImageJ (17) plugin. Both versions of Adiposoft (MATLAB-based and Java-based), together with instructions for installation and setup are available at http://sw.wikkii.com/wiki/Adiposoft. The plugins used to manually analyze the histological samples and to process the cells in suspension are available at the same site.
Statistical analysis
Three-sided comparison of Adiposoft, manual, and suspension.
To validate Adiposoft, we compared the result [mean of the distribution of adipocyte diameters (MAD)] obtained using Adiposoft (described above) with our manually computed gold standard (described above) and with the analysis of cells in suspension (described above). To this end, we used samples from three different depots: MES, RP, and SC. Because there were three different analysis methods, the comparison was based on three cell populations. The cells analyzed by Adiposoft and manually were exactly the same, whereas the cells analyzed in suspension were different, although the cells were extracted from the same region and animal. We analyzed a total of 54 tissue samples, organized into nine depot method groups.
The statistical differences between the nine groups were analyzed using two-way ANOVA, with one dependent variable (MAD) and two independent variables (depot and method). ANOVA detects whether there are significant differences between groups for the two independent variables, as well as for their interaction (depot*method). A level of probability P < 0.05 was used for statistically significant results, and P < 0.01 for very statistically significant results. For all statistical tests, SPSS v15.0 was used (SPSS, San Diego, CA).
Direct comparison of Adiposoft with the manual method.
To compare the accuracy of Adiposoft with the manual method, we calculated the Pearson correlation between the per-image average adipocyte diameter on 250 images from all rats and depots. Then, we built a Bland and Altman plot, which graphically displayed the agreement and bias between both methods by plotting the per-image difference between the average diameter calculated using both methods. The plot indicates agreement if more than 95% of the data lies within two standard deviations of the average value. Additionally, we estimated the confidence interval with the interimage standard deviation error. For the statistical tests, we used SPSS v15.0 (SPSS, San Diego, CA) and the open-source statistical software package R (Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Three-sided comparison of Adiposoft, manual, and suspension methods
The dependent variable MAD was normally distributed for the groups formed by the combination of depot and method, as assessed by the Shapiro-Wilk test (all P > 0.05). There was also homogeneity of variance between groups as assessed by Levene's test for equality of error variances (0.438).
The two-way ANOVA test (manual versus Adiposoft versus suspension) with the three different depots (MES, RP, SC) gave the expected results: i) we found no interaction between the depot and the method used to analyze the size of the adipocytes (P = 0.566); ii) for a given depot, the election of method did not produce significantly different measurements (P = 0.278); and iii) when we used the same method, we found significant differences between depots (P < 0.001).
We then used a simple main effect analysis of MAD to confirm that the MAD of a given depot is not significantly different when analyzed with different methods (for MES, P = 0.413; for RP, P = 0.163; and for SC, P = 0.984). Also, using this type of analysis, we were able to see very statistically significant differences between depots (P < 0.001) when using the three methods separately.
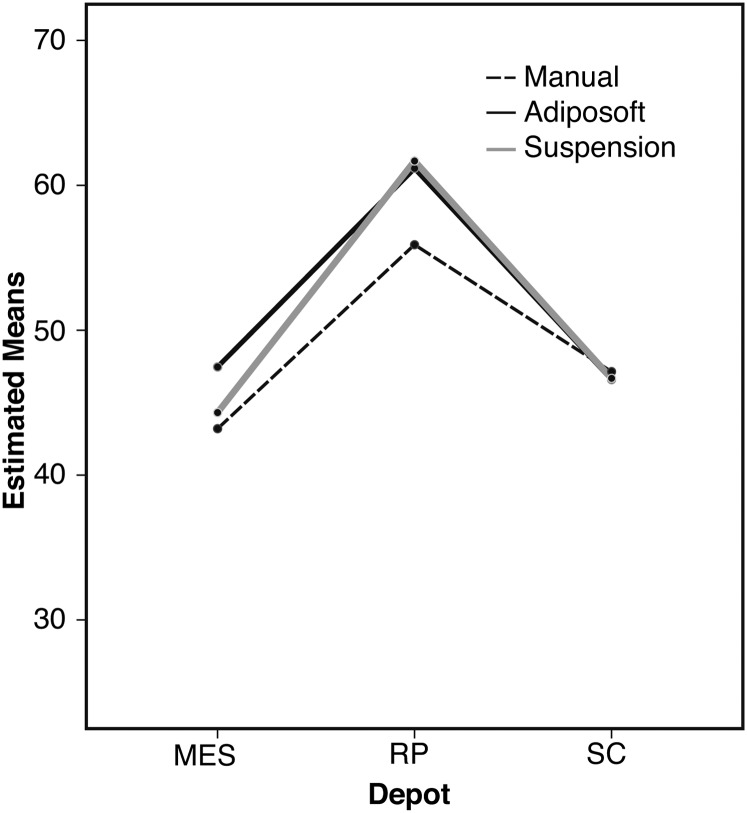
Finally, the Tukey posthoc test showed that adipocytes of the RP depot were significantly larger, which is a well-known feature, than those from MES and SC depots for all three methods used. This can be observed in Fig. 3, which represents the plot of results.
