Abstract
Intestinal apolipoprotein B (apoB) mRNA undergoes C-to-U editing, mediated by the catalytic deaminase apobec-1, which results in translation of apoB48. Apobec1−/− mice produce only apoB100 and secrete larger chylomicron particles than those observed in wild-type (WT) mice. Here we show that transgenic rescue of intestinal apobec-1 expression (Apobec1Int/O) restores C-to-U RNA editing of apoB mRNA in vivo, including the canonical site at position 6666 and also at approximately 20 other newly identified downstream sites present in WT mice. The small intestine of Apobec1Int/O mice produces only apoB48, and the liver produces only apoB100. Serum chylomicron particles were smaller in Apobec1Int/O mice compared with those from Apobec1−/− mice, and the predominant fraction of serum apoB48 in Apobec1Int/O mice migrated in lipoproteins smaller than chylomicrons, even when these mice were fed a high-fat diet. Because apoB48 arises exclusively from the intestine in Apobec1Int/O mice and intestinal apoB48 synthesis and secretion rates were comparable to WT mice, we were able to infer the major sites of origin of serum apoB48 in WT mice. Our findings imply that less than 25% of serum apoB48 in WT mice arises from the intestine, with the majority originating from the liver.
Keywords: RNA binding, cytidine deaminase, hyperediting, apoB48, lipoprotein assembly
The continued epidemic of obesity has focused attention on the metabolic pathways and key players that regulate dietary lipid absorption from the mammalian small intestine. Lipoprotein biogenesis and bulk lipid transport from enterocytes of the small intestine is coordinated through interactions between the structural protein apolipoprotein B (apoB) and a resident endoplasmic reticulum (ER) chaperone, microsomal triglyceride transfer protein (Mttp), that together facilitate complex lipid delivery into the secretory pathway and orchestrate chylomicron assembly and secretion (reviewed in Refs. 1, 2).
Adult mammalian enterocytes synthesize and secrete a truncated apoB isoform, apoB48, produced as a result of posttranscriptional cytidine-to-uridine (C-to-U) RNA editing of the nascent nuclear apoB transcript, via a heteromeric enzymatic complex that contains apobec-1 (the catalytic deaminase) (3) and its obligate RNA binding subunit, apobec-1 complementation factor (ACF) (4, 5). There is a tissue-specific pattern of apoB isoform distribution, in which tissues that express apobec-1 (such as the intestine) produce apoB48, whereas tissues that express apoB but not apobec-1 (such as the human liver) produce only apoB100 (6). Mouse and rat liver express apobec-1 and, as a result, synthesize and secrete both apoB48 and apoB100, but there is wide variation in the extent of C-to-U editing from tissues of other mammalian species (7). For all practical purposes, however, adult murine and human small intestine secrete virtually only apoB48 (8).
Apobec1−/− mice are unable to edit apoB RNA in any tissue and, as a result, synthesize and secrete lipoproteins from both the liver and small intestine that contain exclusively apoB100 (9, 10). Nascent intestinal lipoproteins isolated from Apobec1−/− mice were larger than those from wild-type mice, and as inferred from lymphatic transport studies, Apobec1−/− mice secrete fewer triglyceride-rich particles (11). These findings imply a subtle functional advantage to intestinal apoB48 (versus apoB100) in terms of accommodating efficient lipid transport by optimizing chylomicron assembly (11).
However, several features of mammalian intestinal apoB RNA editing and the functional significance of apobec-1 remain incompletely understood. Earlier studies indicated that transgenic overexpression of apobec-1 caused C-to-U hyperediting at multiple sites (including the canonical nucleotide) in hepatic apoB RNA both in vivo (12) and in hepatoma cell lines (13). These findings raised questions of how C-to-U RNA editing at the canonical site is regulated physiologically and whether hyperediting might be demonstrated upon apobec-1 overexpression in nonhepatic tissues (14, 15). In addition, as alluded to above, the importance of intestinal apoB48 and its contribution to serum apoB48 levels in the mouse has been difficult to discern, at least in part because mouse liver secretes both isoforms (7). Here we use a strategy of transgenic rescue to reveal unanticipated sites of C-to-U editing in intestinal apoB RNA and to characterize apoB-containing lipoproteins in mice expressing only apoB48 in the small intestine.
METHODS
Generation of transgenic animals
The coding region of human apobec-1 was cloned into pGEM-AclI-villin-EGFP-KpnI-BGHpolyA-AclI vector (a gift from Dr. J. Turner, University of Chicago). The 711 bp coding region was inserted into the unique KpnI site downstream of EGFP motif, resulting in the expression of a N-terminal EGFP-tagged human Apobec-1. The expression of the transgene was under the control of a 9 kb villin promoter (16). This construct (Fig. 1A) was microinjected into fertilized eggs from C57BL/6 × CBA mice. Apobec-1 transgenic mice were then mated with C57BL/6 Apobec1−/− mice to generate lines of Apobec-1Int/O animals expressing Apobec-1 only in the intestine.
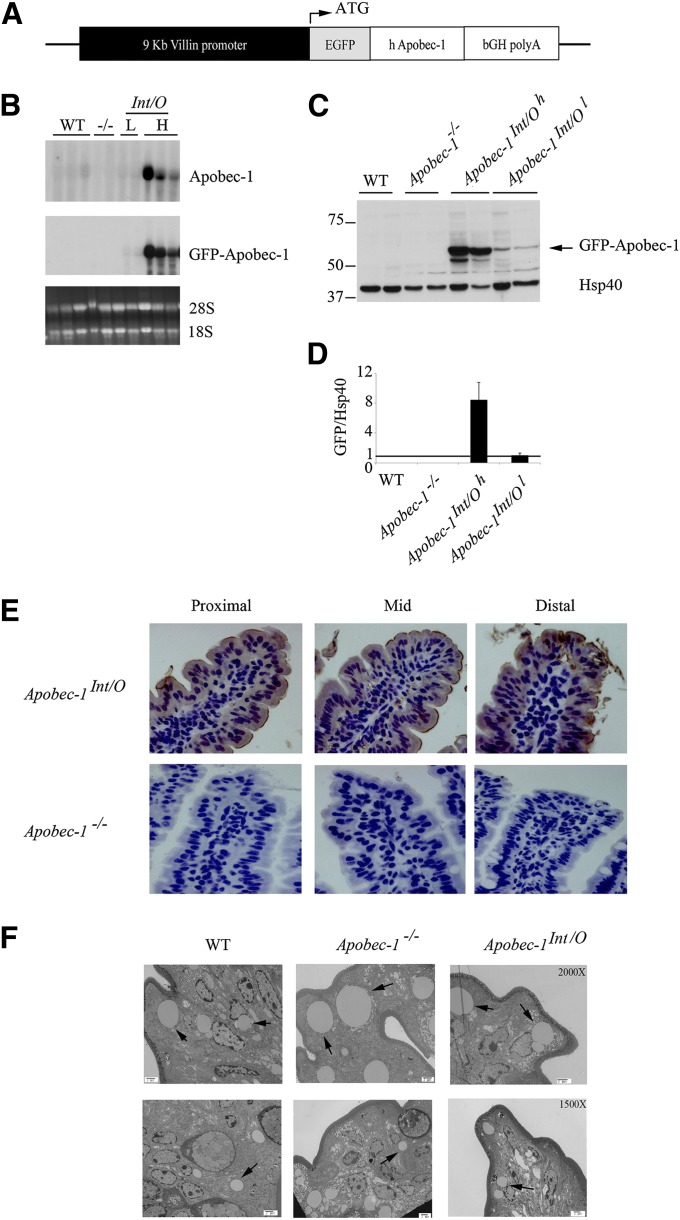
Fig. 1.
Generation of intestine-only Apobec-1 (Apobec-1Int/O) transgenic mice. A. Schematic of transgenic construct. Human apobec-1 was cloned downstream of the coding sequence of green fluorescent protein (EGFP) and expressed as a GFP fusion. The expression of the transgene is under control of the Villin promoter (black box). B. Northern blot analysis. Total RNA was extracted from proximal scraped mucosa of individual WT, Apobec-1−/−, Apobec-1Int/O low-expressor (L), and Apobec-1Int/O high-expressor (H) mice was resolved by electrophoresis and hybridized with either a 445-nt 32P-labeled DNA probe recognizing both murine and human apobec-1 or a 445 nt 32P-labeled DNA probe recognizing the GFP coding sequence. C. Western blot analysis of transgene expression. Equal amounts of protein extracted from scraped proximal mucosa processed from individual animals were resolved by SDS-PAGE and probed with anti-GFP antibody and Hsp40 for loading control. D. Histogram representing protein expression of GFP-Apobec-1 relative to Hsp40. The high-expressor line (Apobec-1Int/Oh) shows ∼8-fold higher level of transgene expression compared with the low-expressor (Apobec-1Int/Ol) line. E. Immunohistochemical analysis. Proximal, mid and distal sections of the small intestine from Apobec-1Int/O and Apobec-1−/− mice were stained with anti-GFP antibody. Note that transgene distributes between nucleus and cytoplasm of epithelial cells. F. Electron microscopy of small intestines harvested from chow-fed WT, Apobec-1−/− and Apobec-1Int/O 4 h after lipid gavage. 10-15 pictures were analyzed per mouse using 3 mice per genotype. The arrows indicate lipid droplets. Note that there is no significant difference in the size of lipid droplets between the 3 genotypes.
