Abstract
A general mass spectrometry-based screen for unusually hydrophobic cellular small-molecule RNA conjugates revealed geranylated RNA in E. coli, Enterobacter aerogenes, Pseudomonas aeruginosa, and Salmonella thyphimurium. The geranyl group is conjugated to the sulfur atom in two 5-methylaminomethyl-2-thiouridine nucleotides. These geranylated nucleotides occur in the first anticodon position of tRNAGluUUC, tRNALysUUU, tRNAGlnUUG at a frequency of up to 6.7% (~400 geranylated nucleotides per cell). RNA geranylation levels can be increased or abolished by mutation or deletion of the selU (ybbB) gene in E. coli, and purified SelU protein in the presence of geranyl pyrophosphate and tRNA can produce geranylated tRNA. The presence or absence of the geranyl group in tRNAGluUUC, tRNALysUUU, and tRNAGlnUUG affects codon bias and frameshifting during translation. These RNAs represent the first reported examples of oligoisoprenylated cellular nucleic acids.
The known roles of RNA in living systems have rapidly expanded over the past three decades. The development and application of methods including bioinformatic analysis, oligonucleotide microarray technology, and high-throughput sequencing have revealed that RNA is an active contributor to many biological processes including gene regulation,1-2 catalysis,3 and viral defense.4 While the known biological repertoire of cellular RNA has grown dramatically, the observed chemical diversity of RNA has remained relatively unchanged over the past several decades. Indeed, approximately two-thirds of all known RNA modifications were reported before 1980.5 The application of modern analytical methods to either a small subset of RNAs or to the study of specific modifications of RNA have enabled the recent discovery of agmatine-modified tRNA in archaea6-7 and to the elucidation of the biosynthesis of several nucleosides.8-10 We recently developed general and highly sensitive chemical screens to reveal cellular small-molecule RNA conjugates in a broad and unbiased manner that is not limited to any particular class of modifications or any specific subset of RNAs. These screens were previously designed to capture chemically reactive11 and hydrophilic12 novel RNA nucleotides, resulting in the discovery of coenzyme A-linked RNA and NAD-linked RNA isolated from several bacteria.11-12
Large, hydrophobic small molecules conjugated to cellular RNAs could potentially influence the structure, function, or subcellular localization of RNA. Such conjugates could also be evolutionary fossils from ancient RNA-mediated lipid synthesis, as has been previously speculated13. We therefore implemented a chemical screen to isolate and identify unusually hydrophobic small molecules conjugated to biological RNA. Here we report the discovery, structural elucidation, and initial biological characterization of geranylated RNAs from bacterial cells. These findings represent the first reported examples of oligoisoprenylated nucleotides from cells.
Results
Screen for hydrophobic small molecule-RNA conjugates
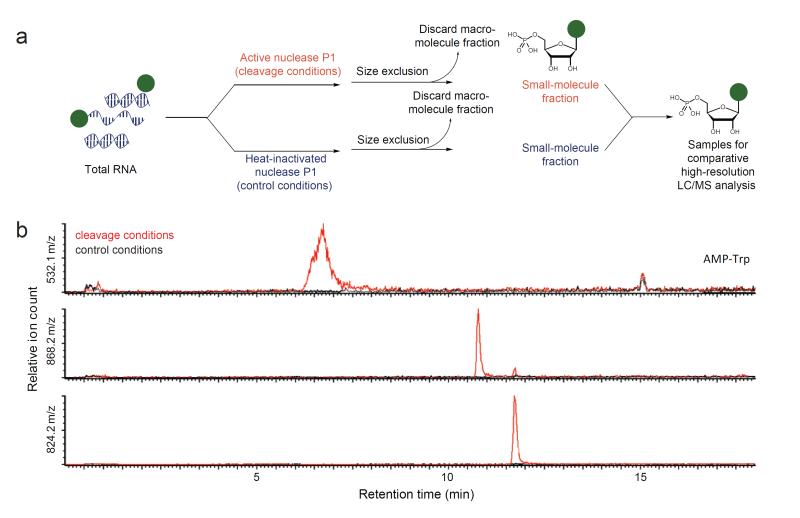
We modified our previously reported small molecule-RNA conjugate screen12 to increase the likelihood of detecting unusually hydrophobic nucleotides. The complete experimental details are provided in the Methods and the Supplementary Information. Briefly, we isolated total cellular RNA isolated in a manner that minimizes contamination with small molecules, DNA, or protein. We digested the resulting RNA pool with nuclease P1 and analyzed the products by high-resolution LC/MS. To ensure that the identified species are covalently attached to RNA, we treated an identical sample of RNA with heat-inactivated nuclease P1 and analyzed by LC/MS in parallel (Fig. 1a, Supplementary Methods). Small molecules conjugated to RNA generate non-canonical nucleotides (or fragments thereof) in the nuclease P1-digested sample that are absent in the control sample treated with inactivated nuclease P1. To focus the screen on hydrophobic RNA modifications, we (i) performed size-exclusion chromatography after digestion in the presence of organic solvents, (ii) implemented a C8 reverse-phase column for LC/MS analysis, and (iii) only considered species eluting later than AMP-Trp, the most hydrophobic of the aminoacyl adenylates, for further evaluation. The application of this screen to RNA isolated from E. coli revealed eight unknown species that are more hydrophobic than AMP-Trp and that are enriched at least three-fold in the active nuclease P1 samples relative to the control samples (Supplementary Results, Supplementary Table 1).
Figure 1.
Discovery of two hydrophobic small molecule-RNA conjugates with [M-H]− m/z = 824.200 and 868.189. (a) Scheme of the general method for discovering biological small molecule-RNA conjugates applied in this work. Total cellular RNA samples are treated with active nuclease P1 or with heat-inactivated nuclease P1 under otherwise identical conditions, then subjected to comparative LC/MS analysis. Nucleotide ions more abundant in the active nuclease sample than in the heat-inactivated nuclease (control) sample represent candidate cellular small molecule-RNA conjugates. (b) Extracted ion chromatogram of AMP-tryptophan and the two unknown nucleotides elucidated in this work.
