Abstract
Proteins in the superfamily of voltage-gated ion channels mediate behavior across the tree of life. These proteins regulate the movement of ions across cell membranes by opening and closing a central pore that controls ion flow. The best-known members of this superfamily are the voltage-gated potassium, calcium (Cav), and sodium (Nav) channels, which underlie impulse conduction in nerve and muscle. Not all members of this family are opened by changes in voltage, however. NALCN (NA+ leak channel nonselective) channels, which encode a voltage-insensitive “sodium leak” channel, have garnered a growing interest. This study examines the phylogenetic relationship among Nav/Cav voltage-gated and voltage-insensitive channels in the eukaryotic group Opisthokonta, which includes animals, fungi, and their unicellular relatives. We show that NALCN channels diverged from voltage-gated channels before the divergence of fungi and animals and that the closest relatives of NALCN channels are fungal calcium channels, which they functionally resemble.
Keywords: NALCN, Cch1, maximum likelihood, pore motif
The eukaryotic supergroup Opisthokonta contains two large kingdoms with very different life styles: fungi and animals (supplementary fig. S1, Supplementary Material online; Parfrey et al. 2011; Torruella et al. 2011). The most obviously distinguishing feature of animals is the elaboration of motile behavior in adults, facilitated by the evolution of nerves and muscle. Recent studies have used comparative genomics and phylogenetics to examine the history of nervous system genes and show how this history bears on eukaryotic diversity (Cai and Clapham 2011; Emes and Grant 2011; Liebeskind et al. 2011; Cai 2012). We continue this project, focusing here on the evolution of opisthokont four-domain ion channels.
The voltage-gated ion channel family includes the potassium, calcium (Cav), and sodium (Nav) channels that mediate the neural code by creating action potentials (Hille 2001). Cav and Nav channels have four domains, each with six transmembrane segments. Each domain has a pore loop between the fifth and sixth segments, forming a pore motif of four amino acids that determines ion selectivity. It is hypothesized that Cav channels arose from single domain potassium channels by internal duplication at the base of eukaryotes (Hille 2001) and that Nav channels arose from the Cav family just before the origin of opisthokonts (Cai 2012).
Sodium leak channels, or NALCN (NA+ leak channel nonselective), are four-domain channels that are built on the same six trans-membrane segment domain as their better studied relatives, the Cav and Nav channels, but are voltage insensitive. NALCN channels have been implicated in numerous rhythmic behaviors (Ren 2011) such as breathing in mice (Lu et al. 2007), crawling in Caenorhabditis elegans (Pierce-Shimomura et al. 2008), and circadian rhythms in flies (Nash et al. 2002).
NALCN channels maintain and regulate firing rates in rhythmically firing neurons by modulating neuronal resting potential (Lu et al. 2007). As they leak sodium into the cell, the membrane becomes depolarized from the very negative potential set by the efflux of potassium and moves toward the threshold at which Nav channels begin to open. NALCN channels may therefore be thought of as affecting the gain of the neuron: the more NALCN channels are open, the more likely an input is to initiate firing (or that a rhythmic neuron will continue to fire). Although they are insensitive to voltage, their open state can be affected by the presence of various neurotransmitters and by calcium, and they rely on accessory proteins for their function (Lu et al. 2009; Swayne et al. 2009; Lu et al. 2010).
It has been shown previously that NALCN channels diverged from voltage-gated channels before the diversification of Cav and Nav channels (Lee et al. 1999) and have some similarities to the lone family of fungal four-domain channels (Hong et al. 2010; Ren 2011). These fungal channels are strongly selective for calcium but like NALCN are voltage insensitive and rely on an accessory protein for gating (Hong et al. 2010). Fungal calcium channels have been implicated in mating in yeast (Paidhungat and Garrett 1997), calcium-store restoration in the meningitis-causing fungus Cryptococcus neoformans (Liu et al. 2006; Hong et al. 2010), and ascospore discharge in the plant pathogen Gibberella zeae (Hallen and Trail 2008). These channels will be called fungal calcium channels here for simplicity, but there are other calcium channels in fungi that are not homologous to animal four-domain channels (Zelter et al. 2004).
