Abstract
Genome reduction in obligately intracellular bacteria is one of the most well-established patterns in the field of molecular evolution. In the extreme, many sap-feeding insects harbor nutritional symbionts with genomes that are so reduced that it is not clear how they perform basic cellular functions. For example, the primary symbiont of psyllids (Carsonella) maintains one of the smallest and most AT-rich bacterial genomes ever identified and has surprisingly lost many genes that are thought to be essential for its role in provisioning its host with amino acids. However, our understanding of this extreme case of genome reduction is limited, as genomic data for Carsonella are available from only a single host species, and little is known about the functional role of “secondary” bacterial symbionts in psyllids. To address these limitations, we analyzed complete Carsonella genomes from pairs of congeneric hosts in three divergent genera within the Psyllidae (Ctenarytaina, Heteropsylla, and Pachypsylla) as well as complete secondary symbiont genomes from two of these host species (Ctenarytaina eucalypti and Heteropsylla cubana). Although the Carsonella genomes are generally conserved in size, structure, and GC content and exhibit genome-wide signatures of purifying selection, we found that gene loss has remained active since the divergence of the host species and had a particularly large impact on the amino acid biosynthesis pathways that define the symbiotic role of Carsonella. In some cases, the presence of additional bacterial symbionts may compensate for gene loss in Carsonella, as functional gene content indicates a high degree of metabolic complementarity between co-occurring symbionts. The genomes of the secondary symbionts also show signatures of long-term evolution as vertically transmitted, intracellular bacteria, including more extensive genome reduction than typically observed in facultative symbionts. Therefore, a history of co-evolution with secondary bacterial symbionts can partially explain the ongoing genome reduction in Carsonella. However, the absence of these secondary symbionts in other host lineages indicates that the relationships are dynamic and that other mechanisms, such as changes in host diet or functional coordination with the host genome, must also be at play.
Keywords: amino acid biosynthesis, Carsonella, endosymbiont, gene loss, purifying selection
Introduction
Genome reduction is pervasive in obligately intracellular bacteria and has been observed in both pathogens and mutualists across a wide range of bacterial and host taxa (Andersson and Kurland 1998; Moran et al. 2008). The most extreme examples of bacterial genome reduction are found in insect endosymbionts that provide essential nutrients to their hosts. These include well-known systems such as the symbiosis between Buchnera and aphids (Shigenobu et al. 2000), but, in recent years, far more extreme cases have been identified in the bacterial symbionts of other host insects (reviewed in McCutcheon and Moran 2011). The first such case was discovered with the complete genome sequencing of Candidatus Carsonella ruddii (hereafter referred to as Carsonella) from the hackberry petiole gall psyllid, Pachypsylla venusta (Nakabachi et al. 2006). Carsonella is a member of the Gammaproteobacteria and appears to be universally present in psyllids, with phylogenetic evidence indicating an ancient history of vertical transmission and co-speciation between host and symbiont (Thao et al. 2000a; Spaulding and von Dohlen 2001; Thao et al. 2001). Carsonella inhabits specialized cells (bacteriocytes) inside an abdominal organ (the bacteriome), where the bacteria are believed to play an essential role in synthesizing amino acids that are lacking from the host’s diet of phloem sap (Buchner 1965; Fukatsu and Nikoh 1998; Nakabachi et al. 2006).
When it was first sequenced, the Carsonella genome represented by far the smallest (160 kb) and most AT-rich (83.4%) bacterial genome yet identified, with a highly reduced gene content that fell well short of predicted requirements for a minimal cellular genome (Nakabachi et al. 2006). Furthermore, many of its genes were proposed to be non-functional based on the loss of key domains or catalytic residues (Tamames et al. 2007). For many other genes, homologs could not be identified in other species, probably due to the extreme sequence divergence, adding further uncertainty to the characterization of functional elements. The extreme nature of genome reduction and sequence divergence in Carsonella raises questions about how it is capable of performing basic cellular functions and fulfilling its inferred role as a nutritional symbiont.
Similar examples of highly reduced genomes have been discovered in other insect symbionts, including Tremblaya in mealybugs (139 kb), Hodgkinia in cicadas (144 kb), and Zinderia in spittlebugs (209 kb) (McCutcheon et al. 2009; McCutcheon and Moran 2010; McCutcheon and von Dohlen 2011). In all three of these cases, the bacteria co-exist with a second intracellular species, with the pair of symbionts exhibiting evidence of metabolic complementarity. In the most striking example, the primary bacterial symbiont of mealybugs (Tremblaya) has acquired its own intracellular bacteria (Moranella), creating a nested structure of cells-within-cells-within-cells (von Dohlen et al. 2001). Although the structure of the mealybug–Tremblaya–Moranella relationship is unprecedented, the co-occurrence of multiple bacterial endosymbionts in the same insect host is common. Such bacteria are typically classified as either primary or secondary symbionts. Primary symbionts are generally restricted to bacteriocytes and characterized by a mutually obligate relationship with the host and an ancient history of vertical transmission and co-speciation. In contrast, secondary symbionts are evolutionarily younger. They engage in a diverse range of (often facultative) relationships and can show evidence of horizontal transfer both within and among host species (Oliver et al. 2010).
Many psyllids harbor a second bacteriome-associated symbiont that inhabits the syncytium found between bacteriocytes in the bacteriome (Buchner 1965; Fukatsu and Nikoh 1998; Subandiyah et al. 2000). Like Carsonella, these bacteria are vertically transmitted from mother to offspring, but their relationships with psyllids originated more recently and from multiple independent events (Buchner 1965; Subandiyah et al. 2000; Thao et al. 2000b). Despite the prevalence of these secondary symbionts in psyllids, they appear to be absent from P. venusta (Nakabachi et al. 2006). Therefore, it is not clear whether this host species (and the lone sequenced Carsonella genome) are broadly representative of psyllid diversity. For example, it has been hypothesized that the gall-forming lifestyle of hackberry psyllids in the genus Pachypsylla may provide a more robust nutritional environment, thereby reducing their dependence on intracellular bacteria (Spaulding and von Dohlen 2001). The lack of genomic data from secondary symbionts or Carsonella strains in other psyllids is a significant impediment to understanding the evolutionary mechanisms responsible for extreme genome reduction in Carsonella.
In this study, we compared complete Carsonella genome sequences from closely related pairs of host species in three different psyllid genera. This nested sampling approach allowed us to examine the patterns of recent sequence divergence, while still sampling Carsonella diversity from distantly related lineages within the family Psyllidae. These lineages include hosts that harbor secondary symbionts, and we report complete genomes from two of these symbionts. On the basis of these data, we address the following questions: 1) To what extent is the Carsonella genome conserved and evolving under functional constraint across host lineages? 2) Is gene loss and genome reduction an ongoing process in Carsonella? and 3) What role do secondary symbionts in psyllids play in Carsonella genome reduction?