Two novel hydrophobic small molecule-RNA conjugates
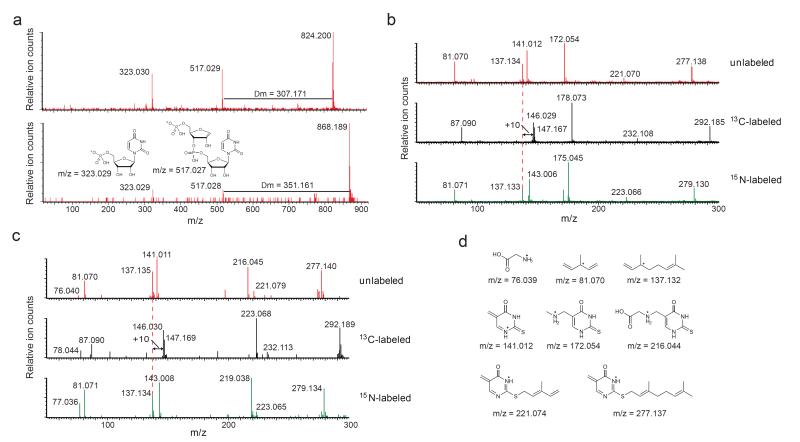
We chose two of these species, with [M-H]− m/z = 824.200 and [M-H]− m/z = 868.189, for further investigation (Fig. 1b) based on their hydrophobicity, their high degree of enrichment, and their relatedness; MS/MS fragmentation of either species resulted in ions consistent with uridine monophosphate, an unmodified ribose, and an unknown group (307.171 or 351.161 Da) (Fig. 2a). We hypothesized that the unknown groups were two novel nucleobases, and that breakage of the glycosidic bond between the novel nucleobase and a ribose gives rise to the observed UMP-ribose adduct ([M-H]− m/z = 517.029). Further MS/MS/MS fragmentation of the UMP-ribose adduct demonstrates that the dinucleotides are of the sequence 5′-X-U-3′ (Supplementary Fig. 1).
Figure 2.
Mass spectrometric characterization of two novel hydrophobic small molecule-RNA conjugates. (a) Negative ion mode MS/MS of the two unknown nucleotides reveals a dinucleotide structure with uracil and two unknown nucleobases of 307.171 and 351.161 Da. (b and c) Positive mode MS/MS of the unknown nucleobases of 307.171 Da (b) and 351.161 Da (c) from unlabeled as well as 13C- and 15N-labeled RNA. The spectra of the unlabeled RNA are shown in red, the 13C-labeled samples are shown in black, and the 15N-labeled samples are shown in green. The +10 Da shifts, indicating 10 carbon atoms, of the geranyl fragment in the 13C-labeled RNA spectra are shown. (d) Proposed structures for the individual ion fragments observed in the MS/MS experiments.
To determine the molecular formula of the unknown nucleotides, we isolated RNA from E. coli grown with 13C-glucose as the sole carbon source or 15N-ammonium chloride as the sole nitrogen source. The resulting increase in mass of the parent and the daughter ions revealed the molecular formulas of the unknown dinucleotides and the corresponding unknown nucleobases: 825.2 Da dinucleotide = C30H45N5O16P2S and 307.2 Da nucleobase = C16H25N3OS; 869.2 Da dinucleotide = C31H45N5O18P2S and 351.2 Da nucleobase = C17H25N3O3S (Supplementary Fig. 2). These empirical formulas are consistent with 5′-X-U-3′ structures. Based on the high degree of hydrophobicity of the parental species, we reasoned that the unknown nucleobases likely contain a lipid-like group. The empirical formulas also suggested that the two nucleobases likely differ by the presence or absence of a carboxylic acid.
To further characterize the structures of the unknown nucleobases, we performed MS/MS experiments in positive and negative ion mode on both unlabeled and isotopically labeled samples (Fig. 2b,c, Supplementary Fig. 3). Based on the isotope labeling data, a key fragment, [M+H]+ m/z = 137.14, had a predicted neutral molecular formula of C10H16, corresponding to three degrees of unsaturation (Fig. 2d). We hypothesized this ion might arise from the elimination of a geranyl group attached to the novel nucleobases (Supplementary Fig. 4a). The presence of a characteristic ion fragment with [M+H]+ m/z = 81.07 in the MS/MS of both nucleobases which we also observed in the MS/MS spectra of geraniol, geranyl acetate, farnesol, and geranylgeraniol, four potentially related lipid-like molecules analyzed for comparison, supports this hypothesis (Supplementary Fig. 4b).
The removal of a geranyl group (C10H16) from the empirical formulae of the unknown nucleobases resulted in molecular formulas matching two known modified nucleobases 5-methylaminomethyl-2-thiouridine (mnm5s2U) and 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U). We therefore speculated that the unknown nucleobases could be geranylated mnm5s2U and geranylated cmnm5s2U. These structures are consistent with all ion species arising from the fragmentation of the nucleobases (Supplementary Table 2). Importantly, the [M+H]+ m/z = 277.139 fragment observed in the positive mode MS/MS of both nucleobases, a fragment consistent with the presence of the geranyl group and the absence of the (carboxy)methylamino group, strongly suggests that the proposed geranyl group is not linked to the exocyclic amine of either novel nucleobase (Fig. 2d).