Northern blot analysis and semiquantitative PCR
Intestinal segments were flushed and scraped mucosa was flash-frozen in liquid nitrogen for total RNA extraction. Total RNA (20 µg) prepared in 1× MOPS, 6.7% formaldehyde, 72% formamide, and 40 μg/ml ethidium bromide was subjected to denaturing formaldehyde/1.2% agarose electrophoresis and transferred to Imobilon-NY+ membrane (Millipore, Billerica, MA). Membranes were prehybridized and hybridized in ExpressHyb hybridization solution (Clontech Laboratories, Mountain View, CA) at 70°C when probing for apobec-1 or 76°C when probing with GFP. Apobec-1 and GFP probes (445 nt) generated by PCR were labeled by random priming using [32P]dATP and Klenow (NEB Biolabs, Ipswich, MA). Apobec-1 probe was generated using the following primers: Fwd H/M 34: 5′-CCCACTCTGAGGAGAAG-3′ and Rev H/M 479: 5′-GGTAGTTGACGAAATTCCTCCAGCAG-3′. Washes were performed at room temperature for 40 min in 2× SSC, 0.05% SDS, followed by two washes at 50°C in 0.1× SSC, 0.1% SDS. Membranes were exposed to PhosphorImager SI (Molecular Dynamics, Sunnyvale, CA). Integrity and loading were monitored by visualization of ribosomal subunits. For semiquantitative evaluation of human Apobec-1 RNA expression, total RNA from wild-type, Apobec-1−/−, Apobec-1 Int/Ohigh-expressor (H), and Apobec-1 Int/Olow-expressor (L) animals was treated with DNase (Ambion, Foster City, CA) and used to synthesize cDNA. A 220-nt motif targeting exon 6 of Apobec-1 (supplementary Fig. I) was amplified using the following primers: Fwd H/M 64 Apobec-1: 5′-GAGTTTGAC/AGTCTTCTA/TTGACCC-3′; Rev H/M 284 Apobec-1: 5′-TCCCC/AGCAGGGACTCCAGGAC-3′. Semiquantitative RT-PCR was optimized (supplementary Fig. II) and performed as follows: 94°C 2 min, 29 cycles [95°C 30 s, 55°C 45 s, 72°C 1 min], 72°C 10 min. PCR products were resolved on a 1% agarose gel and quantitated using Kodak/Image Digital Science and Carestream Molecular Imaging System software. Apobec-1 PCR products were normalized to GAPDH. Quantitative PCR was performed using individual samples (3–4 per genotype). For each sample, reactions were performed in triplicate on an ABI Prism 7000 sequence detection system using SYBR GreenER PCR Master Mix (Invitrogen, Grand Island, NY) and primers designed by Primer Express software (Applied Biosystems). Murine Apobec-1-specific primers were as follows: Fwd: 5′-TGTTTATTTACATAGCACGGCTTTATC-3′ and Rev: 5′-TGCTAATAAGGTCCCTGAGTCCTT-3′. Degenerate human Apobec-1 primers: H/M Fwd: 5′-AGAATC/TGAA/GCCCT/CG/AG/CGAGTTT-3′ and H/M Rev: 5′-GATC/TTCA/GTAC/GAGCAGACAGGT/CCTCTT-3′. RNA levels are expressed as fold change compared with WT intestinal mRNA, normalized to GAPDH RNA.
Protein extraction, Western blot, and histologic analysis
Scraped mucosa was homogenized in 0.5 ml of tissue lysis buffer containing 20 mM Tris (pH 8); 0.15 M NaCl; 2 mM EDTA; 1 mM sodium vanadate; 0.1 M sodium fluoride; 50 mM βeta-glycerophosphate; 5% glycerol, 2× protease inhibitor (Roche Applied Science, Indianapolis, IN); 1% Triton; and 0.1% SDS. Aliquots of homogenate (100 µg protein) were resolved by 10% SDS-PAGE, transferred to PVDF membrane, and probed with anti-GFP antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Equal loading was verified using anti-Hsp40 antibody (Assay Designs, Ann Arbor, MI). For detection of isoforms of apoB, homogenates were resolved through a 4–15% gradient SDS-PAGE and probed with rabbit anti-murine apoB IgG using antisera previously characterized in our laboratory (17). Histological analysis was conducted on formalin-fixed tissues (proximal, mid, and distal intestine) in which 4 μm sections were stained with anti-GFP antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
ApoB RNA editing analysis by primer extension and sequencing
RNA was isolated from scraped mucosa using TRIzol (Invitrogen, Grand Island, NY). DNase-treated RNA (1 µg) was used for primer extension analysis of apoB RNA editing as previously detailed (17). Sequence analysis of apoB RNA hyperediting was conducted on a 738-nt region spanning nucleotides 6508 to 7246, amplified by RT-PCR using total RNA isolated from scraped mucosa of WT, Apobec-1−/−, and Apobec-1Int/O mice. PCR products were purified and subcloned into pPCR-Script SK(+) plasmid (Agilent Technologies/Stratagene, La Jolla, CA). Twenty individual clones were sequenced using Big Dye terminator mix. Sequences were aligned with SerialCloner 3-1 software.
Enterocyte isolation and pulse chase analysis of apoB secretion
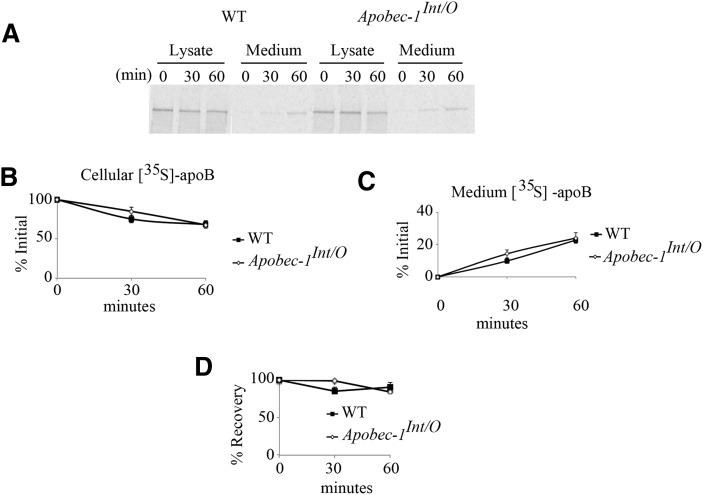
Enterocytes were isolated using EDTA chelation from four individual mice per genotype and used for pulse-chase experiments as described (18). In brief, freshly isolated enterocytes were pulse labeled for 30 min with 250 µCi [35S]-protein labeling mix (Cat# NEG072, PerkinElmer, Boston, MA) and chased for 60 min in DMEM supplemented with 10 mM methionine and 5 mM cysteine. A mixed micellar lipid solution (0.4 mM sodium taurocholate, 0.54 mM sodium taurodeoxycholate, 0.3 mM phosphatidycholine, 0.45 mM oleic acid, 0.26 mM monoolein) was added to both pulse and chase medium for all the experiments (18). At the end of chase, medium and cells were collected, and aliquots of lysate and medium were immunoprecipitated with rabbit anti-mouse apoB IgG. Quantitation of [35S]incorporation into apoB48 was conducted using 4–15% SDS-PAGE gel separation of the immunoprecipitation reactions, which were then quantified using PhosphorImager analysis (Molecular Dynamics, Sunnyvale, CA).
Lipoprotein fractionation by fast-pressure liquid chromatography
Animals were fed chow or a Western diet (Harlan Teklad #TD 88137, Madison, WI) as indicated for 10–12 weeks before study. After an overnight fast, animals were weighed and injected intravenously with Pluronic F127 (19), followed 30 min later by an intragastric bolus of 0.5 ml of a lipid mixture containing 100 µl corn oil and 400 µl 20% intralipid. Blood was collected before Pluronic F127 injection (time 0) and 4 h after lipid gavage. Serum was recovered by centrifugation at 4,000 rpm for 20 min at 4°C. Pooled serum (100 µl) was analyzed by fast-pressure liquid chromatography (FPLC) using two Superose 6 columns connected in series (GE Healthcare Life Sciences, Pittsburgh, PA). Each FPLC run yielded 56 fractions of 500 µl each. Aliquots of each individual fraction (30 µl) were resolved by 4–15% SDS-PAGE, and the distribution of apoB, apoA1, apoA4, and apoE was determined by Western blot. Cholesterol and triglyceride content in each fraction was determined enzymatically using Cholesterol E and L-Type Triglyceride M kits (Wako Chemicals, Richmond, VA).