Materials and Methods
Sampling and DNA Extraction
Two different methods were used to generate DNA samples for genome sequencing. Total insect DNA from a pool of multiple host individuals was used for four psyllid species (Ctenarytaina eucalypti, Ctenarytaina spatulata, Heteropsylla cubana, and Heteropsylla texana). These samples were originally produced as part of a larger phylogenetic study, and the methods for sample collection and DNA extraction have been described previously (Thao et al. 2000a). In addition, nipple galls were collected from the leaves of a single hackberry tree in August 2011 in New Haven, CT. Hackberry nipple galls can be inhabited or colonized by a complex of very closely related psyllid species that are difficult to distinguish (Lewis and Walton 1964; Yang and Mitter 1994; Yang et al. 2001). Therefore, following previous convention (Thao et al. 2000a; Straka et al. 2010), we refer to the hackberry nipple gall psyllids generically as Pachypsylla celtidis. Psyllid nymphs were removed from these galls and dissected in buffer containing 35 mM Tris–HCl, pH 7.4, 25 mM KCl, 100 mM ethylenediaminetetraacetic acid (EDTA), and 250 mM sucrose. DNA was extracted from a pool of bacteriomes from 20 dissected nymphs, using a Qiagen QiaAmp DNA Micro Kit. Approximately 10 ng of extracted DNA was then used as a template for whole genome amplification with the Qiagen REPLI-g Mini Kit.
DNA Sequencing
DNA samples were used to construct paired-end libraries with insert sizes of approximately 400 bp, which were sequenced on an Illumina HiSeq 2000. An initial run was performed with a pair of libraries (from C. eucalypti and P. celtidis) generated with eight cycles of polymerase chain reaction (PCR) amplification, using Agilent PFU Ultra II polymerase. These libraries were sequenced as part of a larger multiplexed pool in a single 2 × 76 bp lane. Preliminary analysis of the resulting data found that coverage of the Carsonella genomes was highly uneven (supplementary fig. S1, Supplementary Material online). As observed previously with Illumina sequencing (Aird et al. 2011), read depth was strongly related to nucleotide composition, with little or no coverage of the most AT-rich portions of the Carsonella genomes (supplementary fig. S1, Supplementary Material online). In contrast, the data provided much more even coverage of the secondary symbiont genome in C. eucalypti, which has a relatively neutral GC content (table 1). Therefore, data from the initial C. eucalypti library were used to assemble the secondary symbiont genome, but new libraries were constructed for all species, using fewer PCR cycles (four) and a different polymerase (KAPA Bio HiFi) to reduce nucleotide composition bias. The resulting libraries were sequenced as part of a larger multiplexed pool in a single 2 × 101 bp lane. The modified library construction methods produced dramatically improved depth and evenness of Carsonella sequencing coverage (supplementary fig. S1, Supplementary Material online), facilitating complete genome assembly. All Illumina library constructions and sequencing were performed at the Yale Center for Genome Analysis.
Table 1.
Summary of Sequenced Symbiont Genomes.
| Genome Size (bp) | G + C Content (%) | Protein Genes | rRNA Genes | tRNA Genes | GenBank | |
|---|---|---|---|---|---|---|
| Primary symbionts (Carsonella) | ||||||
| Ctenarytaina eucalypti | 162,589 | 14.0 | 190 | 3 | 28 | CP003541 |
| Ctenarytaina spatulata | 162,504 | 14.2 | 190 | 3 | 28 | CP003542 |
| Heteropsylla cubana | 166,163 | 14.2 | 192 | 3 | 28 | CP003543 |
| Heteropsylla texana | 157,543 | 14.6 | 180 | 3 | 28 | CP003544 |
| Pachypsylla celtidis | 159,923 | 15.6 | 183 | 3 | 28 | CP003545 |
| Pachypsylla venusta | 159,662 | 16.6 | 182 | 3 | 28 | AP009180 |
| Secondary symbionts | ||||||
| Ctenarytaina eucalypti | 1,441,139 | 43.3 | 918 | 3 | 40 | CP003546 |
| Heteropsylla cubana | 1,121,596 | 28.9 | 576 | 3 | 38 | CP003547 |
Genome Assembly and Annotation
Initial assembly of bacterial symbiont genomes was performed with Velvet v1.1.06 (Zerbino and Birney 2008), using a k-mer of either 41 or 51 bp (for 76 and 101 bp read lengths, respectively). Because sequencing produced exceptionally high Carsonella coverage (>>100×), only a subset of reads corresponding to an average coverage of 50–100× was used for each initial assembly. Coverage levels for the secondary symbiont genomes of C. eucalypti and H. cubana were significantly lower (but still suitable for complete genome assembly), so the full sequencing datasets were used for these genomes. Assembled contigs corresponding to the target bacterial genomes were distinguished from contigs from the host and other bacteria based on a combination of homology to published sequences, GC content, and read depth. For each genome, these contigs were oriented into a single circular scaffold using connectivity data from Illumina read pairs.
Illumina read pairs mapping to each circular scaffold were then identified with SOAP v2.21 (Li et al. 2009) and re-assembled de novo with MIRA v3.4.0 (Chevreux et al. 1999). Read pairs were included even if only one of the two sequences mapped to the original scaffold. MIRA was run with quality set to normal, paired-end insert size constrained to 100–600 bp, and the uniform read distribution setting disabled. MIRA and Velvet assemblies were subsequently merged to eliminate any gaps that were unique to one of the two assemblies. MIRA generally produced assemblies with fewer gaps than Velvet, but the initial Velvet assembly and SOAP read mapping were essential, because MIRA performed poorly when provided with unfiltered sequence data, consisting mostly of host reads.
Remaining assembly gaps typically coincided with regions of reduced read quality and coverage (e.g., long homopolymers). Most of these gaps could be closed by mapping the full set of reads to these regions and performing local reassemblies. In some cases, automated assembly failed to close gaps, but inspection of contig ends in Tablet v1.12.03.26 (Milne et al. 2010) showed clear regions of overlap. In such cases, contig ends were aligned and joined manually using MEGA v5.0 (Tamura et al. 2011). This approach allowed for the closing of all but three gaps (two in Carsonella from C. spatulata and one in Carsonella from P. celtidis), which all coincided with extremely long homopolymers (>25 bp) and were closed by Sanger sequencing of cloned PCR products (Promega pGEM-T Easy Vector Kit).
To identify and correct any small indel or base substitution errors introduced in the assembly process, the full set of Illumina reads were mapped onto each closed genome with SOAP as described previously (Sloan et al. 2012). Mapping coverage data were also analyzed to identify signatures of misassemblies. We found one region spanning the 3′ end of ileS and 5′ end of rluD with abnormally high read depths in the Carsonella genomes of both Heteropsylla species (more than 5-fold above average). In both cases, the region also exhibited a reduction in the average paired-end span and a large number of paired-end reads mapping in inverted orientation, suggesting the presence of a tandem repeat that was collapsed during assembly. In H. cubana, the repeating unit (152 bp) could be inferred directly from the assembled Illumina reads, whereas the longer repeating unit (412 bp) in H. texana had to be confirmed by Sanger sequencing of a PCR product generated with primers designed in inverted orientation. The reported genome sequences include two tandem copies of these repeats, but given the high read depths, it is likely that the actual copy number is higher (and possibly variable). The repeating units correspond to Carsonella nucleotide positions 128,103–128,254 and 119,600–120,011 in H. cubana and H. texana, respectively.
Because our DNA samples were generated from pools of multiple individuals, read mapping was used to identify polymorphic sites within each genome. Read pairs were mapped to their respective genomes using SOAP (parameters: m = 1, x = 1,000, g = 3, r = 2), and single-nucleotide polymorphisms (SNPs) were identified by parsing the resulting mapping output with a custom Perl script.
Finished genome sequences were annotated with the Joint Genome Institute IMG ER pipeline (Markowitz et al. 2009). The Carsonella annotations were refined based on the results of HMMER v3.0 hmmscan (Eddy 2011) searches against the Pfam 26.0 (Punta et al. 2012) and TIGRFAMs 12.0 (Selengut et al. 2007) databases as well as comparisons among the five new genomes and the one previously annotated genome (Nakabachi et al. 2006; Tamames et al. 2007). The identification and reconstruction of functional metabolic pathways were further aided by comparisons with the EcoCyc database (Keseler et al. 2011) and the KAAS annotation server (Moriya et al. 2007). Annotated genome sequences were submitted to GenBank (table 1), and chromosome maps were generated with Circos v0.56 (Krzywinski et al. 2009).