In this study, we sought to clarify the phylogenetic relationships between the major lineages of opisthokont four-domain ion channels. We use this phylogenetic information to infer the historical timing of key amino acid replacements that may have had large-scale effects on opisthokont evolution. This study builds upon and synthesizes previous work, which identified the unique place of voltage-insensitive ion channels but did not place this information in the context of opisthokont evolution (Paidhungat and Garrett 1997; Lee et al. 1999; Ren 2011).
We used basic local alignment search tool searches to identify NALCN homologs in some of the oldest animal lineages, including cnidarians (Nematostella vectensis), placozoans (Trichoplax adhaerens), and sponges (Amphimedon queenslandica). We also found four-domain fungal channels in diverse fungal lineages, including the early-branching Zygomycota (Phycomyces blakesleeanus, and Mucor circinelloides) and Blastocladiomycota (Allomyces macrogynus). Fungal calcium channels and NALCN homologs were notably absent in single-celled opisthokont genomes. We aligned the new sequences with previously identified Cav and Nav channels from animals, choanoflagellates, and the apusozoan protist Thecamonas trahens (Liebeskind et al. 2011; Cai 2012), thought to be the sister group to opisthokonts (Torruella et al. 2011). Support for a monophyletic apusozoan clade is weak (Cavalier-Smith and Chao 2003), but we will refer to the apusomonad Thecamonas as an apusozoan to be consistent with the online database from which the sequence came (supplementary materials, Supplementary Material online) and with recent literature (Torruella et al. 2011).
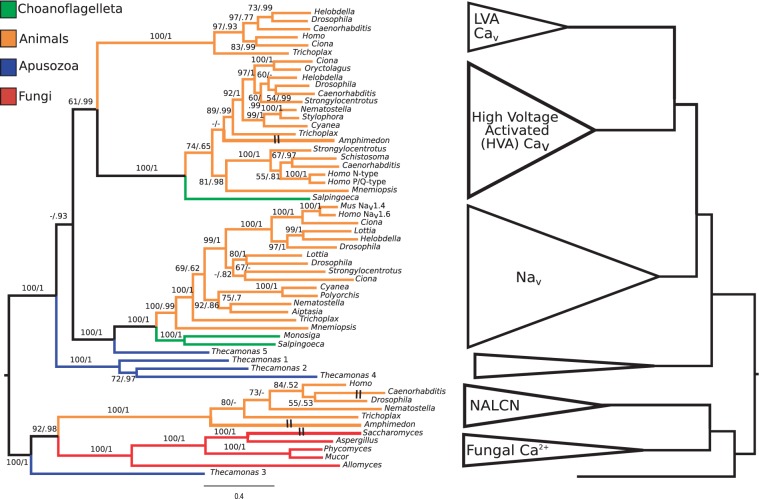
Phylogenetic analysis using maximum likelihood and Bayesian methods places fungal calcium channels and NALCN-like sequences within a well-defined clade to the exclusion of voltage-gated Cav and Nav sequences (fig. 1, supplementary methods, Supplementary Material online). The topology was robust to model choice and estimation method and is consistent with known species trees (Torruella et al. 2011). The voltage-insensitive clade split from the voltage-gated group that includes animal Cav and Nav channels before the divergence of the fungal and animal lineages.
Fig. 1.
Phylogenetic tree of opisthokont four-domain ion channels. NALCN and four-domain fungal calcium channels group together in a well-supported clade. Bootstrap proportions and posterior probabilities are reported for each branch. The cartoon on the right shows major groups of channels, including low- and high-voltage-activated Cav channels (LVA and HVA, respectively), Nav channels, NALCN, and four-domain fungal calcium channel. Hash marks on the left-hand tree denote branches that have been shortened for ease of display.