Electron microscopy
Pooled serum (200 µl) collected 4 h after lipid gavage was layered under 600 µl 0.15 M NaCl and centrifuged at 50,000 rpm for 30 min in the MLA-130 rotor in a tabletop ultracentrifuge (Beckman Instruments). Chylomicrons were collected and submitted to negative staining electron microscopy, and then visualized using a Zeiss 902 electron microscope. At the time of sacrifice (4 h post-lipid gavage), 2 mm segments of the jejunum were flushed with PBS followed by immersion in fixative solution containing 3% Glutaradehyde (EM grade); 1% formaldehyde (EM grade) in 0.1 M sodium cacodylate (pH 7.4); 2 mM Cacl2; and 2 mM MgCl2. Specimens were kept at 4°C in 1.5 ml fixative, stained in 2% aqueous uranyl acetate, and viewed on a Hitachi H-600 electron microscope (Hitachi High Technologies, Schaumburg, IL). Three animals per genotype were examined to obtain representative images.
Statistical comparisons
All data are shown as mean ± SE unless otherwise noted. Statistical comparisons were made using GraphPad Prism 4.0 (GraphPad, San Diego, CA) with one-way ANOVA for multiple group comparisons and t-testing as a posthoc test for comparing different pairs. A value of P < 0.05 was considered significant.
RESULTS
Generation of intestine-only Apobec-1 (Apobec-1Int/O) transgenic mice
Three founder transgenic lines were generated using the construct shown in Fig. 1A. For the purposes of this report, we focused on two lines in which human apobec-1 was expressed homogeneously in the small intestine at either a relatively high or low level (defined below) when compared with endogenous murine apobec-1. The apobec-1 intestinal transgenic founders were then crossed into the Apobec1−/− line to generate lines of Apobec1Int/O mice. Northern blot analysis revealed a strong signal only with the high-expressor line (Fig. 1B). The relative expression levels of transgenic apobec-1 mRNA were determined in comparison to endogenous murine apobec-1 by semiquantitative PCR targeting a conserved region within murine exon 6 (supplementary Figs. I and II). These data indicate a range of ∼5-fold overexpression of the high-overexpressor line and ∼2-fold overexpression with the low-overexpressor line, both compared with WT mice (supplementary Fig. IIC). Quantitative RT-PCR using species-specific primers suggested a much greater range of overexpression within the lines (supplementary Fig. IID). However, relative expression levels of the GFP-apobec-1 fusion protein in extracts from these lines of mice (Fig. 1C) indicated ∼8-fold higher levels in the high-expressor line compared with the low-expressor line (Fig. 1D), which was more in line with the semiquantitative RT-PCR data. Transgenic rescue in these lines was confirmed by immunohistochemical localization of the fusion protein, which indicated homogeneous expression in villus enterocytes in both nuclear and cytoplasmic compartments (Fig. 1E). Electron micrographs of jejunal enterocytes from chow-fed mice revealed comparable morphology between WT, Apobec1−/−, and Apobec1Int/O mice without changes in lipid droplet accumulation (Fig. 1F). Other parameters of intestinal lipid metabolism are detailed in Table 1 and discussed later.
TABLE 1.
Basic physiological parameters in WT and Apobec-1Int/O mice fed chow or Western diet
| Parameter | Chow | Western Diet |
| Body weight (g) | ||
| WT | 24.4 ± 0.6 | 24.9 ± 1.9 |
| Apobec-1Int/O | 21.8 ± 0.8 | 29.5 ± 1.6 |
| Serum cholesterol (mg/dl) | ||
| WT | 79 ± 2 | 166 ± 6 |
| Apobec-1Int/O | 79 ± 7 | 221 ± 19 |
| Serum triglycerides (mg/dl) | ||
| WT | 65 ± 9 | 8.1 ± 1 |
| Apobec-1Int/O | 52 ± 7 | 11 ± 4 |
| Intestinal cholesterol (μg/mg protein) | ||
| WT | 64 ± 19 | 45 ± 6 |
| Apobec-1Int/O | 52 ± 9 | 68 ± 12 |
| Intestinal triglycerides (μg/mg protein) | ||
| WT | 118 ± 30 | 526 ± 104 |
| Apobec-1Int/O | 141 ± 47 | 426 ± 121 |
n= 3 WT and 6 Apobec-1Int/O on chow diet. n = 5 WT and 6 Apobec-1Int/O on Western diet. Data represent mean ± SE. No significant difference was observed between genotypes for any parameter.
ApoB C-to-U RNA editing in the intestine of Apobec-1−/− mice
Intestinal RNA was examined by primer extension assay (17) to determine the extent of C-to-U editing of endogenous murine apoB RNA at the canonical site. WT mice showed greater than 90% C-to-U editing, whereas Apobec1−/− mice demonstrated no RNA editing, as expected (Fig. 2A, B). Transgenic rescue of apobec-1 in both lines of Apobec1Int/O mice also revealed greater than 90% C-to-U editing at the canonical site, indicating that the transgene is fully functional with regard to its physiological target (Fig. 2A, B).
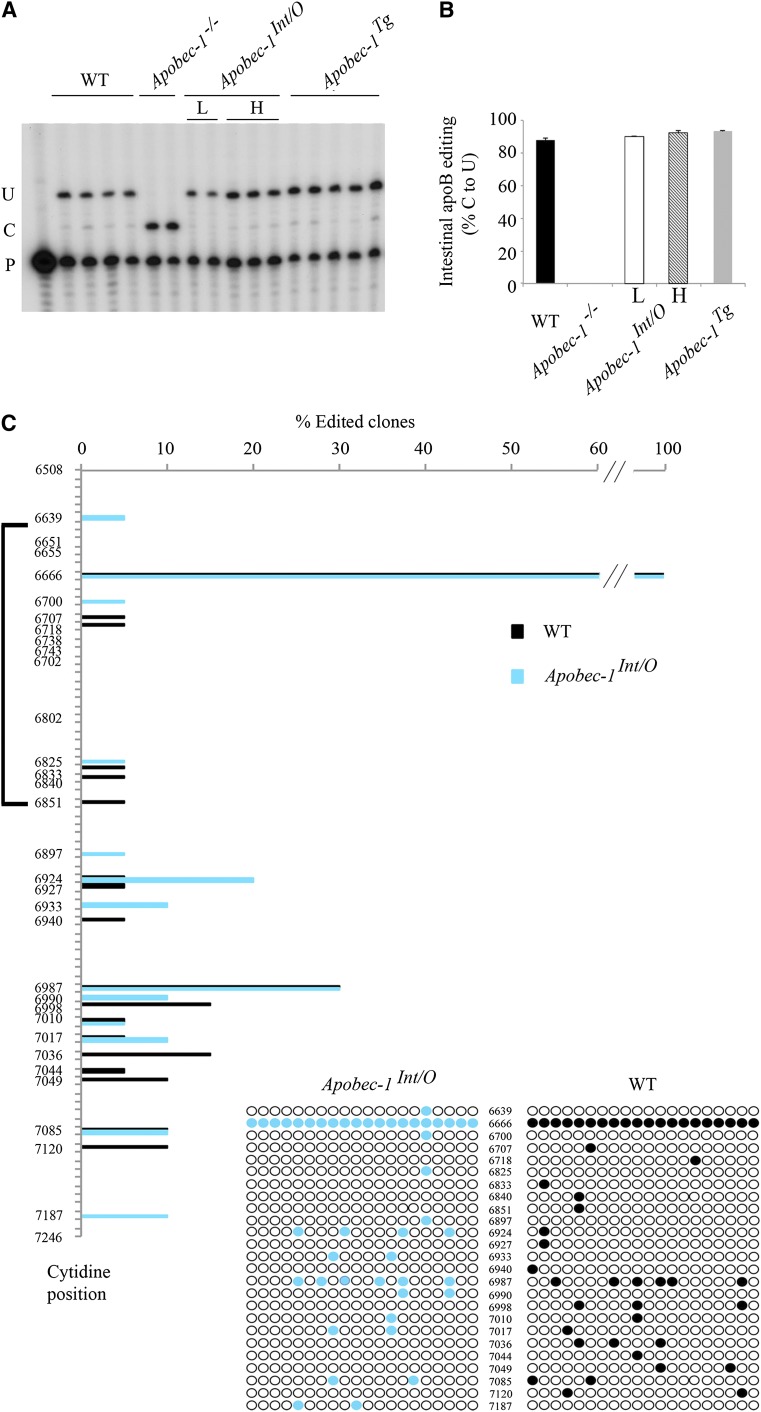
Fig. 2.
