Abstract
Transforming growth factor-β (TGFβ) is activated as a result of liver injury, such as cholestasis. However, its influence on endogenous metabolism is not known. This study demonstrated that TGFβ regulates hepatic phospholipid and bile acid homeostasis through MAD homolog 3 (SMAD3) activation as revealed by lithocholic acid-induced experimental intrahepatic cholestasis. Lithocholic acid (LCA) induced expression of TGFB1 and the receptors TGFBR1 and TGFBR2 in the liver. In addition, immunohistochemistry revealed higher TGFβ expression around the portal vein after LCA exposure and diminished SMAD3 phosphorylation in hepatocytes from Smad3-null mice. Serum metabolomics indicated increased bile acids and decreased lysophosphatidylcholine (LPC) after LCA exposure. Interestingly, in Smad3-null mice, the metabolic alteration was attenuated. LCA-induced lysophosphatidylcholine acyltransferase 4 (LPCAT4) and organic solute transporter β (OSTβ) expression were markedly decreased in Smad3-null mice, whereas TGFβ induced LPCAT4 and OSTβ expression in primary mouse hepatocytes. In addition, introduction of SMAD3 enhanced the TGFβ-induced LPCAT4 and OSTβ expression in the human hepatocellular carcinoma cell line HepG2. In conclusion, considering that Smad3-null mice showed attenuated serum ALP activity, a diagnostic indicator of cholangiocyte injury, these results strongly support the view that TGFβ-SMAD3 signaling mediates an alteration in phospholipid and bile acid metabolism following hepatic inflammation with the biliary injury.
Keywords: cholestasis, hepatitis, transforming growth factor, MAD homolog
Bile acids (BA) are required for the absorption and excretion of lipophilic metabolites such as cholesterol. BA production is initiated by CYP7A1-catalyzed cholesterol oxidation in liver (1). BA are transported to the bile by the bile salt export pump (BSEP, also called ATP-binding cassette B11; ABCB11) (2) and multidrug resistance protein 2 (MRP2, also called ABCC2) (3, 4). Most BA are reabsorbed from the small intestine and are taken up by the liver via the BA uptake transporter Na+-taurocholate cotransporting polypeptide (NTCP, also termed solute carrier 10A1; SLC10A1) (5, 6) and organic anion-transporting peptides (OATP, also named SLCO family) (7). BA homeostasis is maintained by enterohepatic circulation in which greater than 90% are reabsorbed from the small intestine and transported to the liver.
Interruption of BA excretion from the liver can enhance hepatic BA levels, leading to cholestatic liver disease. Accumulation of hepatic BA results in activation of the farnesoid X receptor (FXR). A critical role for FXR in the regulation of BA homeostasis was established using Fxr-null mice (8). FXR binds to BA and directly or indirectly contributes to the downregulation of CYP7A1 (BA synthesis), SLC10A1 and SLCO family (BA uptake) expression (9–11), and upregulation of ABCB11 and ABCC2 (BA export) expression (12–14). In addition, FXR activation enhances expression of the organic solute transporter α/β (OSTα/β) genes in the liver (15) and accelerates BA excretion to the bloodstream. Thus, FXR functions as the chief sensor of intracellular BA levels and contributes to BA homeostasis (16).
When hepatic BA accumulate in liver, hepatic inflammation is induced by tumor necrosis factor (TNF)α and transforming growth factor (TGF)β. TNFα signaling is known to downregulate SLCO1A1 and SLC10A1 expression in the liver (17) and reduce bile canalicular contraction (18). In addition, TNFα upregulates multidrug resistance-associated protein 3 (MRP3; basolateral bile salt export transporter, also called ABCC3) and can protect from liver injury that results from obstructive cholestasis (19). Kupffer cell activation mediates induction of hepatic multidrug resistance-associated protein 4 (MRP4; basolateral bile salt export transporter, also called ABCC4) expression (20), which is involved in basolateral BA export. The TGFβ signal regulates expression of CYP7A1 gene (21), the rate-limiting enzyme of the BA synthesis. Proinflammatory cytokines can also influence BA homeostasis.
Among the BA, lithocholic acid (LCA) is the most potent chemical causing liver toxicity. LCA levels are elevated in patients with liver disease (22) and intrahepatic cholestasis (23). Experimental interventions to protect against LCA toxicity were investigated using animal models, revealing that the nuclear receptor pregnane X receptor (PXR) protects against LCA toxicity through upregulation of cytochrome P450 3A (CYP3A) and sulfotransferase 2A (SULT2A) that metabolize LCA for elimination (24–26). A variety of LCA metabolites were reported to be associated with this protection (27–31). Recently, endogenous bile acid metabolism associated with LCA toxicity was investigated (27), and LCA exposure was reported to change serum chemistry, such as phospholipids, cholesterol, free fatty acids, and triglycerides (32). Furthermore, a comprehensive view of LCA-induced alterations in endogenous metabolites was investigated by use of metabolomics for the detection and characterization of small organic chemicals in biological matrices. LCA exposure decreased serum lysophosphatidylcholine (LPC) levels, leading to cholestasis (33). The change in serum LPC was associated with increased serum alkaline phosphatase (ALP), a marker of cholangiocyte injury, and increased hepatic lysophosphatidylcholine acyltransferase (LPCAT)4 expression. In addition, the TGFβ-MAD homolog 3 (SMAD3)-dependent LPCAT4 induction was observed in mouse primary hepatocytes (33). TGFβ is ubiquitously expressed as latent type, and it is transformed to active type by a variety of agents. Active TGFβ binds to the TGFβ receptors 1 and 2, resulting in enhanced SMAD3 phosphorylation (34). Phosphorylated SMAD3 can lead to dynamic alterations in gene expression patterns. Thus, although TGFβ-SMAD3 signaling is expected to mediate the metabolic alteration in cholestasis, in vivo studies are required to confirm this hypothesis. In the current study, LCA-induced metabolic alterations in Smad3-null mice compared with wild-type mice was investigated, revealing that TGFβ-SMAD3 signaling is involved in BA and LPC metabolism in LCA-induced liver injury.
MATERIALS AND METHODS
Materials
Bile acids (tauromurideoxycholate and tauro-5β-cholanic acid-3-one) were synthesized as described in the supplementary data. The other BA and fatty acids were purchased from Sigma-Aldrich (St. Louis, MO) or Steraloids (Newport, RI). Lysophosphatidylcholines ware purchased form Avanti Polar Lipids (Alabaster, AL).
Animals and diets
Male mice (C57BL/6), MAD homolog 3 (Smad3)-null mice (35), and background-matched wild-type mice were housed in temperature- and light-controlled rooms and given water and pelleted NIH-31 chow ad libitum. For the LCA studies, mice were given 0.6% LCA-supplement diet with the AIN93G diet as a control (Dyets, Bethlehem, PA). Three wild-type and three Smad3-null mice were fed the control diet, and five wild-type and four Smad3-null mice were given the LCA diet. All animal studies were carried out in accordance with Institute of Laboratory Animal Resources (ILAR) guidelines and protocols approved by the National Cancer Institute Animal Care and Use Committee.
Serum chemistry
Serum was prepared using Serum Separator Tubes (Becton, Deckinson and Co., Franklin Lakes, NJ). The serum catalytic activity of alanine aminotransferase (ALT) and alkaline phosphatase (ALP) was measured with ALT and ALP assay kit, respectively (Catachem, Bridgeport, CT).