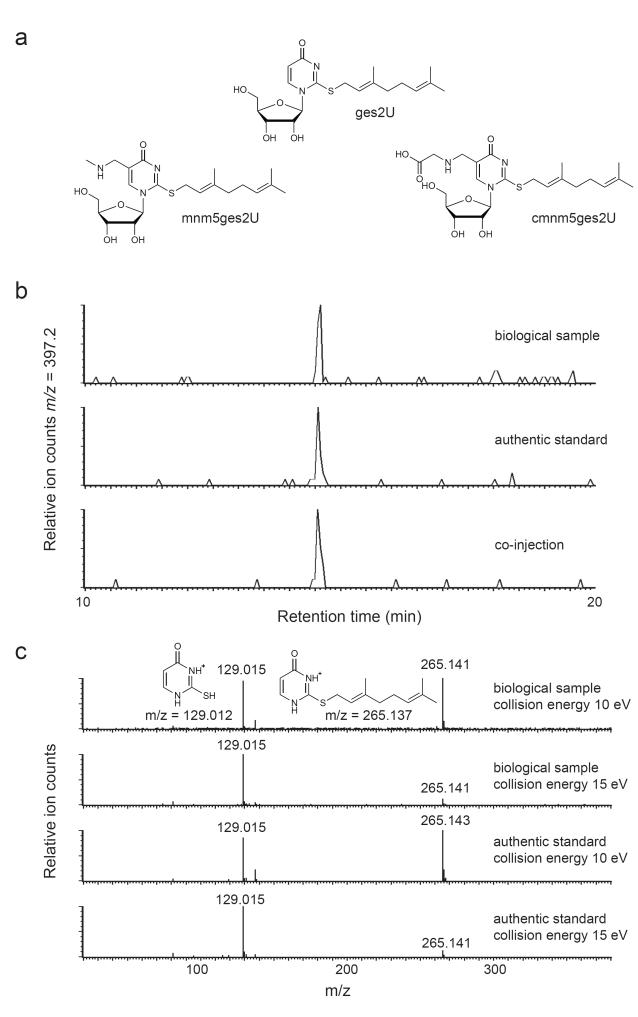
To confirm that the unknown nucleotides were derivatives of mnm5s2U and cmnm5s2U and to verify the regiochemistry of the geranyl group, we prepared RNA from E. coli strains lacking genes involved in the biosynthesis of mnm5s2U and cmnm5s2U. E. coli ΔmnmA or ΔmnmE lack the ability to install sulfur at the C2 of uridine or to incorporate the methylaminomethyl group at the C5 position, respectively.14-15 Neither sample contained detectable quantities of the previously observed unknown nucleotides (Supplementary Fig. 5), suggesting that the geranyl group is bound to either the sulfur atom or to the methylaminomethyl group. While RNA isolated from E. coli ΔmnmA did not contain any corresponding geranylated product (e.g., geranyl-mnm5U-U), a geranyl-2-thiouridine containing dinucleotide (ges2U-U) was present in RNA from E. coli ΔmnmE strains. These results indicate that the 2-thio position is either the position of geranylation (Fig. 3a), or is necessary for recognition by enzyme(s) responsible for installing the geranyl group.
Figure 3.
Structural elucidation of two geranylated nucleosides. (a) Structures of geranylated 2-thiouridine (ges2U), geranylated 5-methylaminomethyl-2-thiouridine (mnm5ges2U), and geranylated 5-carboxymethylaminomethyl-2-thiouridine (cmnm5ges2U). (b) LC comparison of biologically generated and authentic synthetic ges2U. (c) Comparison of biologically generated and authentic ges2U by MS/MS fragmentation. Proposed structures of ion fragments are shown.
To test the hypothesis that geranylation occurs at the 2-thio position, we developed a synthetic route to geranyl-2-thiouridine (ges2U, 1) from 2-thiouridine (Supplementary Fig. 6). Due to the challenge of discriminating the desired S-alkylation product from undesired N- and O-substituted adducts, we used 2-dimensional NMR of synthetic ges2U as well as X-ray crystallography of the nucleobase (geranyl-2-thiouracil, Supplementary Data Set 1) derived from synthetic ges2U to unambiguously confirm the structure of the synthetic ges2U (Supplementary Figs. 15-22). To generate a biological sample for comparison, we subjected RNA from E. coli ΔmnmE to P1 digestion, base hydrolysis, and dephosphorylation. The resulting biological sample and the synthetic ges2U exhibited identical retention times and eluted as a single peak upon co-injection and HPLC (Fig. 3b). In addition, side-by-side MS/MS comparison at varying collision energies16 revealed an identical ion fragmentation pattern for both molecules (Fig. 3c). Collectively, these results unambiguously indicate that these novel E. coli nucleotides contain a geranyl group that is linked to the 2-thio group, and confirm that our proposed structures for cellular mnm5ges2U (2) and cmnm5ges2U (3) are correct (Fig. 3a). We further used the synthetic standard of ges2U to estimate the amount of cellular ges2U in E. coli ΔmnmE cells by generating a standard curve with known quantities of authentic ges2U. Ion counts of ges2U in digested E. coli ΔmnmE RNA correspond to 398±81 molecules of ges2U per E. coli cell under the growth conditions used (Supplementary Fig. 7).
Geranylated nucleotides are in anticodon loops of tRNAs
Next we sought to identify the RNA sequence(s) that contained the geranylated nucleotides. We fractionated total RNA by size and analyzed individual fractions by P1 digestion followed by LC/MS. Both modifications were present on RNAs of length 50 to 80 nucleotides (Supplementary Fig. 8), suggesting their possible attachment to tRNA. Considering that mnm5s2U is known to be present at the first uridine (U34) in the anticodon loop of the tRNAGluUUC and tRNALysUUU, and that tRNAGlnUUG contains a potentially related s2U derivative at the same position17-19, we speculated that the geranylated nucleotides might also be present at U34 of these tRNAs. Because these three tRNAs all contain a uridine at position 35 as well, this hypothesis is consistent with our observation of geranylated bases in 5′-X-U-3′ dinucleotides.