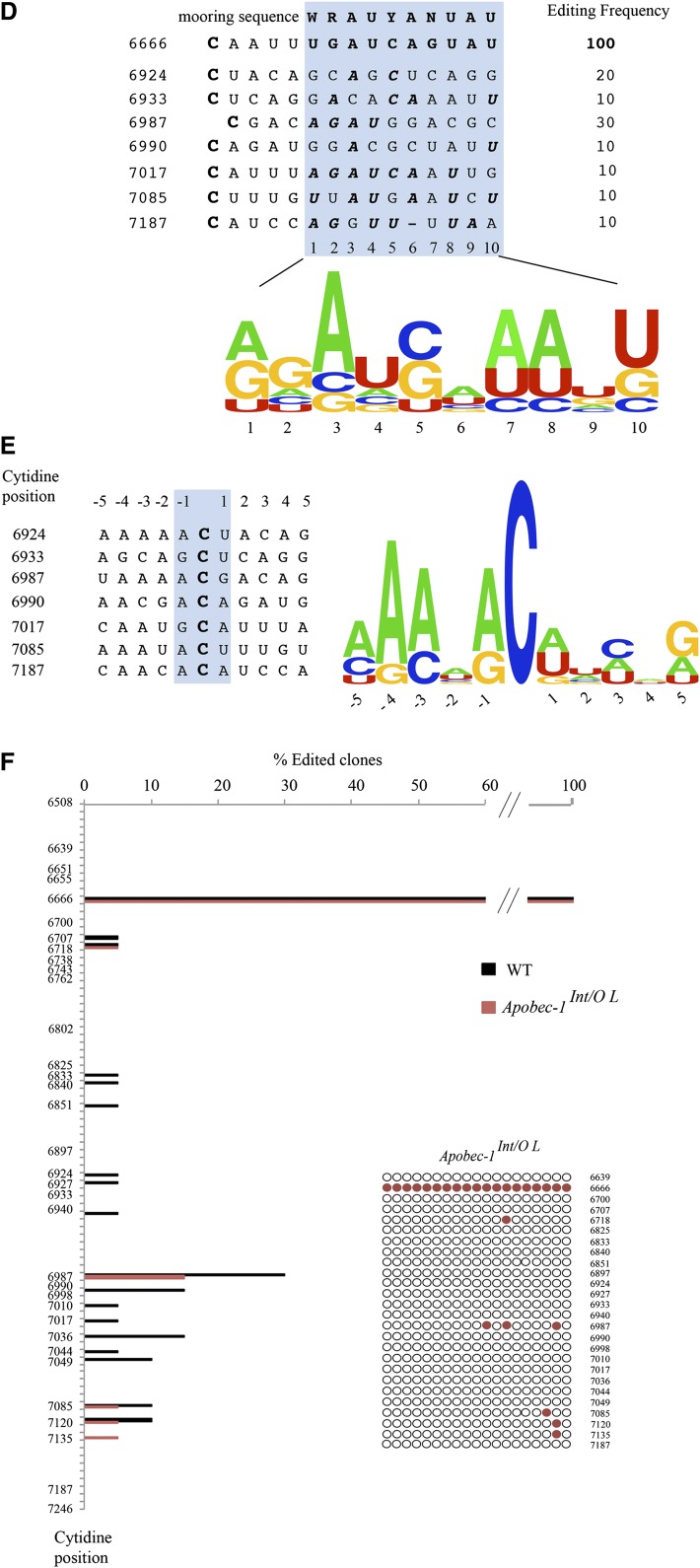
Transgenic rescue of apobec-1 in the intestine of Apobec-1−/− mice restores ApoB C-to-U RNA editing. A: Endogenous apoB mRNA editing was determined by primer extension. The relative mobility of the unedited (C) and edited (U) products is indicated on the left. B: Bar graph representation of percentage C-to-U editing for each group as mean ± SE. (n = 3–5). C, D: Hyperediting of apoB mRNA analyzed by DNA sequencing. A 738 bp fragment (6508–7246 nt) of apoB mRNA overlapping the C6666 canonical editing site was amplified by RT-PCR subcloned into pPCR-Script vector and sequenced. Twenty clones per genotype (from two mice per genotype) were analyzed. Targeted cytidines identified in Apobec-1Int/O clones are indicated by blue circles, aligned with the nucleotide position. Edited cytidines identified in WT clones are indicated with black circles, aligned with the nucleotide position. The bracket on the left side of the vertical panel indicates the region of apoB mRNA found by Innerarity and colleagues to be promiscuously edited in hepatic apobec-1 transgenic mice (20). D: Alignment of a 14 nt sequence downstream of C-to-U editing sites with ≥ 10% editing identified in RNA from Apobec-1Int/Oh mice [see (C)]. The sequences were compared with the apoB RNA canonical mooring sequence shown on the top. W, A/U; R, A/G; Y, U/C. The edited C is indicated in bold font at the beginning of each sequence. The nucleotides matching the mooring sequence are indicated in bold italic letters. Below is shown the frequency plot of nucleotides in the 10 nt motif containing matching residue aligned with the consensus mooring sequence (blue box). E: Alignment and frequency plot of nearest neighbor nucleotides flanking each C-to-U RNA editing site. Positions are indicated relative to the targeted cytidine. Frequency plots were created using Weblogo software (Berkeley, CA). F: Hyperediting of intestinal apoB RNA isolated from Apobec-1Int/O low-expressor mice and analyzed by sequencing as in (C). Edited cytidines identified in 20 independent clones are indicated by red circles.
We next asked whether transgenic rescue of apobec-1 was associated with changes in cytidine deamination of other targets in apoB RNA, as prior studies in transgenic mouse and rabbit liver had indicated that overexpression of apobec-1 induced promiscuous hyperediting (12, 20). We undertook Sanger sequencing of cDNA clones from a 738 nt fragment spanning nt 6508–7246, including the region from nt 6643–6851 examined in that earlier study (20). Our findings demonstrate that C-to-U RNA editing is detectable in many sites 3′ to the canonical target at position 6666 and, importantly, that these RNA modifications were found in both WT and Apobec1Int/O mice (high expressor) (Fig. 2C). The data show that the extent of C-to-U editing varies across the sites, with a maximum of 30% editing at 6987 and generally lower levels (∼10%) at other sites (Fig. 2C). The findings reveal sites at which C-to-U RNA editing was found only in the Apobec1Int/O mice (e.g., 6933, 6990, and 7187) and also sites in which C-to-U RNA editing was only found in the WT intestine (6998, 7036, and 7120). However, for the most part, there was concordance in the patterns observed between WT and Apobec1Int/O mice. The observed pattern of C-to-U RNA editing of these additional sites does not appear to be processive (5′→3′ or 3′→5′) based on the finding that none of the sites appear linked to one another. One additional editing site (1/20 clones) was identified (6639) 5′ to the canonical base, changing a CUC (Leu) to UUC (Phe), but all the other changes within the 738 nucleotides examined were found 3′ to the canonical site, which was quantitatively edited. Whether additional sites for C-to-U RNA editing exist in exon 26 upstream of nucleotide 6508 or in other 5′ exons remains to be determined, but we will return to this question later.
The findings with regard to the sites of hyperediting of intestinal apoB RNA contrast with those demonstrated earlier for hepatic apoB RNA in liver-specific apobec-1 transgenic mice (20), in which a major cluster of cytidine residues at positions 6738-–6762 and another cluster at 6802–6812 were each observed to undergo C-to-U hyperediting. None of those sites were found to undergo C-to-U editing in mouse intestine in either WT or Apobec1Int/O mice (Fig. 2C). These findings strongly suggest that site selectivity of apoB RNA modifications is influenced in a tissue-specific fashion. We checked whether targeting of these additional sites was mooring sequence-dependent (13, 20). When compared with the canonical mooring sequence, we identified only one site (7017) with an almost perfect match with the consensus motif (Fig. 2D). However, editing efficiency at this site (∼10%) was similar to other sites (6933, 6990, 7085, and 7187) with more mismatches to the mooring sequence, suggesting that the presence of this motif is not a critical determinant of site selection (Fig. 2D). We also examined the nearest neighbor nucleotide preference surrounding editing sites with frequency higher than 10%. The nucleotide immediately upstream of the targeted cytidine (position −1) is an adenosine (70%) or guanosine (28%), compared with the immediate downstream nucleotide, adenosine (43%), uridine (43%), or guanosine (14%) (Fig. 2E). These findings contrast with the nearest neighbor preferences identified for liver hyperediting in which the upstream preference was for thymidine (70%) or adenosine (30%) (20). The findings together suggest that sequence-specific (contextual) elements interact with tissue-specific factors in the specificity of apobec-1 mediated C-to-U RNA editing of apoB.
We also examined these patterns of C-to-U RNA editing in the low-expressor line of Apobec1Int/O mice since earlier work indicated a dose response for hyperediting in apobec-1 transgenic liver (21, 22). Our findings demonstrate that low-expressor Apobec1Int/O mice exhibit reduced levels of hyperediting compared with both WT and the high-expressor Apobec1Int/O line, with the dominant location again at the 6987 site (Fig. 2F).
Taken together, the findings reveal an unanticipated array of targets for C-to-U RNA editing in WT murine intestine that is eliminated in Apobec1−/− mice (data not shown) and largely restored in a dose-dependent manner by transgenic rescue in Apobec1Int/O mice. For the purposes of the functional characterization (detailed below), we used Apobec1Int/O mice expressing high levels of the transgene.
