Summary
Poly(ADP-ribose) polymerases (PARPs) (also known as ADP-ribosyl transferase D proteins) modify acceptor proteins with ADP-ribose modifications of varying length (reviewed in refs 1–3). PARPs regulate key stress response pathways, including DNA damage repair and the cytoplasmic stress response2,3,4,5,6. Here, we show that PARPs also regulate the unfolded protein response (UPR) of the endoplasmic reticulum (ER). Human PARP16/ARTD15 is a tail-anchored ER transmembrane protein required for activation of the functionally related ER stress sensors PERK and IRE1α during the UPR. The third identified ER stress sensor, ATF6, is not regulated by PARP16. Similar to other PARPs that function during stress, PARP16 enzymatic activity is up-regulated during ER stress when it (ADP-ribosyl)ates itself, PERK and IRE1α. (ADP-ribosyl)ation by PARP16 is sufficient for activating PERK and IRE1α in the absence of ER stress, and is required for PERK and IRE1 α activation during the UPR. Modification of PERK and IRE1α by PARP16 increases their kinase activities and the endonuclease activity of IRE1α. Interestingly, the C-terminal luminal tail of PARP16 is required for PARP16 function during ER stress, suggesting that it transduces stress signals to the cytoplasmic PARP catalytic domain.
Keywords: PARP16, (ADP-ribosyl)ation, (ADP-ribose), tail-anchored protein, C-tail, transmembrane protein, endoplasmic reticulum, unfolded protein response, ER stress, PERK, IRE1α, ATF6, eIF2α, XBP-1
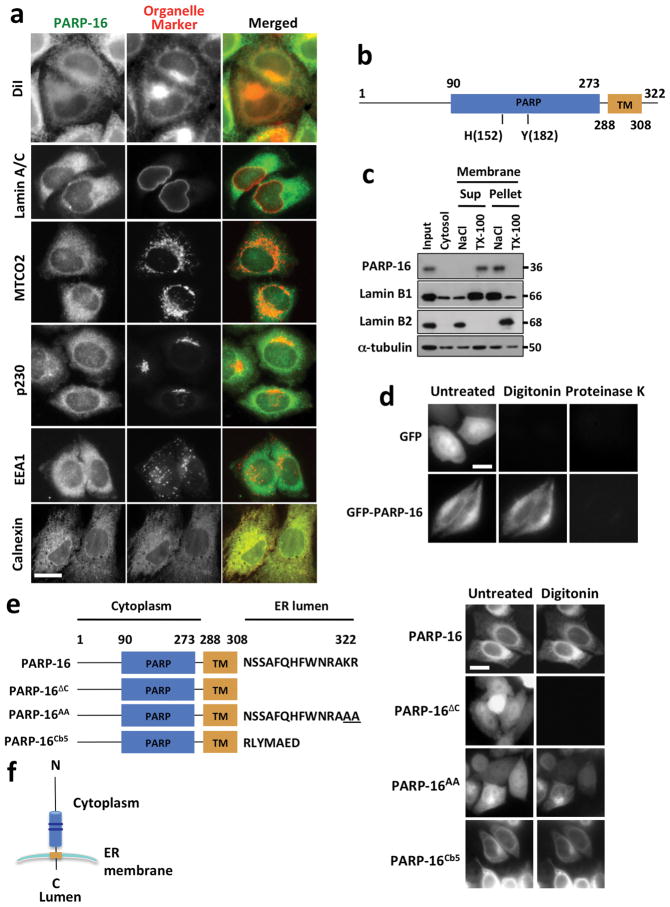
We previously identified a reticular membrane localization for uncharacterized PARP16 in a screen analyzing PARP function using lypophilic dye DiI (personal communication, Sejal Vyas; Fig. 1a). To identify organelles to which it localizes, HeLa cells (utilized in all subsequent experiments) were stained with antibodies against PARP16 and markers for membrane bound organelles–Calnexin, Lamin A/C, MTCO2, p230, and EEA1. Of these, PARP16 and Calnexin localization strongly overlapped, suggesting that PARP16 is an ER protein (Fig. 1a).
Figure 1. PARP16 is a tail-anchored ER transmembrane protein.
HeLa cells used in all figures. a, Cells stained for PARP16 (green) and organelle markers (red). b, PARP16 domain structure. TM: transmembrane domain; PARP: PARP catalytic domain. c, Membrane extraction assay. Immunoblots of input, cytosol, supernatant and pellet of 1 M NaCl or 1% Triton X-100 (TX-100) treated membrane fractions. Molecular weight (MW) (kD) at right of blot. d, Protease protection assay using Untreated, Digitonin- or Proteinase K-treated cells expressing GFP-PARP16 or GFP. e, Left: PARP16 mutants. Right; Untreated or digitonin-treated cells expressing GFP fusions of indicated proteins. f, PARP16 topology within the ER. Dark blue bands mark conserved histidine and tyrosine residues within the PARP catalytic domain. Bars, 10 μm.