Phylogenetic Analysis
To assess the phylogenetic relationships among the six available Carsonella genomes, protein-coding gene sequences from each genome were aligned with orthologs from the outgroups Halomonas elongata (FN869568), Pseudomonas aeruginosa (AE004091), and Escherichia coli (U00096). We excluded all genes that were previously annotated as “hypothetical protein” or “putative” (Tamames et al. 2007), as these tend to be more divergent and difficult to align reliably. Amino acid sequences were aligned with MUSCLE v3.7 (Edgar 2004), and regions with poor alignment quality were trimmed with Gblocks v0.91 b using the following parameter settings: b1 = 7, b2 = 8, b3 = 8, and b4 = 20 (Castresana 2000). After trimming, a total of 82 genes representing 13,962 amino acids positions were concatenated. A maximum likelihood phylogenetic analysis was performed on the resulting dataset with RAxML v7.2.6 (Stamatakis 2006). The search included 1,000 bootstrap replicates and employed an LG+G+F model of evolution, which was chosen based on a model selection analysis in ProtTest v2.4 (Abascal et al. 2005). In addition, 43 gene alignments greater than 100 amino acids in length (after Gblocks trimming) were subjected to individual phylogenetic analysis in RAxML. The resulting set of tree topologies was summarized with PhyloSort v1.3 (Moustafa and Bhattacharya 2008).
Preliminary analysis of the secondary symbiont genomes from C. eucalypti and H. cubana suggested that they were closely related to a clade of insect-associated endosymbionts within the Enterobacteriaceae. To better assess the phylogenetic placement of these bacteria, we used AMPHORA2 (Wu and Scott 2012) to generate trimmed amino acid alignments for a conserved set of 31 genes from a broad sampling of completely sequenced genomes from the Enterobacteriaceae and related outgroups. After excluding two genes (pyrG and smpB) that were not identified by AMPHORA2 in all species, the remaining 29 alignments (5,855 amino acid positions) were concatenated, and a phylogenetic analysis was conducted with RAxML with 500 bootstrap replicates and an LG+G+I model of evolution, as identified by ProtTest.
Synonymous and Non-synonymous Divergence in Carsonella
To assess the extent of purifying selection acting on amino acid sequences in Carsonella, protein-coding genes were aligned for each of the three pairs of Carsonella genomes from congeneric host species and used to estimate divergence at both synonymous (dS) and non-synonymous (dN) sites. Gene sequences were aligned in frame by generating amino acid alignments with MUSCLE and then converting them back into nucleotide sequences. Pairwise sequence divergence was estimated in HyPhy v2.1.0, using an MG94×HKY85 codon-based model of evolution (Pond et al. 2005). To account for the extreme nucleotide composition bias in Carsonella, codon frequencies were estimated with an F3×4 model. In a small number of genes, synonymous divergence appeared to be at or near saturation, resulting in unrealistically high estimates of dS. For the purposes of calculating dN/dS, we constrained dS to a maximum value of 3 in these instances. For statistical comparisons of dN/dS estimates among genes or among lineages, values were first log-transformed. All reported statistical tests were performed in R v2.8.0.
Results
Conservation of Carsonella Genome Size, Structure, and GC Content
Illumina sequencing of psyllid DNA samples generated 14.7–23.7 million quality-filtered read pairs per library, yielding sufficient Carsonella coverage for complete genome assembly. In four libraries generated from total insect DNA, Carsonella sequences accounted for 1.11–1.96% of all reads, resulting in median genome coverages of 257–589×. As expected, Carsonella sequences represented a larger proportion of reads (8.93%) in a fifth library generated from DNA extracted and amplified from isolated bacteriome tissue, corresponding to a median genome coverage of 1532×.
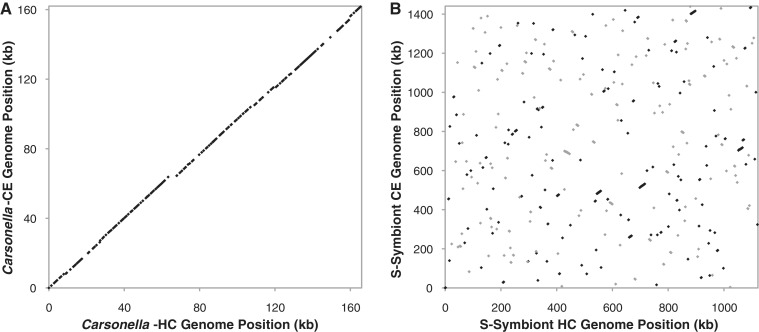
We found that Carsonella strains from divergent host lineages are generally conserved in genome architecture (table 1; supplementary fig. S2, Supplementary Material online). The six sequenced Carsonella genomes (including the originally published genome from P. venusta) vary by less than 6% in size (157.5–166.2 kb), and all exhibit extremely low GC content (14.0–16.6%). In addition, all six show perfect conservation of synteny with no large inversions or structural rearrangements (fig. 1A).
Fig. 1.
Conservation of synteny between Carsonella strains (A) but not secondary symbionts (B) from Ctenarytaina eucalypti (“CE”) and Heteropsylla cubana (“HC”). Black and gray dots indicate the genes shared between the two genomes in forward and reverse orientation, respectively. All other sequenced Carsonella genomes also exhibited perfect conservation of synteny (data not shown).
For all psyllid species, DNA samples were derived from multiple field-collected individuals and therefore harbor genetic variation. With the exception of P. celtidis, however, the amount of polymorphism within each sample was relatively low, as SNPs with a minor allele frequency of at least 10% accounted for less than 0.1% of sites. In contrast, such SNPs were found at 0.3% of sites in P. celtidis. This higher level of variation may reflect natural differences in polymorphism among host species, but it could alternatively indicate the presence of multiple closely related species inhabiting hackberry leaf nipple galls (see Materials and Methods).