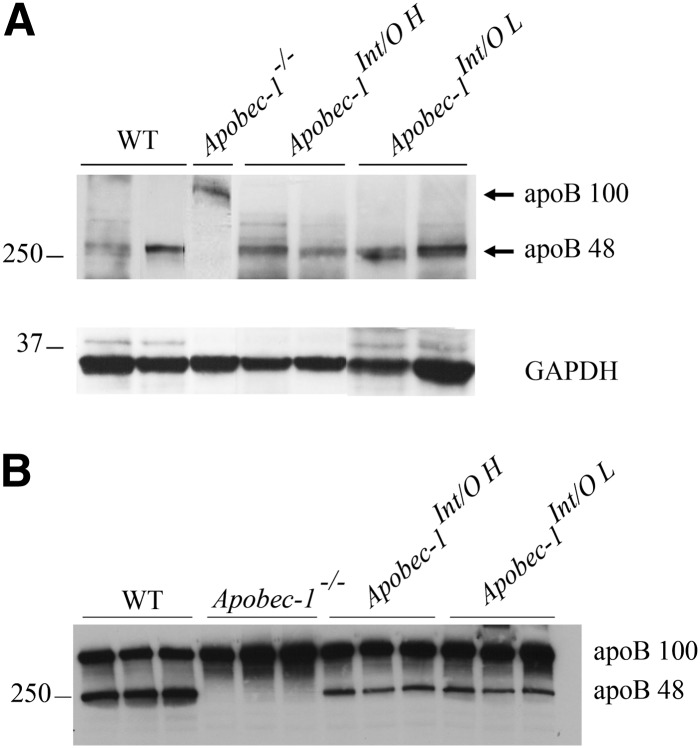
Intestinal apoB48 production in Apobec1Int/O mice
We examined apoB isoform expression in mice of the indicated genotypes, which revealed that apoB48 was the dominant intestinal isoform expressed in Apobec1Int/O mice and that there were no other truncated species detectable either in serum or intestinal mucosa (Fig. 3A, B). We next examined apoB48 synthesis and secretion in isolated enterocytes from WT and Apobec1Int/O mice, the findings demonstrating comparable synthesis and secretion rates with no differences in the overall recovery of apoB (Fig. 4A–D). These findings demonstrate that intestine-specific apobec-1 transgenic expression rescues the patterns of synthesis and secretion of intestinal apoB48 in Apobec1Int/O mice. In addition, there was no discernable difference between WT and Apobec1Int/O mice in terms of weight gain or lipid absorption (data not shown) or in terms of intestinal or fasting serum cholesterol or triglyceride concentrations either on a chow or Western diet (Table 1). These findings would be predicted based on the predominant pattern of intestinal apoB48 production in WT mice.
Fig. 3.
Intestinal and serum apoB48 protein expression is restored in Apobec-1Int/O mice. A: Protein from scraped proximal mucosa was resolved by 4–15% SDS-PAGE and probed with anti-apoB and anti-GAPDH antibody. B: Serum from chow-fed mice was separated by 4–15% SDS-PAGE and analyzed with anti-apoB antibody.
Fig. 4.
ApoB48 synthesis and secretion in Apobec-1Int/O and WT enterocytes. Enterocytes from WT and Apobec-1Int/O chow-fed mice were pulse labeled for 30 min and chased for 30–60 min. A–D: Lysates and media were collected at the indicated times. [35S]ApoB isoforms were immunoprecipitated from cell lysates and media, separated by SDS-PAGE, and quantitated by PhosphorImager. [35S]-ApoB48 is expressed as a percentage of the initial label incorporated at the beginning of the chase. Four separate experiments were performed for each genotype using enterocytes isolated from four individual animals. Data are represented as mean ± SE.
Serum apoB isoform and lipoprotein distribution in Apobec1Int/O mice
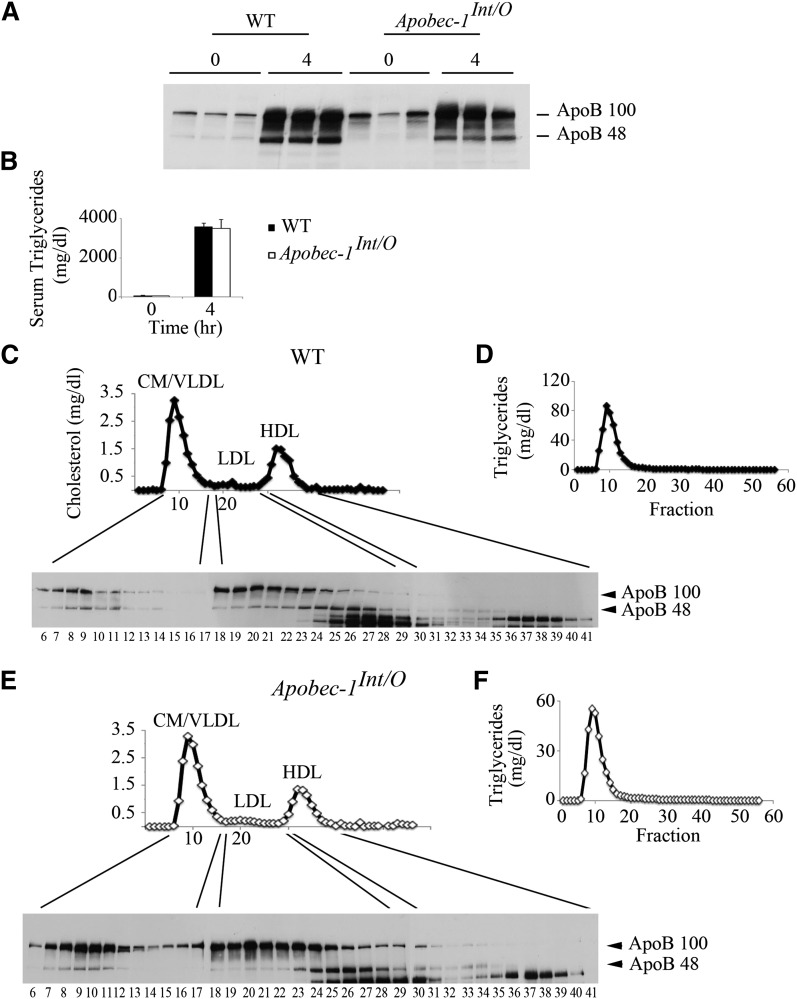
We next turned our attention to the distribution pattern of apoB isoforms in Apobec1Int/O mice, as these mice produce apoB48 exclusively in the intestine and apoB100 exclusively in the liver. Following Pluronic F127 administration, we found that the net (representing both hepatic plus intestinal) secretion rate of triglyceride-rich lipoproteins was comparable but with reduced abundance of apoB48 in Apobec1Int/O mice compared with WT 4 h following a lipid bolus (Fig. 5A, B; see also Fig. 3B). The distribution of isoforms across the lipoprotein profile revealed a subtle change in apoB48 distribution by genotype. ApoB48 fractionated into chylomicron/VLDL-sized particles in both WT and Apobec-1Int/O mice (Fig. 5C, E), but apoB48 distributed into a more homogeneous population of LDL particles in Apobec-1Int/O mice (Fig. 5E, fractions 24–28) compared with WT mice, where apoB48 appeared in broad population of LDL-sized particles (Fig. 5C, fractions 18–29). Triglyceride distribution was indistinguishable between the genotypes, almost exclusively within chylomicron and VLDL fractions, as expected (Fig. 5D, F).
Fig. 5.
Serum apoB48 is distributed in smaller LDL and HDL-size particles in Apobec-1Int/O mice. Following an overnight fast, chow-fed Apobec-1Int/O and WT mice were administered Pluronic F127 and received an intragastric bolus of lipid. Serum was collected before (time 0) and 4 h after gavage. A: Serum (2 µl) were separated by 4–15% SDS-PAGE and probed with apoB antibody. B: Serum triglyceride levels were evaluated enzymatically at indicated times. Data represent mean ± SE from three WT and six Apobec-1Int/O mice. C, E: Pooled serum from three WT and six Apobec-1Int/O animals collected 4 h after lipid gavage was fractionated by FPLC, and 56 (500 µl) fractions were collected. Aliquots of individual fractions were separated by SDS-PAGE and probed with apoB antibody. This is a representative of two separate experiments. Cholesterol (C, E) and triglycerides (D, F) content was enzymatically determined in individual fractions. The reactive bands migrating faster than apoB48 are nonspecific.
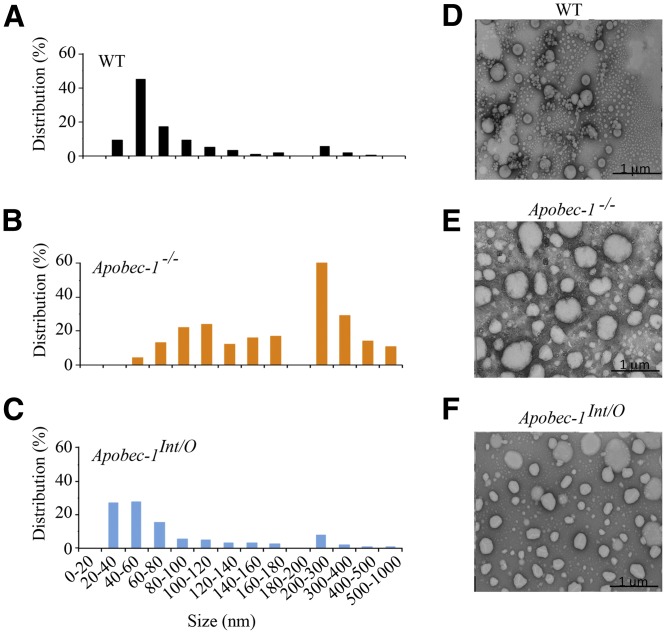
We next evaluated the size distribution of chylomicron and VLDL particles from the indicated genotypes using negative-stain electron microscopy, which revealed a shift to a smaller population of particles in Apobec1Int/O mice (Fig. 6), again suggesting that intestinal lipoprotein production is altered upon rescue of intestinal apoB48 production.