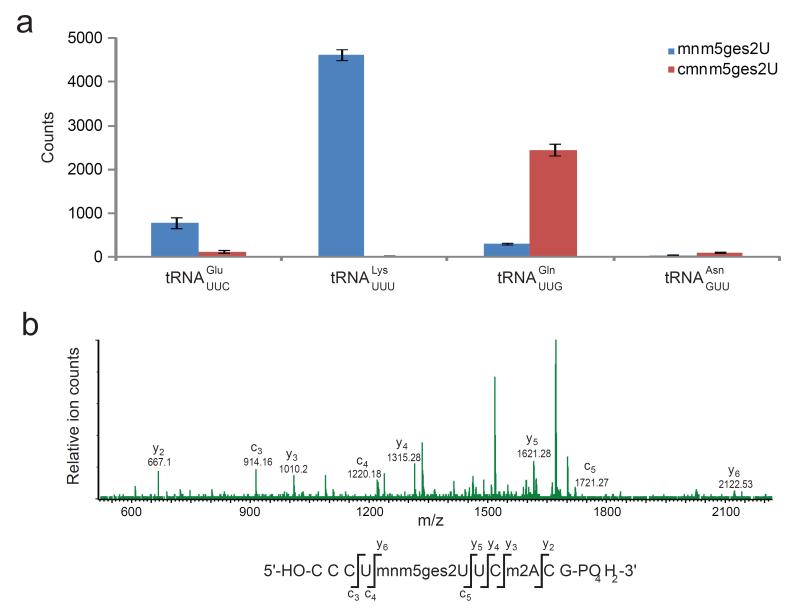
To test this hypothesis, we used complimentary oligonucleotides specific for tRNAGluUUC, tRNALysUUU, and tRNAGlnUUG to isolate these tRNAs from whole E. coli tRNA. In addition, we isolated tRNAAsnGUU, which does not contain any 2-thiouridine derivatives and therefore should not contain any geranylated nucleotides, as a negative control.20 P1 nuclease digestion and LC/MS analysis revealed that geranylated mnm5s2U is primarily present in tRNAGluUUC and tRNALysUUU, and that geranylated cmnm5s2U is present on tRNAGlnUUG (Fig. 4a). We observed that tRNAGlnUUG also contains mnm5ges2U to a lower extent, suggesting that this tRNA contains either mnm5s2U or cmnm5s2U at U34. As expected, we observed no geranylated nucleotides in the isolated tRNAAsnGUU.
Figure 4.
Characterization of geranylated cellular RNAs. (a) MS analysis of individual tRNAs isolated from cells reveals mnm5ges2U to be present on tRNAGluUUC, tRNALysUUU, and (to a lesser extent) tRNAGlnUUG, and cmnm5ges2U to be present on tRNAGlnUUG. The same analysis is shown from tRNAAsnGUU as a negative control. (b) MS/MS fragmentation of the T1 digestion product of tRNAGluUUC reveals that the geranylated nucleotide is U34.
To probe the position of the geranylated nucleotide within these tRNAs, we digested these isolated tRNAs with RNase T1, which specifically cleaves after G. We observed fragments corresponding to both (c)mnm5s2U and (c)mnm5ges2U at position U33 or U34 (Supplementary Fig. 9). Samples digested with RNase A, which cleaves specifically after pyrimidines,21 contained the dinucleotides mnm5ges2U-U and cmnm5ges2U-U. These results are consistent with geranylation of the anticodon loop since uridine is immediately 5′ of the anticodon in the tRNAs in question. MS/MS analysis of RNase T1 digestion products provided a partial sequence22 of the anticodon loop and definitively confirmed U34 as site of geranylation in tRNAGluUUC, tRNALysUUU and tRNAGlnUUG. (Fig. 4b, Supplementary Fig. 10). Collectively, these findings suggest that geranylated (c)mnm5s2U co-exists with (c)mnm5s2U at position U34 in tRNAGluUUC, tRNALysUUU, and tRNAGlnUUG. Based on previously reported abundances of tRNAs in E. coli cells23-24, we estimate that approximately 2.8-6.7% of these three tRNAs are geranylated.
SelU geranylates mnm5s2U and cmnm5s2U
To characterize the evolutionary conservation of these geranylated nucleotides, we analyzed whole cellular RNA or tRNA from eleven different organisms. Both geranylated nucleotides were present in Enterobacter aerogenes, Pseudomonas aeruginosa, and Salmonella thyphimurium but were undetectable in Vibrio fischeri, Bacillus subtilis, and several samples of eukaryotic RNA (Supplementary Table 3). These results are consistent with a previous report describing the presence of an uncharacterized C10H17 group, noted by the authors to have the same molecular formula as a geranyl group, on mnm5s2U in S. typhimurium carrying a mutation (G67E) in the sufY gene.17 SelU, the E. coli homolog of sufY, is known to catalyze the substitution of the sulfur at the C2 position of (c)mnm5s2U with selenium when selenium is present at high concentrations25. Notably, the presence or absence of homologs of sufY in each of the eleven organisms tested perfectly predicts the observed presence or absence of geranylated RNA (Supplementary Table 3), suggesting the potential role of this gene in RNA geranylation.
