Abstract
In bacteria, physiological change may be effected by a single gene acquisition, producing ecological differentiation without genetic isolation. Natural selection acting on such differences can reduce the frequency of genotypes that arise from recombination at these loci. However, gene acquisition can only account for recombination interference in the fraction of the genome that is tightly linked to the integration site. To identify additional loci that contribute to adaptive differences, we examined orthologous genes in species of Enterobacteriaceae to identify significant differences in the degree of codon selection. Significance was assessed using the Adaptive Codon Enrichment metric, which accounts for the variation in codon usage bias that is expected to arise from mutation and drift; large differences in codon usage bias were identified in more genes than would be expected to arise from stochastic processes alone. Genes in the same operon showed parallel differences in codon usage bias, suggesting that changes in the overall levels of gene expression led to changes in the degree of adaptive codon usage. Most significant differences between orthologous operons were found among those involved with specific environmental adaptations, whereas "housekeeping" genes rarely showed significant changes. When considered together, the loci experiencing significant changes in codon selection outnumber potentially adaptive gene acquisition events. The identity of genes under strong codon selection seems to be influenced by the habitat from which the bacteria were isolated. We propose a two-stage model for how adaptation to different selective regimes can drive bacterial speciation. Initially, gene acquisitions catalyze rapid ecological differentiation, which modifies the utilization of genes, thereby changing the strength of codon selection on them. Alleles develop fitness variation by substitution, producing recombination interference at these loci in addition to those flanking acquired genes, allowing sequences to diverge across the entire genome and establishing genetic isolation (i.e., protection from frequent homologous recombination).
Keywords: codon usage bias, codon selection, speciation, recombination interference
Introduction
The hallmark of bacterial adaptation to novel environments is physiological differentiation, whereby evolved organisms interact with their environments differently than did their ancestors. Such physiological differentiation often involves change in biochemical activities as the result of gene gain, gene loss, or the occurrence of mutations that change the biochemical activities of existing gene products. These adaptive shifts can be readily identified as changes in gene inventory (Ochman et al. 2000; Hacker and Carniel 2001) or as sites showing evidence of positive selection for change (Nielsen and Yang 1998; Suzuki and Gojobori 1999). However, exploration of the novel ecological niches afforded by these changes may also demand expression changes among genes not involved in qualitative physiological adaptations. For example, changes in the abundance of a familiar nutrient will result in a concomitant change in the demand for the enzymes to metabolize that nutrient. Here, adaptation can occur through synonymous changes affecting the nature of mRNA/tRNA interactions. Such codon selection is common in genes of both prokaryotic and eukaryotic taxa (Sharp et al. 1988), most likely due to the influence of codon identity on the duration for which a ribosome is occupied synthesizing a particular polypeptide and/or its influence on the accuracy of translation (Plotkin and Kudla 2010).
Selection on synonymous codons produces systemic biases in codon usage among the open reading frames (ORFs) found in a genome, where the frequencies of certain codons increase relative to their synonyms. Although codon selection is not the only selective force that affects the nucleotide identity of synonymous sites, it is the primary selective force in many bacteria, with the less-preferred codons existing as a result of mutation and genetic drift (Bulmer 1991). This bias increases in tandem with the expression level of the gene (Ikemura 1981), indicating stronger selection in these ORFs (Sharp and Li 1987a,b). The genes encoding core physiological processes often exhibit high frequencies of preferred codon usage (Sharp and Li 1987b; Karlin and Mrazek 2000). Aside from widely conserved, highly expressed genes (e.g., those encoding ribosomal proteins), enrichment for preferred codon usage is also seen in genes that are distinctive to particular groups of bacteria (e.g., photosynthesis genes in cyanobacteria [Mrazek et al. 2001]), indicating that codon selection acts beyond those genes that are essential for all organisms. Although differences in preferred codon usage have been noted among orthologous genes (Karlin and Mrazek 2000), these differences have not been examined quantitatively; therefore, the extent to which change in selection is responsible for such differences is unknown. However, such changes are likely to be common as differences in gene expression among lineages may arise from either regulatory changes or simple environmental changes, thereby resulting in different levels of codon optimization in the orthologous ORFs.
We posit that the ecological changes resulting from gene gain, loss, or modification will result in expression changes among otherwise conserved genes, thereby altering the strength of codon selection among them. Such changes in codon selection have been difficult to evaluate since previous statistics lacked a theoretical framework to evaluate the significance of the differences in codon usage. We have developed a statistical technique that permits the comparison of codon selection between orthologous genes (Retchless and Lawrence 2011). This statistic, Adaptive Codon Enrichment (ACE), scores each gene based on its codon composition, with codons enriched in highly expressed genes having a higher score. ACE incorporates information about the codon frequencies of genes that experience little-to-no codon selection, thus allowing the significance of ACE values to be evaluated in the context of a null model of stochastic codon usage. Genes showing no enrichment for the adaptive codons have an ACE value of 0, and enrichment can be reported either as a z statistic based on the standard deviation of the entire gene (ACEz) or a length-normalized statistic that treats each codon as a separate unit (ACEu). Critically, ACE places codon usage bias in the context of a probabilistic distribution, measuring the extent to which preferred codons are over-represented relative to that expected from stochastic sampling of codons. This method permits normalization both within and across genomes, so that differences in codon usage bias between orthologs can be examined robustly in light of the variance expected from stochastic factors. Therefore, unlike other metrics of codon usage bias, ACE incorporates a method for separating neutral variation in codon usage from potential adaptations to ecological differences when comparing values of different genes. In this way, we can evaluate how codon selection has changed as bacteria evolved and identify those genes for which relative expression level has increased or decreased during organismal diversification.
In this study, we use the Enterobacteriaceae as a model group to examine how the changes in codon adaptation among genes may reflect changes in gene deployment during adaptation to different environments. The Enterobacteriaceae are a well-studied group of organisms that includes species with lifestyles as different as commensals of poikilotherms, commensals of mammals and birds, pathogens of mammals, pathogens of plants, and environmental detritovores.
Materials and Methods
Genomes
Genome sequences for Citrobacter koseri ATCC BAA-895, Citrobacter rodentium ICC168, Cronobacter turicensis z3032, Dickeya zeae Ech1591, Enterobacter cloacae ATCC 13047, Enterobacter sp. 638, Erwinia amylovora ATCC 49946, Erwinia tasmaniensis Et1/99, Escherichia coli MG1655, Escherichia fergusonii ATCC 35469, Klebsiella pneumoniae 78578, Klebsiella variicola At-22, Pectobacterium wasabiae WPP163, Salmonella enterica Typhimurium LT2, Salmonella enterica Arizonae 62:z4, Serratia proteamaculans 568, and Yersinia enterocolitica 8081 were downloaded from NCBI RefSeq; genes were identified using the annotation provided. Sequences from the Human Microbiome Project (HMP) were obtained from the HMP database at http://www.hmpdacc.org.
Identification of Orthologs and Genes within Operons
Orthologous proteins were identified as reciprocal best Basic Local Alignment Search Tool hits, which, when aligned, showed greater than 70% amino acid identity across more than 60% of their length. Genes unique to a genome were identified as those lacking any homolog with greater than 40% amino acid identity. Operons in Escherichia coli were delineated as described in the Database of Prokaryotic Operon, version 2 (Dam et al. 2007). Conserved operons between species were identified as those shared more than 50% of their genes.
Calculation of ACE
ACE was calculated as described (Retchless and Lawrence 2011). A reference set of genes reflecting little to no codon selection was assembled from that 80% of each genome that had the most typical di- and trinucleotide compositions; the set of genes reflecting strong codon selection was assembled from orthologs of 40 translation genes—tufA, tsf, fusA, rplA-rplF, rplI-rplT, and rpsB-rpsT (Sharp et al. 2005). The value of each codon is the logarithm of the ratio of its frequency (relative to its synonyms) in the set of translation genes to its frequency in the reference set; the value for each gene is the sum of the values of the constituent codons, normalized for variation among synonymous codons. The stochastic variation in codon composition was modeled as though the codons in each gene were sampled (with replacement) from a pool of codons with a composition identical to that of all genes with orthologs in each of the genomes in the analysis, as described previously (Retchless and Lawrence 2011). ACEz is normalized to the stochastic variation for the entire gene, whereas ACEu is length normalized by accounting for the variation of each codon individually.
Comparison of ACE across Genomes
The distributions of ACEu values were scaled to the distribution in E. coli by way of a second-order polynomial regression among orthologs, which accounted for the nonlinearity of the crossgenome relationship without overfitting (supplementary table S1, Supplementary Material online). When ACEu values were scaled for crossgenome comparisons with second-order polynomial regressions, the stochastic variance of the null model was scaled according to the slope of the tangent line at the point defined by that ACEu value. When ACE values for operons were compared between species, only the values for orthologous genes were considered. Differences between the ACE values of genes were calculated using a two-tailed z test.