Based on primary sequence, PARP16 is predicted to be a tail-anchored (TA) protein with a hydrophobic transmembrane domain at amino acid 288–308 (Fig. 1b; UniProtKB)7. TA proteins are single-spanning transmembrane proteins that contain cytoplasmic N-termini, short transmembrane domains (< 30 a.a.) and C-terminal domains called C-tails (~10–15 amino acids) positioned within the lumen of target organelles. C-tails target to the ER via net positive charge rather than specific amino acid composition and are inserted post-translationally into ER membrane via the GET (Golgi-ER trafficking) complex8,9. To determine if PARP16 is a TA protein, we performed membrane extraction assays to confirm that PARP16 is transmembrane, protease protection assays to determine if the N-terminus is cytoplasmic, and truncation/mutation assays to determine if the C-terminus acts as a C-tail10. Treatment of purified membrane fractions with 1 M NaCl released peripherally associated membrane protein Lamin B2 but not PARP16, while treatment with 1% Triton X-100 resulted in the release of transmembrane protein Lamin B1 and PARP16, identifying PARP16 as a transmembrane protein (Fig. 1c). A N-terminal GFP-fusion to PARP16, GFP-PARP16 remained membrane associated in response to Digitonin treatment (in contrast to GFP only controls), while subsequent Proteinase K treatment resulted in loss of fluorescence, suggesting that the N-terminus or PARP16 is cytoplasmic (Fig. 1d). Finally, a C-tail truncation (PARP16ΔC) and a PARP16AA mutant failed to localize to the ER (a small portion of PARP16AA remained ER associated) while a Cytochrome b5 chimera (PARP16Cb5) with PARP16 C-tail replaced with ER-associated Cytochrome b5 C-tail retained ER localization even upon Digitonin treatment, demonstrating that the C-terminus of PARP16 functions as a C-tail (Fig. 1e, f).
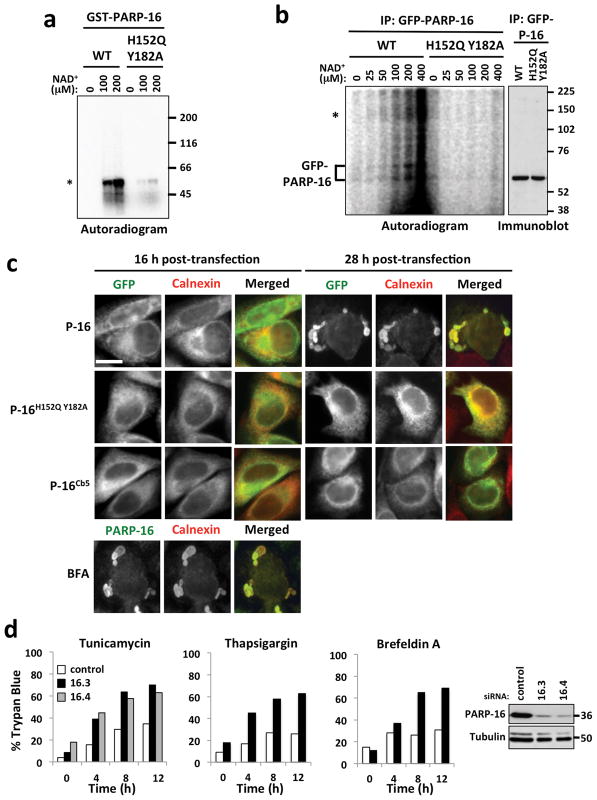
Human PARP16 (ADP-ribosyl)ates itself (personal communication, Sejal Vyas) and contains histidine and tyrosine residues at amino acid 152 and 182 within its catalytic domain, residues thought to be critical for enzymatic activity (Fig. 1b)11. To determine if these residues are required for enzymatic activity, GST-PARP16 and GST-PARP16H152Q Y182A were expressed and purified in E. coli, and 32P-NAD+ incorporation assays performed. Self-modification of GST-PARP16 was detected at its molecular weight in a NAD+ dose-dependent manner, while GST-PARP16H152Q Y182A exhibited incorporation activity at ~ 6% of wild-type (Fig. 2a).
Figure 2. (ADP-ribosyl)ation activity of PARP16 is required for the ER stress response.
MW at right of blots. a, 32P-NAD + incorporation by recombinant GST-PARP16 and GST-PARP16H152Q Y182A. Asterisk= MW of GST-PARP16. b, EMAA using GFP-PARP16 or GFP-PARP16H152Q Y182A containing microsomes. Asterisk= high MW incorporation of 32P-NAD. c, Cells expressing GFP-PARP16, GFP-PARP16H152Q Y182A, or GFP-PARP16Cb5 for 16h or 28 h, or untransfected cells treated with Brefeldin A (BFA), stained for PARP16 (green) and Calnexin (red). d, Trypan blue staining of control or PARP16 knock-downs at indicated time points after Tunicamycin, Thapsigargin or Brefeldin A treatment. 16.3 and 16.4 are different siRNAs against PARP16 (n=4 for siRNA 16.3, and 2 for siRNA 16.4). For Tunicamycin, Thapsigargin, or Brefeldin A-treated PARP16 knock down cells, 0.001 < p < 0.05. Immunoblots of knock-down shown at right. Bar, 10 μm.