UPLC-ESI-QTOFMS analysis
Serum was prepared using Serum Separator Tubes (Becton, Dickinson and Co.). The serum was diluted with 19 vol of 66% acetonitrile and centrifuged twice at 18,000 g for 20 min to remove insoluble materials. UPLC-ESI-QTOFMS was preformed as previously reported (27). In brief, the aliquots (5 μl) were injected into a reverse-phase 50 × 2.1 mm ACQUITY 1.7 μm C18 column (Waters, Milford, MA) using an ACQUITY UPLC system (Waters) with a gradient mobile phase comprising 0.1% formic acid and acetonitrile containing 0.1% formic acid. The eluant was introduced by electrospray ionization into the mass spectrometer [Q-TOF Premier (Waters)] operating in negative electrospray ionization mode. The capillary and sampling cone voltages were set to 3000 and 30 V, respectively. The desolvation gas flow was set to 650 l/h, and the temperature was set to 350°C. The cone gas flow was 50 l/h, and the source temperature was 120°C. Data were acquired in centroid mode from m/z 50 to 800 in MS scanning. Tandem MS collision energy was scanned from 5 to 35 V.
Data processing and multivariate data analysis
Data processing and multivariate data analysis were conducted as previously reported (27). Partial least squares (PLS) and contribution analyses were performed using SIMCA-P+12 (Umetrics, Kinnelon, NJ).
Quantification of serum bile acid and lysophosphatidylcholine
Serum bile acid levels were determined with standard curves using authentic metabolites. Quantification of serum lysophosphatidylcholine was performed according to a previously reported method (33).
RNA analysis
RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA), and qPCR was performed using cDNA generated from 1 µg total RNA with a SuperScript II Reverse Transcriptase kit and random oligonucleotides (Invitrogen). Primers were designed using qPrimerDepot. All sequences are listed in supplementary Table I. Quantitative PCR reactions were carried out using SYBR green PCR master mix (Applied Biosystems, Foster City, CA) in an ABI Prism 7900HT Sequence Detection System. Values were quantified using comparative CT method, and samples were normalized to 18S rRNA.
Western blotting
For whole-cell extracts, a culture cell was lysed with sample buffer and subjected to Western blotting. The samples were boiled for 5 min and then separated and transferred to PVDF membranes using standard Western blotting techniques. The membranes were incubated with an antibody against phospho-SMAD3 at a dilution of 1:1,000, ab52903 (Abcam, Cambridge, UK) or SMAD3 at a dilution of 1:1,000 (ab28379, Abcam). The signals were normalized to signals obtained with a GAPDH Ab used at a dilution of 1:10,000 (MAB374, Millipore, Billerica, MA).
Histological analysis
Small blocks of liver tissue were immediately fixed in 10% neutral formalin and embedded in paraffin. Sections (4 µm thick) were stained with hematoxylin and eosin. At least three discontinuous liver sections were evaluated for each mouse. Immunolocalization of TGFβ1 and phospho-Smad3 were performed as described (36) with the addition of antigen retrieval in 1 mM EDTA (pH 8) at 95°C for 10 min for sections stained with the pSmad3 antibody. TGFβ1 was detected with the primary antibody LC-1-30-1 at 3 µg/ml, and pSmad3 was detected with a rabbit monoclonal antibody from Epitomics (Cat. No. 1880-1) at a dilution of 1:500.
Generation of Smad3-expressing adenovirus with Cre/LoxP system
A Smad3-expressing adenovirus with Cre/LoxP system (Ad-L-Smad3) was generated by using the Adenovirus Cre/LoxP-Regulated Expression Vector Set (Takara, Tokyo, Japan). A Cre-expressing adenovirus (Ad-Cre) and a GFP-expressing adenovirus (Ad-L-GFP) with Cre/LoxP system were previously generated (37). The titer of these adenoviruses was measured by the 50% tissue culture infectious dose method.
Culture of primary hepatocytes and HepG2 cells
Primary hepatocytes were prepared as previously reported (33). After starvation with FBS-negative Williams’ Medium E for 2 h, the hepatocytes were exposed to 5 ng/ml of TGFβ (R and D Systems, Minneapolis, MA) for 12 h, and then collected and lysed for gene expression analysis by use of qPCR. HepG2 cells were seeded on a 12-well plate and infected with recombinant adenovirus. Two days later, the HepG2 cells with the starvation were exposed to 5 ng/ml of TGFβ. The cells were subjected to the Western blotting (treatment with TGFβ for 1 h) or the qPCR (treatment with TGFβ for 12 h).
Statistical analysis
Statistical analysis was performed using Prism version 5.0c (GraphPad Software, San Diego, CA). A P-value of less than 0.05 was considered as significant difference.
RESULTS
Lithocholic acid exposure enhanced TGFβ and the receptors mRNA level in the liver
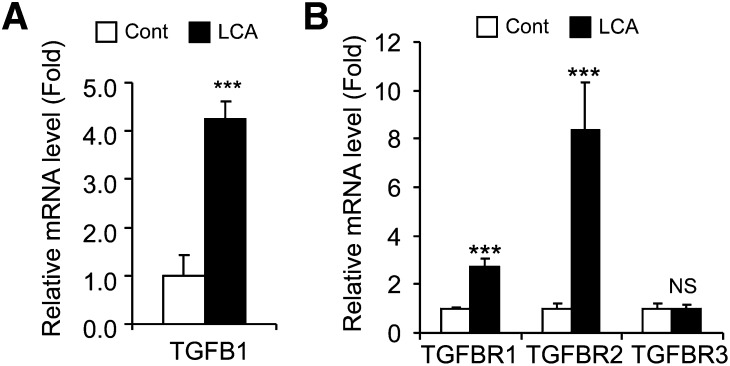
The influence of lithocholic acid (LCA) exposure on TGFβ signaling was investigated using C57BL/6 mice treated with the synthetic AIN93G diet (Cont) and 0.6% LCA-supplemented AIN93G diet (LCA) for 7 days. Hepatic TGFB1, TGFBR1, and TGFBR2 mRNA levels increased after LCA exposure, although TGFBR3 mRNA level did not changed in the livers (Fig. 1). These results suggest that LCA exposure stimulates TGFβ signaling in the livers.
Fig. 1.
Hepatic TGFβ-SMAD3 signaling after LCA exposure. qPCR analysis of TGFB1 (A) and TGFBR (B) mRNA expression in the livers of C57BL/6 mice. Significance was determined by unpaired t-test (***P < 0.001).