Rapid Evolution and Widespread Functional Constraint in Carsonella Protein Genes
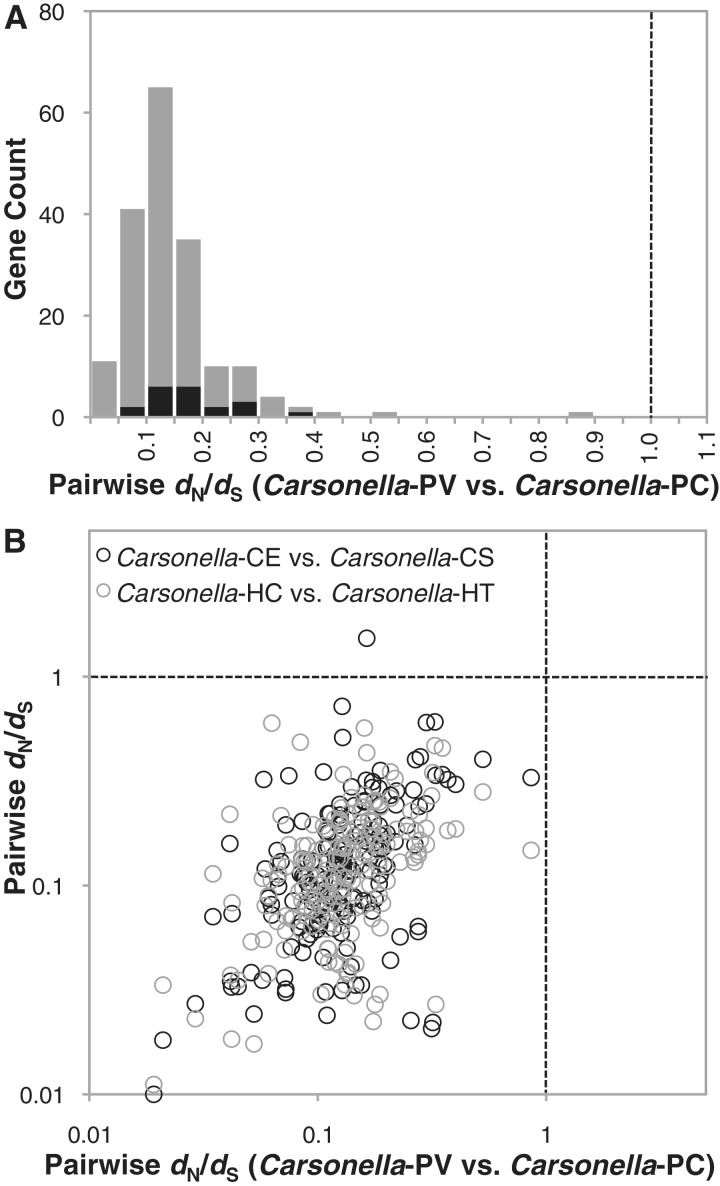
We analyzed patterns of sequence divergence between pairs of Carsonella genomes from congeneric hosts to assess the effect of selection on protein-coding genes. Median estimates for the number of substitutions per synonymous site (dS) within Ctenarytaina, Heteropsylla, and Pachypsylla were 0.40, 0.70, and 0.75, respectively. Within Pachypsylla, the majority of protein-coding genes appears to be evolving under purifying selection. For every gene, estimates of dN/dS were below the neutral expectation of 1, indicating selection against changes in amino acid sequence (fig. 2A). A set of 20 Carsonella genes previously identified as potentially non-functional in P. venusta (Tamames et al. 2007) exhibited evidence for a relaxation in purifying selection, with significantly higher dN/dS than genes with annotated functions (t-test; P = 0.0001), but their values were still consistently below 1 with a mean of 0.18 (fig. 2A). Carsonella sequence divergence within each of the other two host genera exhibited very similar patterns of constraint, and gene-specific dN/dS values were significantly correlated among all three genera (P < 0.0001 for all pairwise comparisons; fig. 2B).
Fig. 2.
Purifying selection on Carsonella protein-coding genes. (A) Distribution of gene-specific dN/dS values based on Carsonella divergence within Pachypsylla hosts. (B) Correlation between gene-specific dN/dS values across the three host genera. Dotted lines show that the neutral expectation of dN/dS = 1. Carsonella genomes are specified by host name initials.
Ongoing Gene Loss in Carsonella
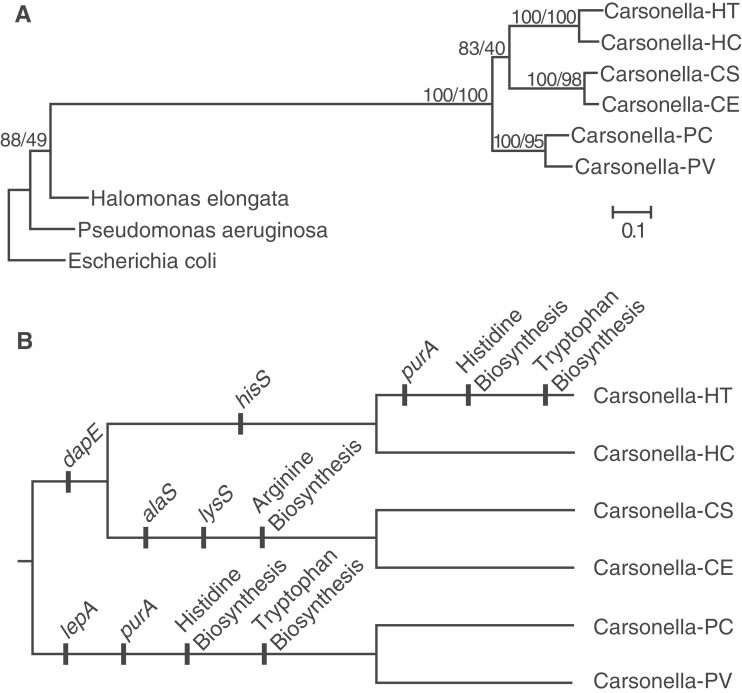
Despite the conservation in Carsonella genome structure and evidence for purifying selection on protein-coding sequences throughout the genome, gene loss remains an active process in these highly reduced genomes. To infer the timing of gene loss events, we first performed a phylogenetic analysis to assess the relationships among the sequenced Carsonella genomes from six different host species. As expected, this analysis provided strong support for the sister relationship between each pair of Carsonella genomes from congeneric hosts (fig. 3A). However, the extensive sequence divergence posed a challenge for rooting the Carsonella tree and created ambiguity with respect to the relationships among the three host genera. We found weak support for a sister relationship between the Carsonella lineages in Ctenarytaina and Heteropsylla (fig. 3A). This would be consistent with the shared absence of dapE from both of those lineages (fig. 3B). However, this topology is in conflict with some morphology-based host classifications, which group Ctenarytaina and Pachypsylla as members of the subfamily Spondyliaspidinae (Thao et al. 2000a). Therefore, it is possible that the inferred placement of the root is incorrect and that dapE has been lost at least twice independently.
Fig. 3.
Phylogenetic relationships and ongoing gene loss in Carsonella. (A) Maximum likelihood phylogeny of Carsonella lineages inferred from 82 concatenated protein sequences. Values indicate the percentage of bootstrap replicates (left) and individual gene trees (right) supporting the corresponding node. (B) Inferred history of gene loss events since divergence of Carsonella lineages. “Arginine biosynthesis” includes argF, carA-1, carA-2, and carB. “Tryptophan biosynthesis” includes trpE and trpG. “Histidine biosynthesis” includes hisA, hisB (lost in Pachypsylla only), hisC, hisD, hisE, hisF, hisG, hisH, hisI, and prs. Losses of hypothetical protein genes are not shown. Carsonella strains from different host species are designated using the first letters of the host genus and species names, as listed in table 1.
Comparisons of gene content among the six Carsonella genomes revealed numerous examples of gene loss, including many that have occurred since the divergence within a single host genus (Heterospylla; fig. 3B). These losses have had an especially large impact on amino acid biosynthesis pathways. The original Carsonella genome sequence lacked entire gene sets underlying histidine and tryptophan biosynthesis. In contrast, the Carsonella genomes from both Ctenarytaina species and H. cubana have retained a set of 10 genes that code for an intact histidine biosynthesis pathway and two genes (trpE and trpG) responsible for converting chorismate to anthranilate, a key step in tryptophan biosynthesis. The histidine biosynthesis genes are distributed across five regions in the Carsonella genome, indicating the occurrence of numerous independent deletion events. All the histidine and tryptophan biosynthesis genes were likely lost before the divergence of the Pachypsylla hosts, because they are also absent from P. celtidis. In addition, all but one of these genes (hisB) are absent from the Carsonella genome in H. texana, indicating a striking example of parallel gene loss. The independent loss of the purine biosynthesis gene purA and an adjacent gene of unknown function in Carsonella within both Pachypsylla and H. texana further supports the parallelism between these lineages (fig. 3B). Amino acid biosynthesis pathways have also been affected by the loss of many of the arginine biosynthesis genes in the Ctenarytaina lineage and the absence of dapE (a component of the lysine biosynthesis pathway) outside the Pachypsylla lineage (fig. 3).