Analysis of PARP16 membrane topology suggests its catalytic domain is cytoplasmic (Fig. 1f). To determine if PARP16 ADP-ribosylation activity is cytoplasmic, and examine its function in the context of ER membrane, we developed an ER microsome assay to monitor NAD+ incorporation called the ER microsome (ADP-ribosyl)ation Assay or EMAA. Microsomes were purified from cells expressing GFP-PARP16 or GFP-PARP16H152Q Y182A, incubated with 32P-NAD+, dissolved to extract and purify GFP-PARP16, then 32P-NAD+ incorporation into GFP-PARP16 assayed via autoradiography12. Microsomes purified for this purpose stained positive for ER tracker, were highly enriched in ER proteins, and were intact since they did not contain protein from other cellular compartments (Fig. S1a, b). Since intact ER microsomes are impermeable to NAD+, any incorporation of 32P-NAD+ occurs outside of the microsome lumen13. Self-modification of GFP-PARP16 was detected at its molecular weight in a NAD+ dose-dependent manner while GFP-PARP16H152Q Y182A failed to incorporate NAD+ (Fig. 2b, Left), suggesting that PARP16 (ADP-ribosyl)ation activity is cytoplasmic and requires His152 and Tyr182. Multiple migrating forms of GFP-PARP16 were detected. In addition, 32P-NAD+ was incorporated at higher molecular weight, suggesting the presence of binding proteins that are modified by PARP16 (Fig. 2b, Asterisk).
Interestingly, prolonged PARP16 over-expression (> 28 h) resulted in abnormal ER morphology with >80% of PARP16 overexpressing cells containing abnormal globular ER structures (Fig. 2c, S2). This phenotype was time and/or protein concentration dependent as ER appeared normal at 16 h of expression, and required PARP16 enzymatic activity and an intact C-tail as only ~ 5% of cells expressing PARP16H152Q Y182A or PARP16Cb5 at levels similar to wild type PARP16, contained abnormal ER (Fig. 2c, S2). Since GFP-PARP16 and GFP-PARP16H152Q Y182A both localized to the ER, enzymatic activity is not required for ER localization.
Abnormal ER structures resulting from prolonged PARP16 over-expression resemble ER from stressed cells14, leading us to examine PARP16 function in the unfolded protein response (UPR) (Fig. 2c). The UPR is an ER stress response activated by an increase in unfolded proteins within the ER lumen. In mammals, three transmembrane ER stress sensors, PERK and IRE1α, kinases with functionally interchangeable luminal domains, and the transcription factor ATF6, regulate separate but interconnected UPR signaling pathways (reviewed in refs 15 – 19). Under non-stress conditions, each sensor is bound to and inhibited by BiP, an ER specific chaperone. To determine if PARP16 functions in the UPR, we knocked it down with two siRNAs generated against distinct sequences, then activated the UPR using Tunicamycin, Brefeldin A, and Thapsigargin, and examined the effects. Consistent with perturbed UPR function, PARP16 knock-down rendered cells highly sensitive to ER stress, resulting in increased cell death (Fig. 2d).
The high sensitivity to ER stress in PARP16 knock-downs could also be explained by ER dysfunction or a general misregulation of cellular stress responses. However, ER function was intact in PARP16 knock-downs as measured by intracellular concentration of ROS (reactive oxygen species) and Ca2+. PARP16 and control knock-down cells exhibited similar ROS generation, measured via CM-H2DCFDA in the presence or absence of H2O2, and similar Ca2+ leakage to the cytoplasm assayed via Fura-4F 340/380 nm fluorescence upon Thapsigargin treatment (Fig. S3a, b). Non-UPR related cellular stress responses, such as DNA damage repair and the cytoplasmic stress response, both known to require PARP activity, were also intact in PARP16 knock-downs. PARP16 and control knock-down cells exhibited a similar response to DNA damage induced by Cisplatin, with similar levels of γ-H2AX foci formation, and cytoplasmic stress induced by Arsenite, although the number of cells positive for TIA-1 staining stress granules were slightly reduced relative to controls (Fig. S3c, d).