LCA-induced liver injury was alleviated in Smad3-null mice
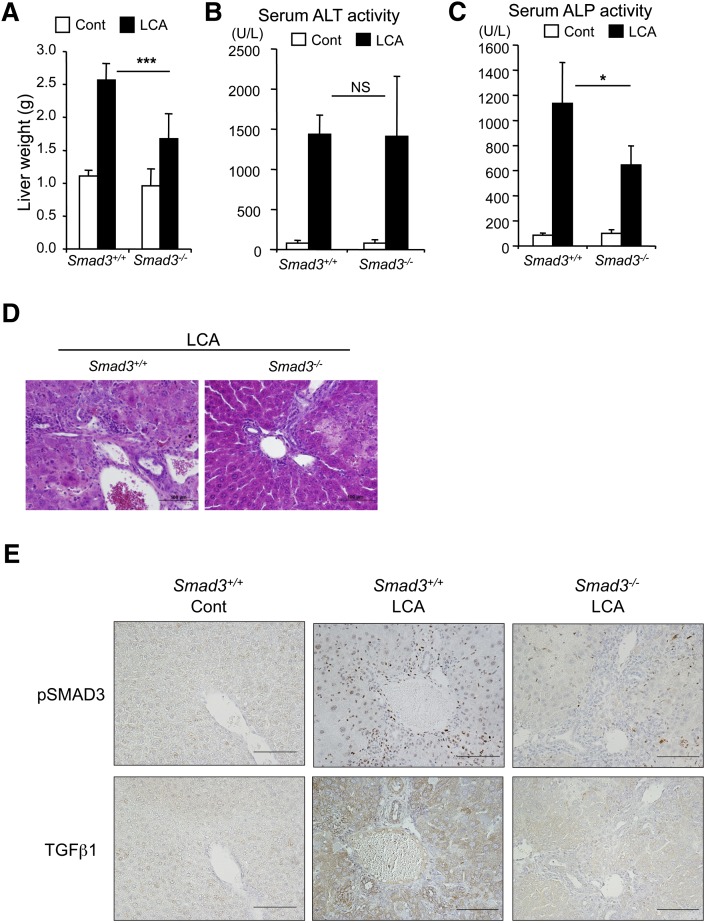
TGFβ activates SMAD3 via the TGFβ receptors. Thus, to investigate whether SMAD3 was involved in LCA-induced liver injury, Smad3-null mice were treated with control diet and LCA diet for 6 days. After LCA exposure, the liver mass of Smad3-null mice was smaller than that of LCA-treated wild-type mice (Fig. 2A). In addition, LCA-increased serum ALP activities were significantly attenuated in the Smad3-null mice, although serum ALT activities were not changed (Fig. 2B, C). Furthermore, liver histology showed mild features of inflammatory cell infiltration around the portal vein in Smad3-null mice, which was not observed in similarly treated wild-type mice (Fig. 2D). Immunohistochemistry revealed TGFβ protein around the portal vein with lower expression of the TGFβ in Smad3-null mice compared with wild-type mice (Fig. 2E), suggesting lower TGFβ stimulation of the SMAD3 activation in the liver (supplementary Fig. I). In addition, a dramatic attenuation of the SMAD3 phosphorylation signal was observed in the hepatocyte nuclei of Smad3-null mice (Fig. 2E). These results suggest that TGFβ-SMAD3 signaling is associated with the LCA-induced biliary injury and raise the possibility that TGFβ-SMAD3 signaling alters hepatic metabolism.
Fig. 2.
Difference between wild-type and Smad3-null mice in LCA-induced liver injury. (A) Liver mass of wild-type and Smad3-null after LCA exposure. Serum ALT (B) and ALP (C) activities. Significance was determined by one way-ANOVA with Bonferroni's test (*P < 0.05; ***P < 0.001). NS, no significance. Liver histology with hematoxylin and eosin staining (D) and with immunostaining for TGFβ and phosphorylated SMAD3 (E).
Difference between wild-type and Smad3-null mice in the serum metabolome after LCA exposure
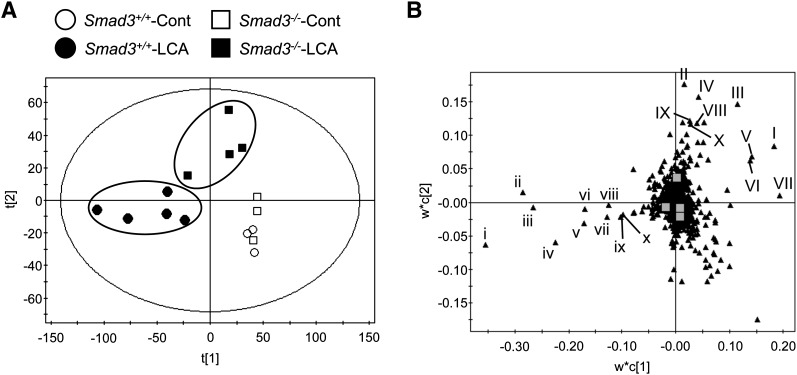
To examine serum metabolites, PLS and contribution analyses were performed with UPLC-ESI-QTOFMS negative mode data derived from serum of mice fed LCA or control diet. PLS analysis showed a separation between the LCA-treated wild-type (Fig. 3A) and the LCA-treated Smad3-null group that was further examined with a loadings plot (Fig. 3B). Contribution analysis revealed 10 enhanced and 10 attenuated ions as the top-ranking ions giving rise to the separation. Lysophosphatidylcholine (LPC) and fatty acid fragments were determined as raised ions in Smad3-null mice compared with the wild-type mice (Table 1). The most lowered ions were derived from bile salts (Table 2). After LCA feeding, the serum metabolome of Smad3-null mice was much different from that of the wild-type mice in LPC and bile salts.
Fig. 3.
Difference in LCA-altered serum metabolites between wild-type and Smad3-null mice. (A) PLS of the serum metabolome among wild-type with control diet (Smad3+/+-Cont, open circle); wild-type with LCA diet (Smad3+/+-LCA, filled circle); Smad3-null with control diet (Smad3−/−-Cont, open square); and Smad3-null mice (Smad3−/−-LCA, filled square). (B) Loading plot of the PLS. Lowercase and uppercase Roman numerals represent enhanced and attenuated ions in the wild-type mice fed LCA diet, determined by the contribution analysis (compared with that of the Smad3-null mice fed LCA diet), respectively. Identified ions are listed in Tables 1 and 2.
TABLE 1.
Top ten of serum metabolite ions that were of higher intensity in Smad3-null group with LCA diet than in wild-type group with LCA diet
| Rank | Mark | RT (min) | Found (m/z) | Candidate | Mass Error (ppm) |
| 1 | I | 5.37 | 568.3585 | Stearoyl-LPC (18:0 LPC) | 5.10 |
| 2 | II | 5.94 | 279.2310 | Linoleic acid | 5.01 |
| 3 | III | 4.90 | 566.3436 | Oleoyl-LPC (18:1 LPC) | 3.88 |
| 4 | IV | 5.78 | 327.2324 | Docosahexaenoic acid | 0.00 |
| 5 | V | 4.63 | 540.3279 | Palmitoyl-LPC (16:0 LPC) | 4.07 |
| 6 | VI | 5.23 | 568.3582 | Stearoyl-LPC (18:0 LPC) | 5.63 |
| 7 | VII | 4.75 | 540.3275 | Palmitoyl-LPC (16:0 LPC) | 4.81 |
| 8 | VIII | 5.88 | 303.2313 | Arachidonic acid | 3.63 |
| 9 | IX | 6.42 | 381.1707 | Not determined | |
| 10 | X | 6.42 | 281.2467 | Oleic acid | 4.98 |
The ion ranking, based on the PLS-DA analysis, indicates the highest confidence and greatest contribution to separation between the wild-type and the Smad3-null mice after LCA exposure. RT, retention time.
TABLE 2.
Top ten of serum metabolite ions that were lower intensity in Smad3-null group with LCA-diet than in wild-type group with LCA-diet
| Rank | Mark | RT (min) | Found (m/z) | Candidate | Mass Error (ppm) |
| 1 | i | 3.19 | 498.2868 | Taurochenodeoxycholate (TCDC) | 4.21 |
| 2 | ii | 3.72 | 482.2913 | Taurolithocholate (TLC) | 5.60 |
| 3 | iii | 3.72 | 480.2764 | Tauro-5β-cholanic acid-3-one (T3KL) | 4.16 |
| 4 | iv | 3.17 | 496.2722 | Not determined | |
| 5 | v | 2.49 | 514.2813 | Tauro-α/β-muricholatea (TαMC/TβMC) | 4.86 |
| 6 | vi | 2.61 | 498.2856 | Tauromurideoxycholate (TMDC) | 6.62 |
| 7 | vii | 3.09 | 498.2860 | Not determined | |
| 8 | viii | 2.77 | 498.2852 | Tauroursodeoxycholate/Taurohyodeoxycholateb (TUDC/THDC) | 7.43 |
| 9 | ix | 2.83 | 496.2700 | Not determined | |
| 10 | x | 2.74 | 496.2703 | Not determined |
The ion ranking, based on the PLS analysis, indicates the highest confidence and greatest contribution to separation between the wild-type and the Smad3-null mice after LCA exposure. RT, retention time.