Correlations of ACE Values within Operons
We tested the null hypothesis that ACEu values are independent among genes within operons by means of analysis of variance (ANOVA) between groups, with the F statistic representing the extent that variance between operons exceeded the variance within operons. The likelihood of obtaining the observed F value was evaluated with the F distribution. An alternative evaluation was performed by randomly assigning ACEu values (or the ACEu differences between orthologs) to operons and testing whether the observed F value exceeded the value resulting from random assignment; this method confirmed the significant results obtained with the F distribution.
Phylogeny Construction
Phenetic relationships between 17 enteric species were constructed on orthologous genes shared among all taxa using the neighbor-joining algorithm (Saitou and Nei 1987). Protein sequence difference was measured as the average divergence at nonsynonymous sites (Yang and Nielsen 2000), weighted by gene length. Difference in patterns of codon selection was measured as the Euclidian distance between ACEu values as represented by the first nine principal components of a PCA performed on all 17 genomes. Bootstrap support for nodes on the dN and ACE dendrograms was calculated using 1,000 and 100 resamples by genes, respectively.
Cladistic relationships between eight enteric taxa were constructed using maximum likelihood by PhyML (Guindon and Gascuel 2003). The similarity of individual gene phylogenies to the consensus species phylogeny was assessed using the Shimodaira–Hasegawa test (Shimodaira and Hasegawa 1999) implement by PAML (Yang 2007). The species phylogeny was constructed using a polychotomy of the Salmonella, Escherichia, and Citrobacter lineages as these nodes are not resolved by virtue of intraspecific recombination during speciation (Retchless and Lawrence 2010).
Tetrad Analysis of Human Microbiome Genomes
In this analysis of the relationship between bacterial niche and ACEu profile, informative tetrads are limited to those where the phylogenetic pairing is different from the pairing that arises from either the site of isolation or the ACEu profile. The alternative outcomes are that the pairing from the ACEu profile either matches or conflicts with the pairing based on isolation site. This reflects the fact that there are three possible pairings of genomes within a tetrad, and one of those configurations is excluded as noninformative due to it being occupied by the pairing that reflects phylogeny. Significance is assessed by a one-tailed binomial test.
Results
Codon Selection Reflects Environment of Expression
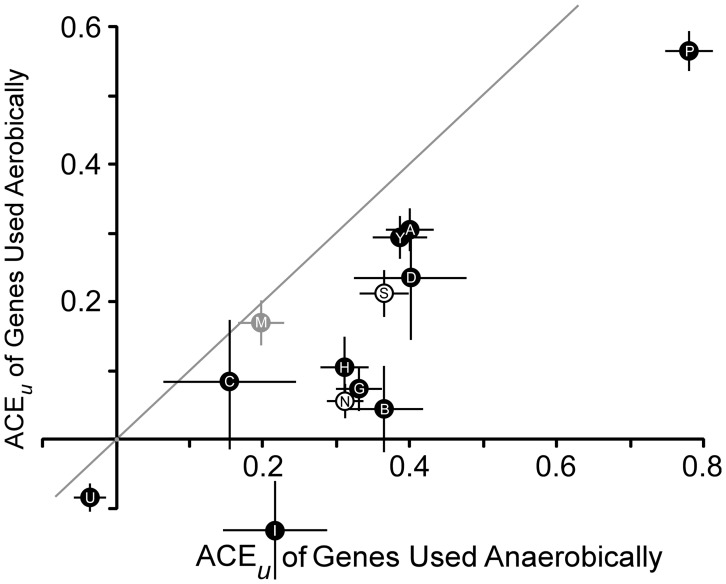
Codon compositions of genes may reflect their roles in cellular metabolism, whereby genes with certain functions use preferred subsets of codons. Alternatively, codon composition may reflect adaptations to their degree of expression in the specific environments wherein they are used. If so, then difference in codon composition among metabolically analogous genes within a genome should be correlated based on the environmental conditions that stimulate their expression. To test this prediction, we compared pairs of genes and operons in the E. coli chromosome whose products perform the same physiological function but are expressed preferentially under either aerobic or anaerobic conditions (fig. 1).
Fig. 1.
ACEu values for Escherichia coli gene pairs with similar functions expressed under aerobic or anaerobic conditions. Homologous gene pairs (anaerobic gene first): frdA/sdhA, frdB/sdhB, frdC/sdhC, frdD/sdhD, narG/narZ, narH/narY, and narI/varV; analogous operon pairs: cydAB/cyoABCD, plfB/aceEF, menABCDEFH/ubiABCDEFGH, and metH/metE. All values are for E. coli genes (black points) except the Salmonella enterica Typhimurium metHE genes (gray point). Error bars represent 1 standard deviation.
We examined the gene pairs encoding subunits of succinate dehydrogenase (sdh) or fumarate reductase (frd), cytochrome oxidase bo (cyo) or bd (cyd), pyruvate dehydrogenase (ace) or pyruvate formate lyase (pfl), proteins responsible for ubiquinone (ubi) or menaquinone (men) biosynthesis, or the constitutive (narZWY) or inducible (narGHI) respiratory nitrate reductase. In addition, we consider the alternative metE and metH methionine synthase genes in the closely related bacterium S. enterica; unlike, E. coli, Salmonella synthesizes coenzyme B12 de novo under anaerobic conditions, allowing greater use of the B12-dependent MetH enzyme anaerobically. This comparison was performed using the ACEu statistic, which is normalized to the length of the ORF.
In all cases, codon usage bias was more pronounced in the genes expressed primarily under anaerobic conditions, whose relatively larger ACEu values lay significantly to the right of the diagonal in figure 1. As the same physiological function is performed by each encoded protein within any pair, we surmise that the influence of ecological conditions on gene expression—rather than simply the cellular roles—shapes codon usage bias in their cognate genes. Therefore, changes in these conditions among bacterial species could lead to changes in codon selection among orthologs.
Degree of Codon Selection on Orthologous Genes Is Not Uniformly Conserved across Genomes
ACE was designed to respond to the magnitude of codon selection acting on an ORF, being normalized to both the codon frequencies that are typical of each genome and the amino acid composition of individual genes. Consequently, it is well suited for crossgenome comparisons, and the ACE values of orthologs should be strongly correlated if orthologous genes in two genomes experience similar levels of codon selection. Weak correlation between genomes may indicate a reallocation of selective power among ORFs, and large differences between the values of an orthologous pair (after accounting for genome-wide differences in total codon selection) may reflect differences in the relative magnitude of codon selection acting on those two orthologs.
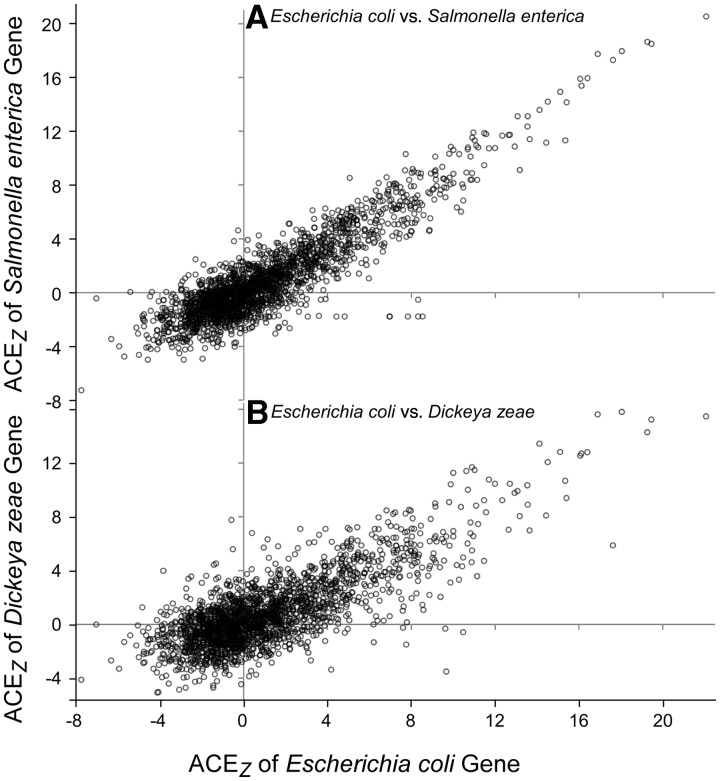
For an initial examination of these relationships, we identified genes shared between E. coli, S. enterica, and D. zeae and compared the ACEz values of the orthologs (fig. 2). ACEz reports the ACE in terms of the standard normal distribution that would be expected for each gene if the codons for its amino acids were sampled from the genome-wide relative frequencies of synonymous codons (Retchless and Lawrence 2011). Strong correlations of the ACEz values are observed when E. coli genes are compared with their orthologs in Salmonella (R = 0.904, fig. 2A) or Dickeya (R = 0.791, fig. 2B). Thus, highly biased genes that experience strong codon selection in one genome are generally likely to be highly biased in related genomes. This is not surprising, as the expression patterns of most genes would be constrained by their cellular roles (e.g., ribosomal proteins or two-component sensor proteins) and general ecological similarity between taxa (Karlin and Mrazek 2000). However, there are also genes which differ in their degree of codon usage bias, evident by genes which lie significant distances from the central trend. Such relationships are observable both in the closely related and ecological similar pair of E. coli and S. enterica (fig. 2A) and in the more distantly related and ecologically different pair of E. coli and D. zeae (fig. 2B), which have a lesser overall correlation among orthologs. So, similar to analogous genes in the same species (fig. 1), homologous genes in different species show evidence for differences in codon selection.