Unlike Nav and Cav channels, which underwent several rounds of duplication in animals (Liebeskind et al. 2011; Zakon et al. 2011), NALCN channels were found in single copy in most species examined (fig. 1). The sponge A. queenslandica, the cnidarian N. vectensis, and the nematode C. elegans (Pierce-Shimomura et al. 2008) are exceptions to this rule and each have two genes. This is notable because neither sponges nor nematodes have Nav channels. The presence of NALCN in all examined species has been noted previously in bilaterians (Ren 2011), and this finding extends this trend to nonbilaterians.
Nonbilaterian NALCN channels do not have the same pore sequence as previously identified NALCN channels (E/E/K/E or E/K/E/E). NALCN channels in Amphimedon, Trichoplax, and Nematostella have E/E/E/E in the pore, identical to high-voltage-activated Cav channels (fig. 2). The lysine (“K”) in the third domain of NALCN channels is thought to render the channels nonselective among cations (Lu et al. 2007) because single lysine substitutions in wild-type (E/E/E/E) Cav channel pores eliminate selectivity for calcium over monovalent cations (Yang et al. 1993). It is therefore likely that nonbilaterian NALCN channels actually function as calcium-permeable channels. These findings reinforce the view that changes in ion channel selectivity were major steps in the evolution of complex nervous systems in animals (Hille 2001; Liebeskind 2011; Liebeskind et al. 2011).
Fig. 2.
Pore states mapped onto the voltage-insensitive subtree. We show two fixations for a polar, uncharged amino acid along the branches leading to ascomycetes and zygomycetes, but it is equally possible that either an asparagine (N) or a glutamine (Q) could have fixed in the common ancestor of these lineages (black circle) and changed to the other amino acid along one of the branches. Early branching lineages have pores with acidic residues (D or E).
The earliest branching fungal calcium channel (Allomyces) also had an acidic pore motif (fig. 2), which suggests that the common ancestor of all voltage-insensitive channels had an acidic pore and was permeable to calcium. The most diverse lineages of fungi (ascomycetes and zygomycetes) then fixed for polar uncharged amino acids (N or Q) in the first domain pore loop (fig. 2). Basidiomycetes are another diverse fungal clade that was not sampled here but have an identical pore to their sister group, the ascomycetes (data not shown). Unlike animal Cav channels, fungal calcium channels with an N/E/E/E pore are not permeable to sodium even in the absence of calcium (Hong et al. 2010). Because fungal calcium channels are necessary for survival in low-calcium environments in several fungal lineages, this may be adaptive (Liu et al. 2006; Hong et al. 2010). The fixation for N or Q in the pore accompanied a loss of swimming zoospores in fungi at the blastocladiomycete/zygomycete boundary. This is notable because calcium channels underlie mating in yeast and ascospore bursting in Gibberella zeae (Fischer et al. 1997; Hallen and Trail 2008) and may therefore be involved in mating behavior in many other fungi.
Thus, the early branching lineages of both NALCN and fungal channels retained acidic motifs, but the most diverse groups of sampled animals and fungi (bilaterians in animals, and ascomycetes and zygomycetes in fungi) evolved different pore motifs early in their diversification. How these changes may have affected the evolution of animals and fungi will require characterization of channels that group close to the roots of these groups.
Characterized NALCN and fungal calcium channels are voltage insensitive (Lu et al. 2007; Hong et al. 2010), and all channels in this clade had reduced numbers of voltage-sensing residues relative to voltage-gated channels (supplementary fig. S2, Supplementary Material online). This suggests that the homolog in the common ancestor of animals and fungi was not voltage gated and that voltage insensitivity is therefore a shared, derived character of this clade. Because both fungal calcium channels and NALCN rely on accessory proteins for their function, it is also likely that this characteristic evolved at the base of the clade. Although we found no obvious sequence similarity between the known accessory proteins of NALCN and fungal calcium channels, it seems likely that modulation by other proteins facilitated the loss of voltage sensitivity in an ancestral channel and that the modulating proteins themselves have changed over time.