No clear separation between tauro-α-muricholate and tauro-β-muricholate.
No clear separation between tauroursodeoxycholate and taurohyodeoxycholate.
Role of TGFβ-SMAD3 signal in bile acid homeostasis
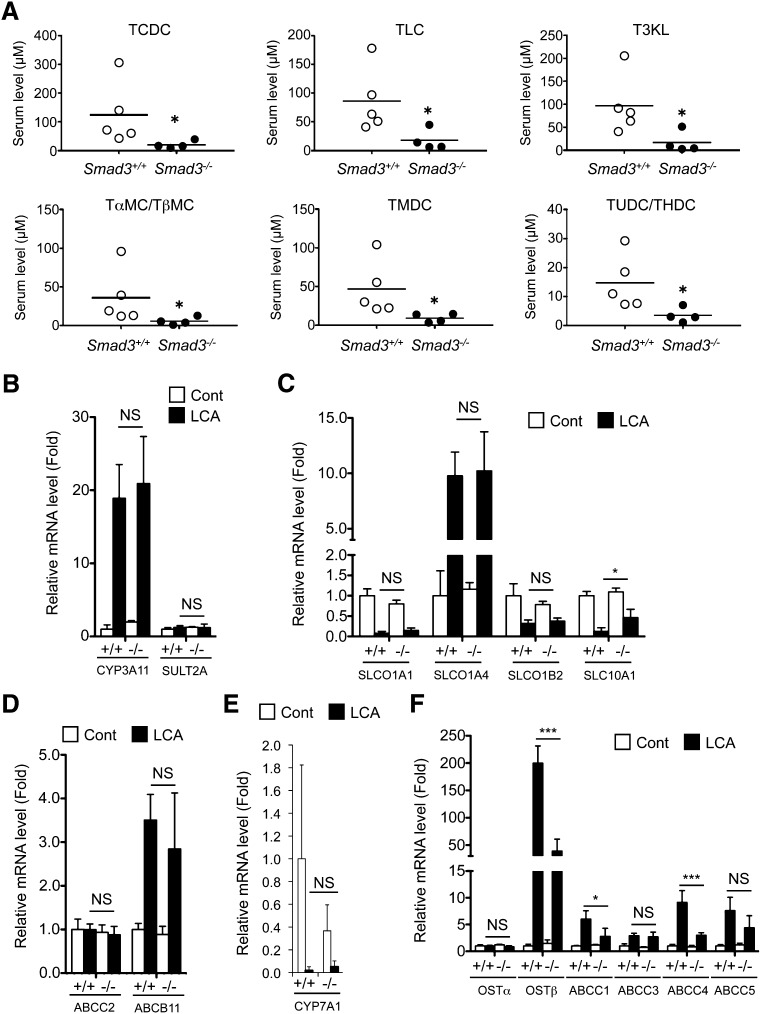
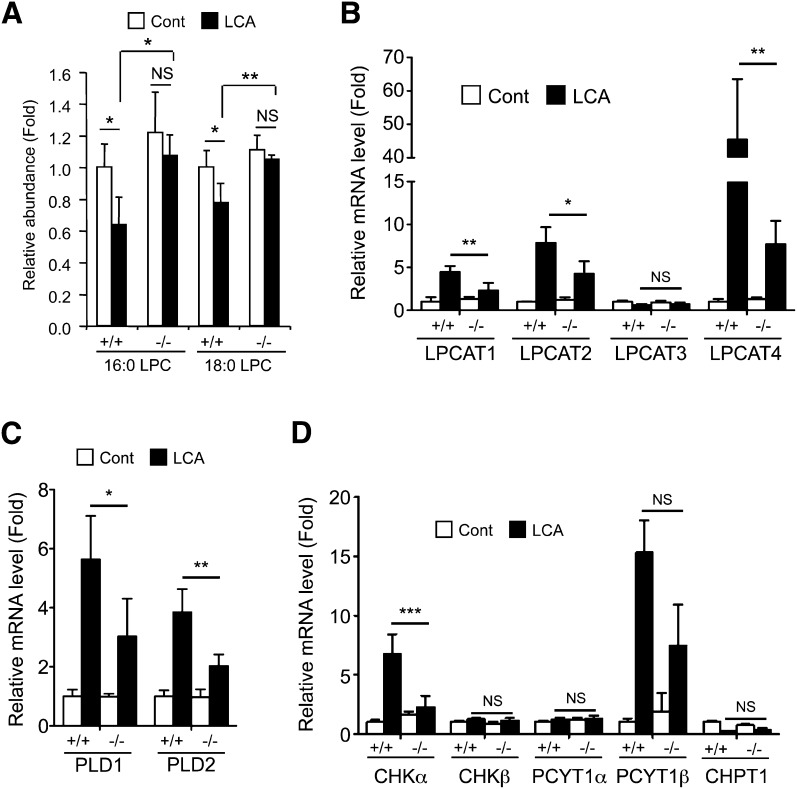
Quantification of serum bile salts was performed with authentic compounds. All of the tested bile acid levels were lower in Smad3-null mice after LCA exposure than those in the wild-type mice (Fig. 4A). As bile acid metabolites can be produced from LCA (supplementary Fig. II) in the liver, the expression of the major metabolic enzymes CYP3A11 and sulfotransferase 2A (SULT2A) was investigated. CYP3A11 and SULT2A expression was not, however, different between the wild-type and the Smad3-null mice after LCA feeding (Fig. 4B). Thus, hepatic expression of bile acid-related transporter genes was investigated, including mRNAs encoding ABCB11, ABCC1, ABCC2, ABCC3, ABCC4, ABCC5, solute carrier (SLC)10A1, solute carrier organic anion transporter (SLCO)1A1, SLCO1A4, SLCO1B2, organic solute transporter (OST)α, and OSTβ. Differences in expression of the bile salt uptake transporters SLCO1A1, SLCO1A4, and SLCO1B2 (intake to hepatocyte, Fig. 4C), major bile salt exporters ABCC2 and ABCB11 (export to bile duct, Fig. 4D), and bile acid synthesis enzyme CYP7A1 (Fig. 4E) were not observed in the liver. However, expression of the basolateral exporting transporter OSTβ and ABCC4 was much lower in the Smad3-null liver than that in the wild-type mice (Fig. 4F). SMAD3 may be involved in the induction of the Ostβ and Abcc4 genes, as LCA exposure decreased hepatic expression of FXR that upregulates the genes (supplementary Fig. III). In addition to the bile acids, quantification of serum palmitoyl-LPC (16:0 LPC) and setearoyl-LPC (18:0 LPC) was performed with use of the authentic compounds. Wild-type mice showed attenuated serum 16:0 LPC and 18:0 LPC after LCA feeding, but Smad3-null mice did not (Fig. 5A). Induction of the related gene expression, as previously reported, was attenuated in Smad3-null mice (Fig. 5B–D). Notably, LPCAT4 expression was lower in the Smad3-null mice compared with the wild-type mice. Thus, Ostβ, Abcc4, and Lpcat4 were expected to be TGFβ-SMAD3 signal-related genes in the liver.
Fig. 4.
Difference in bile acid metabolism between wild-type and Smad3-null mice. (A) Quantification of serum BA levels after LCA exposure. (B–F) qPCR analysis of hepatic bile acid-related gene expression: bile acid metabolism (B), bile acid uptake (C), bile acid excretion to bile (D), bile acid synthesis (E), and bile acid export to vein (F). (A) Significance was determined by Mann-Whitney test (*P < 0.05) or (B–F) one way-ANOVA with Bonferroni's test (*P < 0.05; ***P < 0.001).