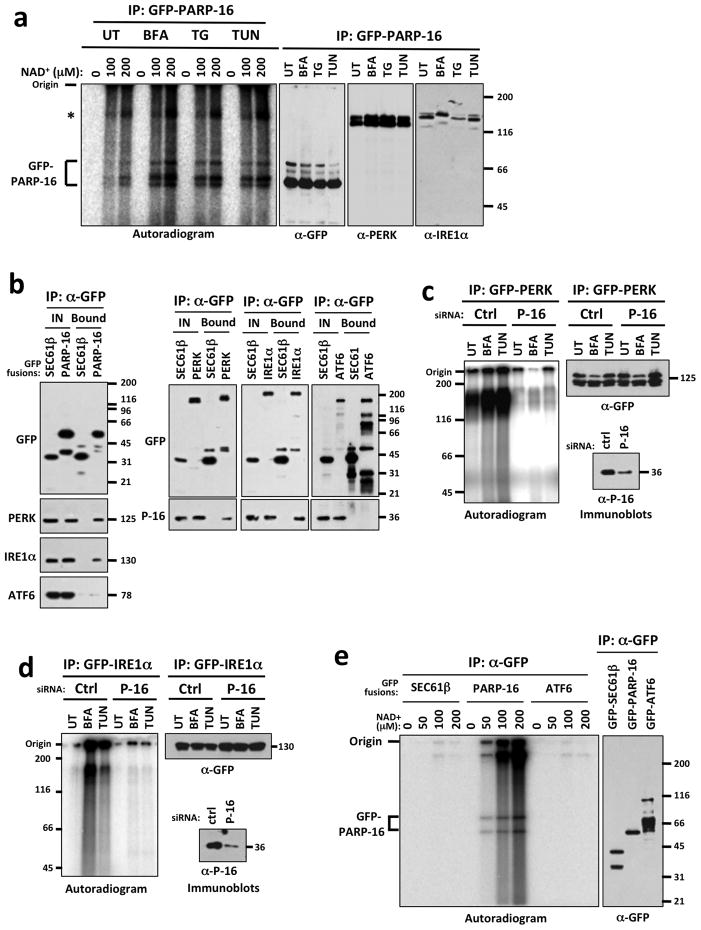
All known PARP-dependent stress responses result in up-regulation of PARP enzymatic activity3,6. We examined PARP16 enzymatic activity during the UPR via EMAA using cells expressing GFP-PARP16 for 16 h, a condition that did not affect ER organization (Fig. 2c). All subsequent EMAAs were performed in this manner. Cells expressing GFP-PARP16 were treated +/− ER stress inducing agents, and PARP16 activity assayed. ER stress resulted in significant increases in GFP-PARP16 self-modification in a NAD+ dose-dependent manner (5~8 fold increase at 100 μM NAD+ and 8~13 fold increase at 200 μM NAD+ depending on stressor), and a dramatic electrophoretic mobility shift of GFP-PARP16 was detected via immunoblot and autoradiogram (Fig. 3a). Additional higher molecular weight bands were also observed on the autoradiogram, migrating at the molecular weight of PERK (125 kD) and IRE1α (130 kD), but not ATF6 (75 kD). PERK and IRE1α were found in these GFP-PARP16 precipitates via immunoblot under 450 mM NaCl conditions, demonstrating a robust association between PARP16, PERK and IRE1α (Fig. 3a, Right panels). These high molecular weight bands of NAD+ incorporation could represent (ADP-ribosyl)ation of PERK and IRE1α.
Figure 3. PERK and IRE1α are (ADP-ribosyl)ated in a PARP16-dependent manner during the UPR.
MW (kD) at right of blots. (UT)= untreated; (BFA)= Brefeldin A treated; (TG)= Thapsagargin treated; (TUN)= Tunicamycin treated. a, Autoradiogram of EMAA showing ADP-ribose incorporation. Immunoblots of GFP-PARP16 precipitates are shown at right. Asterisk= high MW NAD+ incorporation. n = 5; 0.01 < p of fold increase < 0.05 for all stressors. b, ER microsome based co-immunoprecipitation assays of GFP-fusion proteins. Shown are immunoblots of precipitated GFP fusions. c–d, EMAA using control or PARP16 knock-downs. Shown are autoradiogram and immunoblots of GFP-PERK (c) or GFP-IRE1α immunoprecipitates (d). For both (c) and (d), n=4; 0.005 < p of fold increase < 0.05 for all stressors. PARP16 immunoblots of control and PARP16 knock-down lysates are shown. e, EMAA for SEC61β, ATF6 and PARP16. Shown are autoradiogram and immunoblot of the immunoprecipitated GFP fusions. n=2.
To determine if PARP16 binds to ER stress sensors in the absence of ER stress, GFP fusions to PARP16, PERK, IRE1α, ATF6 or SEC61β, an UPR-unrelated ER transmembrane protein, were expressed and co-immunoprecipitation assays performed. While PERK and IRE1α were present in GFP-PARP16 precipitates, and PARP16 was identified in GFP-PERK and IRE1α precipitates, no significant binding was identified between ATF6, SEC61β and PARP16 (Fig. 3b). Thus, PARP16 selectively binds to PERK and IRE1α but not ATF6 in the presence and absence of ER stress.
Our data suggested that PERK and IRE1α could be substrates of PARP16. We examined NAD+ incorporation onto GFP-PERK or GFP-IRE1α via EMAA in cells transfected with control or PARP16 siRNA, treated with or without ER stress inducing drugs. Low level (ADP-ribosyl)ation of GFP-PERK and GFP-IRE1α was detected in control knock-down cells in the absence of drug (Fig. 3c, d), likely due to the previously described UPR induction upon PERK or IRE1α expression20,21. (ADP-ribosyl)ation of GFP-PERK and GFP-IRE1α increased under ER stress (5 fold and 4–11 fold, respectively with differences dependent on stressor). In both cases this increase required PARP16, as modification was dramatically reduced in PARP16 knock downs (Fig. 3c, d). Neither GFP-SEC61β nor GFP-ATF6 were (ADP-ribosyl)ated in similar assays (Fig. 3e).