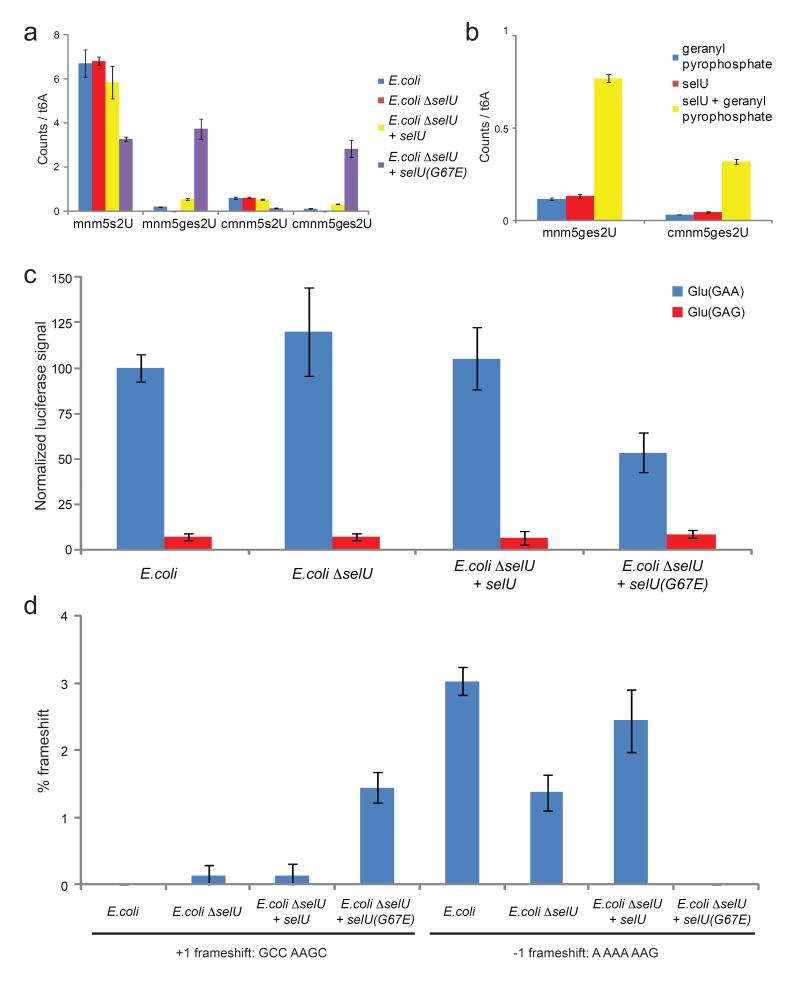
To test the role of selUin producing geranylated RNA, we isolated RNA from a selU-deficient E. coli strain (E. coli ΔselU), as well as E. coli ΔselU bearing a plasmid with a complementary copy of either selU(wt) or selU(G67E). In E. coli ΔselU we observed neither mnm5ges2U nor cmnm5ges2U. E. coli ΔselU complemented with selU(wt) contained similar levels of geranylated RNA as in E. coli, while complementation with selU(G67E) resulted in increased amounts of mnm5ges2U and cmnm5ges2U (Fig. 5a). Collectively, these results confirm that selU is involved in the production of geranylated RNA. We obtained similar results when complementing E. coli ΔselU with S. typhimurium sufY(G67E), S. typhimurium sufY(G67R) (a second mutation with similar phenotype as G67E17), and E. coli selU(G67R) (Supplementary Fig. 11). Since selU is part of the biosynthetic pathway of 5-methylaminomethyl-2-selenouridine (mnm5se2U) and 5-carboxymethylaminomethyl-2-selenouridine (cmnm5se2U), we analyzed RNA from E. coli grown under varying selenium concentrations.25 Interestingly, selenation occurs at the expense of geranylation at selenium concentrations above 10 nM (Supplementary Fig. 12).
Figure 5.
Biological abundance and properties of geranylated RNA. (a) E. coli selU mutations affect geranylated nucleotide levels. No geranylated nucleotides were detected in E. coli cells lacking selU (ΔselU). Complementation with wild-type selU restores geranylation. Complementation with mutant selU(G67E) results in high levels of geranylation. Nucleoside levels are shown as relative ratios of MS counts of the individual nucleosides versus t6A. Error bars represent the standard deviation of three independent biological replicates. (b) SelU geranylates mnm5s2U and cmnm5s2U on tRNA in vitro in a geranyl pyrophosphate-dependant manner. Nucleoside levels are shown as relative ratios of MS counts of the individual nucleosides versus t6A. Error bars represent the standard deviation of three analytical replicates. (c) Luciferase reporter assay of glutamate codon translation efficiency for GAA and GAG reveals a strong bias favoring GAA translation under low geranylation conditions and a significantly lower preference under high geranylation conditions due to reduced efficiency of GAA decoding. Error bars represent the standard deviation of three independent biological replicates. (d) +1 and −1 frameshift efficiency at the sequences GCC AAGC and A AAA AAG inserted between GST and MBP reading frames. The Y-axis indicates the amount of GST-MBP fusion protein detected by western blot as a percentage of total GST-containing proteins produced. Error bars reflect the range of two technical replicates.
To determine whether SelU directly catalyses geranylation or plays a regulatory role in the reaction, we expressed and purified E. coli SelU. Upon incubation of E. coli tRNA in the presence or absence of purified SelU and geranyl pyrophosphate, we observed a 6- to 7-fold increase of mnmges2U and cmnm5ges2U in samples containing SelU and geranyl pyrophosphate (Fig. 5b), thus confirming that SelU geranylates tRNA containing (c)mnm5s2U using geranyl pyrophosphate as co-substrate.
Geranylation affects codon bias and frameshifting
We hypothesized that a large hydrophobic modification at the first position of an anticodon could affect recognition of the third position in the corresponding codon during translation. tRNAGluUUC, tRNALysUUU and tRNAGlnUUG recognize the codons GAA/GAG, AAA/AAG, and CAA/CAG, respectively. Although CAG is further decoded by a separate tRNAGlnCUG, only tRNAGluUUC and tRNALysUUU decode GAA/GAG and AAA/AAG, respectively. Therefore, we compared the translation efficiency of the glutamate codons GAA and GAG at varying geranylation levels using a luciferase reporter containing a six-codon leader sequence of either (GAA)6 or (GAG)6 after the ATG start codon. Consistent with literature reports, we observed that E. coli tRNAGluUUC decodes GAA 10-fold more efficiently than GAG.26-28 While this ratio did not change significantly for E. coli ΔselU and E. coli ΔselU complemented with selU(wt), complementation with selU(G67E) strongly reduced the translation efficiency of GAA codons, altering the relative GAA:GAG translation efficiency ratio from 13:1 to 6:1 (Fig. 5c). We verified that these differences do not arise from changes in transcript levels, which did not differ significantly among all strains assayed (Supplementary Fig. 13). Collectively, these findings indicate that tRNA geranylation affects translational efficiency in a codon-dependent manner.