Figure 1 is rooted at the midpoint. To get a more reliable rooting, we used voltage-insensitive and voltage-gated channels as queries to search nonopisthokont genomes for a channel that diverged before the diversification of the channels represented in figure 1. Most major eukaryote lineages have four-domain channels, many of which are hypothesized to be calcium channels on the basis of their pore motifs (Verret et al. 2010; Prole and Taylor 2011). We added 11 nonopisthokont sequences that had good coverage of taxa and channel types to the phylogeny in figure 1.
These sequences could not be reliably placed within the phylogeny, however, making root placement and the status of the Thecamonas channels uncertain (supplementary fig. S3b, Supplementary Material online). However, rooting between voltage-gated and voltage-insensitive channels, as in figure 1, produces a more parsimonious pattern of gene loss in fungi than rooting with either voltage-gated group (one loss instead of two). Some nonopisthokont channels were identical to animal Cav channels in their pore sequence but had Nav-like inactivation loop motifs (supplementary fig. S4, Supplementary Material online) (Smith and Goldin 1997). These enigmatic similarities cannot be adequately explained at present but suggest a complicated evolutionary history.
Our phylogeny suggests that an ancient loss of voltage sensitivity in a lineage of four-domain ion channels and key amino acid replacements affecting ion selectivity in this lineage were both factors in the diversification of fungi and animals. This phylogenetic information clarifies the evolution of voltage-insensitive four-domain channels and suggests fungal calcium channels as possible models for future NALCN research.
Supplementary Material
Supplementary methods and figures S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Emily McTavish, Thomas Keller, April Wright, and Ammon Thompson for helpful comments on the initial manuscript, and associate editor Dr. Andrew Roger and two anonymous editors for thoughtful reviews. They also acknowledge the Broad Institute and their Origins of Multicellularity project for making the genomes of several taxa used in this study available to the public. Research was performed at the University of Texas at Austin. This work was supported by National Institute of Health Grant R01GM084879.
References
- Cai X. Ancient origin of four-domain voltage-gated Na+ channels predates the divergence of animals and fungi. J Membr Biol. 2012;245:117–123. doi: 10.1007/s00232-012-9415-9. [DOI] [PubMed] [Google Scholar]
- Cai X, Clapham DE. Ancestral Ca2+ signaling machinery in early animal and fungal evolution. Mol Biol Evol. 2011;29:91–100. doi: 10.1093/molbev/msr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE-Y. Phylogeny of choanozoa, apusozoa, and other protozoa and early eukaryote megaevolution. J Mol Evol. 2003;56:540–563. doi: 10.1007/s00239-002-2424-z. [DOI] [PubMed] [Google Scholar]
- Emes RD, Grant SGN. The human postsynaptic density shares conserved elements with proteomes of unicellular eukaryotes and prokaryotes. Front Neurosci. 2011;5 doi: 10.3389/fnins.2011.00044. Available from: http://www.frontiersin.org/Journal/Abstract.aspx?s=750&name=neurogenomics&ART_DOI=10.3389/fnins.2011.00044 (accessed July 31, 2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Schnell N, Chattaway J, Davies P, Dixon G, Sanders D. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Lett. 1997;419:259–262. doi: 10.1016/s0014-5793(97)01466-x. [DOI] [PubMed] [Google Scholar]
- Hallen HE, Trail F. The L-type calcium ion channel Cch1 affects ascospore discharge and mycelial growth in the filamentous fungus Gibberella zeae (anamorph fusarium graminearum) Eukaryot Cell. 2008;7:415–424. doi: 10.1128/EC.00248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. Ion channels of excitable membranes. 3rd ed. Sunderland (MA): Sinauer Associates; 2001. [Google Scholar]
- Hong M-P, Vu K, Bautos J, Gelli A. Cch1 restores intracellular Ca2+ in fungal cells during endoplasmic reticulum stress. J Biol Chem. 2010;285:10951–10958. doi: 10.1074/jbc.M109.056218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Cribbs LL, Perez-Reyes E. Cloning of a novel four repeat protein related to voltage-gated sodium and calcium channels. FEBS Lett. 1999;445:231–236. doi: 10.1016/s0014-5793(99)00082-4. [DOI] [PubMed] [Google Scholar]