Fig. 3.
Plot of Tukey posthoc test results for two-way ANOVA tests for the mean diameter.
Direct comparison between Adiposoft and manual analysis
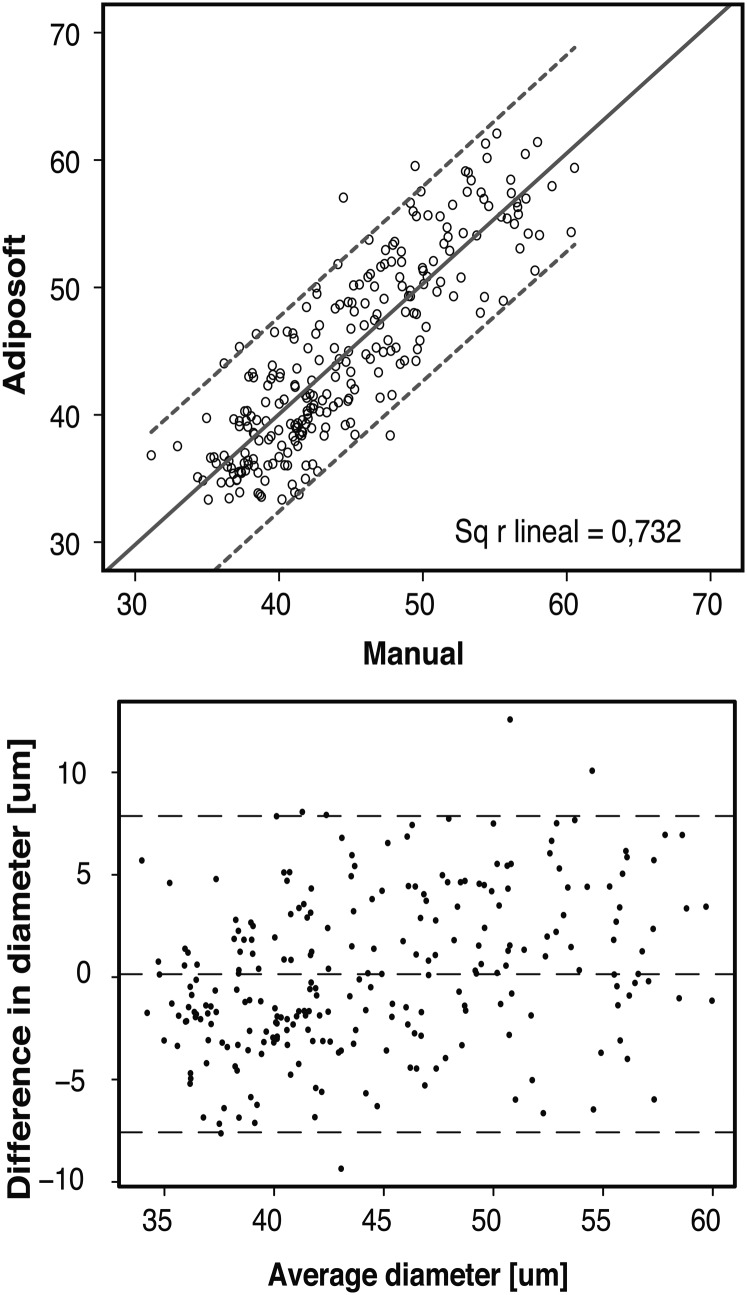
We found high positive correlation between the per-image average adipocyte diameter provided by Adiposoft and the manual methods (Pearson correlation = 0.856, with 95% confidence interval, low = 0.73 and high = 0.83).