Next, we investigated whether tRNA geranylation levels can affect the efficiency of frameshifting during translation. In S. thyphimurium, the sufY(G67E) mutant resulting in the uncharacterized C10H17 fragment has been previously reported to enable the readthrough of a single C insertion in the his operon17. We studied the effect of varying tRNA geranylation levels on frameshift of the nucleotide sequences GCC AAGC, which promotes +1 frameshifting, and A AAA AAG, which promotes −1 frameshifting, at the lysine codon AAG.29-30 The sequences were inserted between the genes of glutathione S-transferase (GST) and maltose-binding protein (MBP). The inserted sequence was followed by a stop codon in frame with the GST-encoding sequence, while the MBP-encoding sequence was in the +1 or −1 frame, such that production of MBP requires a +1 or −1 translational frameshift. High levels of geranylation in E. coli ΔselU complemented with selU(G67E) induced the +1 frameshift (GCC AAGC) by >10-fold, and inhibited the −1 frameshift (A AAA AAG) by >10-fold relative to wild-type E. coli. For the +1 frameshift, we observed no detectable change in frameshift efficiency relative to wild-type E. coli for E. coli ΔselU or E. coli ΔselU complemented with selU, while for the −1 frameshift we observed a two-fold reduction in frameshift efficiency for E. coli ΔselU and no significant change for E. coli ΔselU complemented with selU (Figure 5d). +1 frameshifting at GCC AAGC has been reported to be induced by uncharged lysyl-tRNAs due to ribosome stalling at lysine codons in lysine-limiting conditions.30 However, the amount of lysyl-tRNA or tRNALysUUU does not change as a function of geranylation levels (Supplementary Fig. 14), suggesting that geranylation either results in ribosome stalling by affecting a different step of protein synthesis or induces frameshifting by a completely different mechanism.
Discussion
Posttranscriptional modifications are ubiquitous on all tRNA molecules and are known to affect their function.31 Among the most frequently modified nucleotides in tRNAs are the anticodon wobble position (nucleotide 34) and the purine adjacent to the anticodon (nucleotide 37). These modifications ensure binding to the correct codon and are required for the efficient recognition of several codons by the same tRNA.32 Modifications at U34 are of particular importance for wobble base pairing. In bacteria, the 5-methylaminomethyl group on tRNALysUUU and tRNAGluUUC is important for efficient recognition of AAG and GAG, respectively.33-34 The 2-thio group in modified U34 of tRNAGluUUC has been reported to increase recognition of GAA33 and to be required for efficient binding to the tRNA synthetase.35 Moreover, in tRNALysUUU the 2-thio modification enables binding to the ribosome.36-37
Previously reported RNA modifications are predominantly small or hydrophilic.5 Our identification of two lipid-linked nucleotides mnm5ges2U and cmnm5ges2U represent to our knowledge the first characterized oligoisoprenylated nucleotides. These modifications are present in E. coli, E. aerogenes, P. aeroginosa, and S. thyphimurium at U34 in the anticodon of tRNAGluUUC, tRNALysUUU and tRNAGlnUUG. It is tempting to speculate that the unusual physical properties of very hydrophobic RNA modifications may have obscured their discovery using previous methods. The conditions traditionally used for thin layer chromatography and HPLC optimized for hydrophilic mononucleotides often do not allow the resolution of very hydrophobic molecules.38 Further, while mass spectrometry has previously been used to characterize novel nucleotides, the use of mass spectrometry in the primary screening method in this work enabled the detection and analysis of low-abundance species.
Our findings suggest that RNA geranylation is an alternative to selenation at low selenium concentrations. Interestingly, our results indicate that selU can directly catalyze geranylation of (c)mnm5s2U, in addition to its previously known role in selenation. That two such disparate chemical alternatives are mediated by a single enzyme (SelU in E. coli) may seem surprising, but we speculate that the active site can accommodate either selenophosphate, the substrate for 2-selenouridine synthesis,25 or geranyl pyrophosphate, the substrate for geranylation. Indeed, in Salmonella mutation of Cys 97, the homologue of the active site residue for selenation in selU, to Ala in the rhodanese domain of sufY(G67E) strongly reduces the level of the previously uncharacterized C10H17 modification.17 Even if the same residues on the protein mediate both selenation and geranylation, the two reactions are likely to proceed through distinct pathways since for selenation the sulfur at 2-thio position must be converted into a leaving group as selenium acts as nucleophile, while for geranylation the 2-thio group acts as nucleophile. Further studies on this enzyme are necessary to reveal the detailed mechanism of RNA geranylation.
High geranylation levels can affect codon bias and frameshift efficiency during translation. The exact mechanisms by which geranylation modulates codon bias and frameshifting remains to be elucidated. Frameshifting at A AAA AAG is thought to be based on the preferential recognition of AAA over AAG by the anticodon mnm5s2UUU.39 Hypomodification of tRNALysUUU at U34 reduces −1 frameshifting at NNA AAG sites, possibly due to improved wobble pairing.40 Likewise, high geranylation levels may also reduce the preference of tRNALysUUU for AAA over AAG, thereby inhibiting frameshifting. This hypothesis is supported by our observation of decreased codon bias of tRNAGluUUG upon geranylation. Considering the importance of position of U34 in codon recognition as well as tRNA-synthetase and ribosome binding,35-37 it is possible that a large hydrophobic modification at this tRNA position could have a broad impact on translation.
Recent studies in S. cerevisiae suggest that the large-scale coordination of tRNA modification is a translational response to cellular stress.41-42 Modification of RNA with unusually hydrophobic groups could also affect subcellular localization by inducing association with proteins or the lipid bilayer. Small molecule-RNA conjugates may therefore represent an additional basis for translation-independent RNA localization.43-44 Geranylation and other hydrophobic modifications of RNA may also play even broader roles in cellular function. Boxer and coworkers recently reported that lipid-linked DNA oligonucleotides enable vesicle fusion in vitro by a process resembling SNARE-mediated exocytosis.45 While lipid-oligonucleotide conjugates that induce membrane association in living systems have yet to be discovered, analogous processes enabled by lipid-linked cellular RNA may exist in nature.
Methods
RNA digestion with nuclease P1
For negative ion mode mass spectrometric analysis, 500 μg of isolated total RNA or commercial RNA (wheat tRNA or bovine total RNA (Sigma)) were digested with 10 U nuclease P1 (Wako Chemicals) in 500 μL of 50 mM NH4OAc, pH 4.5 at 37 °C for 40 min. The control samples were prepared by incubating 500 μg of RNA with 10 U heat-inactivated nuclease P1 (pre-incubated at 95 °C for two hours) in 500 μL of 50 mM NH4OAc, pH 4.5 at 37 °C for 40 min. The digestion products were purified by size-exclusion chromatography (NAP5, GE Healthcare) and the small-molecule fraction was collected and lyophilized. The size-exclusion chromatography step during the screen was performed in the presence of 30-50% methanol.