- Liebeskind BJ. Evolution of sodium channels and the new view of early nervous system evolution. Commun Integr Biol. 2011;4:679–683. doi: 10.4161/cib.17069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeskind BJ, Hillis DM, Zakon HH. Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci U S A. 2011;108:9154–9159. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Du P, Heinrich G, Cox GM, Gelli A. Cch1 mediates calcium entry in cryptococcus neoformans and is essential in low-calcium Environments. Eukaryot Cell. 2006;5:1788–1796. doi: 10.1128/EC.00158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Su Y, Das S, Liu J, Xia J, Ren D. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell. 2007;129:371–383. doi: 10.1016/j.cell.2007.02.041. [DOI] [PubMed] [Google Scholar]
- Lu B, Su Y, Das S, Wang H, Wang Y, Liu J, Ren D. Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature. 2009;457:741–744. doi: 10.1038/nature07579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Zhang Q, Wang H, Wang Y, Nakayama M, Ren D. Extracellular calcium controls background current and neuronal excitability via an UNC79-UNC80-NALCN cation channel complex. Neuron. 2010;68:488–499. doi: 10.1016/j.neuron.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash HA, Scott RL, Lear BC, Allada R. An unusual cation channel mediates photic control of locomotion in Drosophila. Curr Biol. 2002;12:2152–2158. doi: 10.1016/s0960-9822(02)01358-1. [DOI] [PubMed] [Google Scholar]
- Paidhungat M, Garrett S. A homolog of mammalian, voltage-gated calcium channels mediates yeast pheromone-stimulated Ca2+ uptake and exacerbates the cdc1(Ts) growth defect. Mol Cell Biol. 1997;17:6339–6347. doi: 10.1128/mcb.17.11.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Chen BL, Mun JJ, Ho R, Sarkis R, McIntire SL. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc Natl Acad Sci U S A. 2008;105:20982–20987. doi: 10.1073/pnas.0810359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prole DL, Taylor CW. Identification of intracellular and plasma membrane calcium channel homologues in pathogenic parasites. PLoS One. 2011;6:e26218. doi: 10.1371/journal.pone.0026218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D. Sodium leak channels in neuronal excitability and rhythmic behaviors. Neuron. 2011;72:899–911. doi: 10.1016/j.neuron.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR, Goldin AL. Interaction between the sodium channel inactivation linker and domain III S4-S5. Biophys J. 1997;73:1885–1895. doi: 10.1016/S0006-3495(97)78219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne LA, Mezghrani A, Varrault A, et al. (11 co-authors) The NALCN ion channel is activated by M3 muscarinic receptors in a pancreatic [beta]-cell line. EMBO Rep. 2009;10:873–880. doi: 10.1038/embor.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torruella G, Derelle R, Paps J, Lang BF, Roger AJ, Shalchian-Tabrizi K, Ruiz-Trillo I. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single copy protein domains. Mol Biol Evol. 2011;29:531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verret F, Wheeler G, Taylor AR, Farnham G, Brownlee C. Calcium channels in photosynthetic eukaryotes: implications for evolution of calcium-based signalling. New Phytol. 2010;187:23–43. doi: 10.1111/j.1469-8137.2010.03271.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Elllnor PT, Sather WA, Zhang J-F, Tsien RW. Molecular determinants of Ca2+ selectivity and ion permeation in L-type Ca2+ channels. Nature. 1993;366:158. doi: 10.1038/366158a0. [DOI] [PubMed] [Google Scholar]
- Zakon HH, Jost MC, Lu Y. Expansion of voltage-dependent Na+ channel gene family in early tetrapods coincided with the emergence of terrestriality and increased brain complexity. Mol Biol Evol. 2011;28:1415–1424. doi: 10.1093/molbev/msq325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelter A, Bencina M, Bowman BJ, Yarden O, Read ND. A comparative genomic analysis of the calcium signaling machinery in Neurospora crassa, Magnaporthe grisea, and Saccharomyces cerevisiae. Fungal Genet Biol. 2004;41:827–841. doi: 10.1016/j.fgb.2004.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.