Fig. 6.
Electron microscopy of serum chylomicrons. Chylomicrons (density < 1.006) were isolated from pooled serum collected 4 h after Pluronic F127 injection and lipid gavage and submitted for negative staining electron microscopy. The histograms (A–C) representing the size distribution were generated by measuring 400 particles per sample. Representative images from three animals per genotype are shown (D, WT; E, Apobec-1−/−; F, Apobec-1Int/O).
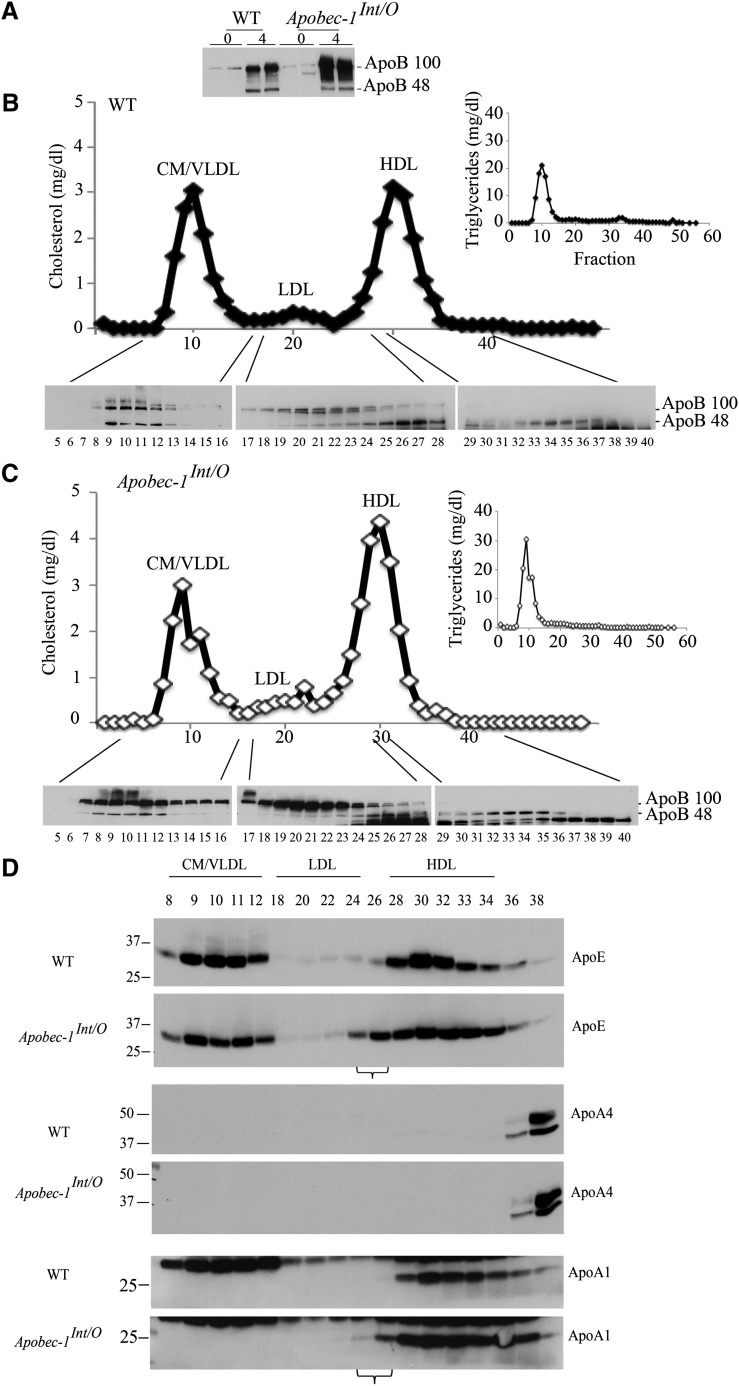
The findings to this point suggest that there is a shift in the profile of intestinal lipoproteins in Apobec1Int/O mice. We extended this characterization to mice fed a high-fat Western diet (see Table 1 for lipid data) in which the FPLC profile again revealed that only a fraction of apoB48 from Apobec1Int/O mice distributed into chylomicron/VLDL-sized particles (Fig. 7A–C). As noted above in chow-fed mice, triglycerides distributed virtually exclusively in chylomicron and VLDL fractions. We also examined the distribution of apoE and apoA1, which revealed comparable distribution in chylomicron and VLDL fractions of both genotypes (Fig. 7D). By contrast, apoE and apoA1 were found in the small LDL/large HDL-sized range in Apobec1Int/O mice (Fig. 7D, bracketed fractions), suggesting again a distinctive pattern of particle distribution in association with intestinal apoB48 in these mice.
Fig. 7.
Serum lipoprotein distribution of apoB48 in Western diet-fed Apobec-1Int/O and WT mice. Animals were fed a Western diet for 12 weeks and overnight fasted mice received Pluronic 127 followed by an intragastric lipid bolus. Animals were euthanized 4 h later. A: Serum was collected and 2 µl was separated by SDS-PAGE and analyzed with apoB antibody. B, C: Pooled serum from five WT and six Apobec-1Int/O mice were fractionated by FPLC, and individual fractions were resolved by SDS-PAGE. Distribution of apoB48 was determined by Western blot using anti-apoB antibody. Cholesterol and triglyceride content were determined enzymatically. Note that HDL-sized particles from Apobec-1Int/O mice are enriched in apoB48 compared with WT controls. D: Individual FPLC fractions were resolved by SDS-PAGE, and lipoprotein distribution of apoE, apoA4, and apoA1 was analyzed by Western blot. Note that small LDL-sized particles are enriched in apoE and apoAI in Apobec-1Int/O mice, as indicated by the horizontal brackets underlining fractions 24 and 26.
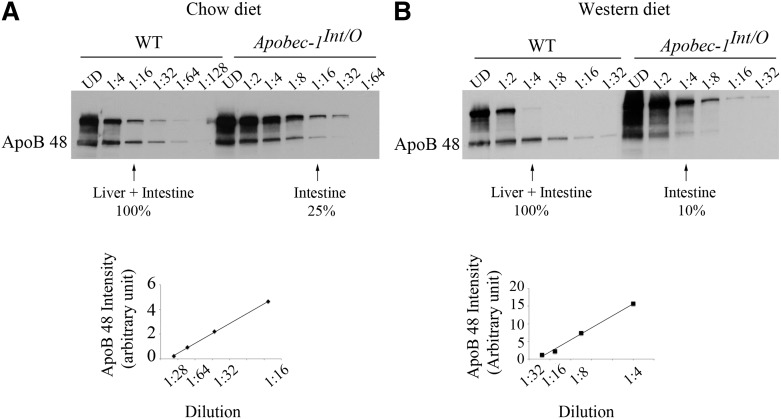
Finally, because apoB48 arises exclusively from the intestine of Apobec1Int/O mice and because intestinal apoB48 synthesis and secretion rates (as inferred from our in-vitro studies in isolated enterocytes; see Fig. 4) were comparable between the genotypes, we reasoned that it would be feasible to infer the contribution of intestinal apoB48 production to serum apoB48 levels in WT mice under conditions where clearance of intestinal and hepatic apoB48 particles was eliminated (i.e., following Pluronic F127 administration). Accordingly, we constructed a semiquantitative standard curve of serum apoB48 in both chow- (Fig. 8A) and Western diet-fed mice (Fig. 8B) using serial dilutions to obtain comparable apoB48 abundance under each dietary condition. The findings (Fig. 8A, B) imply that the intestine contributes ∼10–25% of total serum apoB48, with the majority of apoB48 production reflecting hepatic secretion.
Fig. 8.
Relative contribution of liver and intestine to serum apoB48. Pooled serum collected 4 h following lipid gavage from chow-fed (A) and Western diet-fed animals (B) were diluted 1:4 to 1:128 (A) and 1:2 to 1:32 in PBS (B). One microliter of diluted serum was resolved by 4–15% SDS-PAGE and probed using anti-apoB antibody. ApoB48 band intensity was quantitated using Kodak/Image Digital Science and the Carestream Molecular Imaging System software. Band intensities from WT apoB48 were used to create a standard curve. ApoB48 band intensity within the WT standard curve was used to evaluate the relative contribution of intestine to apoB48 production. As an example, in (A), the intensity of the band from Apobec-1Int/O serum diluted 1:16 represents 25% of the band intensity from WT serum of the same dilution.