Fig. 5.
Difference in phospholipid metabolism between wild-type and Smad3-null mice. (A) Quantification of serum LPC levels after LCA exposure. (B–D) qPCR analysis of hepatic gene expression: lysophosphatidylcholine transferase 1–4 (B), phospholipase D (C), and de novo synthesis of phosphatidylcholine (D). Significance was determined by one way-ANOVA with Bonferroni's test (*P < 0.05; **P < 0.01; ***P < 0.001).
TGFβ-SMAD3 signal regulated the genes encoding OSTβ and LPCAT4 in mouse hepatocytes and human cells
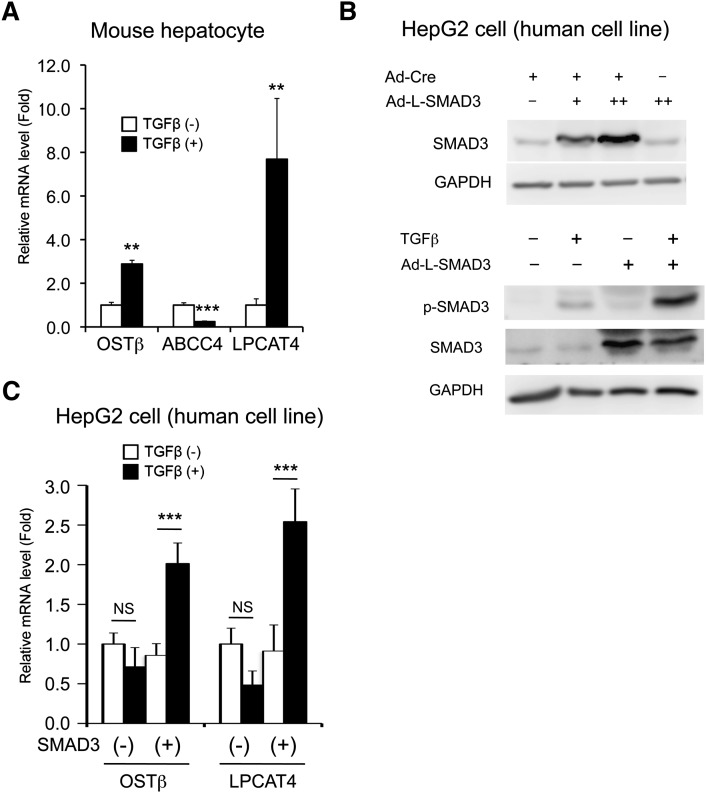
To investigate whether TGFβ signaling directly induced the expression of Ostβ, Abcc4, and Lpcat4 genes in the hepatocyte, the respective mRNAs were quantitated with isolated and cultured mouse primary hepatocytes. TGFβ-elevated OSTβ and LPCAT4 mRNA levels were observed in hepatocytes, while ABCC4 expression was decreased (Fig. 6A). In addition, with Cre recombinase-dependent human SMAD3 expression (Fig. 6B), the induction was also observed in the human hepatocarcinoma cell line HepG2 (Fig. 6C). These results strongly indicate that TGFβ-SMAD3 signaling mediates induction of Ostβ and Lpcat4 gene expression.
Fig. 6.
TGFβ stimulates both mouse and human OSTβ and LPCAT4 gene transcription. (A) qPCR analysis for OSTβ, ABCC4, and LPCAT4 mRNA expression in mouse primary hepatocytes after TGFβ exposure. Significance was determined by unpaired t-test (**P < 0.001; ***P < 0.001). (B) Western blotting for human SMAD3 expression and TGFβ-stimulated SMAD3 phosphorylation in HepG2 cells infected with SMAD3-expressing adenovirus. Two days after the infection (Ad-L-GFP, Ad-Cre, and/or Ad-L-SMAD3), the HepG2 cells (seeded on a 12-well plate) were subjected to Western blotting. Adenovirus (13 MOI) was adjusted with Ad-L-GFP. −, +, and ++ is presented as 0, 3, and 10 MOI, respectively. TGFβ (2.5 ng/ml) exposure was performed with 2 h starvation (FBS negative) after the SMAD3 introduction with Ad-Cre. One hour later, the TGFβ-exposed HepG2 cells were subjected to Western blotting. (C) qPCR analysis for OSTβ and LPCAT4 gene expression in the SMAD3-introduced HepG2 cells after TGFβ exposure. Significance was determined by one way-ANOVA with Bonferroni's test (***P < 0.001). NS, no significance.
DISCUSSION
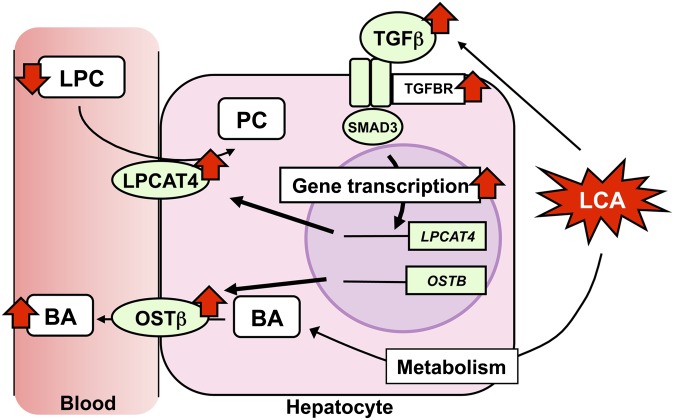
The current study demonstrated that TGFβ-SMAD3 signaling was involved in bile acid and phospholipid homeostasis in part through inducing hepatic expression of Ostβ and Lpcat4 genes (Fig. 7). UPLC-ESI-QTOFMS, in conjunction with a PLS analysis, illustrated a clear difference between wild-type and Smad3-null mice in serum metabolites after LCA exposure, with BA metabolites being markedly elevated while LPC were decreased. Alteration in the saturated LPC tightly associates not only with LCA-induced liver injury but also with the other cholestasis model (38), which is related to the increased serum BA. Interestingly, Smad3-null mice showed lowered serum ALP activity (a cholangiocyte injury marker) associated with acute liver injury compared with wild-type mice, but they did not have altered serum ALT activity (a hepatocyte injury marker). In chronic liver disease, such as cholestasis and primary biliary cirrhosis, chronic autoimmune-mediated damage to bile duct epithelial cell (BDEC) can cause inflammation with activation of wound healing in the liver (39). In bile duct ligation (BDL) model of obstructive cholestasis, the BDEC-expressed β6 integrin activates the latent TGFβ (40, 41). In another biliary injury model, the BDEC-selective toxicant α-naphthylisothiocyanate-exposed mouse, β6 integrin-mediated TGFβ activation was observed (42). In addition, TGFβ expression was observed in metaplastic bile duct epithelium (43), as was found in the present study. These observations strongly support the view that the serum metabolic alteration reflects the severity of biliary injury. This finding, together with ALT, ALP, and γ-glutamyltransferase, may contribute to an accurate diagnosis of liver injury.
Fig. 7.
Proposed mechanism of TGFβ-SMAD3-mediated metabolic alteration in LCA-induced liver injury. LCA-stimulated TGFβ-SMAD3 signal is involved in serum metabolic alteration (increased BA and decreased LPC) via induction of LPCAT4 and OSTβ expression.