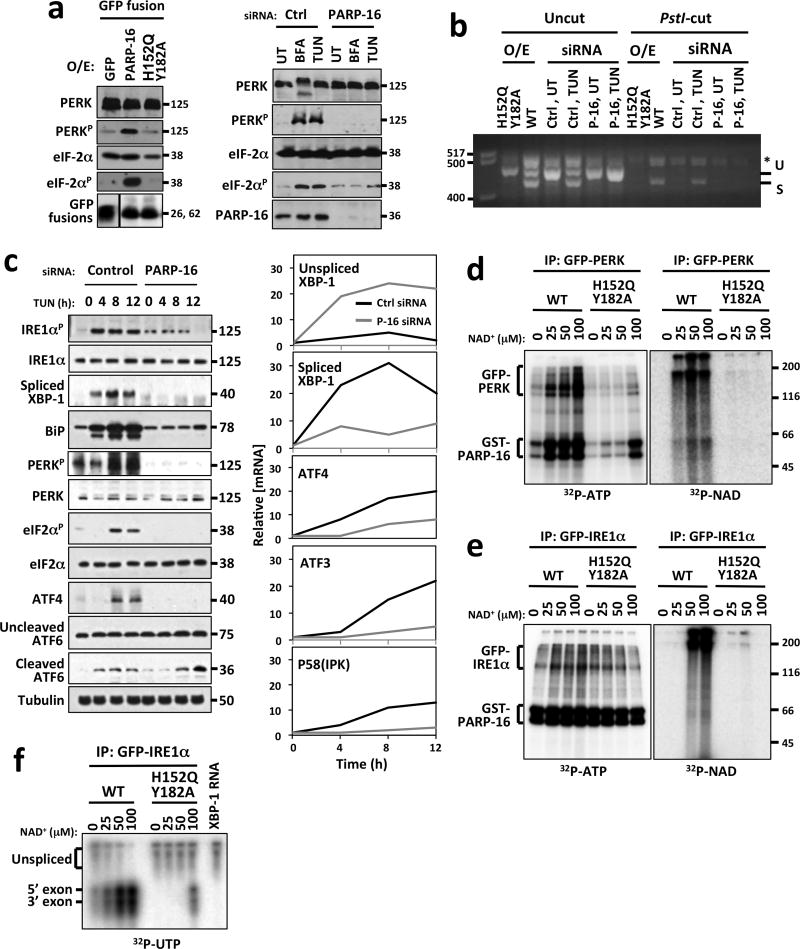
To determine the effects of (ADP-ribosyl)ation on PERK and IRE1α signaling, GFP-PARP16, GFP-PARP16H152Q Y182A, or GFP alone were over-expressed at similar concentrations, and PERK and IRE1α activation examined using two standard assays; (i) detection of PERK phosphorylation at Thr 981 and phosphorylation of its substrate eIF2α at Ser 51 using phospho-specific antibodies, and (ii) monitoring splicing of the IRE1α substrate XBP-1 mRNA. Over-expression of GFP-PARP16, but not GFP-PARP16H152Q Y182A or GFP resulted in PERK and eIF2 α phosphorylation, and XBP-1 splicing (Fig. 4a, b, S4a), suggesting that (ADP-ribosyl)ation by PARP16 is sufficient to activate PERK and IRE1α.
Figure 4. Enzymatic activity of PARP16 is required for activation of PERK- and IRE1α–mediated UPR.
MW (kD or bp) at right or left of blots or gels. (UT)= untreated; (BFA)= Brefeldin A; (TG)= Thapsagargin; (TUN)= Tunicamycin; PERKp= phospho-PERK; eIF2αp= phospho-eIF2 α. Asterisk= hybrid amplicons. a, Left, Immunoblots of cells overexpressing GFP or GFP fusions to PARP16 or PARP16H152Q/Y182A. Right, Immunoblots of cells transfected with control or PARP16 siRNA. b, XBP-1 mRNA splicing assay from control, GFP-PARP16 or GFP-PARP16H152Q Y182A expressing cells, or cells transfected with control or PARP16 siRNA. Unspliced (U) and spliced (S) XBP-1 cDNA were amplified via RT-PCR then cut with Pst1 restriction enzyme. Only unspliced XBP-1 is cut by PstI. c, Left, Immunoblots of cell lysates. Right, RT-qPCR analysis of UPR-dependent transcription in control or PARP16 knock-downs treated with Tunicamycin. d and e, ER microsome based (ADP-ribosyl)ation and kinase assays. Microsomes containing GFP-PERK (d) or GFP-IRE1α (e) were (ADP-ribosyl)ated via addition of 0.5 μg GST-PARP16 or GST-PARP16H152Q Y182A in the presence of 32P-NAD+. Duplicate NAD+ incorporation reactions performed under identical conditions using unlabeled NAD+, then kinase activity assayed via γ32P-ATP incorporation. For d and e, n=4. f, ER microsome based (ADP-ribosyl)ation and IRE1α endonuclease assays. (ADP-ribosyl)ation of GFP-IRE1α was performed in a similar manner to (d) using unlabeled NAD+. In vitro transcribed 32P labeled XBP-1 transcript was incubated with the (ADP-ribosyl)ated GFP-IRE1α immunoprecipitates and assayed for splicing as indicated via presence of 5′ and 3′ exons. n=4.