One surprising finding from the original Carsonella genome sequence was that it lacked a complete set of tRNA synthetases (Nakabachi et al. 2006; Tamames et al. 2007). The loss of these genes is even more extensive in some of the other Carsonella genomes, as the alanyl (alaS), histidinyl (hisS), and lysyl (lysS) tRNA synthetases are absent from either the Ctenarytaina or the Heteropsylla lineages (fig. 3). In addition, the translation elongation factor lepA has been lost from the Pachypsylla lineage, and a small number of hypothetical protein genes are absent from various lineages.
Gene losses were generally associated with deletion of most or all the locus, but there are also a few examples of potential pseudogenes retained in the genome. In particular, the Carsonella genes for phenylalanyl tRNA synthetase (pheS) and the translation initiation factor IF-3 (infC) lack identifiable start codons in H. cubana and Ctenarytaina, respectively.
Secondary Symbiont Genomes
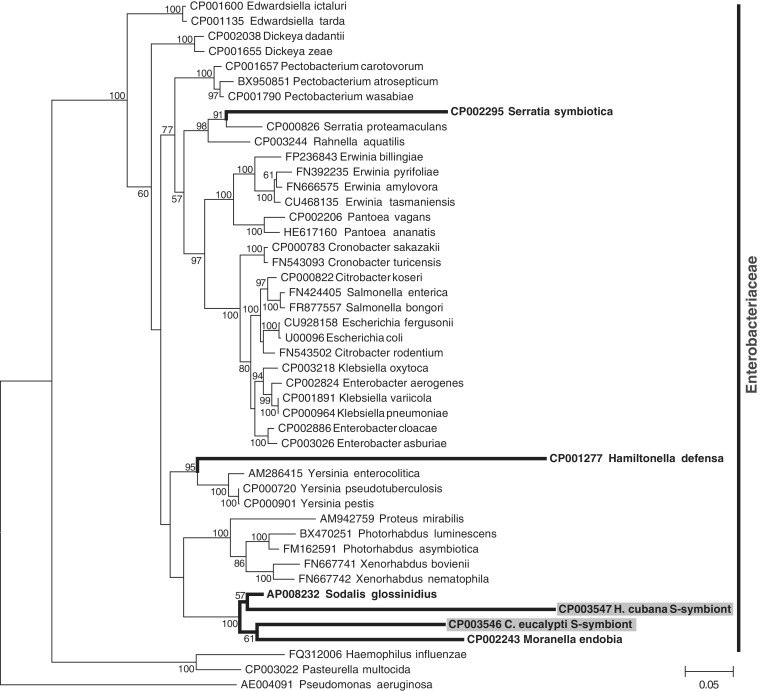
In addition to the Carsonella genomes, the assemblies from each psyllid species contained other contigs of bacterial origin. Ctenarytaina eucalypti, C. spatulata, and H. cubana contained contigs with homology to a clade of insect-associated endosymbionts in the Enterobacteriaceae (fig. 4). The only members of this group with fully sequenced genomes are the tsetse fly secondary symbiont Sodalis glossinidius (Toh et al. 2006) and the mealybug secondary symbiont Candidatus Moranella endobia (McCutcheon and von Dohlen 2011), but related bacteria have been previously identified in psyllids (Fukatsu and Nikoh 1998; Thao et al. 2000b; Spaulding and von Dohlen 2001) and other insect hosts, for example, weevils (Lefevre et al. 2004; Toju et al. 2010). A phylogenetic analysis of 29 protein-coding genes did not support monophyly of the C. eucalypti and H. cubana symbionts (fig. 4), which is further supported by the almost complete lack of conserved synteny between these genomes (fig. 1B) and previous findings of independent origins of secondary symbionts in psyllids (Subandiyah et al. 2000; Thao et al. 2000b; Spaulding and von Dohlen 2001).
Fig. 4.
Maximum likelihood phylogeny of insect secondary symbionts (bold) and free-living members of the Enterobacteriaceae based on 29 concatenated protein sequences. Gray boxes indicate psyllid secondary symbionts. Values at each node indicate percentage bootstrap support.
Although we did not find any sequences corresponding to this clade of secondary symbionts in the H. texana or P. celtidis assemblies, these hosts did yield contigs with clear homology to Arsenophonus and Wolbachia, respectively. Both of these intracellular bacteria are widely associated with insects and can be involved in the manipulation of host reproductive systems (Werren et al. 2008; Novakova et al. 2009). The presence of Arsenophonus is also correlated with the incidence of parasitoid infection in the psyllid Glycaspis brimblecombei, suggesting a possible role in defense (Hansen et al. 2007). Wolbachia contigs were also found in the H. cubana assembly in addition to sequences from two other Alphaproteobacteria (tentatively assigned to the Acetobacteraceae and Rhizobiales).
Of the identified bacterial sequences, only those corresponding to the Enterobacteriaceae secondary symbionts of H. cubana and C. eucalypti had sufficient coverage and contiguity for complete genome assembly. The H. cubana assembly produced a 1.12 Mb chromosome with a median coverage of 39×, and the C. eucalypti assembly produced a 1.44 Mb chromosome with a median coverage of 60×.
This pair of secondary symbiont genomes shows signatures of reduction and long-term evolution as vertically transmitted endosymbionts. Both exhibit accelerated rates of sequence evolution (fig. 4) and are noticeably devoid of large repeats and mobile genetic elements, with no duplicated sequences greater than 400 bp in length. The genomes are also greatly reduced in size relative to those of most free-living bacteria and many facultative insect endosymbionts (table 2) and have an unusually low gene density (supplementary fig. S3, Supplementary Material online), a hallmark of genomes experiencing ongoing gene loss and pseudogenization (McCutcheon and Moran 2011).
Table 2.
Examples of Insect Secondary Symbiont Genome Sizes from the Enterobacteriaceae.
| Endosymbiont | Host | Chromosome Size (kb) | Reference |
|---|---|---|---|
| Seconday symbiont | Heteropsylla texana | 1122 | This study |
| Secondary symbiont | Ctenarytaina eucalypti | 1441 | This study |
| Serratia symbiotica | Cinara cedri | 1763 | Lamelas et al. 2011 |
| Regiella insecticola LSR1 | Acyrthosiphon pisum | 2035 | Degnan et al. 2010 |
| Hamiltonella defensa | Acyrthosiphon pisum | 2110 | Degnan et al. 2009 |
| Serratia symbiotica | Acyrthosiphon pisum | 2789 | Burke and Moran 2011 |
| Sodalis glossinidius | Glossina morsitans | 4171 | Toh et al. 2006 |
Reduced size and low gene density combine to yield very gene-poor genomes, with a history of gene loss evident across all functional categories (supplementary fig. S4, Supplementary Material online). The apparent loss of many metabolic capabilities offers further support for the evolution of an obligate intracellular lifestyle. For example, both secondary symbionts lack most essential amino acid biosynthesis pathways (see below). Although they retain basic gene sets necessary for glycolysis and the pentose phosphate pathway, they both appear to have lost genes necessary for a complete tricarboxylic acid (TCA) cycle. The pair of genomes also shows examples of differential gene loss. For example, the C. eucalypti secondary symbiont has retained genes for major oxidative phosphorylation complexes such as NADH dehydrogenase and ATP synthase, whereas these have been lost from the H. cubana secondary symbiont. As typically observed in intracellular bacteria (Moran et al. 2008), both genomes have also lost numerous genes involved in DNA recombination and repair (supplementary fig. S5, Supplementary Material online), which may contribute to accelerated rates of sequence evolution and biased nucleotide compositions. Unlike other secondary symbionts such as Sodalis glossinidius, Hamiltonella defensa, and Regiella insecticola, the psyllid secondary symbionts appear to lack a type III secretion system, which can be involved in host cell invasion (Dale et al. 2001). The loss or degeneration of such systems may indicate evolution toward a more mutualistic and obligate relationship (Dale et al. 2005).