TGFβ was reported to be a controller of BA synthesis through regulation of the human CYP7A1 gene (21), whereas no difference in hepatic CYP7A1 mRNA level was observed between wild-type and Smad3-null mice in the current study, thus suggesting that other factors were important for regulating mouse Cyp7a1 gene transcription. On the other hand, the present study revealed that TGFβ-SMAD3 was a mediator of basolateral export transporter OSTβ expression. OSTβ together with OSTα are expressed in hepatocytes and cholangiocytes, and they mediate BA movement from hepatocyte and cholangiocyte to the circulation. In addition, decreased BA pool size and attenuated serum BA levels were observed in mice lacking OSTα/β (44). FXR, which is a chief sensor of intracellular BA levels, is believed to control the OSTα/β gene transcription in human and rodents that regulates hepatic BA levels via controlling the enterohepatic system. LCA attenuated hepatic FXR expression, but not PXR expression, that regulates expression of the BA metabolic enzymes CYP3A and SULT2A. Despite hepatic FXR attenuation, LCA induction of OSTβ expression was observed in the liver. Considering the enhancement of hepatocyte SMAD3 activation and SMAD3-dependent elevation of serum BA levels, these observations may suggest that TGFβ-SMAD3-OSTβ signaling is an important factor for controlling the hepatocyte BA levels under conditions of reduced hepatic FXR, whereas TNFα and nuclear factor (erythroid-derived 2)-like 2 (NRF2) mediate elevation of serum BA levels via hepatic ABCC3 and ABCC4 induction (19, 45). Thus, proinflammatory cytokine signaling (with oxidative stress) was considered to turn the direction of hepatic bile acid flow (from vein to hepatocyte, and bile duct). Hepatic BA accumulation can stimulate enhanced oxidative stress and accelerate the proinflammatory response in the liver. Proinflammatory cytokines may play an important role in the bile acid-induced acute liver injury. Future studies will be required to establish the pathophysiological significance of these findings.
Cholic acid (CA) challenge is frequently used as a cholestasis model, as well as LCA. Interestingly peroxisome proliferator-activated receptor (PPAR)α-deficient mice showed liver injury after cholic acid CA challenge, whereas the corresponding wild-type mice did not (38). As PPARα is activated by fatty acid metabolites and controls lipid metabolism, this observation may suggest that lipid is associated with BA homeostasis. Synthetic PPARα activators, such as [4-chloro-6-(2,3-xylido)-2-pyrimidinylthio)] acetic acid (also called Wy-14,643), ciprofibrate, and bezafibrate, are known to downregulate the human CYP7A1 and rodent Cyp7a1 gene promoters (46–48). In addition, PPARα was reported to mediate clofibrate-induced hepatic MRP3 (ABCC3) and MRP4 (ABCC4) expression (49), and ciprofibrate reduces hepatic NTCP (SLC10A1), OATP1 (SLCO1A1), and BSEP (ABCB11) expression in mice (50). However, Ppara-null mice did not diminish altered hepatic expression of these genes after CA challenge (38), and thus, the pathophysiological significance of PPARα on bile acid homeostasis remains unclear. Therefore, the PPARα-mediated protection against CA toxicity may be accomplished through the other gene function. The hydrophobic BA, such as LCA, deoxycholic acid, and chenodeoxycholic acid, can lead to mitochondria dysfunction and induce hepatic apoptosis and necrosis. CA challenge elevates the hydrophobic BA levels in the liver. PPARα activation protects from liver injury due to mitochondria dysfunction, such as acetaminophen-induced liver injury (51), and the PPARα protection against mitochondria damage may contribute to alleviation of the CA-induced cholestasis. However, a detailed understanding requires further study. As we observed that Fxr-null mice showed increased hepatic lipid levels with hepatic accumulation of bile acid (52), lipid metabolism may be associated with bile acid-induced liver injury or bile acid homeostasis.
Recently, the number of patients with fat accumulation-related liver disease (including hepatic steatosis and nonalcoholic steatohepatitis; NASH) is increasing not only in middle agers but also in teenagers (53, 54). NASH, with chronic hepatic inflammation, can lead to fibrogenesis and development of cirrhosis and cancer. Some patients with NASH were reported to show increased plasma bile acid levels (55). In a previous report, methionine- and choline-deficient diet-fed mice (experimental NASH model) also showed increased serum bile acids (56). In addition, experimental NASH was associated with decreased serum LPC, similar to the LCA-exposed mice (56). Furthermore, elevated hepatic TGFβ levels were observed in the NASH model (56). These observations suggest a metabolic link, in which TGFβ is involved in lipid and BA metabolism following a variety of liver diseases. Understanding the molecular mechanism of bile acid-induced liver injury (cholestasis) that is common with fat accumulation-related liver disease (NASH) would contribute to the development of clinical therapies.
In conclusion, the present study revealed that TGFβ-SMAD3 signaling was involved in hepatic bile acid and phospholipid homeostasis regulating LPCAT4 and OSTβ gene expression. Serum LPC levels following TGFβ-SMAD3 signaling may be involved in biliary injury. Proinflammatory cytokines, such as TGFβ, are mainly secreted from nonparenchymal cells. Thus, a further understanding of liver diseases requires additional studies on the role of hepatocyte-nonparenchymal cell interactions in hepatic metabolism. Knowledge of metabolic linkage between proinflammatory signal and hepatic lipid and bile acid homeostasis may contribute to further understanding of chronic liver diseases, such as steatohepatitis, liver fibrosis and cirrhosis, as well as cholestasis.
Supplementary Material
Acknowledgments
The authors thank John Buckley and Linda Byrd (National Cancer Institute) for technical assistance. T.M. was supported by a fellowship from the Japan Society for the Promotion of Science.
Footnotes
Abbreviations:
- ALP
- alkaline phosphatase
- ALT
- alanine aminotransferase
- BA
- bile acid
- CA
- cholic acid
- CHK
- choline kinase
- CHPT1
- choline phosphotransferase 1
- CYP
- cytochrome P450
- FXR
- farnesoid X receptor
- LCA
- lithocholic acid
- LPC
- lysophosphatidylcholine
- LPCAT
- lysophosphatidylcholine acyltransferase
- OST
- organic solute transporter
- PCYT1
- phosphate cytidylyltransferase 1
- PLD
- phospholipase D
- PLS
- partial least squares
- PXR
- pregnane X receptor
- SLC
- solute carrier
- SLCO
- solute carrier organic anion transporter
- SMAD3
- MAD homolog 3
- SULT
- sulfotransferase
- T3KL
- touro-5β-cholanic acid-3-one
- TC
- taurocholate
- TCDC
- taurochenodeoxycholate
- TGFβ
- transforming growth factor-β
- THDC
- taurohyodeoxycholate
- TLC
- taurolithocholate
- TMDC
- tauromurideoxycholate
- TNFα
- tumor necrosis factor α
- TUDC
- tauroursodeoxycholate
- UPLC-ESI-QTOFMS
- ultra-performance liquid chromatography coupled with electrospray ionization quadruple time-of-flight mass spectrometry
This work was supported by the National Cancer Institute Intramural Research Program, Center for Cancer Research, National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table, four figures, and methods.