To determine if PARP16 is required for PERK or IRE1α activation, we compared activation in PARP16 knock-downs to controls. Control cells treated with Brefeldin A or Tunicamycin resulted in robust phosphorylation of PERK and eIF2α, and XBP-1 splicing, while PARP16 knock-downs similarly treated failed to activate PERK or IRE1α (Fig. 4a, b, S4a. Since PERK and IRE1 α activity result in the time-dependent activation of downstream transcriptional programs regulated by ATF4 and spliced XBP-1, respectively, we analyzed PERK and IRE1α signaling every 4 h over a 12 h period in PARP16 knock-downs and controls treated with Tunicamycin. Components of each pathway were analyzed via immunoblot or RT-qPCR analysis. While IRE1α activation, detected by phosphorylation of IRE1α, occurred 4 h post treatment in controls, such phosphorylation was barely detectable in PARP16 knock-downs at any time (Fig. 4c, Left). At 4 h, spliced XBP-1 protein began to accumulate in controls, but was undetectable in PARP16 knock-downs (Fig. 4c, Left). IRE1α dependent transcriptional programs were also defective in PARP16 knock-downs; In control cells, unspliced XBP-1 mRNA decreased and spliced XBP-1 mRNA increased (Fig. 4c, Right), whereas in PARP16 knock-downs, unspliced XBP-1 mRNA increased, due to ATF6 activation, and spliced XBP-1 mRNA induction was reduced 5-fold at 4h and 15-fold at 8 h (Fig. 4c, Right). P58(IPK) mRNA was induced in control but not PARP16 knock downs (Fig. 4c, Right). As expected, BiP concentrations increased at 4 h and plateaued at 8 h in control knock-downs due to increased transcription of BiP mRNA by spliced XBP1. A minor increase in BiP appeared at 12 h in PARP16 knock-downs (Fig. 4c, Left).
Activation of the PERK branch appeared at 8 h in controls as determined by PERK and eIF2α phosphorylation, and ATF4 synthesis. Such phosphorylation was barely detectable in PARP16 knock-downs at this time-point, and PERK-dependent transcriptional programs were defective; while ATF3 and ATF4 mRNA began to accumulate at 8 h in controls, accumulation was reduced 5-fold at 8 h and 10-fold at 12 h in PARP16 knock downs (Fig. 4c). ATF6 activation was also monitored by examining cleavage to its active transcription factor. Cleavage appeared at 4 h in control and PARP16 knock downs, confirming that ATF6 activation is intact in the PARP16 knock downs (Fig. 4c, Left).
While our data strongly point to direct effects of PARP16 on PERK and IRE1α signaling, compromised ERAD (ER-associated degradation) and/or chaperone capacities of the ER in PARP16 knock-downs could also affect UPR activation. ERAD activity in PARP16 knock-downs was examined by measuring clearance of CD3δ-YFP, a model substrate of ERAD machinery. CD3δ-YFP degradation kinetics were similar in PARP16 knock downs and controls as assayed by cycloheximide chase. Inhibition of the proteasome via MG132 rescued degradation (Fig. S5a), suggesting that ERAD activity is similar in control and PARP16 knock-downs. Cells overexpressing intermediate amounts of mCherry-PARP16 also exhibited similar kinetics of CD3δ-YFP clearance (Fig. S5a), suggesting that overexpression of PARP16 does not perturb ERAD activity. The protein-folding capacity of the ER in PARP16 knock-downs appear to be similar to controls as the protein concentrations of ER chaperones BiP and Calnexin, and disulfide isomerases PDI and ERp57, were similar (Fig. S5b).