Fig. 4A shows the dispersion of the per-image average diameter as measured by the Adiposoft and manual methods. Fig. 4B shows the Bland and Altman plot that displays the agreement between methods. The middle line corresponds to the existing bias, which in our case is very low (0.17 µm). The limits of agreement are shown as dashed lines. A value above the upper limit (+3.82 µm) or below the lower limit (−3.82 µm) has 95% likelihood of representing a real difference between the quantification obtained using the two methods. The calculated intra-image variability is 0.159 ± 0.345.
Fig. 4.
(Top) Plot showing the agreement between the average diameters calculated using Adiposoft or the manual method. The plot is based on the analysis of 250 images. The line shows the linear regression with the 95% confidence interval. (Bottom) Bland Altman plot of diameter measures.
Regarding time consumption, Adiposoft can analyze 40 fields in less than 5 min in a regular computer without any user intervention, whereas a manual operator, depending on the skills, could use an average of 5 min per image. Therefore, Adiposoft is between one and two orders of magnitude faster than the average manual operator.
DISCUSSION
In this article, we have presented Adiposoft, fully automated open-source software for the quantification of adipocyte cellularity in histological sections. We have described the sequence of image analysis routines implemented by the program and have compared the performance of the software with our gold standard of manually segmenting the cells and with the analysis of cells in suspension. The comparison was based on the frequency size distribution of adipose cells in three different rat fat depots.
Our results show significant concordance between Adiposoft and the other two methods, along with the expected ability to distinguish the cellular composition of all three different rat fat depots. Furthermore, we found high correlation and low disagreement between Adiposoft and the manual delineation of cells. In terms of time consumption, Adiposoft is between one and two orders of magnitude faster than the manual method.
Therefore, Adiposoft provides similarly accurate results while being less time and effort consuming than the other two methods. Namely, Adiposoft requires a single click to analyze a whole set of images. This is significantly more effortless than manually segmenting the cells, which in its less effort consuming version requires manually planting a seed inside all the cells of each image and manually correcting inaccurate results. Adiposoft works with standard histological sections, whereas the analysis of cells in suspension requires collagenase digestion of the adipose tissue sample, adipocyte filtration and washes, and subsequent microscopic measurement of the diameter (19). The disadvantages of this method include the need to extract and separate the adipocytes, which can easily break and divide during the process (thus altering the results), and the need for qualified staff to perform this protocol. Furthermore, in animal models with diet-induced obesity, hypertrophic adipocytes are more susceptible to breaking by treatment with collagenase. In summary, the automated analysis of histological sections by Adiposoft provides similar results to the analysis of cells in suspension, with the added value that the results are obtained without losing contextual histological information. The results should be more accurate in cases with diet-induced obesity due to the problems associated with the preparation protocol.
A reasonable concern about the method used to evaluate Adiposoft stems from the fact that we used collagenase digestion to create the cell suspensions to evaluate our software. This procedure is usually problematic when handling large adipocytes, as they tend to break apart and coalesce into fat droplets (20). Distinguishing these fat droplets from intact fat cells is not an easy task on unstained samples. Perilipin immune staining is commonly used to distinguish between intact adipocytes and lipid droplets. Our samples were not stained with Perilipin, which allowed for a small statistical error. However, fresh tissue from healthy normal rats was treated by highly experienced staff, thus minimizing the rupture of adipocytes and consequent coalescence into fat droplets.
Two limitations of Adiposoft are the relatively time-consuming task of acquiring the images and the analysis of nonfatty tissue and low-quality images.