RNA digestion with nuclease P1 and alkaline phosphatase
For positive ion mode mass spectrometric analysis, 500 μg of isolated total RNA or commercial RNA were digested with 10 U nuclease P1 and 100 U alkaline phophatase (Sigma) in 500 μL of 50 mM NH4OAc, pH 6 at 37 °C for 4 hours. The digestion products were purified and lyophilized as described above.
LC/MS analysis
LC/MS was performed using a Waters Aquity UPLC Q-TOF Premier instrument. For negative ion mode analysis of the small molecule-RNA conjugate screen and characterization of nuclease P1-digested RNA, samples were dissolved in 50 μL of 0.05% NH3HCOOH in 1:1 deionized water:DMSO and LC was performed using a gradient from 0.1% aqueous ammonium formate (A1) to methanol (B1) on an Aquity UPLC BEH C8 column (1.7 μm, 2.1 mm x 50 mm, Waters) at a constant flow rate of 0.3 mL/min. The mobile phase composition was as follows: 100% A1 for 1 min; linear increase over 21 min to 100% B1; maintain at 100% B1 for 8 min; return to 100 % A1 over 5 min. The column was washed several times using short gradients from A1 to B1 between subsequent LC runs. Electrospray ionization was used with a capillary voltage of 3 kV, a sampling cone voltage of 25 V and 40 V, and a LM resolution of 4.7. The desolvation gas temperature was 300 °C, the flow rate was 800 L/hour, the source temperature was 150 °C, and the detector was operated in negative ion mode. LC/MS/MS experiments of the identified dinucleotides were performed at collision energies of 20 and 30 eV. LC/MS/MS/MS for fragmentation of the nucleobase of mnm5ges2U was performed with the cone voltage at 180 V and collision energies of 10 and 20 eV.
Positive ion mode analysis for the characterization of RNA digested with nuclease P1 and alkaline phosphatase (Sigma) was performed by dissolving the samples in 50 μL of 0.1% aqueous NH3HCOOH and performing LC using a linear gradient from 0.1% aqueous formic acid (A2) to acetonitrile (B2) on an Aquity UPLC BEH C8 column (1.7 μm, 2.1 mm x 50 mm) at a constant flow rate of 0.3 mL/min. The mobile phase composition was as follows: 100% A2 for 1 min; linear increase over 21 min to 100% B2; maintain at 100% B2 for 8 min; return to 100% A2 over 5 min. Electrospray ionization conditions were as described above with the detector operating in positive ion mode. LC/MS/MS experiments of the identified dinucleotides were performed at collision energies of 10, 20, and 30 eV. LC/MS/MS/MS analysis of the nucleobase of mnm5ges2U was performed with a cone voltage of 70 V and collision energies of 10, 20, 30, and 40 eV. LC/MS/MS/MS analysis of the nucleobase of cmnm5ges2U was performed with the cone voltage at 40 V and collision energies of 10, 20, 30, and 40 eV.
Synthesis of ges2U (1-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-2-(((E)-3,7-dimethylocta-2,6-dien-1-yl)thio)pyrimidin-4(1H)-one)
A solution of 2-thiouridine (26.0 mg, 0.1 mmol, Santa Cruz Biotechnology), geranyl bromide (57 μL, 0.3 mmol) and N,N-diisopropylethylamine (52.2 μL, 0.3 mmol) in methanol (1 mL) was stirred at 25 °C for 12 h. The resulting reaction mixture was concentrated in vacuo and directly subjected to silica gel chromatography. A white solid (38.9 mg, 98% yield) was obtained after flash chromatography: TLC Rf = 0.10 (10% MeOH/EtOAc); 1H NMR (600 MHz, CDCl3) δ 8.25 (d, J = 7.2 Hz, 1H), 5.98 (d, J = 7.2 Hz, 1H), 5.91 (d, J = 5.4 Hz, 1H), 5.28 (t, J = 7.2 Hz, 1H), 5.05 (t, J = 7.2 Hz, 1H), 4.45 (m, 1H), 4.37 (m, 1H), 4.18 (s, 1H), 3.88-3.96 (m, 2H), 3.78-3.86 (m, 2H), 2.04-2.12 (m, 2H), 1.98-2.04 (m, 2H), 1.72 (s, 3H), 1.68 (s, 3H), 1.59 (s, 3H); 13C NMR (125 MHz, CDCl3) δ 170.4, 164.9, 143.8, 141.2, 132.1, 123.9, 115.9, 109.0, 92.2, 86.1, 75.7, 70.8, 61.5, 39.9, 31.7, 26.6, 25.9, 18.0, 17.0; HRMS-EI (m/z): [M+H]+ calculated for C19H28N2O5S = 397.1792, observed = 397.1800.
Supplementary Material
Acknowledgements
This work was supported by the Howard Hughes Medical Institute and the NIH/NIGMS (R01GM065865). C.E.D. acknowledges fellowship from the Novartis Foundation. A.M.L. was supported by a NIH National Research Service Award Postdoctoral Fellowship (F32GM095028). We thank Dr. Shao-Liang Zheng for his help with X-ray diffraction and structural determination of synthetic geranyl-2-thiouracil. We thank Dr. Alan Saghatelian for providing Pseudomonas aeruginosa and Dr. Irene Chen for providing Salmonella typhimurium and BL2 facilities. We are also grateful to Jacob Carlson for his assistance, and to Professor Michael Ibba for helpful discussions.
Footnotes
Competing financial interest The authors declare no competing financial interests.
Author contributions C.E.D., Y.C., A.M.L., and Y.G.C. designed, and performed the experiments. All authors analyzed the results and wrote the manuscript.