The increase in PARP16 enzymatic activity, and (ADP-ribosyl)ation of PERK and IRE1α during the UPR raised the possibility that (ADP-ribosyl)ation directly regulates PERK and IRE1α enzymatic activity. We examined PERK and IRE1α kinase activity in response to (ADP-ribosyl)ation by PARP16 via self-phosphorylation assays. ER microsomes purified from GFP-PERK or GFP-IRE1α expressing cells were washed with 1M NaCl to remove bound PARP16, returned to physiological salt buffer, split into duplicate reactions, and incubated with unlabeled NAD+ plus either GST-PARP16 or GST-PARP16H152Q Y182A, or 32P-NAD+ plus either recombinant protein. (ADP-ribosyl)ated GFP-PERK or GFP-IRE1α were extracted from the microsomes and purified under 1 M NaCl conditions to remove added recombinant PARP16 proteins. Reactions containing unlabeled NAD+ were incubated with 32P-ATP, and reactions containing 32P-NAD+ were incubated with unlabeled ATP. (ADP-ribosyl)ation of GFP-PERK and GFP-IRE1α increased in a GST-PARP16 and NAD+ dose dependent manner (32P-NAD+ autoradiograms in Fig. 4d, e, S4c). 32P-NAD+ incorporation at the molecular weight of GST-PARP16 was also observed (32P-NAD+ autoradiograms in Fig. 4d, e, S4c), representing residual binding of GST-PARP16 with GFP-PERK or GFP-IRE1α even after 1 M NaCl washes. As shown in 32P-ATP autoradiograms in Fig. 4d, e and S4c, increased (ADP-ribosyl)ation of GFP-PERK or GFP-IRE1α resulted in a dose-dependent increase in kinase activity (for GFP-PERK a 4–18 fold increase depending on NAD+ concentration, and for IRE1α, a2–5 fold increase depending on NAD+ concentration), suggesting that (ADP-ribosyl)ation by PARP16 directly up-regulates GFP-PERK and GFP-IRE1α kinase activity. Phosphorylation by PERK at the molecular weight of GST-PARP16 was detected in a GST-PARP16 and NAD+ dose dependent manner, indicating that PARP16 is likely a substrate of PERK. Such phosphorylation was dramatically reduced (5–10 fold reduction depending on NAD+ concentration) in GST-PARP16H152Q Y182A samples (Fig. 4d and S4c). Phosphorylation by GFP-IRE1α at the molecular weight of GST-PARP16 and GST-PARP16H152Q Y182A was also detected (Fig. 4e), suggesting that PARP16 is an IRE1α substrate and that PARP activity is not required for phosphorylation by IRE1α. Such phosphorylation does not appear NAD+ concentration dependent.
Next, we examined the effects of (ADP-ribosyl)ation on IRE1α endonuclease activity. GFP-IRE1α purified as in Fig. 4e was incubated with 32P labeled mouse XBP-1 mRNA containing the intron flanked by truncated exons. Increased (ADP-ribosyl)ation of GFP-IRE1α resulted in a NAD+ dose-dependent cleavage of XBP-1 mRNA indicated by the appearance of 5′ and 3′ exons (5–12 fold increase depending on NAD+ concentration; Fig. 4f), suggesting that (ADP-ribosyl)ation of IRE1α by PARP16 directly up-regulates endonuclease activity.
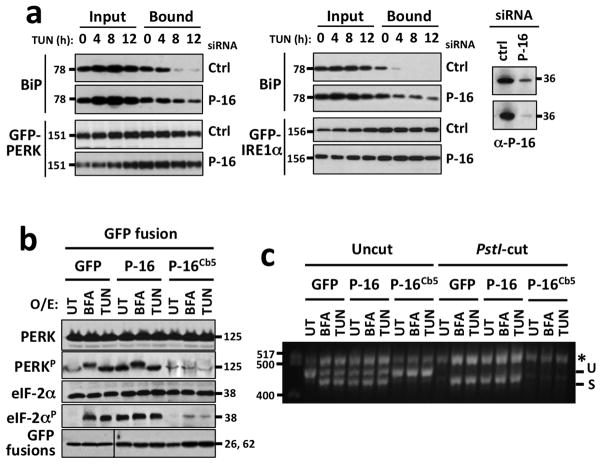
One potential mechanism by which PARP16 regulates PERK and IRE1α is via BiP binding. We examined BiP dissociation from PERK and IRE1α during the UPR to determine if it is affected in PARP16 knock downs. ER microsomes were purified from control or PARP16 knock-down cells expressing either GFP-PERK or GFP-IRE1α and treated with Tunicamycin. GFP fusions were purified from the microsomes every 4 h for 12h, and immunoprecipitates analyzed for the presence of BiP. In controls, BiP dissociated from GFP-IRE1α and GFP-PERK at 4 and 8 h respectively (Fig. 5a). In PARP16 knockdowns, BiP remained bound to GFP-PERK and GFP-IRE1α throughout the time course with a slight reduction in binding, suggesting that BiP dissociation was impaired (Fig. 5a). Since BiP displacement from PERK and IRE1α occurs inside the ER lumen, these data indicate a potential function for the PARP16 C-tail in facilitating BiP dissociation from the luminal domains of PERK and IRE1α.
Figure 5. The C-tail of PARP16 is required for activation of PERK- and IRE1α–mediated UPR.
a, ER microsome based co-immunoprecipitation assays. GFP-PERK or GFP-IRE1α purified from ER microsomes from cells treated with control (Ctrl) or PARP16 (P-16) siRNA, then analysed for BiP binding via immunoblot. PARP16 blots from each assay are shown. b, Immunoblots of cell expressing GFP, or GFP fusions to PARP16 or PARP16Cb5. c, XBP-1 mRNA splicing assay, similar to (b) but performed on PARP16 or PARP16Cb5 overexpressing cells.
The requirement of PARP16 for PERK and IRE1α activation suggests that PARP16 functions upstream of PERK and IRE1α and that the C-tail of PARP16 might transduce stress signals from the ER lumen to the cytoplasmic PARP domain. To determine if this is the case, we expressed GFP-PARP16Cb5 and treated with ER stress inducing drugs. Cells expressing GFP-PARP16Cb5 were unable to activate PERK and IRE1α (Fig. 5b, c and S4b), suggesting that the luminal C-tail of PARP16 is necessary for PARP16 function in the UPR, and that PARP16Cb5 acts as a dominant negative for PARP16 function in the UPR.