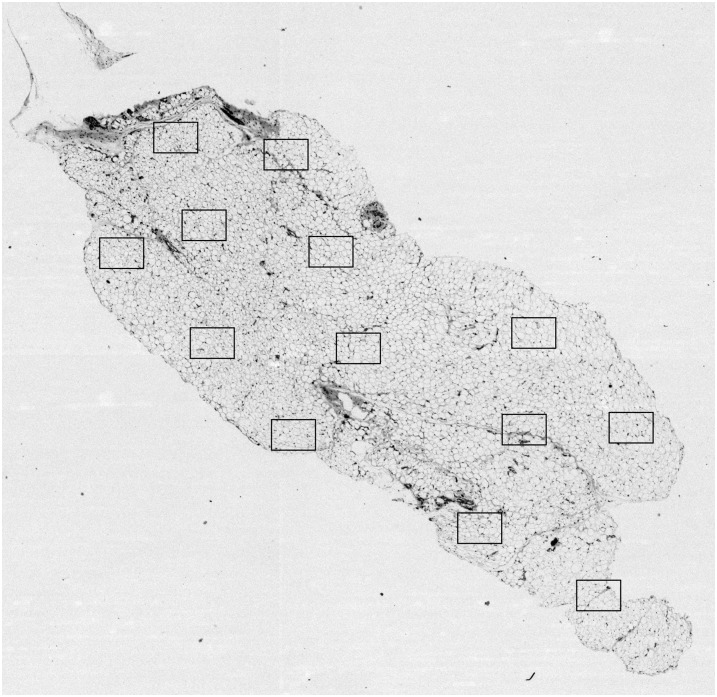
To address the first issue, to automate the microscopy acquisition of the samples, we have implemented a Metamorph macro (Molecular Devices). (The macro is available at http://sw.wikkii.com/wiki/Adiposoft.) The macro first acquires the entire tissue section at low magnification (1.25× in Fig. 5). Then, the areas occupied by the tissue are detected. Finally, a predefined number of high-resolution, randomly located images are acquired at high magnification (20×, at the squares overlayed in Fig. 5). To that end, the system automatically focuses each field of view and then applies a white balance and shadow correction on every acquired image. The high-magnification images are then automatically analyzed by Adiposoft as described in Materials and Methods.
Fig. 5.
Sample H&E-stained tissue section acquired at low magnification (1.25×). The squares indicate the positions and the size of some random high-resolution (20×) acquisitions.
Regarding the analysis of nonfatty tissue, the tissue depots used were fairly homogenous, mostly containing only fatty cells. Therefore, most high-resolution images contain only adipocytes. However, it is true that sometimes high-resolution images may contain areas with background because the images were taken at the periphery of the tissue or in areas of the tissue containing fissures. Sometimes there might also appear blood vessels or connective tissue structures. In those cases, our software is able to remove those parts from the images, based on the intensity, size, and shape of the structures that they form. In summary, very rarely is one of those nonfatty structures confused with a cell, and when it does happen, the effect of this error on the final result of the analysis, based on hundreds or thousands of cells, is meaningless.
Related to this last issue, during the preparation of histological sections, adipocytes can break, thus complicating the segmentation of the cells. Low-quality images with many broken adipocytes can be discarded; alternatively, Adiposoft can be run in semi-automatic mode. In this mode, the user can manually intervene in some of the steps shown in Fig. 1. Namely, the results can be edited by deleting false positives, dividing adipocytes clusters, or restoring incorrectly deleted regions. This option was not used in the evaluation of Adiposoft presented above, which was based on the fully automated mode. Furthermore, our data show that the diameter of adipocytes in the histological sections is on average smaller than in freshly isolated adipocytes (cells in suspension). This result is reasonable because the fixation procedure produces approximately a 20% tissue volume reduction. This issue must be taken into account when using the absolute size values provided by Adiposoft, but it is not a significant problem when comparing different adipose depots or treatment groups, as they are all equally affected by this effect.
In summary, Adiposoft provides a good cost-benefit solution to the problem of analyzing adipose cellularity. Adiposoft automates the analysis of histological adipose tissue samples faster and independently of the user, thus providing more objective results than the existing software and manually based methods.
Footnotes
Abbreviations:
- H&E
- hematoxylin and eosin
- MAD
- mean of the distribution of adipocyte diameters
- MD
- mean diameter
- MES
- mesenteric
- RP
- retroperitoneal
- SC
- subcutaneous
This work was supported by the UTE Project CIMA and the Spanish Science and Innovation Ministry (MICINN) under grant DPI2009-14115-C03-03 and by Línea Especial (LE/97) from the University of Navarra. Arrate Muñoz-Barrutia holds a Ramón y Cajal fellowship of MICINN. Noemí Boqué holds a fellowship from Asociación de Amigos de la Universidad de Navarra.
REFERENCES
- 1.DiGirolamo M., Fine J. B., Tagra K., Rossmanith R. 1998. Qualitative regional differences in adipose tissue growth and cellularity in male Wistar rats fed ad libitum. Am. J. Physiol. 274: R1460–1467 [DOI] [PubMed] [Google Scholar]
- 2.Ong K. K.2010. Early determinants of obesity. In Adipose Tissue Development: From Animal Models to Clinical Conditions. Endocrine Development .Vol. 19. C. Lévy-Marchal and L. Pénicaud, editors. Karger, Basel. 53–62.