References
- 1.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 2.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–90. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat Rev Mol Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 4.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–44. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 5.Cantara WA, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeuchi Y, et al. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat Chem Biol. 2010;6:277–82. doi: 10.1038/nchembio.323. [DOI] [PubMed] [Google Scholar]
- 7.Mandal D, et al. Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc Natl Acad Sci U S A. 2010;107:2872–7. doi: 10.1073/pnas.0914869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol Cell. 2006;21:97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Miles ZD, McCarty RM, Molnar G, Bandarian V. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc Natl Acad Sci U S A. 2011;108:7368–72. doi: 10.1073/pnas.1018636108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–54. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowtoniuk WE, Shen Y, Heemstra JM, Agarwal I, Liu DR. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc Natl Acad Sci U S A. 2009;106:7768–73. doi: 10.1073/pnas.0900528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YG, Kowtoniuk WE, Agarwal I, Shen Y, Liu DR. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat Chem Biol. 2009;5:879–81. doi: 10.1038/nchembio.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott AI. How were porphyrins and lipids synthesized in the RNA world? Tetrahedron Letters. 1997;38:4961–4964. [Google Scholar]
- 14.Hagervall TG, Edmonds CG, McCloskey JA, Bjork GR. Transfer RNA(5-methylaminomethyl-2-thiouridine)-methyltransferase from Escherichia coli K-12 has two enzymatic activities. J Biol Chem. 1987;262:8488–95. [PubMed] [Google Scholar]
- 15.Kambampati R, Lauhon CT. MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry. 2003;42:1109–17. doi: 10.1021/bi026536+. [DOI] [PubMed] [Google Scholar]
- 16.Nicolas EC, Scholz TH. Active drug substance impurity profiling part II. LC/MS/MS fingerprinting. J Pharm Biomed Anal. 1998;16:825–36. doi: 10.1016/s0731-7085(97)00132-5. [DOI] [PubMed] [Google Scholar]
- 17.Chen P, Crain PF, Nasvall SJ, Pomerantz SC, Bjork GR. A “gain of function” mutation in a protein mediates production of novel modified nucleosides. EMBO J. 2005;24:1842–51. doi: 10.1038/sj.emboj.7600666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juhling F, et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–62. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yaniv M, Folk WR. The nucleotide sequences of the two glutamine transfer ribonucleic acids from Escherichia coli. J Biol Chem. 1975;250:3243–53. [PubMed] [Google Scholar]
- 20.Yokogawa T, Kitamura Y, Nakamura D, Ohno S, Nishikawa K. Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res. 2010;38:e89. doi: 10.1093/nar/gkp1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volkin E, Cohn WE. On the Structure of Ribonucleic Acids .2. The Products of Ribonuclease Action. Journal of Biological Chemistry. 1953;205:767–782. [PubMed] [Google Scholar]
- 22.Mcluckey SA, Vanberkel GJ, Glish GL. Tandem Mass-Spectrometry of Small, Multiply Charged Oligonucleotides. Journal of the American Society for Mass Spectrometry. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 23.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–63. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 24.Jakubowski H, Goldman E. Quantities of individual aminoacyl-tRNA families and their turnover in Escherichia coli. J Bacteriol. 1984;158:769–76. doi: 10.1128/jb.158.3.769-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfe MD, et al. Functional diversity of the rhodanese homology domain: the Escherichia coli ybbB gene encodes a selenophosphate-dependent tRNA 2-selenouridine synthase. J Biol Chem. 2004;279:1801–9. doi: 10.1074/jbc.M310442200. [DOI] [PubMed] [Google Scholar]
- 26.Looman AC, et al. Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. EMBO J. 1987;6:2489–92. doi: 10.1002/j.1460-2075.1987.tb02530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J Mol Biol. 1991;222:265–80. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- 28.Wittwer AJ, Ching WM. Selenium-containing tRNA(Glu) and tRNA(Lys) from Escherichia coli: purification, codon specificity and translational activity. Biofactors. 1989;2:27–34. [PubMed] [Google Scholar]
- 29.Gurvich OL, et al. Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. EMBO J. 2003;22:5941–50. doi: 10.1093/emboj/cdg561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsley D, Gallant J. On the directional specificity of ribosome frameshifting at a “hungry” codon. Proc Natl Acad Sci U S A. 1993;90:5469–73. doi: 10.1073/pnas.90.12.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T. Biosynthesis and function of tRNA wobble modifications. In: Grosjean H, editor. Fine-Tuning of RNA Functions by Modification and Editing. Vol. 12. Springer; Berlin / Heidelberg: 2005. pp. 23–69. [Google Scholar]
- 32.Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 33.Kruger MK, Pedersen S, Hagervall TG, Sorensen MA. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J Mol Biol. 1998;284:621–31. doi: 10.1006/jmbi.1998.2196. [DOI] [PubMed] [Google Scholar]
- 34.Yarian C, et al. Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem. 2002;277:16391–5. doi: 10.1074/jbc.M200253200. [DOI] [PubMed] [Google Scholar]
- 35.Sylvers LA, Rogers KC, Shimizu M, Ohtsuka E, Soll D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry. 1993;32:3836–41. doi: 10.1021/bi00066a002. [DOI] [PubMed] [Google Scholar]
- 36.Ashraf SS, et al. Single atom modification (O-->S) of tRNA confers ribosome binding. RNA. 1999;5:188–94. doi: 10.1017/s1355838299981529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yarian C, et al. Modified nucleoside dependent Watson-Crick and wobble codon binding by tRNALysUUU species. Biochemistry. 2000;39:13390–5. doi: 10.1021/bi001302g. [DOI] [PubMed] [Google Scholar]
- 38.Lane BG. Historical Perspectives on RNA Nucleoside Modifications. In: Grosjean H, Benne R, editors. Modification and editing of RNA. ASM Press; 1998. pp. 1–20. [Google Scholar]
- 39.Tsuchihashi Z, Brown PO. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev. 1992;6:511–9. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- 40.Licznar P, et al. Programmed translational −1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. EMBO J. 2003;22:4770–8. doi: 10.1093/emboj/cdg465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Begley U, et al. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell. 2007;28:860–70. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan CT, et al. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keiler KC. RNA localization in bacteria. Curr Opin Microbiol. 2011;14:155–9. doi: 10.1016/j.mib.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in E. coli. Science. 2011;331:1081–4. doi: 10.1126/science.1195691. [DOI] [PubMed] [Google Scholar]
- 45.Chan YH, van Lengerich B, Boxer SG. Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides. Proc Natl Acad Sci U S A. 2009;106:979–84. doi: 10.1073/pnas.0812356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.