Fig. 2.
Scatterplot showing the ACEz values for orthologous genes shared among Escherichia coli, Salmonella enterica Typhimurium, and Dickeya zeae.
Changes in Codon Selection Are Shared among Genes within Operons
Changes in codon selection should affect coregulated genes in a similar manner. Bacterial operons represent intimately coregulated clusters of genes, as they are coexpressed from a common promoter. We predict that the degree of codon usage bias (measured by ACEu) will be correlated among genes in the same operon due to their expression patterns being correlated. Moreover, if evolutionary changes in codon usage bias reflect changes in codon selection arising from gene expression, then differences in ACEu values between orthologs should be correlated among genes in the same operon.
For this analysis, we expanded the data set to include eight species of enteric bacteria, for which we could identify 2,235 orthologous genes shared among all eight genomes. This set provides a balance between a large sample of diverse genomes and a large set of orthologous genes present in all genomes. Differences between species in the overall strength of selection result in some genomes exhibiting a greater range of ACEu values than others (Retchless and Lawrence 2011). Therefore, comparisons between genomes required that the values be scaled by regression. As the relationship in ACEu values among orthologs exhibited some curvature, reflecting differences in the overall degree of codon selection within each genome, a second-order polynomial equation was used for regression (supplementary table S1, Supplementary Material online). Following these adjustments, Pearson correlations between genomes ranged from 0.80 to 0.96, with more closely related taxa generally showing stronger correlation (supplementary table S2, Supplementary Material online).
To test the prediction that codon bias of genes in the same operons are correlated, we performed an ANOVA on the 442 E. coli operons that contained two or more genes with orthologs in each of the eight genomes, thereby including 1,285 of the 2,235 orthologous genes present in the eight genomes. In all genomes we examined, variability in ACEu was much smaller within operons than would be expected from randomly chosen gene sets, with P values for the ANOVA ranging from 10−52 to 10−96 (table 1, values on the diagonal). This reflects the relatively similar expression levels of genes within the same operon, despite some genes being transcribed from multiple promoters and the mRNA of cotranscribed genes decaying at different rates.
Table 1.
Likelihood of Observing the Actual Level of Variation between the Mean ACEu Values (or differences between orthologs) of 442 Operons Shared among Eight Genomes if the 1,285 Constituent Genes Were Randomly Distributed among Operons.
| Croa | Ctu | Ecl | Eco | Efe | Sty | Saz | Cko | |
|---|---|---|---|---|---|---|---|---|
| Cro | 3.42E-77b | 5.59E-05c | 1.21E-07 | 1.12E-07 | 2.5E-11 | 4.86E-05 | 3.77E-3 | 4.11E-3 |
| Ctu | 2.39E-96 | 1.48E-05 | 7.59E-22 | 4.24E-27 | 3.83E-14 | 6.34E-10 | 3.08E-13 | |
| Ecl | 3.08E-61 | 1.88E-10 | 1.45E-17 | 1.36E-08 | 2.22E-05 | 5.55E-10 | ||
| Eco | 5.76E-52 | 6.34E-04 | 1.37E-04 | 2.75E-05 | 4.46E-3 | |||
| Efe | 9.14E-52 | 6.21E-05 | 1.07E-05 | 2.17E-06 | ||||
| Sty | 8.58E-78 | 1.81E-01 | 1.51E-01 | |||||
| Saz | 4.24E-74 | 0.53803 | ||||||
| Cko | 2.03E-62 |
aTaxa are Cko, Citrobacter koseri; Cro, Citrobacter rodentium; Ctu, Cronobacter turicensis; Ecl, Enterobacter cloacae; Eco, Escherichia coli; Efe, Escherichia fergusonii; Sty, Salmonella enterica Typhimurium; and Saz, Salmonella enterica Arizonae.
bValues on the diagonal report P values of the ANOVA F statistic testing for similarity among ACEu values for genes in the same operon. Significant values indicate clustering of genes with similar ACEu values.
cValues off the diagonal report P values of the ANOVA F statistic testing for differences in the ACEu values for cognate operons in different species. Significant values indicate a change in the ACEu value between species.
Next, the change in ACEu was examined among the orthologous genes in each pair of genomes. If differences in ACEu between genomes simply reflect stochastic change, then differences among genes within operons would be independent from one another, with equal numbers of genes showing an increase or decrease in ACEu. Alternatively, if change in ACEu reflects change in codon selection, then differences should be correlated among genes in the same operon, such that constituent genes either increase or decrease in ACEu en masse relative to their respective orthologs. An ANOVA shows that changes in ACEu among genes within the same operon are significantly correlated (table 1, off-diagonal values). As expected, values are least significant between closely related species pairs such as E. coli and E. fergusonii (P = 10−4) and Salmonella serovars Typhimurium and Arizonae (P = 10−1), both because ecological differences are only beginning to develop and because few synonymous changes have arisen to produce a signal. In contrast, more distantly related species pairs show strong correlations in differences among genes in the same operon, with P values ranging up to 10−27 (table 1); these correlations were upheld when a larger group of 17 taxa was examined (supplementary table S3, Supplementary Material online).
There are caveats to interpreting the ANOVA results. First, operons have different sizes, ranging from 2 to 15 genes. However, similar results are obtained when examining operons with only two, only three or at least four genes (data not shown), suggesting that this did not influence the results. In addition, groups have different amounts of variance as assessed by Levene’s F (data not shown). To address this issue, we converted ACEu differences to ranks. The resulting rank ANOVA showed comparable results as well (data not shown). Finally, the low likelihoods of these results under the null model were confirmed by randomly reassigning genes to operons 100 times (see Materials and Methods). These data confer high confidence to the conclusion that the degree of codon usage bias changes in parallel among genes in the same operon, indicating that the differences in ACEu values among genomes reflect changes in gene deployment over evolutionary time.