- 3.Galinier A., Carriere A., Fernandez Y., Carpene C., Andre M., Caspar-Bauguil S. 2006. Adipose tissue proadipogenic redox changes in obesity. J. Biol. Chem. 281: 12682–12687 [DOI] [PubMed] [Google Scholar]
- 4.Guilherme A., Virbasius J. V., Puri V., Czech M. P. 2008. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 9: 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Björnheden T., Jakubowicz B., Levin M., Odén B., Edén S., Sjöström L., Lönn M. 2004. Computerized determination of adipocyte size. Obes. Res. 12: 95–105 [DOI] [PubMed] [Google Scholar]
- 6.Tchoukalova Y. D., Harteneck D. A., Karwoski R. A., Tarara J., Jensen M. D. 2003. A quick, reliable, and automated method for fat cell sizing. J. Lipid Res. 44: 1795–1801 [DOI] [PubMed] [Google Scholar]
- 7.Di Girolamo M., Mendlinger S., Fertig J. W. 1971. A simple method to determine fat cell size and number in four mammalian species. Am. J. Physiol. 221: 850–858 [DOI] [PubMed] [Google Scholar]
- 8.Lemonnier D. 1972. Effect of age, sex, and site on the cellularity of the adipose tissue in mice and rats rendered obese by a high-fat diet. J. Clin. Invest. 51: 2907–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H. C., Farese R. V. 2002. Determination of adipocyte size by computer image analysis. J. Lipid Res. 43: 986–989 [PubMed] [Google Scholar]
- 10.Casteilla L., Penicaud L., Cousin B., Denis C.2001. Choosing an adipose tissue depot for sampling. Factors in selection and depot specificity. In Adipose Tissue Protocols (Methods in Molecular Biology). Vol 155. G. Ailhaud, editor. Springer, New York. 1–19.
- 11.Boqué N., Campión J., Paternain I., García-Díaz D. F., Galarraga M., Portillo M. P., Milagro F. I., Ortiz de Solórzano C., Martínez J. A. 2009. Influence of dietary macronutrient composition on adiposity and cellularity of different fat depots in Wistar rats. J. Physiol. Biochem. 65: 387–395 [DOI] [PubMed] [Google Scholar]
- 12.Otsu N. 1978. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9: 62–66 [Google Scholar]
- 13.Sun T., Neuvo Y. 1994. Detail-preserving median based filters in image processing. Pattern Recognition Letters. 15: 341–347 [Google Scholar]
- 14.Vincent L., Soille P. 1991. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE PAMI. 13: 583–598 [Google Scholar]
- 15.Malpica N., Ortiz de Solórzano C., Vaquero J. J., Santos A., Vallcorba I., García Sagredo J. M., del Pozo F. 1997. Applying watershed algorithms to the segmentation of clustered nuclei. Cytometry. 28: 289–297 [DOI] [PubMed] [Google Scholar]
- 16.Bourgeois F., Alexiu A., Lemonnier D. 1983. Dietary-induced obesity: effect of dietary fats on adipose tissue cellularity in mice. Br. J. Nutr. 49: 17–26 [DOI] [PubMed] [Google Scholar]
- 17.Abramoff M. D., Magalhaes P. J., Ram S. J. 2004. Image processing with ImageJ. Biophotonics International. 11: 36–42 [Google Scholar]
- 18.Luengo Hendriks C. L., Van Vliet L. J., Rieger B., Van Ginkel M.1999. DIPimage: a scientific image processing toolbox for MATLAB. Delft University of Technology, Delft, The Netherlands.
- 19.Campión J., Martinez J. A. 2004. Ketoconazole, an antifungal agent, protects against adiposity induced by a cafeteria diet. Horm. Metab. Res. 36: 485–491 [DOI] [PubMed] [Google Scholar]
- 20.Guo W., Bigornia S., Leizerman I., Xie W., McDonnell M., Clemente K., Pirtskhalava T., Kirkland J. L., Gokce N., Corkey B. E., et al. 2007. New scanning electron microscopic method for determination of adipocyte size in humans and mice. Obesity (Silver Spring). 15: 1657–1665 [DOI] [PubMed] [Google Scholar]