In conclusion, we show that the ER-associated tail-anchored protein PARP16 selectively (ADP-ribosyl)ates PERK and IRE1α during the UPR, and that such modification is required for activation of PERK and IRE1α at least in part by increasing their kinase and endonuclease activities. Interestingly, S. cerevisiae lacks PARP proteins and has only one ER stress sensor, Ire1. In the absence of stress, the peptide-binding pocket of yeast Ire1 is fully open, while in humans the peptide binding pockets of IRE1α and PERK are partially closed (reviewed in ref 19). Perhaps the C-tail of PARP16 interacts with the peptide-binding pockets of PERK and IRE1α, opening it. Since ATF6 lacks a peptide binding pocket, this could explain the selective activation of PERK and IRE1α by PARP1620. Further, S. cerevisiae Ire1 appears to be activated primarily by direct binding of unfolded protein to the peptide-binding pocket, while mammalian IRE1α relies on BiP dissociation for activation (reviewed in ref 21). PARP16 could facilitate dissociation of BiP from IRE1α and PERK upon ER stress since BiP dissociation from IRE1α and PERK was impaired in PARP16 knock-downs.
Our work brings the number of stress responses requiring PARP activity to three DNA damage, cytoplasmic stress granule assembly, and the unfolded protein response. The latter two stress responses converge on eIF2α phosphorylation, suggesting that PARPs could have evolved functions in regulating translation as a means to respond to cytoplasmic stresses. Cancer cells often exhibit increased protein folding capacities within the ER due to increased protein synthesis. Since PARPs have been shown to be highly druggable, PARP16 could be an attractive target for therapeutic inhibition for cancers and protein folding diseases.
Methods Summary
Provided in the Supplemental Section
Supplementary Material
Acknowledgments
We thank Nica Borgese for sharing reagents; Tenzin Sangpo for technical assistance; T Jacks, H Lodish, H Ploegh, UL RhajBhandary, and M Chesarone-Cataldo for comments on the manuscript. P.C. is a Rita Allen Foundation Scholar, a Kimmel Foundation for Cancer Research Scholar, and was a Howard S. and Linda B. Stern Career Development assistant professor. This work is partially funded by Cancer Center Support (core grant P30-CA14051), grant 5R01GM087465-02 from the National Institutes of Health (to P.C.), Curt and Kathy Marble, and the Jeptha H and Emily V. Wade Fund (to P.C.), and partially supported by the Ludwig fund for Cancer Research fellowship (to M.J.).
Footnotes
Author Contributions
PC and MJ designed experiments and wrote the manuscript. MJ performed the experiments and data analysis.
Publisher's Disclaimer: This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2–3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
Literature cited
- 1.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribodyltransferases. Trends in Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 3.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 4.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 5.Ménissier-de Murcia J, Molinete M, Gradwohl G, Simonin F, de Murcia G. Zinc-binding domain of poly(ADP-ribose)polymerase participates in the recognition of single strand breaks on DNA. J Mol Biol. 1989;210:229–233. doi: 10.1016/0022-2836(89)90302-1. [DOI] [PubMed] [Google Scholar]
- 6.Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-Ribose) Regulates Stress Responses and MicroRNA Activity in the Cytoplasm. Mol Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitley P, Elin Grahn E, Kutay U, Rapoport TA, Heijne G. A 12-Residue-long Polyleucine Tail Is Sufficient to Anchor. J Biol Chem. 1996;271:7583–7586. doi: 10.1074/jbc.271.13.7583. [DOI] [PubMed] [Google Scholar]
- 8.Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borgese N, Brambillasca S, Colombo S. How tails guide tail-anchored proteins to their destinations. Curr Opin Cell Biol. 2007;19:368–375. doi: 10.1016/j.ceb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 10.Lorenz H, Hailey DW, Lippincott-Schwartz J. Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat Methods. 2005;3:205–210. doi: 10.1038/nmeth857. [DOI] [PubMed] [Google Scholar]
- 11.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Stephens SB, Dodd RD, Lerner RS, Pyhtila BM, Nicchitta CV. Analysis of mRNA partitioning between the cytosol and endoplasmic reticulum compartments of mammalian cells. Methods Mol Biol. 2008;419:197–214. doi: 10.1007/978-1-59745-033-1_14. [DOI] [PubMed] [Google Scholar]
- 13.Hamman BD, Chen JC, Johnson EE, Johnson AE. The aqueous pore through the translocon has a diameter of 40–60 A during cotranslational protein translocation at the ER membrane. Cell. 1997;89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- 14.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- 16.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 17.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 19.Walter P, Ron D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 20.Bertolotti A, Zhang Y, Hendershot L, Harding H, Ron D. Dynamic interaction of BiP and the ER stress transducers in the unfolded protein response. Nature Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 21.Kimata Y, Kohno K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr Opin Cell Biol. 2011;23:135–142. doi: 10.1016/j.ceb.2010.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.