Functional Complementarity between Carsonella and Secondary Symbionts in Amino Acid Biosynthesis
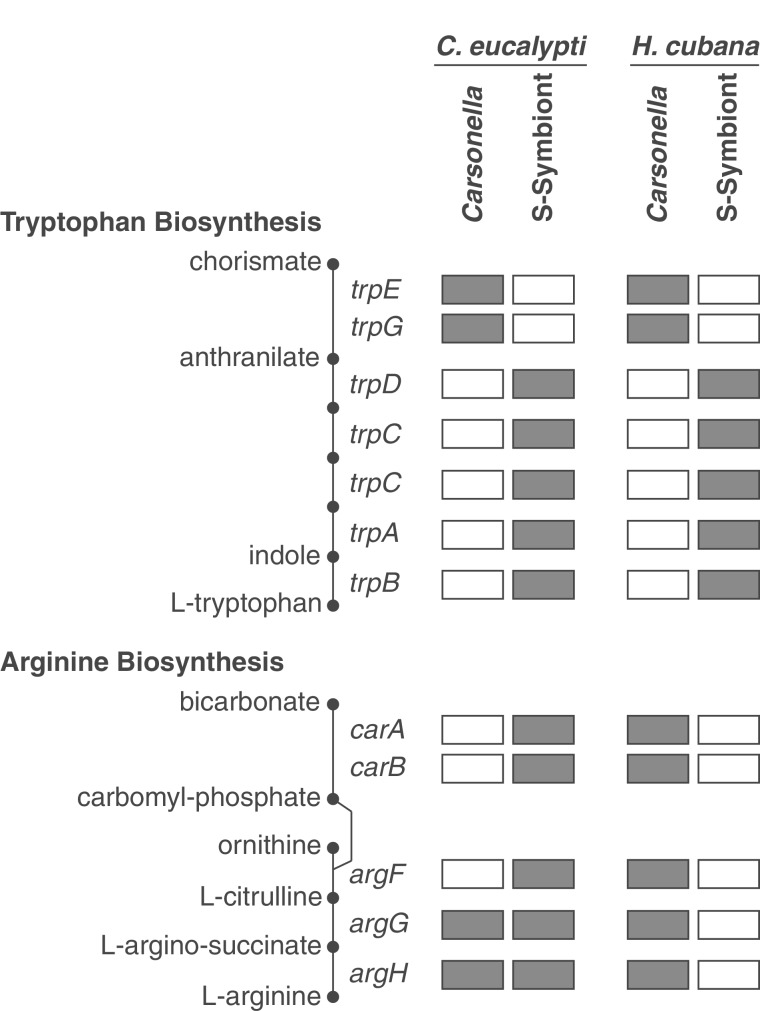
Although Carsonella has retained a disproportionately large number of genes involved in the synthesis of essential amino acids (Nakabachi et al. 2006), these genes have been largely eliminated from the genomes of secondary symbionts in C. eucalypti and H. cubana. However, the few remaining amino acid biosynthesis pathway genes in these secondary symbionts suggest a high degree of functional complementarity with Carsonella (fig. 5). For example, the H. cubana secondary symbiont has lost nearly all essential amino acid biosynthesis genes except the trpDCBA operon, which encodes the genes necessary to catalyze the synthesis of tryptophan from anthranilate (supplementary fig. S6, Supplementary Material online). This neatly complements the tryptophan biosynthesis capacity of Carsonella in H. cubana, which contains the genes necessary to produce anthranilate but lacks those required to complete the final steps of the pathway. Notably, the exact same division of labor appears to have evolved between Carsonella and the secondary symbiont in C. eucalypti (fig. 5). In addition to the trpDCBA operon, the C. eucalypti secondary symbiont also retains the complete set of genes required for synthesis of arginine from ornithine. Although these same genes are present in some Carsonella genomes, the pathway is severely compromised in Carsonella in both Ctenarytaina hosts, suggesting that the responsibility for synthesizing arginine has been shifted to secondary symbionts in this lineage (fig. 3). The Carsonella copies of argF, carA-1, carA-2, and carB have all been deleted in Ctenarytaina. In addition, argG shows evidence of relaxed selection based on an accelerated rate of sequence evolution, an increase in dN/dS, and a decrease in GC content (supplementary fig. S6, Supplementary Material online). The C. eucalypti secondary symbiont genome also contains genes encoding a largely intact lysine biosynthesis pathway. In this case, however, there is no clear indication of complementarity with Carsonella, because both bacteria retain similar sets of genes (supplementary fig. S6, Supplementary Material online). It is possible that lysine biosynthesis genes are retained in the C. eucalypti secondary symbiont for production of the lysine precursor meso-diaminopimelate, which is involved in cell wall biosynthesis (Gerdes et al. 2003). This is consistent with the loss of lysA, which codes for the terminal enzyme in the pathway that converts meso-diaminopimelate into lysine (supplementary fig. S7, Supplementary Material online).
Fig. 5.
Complementarity in amino acid biosynthesis capabilities. Shaded boxes indicate the presence of an intact gene in the respective genome. See supplementary figure S7, Supplementary Material online for a full summary of essential amino acid biosynthesis gene content.
Discussion
Conservation of the Highly Reduced Carsonella Genome
The extreme genome reduction in Carsonella poses fundamental questions about the concept of the minimal cell and the boundary between cellular organisms and organelles (Nakabachi et al. 2006; Tamames et al. 2007). The observed conservation in genome size, structure, and GC content across psyllids demonstrates that the highly derived nature of this genome is broadly characteristic of Carsonella and not a peculiarity resulting from the biology of a particular host lineage (e.g., the gall-forming habit of Pachypsylla). Overall, our analysis indicates that the Carsonella genome experiences very rapid removal of non-functional sequence and conversely that sequence content present in the genome is actively retained by selection. This conclusion is supported by multiple lines of evidence: 1) low dN/dS values in the overwhelming majority of protein genes (fig. 2); 2) the general conservation of open reading frames, which, in the absence of strong selection, would rapidly accumulate stop codons given the AT richness of the genomes; 3) the unusually high gene density in Carsonella, reflecting a paucity of pseudogenes and intergenic sequence (supplementary fig. S3, Supplementary Material online); and 4) the removal of entire pathways by multiple independent deletion events on recent evolutionary timescales (fig. 3).
How can these findings be reconciled with previous observations that many Carsonella genes have lost key domains or catalytic residues and are therefore likely non-functional (Tamames et al. 2007)? One potential explanation is that it is simply difficult to predict functions solely from sequence data in these highly derived genomes and that some genes may have retained their ancestral role even though they are so divergent that conserved functional elements can no longer be recognized. An alternative and perhaps more biologically interesting possibility is that some of these genes have evolved novel functions. This could explain the evidence for ongoing purifying selection despite the loss of domains necessary for ancestral function. For example, Tamames et al. (2007) reported that the DNA primase encoded by dnaG is likely incapable of performing its traditional role in DNA replication, as large deletions have removed most or all of two domains that are required in other bacteria for DNA binding and interaction with the replicative DNA helicase. However, we found that all six Carsonella genomes share these large deletions in dnaG and that the gene has nevertheless continued to evolve under strong purifying selection (dN/dS < 0.2 within all three host genera). Therefore, it is possible that this protein employs novel mechanisms for performing its conserved role in DNA replication or has been retained in Carsonella for an entirely different purpose.