Some Operons Show Strong Change in Codon Selection
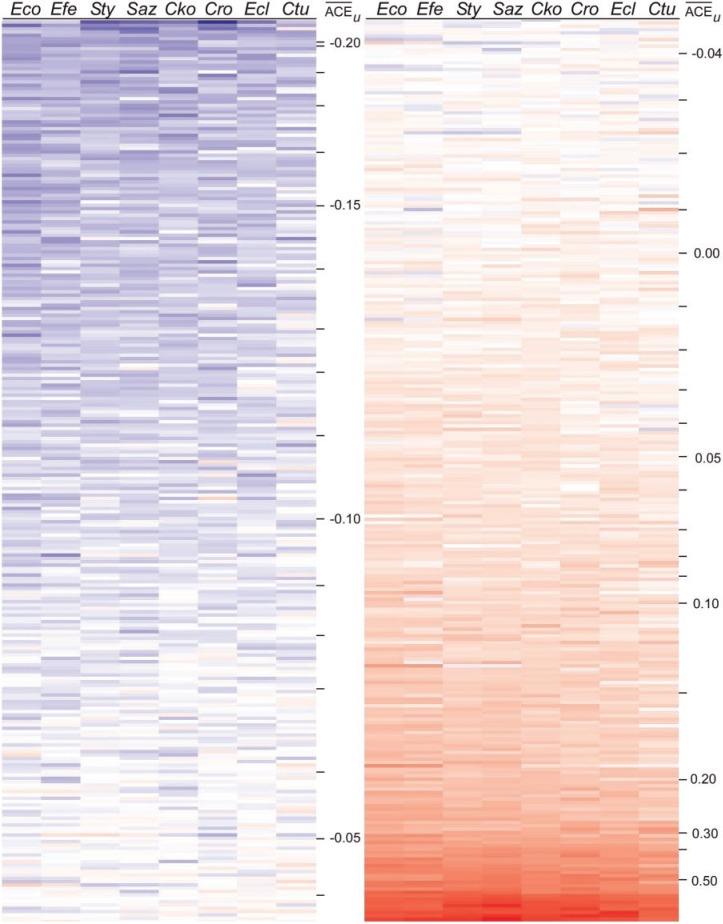
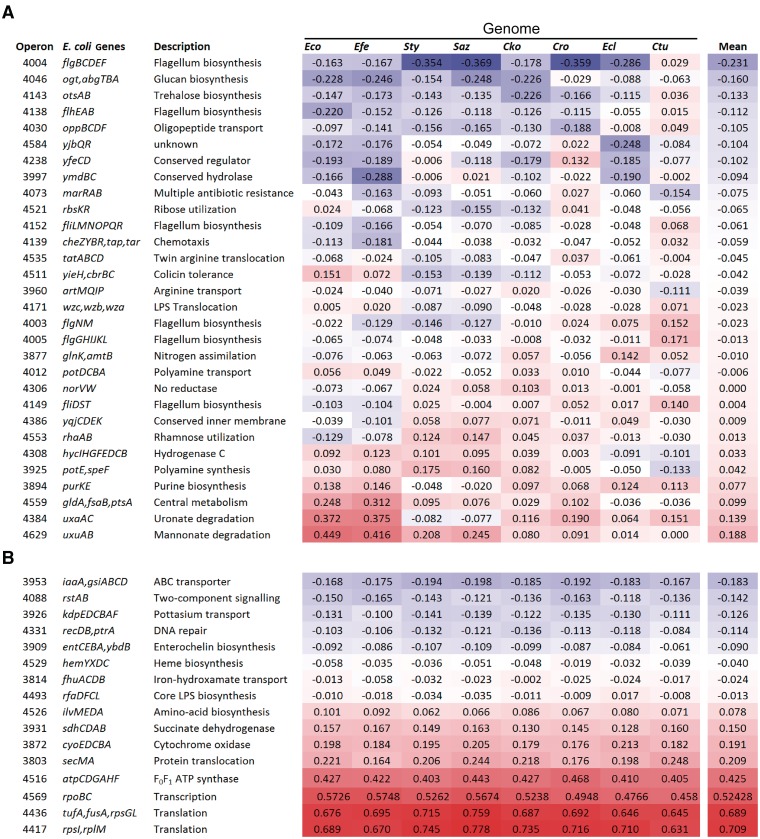
Although table 1 presents that ACEu values for genes in the same operon change in concert, these changes may be modest and may not explain the genes with large differences in ACEu between species (fig. 2). To examine whether operons exhibited significant changes in codon selection as units, we assessed the magnitude of change in ACEu values among 523 orthologous operons in eight genomes of enteric bacteria (figs. 3 and 4); values for operons were calculated as though their constituent genes were a single coding sequence.
Fig. 3.
Heat map of ACEu values for operons containing orthologous genes in eight species of enteric bacteria: Escherichia coli, Escherichia fergusonii, Salmonella enterica Typhimurium, Salmonella enterica Arizonae, Citrobacter koseri, Citrobacter rodentium, Enterobacter cloacae, and Cronobacter turicensis. ACEu values were scaled to E. coli, and operons were sorted by their average ACEu across all eight genomes; values < 0.05 are shaded blue, whereas values > 0.05 are shaded red; darker colors represent more extreme values.
Fig. 4.
A subset of the operons presented in figure 3. (A) Operons with variable levels of codon selection across genomes. (B) Operons with relatively constant levels of codon selection across genomes. Operon numbers correspond to those presented in Dam et al. (2007).
In general, the pattern among orthologous operons mirrors that seen among orthologous genes (fig. 2), with some operons showing substantial differences between genomes (figs. 3 and 4A) even as codon adaptation is broadly conserved (figs. 3 and 4B). The operons showing little change across species (fig. 4B) encode functions that are not expected to change relative importance—e.g., transcription, translation, protein translocation, and ATP generation—among organisms dwelling in different environments. In contrast, others show dramatic changes in the relative degree of codon bias across genomes (fig. 4A). Although the functions of some operons are unknown, several operons showing substantial change in codon usage have known physiological functions. For example, rha genes, responsible for rhamnose degradation, are more highly biased in the two Salmonella genomes. This is not surprising as Salmonella synthesizes coenzyme B12 de novo, allowing it to degrade 1,2-propanediol, a byproduct of rhamnose degradation; therefore, rhamnose utilization may provide a greater benefit in Salmonella than in other bacteria.
No simple rules describe the differences among genomes (figs. 3 and 4); sometimes one operon shows evidence of codon adaptation (red), whereas the cognate operons do not (blue), sometimes the opposite is seen, and sometimes the cognate operons show a more even distribution of high and low ACEu values. The two S. enterica genomes tend to be similar, as do the two Escherichia genomes, yet substantial differences are sometimes apparent even among these pairs of closely related genomes.
Changes in Codon Usage Bias between Genomes Are Significant
The orthologs within some pairs of genomes have strong correlations in their ACEu values (fig. 2A; supplementary table S2, Supplementary Material online), whereas the correlations are weaker for other pairs of genomes (fig. 2B), suggesting that some pairs of genomes have greater differences in their codon adaptation profiles than others. To quantify these differences in terms of discrete locus-specific changes (rather than diffuse variation in all genes), we tested for significant differences in ACEu values between cognate operons in pairs of genomes. The sampling distributions of ACE statistics are approximately normal, and the variance can be expressed analytically (Retchless and Lawrence 2011). The consolidation of genes into operons increases the number of synonymous codons incorporated into each test of statistical significance, while decreasing the overall number of tests performed and summarizing changes according to the shared physiological function of each operon.
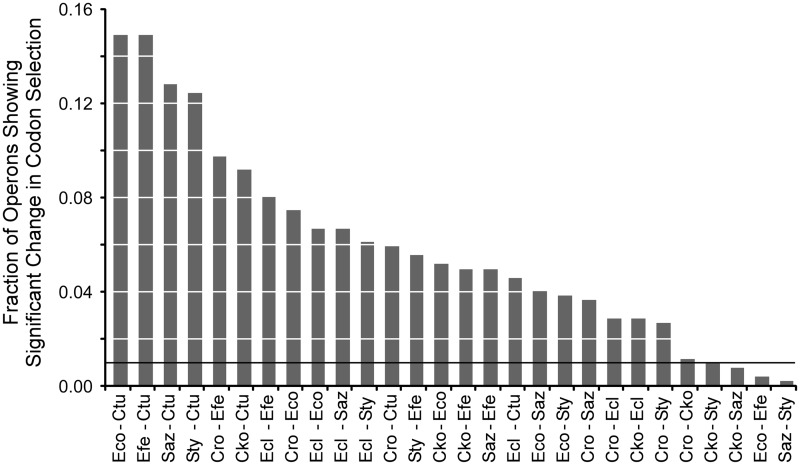
For each of the 28 pairs of genomes that can be drawn from our set of eight taxa, we enumerated the operons with significant differences in ACEu values (P < 0.01, two-tailed Z test; supplementary table S4, Supplementary Material online), normalizing the number of operons as a fraction of the 523 operons examined (fig. 5). Given a significance value of 0.01, we expect 1% of the operons to show this degree of change as the result of simple stochastic variation; this is represented by the dark horizontal line in figure 5. The majority of genome comparisons show a large excess of operons with significant change in ACEu values at P < 0.01. Therefore, we conclude that substantial fractions of these bacterial genomes have experienced changes in codon selection relative to each other, reflected in the significant changes in ACEu values. This conclusion is upheld when one examines changes at either more stringent (P < 0.002) or less stringent (P < 0.05) significance thresholds (supplementary table S4, Supplementary Material online). The exceptions to this trend are not surprising: comparison among closely related genomes (e.g., the two Escherichia species or the two Salmonella species) fail to detect an excess of operons that have changed ACEu values significantly. Here, insufficient time has elapsed for significant differences in codon usage to become manifested.
Fig. 5.
Excess of operons showing significant change in codon selection. The fraction of operons showing differences significant at P < 0.01 is plotted for pairwise comparisons among Escherichia coli, Escherichia fergusonii, Salmonella enterica Typhimurium, Salmonella enterica Arizonae, Citrobacter rodentium, Citrobacter koseri, Enterobacter cloacae, and Cronobacter turicensis. The horizontal line represents the expected fraction that will occur by change alone (1% of the operons). Values were scaled by polynomial regression.
Alternative explanations could account for significant differences in codon usage between orthologous genes, confounding the above examination of differences in codon selection. However, none of the factors that are likely to strongly influence codon composition can explain the widespread observation of significant changes in ACEu, as will be discussed after examining the implications of changes in codon selection among orthologs.
Adjustment to Changes in Codon Selection Saturates within the Enterobacteriaceae
Over time, change in codon selection will lead to change in codon usage, as an increase in selection will result in use of preferred codons or relaxation of selection will allow for fixation of nonpreferred codons. Genes in relatively closely related genomes rarely show significant changes in codon usage (fig. 5). This is expected both because of the ecological similarity in closely related taxa—thus fewer opportunities for a change in expression regimes to produce changes in codon selection—and because there has been less opportunity for adaptive changes to accumulate. One would predict that, at least within bacterial families, the number of genes showing significant changes in codon selection to increase as phylogenetic distance increases.