DISCUSSION
The key findings from this study demonstrate that transgenic expression of human apobec-1 in the intestine of Apobec1Int/O mice rescues apoB RNA editing at the canonical site (6666), restores intestinal apoB48 production, and produces a shift in the spectrum of intestinal lipoproteins. The findings also reveal an unanticipated number of additional sites of C-to-U RNA editing of intestinal apoB, including a newly identified cluster of sites that were present not only in Apobec1Int/O mice in which apobec-1 was overexpressed but also in WT animals. These features together raise important implications for our understanding of the regulation of posttranscriptional C-to-U RNA editing and the functional importance of apoB RNA editing in the assembly and secretion of intestinal lipoproteins. Each of these features merits additional discussion.
Considerable effort has been devoted to understanding the cis-acting elements and trans-acting factors that regulate C-to-U RNA editing of apoB (12, 13, 20, 23). Historically, this work focused predominantly on C-to-U editing of synthetic apoB RNA templates flanking the canonical site and indicated both nearest neighbor preferences (23, 24) and also distinct structural context requirements (25, 26). These RNA elements, together with regulated abundance of apobec-1 and its auxiliary components, including ACF, were viewed as sufficient and necessary to constrain C-to-U editing of apoB to a physiological site at position 6666 (6). However, the finding that forced overexpression of apobec-1 induced hyperediting of apoB RNA at multiple sites suggested that these constraints could be overcome (13, 20). The current findings illustrate several important contrasts from those earlier studies. First, almost none of the sites identified as targets of hyperediting in apobec-1 transgenic mouse and in rabbit liver apoB RNA by Yamanaka et al. (20) were found to be edited in intestinal apoB RNA in Apobec1Int/O mice. This includes the 6802 site, which was identified earlier in human intestinal cDNA as an example of noncanonical C-to-U RNA editing (27). These findings demonstrate that target specificity for C-to-U RNA editing of apoB is regulated in a tissue-specific manner and that distinctive factors constrain and regulate the extent and efficiency of hepatic versus intestinal apoB RNA editing, particularly under conditions where apobec-1 expression may be altered. The second important feature is that noncanonical sites for C-to-U editing, in particular the cluster of targets between 6987 and 7187, were found in both WT and Apobec1Int/O mice albeit at lower efficiency (up to 30%) compared with the canonical site. These findings imply that C-to-U RNA editing of murine intestinal apoB is not confined to a single site, even under circumstances where apobec-1 is expressed at physiological levels. These findings contrast with earlier observations in murine liver, where none of the cDNA clones spanning nt 6643–6851 sequenced from endogenous (murine) or transgenic human apoB were edited (with the exception of 6806) (20). It will be of interest to extend those findings and include the 3′ cluster of C-to-U editing sites identified here in intestinal apoB cDNA. Those studies are currently in progress and will be reported elsewhere.
The predominant overlap in hyperediting site selection and efficiency noted in WT and the high-expressing Apobec1Int/O mice suggest that simply overexpressing human (versus murine) apobec-1 in murine intestine is unlikely to account for the findings. The observation that hyperediting of apoB RNA involved fewer noncanonical sites in the low-expressor line of Apobec1Int/O mice (again, largely overlapping sites observed in WT mice) further supports the conclusion that human and murine apobec-1 exhibit similar functional characteristics with regard to murine intestinal apoB RNA editing in vivo. These conclusions are broadly consistent with the findings from rabbit apobec-1 transgenic overexpression in murine liver and from in-vitro studies in human and rat hepatoma cells (20, 24). However, recent findings using more sensitive amplification methods to detect C-to-U RNA editing events have demonstrated that adenoviral expression of rat and human apobec-1 in hepatoma cell lines exhibits different specificity for introducing hypermutations, suggesting that tissue-specific cofactor interactions may be an important consideration in understanding how site selection is regulated (28).
It is also worth pointing out that virtually all the sites of C-to-U RNA editing were 3′ of the canonical site and that the additional RNA editing events described occur in the 3′ untranslated region (UTR) of the apoB transcript. Recent findings using deep sequencing of intestinal RNA (RNA seq) from WT mice found 32 new (i.e., non-apoB) targets of C-to-U editing, all of which were in the 3′ UTR and all of which were eliminated in the intestine of Apobec1−/− mice (29). These findings, taken together with the current results, indicate that there is a wider range of C-to-U editing targets than previously thought and that these targets reside within the 3′ UTR, raising questions regarding the functional impact of these modifications. It will be important to undertake a comprehensive analysis of the entire apoB transcript from mice of the three genotypes examined in the current study to determine whether there are additional sites of C-to-U editing beyond those identified in the region of 6508–7246. As alluded to above, this analysis will extend to regions both 5′ and 3′ of the regions examined in the current report, and those studies are currently in progress. However, RNA seq studies reported earlier indicated only a low level (up to 10%) of hyperediting of cytidine residues in apoB RNA, downstream and in close proximity to the canonical site (6676 and 6708) (29). It is important to emphasize that Western blots of intestinal mucosal extracts and serum from Apobec1Int/O mice each indicated only apoB48 and no other truncated apoB species, suggesting that there are no new functional translational termination codons created through alternative C-to-U RNA editing. That being said, it is possible that these additional RNA editing events within the 3′ UTR of apoB might influence other aspects of apoB mRNA biology, including transport and translational efficiency. Those possibilities await formal examination.
The current studies demonstrate that intestinal apoB48 synthesis and secretion are restored in Apobec1Int/O mice and reveal no obvious impact on the efficiency of lipid absorption. The findings indicate a subtle shift in apoB48 distribution that includes a population of smaller chylomicron particles (identified by electron microscopy) whose significance remains to be elucidated. In addition, the findings demonstrate that the distribution of apoB48 in Apobec1Int/O mice also includes LDL- and HDL-sized fractions. This observation raises the question of the functional relevance of apoB48 within these fractions. It is worth noting that earlier studies in apoB48-only versus apoB100-only mice demonstrated that cholesterol distributed into predominantly HDL-sized particles whereas TG distributed predominantly into chylomicron/VLDL-sized particles (30). However, those studies left open the relative contribution of intestinal versus hepatic apoB48 in the apoB48-only mice. The current findings demonstrate that, following a lipid bolus, virtually all the triglyceride in serum is transported in the apoB48-containing fractions of chylomicron/VLDL size in both WT and Apobec1Int/O mice. On the other hand, a large fraction of the apoB48 in Apobec1Int/O mice cofractionates with smaller particles, raising the possibility that these apoB48-containing particles in the LDL and HDL range are underlipidated, precursor lipoprotein particles. This possibility will require further experimental validation but is broadly consistent with the two-step hypothesis for triglyceride-rich lipoprotein assembly and secretion as inferred from studies in the liver (31). The lipid cargo of these smaller, apoB48-containing intestinal lipoproteins remains to be defined more completely but our findings (Figs. 5E and 7C) indicate that they contain cholesterol. The possibility that intestinal lipid transport utilizes different secretory pathways (i.e., chylomicron and HDL) is consistent with other work examining intestinal vitamin E (32) and cholesterol secretion (33).
The strategy for generating Apobec1Int/O mice also provided the opportunity to examine the contribution of intestinal apoB48 production to the overall serum apoB48 pool in WT mice. We used the assumption that, because apoB48 synthesis and secretion rates were similar, as inferred from studies in isolated enterocytes, the abundance of serum apoB48 in the respective genotypes would reflect intestinal secretion under circumstances in which apoB uptake was eliminated. The findings suggest that under conditions of either chow or high-fat Western diet feeding, the intestinal contribution of apoB48 in WT mice is only 10–25% of the total. In other words, under both dietary conditions, the majority of apoB48 in WT mice originates from the liver. Following intake of a high-fat Western diet, the contribution of intestinal apoB48 was lower than in chow- fed mice, but these findings should be interpreted with caution as hepatic apoB48 synthesis and secretion rates were not formally analyzed in these experiments. Nevertheless, the results suggest that the intestine accommodates chylomicron assembly and triglyceride mobilization through apoB48 production and secretion of particles in the size range of chylomicrons but in addition secretes a range of smaller particles containing apoB48 whose lipid transport function and metabolic implications are yet to be fully understood.
In summary, the findings from this study illustrate an unanticipated range of targets of C-to-U apoB RNA editing, findings that extend discussion regarding the diversity of DNA versus RNA differences (34–37). Further questions, both in relation to C-to-U RNA editing as a biological process and in relation to apoB48 assembly and secretion as a functional pathway for intestinal lipid export, are the focus of ongoing investigation.
Supplementary Material
Acknowledgments
The authors are very grateful to all members of the Davidson laboratory for their input and to Dr. Steven Farber, Carnegie Institution for Science, Baltimore, for helpful suggestions and comments.
Footnotes
Abbreviations:
- ACF
- apobec-1 complementation factor
- C-to-U
- cytidine-to-uridine
- FPLC
- fast-pressure liquid chromatography
- UTR
- untranslated region
- WT
- wild-type
This work was supported by National Institutes of Health Grants HL-38180, DK-56260, and DK-52574 to N.O.D. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org)contains supplementary data in the form of two figures.
REFERENCES
- 1.Chen Z., Davidson N.O.2006. Genetic regulation of intestinal lipid transport and metabolism. In Physiology of the Gastrointestinal Tract. L. R. Johnson, editor. Elsevier Academic Press, Boston, MA. 1711–1734.