Amino Acid Biosynthesis Pathways in Flux
Given the difficulties with using sequence homology to annotate highly divergent genomes, there are always lingering doubts as to whether genes that are reported as missing are truly absent, or present but simply unidentified. The discovery of histidine and tryptophan biosynthesis genes in some of the newly sequenced Carsonella lineages unambiguously confirms that these pathways were ancestrally present in the symbiont and subsequently lost in some lineages. These and other cases of recent gene loss (fig. 3) demonstrate that Carsonella amino acid biosynthesis pathways are very much in flux and confirm that Carsonella, by itself, is incapable of synthesizing a complete set of essential amino acids for its host.
Secondary symbionts have been found to be involved in a diverse range of mutualistic (and antagonistic) interactions with their hosts. For example, secondary symbionts have been shown to confer resistance to parasitoid infection (Brownlie and Johnson 2009), improve thermotolerance (Montllor et al. 2002), and synthesize important vitamins and co-factors (Lamelas et al. 2011). The role of secondary symbionts in psyllids is not well understood, but genomic evidence from C. eucalypti and H. cubana suggests that they play an important role in complementing amino acid biosynthesis pathways in Carsonella.
The striking complementarity in tryptophan biosynthesis gene content indicates that this pathway is divided between the pair of bacterial symbionts in both C. eucalypti and H. cubana. Interestingly, similar partitions in the tryptophan biosynthetic pathway have also been observed between pairs of symbionts in the mealybug Planococcus citri and the cedar aphid Cinara cedri (Gosalbes et al. 2008; Lamelas et al. 2011; McCutcheon and von Dohlen 2011), suggesting that it is particularly amenable to division among symbiotic compartments.
The specific location of secondary symbionts within C. eucalypti and H. cubana has not been determined, but they likely inhabit the syncytium between the host bacteriocytes that contain Carsonella. Other psyllid secondary symbionts, including those from the same clade in the Enterobacteriaceae, have been localized to this region (Buchner 1965; Fukatsu and Nikoh 1998; Subandiyah et al. 2000). Therefore, metabolic processes within each of the bacterial symbionts are likely separated by a series of host and bacterial membranes, suggesting that the host genome may play an important role in shuttling metabolites among compartments.
The retention of the arginine biosynthesis pathway in the C. eucalypti secondary symbiont offers further support for metabolic complementarity given the corresponding loss of most of these genes from Carsonella in the Ctenarytaina lineage (fig. 3). Similar examples of complementary gene content have provided compelling evidence for mutualistic interdependence between other pairs of insect endosymbionts (Wu et al. 2006; McCutcheon et al. 2009; McCutcheon and Moran 2010; Lamelas et al. 2011), but it is also important to consider alternative evolutionary mechanisms that could produce such patterns. For example, in cases where Carsonella has lost the ability to produce arginine, it may be a more limiting resource in the system, creating stronger selection for secondary symbionts to retain the capacity to synthesize arginine even if they are not serving as a major source of arginine for the host. Therefore, a more definitive confirmation of a shared symbiotic role in amino acid biosynthesis will require showing that the host utilizes both symbionts as sources of amino acid production.
In some cases, such as the loss of the tryptophan and histidine biosynthesis genes in Pachypsylla, deleted Carsonella genes do not appear to be functionally replaced by secondary symbionts. Previous screens have not found evidence of secondary symbionts in this genus, which is largely consistent with our results from deep sequencing. Although we detected Wolbachia in P. celtidis, none of the fragmented contigs from its genome appear to encode genes involved in histidine or tryptophan biosynthesis. Interestingly, the H. texana lineage has experienced the deletion of a nearly identical set of tryptophan and histidine biosynthesis genes, and it is the only other species in our sample that did not contain a secondary symbiont from the Sodalis/Moranella clade.
The loss of Carsonella-encoded pathways in these lineages raises two alternative possibilities. First, gene loss may result from changes in host environment or diet that reduce dependence on particular amino acids. For example, the dramatic changes in host plant morphology elicited by the gall-forming Pachypsylla species may provide an improved source of nutrition with higher concentration of essential amino acids (Price et al. 1987; Spaulding and von Dohlen 2001). Although H. texana is not a gall former, it causes extensive morphological modifications to the leaf and floral shoot structures of its host plants, Prosopis spp. (Donnelly 2002). Future studies should investigate amino acid profiles in the phloem sap from different psyllid host plants to assess whether they can explain the history of differential gene loss in Carsonella. Second, the loss of Carsonella genes could coincide with an increased role of the host genome, resulting from either direct endosymbiotic gene transfer from Carsonella (Nakabachi et al. 2006) or acquisition of homologous genes from some other bacterial source (Nikoh et al. 2010). The role of the host genome in complementing the reduced gene content in Carsonella remains an important area for future investigation.
Secondary Symbiont Acquisition as a Common Driver of Genome Reduction
Although secondary symbionts are often characterized by a facultative relationship with their insect hosts, the sequenced genomes from C. eucalypti and H. cubana bear signatures of reductive genome evolution that are more typical of an obligate relationship. These signatures include more extensive gene loss than normally observed in facultative symbionts, a general lack of repetitive elements, and evidence of metabolic interdependence with Carsonella. On the basis of these observations, we hypothesize that these psyllid secondary symbionts have established long-term, stable associations with their respective hosts, and we predict that broad population sampling within these psyllid species would identify widespread, if not universal, presence of these symbionts.
Coupled with the findings from other recent genomic studies, our results paint a dynamic picture of endosymbiont evolution in insects that is characterized by repeated colonization and establishment of obligate associations, even in hosts that already harbor ancient primary symbionts. The recent characterization of Serratia symbiotica in aphids illustrates the labile boundary between facultative and obligate endosymbiotic relationships. Although facultative in the pea aphid (Montllor et al. 2002; Burke and Moran 2011), this symbiont lineage appears to have evolved an obligate relationship with the related cedar aphid, demonstrating similar patterns of metabolic interdependence between primary and secondary symbiont as we observe in psyllids (Lamelas et al. 2011). This association likely accelerated the process of symbiont genome reduction, as the genomes of both Serratia and the primary symbiont Buchnera are substantially smaller in the cedar aphid than in related host lineages (Perez-Brocal et al. 2006; Lamelas et al. 2011). A similar process appears to have occurred in mealybugs and multiple lineages within the suborder Auchenorrhyncha, including cicadas, sharpshooters, and spittlebugs. These insects demonstrate some of the most extreme examples of bacterial genome reduction yet identified, and, in each case, the host has independently acquired a secondary symbiont that now forms part of an obligate and metabolically complementary relationship (Wu et al. 2006; McCutcheon et al. 2009; McCutcheon and Moran 2010). Therefore, the acquisition of secondary symbionts that can replace primary symbiont functions appears to represent a very common, ratchet-like mechanism that can intensify the process of irreversible reduction of endosymbiont genomes.
Supplementary Material
Supplementary figures S1–S7 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Paul Baumann for providing psyllid DNA samples and John Overton for advice on optimizing Illumina library construction protocols. This work was supported by Yale University and a National Institutes of Health Postdoctoral Fellowship (1F32GM099334).