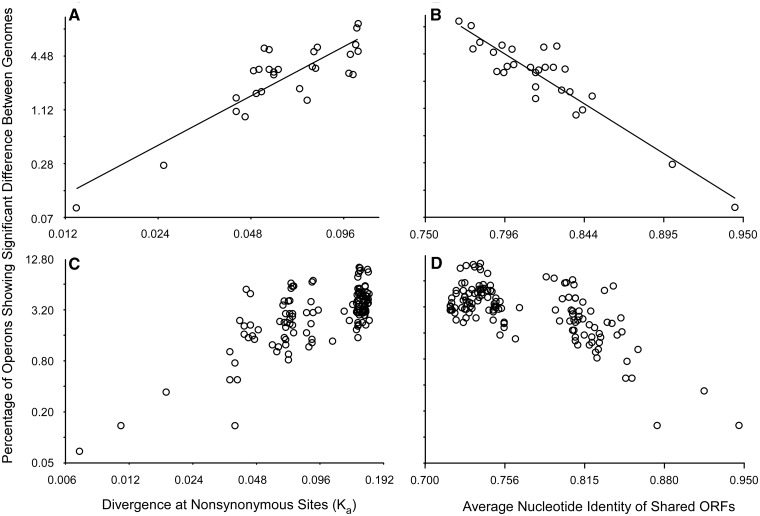
To test the extent to which codon selection can change, we calculated the fraction of 523 operons showing significant differences in codon usage among eight enteric bacteria (supplementary table S4, Supplementary Material online). The overall similarity among these genomes was assessed by either average nucleotide identity or divergence at nonsynonymous sites (Ka) among orthologous genes (Li et al. 1985). As expected, the fraction of operons showing significant changes increased robustly with phylogenetic distance (fig. 6A and B). When one expands the data set to 17 organisms to include more distantly related taxa, smaller numbers of operons are shared among all genomes. Although these data are thus noisier, the fraction of the 391 shared operons that shows significant change in codon usage still increases with phylogenetic distance to a point but then does not increase further (fig. 6C and D); this is evident using either metric of genome distance. This may represent a limit to the extent that codon adaptation profiles can change, which would be expected to occur for two reasons. First, following the initial divergence of the magnitude of codon selection on orthologous operons, the level of selection could subsequently converge, thereby reducing this difference. Second, there may be a limit to the number of operons that can show significant change, as some operons may be consistently highly expressed (e.g., those encoding ribosomal proteins) or weakly expressed.
Fig. 6.
The fraction of operons with significant changes in codon selection increases with phylogenetic distance. (A, B) Pairwise values are reported for Escherichia coli, Escherichia fergusonii, Salmonella enterica Typhimurium, Salmonella enterica Arizonae, Citrobacter rodentium, Citrobacter koseri, Enterobacter cloacae, and Cronobacter turicensis. (C, D) Pairwise values are reported for E. coli, E. fergusonii, S. enterica Typhimurium, S. enterica Arizonae, C. rodentium, C. koseri, E. cloacae, Enterobacter sp. 638, C. turicensis, Dickeya zeae, Klebsiella varicola, Klebsiella pneumoniae, Pectobacterium wasabiae, Erwinia amylovora, Erwinia tasmaniensis, Yersinia enterocolitica, and Serratia proteamaculans.
Relative Contribution of Codon Selection to Speciation
The genetic cohesion of bacterial species may be ascribed to recombination (Dykhuizen and Green 1991), whereby variant alleles are purged and genotypic similarity among strains is maintained. Adaptive changes between lineages contribute to recombination interference, whereby recombinants that lose adaptive changes are counterselected, maintaining the genotypic differences between ecologically distinct classes (Lawrence and Retchless 2009, 2010). This eventually leads to bacterial speciation, where genetic isolation has been imposed at all loci around the chromosome (Lawrence 2002; Retchless and Lawrence 2007, 2010). A major influence on bacterial adaptation has been attributed to genes acquired by lateral gene transfer, whereby introduced genes impart novel physiological functions (Ochman et al. 2000; Hacker and Carniel 2001). Regions first experiencing genetic isolation between incipient species have been associated with the acquisition of genes by lateral transfer (Retchless and Lawrence 2007), suggesting that acquisition of adaptive, physiological differences can drive genetic isolation. Above, we demonstrate that some genes experience significant changes in codon selection. Here, we assess the potential contribution of these adaptive changes to genetic isolation.
To examine the relative contributions of adaptation by gene gain versus adaptive change in codon selection, we compared the genomes of the well-studied enteric bacteria E. coli and S. enterica Typhimurium. We enumerated 125 acquired genes that play likely adaptive roles in E. coli by identifying genes present only in E. coli and E. fergusonii but absent from other enteric bacteria. A similar set of 55 adaptive genes in Salmonella were identified as those restricted to serovars of Salmonella. A set of 665 shared operons was examined for significant changes in codon selection between these two species; dozens of operons were identified with change in ACEu values beyond those predicted by stochastic factors alone (supplementary table S5, Supplementary Material online), with the number dependent on the prescribed significance value. In addition, many individually transcribed genes also showed significant changes in codon selection (supplementary table S5, Supplementary Material online).
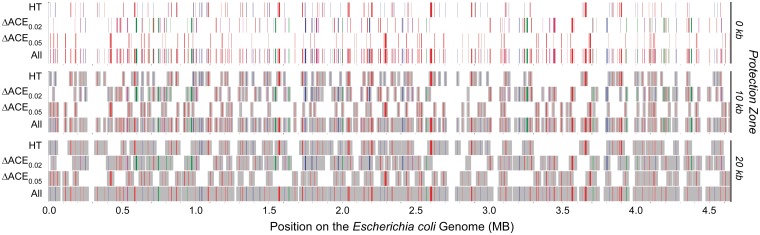
The positions of putative adaptive differences between Escherichia and Salmonella genomes were plotted along the E. coli chromosome (fig. 7); positions of genes gained in Salmonella are assigned to the positions of adjacent, orthologous genes in E. coli. Recombination events are counterselected in the region of DNA around an adaptive locus as these events would affect the adaptive locus itself. Previous analyses have estimated these zones of recombination interference both by finding regions of similar time of divergence (Retchless and Lawrence 2007) and regions with similar phylogenetic history (Retchless and Lawrence 2010); both estimates place the upper boundary of the zone of recombination interference at about 20 kb. When considering a range of sizes for this zone (fig. 7), a large portion of the E. coli chromosome could be protected from recombination by either adaptive gene gains or adaptive changes in codon usage between lineages.
Fig. 7.
Locations of loci inferred to show adaptive change in the divergence of Escherichia coli and Salmonella enterica. Data are presented in three tiers, each with four lanes. Lane 1 shows horizontally transferred genes present in both E. coli and Escherichia fergusonii (red) or both S. enterica Typhimurium and S. enterica Arizonae (blue) but absent from other enterica bacteria; Salmonella loci are plotted by their cognate position on the E. coli chromosome. Lane 2 shows operons that show significant change in ACEu values at P < 0.002 (blue), P < 0.005 (red), P < 0.01 (green), or P < 0.02 (magenta); ACE values were normalized by polynomial regression (see text). Lane 3 shows operons that show significant change in ACEu values at P < 0.05 (red). Lane 4 shows all adaptive loci. Tier 1 shows adaptive loci alone; tier 2 shades 10 kb of flanking DNA on both sides of each adaptive locus; and tier 3 shades 20 kb of flanking DNA.
We have not considered potentially adaptive gene loss events for two reasons. First, these events may be recent, independent events in multiple lineages within a bacterial taxon. Second, deletion events may have been neutral or deleterious. The irreversible nature of gene losses makes their interpretation as adaptive changes more complex than the retention of a protein-coding gene, and subsequent selection against nonsynonymous substitutions following its introduction.