REFERENCES
- 1.Vlahcevic Z. R., Heuman D. M., Hylemon P. B. 1991. Regulation of bile acid synthesis. Hepatology. 13: 590–600 [PubMed] [Google Scholar]
- 2.Gerloff T., Stieger B., Hagenbuch B., Madon J., Landmann L., Roth J., Hofmann A. F., Meier P. J. 1998. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J. Biol. Chem. 273: 10046–10050 [DOI] [PubMed] [Google Scholar]
- 3.Jedlitschky G., Leier I., Buchholz U., Barnouin K., Kurz G., Keppler D. 1996. Transport of glutathione, glucuronate, and sulfate conjugates by the MRP gene-encoded conjugate export pump. Cancer Res. 56: 988–994 [PubMed] [Google Scholar]
- 4.Keppler D., Konig J., Buchler M. 1997. The canalicular multidrug resistance protein, cMRP/MRP2, a novel conjugate export pump expressed in the apical membrane of hepatocytes. Adv. Enzyme Regul. 37: 321–333 [DOI] [PubMed] [Google Scholar]
- 5.Stieger B., Hagenbuch B., Landmann L., Hochli M., Schroeder A., Meier P. J. In situ localization of the hepatocytic Na+/Taurocholate cotransporting polypeptide in rat liver. 1994 Gastroenterology. 107: 1781–1787 [DOI] [PubMed] [Google Scholar]
- 6.Hagenbuch B., Meier P. J. 1994. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter J. Clin. Invest. 93: 1326–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svoboda M., Riha J., Wlcek K., Jaeger W., Thalhammer T. 2011. Organic anion transporting polypeptides (OATPs): regulation of expression and function. Curr. Drug Metab. 12: 139–153 [DOI] [PubMed] [Google Scholar]
- 8.Sinal C. J., Tohkin M., Miyata M., Ward J. M., Lambert G., Gonzalez F. J. 2000. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 102: 731–744 [DOI] [PubMed] [Google Scholar]
- 9.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6: 517–526 [DOI] [PubMed] [Google Scholar]
- 10.Maeda T., Miyata M., Yotsumoto T., Kobayashi D., Nozawa T., Toyama K., Gonzalez F. J., Yamazoe Y., Tamai I. 2004. Mol. Pharm. 1: 281–289 [DOI] [PubMed] [Google Scholar]
- 11.Jung D., Hagenbuch B., Fried M., Meier P. J., Kullak-Ublick G. A. Role of liver-enriched transcription factors and nuclear receptors in regulating the human, mouse, and rat NTCP gene. 2004. Am. J. Physiol. Gastrointest. Liver Physiol. 286: G752–G761 [DOI] [PubMed] [Google Scholar]
- 12.Kast H. R., Goodwin B., Tarr P. T., Jones S. A., Anisfeld A. M., Stoltz C. M., Tontonoz P., Kliewer S., Willson T. M., Edwards P. A. 2002. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J. Biol. Chem. 277: 2908–2915 [DOI] [PubMed] [Google Scholar]
- 13.Plass J. R., Mol O., Heegsma J., Geuken M., Faber K. N., Jansen P. L., Muller M. 2002. Hepatology. 35: 589–596 [DOI] [PubMed] [Google Scholar]
- 14.Ananthanarayanan M., Balasubramanian N., Makishima M., Mangelsdorf D. J., Suchy F. J. 2001. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J. Biol. Chem. 276: 28857–28865 [DOI] [PubMed] [Google Scholar]
- 15.Boyer J. L., Trauner M., Mennone A., Soroka C. J., Cai S. Y., Moustafa T., Zollner G., Lee J. Y., Ballatori N. 2006. Am. J. Physiol. Gastrointest. Liver Physiol. 290: G1124–G1130 [DOI] [PubMed] [Google Scholar]
- 16.Matsubara T., Li F., Gonzalez F. J. FXR signaling in the enterohepatic system. Mol. Cell. Endocrinol. Epub ahead of print. May 17, 2012; doi:10.1016/j.mce.2012.05.004. [DOI] [PMC free article] [PubMed]
- 17.Geier A., Dietrich C. G., Voigt S., Ananthanarayanan M., Lammert F., Schmitz A., Trauner M., Wasmuth H. E., Boraschi D., Balasubramaniyan N., et al. 2005. Cytokine-dependent regulation of hepatic organic anion transporter gene transactivators in mouse liver. Am. J. Physiol. Gastrointest. Liver Physiol. 289: G831–G841 [DOI] [PubMed] [Google Scholar]
- 18.Ikeda S., Mitaka T., Harada K., Sato F., Mochizuki Y., Hirata K. 2003. Tumor necrosis factor-alpha and interleukin-6 reduce bile canalicular contractions of rat hepatocytes. Surgery. 133: 101–109 [DOI] [PubMed] [Google Scholar]
- 19.Bohan A., Chen W. S., Denson L. A., Held M. A., Boyer J. L. 2003. Tumor necrosis factor alpha-dependent up-regulation of Lrh-1 and Mrp3(Abcc3) reduces liver injury in obstructive cholestasis. J. Biol. Chem. 278: 36688–36698 [DOI] [PubMed] [Google Scholar]
- 20.Campion S. N., Johnson R., Aleksunes L. M., Goedken M. J., van Rooijen N., Scheffer G. L., Cherrington N. J., Manautou J. E. 2008. Hepatic Mrp4 induction following acetaminophen exposure is dependent on Kupffer cell function. Am. J. Physiol. Gastrointest. Liver Physiol. 295: G294–G304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T., Chiang J. Y. 2007. A novel role of transforming growth factor beta1 in transcriptional repression of human cholesterol 7alpha-hydroxylase gene. Gastroenterology. 133: 1660–1669 [DOI] [PubMed] [Google Scholar]
- 22.Festi D., Morselli Labate A. M., Roda A., Bazzoli F., Frabboni R., Rucci P., Taroni F., Aldini R., Roda E., Barbara L. 1983. Diagnostic effectiveness of serum bile acids in liver diseases as evaluated by multivariate statistical methods. Hepatology. 3: 707–713 [DOI] [PubMed] [Google Scholar]
- 23.Hofmann A. F. 2004. Detoxification of lithocholic acid, a toxic bile acid: relevance to drug hepatotoxicity. Drug Metab. Rev. 36: 703–722 [DOI] [PubMed] [Google Scholar]
- 24.Kitada H., Miyata M., Nakamura T., Tozawa A., Honma W., Shimada M., Nagata K., Sinal C. J., Guo G. L., Gonzalez F. J., et al. 2003. Protective role of hydroxysteroid sulfotransferase in lithocholic acid-induced liver toxicity. J. Biol. Chem. 278: 17838–17844 [DOI] [PubMed] [Google Scholar]
- 25.Staudinger J. L., Goodwin B., Jones S. A., Hawkins-Brown D., MacKenzie K. I., LaTour A., Liu Y., Klaassen C. D., Brown K. K., Reinhard J., et al. 2001. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. USA. 98: 3369–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie W., Radominska-Pandya A., Shi Y., Simon C. M., Nelson M. C., Ong E. S., Waxman D. J., Evans R. M. 2001. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc. Natl. Acad. Sci. USA. 98: 3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho J. Y., Matsubara T., Kang D. W., Ahn S. H., Krausz K. W., Idle J. R., Luecke H., Gonzalez F. J. 2010. Urinary metabolomics in Fxr-null mice reveals activated adaptive metabolic pathways upon bile acid challenge. J. Lipid Res. 51: 1063–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deo A. K., Bandiera S. M. 2009. 3-ketocholanoic acid is the major in vitro human hepatic microsomal metabolite of lithocholic acid. Drug Metab. Dispos. 37: 1938–1947 [DOI] [PubMed] [Google Scholar]
- 29.Araya Z., Wikvall K. 1999. 6alpha-hydroxylation of taurochenodeoxycholic acid and lithocholic acid by CYP3A4 in human liver microsomes. Biochim. Biophys. Acta. 1438: 47–54 [DOI] [PubMed] [Google Scholar]
- 30.Bodin K., Lindbom U., Diczfalusy U. 2005. Novel pathways of bile acid metabolism involving CYP3A4. Biochim. Biophys. Acta. 1687: 84–93 [DOI] [PubMed] [Google Scholar]
- 31.Miyata M., Watase H., Hori W., Shimada M., Nagata K., Gonzalez F. J., Yamazoe Y. 2006. Role for enhanced faecal excretion of bile acid in hydroxysteroid sulfotransferase-mediated protection against lithocholic acid-induced liver toxicity. Xenobiotica. 36: 631–644 [DOI] [PubMed] [Google Scholar]