- 2.Hussain M. M., Rava P., Walsh M., Rana M., Iqbal J. 2012. Multiple functions of microsomal triglyceride transfer protein. Nutr. Metab. (Lond). 9: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teng B., Burant C. F., Davidson N. O. 1993. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 260: 1816–1819 [DOI] [PubMed] [Google Scholar]
- 4.Mehta A., Kinter M. T., Sherman N. E., Driscoll D. M. 2000. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol. 20: 1846–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lellek H., Kirsten R., Diehl I., Apostel F., Buck F., Greeve J. 2000. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J. Biol. Chem. 275: 19848–19856 [DOI] [PubMed] [Google Scholar]
- 6.Blanc V., Davidson N. O. 2003. C-to-U RNA editing: mechanisms leading to genetic diversity. J. Biol. Chem. 278: 1395–1398 [DOI] [PubMed] [Google Scholar]
- 7.Greeve J., Altkemper I., Dieterich J. H., Greten H., Windler E. 1993. Apolipoprotein B mRNA editing in 12 different mammalian species: hepatic expression is reflected in low concentrations of apoB-containing plasma lipoproteins. J. Lipid Res. 34: 1367–1383 [PubMed] [Google Scholar]
- 8.Powell L. M., Wallis S. C., Pease R. J., Edwards Y. H., Knott T. J., Scott J. 1987. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 50: 831–840 [DOI] [PubMed] [Google Scholar]
- 9.Hirano K., Young S. G., Farese R. V., Jr., Ng J., Sande E., Warburton C., Powell-Braxton L. M., Davidson N. O. 1996. Targeted disruption of the mouse apobec-1 gene abolishes apolipoprotein B mRNA editing and eliminates apolipoprotein B48. J. Biol. Chem. 271: 9887–9890 [DOI] [PubMed] [Google Scholar]
- 10.Nakamuta M., Chang B. H., Zsigmond E., Kobayashi K., Lei H., Ishida B. Y., Oka K., Li E., Chan L. 1996. Complete phenotypic characterization of apobec-1 knockout mice with a wild-type genetic background and a human apolipoprotein B transgenic background, and restoration of apolipoprotein B mRNA editing by somatic gene transfer of Apobec-1. J. Biol. Chem. 271: 25981–25988 [DOI] [PubMed] [Google Scholar]
- 11.Lo C. M., Nordskog B. K., Nauli A. M., Zheng S., Vonlehmden S. B., Yang Q., Lee D., Swift L. L., Davidson N. O., Tso P. 2008. Why does the gut choose apolipoprotein B48 but not B100 for chylomicron formation? Am. J. Physiol. Gastrointest. Liver Physiol. 294: G344–G352 [DOI] [PubMed] [Google Scholar]
- 12.Yamanaka S., Balestra M. E., Ferrell L. D., Fan J., Arnold K. S., Taylor S., Taylor J. M., Innerarity T. L. 1995. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc. Natl. Acad. Sci. USA. 92: 8483–8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sowden M., Hamm J. K., Smith H. C. 1996. Overexpression of APOBEC-1 results in mooring sequence-dependent promiscuous RNA editing. J. Biol. Chem. 271: 3011–3017 [DOI] [PubMed] [Google Scholar]
- 14.Siddiqui J. F., Van Mater D., Sowden M. P., Smith H. C. 1999. Disproportionate relationship between APOBEC-1 expression and apolipoprotein B mRNA editing activity. Exp. Cell Res. 252: 154–164 [DOI] [PubMed] [Google Scholar]
- 15.Galloway C. A., Smith H. C. 2010. The expression of apoB mRNA editing factors is not the sole determinant for the induction of editing in differentiating Caco-2 cells. Biochem. Biophys. Res. Commun. 391: 659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinto D., Robine S., Jaisser F., El Marjou F. E., Louvard D. 1999. Regulatory sequences of the mouse villin gene that efficiently drive transgenic expression in immature and differentiated epithelial cells of small and large intestines. J. Biol. Chem. 274: 6476–6482 [DOI] [PubMed] [Google Scholar]
- 17.Blanc V., Davidson N. O. 2011. Mouse and other rodent models of C to U RNA editing. Methods Mol. Biol. 718: 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Y., Nassir F., Luo J., Buhman K., Davidson N. O. 2003. Intestinal lipoprotein assembly in apobec-1-/- mice reveals subtle alterations in triglyceride secretion coupled with a shift to larger lipoproteins. Am. J. Physiol. Gastrointest. Liver Physiol. 285: G735–G746 [DOI] [PubMed] [Google Scholar]
- 19.Millar J. S., Cromley D. A., McCoy M. G., Rader D. J., Billheimer J. T. 2005. Determining hepatic triglyceride production in mice: comparison of poloxamer 407 with Triton WR-1339. J. Lipid Res. 46: 2023–2028 [DOI] [PubMed] [Google Scholar]
- 20.Yamanaka S., Poksay K. S., Driscoll D. M., Innerarity T. L. 1996. Hyperediting of multiple cytidines of apolipoprotein B mRNA by APOBEC-1 requires auxiliary protein(s) but not a mooring sequence motif. J. Biol. Chem. 271: 11506–11510 [DOI] [PubMed] [Google Scholar]
- 21.Qian X., Balestra M. E., Yamanaka S., Boren J., Lee I., Innerarity T. L. 1998. Low expression of the apolipoprotein B mRNA-editing transgene in mice reduces LDL levels but does not cause liver dysplasia or tumors. Arterioscler. Thromb. Vasc. Biol. 18: 1013–1020 [DOI] [PubMed] [Google Scholar]
- 22.Hersberger M., Patarroyo-White S., Qian X., Arnold K. S., Rohrer L., Balestra M. E., Innerarity T. L. 2003. Regulatable liver expression of the rabbit apolipoprotein B mRNA-editing enzyme catalytic polypeptide 1 (APOBEC-1) in mice lacking endogenous APOBEC-1 leads to aberrant hyperediting. Biochem. J. 369: 255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sowden M., Hamm J. K., Spinelli S., Smith H. C. 1996. Determinants involved in regulating the proportion of edited apolipoprotein B RNAs. RNA. 2: 274–288 [PMC free article] [PubMed] [Google Scholar]
- 24.Sowden M. P., Eagleton M. J., Smith H. C. 1998. Apolipoprotein B RNA sequence 3′ of the mooring sequence and cellular sources of auxiliary factors determine the location and extent of promiscuous editing. Nucleic Acids Res. 26: 1644–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Backus J. W., Smith H. C. 1992. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res. 20: 6007–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith H. C. 1993. Apolipoprotein B mRNA editing: the sequence to the event. Semin. Cell Biol. 4: 267–278 [DOI] [PubMed] [Google Scholar]
- 27.Navaratnam N., Patel D., Shah R. R., Greeve J. C., Powell L. M., Knott T. J., Scott J. 1991. An additional editing site is present in apolipoprotein B mRNA. Nucleic Acids Res. 19: 1741–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z., Eggerman T. L., Bocharov A. V., Baranova I. N., Vishnyakova T. G., Kurlander R. J., Csako G., Patterson A. P. 2012. Hypermutation of ApoB mRNA by rat APOBEC-1 overexpression mimics APOBEC-3 hypermutation. J. Mol. Biol. 418: 65–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg B. R., Hamilton C. E., Mwangi M. M., Dewell S., Papavasiliou F. N. 2011. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 18: 230–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farese R. V., Jr., Veniant M. M., Cham C. M., Flynn L. M., Pierotti V., Loring J. F., Traber M., Ruland S., Stokowski R. S., Huszar D., et al. 1996. Phenotypic analysis of mice expressing exclusively apolipoprotein B48 or apolipoprotein B100. Proc. Natl. Acad. Sci. USA. 93: 6393–6398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sundaram M., Yao Z. 2010. Recent progress in understanding protein and lipid factors affecting hepatic VLDL assembly and secretion. Nutr. Metab. (Lond). 7: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reboul E., Trompier D., Moussa M., Klein A., Landrier J. F., Chimini G., Borel P. 2009. ATP-binding cassette transporter A1 is significantly involved in the intestinal absorption of alpha- and gamma-tocopherol but not in that of retinyl palmitate in mice. Am. J. Clin. Nutr. 89: 177–184 [DOI] [PubMed] [Google Scholar]
- 33.Iqbal J., Hussain M. M. 2005. Evidence for multiple complementary pathways for efficient cholesterol absorption in mice. J. Lipid Res. 46: 1491–1501 [DOI] [PubMed] [Google Scholar]
- 34.Li M., Wang I. X., Li Y., Bruzel A., Richards A. L., Toung J. M., Cheung V. G. 2011. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 333: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickrell J. K., Gilad Y., Pritchard J. K. 2012. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 335: 1302; author reply 1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin W., Piskol R., Tan M. H., Li J. B. 2012. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 335: 1302, author reply 1302 [DOI] [PubMed] [Google Scholar]
- 37.Gu T., Buaas F. W., Simons A. K., Ackert-Bicknell C. L., Braun R. E., Hibbs M. A. 2012. Canonical A-to-I and C-to-U RNA editing is enriched at 3′UTRs and microRNA target sites in multiple mouse tissues. PLoS ONE. 7: e33720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.