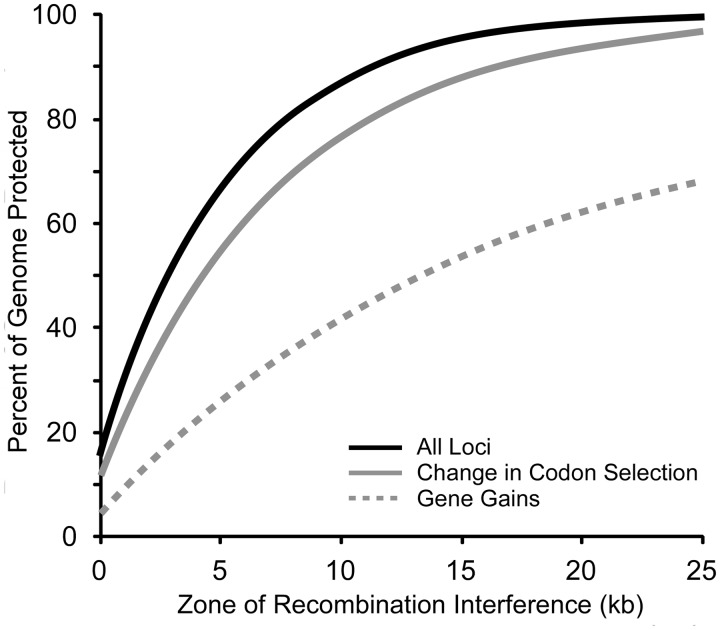
To estimate the relative contributions of adaptive gene gains and adaptive changes in codon selection, we must recognize that some of the loci shown in figure 7 show a large difference in ACEu values by chance alone (fig. 5; supplementary tables S4 and S5, Supplementary Material online). The fraction of genes with large, yet effectively neutral, change increases as the stringency for their identification decreases (supplementary tables S4 and S5, Supplementary Material online). To model recombination interference, we must evaluate only a subset of the genes and operons that are putatively involved in adaptive differences, such that their number corresponds to the number that cannot be accounted for by chance alone (supplementary table S5, Supplementary Material online). To do this, the appropriate numbers of genes and operons were chosen at random, and the percent of the genome affected by recombination interference was calculated; this was repeated 500 times, and the mean fraction of the genome protected was calculated for windows of recombination interference ranging from 0 to 25 kb (fig. 8).
Fig. 8.
Contributions of classes of adaptive loci toward recombination interference. Recombination interference was modeled on both sides of loci of gene insertion and loci showing adaptive changes in codon selection. For the latter, only the fraction of genes beyond those expected by chance are consider; values represent the mean of 500 iterations randomly selecting the identities of genes with potentially adaptive change in codon selection.
We identified 180 potentially adaptive genes acquired by lateral transfer; considered alone, these genes cannot mediate recombination interference across the entirety of the chromosome (fig. 8, dashed line). There are long regions that lack genes recently acquired by lateral transfer. Because the windows for recombination interference are limited to ∼20 kb, genetic isolation of regions far from sites of gene acquisition can be attained by one of two mechanisms. First, the zone of recombination interference could spread from the site of gene acquisition (Lawrence 2002); here, lack of recombination adjacent to sites of insertion leads to the accumulation of substitutions, which themselves provide recombination interference. This would be a very slow process, regardless of the strength of recombination interference adjacent to sites of gene insertion.
Alternatively, recombination interference could be provided by additional adaptive differences, which lie far from sites of gene gain, such as ancestral loci experiencing significant changes in codon selection. Although laterally transferred genes (dashed line) leave large portions of the chromosome initially unaffected by recombination interference, reasonably complete coverage is provided by the combination (black line) of both laterally acquired genes and genes with adaptive changes in codon usage (gray line). Interestingly, the fraction of the genome affected by genes with adaptive change in codon selection far exceeds the fraction affected by laterally acquired genes. Therefore, we conclude that adaptive changes in codon selection could not only play a role in speciation but could be major drivers of recombination interference, and thus genetic isolation, during bacterial speciation. In addition, this disparity may be underestimated for two reasons. First, some foreign genes were likely gained relatively recently, after genetic isolation between E. coli and S. enterica had been achieved, overestimating the contribution of this set of genes to recombination interference. Second, the number of operons with adaptive changes in codon selection is also underestimated, as those arising in the latter stages of genetic isolation between E. coli and S. enterica would share similarity due to their relatively recent separation and would not be recognized as having sufficient numbers of changes.
Codon Selection as a Marker for Ecological Similarity
The degree of codon selection experienced by a gene reflects its relative degree of expression, a measure of its relative importance in the genome—both in terms of nutritional resources dedicated to producing its product and the fitness benefit of optimizing its production. The results above show that the degree of codon selection on individual genes can change between species (fig. 5) and that the number of genes showing significant change in codon selection increases with phylogenetic distance (fig. 6). However, these changes should reflect overall ecology (e.g., fig. 1). Therefore, once synonymous sites have experienced sufficient change, the overall patterns of codon selection should allow insight into the ecological similarities between genomes. Genomes of closely related organisms that acquire dissimilar patterns of codon selection rapidly may have exploited substantially distinct ecologies. In contrast, more distantly related genomes that converge on similar patterns of codon selection among their shared orthologs may be exploiting similar ecologies.
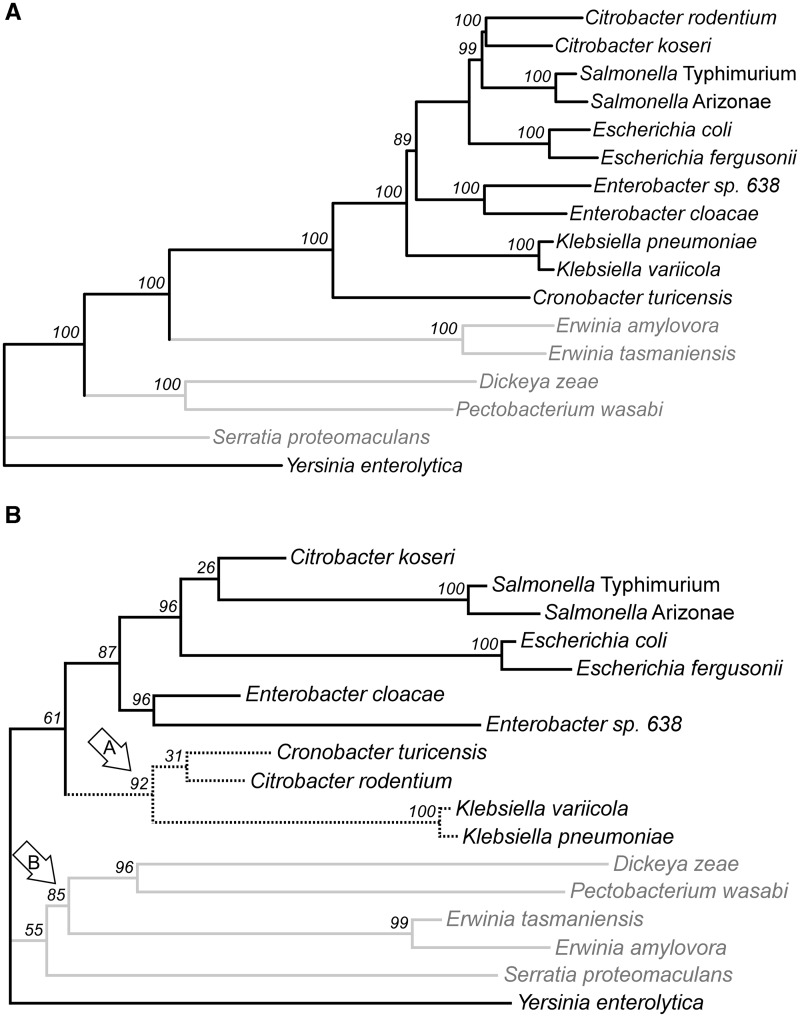
To examine this, we looked at overall similarity in codon selection among 1,460 genes shared among 17 species of enteric bacteria. We examined the similarity in codon selection patterns by principal components analysis; a neighbor-joining tree was constructed using the Euclidean distance between the first nine principal components of ACEu values (fig. 9B); this captured clustering of genes by similarity in codon selection but avoided the noise of the weakly contributing dimensions. To retain methodological similarity, we evaluated phylogenetic relationships as a neighbor-joining tree using average divergence at nonsynonymous sites (fig. 9A). Comparison of these topologies show some expected similarities. Closely related taxa (species of Escherichia, Salmonella, Erwinia, Enterobacter, and Klebsiella) cluster as sister lineages in both analyses. However, two species of Citrobacter are well separated when considering codon selection (fig. 9B, arrow A), and their separation is well supported by bootstrap resampling of genes; this suggests that these lineages are ecologically dissimilar. More dramatically, three separate lineages of plant-associated species (noted in gray in fig. 9) are clustered when considering codon selection patterns, reflecting their ecological similarity; the association of two of these lineages is strongly supported (fig. 9B, arrow B).
Fig. 9.
Relationships among enteric bacteria based on overall similarity. (A) Protein similarity (dN). Difference is calculated as weighted average of divergence at nonsynonymous sites (Yang and Nielsen 2000) among protein-coding genes found among all taxa. Smaller distances reflect greater average similarity of amino acid sequences. Numbers at nodes represent percentage of 1,000 bootstrap replicates bearing that node. (B) Similarity in codon selection (ACEu) is calculated as Euclidean distance of the first nine principal components when comparing average degree of codon usage bias among shared protein coding genes. Smaller distances reflect greater congruence in inferred degree of codon selection. Numbers at nodes represent percentage of 100 bootstrap replicates bearing that node.