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C, Jaffe DB, Nusbaum C, Gnirke A. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- Brownlie JC, Johnson KN. Symbiont-mediated protection in insect hosts. Trends Microbiol. 2009;17:348–354. doi: 10.1016/j.tim.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Buchner P. New York (NY): John Wiley & Sons; 1965. Endosymbiosis of animals with plant microorganisms. [Google Scholar]
- Burke GR, Moran NA. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol. 2011;3:195–208. doi: 10.1093/gbe/evr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S. Genome sequence assembly using trace signals and additional sequence information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB) 1999;99:45–56. [Google Scholar]
- Dale C, Jones T, Pontes M. Degenerative evolution and functional diversification of type-III secretion systems in the insect endosymbiont Sodalis glossinidius. Mol Biol Evol. 2005;22:758–766. doi: 10.1093/molbev/msi061. [DOI] [PubMed] [Google Scholar]
- Dale C, Young SA, Haydon DT, Welburn SC. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc Natl Acad Sci U S A. 2001;98:1883–1888. doi: 10.1073/pnas.021450998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Leonardo TE, Cass BN, Hurwitz B, Stern D, Gibbs RA, Richards S, Moran NA. Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol. 2010;12:2060–2069. doi: 10.1111/j.1462-2920.2009.02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci U S A. 2009;106:9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly G. The host range and biology of the mesquite psyllid Heteropsylla texana. BioControl. 2002;47:363–371. [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T, Nikoh N. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera) Appl Environ Microbiol. 1998;64:3599–3606. doi: 10.1128/aem.64.10.3599-3606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes SY, Scholle MD, Campbell JW, et al. (21 co-authors) Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes MJ, Lamelas A, Moya A, Latorre A. The striking case of tryptophan provision in the cedar aphid Cinara cedri. J Bacteriol. 2008;190:6026–6029. doi: 10.1128/JB.00525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Jeong G, Paine TD, Stouthamer R. Frequency of secondary symbiont infection in an invasive psyllid relates to parasitism pressure on a geographic scale in California. Appl Environ Microbiol. 2007;73:7531–7535. doi: 10.1128/AEM.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Collado-Vides J, Santos-Zavaleta A, et al. (21 co-authors) EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamelas A, Gosalbes MJ, Manzano-Marin A, Pereto J, Moya A, Latorre A. Serratia symbiotica from the aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet. 2011;7:e1002357. doi: 10.1371/journal.pgen.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre C, Charles H, Vallier A, Delobel B, Farrell B, Heddi A. Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Mol Biol Evol. 2004;21:965–973. doi: 10.1093/molbev/msh063. [DOI] [PubMed] [Google Scholar]
- Lewis IF, Walton L. Gall-formation on leaves of Celtis occidentalis L. resulting from material injected by Pachypsylla sp. Trans Am Microsc Soc. 1964;83:62–78. [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, von Dohlen CD. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 2011;21:1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. Tablet—next generation sequence assembly visualization. Bioinformatics. 2010;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montllor CB, Maxmen A, Purcell AH. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol. 2002;27:189–195. [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A, Bhattacharya D. PhyloSort: a user-friendly phylogenetic sorting tool and its application to estimating the cyanobacterial contribution to the nuclear genome of Chlamydomonas. BMC Evol Biol. 2008;8:6. doi: 10.1186/1471-2148-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Nikoh N, McCutcheon JP, Kudo T, Miyagishima SY, Moran NA, Nakabachi A. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 2010;6:e1000827. doi: 10.1371/journal.pgen.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova E, Hypsa V, Moran NA. Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 2009;9:143. doi: 10.1186/1471-2180-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Degnan PH, Burke GR, Moran NA. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. 2010;55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- Perez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, Michelena JM, Silva FJ, Moya A, Latorre A. A small microbial genome: the end of a long symbiotic relationship? Science. 2006;314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Price PW, Fernandes GW, Waring GL. Adaptive nature of insect galls. Environ Entomol. 1987;16:15–24. [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, et al. (16 co-authors) The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. TIGRFAMs and genome properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, Taylor DR. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10:e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaulding AW, von Dohlen CD. Psyllid endosymbionts exhibit patterns of co-speciation with hosts and destabilizing substitutions in ribosomal RNA. Insect Mol Biol. 2001;10:57–67. doi: 10.1046/j.1365-2583.2001.00231.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Straka JR, Hayward AR, Emery RJN. Gall-inducing Pachypsylla celtidis (Psyllidae) infiltrate hackberry trees with high concentrations of phytohormones. J Plant Interact. 2010;5:197–203. [Google Scholar]
- Subandiyah S, Nikoh N, Tsuyumu S, Somowiyarjo S, Fukatsu T. Complex endosymbiotic microbiota of the citrus psyllid Diaphorina citri (Homoptera: Psylloidea) Zool Sci. 2000;17:983–989. [Google Scholar]
- Tamames J, Gil R, Latorre A, Peretó J, Silva FJ, Moya A. The frontier between cell and organelle: genome analysis of Candidatus Carsonella ruddii. BMC Evol Biol. 2007;7:181. doi: 10.1186/1471-2148-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao ML, Clark MA, Baumann L, Brennan EB, Moran NA, Baumann P. Secondary endosymbionts of psyllids have been acquired multiple times. Curr Microbiol. 2000a;41:300–304. doi: 10.1007/s002840010138. [DOI] [PubMed] [Google Scholar]
- Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, Baumann P. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol. 2000b;66:2898–2905. doi: 10.1128/aem.66.7.2898-2905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao ML, Clark MA, Burckhardt DH, Moran NA, Baumann P. Phylogenetic analysis of vertically transmitted psyllid endosymbionts (Candidatus Carsonella ruddii) based on atpAGD and rpoC: comparisons with 16S-23S rDNA-derived phylogeny. Curr Microbiol. 2001;42:419–421. doi: 10.1007/s002840010240. [DOI] [PubMed] [Google Scholar]
- Toh H, Weiss BL, Perkin SA, Yamashita A, Oshima K, Hattori M, Aksoy S. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toju H, Hosokawa T, Koga R, Nikoh N, Meng XY, Kimura N, Fukatsu T. "Candidatus Curculioniphilus buchneri," a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl Environ Microbiol. 2010;76:275–282. doi: 10.1128/AEM.02154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dohlen CD, Kohler S, Alsop ST, McManus WR. Mealybug beta-proteobacterial endosymbionts contain gamma-proteobacterial symbionts. Nature. 2001;412:433–436. doi: 10.1038/35086563. [DOI] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H, Tallon LJ, Zaborsky JM, Dunbar HE, Tran PL. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006;4:1079–1092. doi: 10.1371/journal.pbio.0040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Scott AJ. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics. 2012;28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- Yang MM, Mitter C. Biosystematics of hackberry psyllids (Pachypsylla) and the evolution of gall and lerp formation in psyllids (Homoptera: Psylloidea): a preliminary report. In: Price P, Baranchikov Y, Mattson W, editors. The ecology and evolution of gall-forming insects. Proceedings of the first international gall symposium. 1994. August 1993; Krasnoyarsk, Siberia: USDA. p. 172–185. [Google Scholar]
- Yang MM, Mitter C, Miller DR. First incidence of inquilinism in gall-forming psyllids, with a description of the new inquiline species (Insecta, Hemiptera, Psylloidea, Psyllidae, Spondyliaspidinae) Zool Scr. 2001;30:97–113. [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.