- 32.Miyata M., Nomoto M., Sotodate F., Mizuki T., Hori W., Nagayasu M., Yokokawa S., Ninomiya S., Yamazoe Y. 2010. Possible protective role of pregnenolone-16 alpha-carbonitrile in lithocholic acid-induced hepatotoxicity through enhanced hepatic lipogenesis. Eur. J. Pharmacol. 636: 145–154 [DOI] [PubMed] [Google Scholar]
- 33.Matsubara T., Tanaka N., Patterson A. D., Cho J. Y., Krausz K. W., Gonzalez F. J. 2011. Lithocholic acid disrupts phospholipid and sphingolipid homeostasis leading to cholestasis in mice. Hepatology. 53: 1282–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuzaki K. 2012. Smad phosphoisoform signals in acute and chronic liver injury: similarities and differences between epithelial and mesenchymal cells. Cell Tissue Res. 347: 225–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X., Letterio J. J., Lechleider R. J., Chen L., Hayman R., Gu H., Roberts A. B., Deng C. 1999. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 18: 1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Figueroa J. D., Flanders K. C., Garcia-Closas M., Anderson W. F., Yang X. R., Matsuno R. K., Duggan M. A., Pfeiffer R. M., Ooshima A., Cornelison R., et al. 2010. Expression of TGF-beta signaling factors in invasive breast cancers: relationships with age at diagnosis and tumor characteristics. Breast Cancer Res. Treat. 121: 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saika S., Ikeda K., Yamanaka O., Sato M., Muragaki Y., Ohnishi Y., Ooshima A., Nakajima Y., Namikawa K., Kiyama H., et al. 2004. Transient adenoviral gene transfer of Smad7 prevents injury-induced epithelial-mesenchymal transition of lens epithelium in mice. Lab. Invest. 84: 1259–1270 [DOI] [PubMed] [Google Scholar]
- 38.Li F., Patterson A. D., Krausz K. W., Tanaka N., Gonzalez F. J. 2012. Metabolomics reveals an essential role for peroxisome proliferator-activated receptor α in bile acid homeostasis. J. Lipid Res. 53: 1625–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiocchi C., Lund P. K. 2011. Themes in fibrosis and gastrointestinal inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 300: G677–G683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popov Y., Patsenker E., Stickel F., Zaks J., Bhaskar K. R., Niedobitek G., Kolb A., Friess H., Schuppan D. 2008. Integrin alphavbeta6 is a marker of the progression of biliary and portal liver fibrosis and a novel target for antifibrotic therapies. J. Hepatol. 48: 453–464 [DOI] [PubMed] [Google Scholar]
- 41.Wang B., Dolinski B. M., Kikuchi N., Leone D. R., Peters M. G., Weinreb P. H., Violette S. M., Bissell D. M. 2007. Role of alphavbeta6 integrin in acute biliary fibrosis. Hepatology. 46: 1404–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan B. P., Weinreb P. H., Violette S. M., Luyendyk J. P. 2010. The coagulation system contributes to alphaVbeta6 integrin expression and liver fibrosis induced by cholestasis. Am. J. Pathol. 177: 2837–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takiya S., Tagaya T., Takahashi K., Kawashima H., Kamiya M., Fukuzawa Y., Kobayashi S., Fukatsu A., Katoh K., Kakumu S. 1995. Role of transforming growth factor beta 1 on hepatic regeneration and apoptosis in liver diseases. J. Clin. Pathol. 48: 1093–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ballatori N., Fang F., Christian W. V., Li N., Hammond C. L. 2008. Ostalpha-Ostbeta is required for bile acid and conjugated steroid disposition in the intestine, kidney, and liver. Am. J. Physiol. Gastrointest. Liver Physiol. 295: G179–G186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aleksunes L. M., Slitt A. L., Maher J. M., Augustine L. M., Goedken M. J., Chan J. Y., Cherrington N. J., Klaassen C. D., Manautou J. E. 2008. Induction of Mrp3 and Mrp4 transporters during acetaminophen hepatotoxicity is dependent on Nrf2. Toxicol. Appl. Pharmacol. 226: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marrapodi M., Chiang J. Y. 2000. Peroxisome proliferator-activated receptor alpha (PPARalpha) and agonist inhibit cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J. Lipid Res. 41: 514–520 [PubMed] [Google Scholar]
- 47.Patel D. D., Knight B. L., Soutar A. K., Gibbons G. F., Wade D. P. 2000. The effect of peroxisome-proliferator-activated receptor-alpha on the activity of the cholesterol 7 alpha-hydroxylase gene. Biochem. J. 351: 747–753 [PMC free article] [PubMed] [Google Scholar]
- 48.Post S. M., Duez H., Gervois P. P., Staels B., Kuipers F., Princen H. M. 2001. Fibrates suppress bile acid synthesis via peroxisome proliferator-activated receptor-alpha-mediated downregulation of cholesterol 7alpha-hydroxylase and sterol 27-hydroxylase expression. Arterioscler. Thromb. Vasc. Biol. 21: 1840–1845 [DOI] [PubMed] [Google Scholar]
- 49.Moffit J. S., Aleksunes L. M., Maher J. M., Scheffer G. L., Klaassen C. D., Manautou J. E. 2006. Induction of hepatic transporters multidrug resistance-associated proteins (Mrp) 3 and 4 by clofibrate is regulated by peroxisome proliferator-activated receptor alpha. J. Pharmacol. Exp. Ther. 317: 537–545 [DOI] [PubMed] [Google Scholar]
- 50.Kok T., Bloks V. W., Wolters H., Havinga R., Jansen P. L., Staels B., Kuipers F. 2003. Peroxisome proliferator-activated receptor alpha (PPARalpha)-mediated regulation of multidrug resistance 2 (Mdr2) expression and function in mice. Biochem. J. 369: 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patterson A. D., Shah Y. M., Matsubara T., Krausz K. W., Gonzalez F. J. 2012. Peroxisome proliferator-activated receptor alpha induction of uncoupling protein 2 protects against acetaminophen-induced liver toxicity. Hepatology. 56: 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyata M., Sakaida Y., Matsuzawa H., Yoshinari K., Yamazoe Y. 2011. Fibroblast growth factor 19 treatment ameliorates disruption of hepatic lipid metabolism in farnesoid X receptor (Fxr)-null mice. Biol. Pharm. Bull. 34: 1885–1889 [DOI] [PubMed] [Google Scholar]
- 53.Vajro P., Lenta S., Socha P., Dhawan A., McKiernan P., Baumann U., Durmaz O., Lacaille F., McLin V., Nobili V. 2012. Diagnosis of nonalcoholic fatty liver disease in children and adolescents: position paper of the ESPGHAN Hepatology Committee. J. Pediatr. Gastroenterol. Nutr. 54: 700–713 [DOI] [PubMed] [Google Scholar]
- 54.Alisi A., Feldstein A. E., Villani A., Raponi M., Nobili V. 2012. Pediatric nonalcoholic fatty liver disease: a multidisciplinary approach. Nature Reviews Gastroenterol. Hepatol. 9: 152–161 [DOI] [PubMed] [Google Scholar]
- 55.Kalhan S. C., Guo L., Edmison J., Dasarathy S., McCullough A. J., Hanson R. W., Milburn M. 2011. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism. 60: 404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanaka N., Matsubara T., Krausz K. W., Patterson A. D., Gonzalez F. J. 2012. Disruption of phospholipid and bile acid homeostasis in mice with nonalcoholic steatohepatitis. Hepatology. 56: 118–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.