To examine the relative impact of ecology versus phylogeny in determining similarities of codon selection among orthologs, we selected sets of four bacterial genomes that were sequenced as part of the Human Microbiome Project (Group et al. 2009). The organisms represented four different families from the same bacterial division, with two species each colonizing one of two anatomical locations (among oral cavity, skin, gastrointestinal tract, or urogenital tract). As above, we assessed affinity of strains based on protein similarity or based on similarity of patterns of codon usage as assessed by ACEu. In all cases, patterns of codon usage bias were most similar between organisms that were isolated from the same body location. In the majority of these cases, bacteria from the same environment were also most closely related based on protein similarity; therefore, the similarity of patterns of codon usage could simply reflect phylogenetic similarity. However, in three of these tetrads, codon usage similarity was most similar between the bacteria that were isolated from the same body site, even though these bacteria were not the most closely related (supplementary table S6, Supplementary Material online). Although small sample sizes does not allow for statistical support (P = 0.13), the data trend shows that the similarity in patterns of codon usage bias among shared genes reflects convergence driven by adaptation to a particular habitat, not phylogenetic history. These data are consistent with the hypothesis that genes shared among organisms alter their patterns of codon usage to reflect changes in codon selection accorded by their current ecological niche.
Potential Nonadaptive Sources for Differing Codon Usage
More genes and operons than expected show significant differences in ACEu values between species (fig. 5). We interpreted this excess as evidence for changes in the degree of codon selection between species. However, it is possible that other factors have led to changes in codon usage. Here, we address three possible sources of such differences.
Change in Strand Identity
First, mutational biases are not equivalent between genes transcribed from leading and lagging strands (Lobry 1996; Rocha 2004). Therefore, genes changing orientation relative to the replication origin would experience slightly different mutational pressures, which could lead to greater changes in codon usage than expected. To examine this possibility, we identified those orthologs which were not orientated in the same direction relative to their respective replication origins. Replication origins and termini were identified by homology to the E. coli oriC and dif regions (Kono et al. 2011); in all cases, these corresponded to regions where strand-specific nucleotide patterns invert, as measured by cumulative GC-skew and octameric skew (Hendrickson and Lawrence 2007).
Of the 2,235 genes shared among eight enteric species, between 2 and 171 genes were encoded on different strands in comparisons of two genomes (supplementary table S7, Supplementary Material online). Most comparisons showed between 2 and 10 genes changing strand identity; a large inversion in Citrobacter rodentium led to greater numbers changing strand identity in comparisons involving this taxon. When considering genes with significant changes in ACEu between species, only between 0 and 11 of these genes changed strand identity (supplementary table S7, Supplementary Material online). As a result, the fractions of genes showing significant changes in ACEu do not change significantly when restricting the analysis to genes that retain strand identity (supplementary table S7, Supplementary Material online). Therefore, we conclude that change in strand identity is not responsible for the large number of genes showing significant changes in codon usage.
Xenologous Genes
Alternatively, large differences in ACEu values for some orthologs could reflect a history of gene transfer. ACEu values are calculated relative to the mutational biases of the resident genome; newly acquired xenologs could show unexpected changes in codon usage due to differences in mutational biases between donor and recipient genomes, not changes in selection. Although our data set is stringent in its requirement for strong similarity among genes shared among all taxa, and putative orthologs are largely syntenic (data not shown), it is possible that some xenologous genes are present in the data set.
To determine whether large portions of those genes with significant changes in ACEu values are simply of foreign origin, we inferred the phylogeny of each gene using maximum likelihood and then compared it with the species phylogeny. Using a Shimodaira–Hasegawa test (Shimodaira and Hasegawa 1999), 25 of 2,235 genes rejected the species tree at P < 0.01. Given that 1% of genes should reject at this confidence level, this is not greater than expected. Even if we accept that the 25 genes with P values less than 0.01 are potentially xenologous, only between 0 and 3 of these genes showed significant changes in ACEu values between species (supplementary table S8, Supplementary Material online). As a result, the fraction of genes showing significant changes in ACEu between species does not change when considering only genes whose phylogeny does not reject the species phylogeny (supplementary table S8, Supplementary Material online). Therefore, we conclude that foreign origin is not responsible for the large number of genes showing significant changes in codon usage.
Selection for Change in Protein Sequence
Finally, significant changes in codon usage may be driven by large numbers of changes in amino acid sequences; this may occur in genes whose protein products are exposed on the cell surface and experience pressure to change due to exposure to hosts’ immune systems (Salazar-Gonzalez and McSorley 2005) or natural predators (Wildschutte et al. 2004). Although genes known to experience diversifying selection (e.g., those encoding flagellins) are not included in our data set, this does not remove the possibility that other have escaped our attention. Molloy et al. (2000) identified 58 proteins resident in E. coli’s outer membrane, thus potentially experiencing accelerated rates of protein change. Orthologs of 28 of these genes were found in eight enteric bacterial genomes and are thus present in our data set (supplementary table S9, Supplementary Material online). Of the genes showing significant change in ACEu between species, only between 0 and 5 encoded outer-membrane proteins (supplementary table S9, Supplementary Material online). As a result, the fraction of genes showing significant changes in ACEu between species does not change when considering only genes whose products are not surface exposed. Therefore, we conclude that selection for change in protein sequence is not responsible for the large number of genes showing significant changes in codon usage.
Discussion
Speciation Cascades
Speciation typically involves two classes of changes within descendent lineages. First, ecological differences allow for coexistence; without these differences, competition among sympatric taxa will eliminate the less fit group (Van Valen 1976). Second, genetic isolation both privatizes adaptive mutations within species and prevents the formation of less fit, cross-species progeny (Gevers et al. 2005). In bacteria, these changes are somewhat decoupled at the organismal level. Significant ecological changes may arise by very few genomic changes, such as the acquisition of genomic islands conferring complex traits after their acquisition (Lawrence 1997; Hacker and Carniel 2001). These adaptive differences lead to recombination interference at their underlying loci. However, because bacterial recombination operates on small portions of the genome, the majority of the genome may still experience allelic exchange (Lawrence 2002). Among recombinogenic taxa, then, genetic isolation of the entire chromosome takes place over a long period of time as adaptive changes that catalyze recombination interference must occur throughout the chromosome (Retchless and Lawrence 2007). The lack of recombination leads to the accumulation of neutral genetic differences (Lawrence 2002), which themselves provide a further barrier to recombination by disrupting the formation of heteroduplexes (Vulic et al. 1997, 1999).
Horizontally transferred genes are good candidates for the motivators of ecological change. Acquired genes can provide novel functions allowing for rapid adaptation and effective competition within novel ecological niches. However, changes in gene inventory are not observed across the entire chromosome (fig. 7). Moreover, because an adaptive locus catalyzes recombination interference over a distance of ∼20 kb, more than 200 evenly spaced gene acquisitions would be required to confer genetic isolation across the entire genome. This is more than are observed; therefore, we conclude that there must be another source of adaptive change that drives genetic isolation. Here, we show that another class of adaptive mutations—changes in codon usage driven by shifts in the degree of codon selection—likely play a key role in speciation by initiating recombination interference at otherwise conserved loci.
We envision speciation as a cascade of adaptive events leading from ecological differentiation through genetic isolation. First, the acquisition of genes by lateral transfer initiates ecological differentiation. This may occur among numerous populations within a freely recombining bacterial species by independent acquisitions. Although these adaptive changes promote recombination interference at their sites of insertion (fig. 7), other regions will recombine freely, leading to their genotypic similarity between ecologically distinct populations. However, the ecological changes initiated by the gene gains will lead to changes in relative levels of gene expression as organisms experience a different suite of environments than those experienced by the ancestral taxon. The differences in gene expression promote subsequent changes in codon selection. Over time, the accumulation of adaptive changes at synonymous sites will also provide recombination interference due to natural selection against the recombinant genotype. This additional constraint on gene exchange ultimately leads to complete genetic isolation and the formation of genetically independent species in the Mayrian sense (Mayr 1942, 1963).
Inspection of figure 8 shows that, at least in the case of the divergence of E. coli and Salmonella, adaptive changes in codon selection alone can mediate recombination interference across nearly the entirely of the bacterial chromosome. That is, it is not necessary to invoke numerous adaptive gene gains to provide substrates for recombination interference and hence genetic isolation. Rather, a single gene gain—if it allows for exploitation of a sufficiently distinct ecological niche—may lead to genetic isolation across the entirely of the bacterial chromosome by virtue of the secondary cascade of adaptive changes in codon selection.
Two Stages of Speciation
Genes acquired by lateral transfer play a major role in changing organisms’ favored ecological niches; acquired genes can confer novel functions and allow effective competition in new environments by virtue of the novel biochemical processes they encode. In contrast, although changes in codon selection can lead to adaptive changes in codon usage, these adaptations do not underlie physiological differences between strains. Although adaptive changes in codon usage may drive the majority of genetic isolation between bacterial species, they do not contribute to the physiological differences between species.
When we consider the two aspects of species formation—acquisition of ecological differences and achievement of genetic isolation—it is clear that ecological differences arise first. Ecologically distinct strains are readily created by gene transfer, given the high rate of influx of alien genes (Lawrence and Ochman 1997, 1998), any one of which may significantly alter the recipient’s physiology. Ecologically distinct strains are abundantly evident in collections of isolates of nearly any named species (Walk et al. 2009; Luo et al. 2011). Although it is not easy to predict which genetic variants have led to significant changes in underlying ecology, likely candidates can be identified as those with significantly different distributions in the environment (Gordon et al. 2002; Gordon and Cowling 2003; Luo et al. 2011). It is for this reason that such variant lineages, termed ecotypes (Cohan 2002), are often viewed as bacterial "species," as they exhibit one of the two hallmarks of eukaryotic species.
However, even ecologically distinct strains may continue to recombine at loci that are unencumbered by adaptive change. For groups experiencing little recombination, genetic isolation arises by the accumulation of neutral mutations alone (Hanage et al. 2006; Fraser et al. 2007). However, many groups experience relatively high rates of gene exchange; here, the likelihood of inheriting a novel allele by recombination can exceed that of acquiring a variant allele by mutation (Feil et al. 1999, 2000; Maynard Smith et al. 2000). In these recombining groups, genetic isolation is achieved only after adaptive changes have arisen at a large number of loci around the bacterial chromosome (Lawrence 2002; Retchless and Lawrence 2007). The differences in these time scales had led to confusion and discussion as to the nature of bacterial species (Luo et al. 2011). Confusion arises because the long time frame for genetic isolation of bacterial species would seemingly ignore the potentially dramatic ecological and physiology distinctions between closely related taxa. As a result, the term "speciation" has been used to reflect two different processes: the acquisition of ecological differences and the establishment of genetic isolation (Gevers et al. 2005).
The work presented in this study provides a comprehensive model for bacterial speciation that integrates the processes of ecological differentiation and genetic isolation. Rapid ecological differentiation is provided by gene acquisition events or biochemical adaptations of existing genes, and long-term genetic isolation is conferred by the slow acquisition of adaptive changes in codon usage. Because the rate of accumulation of adaptive changes in codon usage is a function of the overall substitution rate, we expect this to take a significant period of time. Estimates of the rates of speciation, the numbers of species, the precise placement of speciation boundaries, or other quantitative metrics of bacterial relationships are only useful when placed in the context of ecological differentiation, genetic isolation, or both processes.
Supplementary Material
Supplementary tables S1–S9 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by grant GM078092 from the NIH to J.G.L. and a Mellon Fellowship to A.C.R.
References
- Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan FM. What are bacterial species? Annu Rev Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- Dam P, Olman V, Harris K, Su Z, Xu Y. Operon prediction using both genome-specific and general genomic information. Nucleic Acids Res. 2007;35:288–298. doi: 10.1093/nar/gkl1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE, Green L. Recombination in Escherichia coli and the definition of biological species. J Bacteriol. 1991;173:7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Maiden MC, Achtman M, Spratt BG. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol Biol Evol. 1999;16:1496–1502. doi: 10.1093/oxfordjournals.molbev.a026061. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Smith JM, Enright MC, Spratt BG. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics. 2000;154:1439–1450. doi: 10.1093/genetics/154.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Hanage WP, Spratt BG. Recombination and the nature of bacterial speciation. Science. 2007;315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Cohan FM, Lawrence JG, et al. (11 co-authors) Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- Gordon DM, Bauer S, Johnson JR. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology. 2002;148:1513–1522. doi: 10.1099/00221287-148-5-1513. [DOI] [PubMed] [Google Scholar]
- Gordon DM, Cowling A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology. 2003;149:3575–3586. doi: 10.1099/mic.0.26486-0. [DOI] [PubMed] [Google Scholar]
- Group NHW, Peterson J, Garges S, et al. (40 co-authors) The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001;2:376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanage WP, Spratt BG, Turner KM, Fraser C. Modelling bacterial speciation. Philos Trans R Soc Lond B Biol Sci. 2006;361:2039–2044. doi: 10.1098/rstb.2006.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson H, Lawrence JG. Mutational bias suggests that replication termination occurs near the dif site, not at Ter sites. Mol Microbiol. 2007;64:42–56. doi: 10.1111/j.1365-2958.2007.05596.x. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrance of the respective codons in its protein genes. J Mol Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Karlin S, Mrazek J. Predicted highly expressed genes of diverse prokaryotic genomes. J Bacteriol. 2000;182:5238–5250. doi: 10.1128/jb.182.18.5238-5250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono N, Arakawa K, Tomita M. Comprehensive prediction of chromosome dimer resolution sites in bacterial genomes. BMC Genomics. 2011;12:19. doi: 10.1186/1471-2164-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG. Gene transfer in bacteria: speciation without species? Theor Popul Biol. 2002;61:449–460. doi: 10.1006/tpbi.2002.1587. [DOI] [PubMed] [Google Scholar]
- Lawrence JG. Selfish operons and speciation by gene transfer. Trends Microbiol. 1997;5:355–359. doi: 10.1016/S0966-842X(97)01110-4. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci U S A. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Retchless AC. The interplay of homologous recombination and horizontal gene transfer in bacterial speciation. Methods Mol Biol. 2009;532:29–53. doi: 10.1007/978-1-60327-853-9_3. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Retchless AC. The myth of bacterial species and speciation. Biol Phil. 2010;25:569–588. [Google Scholar]
- Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci U S A. 2011;108:7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Feil EJ, Smith NH. Population structure and evolutionary dynamics of pathogenic bacteria. BioEssays. 2000;22:1115–1122. doi: 10.1002/1521-1878(200012)22:12<1115::AID-BIES9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Mayr E. Systematics and the origin of species. New York: Columbia University Press; 1942. [Google Scholar]
- Mayr E. Animal species and evolution. Cambridge: Harvard University Press; 1963. [Google Scholar]
- Molloy MP, Herbert BR, Slade MB, Rabilloud T, Nouwens AS, Williams KL, Gooley AA. Proteomic analysis of the Escherichia coli outer membrane. Eur J Biochem. 2000;267:2871–2881. doi: 10.1046/j.1432-1327.2000.01296.x. [DOI] [PubMed] [Google Scholar]
- Mrazek J, Bhaya D, Grossman AR, Karlin S. Highly expressed and alien genes of the Synechocystis genome. Nucleic Acids Res. 2001;29:1590–1601. doi: 10.1093/nar/29.7.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman E. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2010;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retchless AC, Lawrence JG. Temporal fragmentation of speciation in bacteria. Science. 2007;317:1093–1096. doi: 10.1126/science.1144876. [DOI] [PubMed] [Google Scholar]
- Retchless AC, Lawrence JG. Phylogenetic incongruence arising from fragmented speciation in enteric bacteria. Proc Natl Acad Sci U S A. 2010;107:11453–11458. doi: 10.1073/pnas.1001291107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retchless AC, Lawrence JG. Quantification of codon selection for comparative bacterial genomics. BMC Genomics. 2011;12:374. doi: 10.1186/1471-2164-12-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004;150:1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salazar-Gonzalez RM, McSorley SJ. Salmonella flagellin, a microbial target of the innate and adaptive immune system. Immunol Lett. 2005;101:117–122. doi: 10.1016/j.imlet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 2005;33:1141–1153. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilus, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster, and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988;16:8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li W-H. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 1987a;4:222–230. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li W-H. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987b;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- Suzuki Y, Gojobori T. A method for detecting positive selection at single amino acid sites. Mol Biol Evol. 1999;16:1315–1328. doi: 10.1093/oxfordjournals.molbev.a026042. [DOI] [PubMed] [Google Scholar]
- Van Valen L. Ecological species, multispecies, oaks. Taxon. 1976;25:223–239. [Google Scholar]
- Vulic M, Dionisio F, Taddei F, Radman M. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in Enterobacteria. Proc Natl Acad Sci U S A. 1997;94:9763–9767. doi: 10.1073/pnas.94.18.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulic M, Lenski RE, Radman M. Mutation, recombination, and incipient speciation of bacteria in the laboratory. Proc Natl Acad Sci U S A. 1999;96:7348–7351. doi: 10.1073/pnas.96.13.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walk ST, Alm EW, Gordon DM, Ram JL, Toranzos GA, Tiedje JM, Whittam TS. Cryptic lineages of the genus Escherichia. Appl Environ Microbiol. 2009;75:6534–6544. doi: 10.1128/AEM.01262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildschutte H, Wolfe DM, Tamewitz A, Lawrence JG. Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. Proc Natl Acad Sci U S A. 2004;101:10644–10649. doi: 10.1073/pnas.0404028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.